Abstract
We describe methods for live-cell imaging of yeast cells that we have exploited to image yeast polarity establishment. As a rare event occurring on a fast time-scale, imaging polarization involves a trade-off between spatiotemporal resolution and long-term imaging without excessive phototoxicity. By synchronizing cells in a way that increases resistance to photodamage, we discovered unexpected aspects of polarization including transient intermediates with more than one polarity cluster, oscillatory clustering of polarity factors, and mobile “wandering” polarity sites.
Keywords: Microscopy, Imaging, Cell polarity, Yeast, Polarity establishment, Cdc42, GFP
1 Introduction
The budding yeast Saccharomyces cerevisiae has served as an extraordinarily tractable model system in which to develop a molecular understanding of cell biological phenomena. Despite its small cell size (diameter ~4–6 µm, volume ~40–80 fL depending on whether the cell is haploid or diploid [1]), live-cell imaging approaches have revealed critical features of many cell biological processes in yeast cells. Combining live-cell imaging with genetic perturbations provides a powerful strategy for elucidating the molecular design principles underlying the phenomena under investigation. Here, we provide detailed protocols for imaging polarity establishment in yeast.
Polarity establishment is central to cell migration and to the function of many differentiated cell types. Yeast cells polarize to form a bud during vegetative growth and to form a mating projection during conjugation. Polarity establishment occurs in the G1 phase of the cell cycle and is triggered by activation of G1 cyclin/cyclin-dependent kinase complexes [2]. The location of polarity establishment is influenced by inherited landmark proteins and often occurs near to the preceding cytokinesis site [2]. Because many of the same proteins concentrate at both the cytokinesis site and the polarity site, it can be difficult to clearly distinguish polarity establishment from the completion of the prior cytokinesis. To circumvent this issue, we often image cells in which RSR1, encoding a key transducer of spatial information from the landmarks to the polarity machinery, has been deleted. In cells lacking Rsr1, polarity establishment occurs at more-or-less random locations.
A central challenge for imaging polarity establishment concerns phototoxicity [3]. Because polarity establishment occurs at low frequency (only once per cell cycle or approximately once per 100 min in rapid growth conditions), cells must be imaged for prolonged periods in order to detect significant numbers of events, and imaging must be performed at high temporal resolution because the process itself is rapid, occurring in just 1–2 min. Because the location of polarity establishment cannot be accurately predicted, cells must also be imaged at high spatial resolution, with multiple z-plane acquisition so that polarity site s can be detected wherever they occur over the cell surface. In combination, the requirements for prolonged imaging at high spatiotemporal resolution translate to high fluorescence excitation exposure, which can easily lead to unacceptable phototoxicity. To make matters worse, it appears that cells are more sensitive to phototoxic stress in G1 than they are later in the cell cycle, and imaging conditions that are normally considered to be “low-light” can delay or block polarity establishment [4].
To lower the amount of light needed, we use widefield fluorescence or spinning disk confocal microscope s equipped with sensitive EM-CCD cameras. In addition, we image yeast cells on agarose slabs because these cells are less photosensitive than those imaged in more stressful conditions (e.g., in microfluidic devices). The total imaging time can be reduced by synchronizing the cells so that many undergo polarity establishment within a short time interval. We tested several synchrony protocols and fortuitously discovered that hydroxyurea arrest-release had the unanticipated side-benefit of making cells more photoresistant. We speculate that this treatment (which strongly induces the DNA damage response [5]) prepares cells to mitigate the damaging effects of light.
If yeast cells in G1 are exposed to sufficient amounts of mating pheromone, they arrest with low G1 cyclin/CDK activity, but they nevertheless polarize to form a mating projection. The polarity site in mating cells is not static, but rather wanders around the cortex [6]. Wandering is suppressed by high doses of pheromone, and this process probably contributes to the successful tracking of pheromone gradients during chemotropism [6]. To characterize polarity patch wandering behavior in the absence of suppression by added pheromone, we exploit the fact that pheromone-induced MAPK activation is sufficient to trigger cell-cycle arrest and polarization but does not constrain wandering (pheromone constrains wandering via a separate pathway). Thus, artificial activation of the MAPK using an artificial scaffold protein, Ste5-CTM [7], leads to a uniform population of polarized cells in which wandering of the polarity site is prominent. We induce expression of Ste5-CTM using an orthologous promoter system [8] to avoid affecting cell metabolism by changing the media.
In this chapter, we provide detailed protocols for imaging and quantification of polarity establishment and polarity site wandering in budding and mating yeast.
2 Materials
2.1 Yeast Strains and Fluorescent Probes
The following protocols have been developed using two yeast strains: YEF-473 [9] and BF264-15Du [10]. We expect the protocols to apply to other lab yeast strains, though some small changes especially to imaging parameters (e.g., exposure time) might need to be applied.
While Cdc42 is considered the master regulatory of polarity, we do not usually use Cdc42 fluorescent probes because they are not fully functional [4]. Instead, we use Bem1-GFP and Spa2-mCherry as our primary markers for the polarity patch. These probes are very functional and bright, making them ideal for live cell microscopy.
2.2 Medium
For live cell imaging, grow cells and prepare samples with complete synthetic medium (CSM). YEP medium is not recommended because it is auto-fluorescent, leading to high background fluorescence intensity.
Complete synthetic medium (CSM): Add 950 ml deionized water to a 1 l glass bottle. Add 6.7 g yeast nitrogen base without amino acids (Difco™, Becton Dickinson and Company), 0.74 g CSM (MP Biomedicals), and 0.1 g adenine (see Note 1) to the bottle and mix them together. Autoclave the liquid at 121 °C for 20 min.
40 % dextrose solution: Dissolve 400 g dextrose (Macron Fine Chemicals™) in 1 l deionized water. Sterilize the solution by filtration using membrane filters with pore size 0.2 µm.
2.3 Reagents for Synchronizing/Arresting Cells
2 M Hydroxyurea (HU) solution: Add 1.52 g HU (Sigma-Aldrich) in about 5 ml deionized water. Make up to 10 ml with water and vortex vigorously to dissolve HU. Aliquot 0.5 ml HU solution in eppendorf tubes for individual uses. Store at −20 °C.
α-factor solution: Dilute 10 mg of α-factor (Genesee Scientific) in 5.938 ml water to make 1 mM stock solution and aliquots. For bar1 cells, make up 25 µM stock solution aliquots. Store at −20 °C, and only freeze-thaw once.
β-estradiol solution: Dilute 13.6 mg of β-estradiol (Sigma-Aldrich) in 5 ml ethanol to make 10 mM stock solution. Make up 50 µM stock solution and store at −20 °C.
2.4 Mounting Slab Components
CSM containing 2 % dextrose (see Subheading 2.2).
Agarose (Denville Scientific Inc.).
75 × 25 mm Gold Seal Micro Slides (Gold Seal Products).
18 × 18 mm No. 1 (0.14 mm thickness) microscope cover glass (Globe Scientific Inc.).
Vaseline for sealing the slab: Scoop Vaseline in a 50 ml conical tube and place in a boiling water bath. Once Vaseline is melted, pour into a 10 ml syringe with a 16 G needle.
0.2 µm Tetraspeck beads (Life Technologies), diluted 1:5 in water.
2.5 Microscopes and Imaging Settings
We have optimized our protocols using two microscopes that we describe here. There are many parameters to consider when imaging live yeast cells, and we discuss the trade-offs between parameters to help when using other microscopes, fluorescent probes, conditions, etc.
Widefield fluorescence microscope: Axio Observer Z1 (Carl Zeiss, Thornwood, NY) with an X-CITE 120XL metal halide fluorescence light source, a 100×/1.46 Plan Apochromat oil immersion objective, and a QuantEM backthinned EM-CCD camera (Photometrics, Tucson, AZ). The microscope is equipped with a XL S1 incubator and heating system (PeCon GmbH). The GFP and RFP filter cubes have the following filter sets (excitation/dichroic/emission): 470/495/525 nm and 560/585/630 nm, respectively.
Spinning disk confocal microscope: Andor Revolution XD (Olympus) with a CSU-X1 spinning disk unit (Yokogawa Electric Corporation), a 100×/1.4 UPlanSApo oil immersion objective, and an iXon3 897 EM-CCD camera (Andor Technology). The microscope is equipped with a PZ-2300FT automated stage, which contains a temperature-containing chamber (Applied Scientific Instrumentation Inc.).
Temperature control. Our live cell imaging experiments require temperatures ranging from 24 to 37 °C. Microscopes using small heaters (e.g., Andor Spinning Disk) require ~10 min to heat, and microscopes that are fully enclosed (e.g., Axio Observer Widefield) requires >30 min to heat (see Note 2).
Choosing imaging fields. The number of fields used and the number of cells per field depends on the experimental question, the length of the experiment, the cell size, among other considerations. We have found that when imaging multiple fields in an experiment, the cells from fields not currently being imaged are still exposed to indirect light. Therefore, for certain experiments we only use one field. When finding a suitable field with the desired density of cells, we have found that the cell density is often low in the middle of the slab where the cells have been placed, but the cell density is higher near the edge.
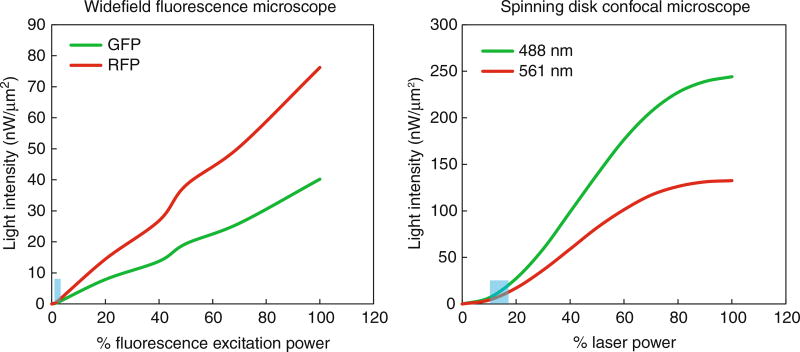
Light exposure. The phototoxicity caused by excitation light has been measured previously [3]. The maximum photon flux that did not cause a delay in cell division was 3.0 × 1011 photons/µm2 per 3D image. Since the light source of different microscopes varies (e.g., laser power or % fluorescence excitation power), we quantified the light intensity from our imaging conditions. The excitation light intensity at different laser power settings (spinning disk microscope) or percent of fluorescence excitation power settings (widefield fluorescence microscope) is shown in Fig. 1. The range of light used in our experiments is marked in blue boxes. Based on our imaging settings, the range of the photon flux shone on cells is listed in Table 1. In the calculations, we assume that the entire area is evenly illuminated by the excitation light; however, in reality, the illumination is uneven with higher intensity in the center than at the edge. Thus, the field we imaged may have a higher photon flux than that shown in the table, but the actual value should be below the 3.0 × 1011 photons/µm2 limit.
Z-stack. Cell structures, including the polarity site, are not always in the medial plane of the cell. Therefore, it is usually necessary to image cells with multiple z-plane acquisition. We used the Nyquist rate calculator (http://www.svi.nl/NyquistCalculator) to determine the minimal sampling density that is required to capture all information of the objects. Based on the microscopic parameters we used, the critical sampling distance (or Nyquist rate) for the widefield fluorescence microscope is 235 nm. With this distance, we took 30 z-plane images to cover the entire cell (HU-treated cells are larger, with diameter ~6–7 µm). The Nyquist rate for spinning disk microscope is 131 nm, which requires 54 z-planes to cover the same depth, but the increased light exposure would lead to more phototoxicity. Therefore, we use 30 z-planes (0.24 µm apart) for live cell imaging on both the widefield and confocal microscopes. When imaging multiple fields and multiple fluorescent channels, especially with small time intervals, sometimes the acquisition is reduced to 15 z-planes (0.5 µm apart) to reduce phototoxicity and reduce the imaging time. Additionally, the z-stack for each fluorescent channel is acquired separately rather than alternating between the fluorescent channels at each z-plane to reduce motor movements and decrease time of acquisition (see Note 3).
Time interval between image acquisitions. We usually use 45 s or 1.5 min intervals for live cell imaging. Shorter time intervals result in more phototoxicity, and it is important to keep in mind the trade-off between temporal resolution, spatial resolution, and phototoxicity.
Fig. 1.
Measured light intensity by microscope. Light intensity was measured using an X-cite power meter (Excelitas Technologies) over different settings of transmitted light for the widefield fluorescence microscope and laser power for the spinning disk confocal microscope. The blue boxes represent the range of settings used during live-cell yeast imaging
Table 1.
Estimated photon flux by microscope
| Microscope | Light intensity (nW/µm2) | Photon flux (photons/µm2 per 3D image) |
|---|---|---|
| Widefield | GFP: 0.15–0.4 | 2.7×109–1.1×1010 |
| RFP: 0.27–0.75 | ||
|
| ||
| Spinning disk | 488: 6.7–17.0 | 8.0×1010–2.5×1011 |
| 561: 4.2–10.0 | ||
3 Methods
3.1 Preparing Agarose Slabs
Combine 0.5 g agarose with 2.5 ml CSM + 2 % dextrose in a 15 ml conical tube. Add α-factor or β-estradiol if needed.
Loosen the cap of the tube and microwave (900 W) for 35–45 s. Forcefully invert or vortex every time the media boils to the top of the tube (about four times). Make sure the agarose completely dissolves. The final solution will be very viscous (see Note 4).
Using a cut pipette tip, quickly pipette 90–180 µl of the agarose solution onto a glass slide and immediately drop a coverslip on the agarose droplet. Repeat this procedure to make several slabs in order to ensure one is level with the glass. The slabs will solidify within 1 min.
Store the prepared slabs in a box (e.g., empty pipette tip box) with some water to prevent the slab from drying out. Use the slabs within a few hours of preparing.
3.2 Mounting Cells onto Agarose Slabs
Centrifuge sample (1 ml) at 16,000 × g for 1 min (see Note 5). Aspirate most of the media, leaving ~10–20 µl media to resuspend cells.
Pipette ~0.7 µl cells into tip and set pipette aside.
If the agarose slab extends beyond the coverslip, remove with a razor blade prior to loading cells.
Gently lift a corner of the coverslip away from the slab using a razor blade, and hold the coverslip in one hand.
Use other hand to pick up the pipette and drop cells onto slab.
Replace coverslip on slab.
Gently press sides of coverslip to ensure proper contact and reduce air pockets.
Seal slabs with Vaseline. Heat Vaseline in needle over flame prior to sealing each edge of coverslip.
3.3 Synchronizing Cells with HU Treatment Prior to Imaging
Grow cells overnight at 30 °C (or 24 °C if the strain is temperature sensitive) in CSM + 2 % dextrose to a log phase culture (OD600 = 0.1–0.7) (see Note 6).
The next morning, dilute the cell culture OD600 = 0.15. Mix 4.5 ml of the diluted cell culture and 0.5 ml 2 M HU solution (final working concentration 200 mM) in a 50 ml flask or glass test tube. Agitate the sample using a rotary shaker or roller drum for 3 h at 30 °C or 4 h at 24 °C to arrest cells in S phase (see Note 7).
Centrifuge 5 ml treated cells in eppendorf tubes at 16,000 × g (or in conical tubes at 2000 × g) and discard supernatant. Wash the cells twice with 1 ml CSM + 2 % dextrose to remove any remaining HU. Resuspend the cells in 5 ml CSM + 2 % dextrose in a 50 ml flask or a glass test tube. Agitate the sample for 1 h at 30 °C or 2 h at 24 °C to release the cells from HU arrest. At 30 °C, most strains take 75–90 min after releasing from HU arrest to enter G1 of the next cell cycle.
Mount cells onto an agarose slab and find a good field for imaging (see Subheading 2.5). We recommend using a field with 10–15 large-budded cells, which are not touching each other to prevent overlapping signal from neighboring cells during quantification. Also, most polarity markers localize to the bud neck during cytokinesis, so choose a field with cells that show signal at the bud neck. These cells are finishing their cell cycle and will polarize in the following G1 phase soon.
3.4 Imaging Polarity Establishment
Ensure that the temperature control chamber has reached a stable temperature at least 30 min prior to imaging.
Image cells (see Subheading 2.5) using settings listed in Table 2.
Table 2.
Settings for imaging polarity establishment
| Widefield | Channel | Transmitted light | EM gain | Exposure (ms) |
| DIC | 100 % | 150 | 50 | |
| GFP (470 nm) | 2 % | 750 | 250 | |
| RFP (560 nm) | 2 % | 750 | 250 | |
| Spinning disk | Channel | Laser power | EM gain | Exposure (ms) |
| DIC | N/A | 200 | 100 | |
| 488 nm | 10 % | 200 | 200 | |
| 561 nm | 10 % | 200 | 200 |
If using one fluorescence channel, use a 30 z-plane stack with images 0.24 µm apart (see Note 10). If using two channels, use a 15 z-plane stack with images 0.5 µm apart. Image at 45 s intervals for 45–90 min total (see Subheading 2.5 for more details)
3.5 Image Deconvolution and Analysis of Polarity Establishment
Deconvolve the z-stack images taken by a widefield fluorescence microscope using deconvolution software (e.g., Huygens Essential) to reduce the blur from out-of-focus light. The classic maximum-likelihood estimation and predicted point spread function method with a signal-to-noise ratio of 10 (see Note 8) is used with a constant background across all images from the same channel on the same day.
Images taken by a spinning disk confocal microscope are denoised by the Hybrid 3D Median Filter plugin in ImageJ (http://rsb.info.nih.gov/ij/plugins/hybrid3dmedian.html). Alternatively, the raw images can be further enhanced by deconvolution with a signal-to-noise ratio of 3 (see Note 8).
Using image quantification software (e.g., Volocity), set a pixel intensity (fluorescence) threshold that selects the pixels corresponding to the polarity patch but not the rest of the cell. If the software is unable to import 3D information for quantification, use the sum projection of the z-stack.
The sum intensity of the polarity patch is recorded from its first detection (we generally focus on early times before bud emergence). The intensity at each time point is normalized to the peak intensity value within the imaging period, and plotted against time.
3.6 Arresting Cells with Ste5-CTM Fusion and Imaging Polarity Patch Wandering
Grow cells overnight at 30 °C in CSM + 2 % dextrose to a log phase culture (OD600 = 0.1–0.7) (see Note 6).
Dilute cells to 1.5 ml of OD600 = 0.1, and treat with 20 nM β-estradiol for 4 h (see Note 9).
Centrifuge 1 ml of culture at 16,000 × g for 1 min, and aspirate most of the liquid. Add 1 µl of Tetraspeck beads (diluted 1:5 in water), and resuspend pelleted cells.
Mount approximately 0.7 µl onto agarose slabs and seal with Vaseline. If using α-factor slabs, let cells sit on slabs for 20 min prior to imaging.
Image cells (see Subheading 2.5) using settings listed in Table 3.
Table 3.
Settings for imaging polarity patch movement
| Spinning disk | Channel | Laser power | EM gain | Exposure (ms) |
|---|---|---|---|---|
| DIC | N/A | 200 | 100 | |
| 488 nm | 15–18 % | 200 | 200 | |
| 561 nm | 15 % | 200 | 200 |
Use a 30 z-plane stack with images 0.24 µm apart. Image at 1.5 min intervals for 30–45 min (see Subheading 2.5 for more details)
3.7 Tracking Polarity Patch Wandering and Calculating Mean Squared Displacement (MSD)
Using image processing software (e.g., Volocity), set a pixel intensity (fluorescence) threshold that selects the pixels that correspond to the polarity patch but not the rest of the cell. Use the same threshold for all the samples being compared.
Remove any objects detected with an area <0.1 µm2.
Determine the centroid of a Tetraspeck bead in the field of view for each movie. This will serve to control for stage drift during imaging.
Determine the location of the centroid of the polarity patch (in three dimensions) for each cell at each time point.
If there are multiple polarity patches for a given time point for a given cell, remove all centroid measurements for that cell at that time point.
From each polarity patch centroid location, subtract the Tetraspeck bead centroid location.
Combine centroids from continuous trajectories: patches that have been continuously measured in a given cell.
Calculate the MSD for each trajectory. For all pairs of time points ti, tj and corresponding centroid positions pi,pj, calculate the squared distance between pi and pj. Then, average all squared distances for each time interval T = tj − ti per cell.
For each cell, average the MSD of all trajectories. We consider each cell an observation unit for statistical analysis, and we typically measure approximately 50 cells per condition in an experiment.
4 Notes
One of the yeast strains (BF264-15D) used in our lab carries a mutation (ade1) that can lead to the accumulation of a fluorescent red metabolite under low-adenine conditions, which interferes with imaging. To prevent this, 0.5 mM adenine is added to the CSM. For strains with a wild-type adenine synthesis pathway, extra adenine is not needed.
Sometimes the temperature recorded by the microscope is not accurate. We recommend determining the temperature that the sample is exposed to rather than relying on the microscope’s thermometer.
When comparing the appearance or movement of two fluorescent probes, note that the second fluorescent channel will be imaged with a few seconds delay compared to the first channel.
The 15 ml conical tube can be kept in a 95 °C heat block to prevent the agarose solution from solidifying after microwaving.
Certain plastics are better at pelleting dilute cell samples. If the cells are not well-pelleted after a 1 min centrifuge spin at 16,000 × g, aspirate the media, scrape the sides of the tube using a pipette tip, and spin again.
We have found that growing multiple dilutions of starter cultures overnight is useful for ensuring one culture will be the appropriate density.
Some strains take longer to respond to HU so longer treatment is needed to obtain a synchronized population. Check cells under microscope after 3 h HU treatment to ensure good synchrony (>70 % large-budded cells).
The signal-to-noise ratio listed here is not the ratio on the raw images; instead, it is the ratio by which the raw images will be enhanced after deconvolution. The signal-to-noise ratio of raw images can be influenced by excitation light intensity, the brightness of fluorescence probes, the sensitivity of camera, etc., so the enhancing ratio we suggest here may not apply to images generated from other microscopes. We suggest choosing a signal-to-noise ratio by deconvolving the same image with different ratios and choosing the one that effectively reduces the blur without introducing erroneous signals not seen on the raw image.
Because the cells are asynchronous when we initially treat them with β-estradiol, the cells arrest at different times. 3–4 h treatment is usually sufficient for >95 % cells to arrest. As the arrested cells continue to grow, larger size yields higher MSD, so it is essential to maintain the same β-estradiol treatment time for all samples.
Bud emergence can occur outside the current focal plane. If the timing of bud emergence is important for analysis, image using a z-stack in the DIC channel.
Acknowledgments
We thank Audrey Howell and Jayme Dyer for their role in developing the protocols discussed here. A.W.M. and C.-F.W. contributed equally to this work. Work in the Lew lab was supported by NIH/NIGMS grants GM62300 and GM103870 to D.J.L.
References
- 1.Klis FM, de Koster CG, Brul S. Cell wall-related bionumbers and bioestimates of Saccharomyces cerevisiae and Candida albicans. Eukaryot Cell. 2014;13:2–9. doi: 10.1128/EC.00250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howell AS, Lew DJ. Morphogenesis and the cell cycle. Genetics. 2012;190:51–77. doi: 10.1534/genetics.111.128314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlton PM, Boulanger J, Kervrann C, et al. Fast live simultaneous multiwavelength four-dimensional optical microscopy. Proc Natl Acad Sci U S A. 2010;107:16016–16022. doi: 10.1073/pnas.1004037107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell AS, Jin M, Wu CF, et al. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 2012;149:322–333. doi: 10.1016/j.cell.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 6.Dyer JM, Savage NS, Jin M, et al. Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr Biol. 2013;23:32–41. doi: 10.1016/j.cub.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louvion JF, Havaux-Copf B, Picard D. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 1993;131:129–134. doi: 10.1016/0378-1119(93)90681-r. [DOI] [PubMed] [Google Scholar]
- 9.Bi E, Pringle JR. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson HE, Wittenberg C, Cross F, Reed SI. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]