Abstract
Human PrimPol is a monomeric enzyme whose DNA primase activity is required to rescue stalled replication forks during nuclear and mitochondrial DNA replication. PrimPol contains an Archeal-Eukaryotic Primases (AEP) core followed by a C-terminal Zn finger-containing domain (ZnFD), that is exclusively required for primer formation and for PrimPol function in vivo. The present study describes the sequential substrate interactions of human PrimPol during primer synthesis, and the relevance of the ZnFD at each individual step. Both the formation of a PrimPol:ssDNA binary complex and the upcoming interaction with the 3′-nucleotide (pre-ternary complex) remained intact when lacking the ZnFD. Conversely, the ZnFD was required for the subsequent binding and selection of the 5′-nucleotide that will become the first nucleotide of the new primer strand. Providing different 5′-site nucleotides, we can conclude that the ZnFD of PrimPol most likely interacts with the γ-phosphate moiety of the 5′-site nucleotide, optimizing formation of the initial dimer. Moreover, the ZnFD also contributes to recognize the cryptic G at the preferred priming sequence 3′GTC5′. Dimer elongation to obtain long DNA primers occurs processively and is facilitated by the 5′-terminal triphosphate, indicating that the ZnFD is also essential in the subsequent translocation/elongation events during DNA primer synthesis.
INTRODUCTION
Most DNA polymerases are unable to initiate DNA synthesis, as they cannot use a dNTP as the initiating/priming source of the hydroxyl group required to transfer elongating nucleotides during DNA replication. The most common mechanism to resolve this issue relies on a specialized RNA polymerase, a primase, which can initiate synthesis from a single ribonucleoside triphosphate (NTP) to produce short RNA primers whose 3′-hydroxyl group is used for subsequent elongation by DNA polymerases (1,2). The molecular mechanism of primer synthesis in conventional primases begins when the primase binds the single-stranded DNA (ssDNA) template, without the strict need to recognize a specific sequence. That said, some primases, especially those from bacterial origin, start primer synthesis at preferred template initiation sites: DnaG from Escherichia coli starts synthesis at 3′-GTC-5′ (3), T7 primase initiates at 3′-CTG- 5′ (4,5), and herpes virus recognizes 3′-GTCC-5′ (6,7). Conversely, eukaryotic primases usually do not show a strict sequence preference, although pyrimidine-rich template regions are favored (8), given that primers generally start with purines (9). Primases can initiate de novo RNA synthesis as they have two NTP binding sites–the 5′-site or initiation site that establishes the NTP at the 5′-end of the primer, and the 3′-site or elongation site that accommodates incoming NTPs at the 3′end of the growing primer (10). Formation of the initial dimer is the rate-limiting step in which the two NTPs, complementary to the template initiation site, are not bound simultaneously to their sites: binding of the 3′ NTP is assumed to occur first, followed by binding of the 5′-site NTP and catalysis (11). The initiating dimer (pppN-p-N) is then translocated to make room for the next incoming nucleotide, and elongation of the primer continues processively usually up to a determined length (5–12 nt).
Based on their structure, primases can be divided into two groups: the DnaG superfamily with a TOPRIM-fold domain (12), and the AEP superfamily, which contains an RNA recognition motif fold (13,14). Iyer et al. (15) have identified more than 10 AEP families, including a novel family termed PrimPol (primase-polymerase), which have been the subject of intensive study since its recent discovery. The first described PrimPol was Orf904 from the Sulpholobus islandicus archaeal plasmid pRN1 (16), but PrimPols are encoded also in the genomes of many bacteria, archaea, eukaryotes, viruses, plasmids and bacteriophages. Some recently characterized members of the PrimPol family include BcMCM from Bacillus cereus (17), TthPrimPol from Thermus thermophilus (18), and human PrimPol, the first to be described in eukaryotes (19–21). Unlike conventional primases, PrimPols use dNTPs to build-up the primers they make, with the exception of the initiating nucleotide that is likely an NTP. Moreover, PrimPols can also use dNTPs to extend pre-existing DNA or RNA primers (22), behaving like conventional DNA polymerases. Because of this dual function, this group of enzymes were originally named PrimPols (16).
Human PrimPol is a monomeric enzyme that contains motifs A, B and C, shared with other AEP primases, but also an extra C-terminal Zn finger-containing domain (ZnFD) that has three conserved cysteines and a histidine (Cys-His-Cys-Cys), with the potential to coordinate a Zn2+ atom to form a Zn-finger, very similar to that present in AEP primases from the herpesvirus family (15). Zn-fingers are generally involved in the maintenance of protein integrity and/or protein-protein interactions, but also in recognizing specific template features at the initiation site of primases (23,24). Interestingly, herpes virus primase (UL52), which belongs to the same AEP subfamily as human PrimPol (15), has been shown to require its C-terminal ZnFD for primase activity and even DNA binding (25). The ZnFD in human PrimPol was also shown to be essential for primase activity both in vivo and in vitro (26,27). A double point mutation in two of the Zn2+ ligands (C419G, H426Y) and a deletion mutant lacking the C-terminal domain (ΔZnFD; lacking aa 410–560) were instrumental in demonstrating that this domain is specifically involved in DNA priming by PrimPol; and that this activity is essential to rescue stalled replication forks challenged by UV or hydroxyurea treatment, and even to maintain the normal replication rate in the absence of exogenous stress (26).
In this work, we further investigated the role of the ZnFD at each step of the DNA priming process by human PrimPol. Our hypothesis, based on previous data, is that PrimPol will initially bind the ssDNA template (binary complex), then the 3′-site nucleotide to form a pre-ternary complex, followed by binding of the 5′-site nucleotide to trigger dimer formation and subsequent elongation to build-up a mature DNA primer. For an optimal approach, we used single-stranded oligonucleotides containing a preferred and unique initiation site (3′..GTC..5′) for human PrimPol, which directs the synthesis of 3pAG or 3pAG initiating dimers, using either ATP or dATP at the 5′-site, and dGTP at the 3′-site (19). We also evaluated the importance of the 3′-G at the priming site (3′..GTC..5′), as a putative cryptic nucleotide (not copied) and its significance for both dimer formation and primer elongation.
MATERIALS AND METHODS
Expression and purification of PrimPol, site-directed mutants and ΔZnFD mutant
Human PrimPol, mutant AxA (catalytically-dead mutant in which Asp114 and Glu116 were simultaneously changed to Ala), and deletion mutant ΔZnFD (Δ410–560) were expressed and purified as previously described (19,26).
Oligonucleotides and nucleotides
Conventional DNA oligonucleotides were synthesized by Sigma Aldrich (St Louis, MO, USA). pppAGT and its control pAGT were obtained from IDT (Coralville, IO, USA). Ultrapure dNTPs were supplied by GE (Fairfield, CT, USA). Radiolabeled nucleotides [γ-32P]ATP, [α-32P]dGTP, [γ-32P]GTP and [α-32P]dTTP (250 μCi; 3000 Ci/mmol,) were purchased from Perkin Elmer (Waltham, MA, USA). T4 polynucleotide kinase, used for 5′ oligonucleotide labeling, was supplied by New England Biolabs (Ipswich, MA, USA).
EMSA for enzyme:ssDNA binary complex
Enzyme:ssDNA binding was performed in buffer A [50 mM Tris-HCl pH 7.5, 40 mM NaCl, 2.5% (w/v) glycerol, 2.5% (w/v) PEG-4000, 1 mM DTT, 0.1 mg/ml BSA], using PNK-[γ-32P]-labeled oligonucleotide 3′T20GTCCT365′ or 3′T20ATCCT365′ (G/ATCC; 1 nM), and increasing concentrations of either wild-type (WT) PrimPol or ΔZnFD mutant (2.5, 5, 10, 20, 40, 80 nM) in the absence of metal, or in the presence of MnCl2 (1 mM) or MgCl2 (5 mM). The reaction (in 20 μl) was incubated at 30°C for 10 min. Subsequently, loading buffer (30% glycerol, 1 mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue) was added and the reaction was analyzed in a native 6% polyacrylamide gel run at 150 V for 120 min at 4°C in Tris-glycine pH 8.3 buffer. After electrophoresis, the gel was dried and the mobility shift of free ssDNA versus enzyme:ssDNA complex was analyzed by autoradiography. Complex stability was evaluated by incubation of either WT PrimPol or ΔZnFD mutant (40 nM) in buffer A with 1 nM of labeled GTCC. After 10 min incubation, increasing concentrations of unlabeled GTCC (1.25, 2.5, 5 and 10 nM) were added to compete with the labeled GTCC, and further incubated for an additional 10 min. The result was analyzed as described before.
EMSA for enzyme:ssDNA:dNTP preternary complex
Pre-ternary complex formation of WT PrimPol, ΔZnFD or null AXA mutants (0.5 μM) and labeled nucleotide [α-32P]dGTP, [γ-32P]ATP, [γ-32P]GTP or [α-32P]dTTP (16 nM) was evaluated in buffer B [50 mM Tris pH 7.5, 2.5% (w/v) glycerol, 40 mM NaCl, 1 mM MnCl2, 1 mM DTT and 0.1 μg/μl BSA], supplemented when indicated with ssDNA 3′T20XTCCT365′ (0.5 μM; 60-mer) (X is: A/G/C or T), and MnCl2 (1 mM) or MgCl2 (5 mM). Reactions (20 μl final volume) were incubated for 10 min at 30°C. Then loading buffer was added and the reactions were analyzed as described above.
DNA primase assays
DNA primase activity of PrimPol and ΔZnFD mutant was evaluated using the 60-mer oligonucleotides 3′T20XTCCT365′ or 3′T10XTCAT155′ (X is: A/G/C or T) as template ssDNA. The 3′T20GTCCT365′ oligonucleotide contains a putative herpes virus priming initiation site (GTCC) (6), which is also a preferred priming site for human PrimPol (19). The reaction mixture (20 μl) contained PrimPol (400 nM or as indicated) in buffer B (as above). The nucleotides and concentrations used are indicated in each figure. Usually, the labeled nucleotide ([γ-32P]ATP or [α-32P]dGTP) is provided at 16 nM and the cold nucleotide/s at 10 μM unless otherwise indicated. To evaluate both sugar and phosphate requirements for the 5′-site nucleotide, AMP, ADP, ATP, dADP and dATP (at 10 μM) were separately used. To measure processivity during primase synthesis by PrimPol, a different template was used (3′T29GTCAGACAGCAT20 5′) and the indicated type and concentration of dNTPs. To analyze PrimPol elongation events following dimer synthesis, synthetic miniprimers mimicking PrimPol products (AG, AGG, CA and ACA) were obtained from Sigma Aldrich, and were labeled using PNK and [γ-32P]ATP. Alternatively, miniprimers harboring a 5′-triphosphate or 5′-monophosphate group (pppAGT or pAGT), obtained from IDT, were used. The primase reaction was incubated during 20 min at 30°C, then stopped by addition of formamide buffer (25 mM EDTA, 95% v/v formamide and 0.3% (w/v) bromophenol blue and xylene-cyanol) and loaded on 8 M urea-containing 20% polyacrylamide sequencing gels. After electrophoresis, de novo synthesized polynucleotides or elongation of miniprimers were detected by autoradiography.
RESULTS
PrimPol interaction with ssDNA does not require the Zn-finger domain and activating metal ions
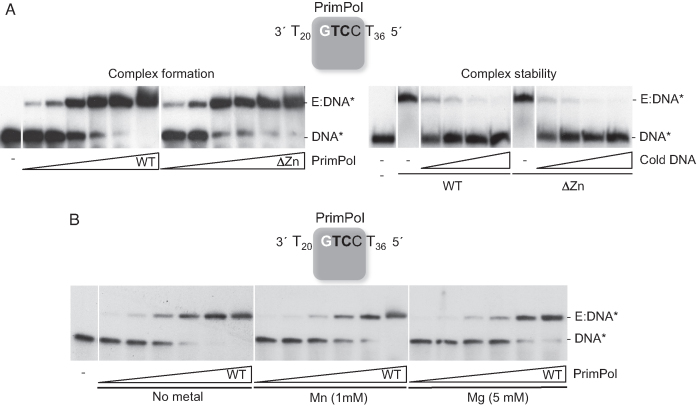
As described (19), human PrimPol efficiently begins primer synthesis at the preferred initiation site 3′GTCC5′, by forming a 3pAG dinucleotide. Hence, this single initiation site, flanked by polydT tracks (3′T20GTCCT365′; abbreviated as GTCC), is an optimal template to test the direct interaction between PrimPol and ssDNA by Electrophoretic Mobility Shift Assay (EMSA). Supporting the concept that PrimPol stably interacts with the 3′GTCC5′ sequence to form a binary complex (E:ssDNA), a single retarded band was detected (Figure 1A, left panel), whose abundance progressively augmented with increasing enzyme concentration. The ZnFD was shown to be irrelevant for this reaction, as the binary complex was formed at a similar protein concentration when using the ΔZnFD mutant, implying that the Zn-finger is not involved in binding ssDNA. We also tested the stability of a pre-formed E:ssDNA binary complex by further addition of an excess of unlabeled ssDNA, and whether the stability was altered by the absence of the ZnFD. The retarded binary complex largely disappeared in the presence of relatively small amounts of competing ssDNA, implying a short-lived interaction, which was not affected by the presence or absence of the ZnFD (Figure 1A, right panel). Furthermore, we examined whether the binary complex formation was affected by the presence of divalent metal ions, observing that the amount of complex formed at different enzyme concentrations did not change when Mn2+ or Mg2+ was added to the reaction (Figure 1B).
Figure 1.
PrimPol binding to ssDNA does not require the Zn-finger domain or activating metal ions. (A) Left panel: EMSA showing the interaction of wild-type PrimPol (WT) or ΔZnFD (ΔZn) mutant (2.5, 5, 10, 20, 40, 80 nM) with [γ-32P]-labeled GTCC oligonucleotide (60-mer; 1 nM) in the absence of metal cofactor; right panel: binary complex stability was assessed by competition of the labeled binary complex (1 nM labeled GTCC and 40 nM WT or ΔZnFD mutant) with increasing concentrations of unlabeled (cold) GTCC (1.25, 2.5, 5 and 10 nM). (B) EMSA using the [γ-32P]-labeled GTCC (60 mer; 1 nM) and WT PrimPol (2.5, 5, 10, 20, 40, 80 nM) in the absence of metal cofactor (left panel), or in the presence of 1 mM MnCl2 (central panel), or 5 mM MgCl2 (right panel). The autoradiographs shown in this figure are representative of at least four independent EMSA experiments.
PrimPol forms a stable pre-ternary complex in the presence of Mn ions, which does not involve the Zn-finger domain
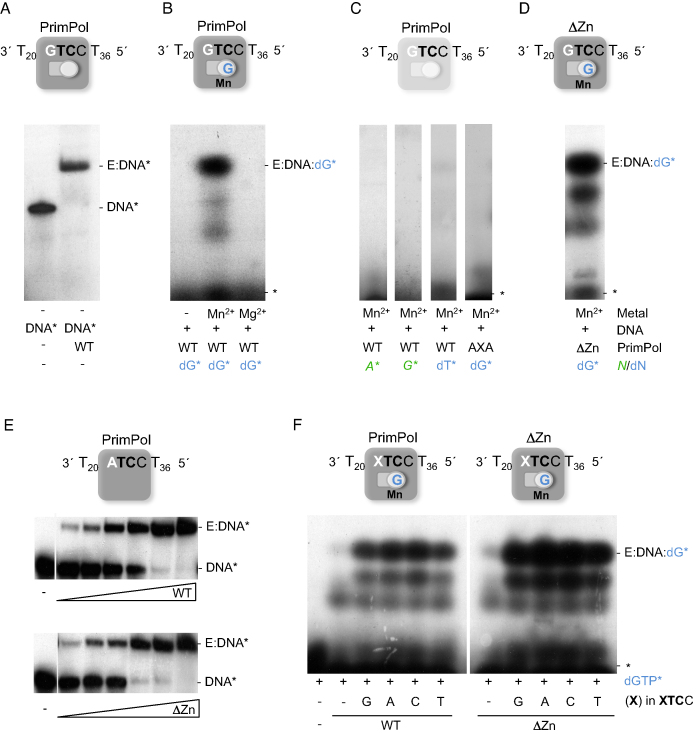
After formation of the enzyme:ssDNA binary complex, a first nucleotide has been proposed to bind at the 3′-site (elongation site) of a primase, followed by binding of the 5′-nucleotide (initiation site), and subsequent dimer formation (11). On the GTCC oligonucleotide, human PrimPol starts primer synthesis by forming a dinucleotide 3pAG, copying TC in the ssDNA template (19). Accordingly, we explored the formation of a pre-ternary complex in PrimPol by assaying the tripartite interaction enzyme:ssDNA:dGTP(3′-site) as an intermediate step preceding catalysis, again using EMSA but in the presence of labeled dGTP. As shown in Figure 2B, we found evidence for the so-called pre-ternary complex (PrimPol:ssDNA:dGTP), which has a similar electrophoretic mobility to that of the binary complex (Figure 2A), but was detected exclusively in the presence of MnCl2 and could not be formed by adding MgCl2 or in the absence of metal. Whereas PrimPol is able to use Mg2+, Mn2+ is largely preferred as a metal cofactor both for priming and elongation (19,20,28), in agreement with the preference for this metal to form a stable pre-ternary complex as shown here. As further support for the physiological relevance of the detected pre-ternary complex (Figure 2C), it could not be formed with: (i) ATP as the 5′-site nucleotide; (ii) GTP as the 3′-site nucleotide, in agreement with the sugar specificity described for PrimPol at the elongation site (19); (iii) dTTP, a non-complementary deoxynucleotide, emphasizing the strict need for templated base selection at this step of pre-ternary complex formation; (iv) the catalytically-dead PrimPol mutant AxA (19), revealing the need for the specific binding of Mn2+ at the catalytic site, contributed by residues Asp114 and Glu116 of human PrimPol. Our results also suggest that the initiating nucleotide (the one to be bound at the 5′-site) binds with a much lower affinity, and that its recruitment likely requires the previous binding of the 3′-nucleotide. Interestingly, the ΔZnFD mutant was able to form a prominent band corresponding to the pre-ternary complex (Figure 2D), indicating that this domain is not required to stabilize the 3′-site nucleotide, in agreement with its dispensability for the polymerase activity (26).
Figure 2.
PrimPol binds the GTCC oligonucleotide and the 3′-site nucleotide dGTP to establish the pre-ternary complex. (A) Binary complex (E:DNA*) of PrimPol (40 nM) with [γ-32P]-labeled oligonucleotide GTCC (1 nM), as detected by EMSA. The scheme above shows the ssDNA and PrimPol, with both 5′-nucleotide (primer; square) and 3′-nucleotide (incoming; circle) binding sites empty. (B) Formation of a pre-ternary complex (E:DNA:dG*) when providing PrimPol (0.5 μM), non-labeled ssDNA GTCC (1 μM), MnCl2 (1 mM), and labeled 3′-nucleotide [α-32P]dGTP (16 nM), which occupies its position as shown in the scheme above. (C) A pre-ternary complex was not detected by EMSA either with [γ-32P]ATP, [γ-32P]GTP, a non-complementary 3′-deoxynucleotide [α32P]-dTTP (16 nM), or with a PrimPol mutant lacking two catalytic metal ligands (AxA). (D) Pre-ternary complex formed with ΔZnFD mutant (ΔZn) (0.5 μM), ssDNA GTCC (1 μM), [α-32P]dGTP (16 nM), and 1 mM MnCl2. (E) EMSA showing the interaction of PrimPol WT or ΔZnFD mutant (2.5, 5, 10, 20, 40, 80 nM) with [γ-32P]-labeled ATCC oligonucleotide (60-mer; 1 nM) in the absence of metal cofactor. (F) Pre-ternary complex of either PrimPol or ΔZnFD mutant (ΔZn) (0.5 μM) with [α-32P]dGTP (16 nM), 1 mM MnCl2 and different oligonucleotides (XTCC), where the cryptic G has been replaced by each of the three other nucleotides A, C and T (depicted as an X in the scheme; 1 μM). The mobility of free labeled ssDNA (DNA*), binary complex (E:DNA*), free labeled nucleotide (*), or pre-ternary complex (E:DNA:dG*) are indicated in the gels. The autoradiographs shown in this figure are representative of at least 3 independent EMSA experiments.
The cryptic G in the template recognition sequence does not determine formation of binary and pre-ternary complexes
Eukaryotic primases do not require a specific DNA sequence to begin primer synthesis in vivo (29), although they do show a preference for pyrimidines that vanishes when higher concentrations of the NTP and Mn2+ are added (30). However, many of them (especially prokaryotic primases) do have an in vitro preference for a short three bases-long recognition motif (4,5,7,31,32) reviewed in (1,2). The 3′-base of the recognition motif is not copied (cryptic), whereas the next two bases, frequently pyrimidines, direct the initiation of primer synthesis that usually begins with two purines (1,33).
We questioned whether the cryptic G in the PrimPol-preferred sequence 3′GTCC5′ is relevant for the sequential interactions with ssDNA and the 3′-incoming nucleotide. PrimPol binding to a ssDNA template, where the cryptic G was replaced by A, produced a PrimPol:ssDNA binary complex (Figure 2E, upper panel) comparable to that shown in Figure 1B, implying that the cryptic G did not affect the overall interaction of PrimPol with ssDNA, as shown even in the absence of the ZnFD (Figure 2E, lower panel).
Next, formation of the pre-ternary complex was evaluated using template variants with the four options at the cryptic base (3′XTCC5′). As shown in Figure 2F, a comparable pre-ternary complex (E:XTCC:dG*) was observed with the four templates, and irrespective of the presence of the ZnFD. Therefore, we conclude that the core AEP domain of PrimPol is responsible for ssDNA binding, template site recognition, and Mn2+-dependent binding of the 3′-site dNTP, and that the cryptic G at the preferred template site does not determine any of these abilities.
The cryptic G-mediated stimulation of dimer synthesis and elongation requires the Zn-finger domain
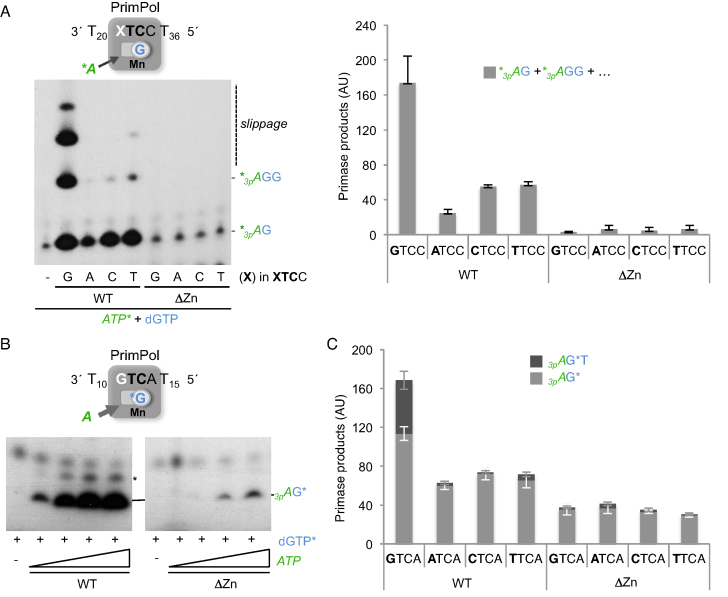
We assessed the effect of the cryptic G at the preferred recognition sequence (GTCC) on the next step after formation of the pre-ternary complex, i.e. dimer synthesis. PrimPol-mediated dimer synthesis was first evaluated on four different templates (XTCC oligonucleotide; where X represents G/A/C or T), providing dGTP at 10 μM to allow formation of the pre-ternary complex, and a low concentration (16 nM) of [γ-32P]ATP as the 5′-site nucleotide. Under these conditions, dimer formation (calculated by integrating all primase products) was significantly higher (4–8-fold) on the template containing G as the cryptic base (see the WT data in Figure 3A). Interestingly, the larger products mainly observed with the cryptic G-containing template also indicated a stimulatory effect of the cryptic G on the translocation and further elongation of the dimer (see later).
Figure 3.
The cryptic G enhances PrimPol primase activity mediated by the Zn-finger domain. (A) Representative primase assay of PrimPol WT or ΔZnFD (400 nM) on either GTCC template or its variants in the cryptic nucleotide (60-mer, 1 μM). The reaction contained [γ-32P]ATP (16 nM) and dGTP (10 μM) as nucleotide substrates. Dimer (*3pAG) and trimer (*3pAGG) products are expected, whereas longer products were the result of slippage-mediated insertion of extra dGTP units. Quantification of primase products (*3pAG+*3pAGG+….) obtained with each template and enzyme in three independent experiments is shown at the right. (B) Representative primase assay (n = 4) of PrimPol WT or ΔZnFD (400 nM) using limiting concentration of the 3′-site nucleotide ([α-32P]dGTP; 16 nM) and increasing concentration of the 5′-site nucleotide (ATP; 0.1, 1, 10, 100 μM), and using a GTCA template to detect the 3pAG* dimer as a main product. A minor product running slower (indicated with an asterisk) likely corresponds to an 2pAG* dimer. (C) Quantification of dimer and trimer products obtained in a primase assay (n = 3) with PrimPol WT or ΔZnFD (400 nM) on the XTCA template variants, providing ATP (10 μM) and [α-32P]dGTP (16 nM) to form the 3pAG* dimer (light gray), and by adding also dTTP (10 μM) to allow formation of the 3pAG*T trimer (dark gray). AU stands for arbitrary units.
We next wondered whether the ZnFD was involved in the recognition of the cryptic G during dimer synthesis. As shown in Figure 3A, the ΔZnFD mutant catalyzed a residual amount of dimer on all of the template variants differing at the cryptic nucleotide, in agreement with previous data indicating its selective role in PrimPol primase activity (26). Considering our previous results showing that the ΔZnFD mutant was able to form a pre-ternary complex (Figure 2D), it was likely that the limiting concentration of the 5′-site nucleotide used in this experiment (16 nM) could be the main factor compromising the primase activity of the ΔZnFD mutant. Hence, we carried out a second primase assay (Figure 3B) maintaining a low and constant concentration of the 3′-site nucleotide ([α-32P]dGTP) and providing increasingly higher concentrations of the 5′-site nucleotide (ATP); moreover, a variant DNA template priming site (GTCA), was used to facilitate a separate analysis of dimer and trimer products, and to avoid slippage-mediated products (like those previously shown in Figure 3A). Under these new experimental conditions, a major band corresponding to the 3pAG* dimer was observed in the presence of the WT PrimPol even at the lowest ATP concentration (Figure 3B, left panel). By contrast, the ΔZnFD mutant required a much higher concentration of ATP to catalyze the 3pAG* dimer (Figure 3B, right panel). Thus, it is likely that the ZnFD is specifically required to stabilize the 5′-site nucleotide.
These new primase assay conditions, especially when providing a high concentration of the 5′-site nucleotide (ATP), allow detection and quantification of a significant amount of dimer even in the absence of the ZnFD. Thus, we re-evaluated the impact of the cryptic G under these conditions, not only during dimer formation (providing ATP+dGTP*), but also for the next elongation step (providing ATP+dGTP*+dTTP to make a trimer). As quantified in Figure 3C, only a G at the cryptic position in the XTCA priming site favors dimer formation (light gray bars) and its rate-limiting elongation (dark gray bars) by WT PrimPol. By contrast, the ΔZnFD mutant was unable to exploit the cryptic G in the template, producing similar amounts of dimers with the four templates, and very inefficient elongation in all cases. We therefore conclude that the ZnFD is involved in the recognition/positioning of the cryptic G, which facilitates not only dimer formation but also its translocation and elongation.
The Zn-finger domain is not required to elongate synthetic primers of minimal size
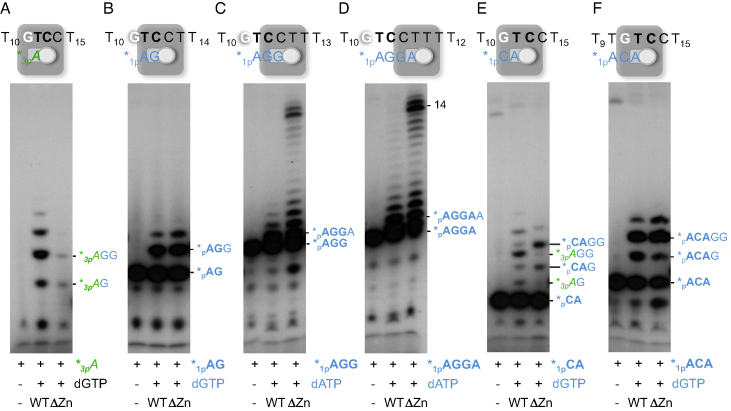
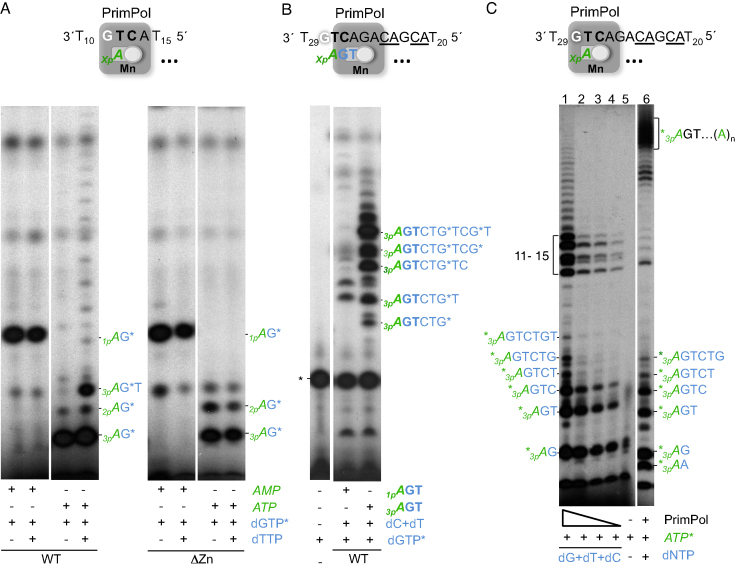
Thus far, our analysis of the importance of the ZnFD of PrimPol during primer synthesis points to a direct role in the stabilization of the 5′-site nucleotide, and in the proper positioning of the cryptic G. A direct effect in catalysis during primer synthesis is much less likely, considering previous data showing that the ZnFD is dispensable for the polymerase and translesion synthesis activities of PrimPol (26,27). To explore this further, a collection of miniprimers that resemble PrimPol products made on the 3′GTCC5′ template (AG, AGG, AGGA…) were commercially synthesized, labeled with [γ-32P]ATP at their 5′-end, and evaluated as valid primers to be extended by WT PrimPol or by the ΔZnFD mutant. Figure 4A shows a positive control in which WT PrimPol and, much less efficiently, the ΔZnFD mutant, synthesize and elongate a dimer when starting from ATP (‘1-nt primer’) and dGTP. However, when a preformed dimer *pAG is provided as primer, it was similarly elongated by the WT and the ΔZnFD mutant, suggesting, at a first glance, that the ZnFD is dispensable once the dimer is formed (Figure 4B). The very same conclusion was drawn when a 3-mer (*pAGG; panel C) or 4-mer oligo (*pAGGA; panel D) was used as a primer, and dATP was added for elongation, although in these cases the ΔZnFD mutant was more efficient during polymerization as previously described (26). This higher efficiency is due to a 50- to 100-fold decrease of the Km for incoming dNTPs measured in DNA polymerization assays (results not shown), in good agreement with the enhanced formation of the pre-ternary complex by the ΔZnFD mutant (Figure 2D and F). Strikingly, we had previously shown that after dimer formation, conversion to trimer was significantly reduced when lacking the ZnFD (Figure 3C), which contradicts the experimental results with the synthetic *pAG primer (Figure 4B). An important difference that could explain this discrepancy is that in the primase assay, the 5′-site nucleotide provided is a triphosphate, whereas the oligonucleotides provided as synthetic miniprimers contain a monophosphate at the 5′-end. This indicates that the ZnFD is also required during elongation, if the primers contain a 5′-terminal triphosphate.
Figure 4.
PrimPol Zn-finger domain is not needed to elongate synthetic mini-primers. PrimPol WT or the ΔZnFD (400 nM) were evaluated in a primase/elongation assay (n = 3) using the 3′T10GTCC T15 5′ oligonucleotide (1 μM) as template, ATP (panel A) or different synthetic/PNK-labeled primers (*pAG, panel B; *pAGG, panel C; *pAGGA, panel D; *pCA, panel E; and *pACA, panel F), each at 16 nM, and the required 3′-incoming nucleotides dGTP or dATP (at 10 μM) for dimer formation or primer elongation. The schemes above show the GTCC template and the position of the different mini-primers provided or the 5′-site initiating ATP. The representative autoradiograms show the position of the different products originated from a single triphosphate as primer (ATP) and those extended from the mini-primers.
Miniprimers covering the cryptic G were also used to evaluate its influence during elongation. When the *pCA primer was used (Figure 4E), the elongation bands obtained with the WT PrimPol and the ΔZnFD mutant differed markedly. The WT PrimPol preferentially used the residual [γ-32P]ATP (used for labeling the CA primer) to make *3pAG dimers and its elongated products (*3pAGG, *3pAGGG…) and poorly used the provided *pCA primer that covers the cryptic G, emphasizing the importance of a free cryptic G to favor primase activity. Conversely, the ΔZnFD mutant preferentially used the *pCA primer for elongation (see the extended products *pCAG and *pCAGG), underlining its inability to prime using a free nucleoside triphosphate at the initiation site. This result reinforces our previous conclusion that the ZnFD domain is involved in the recognition of the G and in the stabilization of the 5′-site nucleotide to initiate primer synthesis. Finally when a 3-mer long primer (*pACA) was used to cover the cryptic G, both WT and ΔZnFD mutant produced the same elongation pattern, corresponding only to the extension of the *pACA primer (Figure 4E). This result suggests that this synthetic primer is more stably bound to the template, fully obliterating the cryptic G. Overall, these data underscore that the cryptic G must be in ssDNA configuration to trigger initiation of primer synthesis by PrimPol.
The Zn-finger domain is required to use a triphosphate as the preferred initiating 5′-nucleotide
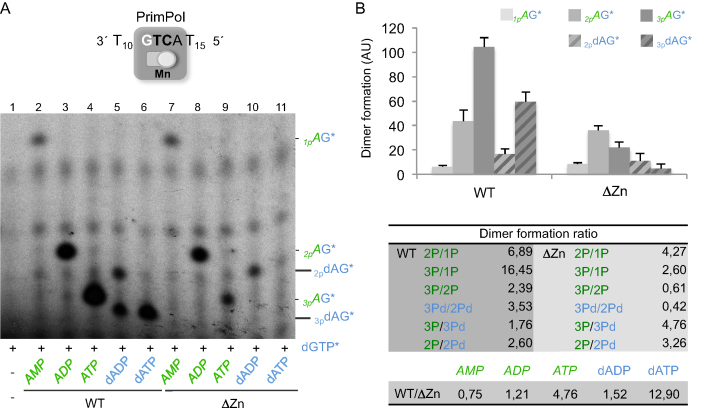
To directly address whether the ZnFD is involved in the interaction with the initiating nucleotide, and particularly the impact of the phosphate moiety, we assayed dimer formation with mono, di, and triphosphate versions of the initiating nucleotide, in both sugar configurations (A and dA). As shown in Figure 5A, formation of AG* dimers by the WT PrimPol proportionally increased with the number of phosphates in the 5′-site ribonucleotide (lanes 2–4): dimer formation was enhanced 7-fold when using ADP instead of AMP, and over 16-fold when using ATP (Figure 5B). This beneficial effect of the phosphates was also tested when using deoxy-A nucleotides, an alternative sugar for PrimPol at the 5′-initiation site but largely preferred at the 3′-elongating site (19). Again, the triphosphate version of the nucleotide was the most efficient to trigger dimer formation (Figure 5A, lanes 5 and 6), with an increase of ∼3.5-fold when using dATP over dADP (Figure 5B). Another conclusion of this experiment is that a ribose is preferred over the deoxyribose at the 5′-site, by a factor of almost 2-fold or 2–5-fold, when comparing ATP with dATP (lanes 4 and 6) or ADP with dADP (lanes 3 and 5), respectively (see also Figure 5B).
Figure 5.
Use of nucleoside triphosphates as the best 5′-nucleotides for dimer formation requires the Zn-finger domain. (A) Representative primase assay using the oligo GTCA to measure dimer formation by wild-type PrimPol WT or its ΔZnFD mutant (400 nM) in the presence of [α-32P]dGTP (16 nM) and different 5′-nucleotides (AMP, ADP, ATP, dADP or dATP, at 10 μM). The different mobility of the dimers formed (1pAG*, 2pAG*…) is shown after autoradiography. Note that the mobility of each dimer is affected by the number of 5′-phosphates, but also by the sugar of the 5′-nucleotide (i.e. 3pAG* vs3pdAG*). (B) Quantification of four different experiments as the one shown in A, representing (with bars) the average of the different dimers formed, according to the 5′-nucleotide used. AU stands for arbitrary units. Dimer formation ratios among selective pair of dimers are indicated in the table below; nomenclature (2P/1P, 3Pd/2Pd….) is abbreviated on the basis of the number of 5′-terminal phosphates.
Unexpectedly, the absence of the ZnFD domain did not significantly alter the efficiency of using AMP (Figure 5A, lane 7), and only modestly affected the efficiency of using a diphosphate, either ADP or dADP (Figure 6A, lanes 8 and 10), thus keeping also the preference for the ribose sugar. However, the ΔZnFD mutant was very inefficient (lane 9) or incapable (lane 11) to use a triphosphate; in fact, the efficiency of forming 3pAG* versus 1pAG* was quite similar (2.5-fold), but very much reduced in comparison with 2pAG* (Figure 5B), suggesting that the extra γ-phosphate makes the nucleotide more unstable in the absence of the ZnFD. Accordingly, a specific defect in binding/stabilizing the γ-phosphate of the 5′-initiating nucleotide is the most likely explanation for the very low primase activity reported for this mutant (26).
Figure 6.
A putative interaction of PrimPol ZnFD with the 5′-end triphosphate is required for dimer translocation and processive elongation. (A) Representative primase assay (n = 3) using the oligo GTCA (1 μM) to measure dimer formation either by wild-type PrimPol (WT) or its ΔZnFD mutant (ΔZn) at 400 nM, in the presence of [α-32P]dGTP (16 nM) and either AMP (100 μM) or ATP (10 μM for the WT; 100 μM for the ΔZn). When indicated, dTTP (10 μM) was added to evaluate elongation of the dimers. (B) Elongation of a synthetic miniprimer (1pAGT or 3pAGT; 10 μM) in the presence of dCTP, dTTP (10 μM) and [α-32P]dGTP (16 nM) by wild-type PrimPol (WT) at 400 nM. *indicates a main labeled product, intrinsic to [α-32P]dGTP that is present in all conditions including the control without enzyme. (C) Lanes 1–4: evaluation of processive primer synthesis by WT PrimPol (400, 200, 100, 50 nM), in the presence of [γ-32P]ATP (16 nM) and 10 μM (dG/dT/dC)TP. A control reaction in the absence of PrimPol and dNTPs (lane 5) shows minor products intrinsic to the batch of labeled ATP. Addition of the four dNTPs at 10 μM, [γ-32P]ATP (16 nM) and WT PrimPol (400 nM) allowed full extension of the primers (lane 6). The autoradiographs shown are representative of 4 independent experiments.
We conclude that the ZnFD of PrimPol is indispensable for the use of a triphosphate as the preferred 5′-initiating nucleotide, probably increasing its stability at the active site to allow dimer synthesis.
A 5′-terminal triphosphate facilitates primer elongation
We have shown that the efficiency of dimer formation by PrimPol strongly depends on the number of phosphates of the 5′-nucleotide, with a triphosphate being the optimal substrate. We next addressed whether the 5′-terminal phosphates also influence the ability to translocate the dimer, and its subsequent elongation, and the impact of the ZnFD. The oligo GTCA was used as the most suitable template to generate dimers using AMP or ATP as the 5′-nucleotide, and labeled dGTP as the first 3′-nucleotide. To obtain comparable amounts of labeled dimers, we adjusted the concentration of the initiating nucleotide, using 100 μM AMP or 10 μM ATP when using the WT PrimPol, and 100 μM of both AMP or ATP when using the ΔZnFD mutant. Elongation of each dimer into a trimer was assayed by addition of the next 3′-nucleotide, dTTP.
As shown in Figure 6A (left panel), the 1pAG* dimer could not be converted into a trimer, whereas the dimer starting with a 5′-end triphosphate (3pAG*) was elongated by the WT PrimPol to form a trimer. Longer elongation products were also obtained from the 3pAG* dimer, very likely due to reiterative incorporation of ATP (provided to form the dimer) opposite the polydT template tail, in spite of the sugar discrimination at the 3′-nucleotide elongations site (19). Thus, when ATP is used as the optimal initiating nucleotide, the 5′-triphosphate that remains after dimer formation is also very important to facilitate translocation and further nucleotide addition, most likely due to a sustained role of the triphosphate in stabilizing the incipient primer. The ΔZnFD mutant was very inefficient in converting the 3pAG* dimer to the extended 3pAG*T primer, suggesting that the Zn-finger domain is responsible for the interaction with the 5′-terminal triphosphate that is required for dimer translocation and subsequent trimer formation.
To further investigate the role of the 5′-terminal triphosphate beyond dimer formation, we compared the elongation efficiency of a synthetic trinucleotide 3pAGT as primer, versus the 5′-monophosphate version 1pAGT. For that, we used a longer template (see Figure 6B), in which the preferred GTC priming site is followed by a heteropolymeric sequence of 8 nt, preceding a polydT20 tail. As expected, the trimer containing the 5′-terminal triphosphate was more efficiently elongated by PrimPol than the one with a single 5′-terminal phosphate (Figure 6B). Moreover, this stimulation by the triphosphate was not observed with the mutant lacking the Zn-finger domain (data not shown). In conclusion, the 5′-terminal triphosphate is essential not only during dimer formation but also during further elongation events.
Considering the importance of the 5′-triphosphate also for elongation by PrimPol, we investigated whether such a ZnFD-mediated interaction with the triphosphate would facilitate the processive synthesis of long primers. Thus, using the same template as in Figure 6B, and providing labeled ATP as the initiating nucleotide and only three dNTPs (dG/dC/dT), elongated primers up to 10 nt were the major expected products. As shown in Figure 6C, the primer elongation pattern showed the accumulation of dimer, trimer and tetramer products, which can be produced either as abortive (dissociated) products or due to a rate-limited elongation at these steps. Moreover, an accumulation of longer extended products (11–15 nt) was also observed, which could be explained by a backwards primer realignment event (22) occurring after copying the CA template sequence that precedes the polydT tail (repetition underlined in the scheme of Figure 6C). Interestingly, the pattern/length of elongated primers remained unaltered even when very low enzyme:template ratios were used, demonstrating the processivity of primer synthesis by PrimPol. When the four dNTPs were provided (Figure 6C, lane 6), much longer primers were synthesized by copying also the polydT tail. Some primases are well known to synthesize primers of defined length, as in the case of the human primase (11). According to our findings, PrimPol is apparently unable ‘to count’, but is competent to synthesize very long primers.
DISCUSSION
PrimPols are unconventional primases whose main role is assisting replication of damaged DNA templates. Early reports have shown that the maintenance of normal DNA replication rates in mammalian cells, and the efficient rescue of replication forks stalled by DNA damage or nucleotide substrate deprivation, exclusively requires the exceptional ability of PrimPol to make DNA primers in response to replication stress (21,26). Accordingly, the stalled replicase can switch to a nearby primer, newly synthesized by PrimPol beyond the blocking lesion, thus resuming normal replication fork progression.
The C-terminal domain of human PrimPol contains a replication protein A (RPA)-interacting domain (21,27,34), and also a conserved Zn-finger that is essential for primase activity, but is fully dispensable for polymerase and translesion synthesis activities (26,27). Expression of human PrimPol versions with a deletion of the ZnFD, or solely with single mutations at Zn2+ ligands, did not rescue normal fork rate and fork stalling in PrimPol-depleted cells, implying that only the primase activity of PrimPol is required during nuclear DNA replication (26), and also during mitochondrial DNA replication (35). However, little was known about the molecular details of the primase reaction in PrimPols, and how the C-terminal Zn-finger contributes to this activity.
Sequential steps during primer synthesis by human PrimPol
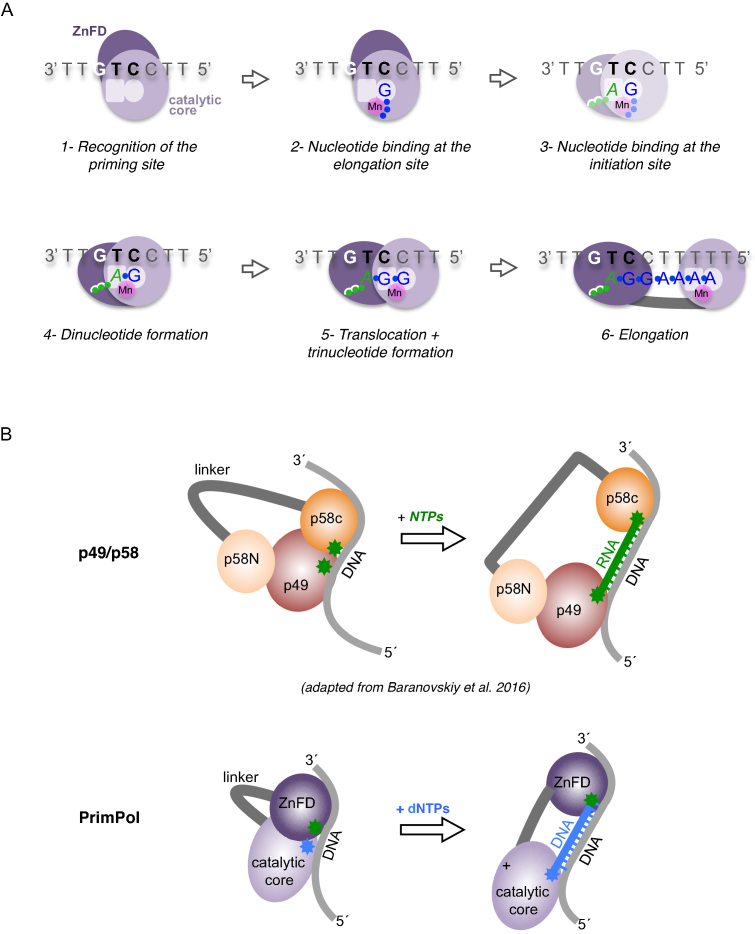
As described here by using templates containing a preferred and unique priming site, we were able to delineate the individual steps and substrate preference of PrimPol when building a primer, and to determine the relevance of the ZnFD at each individual step (Figure 7A). Human PrimPol binds first to any sequence in the ssDNA template, but preferentially to pyrimidine-rich motifs. No divalent metals are required for the stabilization of this enzyme:ssDNA binary complex (Figure 7A, step 1). After PrimPol binding to ssDNA and recognition of a preferred template sequence (GTC in our assays), PrimPol binds a first nucleotide (dGTP) at the elongation site, which is selected by Watson-Crick hydrogen bonding with the templating C at the recognition sequence 3′GTC5′, thus forming a so-called pre-ternary complex (Figure 7A, step 2). Pre-ternary complex formation strictly required the presence of manganese ions, most likely to stabilize the interaction of the incoming nucleotide at the 3′-site. The association of active site motif A (contributing two metal ligands) to the formation of the pre-ternary complex also supports this conclusion. Such a tripartite complex, the pre-ternary complex, was proposed to exist as an early intermediate of the primase reaction (11,36), but was formally demonstrated only in specific members of the AEP superfamily such as LigD-PolDom (36), and now here for the first time in human PrimPol, to our knowledge. Moreover, the capacity to bind both ssDNA template and 3′-nucleotide was shown to reside in the catalytic core of PrimPol, with ZnFD being dispensable at these two steps of the primase reaction.
Figure 7.
Mechanism of DNA priming by human PrimPol. (A) Sequential steps during primer synthesis by human PrimPol. (1) Recognition of the priming site: binding of PrimPol to ssDNA does not rely on the cryptic G of the GTC motif (in bold), and does not require the Zn-finger domain (ZnFD; dark purple). (2) Nucleotide binding at the 3′-elongation site: the 3′-deoxynucleotide (dGTP; in blue) binds to the elongation site without the influence of the ZnFD and the cryptic G, but requires manganese ions (in pink; a single Mn sphere is depicted for simplicity) and template base-complementarity to stabilize such a pre-ternary complex. (3, 4) Nucleotide binding at the 5′-initiation site, and dinucleotide formation: the 5′-ribonucleotide (ATP; in green) binds to the initiation site (step 3 is drawn faint, as it was not experimentally demonstrated) in an optimal configuration facilitated by the cryptic G, to promote dinucleotide 3pA-G formation. The Zn-finger domain plays a key role in positioning the cryptic G and binding the triphosphate moiety of the 5′-site nucleotide, keeping a closed conformation that promotes catalysis. (5) Translocation and trinucleotide formation: the Zn-finger domain maintains the interaction with the cryptic nucleotide and the 5′-end triphosphate, facilitating dimer translocation and next nucleotide addition. (6) Processive elongation: the primer is elongated processively at least up to 10 nucleotides, likely favored by a persisting interaction of the ZnFD with the 5′-triphosphate at the end of the primer, and by its flexible tethering to the catalytic core of PrimPol. (B) Upper scheme: proposed role for the non-catalytic subunit (p58) of the p49/p58 human primase (41); the C-terminal domain (p58C; orange) interacts with the triphosphate of the 5′-initiating nucleotide during dimer synthesis, and also during extension of the RNA primer that will be transferred to Polα. The N-terminal domain (p58N; light orange) interacts with the catalytic subunit p49 (maroon), establishing a limited primer size. Lower scheme: Mirroring the role of p58C, the ZnFD (dark purple) of monomeric PrimPol interacts with the triphosphate of the 5′-initiating nucleotide during dimer synthesis, and also during processive extension of the DNA primer.
The next steps after pre-ternary complex formation are the binding and selection of the initiating nucleotide, which will occupy the PrimPol primer/initiation 5′-site, followed by catalysis and subsequent dimer formation. Attempts to demonstrate the formation of an alternative pre-ternary complex with only the initiating nucleotide (ATP) or even a quaternary complex (enzyme:ssDNA:ATP:dGTP; step 3 at Figure 7A) failed, supporting previous evidence that primases weakly/transiently interact with the initiating nucleotide that will become the 5′-end of the growing primer. Therefore, 5′-nucleotide binding was inferred from the analysis of the first catalytic event, i.e. dimer formation (Figure 7A, step 4), with a special emphasis on the number of phosphates of the 5′-nucleotide provided. Our experiments using different 5′-site nucleotides clearly demonstrated that PrimPol largely prefers a triphosphate in the initiating nucleotide, followed by a diphosphate, and monophosphate as a much less efficient nucleotide substrate. A similar preference for triphosphate-containing initiating nucleotides has been recently described for a PrimPol-like phage enzyme (37). Strikingly, the preference for a triphosphate in human PrimPol is mediated by the ZnFD, explaining the reported reduction in primase activity of a ΔZnFD mutant (26); however, the ZnFD is much less important for 5′-nucleotide substrates with one or two phosphates. Indeed, in the absence of the ZnFD, ADP and dADP were even more efficient than ATP and dATP, respectively, supporting an inhibitory effect of the γ-phosphate. These data imply that the ZnFD of PrimPol is involved in the binding and selection of the 5′-site nucleotide, most likely by establishing interactions with the γ-phosphate moiety of triphosphate-containing nucleotides, making them both valid and optimal substrates for catalysis of the initial dimer. Our analysis also showed a preference for a ribose sugar in the 5′-site nucleotide. That preference was maintained when comparing nucleotides with two or three phosphates, and even in the absence of the ZnFD, suggesting that some residue(s) of the catalytic core of PrimPol can establish stabilizing interactions with the 2′-OH group of the initiating nucleotide. After dimer formation, translocation is needed to copy the next nucleotide of the template (Figure 7A, step 5). Upon translocation, the second nucleotide of the dimer has to move back to be repositioned at the primer/initiation site of the enzyme. Given the preference of PrimPol for inserting dNTPs at the elongation site, also during dimer synthesis (19), the configuration of the initiation site after the first translocation, and also during the next elongation events (Figure 7A, step 6), must be compatible with a deoxyribose, and not only with a ribose as in conventional primases. Interestingly, the efficiency of translocation and further elongation steps also took advantage of a 5′-terminal triphosphate initiating the primer strand, suggesting that the interaction via the ZnFD could serve to facilitate translocation, and/or to avoid enzyme dissociation. In fact, we have observed that PrimPol primer synthesis is highly processive at least until 10–12 nt, in contrast to the distributive pattern of elongation described when starting polymerization from a pre-existing primer (19,27).
We also evaluated the relevance of the so-called ‘cryptic nucleotide’, which precedes the two templating bases that dictate the synthesis of the initiating dimer. Such a cryptic nucleotide (G in the preferred priming site 3′GTC5′; (19)) was irrelevant for the formation of both binary and pre-ternary complexes, but crucial for an efficient dimer formation using triphosphate-containing nucleotides. Interestingly, the cryptic G-mediated stimulation of both dimer synthesis and elongation required the ZnFD. A similar observation was reported for herpes virus primase, where the cryptic G provokes a more efficient polymerization or a decreased rate of primer dissociation (38,39).
Is the role of the ZnFD unique for PrimPols?
Not all primases contain a ZnFD, although they can form a Zn-finger at the catalytic core domain. A relevant example is the catalytic subunit (p49; Prim1) of the dimeric human primase, and AEPs that do not contain a C-terminal ZnFD as in human PrimPol. Instead, p49 interacts with a large regulatory subunit (p58; Prim2) whose N-terminal domain serves to connect the catalytic subunit p49 with Polα (40). Interestingly, the C-terminal domain of the non-catalytic subunit (p58) of human primase, which contains a S-Fe cluster instead of a Zn-finger, was shown to stably interact with a primed-template only if the primer strand contains a 5′-terminal triphosphate (41). Further structural work (42) allowed these authors to propose that the principal role of the non-catalytic subunit is the stabilization of the triphosphate-containing nucleotide at the 5′site of the primase, thus contributing to the formation of the enzyme:ssDNA:NTP:NTP quaternary complex required for dimer synthesis (Figure 7B). The p58 C-terminal domain also interacts with the 3′-DNA template region immediately upstream to the initiation site (41). This interaction is mediated by a specific histidine (His303) that ‘pairs’ with the cryptic base at the priming site, stabilizing the 5′-nucleotide via stacking interactions with the base. Baranovsky and coworkers also proposed that this interaction with the 5′-terminal triphosphate remains during elongation, thus serving to build a primer of a very defined size (Figure 7B) that will be transferred to Polα (42). This mechanism of ‘molecular brake’ has been also proposed for DnaG-like primases (43). A similar dependence on the 5′-triphosphate for primer elongation was also reported for the Deep-sea vent phage DNA polymerase (37), suggesting the need for a specific domain that would be the functional counterpart of the ZnFD of eukaryotic PrimPols. Based on our present data, we propose that the C-terminal extension of the catalytic core of human PrimPol, referred to here as ZnFD, is functionally equivalent to the C-terminal domain of the p58 subunit of the human Pol alpha-primase complex (see Figure 7B). Like p58, the ZnFD of the monomeric human PrimPol is crucial to stabilize the 5′-site nucleotide, especially at preferred PrimPol priming sites, perhaps by mediating the optimal configuration/positioning of the cryptic nucleotide and by directly contributing to the stabilization of the incoming triphosphate-containing nucleotide. Moreover, a sustained interaction with the 5′-triphosphate during elongation could serve to minimize premature dissociation from the growing primer, but allowing the transfer of its 3′-terminus to the elongating polymerase at any time and primer size.
The catalytic core of PrimPol (residues 1 to 354) was recently crystalized with a template/primer and dATP as incoming 3′-nucleotide, thus representing the polymerization mode of PrimPol (44). The structure revealed that PrimPol primarily interacted with the template DNA strand, but showed no contacts with the primer DNA strand, explaining the large distributivity displayed by PrimPol during elongation of a pre-existing primer that does not contain a 5′-terminal triphosphate (34). Additional crystal structures of PrimPol in primase mode will be instrumental to evaluate how the ZnFD contributes to stabilize the incoming 5′nucleotide, which specific residues are involved in the interaction with its sugar and phosphate residues, and whether these interactions are modified or remain during processive elongation to build a mature primer.
FUNDING
Spanish Ministry of Economy and Competitiveness [BFU2012-3769, BFU2014-51672-REDC, BFU2015-65880-P] and Comunidad de Madrid [S2010/BMD-2361] to L.B.; P.A.C. is recipient of a FPI-predoctoral fellowship from Spanish Ministry of Economy and Competitiveness. Funding for open access charge: Spanish Ministry of Economy and Competitiveness [BFU2015-65880-P].
Conflict of interest statement. None declared.
REFERENCES
- 1. Frick D.N., Richardson C.C.. DNA primases. Annu. Rev. Biochem. 2001; 70:39–80. [DOI] [PubMed] [Google Scholar]
- 2. Kuchta R.D., Stengel G.. Mechanism and evolution of DNA primases. Biochim. Biophys. Acta. 2010; 1804:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hiasa H., Sakai H., Tanaka K., Honda Y., Komano T., Godson G.N.. Mutational analysis of the primer RNA template region in the replication origin (oric) of bacteriophage G4: priming signal recognition by Escherichia coli primase. Gene. 1989; 84:9–16. [DOI] [PubMed] [Google Scholar]
- 4. Mendelman L.V., Richardson C.C.. Requirements for primer synthesis by bacteriophage T7 63-kDa gene 4 protein. Roles of template sequence and T7 56-kDa gene 4 protein. J. Biol. Chem. 1991; 266:23240–23250. [PubMed] [Google Scholar]
- 5. Tabor S., Richardson C.C.. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cavanaugh N.A., Kuchta R.D.. Initiation of new DNA strands by the herpes simplex virus-1 primase-helicase complex and either herpes DNA polymerase or human DNA polymerase alpha. J. Biol. Chem. 2009; 284:1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tenney D.J., Sheaffer A.K., Hurlburt W.W., Bifano M., Hamatake R.K.. Sequence-dependent primer synthesis by the herpes simplex virus helicase-primase complex. J. Biol. Chem. 1995; 270:9129–9136. [DOI] [PubMed] [Google Scholar]
- 8. Holmes A.M., Cheriathundam E., Bollum F.J., Chang L.M.. Initiation of DNA synthesis by the calf thymus DNA polymerase-primase complex. J. Biol. Chem. 1985; 260:10840–10846. [PubMed] [Google Scholar]
- 9. Yamaguchi M., Hendrickson E.A., DePamphilis M.L.. DNA primase-DNA polymerase alpha from simian cells: sequence specificity of initiation sites on simian virus 40 DNA. Mol. Cell. Biol. 1985; 5:1170–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frick D.N., Kumar S., Richardson C.C.. Interaction of ribonucleoside triphosphates with the gene 4 primase of bacteriophage T7. J. Biol. Chem. 1999; 274:35899–35907. [DOI] [PubMed] [Google Scholar]
- 11. Sheaff R.J., Kuchta R.D.. Mechanism of calf thymus DNA primase: slow initiation, rapid polymerization, and intelligent termination. Biochemistry. 1993; 32:3027–3037. [DOI] [PubMed] [Google Scholar]
- 12. Aravind L., Leipe D.D., Koonin E.V.. Toprim–a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic. Acids. Res. 1998; 26:4205–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Augustin M.A., Huber R., Kaiser J.T.. Crystal structure of a DNA-dependent RNA polymerase (DNA primase). Nat. Struct. Biol. 2001; 8:57–61. [DOI] [PubMed] [Google Scholar]
- 14. Aravind L., Mazumder R., Vasudevan S., Koonin E.V.. Trends in protein evolution inferred from sequence and structure analysis. Curr. Opin. Struct. Biol. 2002; 12:392–399. [DOI] [PubMed] [Google Scholar]
- 15. Iyer L.M., Koonin E.V., Leipe D.D., Aravind L.. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic. Acids. Res. 2005; 33:3875–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lipps G., Rother S., Hart C., Krauss G.. A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J. 2003; 22:2516–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanchez-Berrondo J., Mesa P., Ibarra A., Martinez-Jimenez M.I., Blanco L., Mendez J., Boskovic J., Montoya G.. Molecular architecture of a multifunctional MCM complex. Nucleic. Acids. Res. 2012; 40:1366–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Picher A.J., Budeus B., Wafzig O., Kruger C., Garcia-Gomez S., Martinez-Jimenez M.I., Diaz-Talavera A., Weber D., Blanco L., Schneider A.. TruePrime is a novel method for whole-genome amplification from single cells based on TthPrimPol. Nat. Commun. 2016; 7:13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garcia-Gomez S., Reyes A., Martinez-Jimenez M.I., Chocron E.S., Mouron S., Terrados G., Powell C., Salido E., Mendez J., Holt I.J. et al. PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell. 2013; 52:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bianchi J., Rudd S.G., Jozwiakowski S.K., Bailey L.J., Soura V., Taylor E., Stevanovic I., Green A.J., Stracker T.H., Lindsay H.D. et al. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell. 2013; 52:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wan L., Lou J., Xia Y., Su B., Liu T., Cui J., Sun Y., Lou H., Huang J.. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. Embo. Rep. 2013; 14:1104–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martinez-Jimenez M.I., Garcia-Gomez S., Bebenek K., Sastre-Moreno G., Calvo P.A., Diaz-Talavera A., Kunkel T.A., Blanco L.. Alternative solutions and new scenarios for translesion DNA synthesis by human PrimPol. DNA Repair (Amst.). 2015; 29:127–138. [DOI] [PubMed] [Google Scholar]
- 23. Laity J.H., Lee B.M., Wright P.E.. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001; 11:39–46. [DOI] [PubMed] [Google Scholar]
- 24. Matthews J.M., Sunde M.. Zinc fingers–folds for many occasions. IUBMB Life. 2002; 54:351–355. [DOI] [PubMed] [Google Scholar]
- 25. Chen Y., Carrington-Lawrence S.D., Bai P., Weller S.K.. Mutations in the putative zinc-binding motif of UL52 demonstrate a complex interdependence between the UL5 and UL52 subunits of the human herpes simplex virus type 1 helicase/primase complex. J. Virol. 2005; 79:9088–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mouron S., Rodriguez-Acebes S., Martinez-Jimenez M.I., Garcia-Gomez S., Chocron S., Blanco L., Mendez J.. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 2013; 20:1383–1389. [DOI] [PubMed] [Google Scholar]
- 27. Keen B.A., Jozwiakowski S.K., Bailey L.J., Bianchi J., Doherty A.J.. Molecular dissection of the domain architecture and catalytic activities of human PrimPol. Nucleic Acids Res. 2014; 42:5830–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zafar M.K., Ketkar A., Lodeiro M.F., Cameron C.E., Eoff R.L.. Kinetic analysis of human PrimPol DNA polymerase activity reveals a generally error-prone enzyme capable of accurately bypassing 7,8-dihydro-8-oxo-2′-deoxyguanosine. Biochemistry. 2014; 53:6584–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirk B.W., Harrington C., Perrino F.W., Kuchta R.D.. Eucaryotic DNA primase does not prefer to synthesize primers at pyrimidine rich DNA sequences when nucleoside triphosphates are present at concentrations found in whole cells. Biochemistry. 1997; 36:6725–6731. [DOI] [PubMed] [Google Scholar]
- 30. Kirk B.W., Kuchta R.D.. Human DNA primase: anion inhibition, manganese stimulation, and their effects on in vitro start-site selection. Biochemistry. 1999; 38:10126–10134. [DOI] [PubMed] [Google Scholar]
- 31. Swart J.R., Griep M.A.. Primase from Escherichia coli primes single-stranded templates in the absence of single-stranded DNA-binding protein or other auxiliary proteins. Template sequence requirements based on the bacteriophage G4 complementary strand origin and Okazaki fragment initiation sites. J. Biol. Chem. 1993; 268:12970–12976. [PubMed] [Google Scholar]
- 32. Tseng T.Y., Frick D.N., Richardson C.C.. Characterization of a novel DNA primase from the Salmonella typhimurium bacteriophage SP6. Biochemistry. 2000; 39:1643–1654. [DOI] [PubMed] [Google Scholar]
- 33. Frick D.N., Richardson C.C.. Interaction of bacteriophage T7 gene 4 primase with its template recognition site. J. Biol. Chem. 1999; 274:35889–35898. [DOI] [PubMed] [Google Scholar]
- 34. Guilliam T.A., Jozwiakowski S.K., Ehlinger A., Barnes R.P., Rudd S.G., Bailey L.J., Skehel J.M., Eckert K.A., Chazin W.J., Doherty A.J.. Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins. Nucleic. Acids. Res. 2015; 43:1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Torregrosa-Munumer R., Forslund J.M.E., Goffart S., Pfeiffer A., Stojkovic G., Carvalho G., Al-Furoukh N., Blanco L., Wanrooij S., Pohjoismaki J.L.O.. PrimPol is required for replication reinitiation after mtDNA damage. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:11398–11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brissett N.C., Martin M.J., Pitcher R.S., Bianchi J., Juarez R., Green A.J., Fox G.C., Blanco L., Doherty A.J.. Structure of a preternary complex involving a prokaryotic NHEJ DNA polymerase. Mol. Cell. 2011; 41:221–231. [DOI] [PubMed] [Google Scholar]
- 37. Zhu B., Wang L., Mitsunobu H., Lu X., Hernandez A.J., Yoshida-Takashima Y., Nunoura T., Tabor S., Richardson C.C.. Deep-sea vent phage DNA polymerase specifically initiates DNA synthesis in the absence of primers. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E2310–E2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramirez-Aguilar K.A., Low-Nam N.A., Kuchta R.D.. Key role of template sequence for primer synthesis by the herpes simplex virus 1 helicase-primase. Biochemistry. 2002; 41:14569–14579. [DOI] [PubMed] [Google Scholar]
- 39. Ramirez-Aguilar K.A., Kuchta R.D.. Mechanism of primer synthesis by the herpes simplex virus 1 helicase-primase. Biochemistry. 2004; 43:1754–1762. [DOI] [PubMed] [Google Scholar]
- 40. Kilkenny M.L., Longo M.A., Perera R.L., Pellegrini L.. Structures of human primase reveal design of nucleotide elongation site and mode of Pol alpha tethering. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15961–15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baranovskiy A.G., Zhang Y., Suwa Y., Gu J., Babayeva N.D., Pavlov Y.I., Tahirov T.H.. Insight into the human DNA primase interaction with template-primer. J. Biol. Chem. 2016; 291:4793–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baranovskiy A.G., Babayeva N.D., Zhang Y., Gu J., Suwa Y., Pavlov Y.I., Tahirov T.H.. Mechanism of concerted RNA-DNA primer synthesis by the human primosome. J. Biol. Chem. 2016; 291:10006–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Corn J.E., Pease P.J., Hura G.L., Berger J.M.. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol. Cell. 2005; 20:391–401. [DOI] [PubMed] [Google Scholar]
- 44. Rechkoblit O., Gupta Y.K., Malik R., Rajashankar K.R., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Structure and mechanism of human PrimPol, a DNA polymerase with primase activity. Sci. Adv. 2016; 2:e1601317. [DOI] [PMC free article] [PubMed] [Google Scholar]