Abstract
Mounting evidence shows that the long non-coding RNA MALAT1 plays a pivotal role in tumorigenesis and metastasis, but the functional significance of MALAT1 in bladder transitional cell carcinoma (BTCC) remains unclear. MALAT1 expression was measured in 56 BTCC patients and 2 BTCC cell lines by real-time PCR. The effects of MALAT1 on BTCC cells were investigated by over-expression approaches in vitro and in vivo. Insights of the mechanism of competitive endogenous RNAs (ceRNAs) were validated through bioinformatic analysis and luciferase assay. MALAT1 up-regulation positively correlated with advanced clinical pathological stage and shorter survival of BTCC patients. Furthermore, MALAT1 over-expression promoted proliferation, migration and invasion of BTCC cells in vitro and in vivo. Particularly, MALAT1 may function as a ceRNA to sponge miR-124, thus modulating the derepression of foxq1, miR-124 target gene, in post-transcriptional levels. The positive MALAT1/foxq1 interaction was confirmed by bivariate correlation analysis, and this positive correlation was of great significance in BTCC tumor growth and metastasis, also accompanied by EMT changes. Overall, this ceRNA regulatory network concerning MALAT1 and the positive MALAT1/foxq1 correlation benefit a better understanding of BTCC pathogenesis and promote the feasibility of lncRNA-directed therapy against this disease.
Keywords: Bladder transitional cell carcinoma (BTCC), MALAT1, competing endogenous RNA(ceRNA), foxq1, proliferation and invasion
Introduction
Bladder cancer ranks the first genitourinary system malignancy in China, with the overwhelming majority being bladder transitional cell carcinomas (BTCC) [1,2]. BTCC is the product of a complicated process associated with genetic and epigenetic abnormalities [3-5]. The biological behavior of this disease exhibits various patterns, for instance, easy relapse, and multiple metastasis. Surgical resection proves to be the most effective treatment for non-invasive BTCC patients. Chemotherapy based on BCG lowers recurrence in patients, but roughly 70% relapse and 25% ultimately progress to distant metastasis [6]. Current strategies to this disease are insufficient and more therapeutic strategies are urgently needed. Probing molecular triggers associated with BTCC diagnosis, progression, metastasis, and prognosis holds potential for BTCC therapy.
In addition to well-known genetic changes in response to regulatory pathways, such as cyclooxygenase (COX-2) [7], β-catenin [8], transforming growth factor (TGF)-β [9], recent researches identified other participators in BTCC progression, including small non-coding RNAs (typically microRNAs) [10] and long non-coding RNAs (lncRNAs) [11]. MicroRNAs (miRNAs) are small ncRNAs that combine to the 3’-untranslated region (3’-UTR) of its target gene to down-regulate mRNAs expression [12,13]. There is a large amount of evidence elucidating the role of miRNAs working as oncogenes or tumor suppressors in tumor progression [13,14]. The functions of lncRNAs in cancers have also been well documented. It has been shown that numerous lncRNAs, such as GAS5 [11], H19 [15], PVT1 [16], HOTAIR [17], etc., were dysregulated in cancers, and associated with tumor progression, opening up a new avenue for exploring tumorigenesis. It should be noted that, lncRNAs can also act as competing endogenous RNAs (ceRNAs), namely miRNA sponge, inhibit miRNAs expression and activities to modulate the derepression of miRNA targets [18,19]. A cardiac hypertrophy-related lncRNA, CHRF, has been reported to serve as an endogenous sponge of miR-489, directly targets miR-489 and modulates its target gene, Myd88 [20]. HOTAIR works as a ceRNA to sink miR-331-3p expression, and regulate the derepression of HER2, miR-331-3p target gene, in gastric cancer [21]. We propose that some lncRNAs may also act as ceRNAs, connecting miRNAs and the post-transcriptional network in bladder pathogenesis.
lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is located on chromosome 11q13, and this region is generally known to be relevant for tumorigenesis and metastasis [22,23]. MALAT1 initially came to light for its detection in early-stage non-small cell lung cancer (NSCLC) samples, wherein MALAT1 promoted the development of NSCLC distant metastasis [24]. Furthermore, MALAT1 up-regulation positively correlates with malignant invasion and poor ending in colorectal cancer, renal cell carcinoma, gastric cancer, etc. [25-28]. Intriguingly, TGF-β-induced up-regulation of MALAT1 promotes bladder cancer metastasis, along with the down-regulation of E-cadherin and up-regulation of N-cadherin [29]. However, the biological significance and molecular mechanism in which MALAT1 functions in BTCC is far from being fully elucidated, although the vast majority of bladder cancer is BTCC.
In this study, we investigated the functional role of MALAT1 in BTCC. We examined the expression of MALAT1 in human BTCC tissues and cell lines, and explored its effects on cell growth, migration and invasion. Its effects on BTCC tumorigenesis and metastasis were also investigated using murine models. Finally, the underlying molecular mechanism of MALAT1 functions in BTCC was explored. This study offers us a better understanding of BTCC pathogenesis, which may benefit BTCC therapy.
Materials and methods
Clinical tissue samples
Bladder tissue samples were obtained from 56 patients (mean age 63 years, range 45-78) with urothelial bladder cancer in The First Affiliated Hospital of Zhengzhou University. Informed consent was obtained from all patients before this study. For each sample, the cancer tissue and its remote normal mucosa were employed and compared. All samples were pathologically confirmed as BTCC by 2 independent pathologists. The TNM classification was in accordance with the 7th edition of the UICC-American Joint Committee on cancer staging. The research protocols were approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Cell culture
The human bladder cancer cell line T24, BIU-87, and human embryo kidney 293T cells (HEK 293T) were supplied by the Institute of Cell Research, Chinese Academy of Sciences, Shanghai, China. All cells were cultured in RPMI-1640 medium (HyClone, Thermo scientific Inc, China) with 10% fetal bovine serum (FBS, HyClone, Thermo scientific Inc, China) at 37°C under 5% CO2.
RNA isolation, reverse transcription, and real-time PCR
Total RNA was extracted from the samples or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol, then reversely transcripted to cDNA in a first-strand cDNA synthesis reaction with PrimeScript RT-PCR kit (Takara Biotechnology Dalian, China), at 37°C for 25 min, then incubated at 85°C for 5 sec in 20 μl of reaction volume. Real-time PCR was carried out using All-in-OneTM qPCR Mix (Applied GeneCopoeia Inc., USA) on an ABI 7500HT System, as described elsewhere [30]. MiR-124 was normalized to snRNA U6. GAPDH was employed as an internal control for MALAT1 and foxq1. Expression fold changes were calculated using 2-ΔΔCt methods [31].
Oligonucleotide transfection
MALAT1 siRNA/Control, miR-124 siRNA/Control, foxq1 siRNA/Control were synthesized from Gene-pharma (Shanghai, China). RNA oligonucleotides were transfected into T24 and BIU-87 cells using lipofectamine2000 reagent (Invitrogen, Carlsbad, CA, USA).
Plasmid construction and stable transfection
To obtain stable cell lines overexpressing MALAT1, pre-MALAT1 was cloned into the pLVTHM lentiviral vector, and the recombinant plasmid was named as MALAT1. The lentivial vectors and packing system were co-transfected in HEK293T cells, and then lentiviral virus was employed to infect the T24 and BIU-87 cell lines.
The lentiviral viruses overexpressing miR-214 were purchased from Genechem (Shanghai, China). Then they were employed to infect T24 and BIU-87 cell lines.
The 3’-UTR of Foxq1 was cloned into the psicheck. 2 vector (Promega, Madison, WI, USA). The mutation of the 3’-UTR of Foxq1 was performed using site-directed mutagenesis kit (Invitrogen, Carlsbad, CA, USA). The resulted vectors were called Foxq1-wildtype (WT) and Foxq1-mutant (MUT), respectively.
The 3’-UTR of MALAT1 was cloned into the psiCHECK-2 vector (Promega, Madison, WI, USA). The mutation of the 3’-UTR of MALAT1 was produced using site-directed mutagenesis kit (Invitrogen, Carlsbad, CA, USA). The resulted vectors were called MALAT1-wildtype (WT) and MALAT1-mutant (MUT), respectively.
Bioinformatics analysis
Binding sites between miRNAs and MALAT1 were predicted with miRanda. The potential target genes of miRNA were predicted using three microRNA target database (PicTar, TargetScan and PITA), and the selected targets gene were validated by real-time PCR and western Blot.
Dual-luciferase assay
T24 cells were cultured in 24-well plates for dual-luciferase report system. Cells were co-transfected with Wild or Mutant type reporter plasmid and miRNA/Ctrl using lipofectamine 2000 reagent (Invitrogen). 24 hours later, cells were harvested to measure the firefly and Renilla luciferase activities using the Dual-Glo luciferase reporter assay kit (Promega, Madison, WI, USA).
Cell proliferation assay
Cell viability was assessed as reported before [32]. Cells at a concentration of 1 × 103 cells/well with five replicate wells were seeded in 96-well plates and cultured for 24 h, 48 h, 72 h, 96 h, respectively. Cell viability was determined using a Cell counting kit-8 (Beyotime, Shanghai, China), according to the manufacturer’s instructions.
Migration and invasion assays in vitro
Transwell migration assay and invasion assay were employed to determine the effects of MALAT1 on cell migration and invasion as described previously [33].
Tumor xenograft model
4-5 week-old female BALB/C nude mice (n=24) were purchased from Centre of Laboratory Animal of Zhengzhou University, and the animal protocol was approved by Institutional Animal Care and Use Committee of Zhengzhou University. Mice were randomly divided into two groups (n=12/group). 1 × 105 MALAT1/T24 or Control/T24 cells were injected subcutaneously into each mouse. Tumor size was measured every other week. 6 weeks later, mice were sacrificed and tumors were dissected. H&E staining was performed for tumor metastasis assay in vivo.
Western blot
Western blotting was performed as reported preciously [34]. Briefly, protein lysates extracted from tissues or cells were separated by 10% SDS-PAGE, and transferred to PVDF (polyvinylidene difluoride) membrane (Millipore, Bedford, MA, USA) for 60 min at 100 V. Then, the membrane was incubated with goat polyclonal antibody against human foxq1 (WuXi PharmaTech Cayman China), followed by HRP (horseradish peroxidase)-labeled goat-anti mouse IgG (Santa Cruz Biotechnology) and detected with chemiluminescence (ECL) reagent (Amersham, Buckinghamshire, UK). GAPDH was used as a protein loading control.
Statistical analysis
The SPSS 15.0 software was employed for general statistical analysis. Data are expressed as the mean ± SEM. The significance of differences between groups was estimated by either Student’s t-test or one-way analysis of variance (ANOVA). Survival rate was analyzed using the Kaplan-Meier method. Relationship between MALAT1 and foxq1 was determined using Spearman’s correlation analysis. For all the tests, the criterion for statistical significance was taken as P < 0.05.
Results
MALAT1 and miR-124 are inversely expressed in BTCC
To examine the expression of MALAT1 in BTCC, tumor tissues and surrounding normal mucosa samples in 56 patients with BTCC were first employed and analyzed by real-time PCR. MALAT1 was significantly up-regulated in tumor tissues compared with normal tissues (n=56) (Figure 1A). Considering the predictable biding site between MALAT1 and miR-124 by bioinformatics analysis, we further tested the expression of miR-124 in tumor tissues and matched normal mucosa samples. In contrast, miR-124 was significantly down-regulated in tumor tissues compared with normal tissues (n=56) (Figure 1A). Besides, significantly higher levels of foxq1, putative target gene of miR-124, were found in tumor tissues with inferior expression of MALAT1, and there was significant positive correlation between MALAT1 and foxq1 expression in BTCC (Figure 1B). We further analyzed the expression patterns in tumor tissues according to clinical stage. High-stage carcinomas (N3) had higher MALAT1 and foxq1 mRNA expression and lower miR-124 mRNA expression than low-stage carcinomas (NO-N2), indicating significant correlations of MALAT1, miR-124, and foxq1 with BTCC clinical pathological stage (Figure 1C). Furthermore, MALAT1 and foxq1 were more abundant and miR-124 was weaker in highly invasive and mesenchymal-like bladder cancer T24 cells than none-invasive epithelial bladder cancer BIU-87 cells (Figure 1D). Kaplan-Meier survival curves revealed that patients with high levels of MALAT1 and foxq1, or weak levels of miR-124 demonstrated an unfavorable clinical outcome (Figure 1E).
Figure 1.
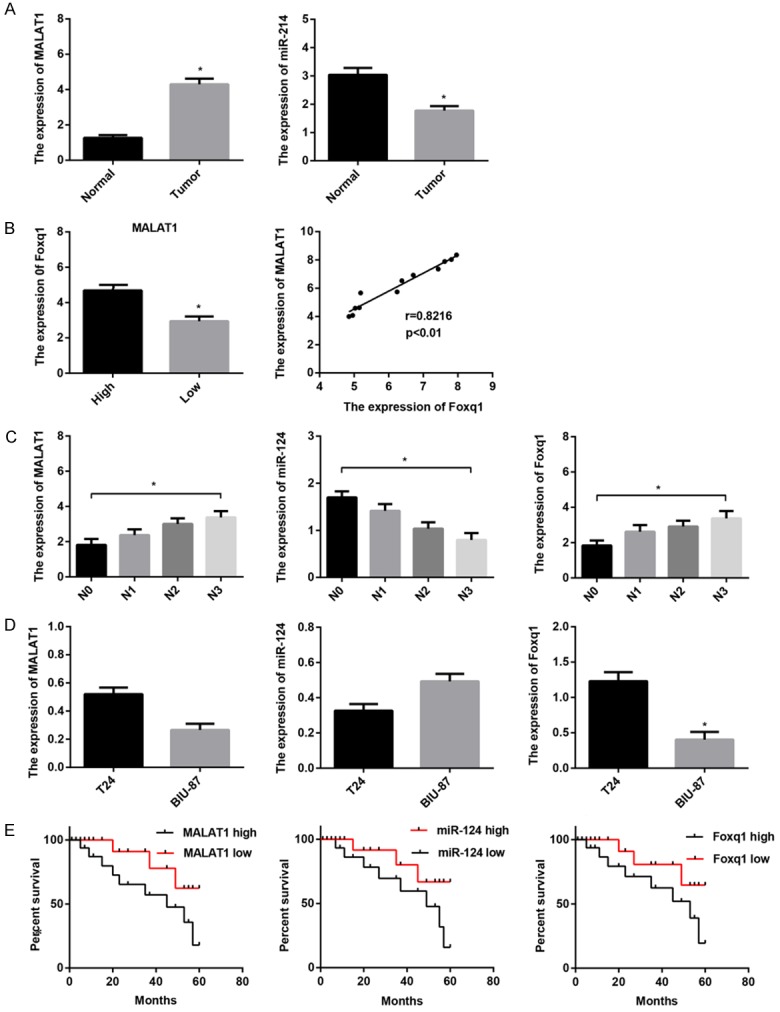
Expression of MALAT1, miR-124, foxq1 in BTCC. The relative expression was examined using real-time PCR in 56 patients with BTCC and 2 BTCC cell lines. Data are shown as mean ± SEM. A. MALAT1 was up-regulated in 56 tumor tissues compared with surrounding normal mucosa tissues; miR-124 was down-regulated in tumor tissues in comparison with matched normal mucosa tissues. B. MALAT1 and foxq1 were positively expressed in BTCC. C. MALAT1 and foxq1 expression positively correlated with BTCC clinical pathological stage; miR-124 expression inversely correlated with clinical pathological stage. D. MALAT1 and foxq1 were more abundant and miR-124 was weaker in T24 cells than BIU-87 cells. E. Kaplan-Meier survival curves revealed that patients with high levels of MALAT1 and foxq1, or weak levels of miR-124 exhibited an unfavorable clinical outcome. Patients were divided into MALAT1-high/low group; miR-124-high/low group; foxq1-high/low group based on the median of their expression in BTCC, respectively. *P < 0.05.
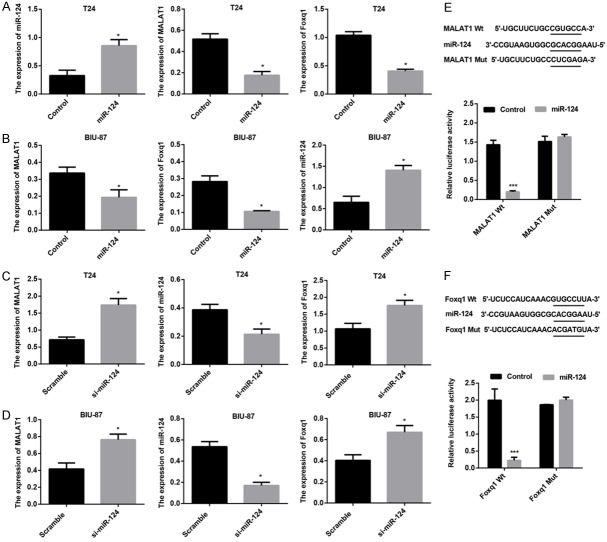
MiR-124 down-regulated the expression of MALAT1/foxq1 by directly targeting the 3’-UTR in BTCC cells
Bioinformatic algorithms have revealed that MALAT1 and foxq1 were putative targets of miR-124. To explore whether MALAT1 and foxq1 expression are regulated by miR-214, T24 and BIU-87 cells were transfected with miR-214 mimics or si-miR-214, and miR-124, MALAT1, foxq1 mRNA expression were measured. The results showed that forced miR-214 expression markedly elevated miR-214 mRNA expression, reduced MALAT1 and foxq1 mRNA expression in both T24 and BIU-87 cells (Figure 2A and 2B); while miR-214 knockdown resulted in a significant decrease in miR-214 mRNA expression and significant increases in MALAT1 and foxq1 mRNA expression in T24 and BIU-87 cells (Figure 2C and 2D). To further determine repression effect of miR-214 on MALAT1/foxq1 expression was associated with the predicted binding sites in 3’UTR region (Figure 2E and 2F), dual-luciferase assay was employed. Co-transfection of miR-214 mimic and MALAT1-wt-3’-UTR in T24 cells markedly down-regulated luciferase activity compared with the control cells, whereas luciferase activity was unaffected in T24 cells cotransfected with miR-214 mimic and MALAT1-mt-3’-UTR (Figure 2E). Similarly, the luciferase activity was dramatically inhibited in T24 cells co-transfected with miR-214 mimic and foxq1-wt-3’-UTR compared with the control cells, while no significant difference in luciferase activity was noted in T24 cells co-transfected with foxq1-mt-3’-UTR and miR-214 mimic (Figure 2F).
Figure 2.
MALAT1 and foxq1 are direct targets of miR-124. Real-time PCR analysis of MALAT1, miR-124, and foxq1 mRNA expression in T24 cells (A) and BIU-87 cells (B) transfacted with Control, miR-124 mimics, respectively. Real-time PCR analysis of MALAT1, miR-124, and foxq1 mRNA expression in T24 cells (C) and BIU-87 cells (D) transfacted with Scramble, si-miR-124, respectively. (E) The putative miR-124-binding site in 3’UTR region of MALAT1. The relative luciferase activity was suppressed in cells transfected with the reporter vector MALAT1-WT, not in cells transfected with MALAT1-MUT. (F) The putative miR-124-binding site in 3’UTR region of foxq1. The relative luciferase activity was down-regulated in cells transfected with the reporter vector foxq1-WT, not in cells transfected with foxq1-MUT. Data are shown as mean ± SEM, *P < 0.05, ***P < 0.001.
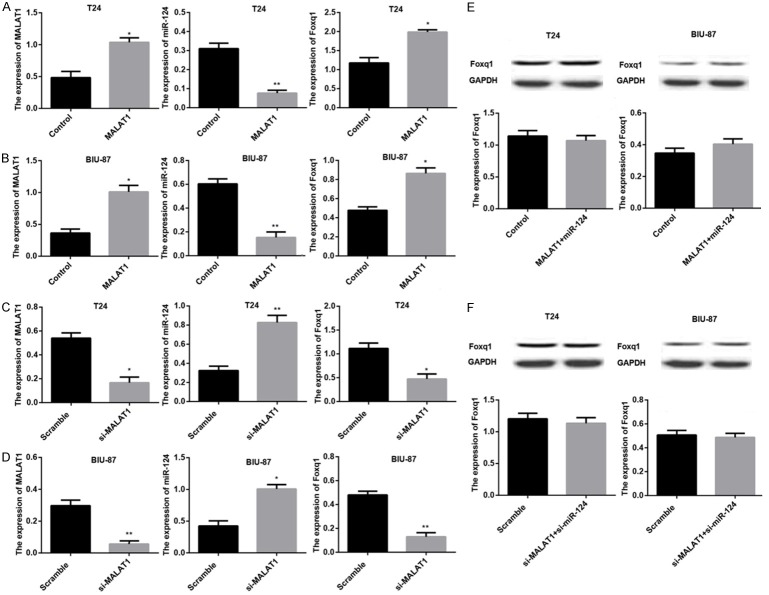
MALAT1 regulates miR-124/foxq1 cascade expression in BTCC cells
It has been well documented that lncRNAs can act as ceRNAs, inhibit miRNAs expression to modulate the derepression of miRNA targets. Based on the results in Figure 2, we wondered whether MALAT1 could work as miR-214 sponge to inhibit miR-214 expression and regulate the derepression of miR-214 target gene, foxq1. The results showed that ectopic MALAT1 expression remarkably raised MALAT1 and foxq1 mRNA expression, down-regulated miR-214 mRNA expression in both T24 and BIU-87 cells (Figure 3A and 3B); while knocking down MALAT1 led to significant decreases in MALAT1 and foxq1 mRNA expression, accompanied by a significant increase in miR-214 mRNA expression in T24 and BIU-87 cells (Figure 3C and 3D). MiR-124 mimic restored foxq1 expression at mRNA and protein levels in T24 and BIU-87 cells both overexpressing MALAT1 (Figure 3E). The similar results were observed in T24 and BIU-87 cells co-transfected with si-miR-124 and si-MALAT1 (Figure 3F).
Figure 3.
MALAT1 regulates miR-124/foxq1 cascade expression in BTCC cells. The relative expression of MALAT1, miR-124, and foxq1 was examined using real-time PCR and western blot in T24 cells and BIU-87 cells. Data are shown as mean ± SEM. Real-time PCR analysis of MALAT1, miR-124, and foxq1 mRNA expression in T24 cells (A) and BIU-87 cells (B) transfacted with Control, MALAT1, respectively. Real-time PCR analysis of MALAT1, miR-124, and foxq1 mRNA expression in T24 cells (C) and BIU-87 cells (D) transfacted with Scramble, si-MALAT1, respectively. (E) Western blot analysis of foxq1 expression in T24 and BIU-87 cells both transfacted with Control, MALAT1+miR-124, respectively. (F) Western blot analysis of foxq1 expression in T24 and BIU-87 cells both transfacted with Scramble, si-miR-124+si-MALAT1, respectively. *P < 0.05.
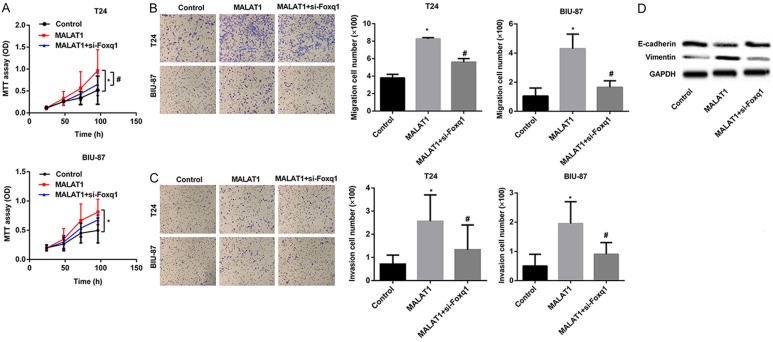
Foxq1 deletion partly inverses the changes caused by MALAT1 in vitro
Figure 3 illustrates the enhancing effect of MALAT1 on foxq1 expression, to further determine the relationship between MALAT1 and foxq1 in BTCC progression, we transfected T24 and BIU-87 cells with MALAT1 or MALAT1+si-foxq1. CCK-8 array was first utilized to examine cell viability. The results showed that forced expression of MALAT1 dramatically promoted cell growth of T24 and BIU-87 cells, whereas foxq1 gene silencing partly abolished T24 and BIU-87 proliferation induced by MALAT1 (Figure 4A). After analyzing the proliferation ability of MALAT1, we tested cell migration and invasion capacities. Migration and invasion arrays demonstrated that MALAT1 enhanced cell migration and invasion, while foxq1 deletion partly eliminated the increased migration and invasion cells induced by MALAT1 in T24 and BIU-87 cells (Figure 4B and 4C). Considering that tumor metastasis always accompanied by epithelial-mesenchymal transition (EMT), to determine whether silencing foxq1 inhibited T24 and BIU-87 motility induced by MALAT1 was concerned with EMT, we examined the expression of epithelial marker E-cadherin and mesenchymal marker Vimentin using western blot in BIU-87 cells. Obviously, up-regulated E-cadherin concomitant with down-regulated Vimentin, as determined at protein levels, were obtained in MALAT1+si-foxq1 cells, compared with MALAT1 cells (Figure 4D).
Figure 4.
Foxq1 deletion partly inverses the changes caused by MALAT1 in vitro. (A) T24 and BIU-87 cells were both transfected with MALAT1 or MALAT1+si-foxq1. Effects of down-regulated foxq1 in MALAT1 overexpressing cells on cell proliferation were measured. Effects of silencing foxq1 in MALAT1 overexpressing cells on cell migration (B) and invasion (C) were investigated. (D) Western blot analysis of foxq1, E-cadherin, and Vimentin expression in BIU-87 cells. Data are shown as mean ± SEM. *P < 0.05, Control vs. MALAT1; #P < 0.05, MALAT1 vs. MALAT1+si-foxq1.
MALAT1 facilitates tumor growth and metastasis in vivo
We finally implanted MALAT1/T24 or Control/T24 cells subcutaneously in nude mice (n=12/group) to investigate the effects of MALAT1 overexpression on tumor growth in vivo. As shown in Figure 5A, tumor volume was measured every other week, and MALAT1 overexpression contributed to a dramatic increase in tumor volume relative to Control cells after 6 weeks. Subsequently, tumors were removed and weighed, tumor weight gain was notably observed in nude mice implanting with MALAT1/T24 cells (Figure 5A). To further explore the metastatic effects of MALAT1 in vivo, livers and lungs were dissected for histology analysis through H&E staining. Mice injecting with MALAT1/T24 cells exhibited more metastases to livers and lungs than mice owning Control cells (Figure 5B). Besides, MALAT1/T24 cells resulted in amazing decline in miR-124 mRNA expression, along with significant up-regulation of foxq1 and Vimentin, and down-regulation of E-cadherin at protein levels in resected tumors (Figure 5C).
Figure 5.
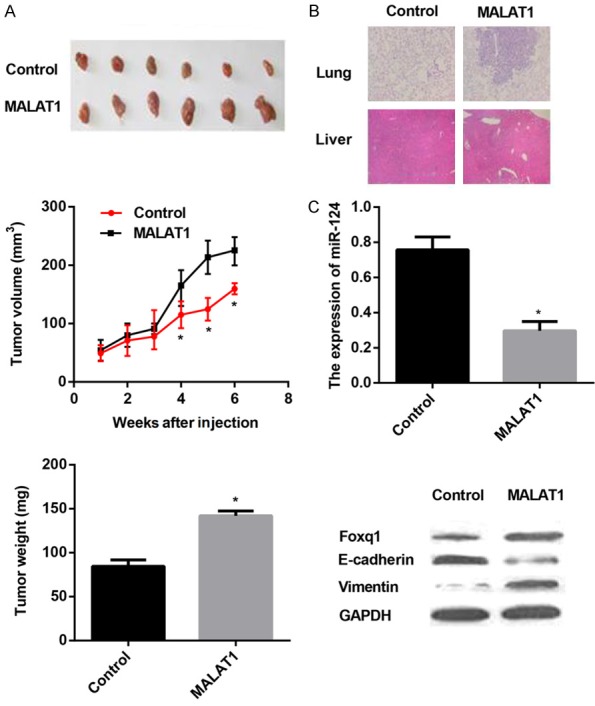
MALAT1 promotes tumor growth and metastasis in vivo. MALAT1/T24 or Control/T24 cells were subcutaneously implanted into nude mice (n=12/group). A. Tumor volume was calculated every one week. 6 weeks later, tumors were removed and tumor weight was measured. B. Livers and lungs were dissected for histology analysis to evaluate distant metastasis. C. Real-time PCR analysis of miR-124 expression and western blot analysis of foxq1, E-cadherin, and Vimentin expression in tissues of resected tumors. Data are presented as mean ± SEM. *P < 0.05.
Discussion
LncRNAs are non-protein coding transcribed RNAs with more than 200 nucleotides, but they can regulate protein-coding genes at both transcriptional and post-transcriptional levels and play key roles in various biological processes. Several recent papers have indicated that lncRNAs dysregulation may influence the regulation of the eukaryotic genome and benefit cellular growth, resulting in progressive tumor growth [35,36]. Hence, lncRNAs may open a missing field differing from other common oncogenic and tumor suppressor networks.
In the present study, we investigated the expression of MALAT1 in BTCC tssues and their surrounding normal mucosa tissues. We also identified the functional role of MALAT1 in BTCC cells by employing gain-of-function approaches. The results showed that MALAT1 was up-regulated in BTCC tissues in comparison with surrounding normal mucosa tissues, and that MALAT1 up-regulation correlated with advanced clinical-pathological stage. Moreover, the survival time of patients with lower MALAT1 expression was significantly longer than that of patients with strong MALAT1 expression. Furthermore, forced expression of MALAT1 facilitated the proliferation, migration and invasion of BTCC cells, both in vitro and in vivo. These results suggest that MALAT1 plays an oncogenic role in BTCC progression, considering MALAT1 as a novel prognostic parameter and therapeutic target in BTCC.
Enlightened by the ceRNAs regulatory network and emerging evidence that suggests that lncRNAs may take part in this regulatory pathway, our hypothesis was that MALAT1 may also act as a ceRNA to sink miRNA expression. In support of this notion, bioinformatics analysis revealed the putative binding site between MALAT1 and miR-124. Mounting evidence indicates that miR-124 is dysregulated in human cancers. So far, several reports have shown that miR-124 was significantly down-regulated, exerted tumor suppressive functions, and correlated with clinical characteristic and prognosis in breast cancer, colorectal cancer, bladder cancer, etc. [37-39]. luciferase assay was further employed to validate the predicted binding site between miR-124 and MALAT1. As expected, we discovered that miR-124 formed complementary base pairing with MALAT1 and induced expression repression of MALAT1 in T24 cells. Real-time PCR analysis showed that miR-124 expression was inversely correlated with MALAT1 expression, also clinical stage in BTCC. Moreover, the survival time of patients with higher miR-124 expression was significantly longer than that of patients with weak miR-124 expression, contrary to the clinical outcomes of patients with different expression of MALAT1. Furthermore, MALAT1 also down-regulated miR-124 expression. Overall, these findings are line with our hypothesis and indicate that MALAT1 may directly interact with miR-124 to link the post-transcriptional network in BTCC pathogenesis.
To investigate whether MALAT1-induced reduction of miR-124 resulted in a derepression to its mRNA targets, we focused on the miR-124 putative target gene, foxq1. Human Foxq1 is first confirmed to encode a protein of 403 amino acids and is part of the family of the Fox transcription factors in 2001 [40]. Previous studies have revealed fairly high expression of foxq1 in the bladder and stomach of murine tissues [41]. Amazing amplification of foxq1 has been noted in several cancers, such as colorectal cancer [42], bladder cancer [43], glioma [44], promoting tumorigenesis and tumor growth in vivo. Interestingly, foxq1 was also implicated in EMT in tumor metastasis [43,45,46]. In several reports, foxq1 expression were directly affected by miRNA-mediated post-transcriptional mechanisms in carcinomas [30,47]. Here, our study determines that foxq1 is a direct target gene of miR-124. Considering the crosstalk of MALAT1/miR-124, we speculate that MALAT1 may regulate foxq1 expression in BTCC, indicating the role of MALAT1 in the tumorigenesis-regulating network.
Contrary to MALAT1 sequestration of miR-124, we found promotion effect of MALAT1 on foxq1 expression. Furthermore, elevated expression of foxq1 induced by MALAT1 was reversed by miR-124. Additionally, real-time PCR assay revealed that foxq1 was mainly up-regulated in advanced clinical-pathological stage BTCC tissues, and associated with high MALAT1 expression. Taken together, the positive correlation between MALAT1 and foxq1 expression and the repression effect of MALAT1 on miR-124 expression are in favor of the theory that ceRNA can sequester miRNA, thereby protecting their target gene from repression. What’s more, foxq1 deletion partly abolished the enhanced proliferation, migration and invasion induced by MALAT1, accompanied by the reversal of EMT, suggesting the functional significance of miR-124/foxq1 cascade in tumorigenesis-regulating network of BTCC associated with MALAT1.
Conclusively, the findings presented in this study show that MALAT1 functions as an oncogene in BTCC, and a novel MALAT1-miR-124/foxq1 regulatory network in BTCC progression is obtained, indicating that targeting MALAT1/foxq1 interaction may be a novel therapeutic strategy against this disease. However, special attention is that there may existing other lncRNAs that work as ceRNAs to regulate key genes expression in BTCC. Undoubtedly, the identification of these ceRNAs will improve our understanding of how lncRNAs function, and eventually benefit lncRNA-directed therapy for BTCC.
Acknowledgements
This study was funded by the national high tech research and development program (863 Program) (grant number: 2015AA020301).
Disclosure of conflict of interest
None.
References
- 1.Li W, Gu M. SULT1A1 Arg213His polymorphism is associated with bladder cancer risk: a meta-analysis. Med Sci Monit. 2014;20:1590–1595. doi: 10.12659/MSM.890822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murta-Nascimento C, Schmitz-Dräger BJ, Zeegers MP, Steineck G, Kogevinas M, Real FX, Malats N. Epidemiology of urinary bladder cancer: from tumor development to patient’s death. World J Urol. 2007;25:285–295. doi: 10.1007/s00345-007-0168-5. [DOI] [PubMed] [Google Scholar]
- 3.Kim WJ, Quan C. Genetic and epigenetic aspects of bladder cancer. J Cell Biochem. 2005;95:24–33. doi: 10.1002/jcb.20412. [DOI] [PubMed] [Google Scholar]
- 4.Wang HT, Chang JW. Molecular pathology of low malignant bladder transitional cell carcinoma: a current perspective. Histol Histopathol. 2005;20:147–153. doi: 10.14670/HH-20.147. [DOI] [PubMed] [Google Scholar]
- 5.Nordentoft I, Lamy P, Birkenkamp-Demtröder K, Shumansky K, Vang S, Hornshøj H, Juul M, Villesen P, Hedegaard J, Roth A. Mutational context and diverse clonal development in early and late bladder cancer. Cell Rep. 2014;7:1649–1663. doi: 10.1016/j.celrep.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 6.Hassen W, Droller MJ. Current concepts in assessment and treatment of bladder cancer. Curr Opin Urol. 2000;10:291–299. doi: 10.1097/00042307-200007000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Zhan J, Tang XD. Expression of cyclooxygenase-2 in human transitional cell bladder carcinomas. Ai Zheng. 2002;21:1212–1216. [PubMed] [Google Scholar]
- 8.Hu X, Ruan Y, Cheng F, Yu W, Zhang X, Larré S. p130Cas, E-cadherin and β-catenin in human transitional cell carcinoma of the bladder: expression and clinicopathological significance. Int J Urol. 2011;18:630–637. doi: 10.1111/j.1442-2042.2011.02793.x. [DOI] [PubMed] [Google Scholar]
- 9.Mcgarvey TW, Tait E, Tomaszewski JE, Malkowicz SB. Expression of transforming growth factor-beta receptors and related cell-cycle components in transitional-cell carcinoma of the bladder. Mol Urol. 1999;3:371–380. [PubMed] [Google Scholar]
- 10.Zhang H, Fan Q, Cao Y, Chen M, Zu X. Down-regulated microRNA-101 in bladder transitional cell carcinoma is associated with poor prognosis. Med Sci Moni. 2014;20:812–817. doi: 10.12659/MSM.890300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Q, Wang N, Qi J, Gu Z, Shen H. Long non-coding RNA-GAS5 acts as a tumor suppressor in bladder transitional cell carcinoma via regulation of chemokine (C-C motif) ligand 1 expression. Mol Med Rep. 2016;13:27–34. doi: 10.3892/mmr.2015.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 17.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:1–10. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallen AN, Xiao-Bo Z, Jie X, Chong Q, Jing M, Lei Y, Lingeng L, Chaochun L, Jae-Sung Y, Haifeng Z. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcella C, Davide C, Ivano L, Tiziana S, Olga S, Mauro C, Anna T, Irene B. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 21.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:2739–2748. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bekri S, Adélaïde J, Merscher S, Grosgeorge J, Carolibosc F, Peruccalostanlen D, Kelley PM, Pébusque MJ, Theillet C, Birnbaum D. Detailed map of a region commonly amplified at 11q13-->q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79:125–131. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- 23.Chakrabarti R, Srivatsan ES, Wood TF, Eubanks PJ, Ebrahimi SA, Gatti RA, Passaro E, Sawicki MP. Deletion mapping of endocrine tumors localizes a second tumor suppressor gene on chromosome 11q13. Genes Chromosomes Cancer. 1998;22:130–137. doi: 10.1002/(sici)1098-2264(199806)22:2<130::aid-gcc7>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 24.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E. MALAT-1, a novel noncoding RNA, and thymosin |[beta] |4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:6087–6097. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 25.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: A long non-coding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 26.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:1–13. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557–564. doi: 10.1016/j.biopha.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 30.Xiao HP, Hao RH, Lu J, Xiong L, Fei PZ, Bao Z, Shao XL, Lu W, Chen HH, Xia X. MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol Cancer. 2014;13:1–13. doi: 10.1186/1476-4598-13-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanguilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 32.Yu BL, Peng XH, Zhao FP, Liu X, Lu J, Wang L, Li G, Chen HH, Li XP. MicroRNA-378 functions as an onco-miR in nasopharyngeal carcinoma by repressing TOB2 expression. Int J Oncol. 2014;44:1215–1222. doi: 10.3892/ijo.2014.2283. [DOI] [PubMed] [Google Scholar]
- 33.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JE, Lee MH, Nam DH, Song HK, Kang YS, Lee JE, Kim HW, Cha JJ, Hyun YY, Han SY, Han KH, Han JY, Cha DR. Celastrol, an NF-κB inhibitor, improves insulin resistance and attenuates renal injury in db/db Mice. PLoS One. 2013;8:e62068–e62068. doi: 10.1371/journal.pone.0062068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. 2010;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang MJ, Li Y, Wang R, Wang C, Yu YY, Yang L, Zhang Y, Zhou B, Zhou ZG, Sun XF. Downregulation of microRNA-124 is an independent prognostic factor in patients with colorectal cancer. Int J Colorectal Dis. 2013;28:183–189. doi: 10.1007/s00384-012-1550-3. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Luo J, Wang B, Wang D, Xie X, Yuan L, Guo J, Xi S, Gao J, Lin X. Microrna-124 targets flotillin-1 to regulate proliferation and migration in breast cancer. Mol Cancer. 2013;12:1–13. doi: 10.1186/1476-4598-12-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Wu Q, Xu B, Wang P, Fan W, Ying C, Gu X, Meng F. miR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1. FEBS J. 2015;282:4376–4388. doi: 10.1111/febs.13502. [DOI] [PubMed] [Google Scholar]
- 40.Bieller A, Pasche B, Frank S, Gläser B, Kunz J, Witt K, Zoll B. Isolation and characterization of the human forkhead gene FOXQ1. DNA Cell Biol. 2001;20:555–561. doi: 10.1089/104454901317094963. [DOI] [PubMed] [Google Scholar]
- 41.Hoggatt AM, Kriegel AM, Smith AF, Herring BP. Hepatocyte nuclear factor-3 homologue 1 (HFH-1) represses transcription of smooth muscle-specific genes. J Biol Chem. 2000;275:31162–31170. doi: 10.1074/jbc.M005595200. [DOI] [PubMed] [Google Scholar]
- 42.Kaneda H, Arao T, Tanaka K, Tamura D, Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y, Yamada Y, Tsurutani J, Okamoto I, Nakagawa K, Nishio K. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010;70:2053–2063. doi: 10.1158/0008-5472.CAN-09-2161. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Z, Zhu Z, Pang Z, Xing Y, Wan F, Lan D, Wang H. Short hairpin RNA targeting FOXQ1 inhibits invasion and metastasis via the reversal of epithelial-mesenchymal transition in bladder cancer. Int J Oncol. 2013;42:1271–1278. doi: 10.3892/ijo.2013.1807. [DOI] [PubMed] [Google Scholar]
- 44.Sun HT, Cheng SX, Tu Y, Li XH, Zhang S. FoxQ1 promotes glioma cells proliferation and migration by regulating NRXN3 expression. PLoS One. 2013;8:268–277. doi: 10.1371/journal.pone.0055693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu F, Ethier SP, Miller F, Wu G. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 2011;71:1292–1301. doi: 10.1158/0008-5472.CAN-10-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jian F, Zhang X, Zhu H, Wang X, Ni S, Huang J. FoxQ1 overexpression influences poor prognosis in non-small cell lung cancer, associates with the phenomenon of EMT. PLoS One. 2012;7:1726–1729. doi: 10.1371/journal.pone.0039937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao B, Liu H, Gu Z, Ji C. Expression of microRNA-133 inhibits epithelial-mesenchymal transition in lung cancer cells by directly targeting FOXQ1. Arch Bronconeumol. 2016;52:505–511. doi: 10.1016/j.arbres.2015.10.016. [DOI] [PubMed] [Google Scholar]