Abstract
MicroRNAs (miRNAs) play an important role in human tumorigenesis as oncogenes or tumor suppressors by directly binding to the 3’-untranslated region of their target mRNAs. MiR-520d-3p has been reported as a tumor suppressor gene in ovarian cancer and gastric cancer, while the function of miR-520d-3p in human breast cancers is still uninvolved. In this study, we initially identified that the expression of miR-520d-3p was significantly reduced in breast cancer specimens and cell lines. The restoration of miR-520d-3p expression not only reduced breast cancer cell viability by causing the accumulation of G2 phase and cell apoptosis, but also inhibited tumorigenicity in vivo. In addition, as a critical target of miR-520d-3p, the activity of spindle and kinetochore associated 2 (SKA2) was greatly inhibited by miR-520d-3p, and overexpression of miR-520d-3p decreased the expression of SKA2. SKA2 downregulation suppressed cell viability, whereas restoration of SKA2 expression significantly reversed the inhibitory effects of miR-520d-3p antitumor activity. Furthermore, SKA2 was frequently overexpressed in clinical specimens and cell lines, and the expression levels were statistically inversely correlated with miR-520d-3p expression. In conclusion, our data demonstrated that miR-520d-3p antitumor activity is achieved by targeting the SKA2 in human breast cancer cells, suggesting that miR-520d-3p may be a potential target molecule for the therapy.
Keywords: miR-520d-3p, breast cancer, antitumor activity, SKA2
Introduction
At present, breast cancer, as one of the most frequently diagnosed types of malignancy, has become the severe threaten to female health [1,2]. Despite much achievements in breast cancer therapy with the development of modern technology have been made in this field, the precise molecular mechanisms behind the development and progression of breast cancer remain enigmatic [3,4].
MicroRNAs (miRNAs), a group of non-coding small RNA consisting of 18-25 nucleotides, can post-transcriptionally regulate gene expression through binding to the 3’-untranslated region (3’-UTR) of their target mRNAs, to result in mRNA degradation or translation inhibition [5,6]. A large number of studies have reported that miRNAs function as onco- or tumor suppressor genes and their aberrant expression are vital in the tumorigenesis and malignant progression [7-10]. To date, extensive studies have identified that miRNAs have essential roles in breast cancer progression [11-14]. Therefore, further exploration of the expression and function of miRNAs will help us further understanding the molecular mechanisms of breast cancer development.
Previous researches have reported that miR-520d-3p expression was down-regulated and inversely correlated with the expression of EphA2 in gaster cancer tissues and cell lines. Lower expression of miR-520d-3p was associated with poorer overall survival. Overexpression of miR-520d-3p dramatically inhibited the proliferation, cell cycle progression, invasion, and migration [15]. MiR-520d-3p was also found to suppress tumor growth and migration/invasion in ovarian cancer [16]. In hepatocellular carcinoma, EGFRvIII expression can induce miR-520d-3p downregulation, which may lead to drug resistance [17]. However, nothing is known about the role of miR-520d-3p in breast cancer.
In this study, we will evaluate the clinical significance of miR-520d-3p as a tumor suppressor in breast cancer tissues, and the potential role of miR-520d-3p in breast cancer cells growth, apoptosis and cell cycle. Otherwise, SKA2 is a potential target gene analyzed by software, we also found that there was a significant inverse correlation between SKA2 and miR-520d-3p. Therefore, these data not only associate miR-520d-3p with tumorigenesis of breast cancer, but also provide a new therapeutic target for treating breast cancer.
Materials and methods
Patients
Both tumor and adjacent normal tissue were collected from patients in Ningbo No. 2 Hospital, none of these patients underwent local or systemic therapy before surgery. The clinical stage was defined according to TNM staging system. The relevant characteristics of these 80 patients are described in Table 1. This research was approved by the Research Ethics Committee of Ningbo No. 2 Hospital and all patients signed the informed consent.
Table 1.
MiR-520d-3p expression and clinicolpathlolgic characteristics of patients
| Characteristic | Patients Number | miR-520d-3p Expression Level | P | |
|---|---|---|---|---|
|
| ||||
| Low (%) | High (%) | |||
| Age (year) | ||||
| ≤ 50 | 36 | 27 (75) | 9 (25) | 0.818 |
| > 50 | 44 | 32 (72.7) | 12 (27.3) | |
| Tumor (cm) | ||||
| < 2 | 33 | 19 (57.6) | 14 (42.4) | 0.022 |
| 2-5 | 34 | 29 (85.3) | 5 (14.7) | |
| > 5 | 13 | 11 (84.6) | 2 (15.4) | |
| Tumor TNM staging | ||||
| I+II | 42 | 27 (64.3) | 15 (35.7) | 0.043 |
| III+IV | 38 | 32 (84.2) | 6 (15.8) | |
| Tumor overall NHS grade | ||||
| 1 | 5 | 6 (60) | 2 (40) | 0.529 |
| 2 | 42 | 33 (78.6) | 9 (21.4) | |
| 3 | 33 | 23 (69.7) | 10 (30.3) | |
| Pathological typing | ||||
| Infiltrative type | 71 | 54 (76.1) | 17 (23.9) | 0.188 |
| No infiltrative type | 9 | 5 (55.6) | 4 (44.4) | |
Cell lines and transfections
Human breast cancer cell lines (MCF-7, MDA-MB-231 and T47D) were purchased from American Type Culture Collection and normal breast cell lines MCF-10A cell was obtained from Institute of Biochemistry and Cell Biology (Shanghai, China). All Cells were grown in the DMEM (HyClone, UT, USA) medium supplemented with 10% fetal bovine serum (FBS, ExCell Bio, Shanghai, China), at 37°C, in 5% CO2 incubator (Memmert, Germany). MCF-7 and T47D lines were transfected with miR-520d-3p mimics, miR-520d-3p NC, siSKA2 and siSKA2 NC (GenePharma, Shanghai, China) with Lipofectamine 2000 (Invitrogen, USA) following the manufacturer’s instructions. The SKA2 cDNA plasmid without 3’-UTR (Addgene, Cambridge, USA) and miR-520d-3p mimics were co-transfected or respective-transfected for MTT assays and Western boltting analyses in MCF-7 cells. MiR-520d-3p overexpression plasmid and negative control plasmid purchased from GenePharma (Shanghai, China) were transfected into MCF-7 cells using Lipofectamine 2000 for the in vivo tumorigenicity assay.
Western blotting analysis
Total protein extracted from cells was quantified by the analysis of bicinchoninic acid (Beyotime, Shanghai, China). Cellular proteins were separated using 12% SDS-polyacrylamide gel, transferred onto PVDF membranes (Millipore, Billerica, MA). Western blot analysis was performed with primary antibodies against Ki-67 (Abcam, Cambridge, UK), cyclin A (Cell Signaling Technology, MA, USA), CDK1 (Cell Signaling Technology, MA, USA), cyclin E (Cell Signaling Technology, MA, USA), Bax (Cell Signaling Technology, MA, USA), Bcl-2 (Cell Signaling Technology, MA, USA), SKA2 (Abcam, Cambridge, UK) and GAPDH (Cell Signaling Technology, MA, USA). Then, membranes were incubated with horseradish peroxidaselabeled secondary antibody (Boster, Wuhan, China). The protein bands on the membrane were visualized using a chemiluminescence imaging system (LI-COR Biosciences, CA, USA).
Quantitative real-time PCR
Total RNA was isolated from the breast cancer cells using Trizol reagent (Invitrogen, USA). CDNA was prepared using a reverse transcription kit (Thermo, USA). The cDNAs were amplified by qRT-PCR using SYBR Green PCR Master Mix (Roche, US) on a LightCycler480 system, and fold changes were calculated by relative quantification (2-ΔΔCt). The PCR primers were as follows: miR-520d-3p, forward: 5’-GGTCTACAAAGGGAAGC-3’ and reverse: 5’-TTTGGCACTAGCACATT-3’; U6, forward: 5’-CTCGCTTCGGCAGCACA-3’ and reverse: 5’-AACGCTTCACGAATTTGCGT-3’. SKA2, forward: 5’-CTGAAACTATGCTAAGTGGGGGAG-3’ and reverse: 5’-TTCCAAACATCCTGACACTCAAAAG-3’; GAPDH, forward: 5’-AAGCCTGCCGGTGACTAAC-3’ and reverse: 5’-GCATCACCCGGAGGAGAAAT-3’.
Cell viability assay
Breast cancer cells transfected with miR-520d-3p mimics, SKA2 siRNA and negative normal control (NC) were plated in 96-well plates at 1 × 104 cells per well. Following incubation for 0, 24, 48, 72 and 96 h, 20 µl of CellTiter96® AQueous One Solution (Promega, WI, USA) was then added to each well. After 3 h of additional incubation at 37°C, the absorbance was measured at 490 nm on a microplate reader (Beckman Counter).
Flow cytometry analysis of apoptosis and cell cycle
For apoptosis analysis, cells were transfected with miR-520d-3p mimics for 24 h. The transfected cells were harvested and washed twice by PBS, and examined using the Annexin V FITC/PI apoptosis detection kit (Multisciences, Hang Zhou, China) according to the manufacturer’s instructions. For the cell cycle analysis, the cells transfected with miR-520d-3p mimics were also collected. After washing with PBS twice, each sample treated with DNA staining solution containing 1 ml and 10 µl RNase A by using the Cell Cycle Stanining Kit (Multisciences, Hang Zhou, China) was incubated for 30 min at 37°C in the dark. Apoptosis analysis and cell cycle were all analyzed by flow cytometry.
Luciferase reporter assay
The predicted miR-520d-3p binding sites on the 3’-UTR of SKA2, together with a corresponding mutanted miR-520d-3p binding sites on the 3’-UTR of SAK2, were synthesized and inserted into the pGL3 vector (Promega, Madison, WI, USA). For the luciferase reporter assay, MCF-7 cells grown in a 24-well plate were co-transfected with wild-type (WT) or mutant (Mut) 3’-UTR vectors and miR-520d-3p mimic or NC using Lipofectamine 2000. After 48 h, the luciferase activity was analyzed by the Dual-Luciferase Reporter Assay System according to the manufacture’s protocols (Promega, Madison, USA). The values were normalized to those obtained for miRNA negative control transfection.
Xenograft assays in nude mice
To assess the inhibitory effect of miR-520d-3p in breast cancer cell, five-week-old male nude mice (BALB/C) (Shanghai laboratory animal center, China) were used for xenograft model with a protocol approved by the Institutional Animal Ethics Committee of Ningbo University. MCF-7 cells transfected with miR-520d-3p expression plasmid and negative control plasmid were injected subcutaneously into the flank of mouse (n = 5 for each group) to establish the tumor xenograft. Tumor size was measured for length and width every 5 d for 30 d. The mice were sacrificed and photographed at 30 d post-implantation. Xenograft tumors were excised, photographed and weighed.
Statistical analysis
All experiments were repeated three times. Statistical comparisons between two data samples were carried out using Students’ t test. Multiple group comparison was analyzed by using ANOVA with a post-test for subsequent individual group comparisons. Pearson correlation analysis was conducted to assess the statistical significance between cases with high or low levels of miR-520d-3p or SKA2. All data were expressed as the mean standard deviation (SD). A value of P < 0.05 was considered to be statistically significant. Statistical analyses were performed by using SPSS 15.0 software.
Result
MiR-520d-3p expression was down-regulated in breast cancer and associated with clinical stage and tumor size
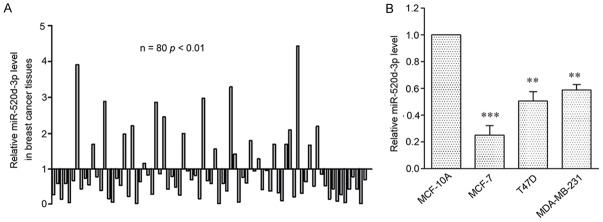
To determine the role of miR-520d-3p in breast cancer, the expression of miR-520d-3p was assessed by qRT-PCR in 80 patient samples. The results showed that miR-520d-3p expression was significantly down-regulated in 59 (73.75%) samples (P < 0.001) (Figure 1A). To further investigate the expression of miR-520d-3p, 3 human breast cancer cell lines (MCF-7, T47D and MDA-MB-231) were evaluated, compared to that of the normal human mammary epithelial cell line MCF-10A, the mRNA levels of miR-520d-3p were significantly inhibited in all 3 cell lines (Figure 1B).
Figure 1.
MiR-520d-3p expression is frequently down-regulated in human breast cancer tissues and cell lines. A. qRT-PCR analysis of miR-520d-3p expression in 80 human breast cancer tissues compared with their matched adjacent non-tumor tissues. B. qRT-PCR analysis of miR-520d-3p expression in three human breast cancer cell lines and a normal human breast epithelial cell line. The level of miR-520d-3p expression was normalized to U6 and compared to the normal cells. Data were shown as mean ± SD from three independent experiments. **P < 0.01, ***P < 0.001 versus MCF-10A cell.
Additionally, clinical characteristics of the patients are listed in Table 1. MiR-520d-3p levels were significantly lower in T3-T4 stage (P < 0.05) and tumor size (P < 0.05) in breast cancer tissues, suggesting that low miR-520d-3p was associated with clinical stage and tumor size.
MiR-520d-3p suppresses breast cancer cells proliferation
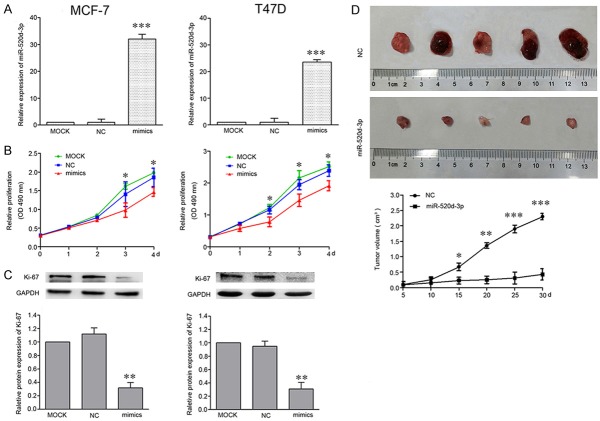
To further characterize the functional importance of miR-520d-3p in breast cancer progression. MCF-7 and T47D cells were transfected with MOCK, 20 µM miR-NC and miR-520d-3p mimics. After 48 h transfection, qPCR was conducted to examine miR-520d-3p expression. Transfection with miR-520d-3p mimics was found to significantly increase miR-520d-3p expression (P < 0.001), compared with the MOCK group (Figure 2A).
Figure 2.
Increased expression of miR-520d-3p inhibits human breast cancer cells growth in vitro and in vivo. A. After cells were transfected with miR-520d-3p mimics, the mRNAs level of miR-520d-3p were significantly increased in MCF-7 and T47D cells, as compared with cells transfected with NC. B. MTT assays were performed to determine the proliferation in MCF-7 and T47D cells compared with miR-520d-3p mimics and NC. C. The expression level of Ki-67 was detected by western blot compared with GAPDH. D. The tumors shown are from the final time point (30 day). miR-520d-3p expression plasmid and negative control plasmid were transfected into MCF-7 cells, which were injected in male athymic mice (n = 5), respectively. The xenograft growth was analyzed after breast cancer cell inoculation and the average size of the tumors was measured every 5 days and shown in the curves. *P < 0.05, **P < 0.01, ***P < 0.001.
Then, we examined the effects of miR-520d-3p overexpression on the proliferation of cells by performing an MTT assay. The data showed that the absorbance in MCF-7 and T47D cells transfected with miR-520d-3p mimics was significantly lower than those in MOCK (P < 0.05; Figure 2B). Previous studies have found that Ki-67 is closely related to cell proliferation [18-20]. Thus, further research showed miR-520d-3p mimics led to the obviously decreased expression of Ki-67 in breast cancer cells (P < 0.05; Figure 2C). Next, breast cancer nude mice were set up to investigate the role of miR-520d-3p on cell growth in vivo, the data demonstrated that miR-520d-3p inhibited the breast cancer cell growth (Figure 2D).
MiR-520d-3p induces cell cycle G2 arrest
To investigate whether the inhibition of miR-520d-3p on cell proliferation of MCF-7 and T47D cells was mediated by cell cycle alteration, cell cycle distribution was analyzed. The data showed that in comparison with the MOCK group, miR-520d-3p expression significantly increased cell population at the G2 phase (Figure 3A) (P < 0.05), indicating that overexpression of miR-520d-3p causes a cell cycle G2 arrest in MCF-7 and T47D cells. We then detected several cycle-related proteins expression by Western blotting. Results showed that over-expression of miR-520d-3p reduced the expression of cyclin A and CDK1, but, the contrary result in cyclin E (Figure 3B).
Figure 3.
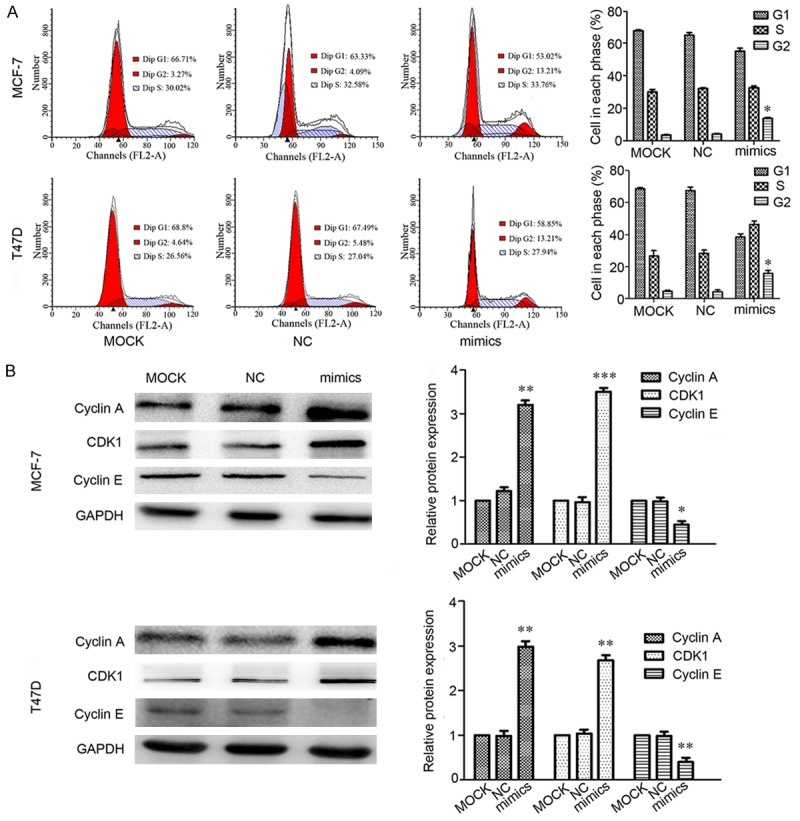
The over-expression of miR-520d-3p causes cell cycle arrest in G2 phase. A. Flow cytometry analysis of cell cycle. At 24 h post-transfection, MCF-7 and T47D cell lines treated with miR-520d-3p mimics showed a remarkable increase at the G2 phase compared with MOCK and NC groups. B. Expression level of cyclin A, CDK1 and cyclin E was determined by Western blotting, using GAPDH as a control. Data are shown as mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
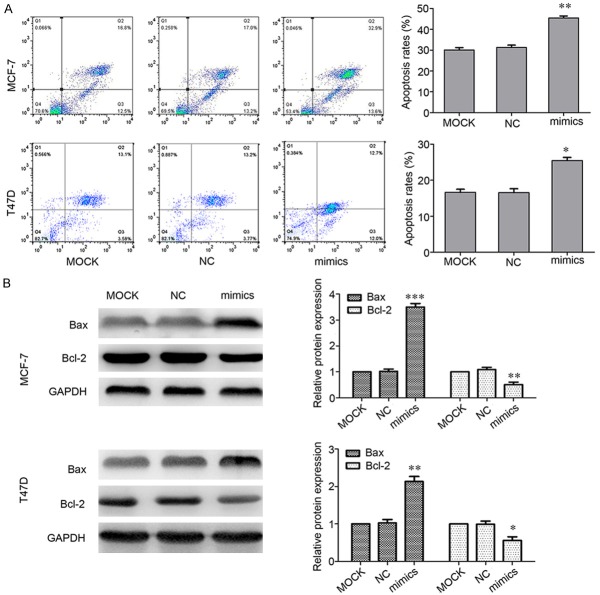
MiR-520d-3p triggers apoptosis in breast cancer cells
Furthermore, the apoptotic levels were examined to elucidate the mechanisms of miR-520d-3p inhibition on MCF-7 and T47D cells proliferation using flow cytometry. It was found that apoptosis rates were changed much in MCF-7 and T47D cells with miR-520a-3p transfection compared to that of the MOCK (Figure 4A). Since down-regulation of Bcl2 and up-regulation of Bax suggested the significant involvement of exposure in the induction of apoptosis [21-23], Western blotting was used to investigate whether miR-520d-3p regulates the expression of Bcl2 and Bax in breast cancer cells. The results revealed that the expression levels of Bcl2 in cells transfected with miR-520d-3p mimics were significantly lower than MOCK. By contrast, miR-520d-3p mimics increased the expression of Bax protein (Figure 4B).
Figure 4.
Up-regulated expression of miR-520d-3p promotes cells apoptosis. A. Flow cytometry analysis of apoptosis. After 24 h treatment, transfection of miR-520d-3p mimics into MCF-7 and T47D cell lines resulted in a significant increase percentage of apoptotic cells compared with the control. B. Expression level of Bax and Bcl-2 was determined by western blotting, using GAPDH as a control. Data are shown as mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
SKA2 is a direct target of miR-520d-3p in breast cancer MCF-7 cell
To elucidate the molecular mechanism of miR-520d-3p on breast cancer cellular functions, the putative target of miR-520d-3p was studied further by conducting a bioinformatical analysis. The predicted results showed that SKA2 is the predicted target gene with high possibility (Figure 5A). Subsequently, we cloned WT or Mut target region sequence of the SKA2 3’-UTR, which was inserted into a luciferase reporter vector (Figure 5B). It was found that miR-520d-3p significantly reduced the activity of the luciferase in the reporter with wild type 3’UTR (Figure 5C).
Figure 5.
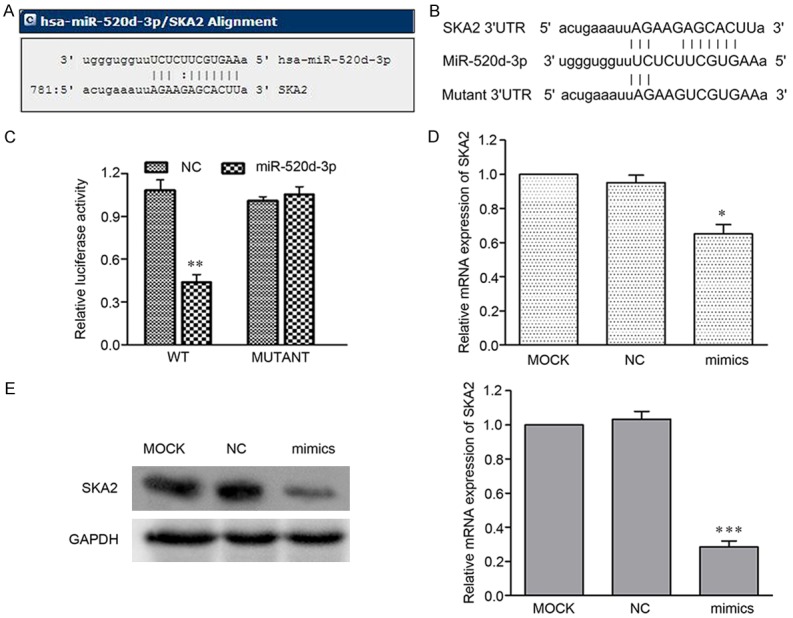
SKA2 is a direct target of miR-520d-3p in breast cancer MCF-7 cell. A. MiRanda prediction of miR-520d-3p targeting of SKA2 at the SKA2 mRNA 3’-UTR. B. The wild-type or mutant type of miR-520d-3p binding sequence in the 3’-UTR of SKA2 were cloned into the pGL3 vector. C. The effect of miR-520d-3p on the activity of firefly luciferase reporter containing SKA2 3’-UTR was suppressed by miR-520d-3p, but not in the mutated construct. D, E. The miR-520d-3p-mediated suppression of SKA2 at both mRNA and protein levels compared to MOCK in MCF-7 cells by qRT-PCR and Western blot, respectively. *P < 0.05, ***P < 0.001.
The effect of miR-520d-3p on SKA2 expressions at the transcriptional and translational levels were investigated further using RT-qPCR and Western blotting analysis. It was found that miR-520d-3p transfection led to a significantly decrease in SKA2 mRNA and protein, compared with the control group (P < 0.05) (Figure 5D and 5E).
Down-regulation of miR-520d-3p highly correlates with upregulation of SKA2 expression in both human breast cancer tissues and cell lines
To further confirm that miR-520d-3p inhibits SKA2 expression in breast cancer, we analyzed the mRNA levels of SKA2 from 20 pairs of breast cancer and normal tissue specimens by qRT-PCR. Compared to the non-tumor counterparts, the expression of SKA2 mRNA was significantly increased in tumor tissues (Figure 6A). The similar result is also true for breast cancer cell lines. SKA2 expression in 3 breast cancer cell lines were markedly higher than that in breast epithelial cell line (Figure 6B and 6C). Then we compared SKA2 and miR-520d-3p expression in these tissues, finally found that there was a statistically significant inverse correlation between SKA2 and miR-520d-3p expression among these 20 pairs of breast cancer tissues (Figure 6D).
Figure 6.
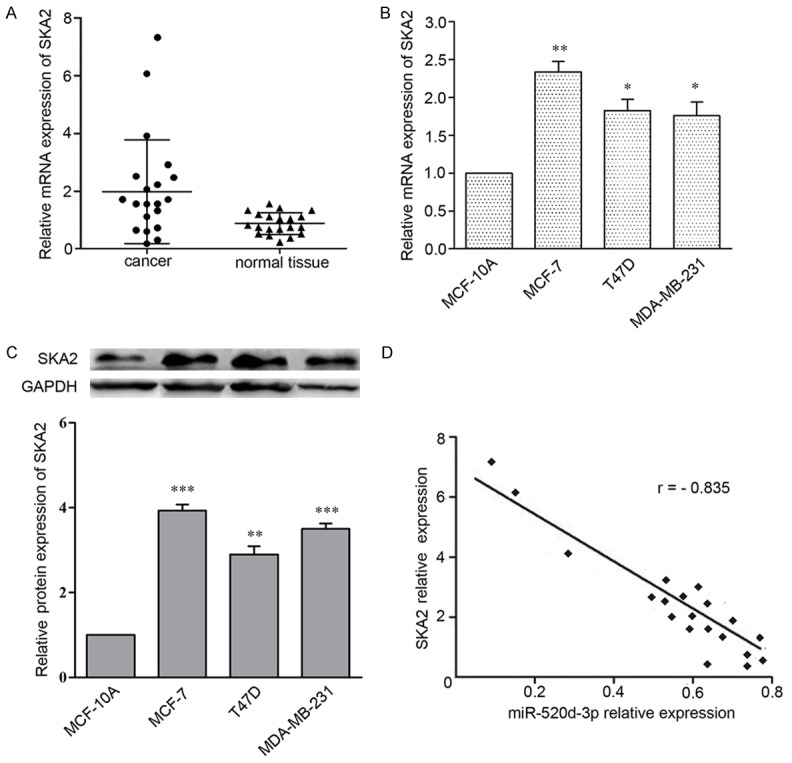
Statistical correlation between the expression levels of miR-520d-3p and SKA2 protein in breast cancer tissues. A. qRT-PCR analysis of SKA2 expression in 20 human breast cancer tissues compared with their matched adjacent non-tumor tissues. B, C. Western blot and qRT-PCR analysis of miR-520d-3p expression in three human breast cancer cell lines and a normal human breast epithelial cell line. Data were shown as mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus MCF-10A cell. D. The inverse correlation in breast cancer tissues was analyzed by Pearson’s correlation method.
MiR-520d-3p-mediated inhibition of SKA2 is involved in tumor suppression of breast cancer MCF-7 cell
To further test whether miR-520d-3p-mediated SKA2 inhibition confers antitumor activity in breast cancer cells, siSKA2 was transfected to assess whether it can mimic the biological effect of 520d-3p. Indeed, siSKA2 caused a significant inhibition of breast cancer cell viability (Figure 7A). While, ectopic expression of SKA2 by SKA2 cDNA plasmid without the 3’-UTR had the contrary effect (Figure 7B). Subsequently, we co-transfected miR-520d-3p mimics and SKA2 cDNA plasmid that lacks the 3’-UTR into breast cancer MCF-7 cell. The results showed that the inhibition of breast cancer cell proliferation by miR-520d-3p mimics were rescued by restoration of SKA2 expression (Figure 7C).
Figure 7.
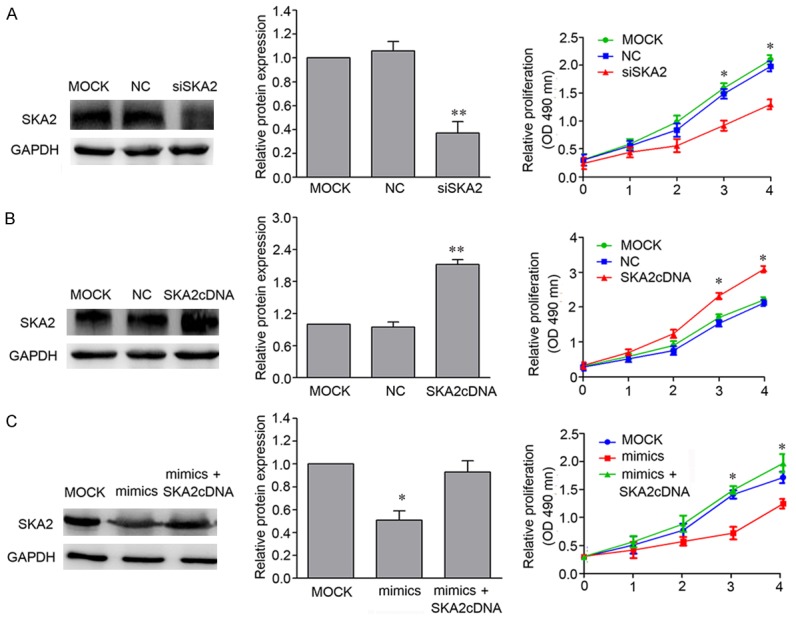
SKA2 mediated miR-520d-3p antitumor activity in breast cancer cells. A. SKA2 depletion-mediated cell viability. MCF-7 cells were transfected with MOCK, NC and SKA2 siRNA respectively. MTT assay was performed at 0, 24, 48, 72 and 96 h. B. Cell survival analysis by overexpression of SKA2 in breast cancer cells. MCF-7 cells were transfected with MOCK, NC and SKA2 cDNA plasmid without the 3’-UTR respectively. C. miR-520d-3p mediated cell survival and SKA2 rescue analysis in breast cancer cells. MCF-7 cells were co-transfected by miR-520d-3p mimics and SKA2 cDNA plasmid without the 3’-UTR followed by MTT assay. Data are shown as mean ± SD from three independent experiments. *P < 0.05, **P < 0.01.
Discussion
MiRNAs, a series of small RNAs, may affect biological features through the regulation expression of their target genes [24-26]. MiR-520d-3p is one of a potential tumor suppressing miRNA [15,16], nevertheless, its function and expression in human breast cancers is not clear.
In the present study, it was demonstrated that miR-520d-3p expression was significantly down-regulated in breast cancer tissues and cells lines. Moreover, lower miR-520d-3p expression was associated with clinical stage and tumor size in breast cancer patients. Data from in vitro and in vivo assays indicated that ectopic expression of miR-520d-3p inhibited proliferation, caused G2 phase arrest and induced apoptosis of MCF-7 and T47D cells, possibly by directly inhibiting SKA2, which was significantly upregulated in breast cancer tissues and cell lines. Further study indicated that miR-520d-3p greatly reduced the SKA2 expression, re-expression of SKA2 protein overcame the inhibitory effect of miR-520d-3p antitumor activity. Otherwise, a statistically inversely correlation was existed between the expressions of miR-520d-3p and SKA2 in breast cancer tissues. Therefore, our current study has further confirmed that miR-520d-3p plays a suppressing role in breast cancer, which is attributed to several biological functions of miR-520d-3p including reduction of breast cancer cell viability and induction of apoptosis and inhibition of tumorigenesis in vivo after over-expression of miR-520d-3p, as well as molecular mechanism of miR-520d-3p-mediated inhibitory effect of SKA2.
SKA2, a conserved protein involved in the kinetochore complex, have been observed in multiple cancer types, including lung cancer [27-29], breast cancer [30], kidney cancer [31], pancreatic cancer [32], gastric cancer [33], Glioma [34] and osteosarcoma [35,36]. Thus, it seemed that aberrant SKA2 expression can affect cell proliferation. In our study, we first found that knockdown of SKA2 expression decreased breast cancer cell viability, this is similar with the effect of miR-520d-3p mimics.
As SKA2 is a potential target gene of miR-520d-3p which predicted by software, we analyzed the relationship between them in cells and tissues.
The data showed that up-regulation of miR-520d-3p attenuated the expression of SKA2 mRNA and protein, the repression of SKA2 expression by miR-520d-3p was rescued by transfection of SKA2 cDNA plasmid that lacks the 3’-UTR. And there was a significantly inverse correlation between them in breast cancer tissues. This findings first supported that miR-520d-3p directly inhibits SKA2 expression in breast cancer via binding to SKA2 3’-UTR.
In conclusion, our study give an evidence that miR-520d-3p is a tumor suppressor gene, which is commonly downregulated in both breast cancer clinic tissues and breast cancer cell lines. Further investigation into the functional and clinical implications of miR-520d-3p/SKA2 may contribute to the development of a novel targeted therapy in breast cancer.
Acknowledgements
Zhouhui Ren and Tong Yang have contributed equally to this work. This work was supported by the National Natural Science Foundation of China (No. 81372209), Hua Mei Research Foundation (No. 2016HMKY35) and Natural Science Foundation of Ningbo (NO. 2013A610224, 2017A610193). Informed consent was obtained from all individual participants in the study.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Libson S, Lippman M. A review of clinical aspects of breast cancer. Int Rev Psychiatry. 2014;26:4–15. doi: 10.3109/09540261.2013.852971. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Zhao Y, Shao N, Ye R, Lin Y, Zhang N, Li W, Zhang Y, Wang S. Overexpression of microRNA-96-5p inhibits autophagy and apoptosis and enhances the proliferation, migration and invasiveness of human breast cancer cells. Oncol Lett. 2017;13:4402–4412. doi: 10.3892/ol.2017.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma F, Li W, Liu C, Li W, Yu H, Lei B, Ren Y, Li Z, Pang D, Qian C. MiR-23a promotes TGF-β1-induced EMT and tumor metastasis in breast cancer cells by directly targeting CDH1 and activating Wnt/β-catenin signaling. Oncotarget. 2017;4:69538–69550. doi: 10.18632/oncotarget.18422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 7.Zheng J, Ge P, Liu X, Wei J, Wu G, Li X. MiR-136 inhibits gastric cancer-specific peritoneal metastasis by targeting HOXC10. Tumour Biol. 2017;39:1010428317706207. doi: 10.1177/1010428317706207. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Gao C, Yang Y, Li G, Dong J, Ai Y, Ma Q, Li W. MiR-424 promotes non-small cell lung cancer progression and metastasis through regulating the tumor suppressor gene TNFAIP1. Cell Physiol Biochem. 2017;42:211–221. doi: 10.1159/000477314. [DOI] [PubMed] [Google Scholar]
- 9.Chang X, Yu C, Li J, Yu S, Chen J. hsa-miR-96 and hsa-miR-217 expression down-regulates with increasing dysplasia in pancreatic intraepithelial neoplasias and intraductal papillary mucinous neoplasms. Int J Med Sci. 2017;14:412–418. doi: 10.7150/ijms.18641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Kong G, Zhang J, Wang T, Ye L, Zhang X. MicroRNA-520b inhibits growth of hepatoma cells by targeting MEKK2 and cyclin D1. PLoS One. 2012;7:e31450. doi: 10.1371/journal.pone.0031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004–8. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 12.Cui W, Zhang Y, Hu N, Shan C, Zhang S, Zhang W, Zhang X, Ye L. miRNA-520b and miR-520e sensitize breast cancer cells to complement attack via directly targeting 3’UTR of CD46. Cancer Biol Ther. 2010;10:232–41. doi: 10.4161/cbt.10.3.12277. [DOI] [PubMed] [Google Scholar]
- 13.Yu H, Xing H, Han W, Wang Y, Qi T, Song C, Xu Z, Li H, Huang Y. MicroRNA-409-5p is upregulated in breast cancer and its downregulation inhibits cancer development through downstream target of RSU1. Tumour Biol. 2017;39:1010428317701647. doi: 10.1177/1010428317701647. [DOI] [PubMed] [Google Scholar]
- 14.Patel N, Garikapati KR, Pandita RK, Singh DK, Pandita TK, Bhadra U, Bhadra MP. MiR-15a/miR-16 down-regulates BMI1, impacting Ub-H2A mediated DNA repair and breast cancer cell sensitivity to doxorubicin. Sci Rep. 2017;7:4263. doi: 10.1038/s41598-017-02800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Yuan W, Mei W, Yang K, Chen Z. MicroRNA 520d-3p inhibits gastric cancer cell proliferation, migration, and invasion by downregulating EphA2 expression. Mol Cell Biochem. 2014;396:295–305. doi: 10.1007/s11010-014-2164-6. [DOI] [PubMed] [Google Scholar]
- 16.Nishimura M, Jung EJ, Shah MY. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013;3:1302–15. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang H, Dong Q, Luo X, Shi B, Wang H, Gao H, Kong J, Zhang J, Li Z. The monoclonal antibody CH12 augments 5-fl uorouracil-induced growth suppression of hepatocellular carcinoma xenografts expressing epidermal growth factor receptor variant III. Cancer Lett. 2014;342:113–20. doi: 10.1016/j.canlet.2013.08.038. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Liu Q, Zhou X, He Y, Guo Q, Shi Q, Eriksson S, Zhou J, He E, Skog S. Thymidine kinase 1 expression in ovarian serous adenocarcinoma is superior to Ki-67: a new prognostic biomarker. Tumour Biol. 2017;39:1010428317706479. doi: 10.1177/1010428317706479. [DOI] [PubMed] [Google Scholar]
- 19.Yalcin SE, Ocal I, Yalcin Y, Selim HS, Caltekin MD, Aydogmus H, Kelekci S. Evaluation of the Ki-67 proliferation index and urocortin expression in women with ovarian endometriomas. Eurasian J Med. 2017;49:107–112. doi: 10.5152/eurasianjmed.2017.17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan M, He T, Zhang P, Zhang J, Li L. Heterogeneity of diffusion-weighted imaging in tumours and the surrounding stroma for prediction of Ki-67 proliferation status in breast cancer. Sci Rep. 2017;7:2875. doi: 10.1038/s41598-017-03122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu M, Cai L, Zheng W, Su Z, Chen Y, Qi S. CD164 regulates proliferation and apoptosis by targeting PTEN in human glioma. Mol Med Rep. 2017;15:1713–1721. doi: 10.3892/mmr.2017.6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Wang R, Chen J, Wu J, Dang Z, Zhang Q, Li B. miR-340 Inhibits proliferation and induces apoptosis in gastric cancer cell line SGC-7901, possibly via the AKT pathway. Med Sci Monit. 2017;23:71–77. doi: 10.12659/MSM.898449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Z, Liu X, Chang K, Liu X, Xiong J. Allyl isothiocyanate inhibits the proliferation of renal carcinoma cell line GRC-1 by inducing an imbalance between Bcl2 and Bax. Med Sci Monit. 2016;22:4283–4288. doi: 10.12659/MSM.897315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Y, Li J, Xu T, Zhou DD, Zhang L, Wang X. MicroRNA-145 induces apoptosis of glioma cells by targeting BNIP3 and Notch signaling. Oncotarget. 2017;37:61510–61527. doi: 10.18632/oncotarget.18604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markopoulos GS, Roupakia E, Tokamani M, Vartholomatos G, Tzavaras T, Hatziapostolou M, Fackelmayer FO, Sandaltzopoulos R, Polytarchou C, Kolettas E. Senescence-associated microRNAs target cell cycle regulatory genes in normal human lung fibroblasts. Exp Gerontol. 2017;96:110–122. doi: 10.1016/j.exger.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Roy S, Bantel H, Wandrer F, Theres Schneider A, Gautheron J, Vucur M, Tacke F, Trautwein C, Luedde T, Roderburg C. miR-1224 inhibits cell proliferation in acute liver failure by targeting the antiapoptotic gene Nfib. J Hepatol. 2017;5:966–978. doi: 10.1016/j.jhep.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Rice L, Waters CE, Eccles J, Garside H, Sommer P, Kay P, Blackhall FH, Zeef L, Telfer B, Stratford I, Clarke R, Singh D, Stevens A, White A, Ray DW. Identification and functional analysis of SKA2 interaction with the glucocorticoid receptor. J Endocrinol. 2008;198:499–509. doi: 10.1677/JOE-08-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia W, Li J, Li S, Chen L, Ding H, Zhao Q, Fan M, Shen B, Shao N. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun. 2010;396:978–82. doi: 10.1016/j.bbrc.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Zhang Y, Zhang C, Weng H, Li Y, Cai W, Xie M, Long Y, Ai Q, Liu Z, Du G, Wang S, Niu Y, Song F, Ozaki T, Bu Y. The gene pair PRR11 and SKA2 shares a NF-Y-regulated bidirectional promoter and contributes to lung cancer development. Biochim Biophys Acta. 2015;1849:1133–44. doi: 10.1016/j.bbagrm.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Shi W, Gerster K, Alajez NM, Tsang J, Waldron L, Pintilie M, Hui AB, Sykes J, P’ng C, Miller N, McCready D, Fyles A, Liu FF. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71:2926–37. doi: 10.1158/0008-5472.CAN-10-3369. [DOI] [PubMed] [Google Scholar]
- 31.Zhuang H, Meng X, Li Y, Wang X, Huang S, Liu K, Hehir M, Fang R, Jiang L, Zhou JX, Wang P, Ren Y. Cyclic AMP responsive element-binding protein promotes renal cell carcinoma proliferation probably via the expression of spindle and kinetochore-associated protein 2. Oncotarget. 2016;7:16325–37. doi: 10.18632/oncotarget.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Z, Li Y, Takwi A, Li B, Zhang J, Conklin DJ, Young KH, Martin R, Li Y. miR-301a as an NF-kappaB activator in pancreatic cancer cells. EMBO J. 2011;30:57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu Y, Gu Q, Zhu Z, Liu B. Overexpressed miR-301a promotes cell proliferation and invasion by targeting RUNX3 in gastric cancer. J Gastroenterol. 2013;48:1023–33. doi: 10.1007/s00535-012-0733-6. [DOI] [PubMed] [Google Scholar]
- 34.Bian EB, Ma CC, He XJ, Wang C, Zong G, Wang HL, Zhao B. Epigenetic modification of miR-141 regulates SKA2 by an endogenous ‘sponge’ HOTAIR in glioma. Oncotarget. 2016;7:30610–25. doi: 10.18632/oncotarget.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang IC, Chiang TI, Lo C, Lai YH, Yue CH, Liu JY, Hsu LS, Lee CJ. Anemone altaica induces apoptosis in human osteosarcoma cells. Am J Chin Med. 2015;43:1031–42. doi: 10.1142/S0192415X15500597. [DOI] [PubMed] [Google Scholar]
- 36.Lin CC, Chao PY, Shen CY, Shu JJ, Yen SK, Huang CY, Liu JY. Novel target genes responsive to apoptotic activity by Ocimum gratissimum in human osteosarcoma cells. Am J Chin Med. 2014;42:743–67. doi: 10.1142/S0192415X14500487. [DOI] [PubMed] [Google Scholar]