Abstract
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths worldwide. Long noncoding RNAs (lncRNAs) are involved in the tumorigenesis and progression of some cancers. However, only a handful of lncRNAs have been functionally identified in HCC. In the present study, we identified a novel functional lncRNA in HCC, termed lncWDR26 (GenBank Accession no. RP11-365O16). Here, we reported that lncWDR26 was significantly downregulated in HCC tissues and cells. Moreover, decreased lncWDR26 expression correlates with larger tumor size, higher clinical stage, and tumor metastasis, and also predicts poor prognosis in patients with HCC. In HCC cells, overexpression of lncWDR26 inhibited growth and metastasis, both in vitro and in vivo. Mechanistically, lncWDR26 suppressed HCC growth and metastasis by inhibiting WDR26 transcription. Notably, lncWDR26 was associated with SIX homeobox 3 (SIX3), and this association was required for the repression of WDR26 transcription. Together, these results indicate that lncWDR26 is a tumor suppressor lncRNA that promotes tumor progression, leading us to propose that lncRNAs may serve as key regulatory hubs in HCC progression.
Keywords: WDR26, SIX3, growth, metastasis
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths worldwide. Liver resection is still the best therapeutic strategy to treat HCC, with a 5-year survival rate of approximately 30% [1]. The prognosis of patients with HCC remains poor despite the advancement in HCC treatment. Thus, understanding the molecular mechanisms of hepatocarcinogenesis and identifying novel prognostic molecular markers can promote the development of appropriate therapeutic strategies earlier in the course of this cancer.
The noncoding RNAs that are the predominant transcripts in the mammalian genome exceed the number of protein-coding genes. It has been reported that abnormal expression of small noncoding RNAs (ncRNAs), especially microRNAs, contributes to the pathogenesis of HCC [2,3]. More recently, a new class of ncRNAs, known as long noncoding RNAs (lncRNA; larger than 200 nucleotides), which are endogenous RNA transcripts in the genome without protein-coding potential, have been reported to be critical regulators in modulating cancer malignancies [4-6]. The mechanisms of actions of well-studied lncRNAs are known to modify chromatin structure and regulate gene expression in cis or trans manner [7]. Dysregulation of lncRNAs has been correlated to tumorigenesis and cancer metastasis [8,9]. In HCC, a handful of lncRNAs have been functionally identified. For example, HOTAIR [10], lncRNA-ATB, UFC1 [11], and MALAT1 [12] were found to be overexpressed in HCC tissues. Overexpression of these lncRNAs is correlated with poor survival of patients, and plays an important role in the development of HCC. Thus, these RNAs may act as “onco-lncRNAs”. In contrast, lncRNA MEG3 [13] and GAS5 [14] were downregulated in HCC tissues compared to normal liver tissues. Restoration of expression of these lncRNAs suppresses the proliferation, migration, and invasion of HCC cells. Therefore, they may function as “tumor suppressor lncRNAs”. Although thousands of lncRNAs have been annotated, only a few lncRNAs have been functionally characterized.
In this study, we characterized the pathological relevance of lncRNA WD repeat domain 26 (lncWDR26) (GenBank Accession no. RP11-365O16) in HCC growth and metastasis. Its expression was analyzed by qRT-PCR using tissue samples from 82 patients with HCC. Further mechanistic investigations into the function of lncWDR26 in HCC were performed through gain-of-function studies, RNA immunoprecipitation (RIP) assays, and RNA pull-down assays, through which we demonstrated that the interaction between lncWDR26 and SIX homeobox 3 (SIX3) was associated with HCC formation and progression.
Materials and methods
Cell culture
Six HCC cell lines (SMMC-7721, PLC/PRF/5, Huh7, SK-Hep-1 and Hep3B) and a normal liver cell line (LO2) were obtained from Cell Bank of Chinese Academy of Sciences. Cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin (Hyclone).
Tissue samples
82 paired HCC and corresponding normal liver tissue samples were collected during HCC surgery. All of the patients gave informed consent for their tissues to be used for this research. Recommendations of the Declaration of Helsinki for biomedical research involving human subjects were also followed. Ethical approval for the study was obtained from Ethics Committee of Cangzhou Central Hospital.
Cell proliferation detection
3×103 cells per well were seeded in the 96-well plate and incubated for different time point, respectively. Cell proliferation was measured with a Cell Counting Kit-8 (CCK-8) (Beyotime, Shanghai, China), following the manufacturer’s instructions. Absorbance was measured at 450 nm using Elx800 Reader (Bio-Tek Instruments, VT, USA).
Western blot
The cells were lysed with RIPA buffer (Beyotime Biothechnology, Beijing, China). Protein lysates were separated by SDS-PAGE and then transferred onto PVDF membranes. The membranes were blocked and incubated with anti-SIX3 (Abcam, Cambridge, MA, USA), or WDR26 (Abcam, Cambridge, MA, USA) or GAPDH antibody (Santa Cruz, CA) at 4°C overnight. After being washed, the membranes were incubated with HRP-conjugated anti-IgG (Santa Cruz, CA). Signal was detected by an ECL system (Amersham Pharmacia, Piscataway, NJ).
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. 1 μg of total RNA was reversely transcribed to cDNA by using EasyScript One-Step gDNA Removal and cDNA Synthesis (Transgen, Beijing, China). qRT-PCR was performed on the Step-One Plus Detection System (Applied Biosystems, USA). Relative expression levels were calculated as ratios normalized against those of GAPDH. Comparative quantification was determined using the 2-ΔΔCt method. Primers were provided as follow: GAPDH-R: 5’-GATTCCACCCATGHCCAAATTC-3’, GAPDH-R: 5’-CTGGAAGATGGTGATGGGATT-3’; lncWDR26-F: 5’-AGACCTGACCTGGAATGAGA-3’, lncWDR26-R: 5’-TGGGCAACAGAGCAAGATT-3’; WDR26-F: 5’-CATGGAAGGAGACTGGGATAAG-3’, WDR26-R: 5’-GATTTCAAGTGCGCCTCTTAC-3’.
Isolation of cytoplasmic and nuclear
Cytoplasmic and nuclear RNA were isolated and purified using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA) according to its manual.
5’ and 3’ rapid amplification of cDNA ends (RACE)
We used the 5’-RACE and 3’-RACE analyses to determine the transcriptional initiation and termination sites of lncWDR26 using a SMARTer™ RACE cDNA Amplification Kit (Clontech, Palo Alto, CA) according to the manufacturer’s instructions.
Cell transfection
LncWDR26 transcript was determined by a RACE assay. Cells were transfected with lentiviral particles expressing full-length human lncWDR26 sequence or an empty lentiviral vector control. After 48 hours, stable clones were selected for 1 week by using puromycin. We constructed lentivirus-based short hairpin RNA (shRNA) expression constructs into pLKO.1 vector (Addgene) to knockdown lncWDR26 expression. Cells were transfected with either of lentiviral vectors encoding specific shRNA sequence or the negative control vector (scramble shRNA, named Con). Following target sequences were used: shlncWDR26: CCTGGAATGAGACTTGCAA; shWDR26: CAGTCAGATGAGGATGTCA; shSIX3: CAAGTAGGCAACTGGTTTA.
Cell cycle assay
The cell cycle was analyzed using an in situ cell proliferation kit FLUOS (Roche) according to the manufacturer’s instruction. The cell cycle distribution was analyzed by flow cytometry. The data were analyzed by FlowJo software (Tree Star).
Apoptosis assay
Cells were stained with fluorescein isothiocyanate-conjugated Annexin V and 7-AAD (Apoptosis Detection Kit, KeyGEN, Nanjing, China) according to the manufacturer’s instruction. Cells were analyzed with flow cytometer, and the data were studied using FlowJo software (Tree Star).
Migration and invasion assays
Cells were seeded on the top side of the membrane pre-coated with Matrigel for invasion assay or without Matrigel for migration assay and incubated for 24 h. Cells inside the upper chamber were obliterated with cotton swabs, while cells on the lower membrane surface were fixed and then stained with 0.5% crystal violet solution. Ten fields were counted randomly in each well.
Chromatin immunoprecipitation (ChIP)
We performed ChIP using the EZ ChIPTM Chromatin Immunoprecipitation Kit (Millipore), as per the manufacturer’s instruction. Briefly, crosslinked chromatin was sonicated into 200 to 1000 bp fragments and immunoprecipitated using anti-SIX3 antibody (Abcam). Normal rabbit immunoglobulin G (IgG) was used as a negative control. qPCR was conducted using SYBR Green Mixture. Primers used for WDR26 promoter regions were F: 5’-GAGTCTCACCGGCTCATTATC-3’ and R: 5’-ATCGGGTTTCACAACCTAC-TT-3’.
RNA pull-down and mass spectrometry assay
RNA pull-down was performed as previously described [15]. In vitro biotin-labeled RNAs (lncWDR26 and its antisense RNA) were transcribed with the biotin RNA labeling mix (Roche) and T7 RNA polymerase (Roche) treated with RNase-free DNase I (Promega) and purified with RNeasy Mini Kit (QIAGEN). Biotinylated RNA was incubated with nuclear extracts of breast cancer cells, and pull-down proteins were run on SDS-PAGE gels. Mass spectrometry followed.
RNA-FISH
Fluorescence-conjugated lncWDR26 probes were used for RNA-FISH. RNA-FISH was performed by using RNA-FISH Kit (Boster, Wuhan, China) according to the standard protocol. Cells were observed with a FV1000 confocal laser microscope (Olympus).
Tumor xenograft
Male nude mice (4-6-week old) were used for xenograft assays. Cells (1×106) were trypsinized, harvested in PBS, and injected subcutaneously into the flank of the animals. Approximately 12 days later, tumors were detectable and tumor size was measured using a vernier caliper. Tumor volumes were calculated by the formula V = ½ (L×W2), where L is the length (longest dimension) and W, the width (shortest dimension). The animal study protocol was reviewed and approved by the local ethics committee at Cangzhou Central Hospital.
Statistical analysis
All the statistical analyses were performed using SPSS software. For comparisons between two groups, two-tailed Student’s t-tests were performed. Survival curve was evaluated using the Kaplan-Meier method, and differences were assessed using the log-rank test.
Results
lncWDR26 expression is decreased in HCC tissues
Firstly, we analyzed the HCC tissue gene profiling data from previous studies [11,16]. lncRNAs act in cis or trans to regulate the expression of neighboring genes [17]. Therefore, we focused on intergenic lncRNAs, which were aberrantly expressed in HCC tissues and located in the nearby coding genes related to HCC development. Among these differentially expressed intergenic lncRNAs, we focused on an uncharacterized lncRNA, termed lncWDR26 (gene symbol: RP11-365O16). lncWDR26 is one of the most decreased lncRNAs in HCC, residing on chromosome 1, upstream of the WDR26 gene. To further validate this result, we investigated lncWDR26 expression in a cohort of 82 paris of HCC tumor and matched normal liver tissue samples using qRT-PCR. The results showed that lncWDR26 expression was significantly lower in the HCC tissues (Figure 1A). Next, we examined lncWDR26 levels in HCC cell lines (SMMC-7721, PLC/PRF/5, Huh7, SK-Hep-1, and Hep3B) and a normal liver cell line (LO2) using qRT-PCR. Compared with LO2 cells, lncWDR26 was present at lower levels in all HCC cell lines (Figure 1B). Collectively, these results showed that lncWDR26 is downregulated in HCC tissues and cells. To further explore the relationship between lncWDR26 expression and clinicopathological features in patients with HCC, patients were divided into two groups: the high lncWDR26 expression group and the low lncWDR26 expression group. Statistical analysis revealed that lower lncWDR26 levels were correlated with larger tumor size, higher clinical stage, and metastasis. However, lncWDR26 expression was not correlated with other factors, including gender, age, liver cirrhosis, and AFP levels in patients with HCC (Table 1).
Figure 1.
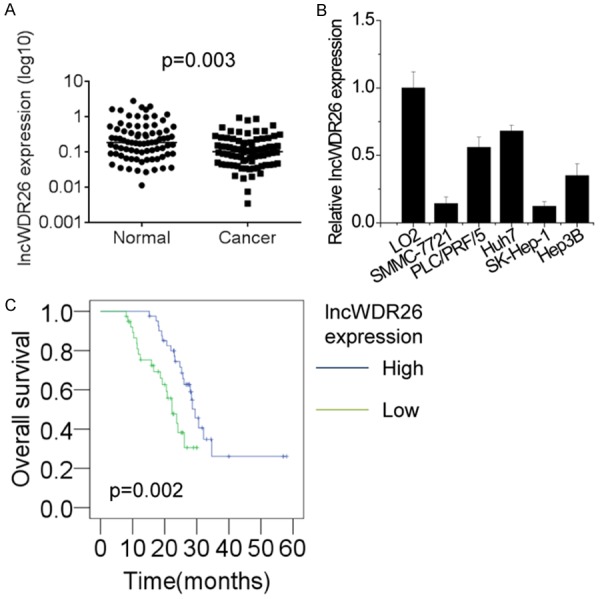
lncWDR26 expression is decreased in HCC tissues. A. The expression levels of lncWDR26 in 82 pairs of HCC and matched normal liver tissues were examined by qRT-PCR. B. The expression levels of lncWDR26 in 5 different HCC cell lines and a normal liver cell line were detected by qRT-PCR. C. The Kaplan-Meier curves for patients with HCC showing low and high lncWDR26 expression.
Table 1.
Relationship between lncWDR26 expression and clinicopathological features of HCC patients
| Clinicopathological features | lncWDR26 expression levels | P value | |
|---|---|---|---|
|
| |||
| Low | High | ||
| Gender | |||
| Male | 25 | 24 | 0.822 |
| Female | 16 | 17 | |
| Age | |||
| ≤55 | 23 | 25 | 0.654 |
| >55 | 18 | 16 | |
| Liver cirrhosis | |||
| With | 22 | 23 | 0.822 |
| Without | 18 | 17 | |
| AFP (ng/mL) | |||
| <20 | 8 | 9 | 0.768 |
| >20 | 34 | 33 | |
| Tumor size (cm) | |||
| ≤5 | 13 | 26 | 0.003 |
| >5 | 29 | 15 | |
| BCLC stage | |||
| A | 14 | 30 | 0.001 |
| B+C+D | 28 | 12 | |
| Metastasis | |||
| Yes | 28 | 15 | 0.005 |
| No | 14 | 27 | |
Abbreviations: AFP, alpha-fetoprotein; BCLC, Barcelona Clinic Liver Cancer. The median expression level was used as cutoff.
Next, we used Kaplan-Meier survival analysis to examine the correlation between lncWDR26 expression and prognosis of patients with HCC. The results showed that lower expression of lncWDR26 predicted a lower rate of 5-year survival of patients with HCC (Figure 1C). These findings suggest that lncWDR26 may exert tumor-suppressive functions in HCC.
Characterization of lncWDR26
A total length of 450 nt of lncWDR26 transcript was determined by RACE assay (Figure 2A). We determined the cellular location of lncWDR26 by performing RNA-FISH and cellular fractionation assays. The results showed that lncWDR26 was mainly localized in the nuclei of HCC cells (Figure 2B and 2C).
Figure 2.
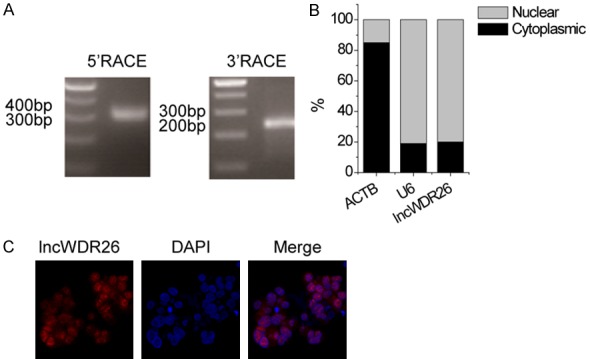
Characterization of lncWDR26. A. Representative images of PCR products from the RACE analysis are shown. B. lncWDR26 RNA expression in different subcellular fractions in SMMC-7721 cells. C. lncWDR26 intracellular localization was visualized in SMMC-7721 cells by FISH assay. Representative images of lncWDR26 staining in SMMC-7721 cells are shown.
Overexpression of lncWDR26 inhibits the growth of HCC cells
To determine the biological functions of lncWDR26 in HCC cells, lncWDR26 was overexpressed in SMMC-7721 and SK-Hep-1 cells by transfection with lncWDR26-overexpressing lentiviral particles (Figure 3A). To assess the role of lncWDR26 in HCC cell phenotype, we performed gain-of-function assays. CCK-8 assay showed that the growth of SMMC-7721 and SK-Hep-1 cells transfected with lncWDR26 was inhibited compared with control cells (Figure 3B). Moreover, colony formation analysis showed that lncWDR26 overexpression significantly suppressed the colony formation capacity of HCC cells (Figure 3C). Because the above results indicate that lncWDR26 exerts a tumor-suppressive effect on HCC cells, we then investigated whether lncWDR26 is involved in regulating cell apoptosis using flow cytometry. Compared with control cells, overexpression of lncWDR26 significantly increased the HCC cell apoptotic rate (Figure 3D). However, lncWDR26 did not have an effect on cell cycle distribution (Figure 3E).
Figure 3.
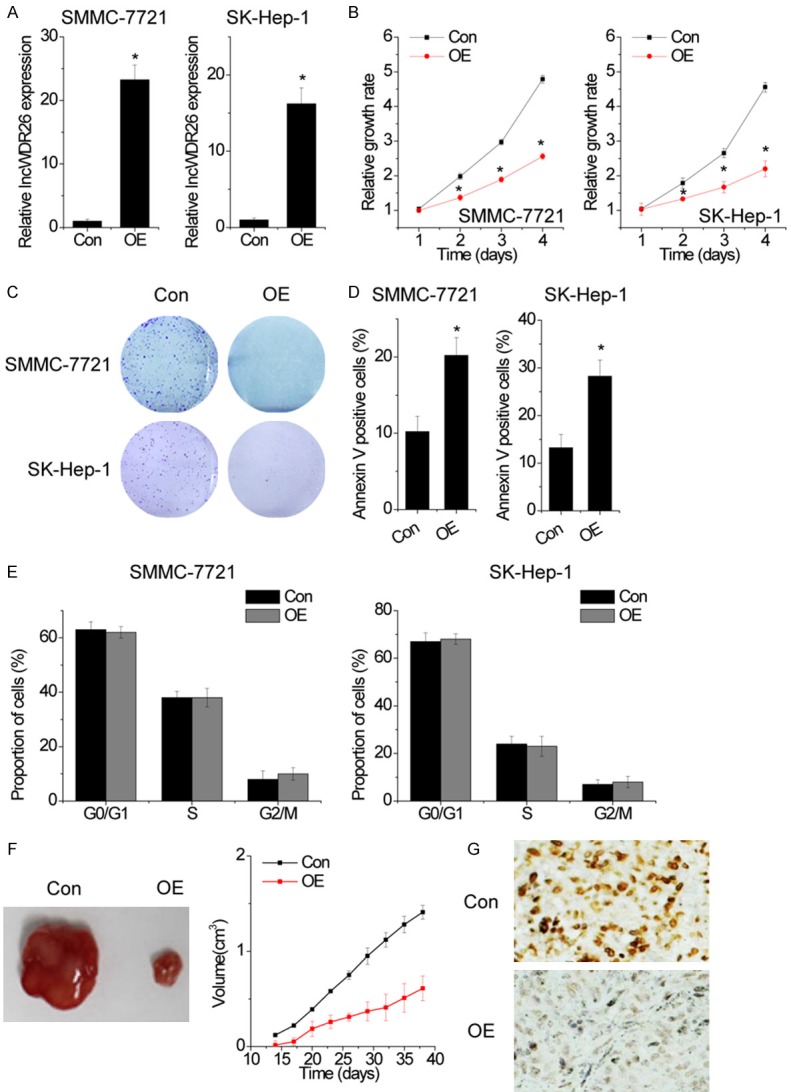
Overexpression of lncWDR26 inhibits the growth of HCC cells. A. HCC cell lines stably overexpressing lncWDR26 were established. The relative expression of lncWDR26 was detected by qRT-PCR. B. The cell proliferation of control and lncWDR26-overexpressing HCC cells was analyzed by CCK-8 assay. C. The effect of lncWDR26 overexpression on colony formation capacity of HCC cells. D. Cells with lncWDR26 overexpression were stained with a combination of annexin V and 7-AAD and analyzed by FACS. Cells positive for annexin V staining were counted as apoptotic cells, and the percentage of apoptotic cells is shown. E. FACS analysis showing no significant changes in cell cycle distribution, in HCC cells overexpressing lncWDR26. F. Effects of lncWDR26 overexpression on tumor growth in vivo. The tumor growth curves are shown. G. Immunohistochemical staining of Ki-67 in xenograft tumor tissues. Data are shown as mean ± standard deviation (SD); *P<0.05.
Next, we injected lncWDR26-overexpressing SMMC-7721 cells or control cells into nude mice to determine whether lncWDR26 could influence HCC tumorigenesis in vivo. The results showed that tumors grown from lncWDR26-overexpressing cells were smaller than those grown from control cells (Figure 3F). Moreover, immunohistochemical analysis confirmed that the tumors formed from lncWDR26-overexpressing SMMC-7721 cells displayed lower Ki-67 staining (Figure 3G). Taken together, our findings demonstrated that lncWDR26 inhibits HCC growth in vitro and in vivo.
Upregulation of lncWDR26 suppresses the metastasis of HCC cells
To evaluate whether lncWDR26 contributes to the progression of HCC, we examined the effect of lncWDR26 on the migratory and invasive behavior of SMMC-7721 and SK-Hep-1 cells. Using transwell assay, we found that the migratory and invasive ability of HCC cells was dramatically inhibited following overexpression of lncWDR26 (Figure 4A).
Figure 4.
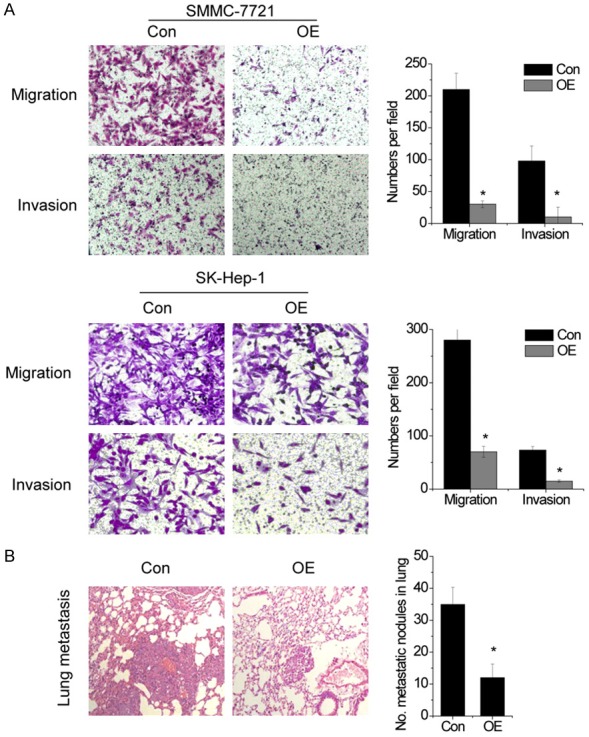
Upregulation of lncWDR26 suppresses the metastasis of HCC cells. A. The effect of lncWDR26 on HCC cell migration and invasion was determined by transwell assay. B. Left: representative hematoxylin and eosin staining of pulmonary metastatic nodules is shown. Right: number of metastatic nodules in the lung of mice. Data are shown as mean ± SD; *P<0.05.
To validate the in vitro results, control and lncWDR26-overexpressing SMMC-7721 cells were injected into the tail vein of nude mice. Overexpression of lncWDR26 resulted in a reduction of metastatic nodules in the mice lungs, as compared with control group (Figure 4B). These results indicate that lncWDR26 is also involved in human HCC progression through affecting cancer cell invasion and metastasis.
lncWDR26 downregulates WDR26 expression
Next, we explored the molecular mechanisms by which lncWDR26 suppressed the growth and metastasis of HCC cells. We detected the expression of the neighboring gene WDR26 in control and lncWDR26-overexpressing HCC cells. Interestingly, lncWDR26 overexpression significantly decreased both mRNA and protein expression of WDR26 (Figure 5A and 5B). We detected the expression levels of WDR26 in the same set of 82 paires HCC tumors and adjacent normal liver tissues by qRT-PCR. We found that WDR26 expression was significantly increased in HCC tissues compared to adjacent normal tissues (Figure 5C). Furthermore, we observed that lncWDR26 expression was negatively correlated with WDR26 expression (r = -0.408, P = 0.005, Figure 5D).
Figure 5.
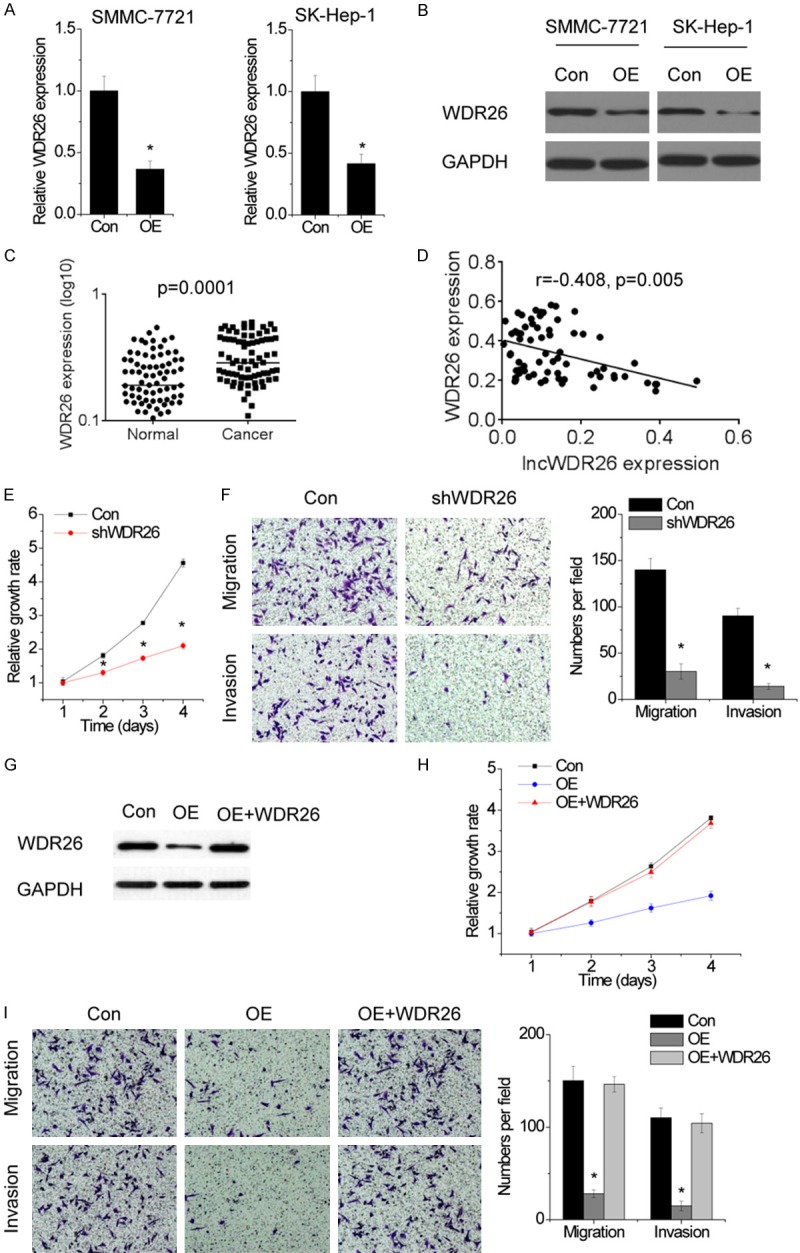
lncWDR26 downregulates WDR26 expression. A. The relative expression of WDR26 in control and lncWDR26-overexpressing cells was detected by qRT-PCR. B. The effect of lncWDR26 overexpression on WDR26 protein level was determined by western blotting. C. The expression levels of WDR26 in 82 pairs of HCC and matched normal liver tissues were examined by qRT-PCR. D. The correlation between lncWDR26 and WDR26 expression. E. The proliferation of control and WDR26-silenced SMMC-7721 cells was analyzed by CCK-8 assay. F. The cell migration and invasion of control and WDR26-silenced SMMC-7721 cells was examined by transwell assay. G. lncWDR26-overexpressing cells were transfected with WDR26. H. Upregulation of WDR26 rescued the suppression of proliferation induced by lncWDR26 overexpression. I. Upregulation of WDR26 rescued the suppression of migration and invasion induced by lncWDR26 overexpression. Data are shown as mean ± SD; *P<0.05.
We silenced WDR26 expression in HCC cells and observed that WDR26 knockdown dramatically suppressed the proliferation (Figure 5E), migration, and invasion of HCC cells (Figure 5F), which was similar to the phenotypes induced by lncWDR26 overexpression. To determine whether lncWDR26 functions upstream of WDR26 in the regulation of malignant phenotypes in HCC, we restored WDR26 expression in lncWDR26-overexpressing SMMC-7721 cells (Figure 5G). Interestingly, upregulation of WDR26 rescued proliferation, migration, and invasion of HCC cells reduced by lncWDR26 overexpression (Figure 5H and 5I). These data suggest that lncWDR26 regulates HCC growth and metastasis through suppression of WDR26.
lncWDR26 interacts with SIX3 to regulate WDR26 transcription
lncRNAs are considered to exert their functions through RNA-interacting proteins that regulate gene expression by various mechanisms. Therefore, we performed an RNA pull-down assay with biotin-labeled lncWDR26 to search for potential lncWDR26-associated proteins. SIX3 was identified to potentially interact with lncWDR26 in HCC cells (Figure 6A). The interaction of lncWDR26 with SIX3 was further validated by RIP assay (Figure 6B). Next, we constructed a series of lncWDR26 truncations to map its binding fragment with SIX3. We found that the 3’-end fragment of lncWDR26 (289 to 450 nt) was essential for binding SIX3 (Figure 6C). However, lncWDR26 overexpression did not influence the protein level of SIX3 (Figure 6D), suggesting that lncWDR26 was not involved in the post-translational regulation of SIX3. Given that SIX3 regulates gene transcription via binding to promoter region, we then evaluated whether lncWDR26 influenced the occupancy of SIX3 on the WDR26 promoter by performing ChIP assay. We found that SIX3 binds to WDR26 promoter region and lncWDR26 overexpression increased its binding level (Figure 6E). In addition, SIX3 depletion restored WDR26 expression reduced by lncWDR26 overexpression (Figure 6F and 6G). Taken together, our data demonstrated that lncWDR26 transcriptionally suppresses WDR26 expression through recruitment of SIX3.
Figure 6.
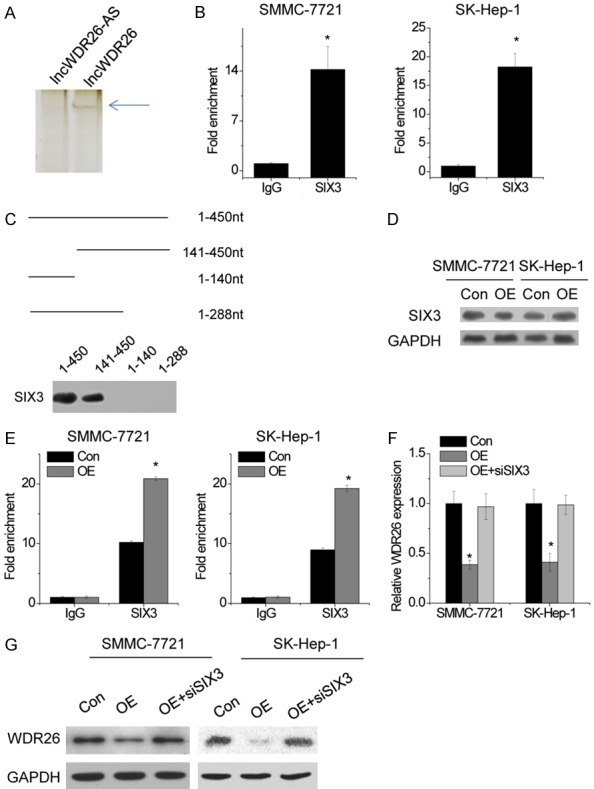
lncWDR26 interacts with SIX3 to regulate WDR26 transcription. A. lncWDR26 antisense (lncWDR26-AS) and lncWDR26 transcripts were labeled with biotin and incubated with SMMC-7721 lysates, followed by silver staining and mass spectrometry. B. The interaction between lncWDR26 and SIX3 was verified by an RIP assay. C. Mapping analysis of SIX3-binding domains of lncWDR26. A schematic diagram of full-length and truncated fragments of lncWDR26 and western blot data of SIX3 in RNA pull-down samples by different lncWDR26 fragments. D. The protein level of SIX3 was analyzed by western blotting in control and lncWDR26-overexpressing HCC cells. E. The binding level of SIX3 at WDR26 promoter region was determined by ChIP assay, followed by qRT-PCR in control and lncWDR26-overexpressing HCC cells. F. Knockdown of SIX3 abolished the decrease in WDR26 mRNA expression induced by lncWDR26 overexpression. G. Downregulation of SIX3 rescued the WDR26 protein level reduced by lncWDR26 overexpression. Data are shown as mean ± SD; *P<0.05.
Discussion
lncRNAs have emerged as critical players in tumorigenesis and cancer progression. However, the potential functions and mechanisms of most lncRNAs in human HCC cells remain unclear. Here, we found that lncWDR26 expression was significantly decreased in HCC tissues and cells. Reduced lncWDR26 expression was associated with poor prognosis in patients with HCC. Using gain-of-function assays, we showed that lncWDR26 negatively regulated HCC cell growth and metastasis in vitro and in vivo. These findings suggest that lncWDR26 functions as a tumor suppressor in HCC, and its underexpression contributes to HCC tumorigenesis and progression.
Evidence from previous studies shows that lncRNAs regulate target gene expression through diverse mechanisms in different cancer cells. Using RIP and RNA pull-down assays, we showed that lncWDR26 directly interacts with the transcription factor, SIX3. Importantly, ChIP assay demonstrated that lncWDR26 could simultaneously recruit SIX3 to WDR26 promoter regions and repress WDR26 transcription. SIX3 acts as a tumor suppressor in some cancers, including astrocytoma and lung adenocarcinoma. SIX3 downregulated a number of genes involved in proliferation and metastasis, such as AURKA/B, S100P, TGFB3, GINS3, and BAG1 [18,19]. Here, we also showed that WDR26 was a direct target gene of SIX3. WDR26 is a member of the WD repeat protein family. Members of this family are involved in a variety of cellular processes, including cell cycle progression, signal transduction, apoptosis, and gene regulation [20]. A previous study suggested that WDR26 contributes to the mitogen-activated protein kinase (MAPK) signaling pathway [21]. In a study of chemo-attractive migration of leukocytes, WDR26 binds to the Gβ-gamma complex and positively controls leukocyte migration [22,23]. A recent study also reported that WDR26 is a negative regulator of the canonical Wnt signaling pathway, and that WDR26 affects β-catenin levels [24]. However, the expression and function of WDR26 in HCC progression remain unclear. In the present study, we demonstrated that WDR26 promotes cellular proliferation, migration, and invasion. Furthermore, our results showed that WDR26 expression is significantly upregulated in HCC tissues compared to that in non-tumor liver tissues. We also observed a negative correlation between lncWDR26 and WDR26 expression in HCC tissues. Interestingly, rescue experiments showed that the tumor-suppressive function of lncWDR26 depends on the repression of WDR26 expression.
In summary, our study is the first to show that the lncRNA, lncWDR26, is downregulated in HCC tissues, is associated with poor prognosis, and may be a positive prognostic factor for patients with HCC. Its effects on HCC growth and metastasis indicate that it exhibits tumor suppressor function in HCC tumorigenesis and progression. lncWDR26 exerts its function through inhibition of WDR26 transcription via association with SIX3. Our findings further the understanding of HCC pathogenesis and facilitate the development of lncRNA-directed diagnostics and therapeutics against HCC.
Disclosure of conflict of interest
None.
References
- 1.Fujiwara N, Friedman SL, Goossens N, Hoshida Y. J Hepatol. 2017 doi: 10.1016/j.jhep.2017.09.016. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Xu J, Li J, Zheng TH, Bai L, Liu ZJ. MicroRNAs in the occurrence and development of primary hepatocellular carcinoma. Adv Clin Exp Med. 2016;25:971–975. doi: 10.17219/acem/36460. [DOI] [PubMed] [Google Scholar]
- 4.Ma Z, Huang H, Xu Y, He X, Wang J, Hui B, Ji H, Zhou J, Wang K. Current advances of long non-coding RNA highly upregulated in liver cancer in human tumors. Onco Targets Ther. 2017;10:4711–4717. doi: 10.2147/OTT.S136915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun W, Yang Y, Xu C, Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017;216-217:105–110. doi: 10.1016/j.cancergen.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Fu LL, Li CJ, Xu Y, Li LY, Zhou X, Li DD, Chen SX, Wang FG, Zhang XY, Zheng LW. Role of lncRNAs as novel biomarkers and therapeutic targets in ovarian cancer. Crit Rev Eukaryot Gene Expr. 2017;27:183–195. doi: 10.1615/CritRevEukaryotGeneExpr.2017019244. [DOI] [PubMed] [Google Scholar]
- 7.Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927–1933. doi: 10.1111/cas.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misawa A, Takayama KI, Inoue S. Long non-coding RNAs and prostate cancer. Cancer Sci. 2017;108:2107–2114. doi: 10.1111/cas.13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, Sugimachi K, Mimori K, Wakabayashi G, Mori M. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 11.Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, Wu D, Liu L. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148:415–426. e418. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Liu D, Zhu Y, Pang J, Weng X, Feng X, Guo Y. Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J Cell Biochem. 2017 doi: 10.1002/jcb.26297. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.He JH, Han ZP, Liu JM, Zhou JB, Zou MX, Lv YB, Li YG, Cao MR. Overexpression of long non-coding RNA MEG3 inhibits proliferation of hepatocellular carcinoma Huh7 cells via negative modulation of miRNA-664. J Cell Biochem. 2017;118:3713–3721. doi: 10.1002/jcb.26018. [DOI] [PubMed] [Google Scholar]
- 14.Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y, Yang X, Shen J, Liu Q, Zhang J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37:2691–2702. doi: 10.1007/s13277-015-4111-x. [DOI] [PubMed] [Google Scholar]
- 15.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, Jean S, Li C, Huang Q, Katsaros D, Montone KT, Tanyi JL, Lu Y, Boyd J, Nathanson KL, Li H, Mills GB, Zhang L. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 17.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Z, Sun Y, She X, Wang Z, Chen S, Deng Z, Zhang Y, Liu Q, Liu Q, Zhao C, Li P, Liu C, Feng J, Fu H, Li G, Wu M. SIX3, a tumor suppressor, inhibits astrocytoma tumorigenesis by transcriptional repression of AURKA/B. J Hematol Oncol. 2017;10:115. doi: 10.1186/s13045-017-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo ML, Okamoto J, Chen Z, Hirata T, Mikami I, Bosco-Clement G, Li H, Zhou HM, Jablons DM, He B. Down-regulation of SIX3 is associated with clinical outcome in lung adenocarcinoma. PLoS One. 2013;8:e71816. doi: 10.1371/journal.pone.0071816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis O, Han F, Adams JC. Molecular phylogeny of a RING E3 ubiquitin ligase, conserved in eukaryotic cells and dominated by homologous components, the muskelin/RanBPM/CTLH complex. PLoS One. 2013;8:e75217. doi: 10.1371/journal.pone.0075217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, Wang Y, Xia C, Li D, Li Y, Zeng W, Yuan W, Liu H, Zhu C, Wu X, Liu M. WDR26: a novel Gbeta-like protein, suppresses MAPK signaling pathway. J Cell Biochem. 2004;93:579–587. doi: 10.1002/jcb.20175. [DOI] [PubMed] [Google Scholar]
- 22.Sun Z, Tang X, Lin F, Chen S. The WD40 repeat protein WDR26 binds Gbetagamma and promotes Gbetagamma-dependent signal transduction and leukocyte migration. J Biol Chem. 2011;286:43902–43912. doi: 10.1074/jbc.M111.301382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Runne C, Chen S. WD40-repeat proteins control the flow of Gbetagamma signaling for directional cell migration. Cell Adh Migr. 2013;7:214–218. doi: 10.4161/cam.22940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goto T, Matsuzawa J, Iemura S, Natsume T, Shibuya H. WDR26 is a new partner of Axin1 in the canonical Wnt signaling pathway. FEBS Lett. 2016;590:1291–1303. doi: 10.1002/1873-3468.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]


