Abstract
Arabidopsis thaliana requires a prolonged period of cold exposure during winter to initiate flowering in a process termed vernalization. Exposure to cold induces epigenetic silencing of the FLOWERING LOCUS C (FLC) gene by Polycomb group (PcG) proteins. A key role in this epigenetic switch is played by transcriptional repressors VAL1 and VAL2, which specifically recognize Sph/RY DNA sequences within FLC via B3 DNA binding domains, and mediate recruitment of PcG silencing machinery. To understand the structural mechanism of site-specific DNA recognition by VAL1, we have solved the crystal structure of VAL1 B3 domain (VAL1-B3) bound to a 12 bp oligoduplex containing the canonical Sph/RY DNA sequence 5′-CATGCA-3′/5′-TGCATG-3′. We find that VAL1-B3 makes H-bonds and van der Waals contacts to DNA bases of all six positions of the canonical Sph/RY element. In agreement with the structure, in vitro DNA binding studies show that VAL1-B3 does not tolerate substitutions at any position of the 5′-TGCATG-3′ sequence. The VAL1-B3–DNA structure presented here provides a structural model for understanding the specificity of plant B3 domains interacting with the Sph/RY and other DNA sequences.
INTRODUCTION
Many plants that grow in a temperate climate zone require a long period of low winter temperature to initiate flowering in a process termed vernalization. In Arabidopsis, prolonged cold exposure promotes flowering in spring through epigenetic silencing of the FLOWERING LOCUS C (FLC) gene, which encodes a potent floral repressor (1,2). It was recently demonstrated that a key role in vernalization is played by plant transcription repressors VAL1 and VAL2 (3,4), which target the Polycomb repressive complex 2 (PRC2) to the region covering the junction of exon I and intron I of FLC, termed the nucleation region, leading to establishment of the H3K27me3 repressive mark. VAL1 and VAL2 possess a unique domain combination, containing the plant-specific B3 DNA-binding domain, EAR transcriptional repressor domain (5), and two potential histone-binding domains, a CW domain and a PHD-like (PHD-L) domain (6,7). The PHD-L domain was demonstrated to read the methylation state of histone H3 via specific interactions with H3K27me2 and K3K27me3 marks (3), while the B3 DNA binding domain is implicated in recognition of the 5′-CATGCA-3′/5′-TGCATG-3′ sequence, termed Sph/RY element (8), two copies of which are found within the FLC nucleation region (3,4). This suggests that VAL1/2 proteins might be recruited to a target gene through multivalent interactions including Sph/RY recognition by the B3 domain and PHD-L-mediated binding of a repressive histone mark (3). Moreover, VAL1/2 proteins were demonstrated to directly interact with Polycomb component LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) (3) and with components of the apoptosis- and splicing-associated protein (ASAP) complex (4), thereby guiding epigenetic silencing machinery to the Sph/RY and H3Kme3 sites. In addition to vernalization, VAL proteins act in other processes, e.g. repression of the embryonic pathway during seed germination (7), and sugar response (5).
To understand the structural basis of DNA target recognition by VAL1, we have solved a 1.9 Å crystal structure of VAL1 B3 domain (VAL1-B3) bound to a 12 bp oligoduplex containing the canonical Sph/RY DNA sequence 5′-CATGCA-3′/5′-TGCATG-3′. Structural comparison of the VAL1-B3–DNA complex with the structure of DNA-bound plant transcription factor ARF1 (9) and structures of bacterial B3-like domains revealed a conserved mode of DNA binding mediated by N- and C-arm structural elements (10–13). VAL1-B3 makes base-specific contacts to all 6 bp of the canonical Sph/RY element and has little tolerance for any single base pair substitution within the canonical Sph/RY sequence.
MATERIALS AND METHODS
DNA oligonucleotides
Oligoduplex substrates used in this study are listed in Table 1. All oligonucleotides were purchased from Metabion (Germany). Radioactive labeling was performed with [γ-33P]ATP (PerkinElmer) and T4 polynucleotide kinase (Thermo Fisher Scientific). Oligoduplexes were assembled by annealing the corresponding radiolabeled and unlabeled strands. Construction of expression vectors, protein expression and purification are described in Supplementary Materials and Methods.
Table 1. Oligoduplex substrates.
| Name | Sequence | Comment |
|---|---|---|
| 12/12NSP | 5′-CGACAGGTGGCT-3′ 3′-GCTGTCCACCGA-5′ | 12 bp oligoduplex lacking the canonical Sph/RY motif. Used as non-cognate DNA in EMSA experiments. |
| 12/12SP | 5′-CGGTGCATGGCT-3′ 3′-GCCACGTACCGA-5′ | 12 bp oligoduplex containing the canonical Sph/RY motif (underlined). Used for crystallization of VAL1-B3 and as cognate DNA in EMSA experiments. |
| 12/12SP_2 | 5′-CGCTGCATGCCT-3′ 3′-GCGACGTACGGA-5′ | As 12/12SP, but contains two G:C to C:G replacements adjacent to the Sph/RY motif (shown in italic). |
| 12/12_1(A|G|C) 12/12_2(C|A|T) 12/12_3(G|A|T) | - - - - - - - -1 2 3 4 5 6- - - - - - - - 5′-CGGTGCATGGCT-3′ 3′-GCCACGTACCGA-5′ | 18 variants. As 12/12SP, but contain 3 possible single bp substitutions at positions 1, 2, 3, 4, 5 or 6. |
| 12/12_4(T|C|G) | ||
| 12/12_5(A|C|G) | ||
| 12/12_6(A|T|C) |
Crystallization
Protein crystallization was performed by sitting drop vapor diffusion method at 277 K. The VAL1-B3 protein in 20 mM Tris–HCl (pH 8.0 at 25 C), 150 mM NaCl and 0.02% NaN3 was concentrated to 350 μM concentration (∼5 mg/ml) and mixed with an equimolar concentration of 12/12SP DNA (Table 1); final concentrations of VAL1-B3 and DNA were 220 μM each. 0.25 μl of the solution was mixed with 0.25 μl of the crystallization solution (#9 solution of the Hampton Research ‘Crystal Screen Cryo’ kit, 0.17 M Ammonium acetate, 0.085 M Sodium citrate tribasic dihydrate pH 5.6, 25.5% w/v Polyethylene glycol 4000, 15% v/v glycerol). Crystals appeared after 3 months.
Data collection and structure determination
Diffraction data were collected at 100 K at the P14 beamline (PETRA III/DESY) on an EIGER 16M detector. No additional cryoprotection was used. Data were processed with XDS (14), SCALA and TRUNCATE (15). The structure was solved by molecular replacement with PHASER (16) as described in Supplementary Materials and Methods. The resultant model was rebuilt in COOT and refined using PHENIX (17) to final Rwork/Rfree of 0.170/0.217. Data collection and refinement statistics are shown in Table 2.
Table 2. Data collection and refinement statistics.
| Data collection | |
| Space group | P 1 21 1 |
| A (Å) | 40.091 |
| B (Å) | 86.577 |
| C (Å) | 65.16 |
| α/β/γ | 90.000/94.135/90.000 |
| Wavelength (Å) | 0.9763 |
| X-ray source | DESY/PETRAIII/P14 |
| Total reflections | 118 797 (12 131) |
| Unique reflections | 34 337 (3404) |
| Resolution range* (Å) | 43.29–1.90 (1.968–1.90) |
| Completeness* (%) | 99 (99) |
| Multiplicity* | 3.5 (3.5) |
| Mean I/σ(I)* | 18.81 (2.69) |
| R(merge)* (%) | 3.294 (52.4) |
| B(iso) from Wilson (Å2) | 39.11 |
| Refinement | |
| Resolution range (Å) | 43.29–1.9 |
| Reflections work (non-anomalous)/test | 34 551/3471 |
| Macromolecule/solvent atoms | 2690/193 |
| R-factor/R-free (%) | 0.1699/0.2174 |
| R.m.s.d. bond lengths (Å)/angles (deg) | 0.013/1.33 |
| PDB ID | 6fas |
Structure and sequence analysis
Structures of VAL1-B3 and related domains were overlaid using Multiprot (18). Structure-based sequence alignments were generated using the Staccato algorithm (19) and ESPRIPT (20). Structures were visualized and figures generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.7 Schrödinger, LLC). Electrostatic potential surfaces were generated using the ‘APBS Tools’ plugin in PyMOL. Phylogenetic trees of the aligned sequences were generated using phylogeny.fr (21).
Electrophoretic mobility shift assay
DNA binding by wt VAL1-B3 was analyzed by the electrophoretic mobility shift assay (EMSA) using 33P-labeled 12 bp oligoduplexes. 1 μM DNA (2–4 nM of radiolabeled DNA mixed with 1 μM of unlabeled oligoduplex) was incubated with the protein (final concentration typically varied from 0.5 to 10 μM) for 15 min in 20 μl of the binding buffer containing 30 mM Mes/30 mM histidine (pH 6.0 at 25°C), 0.1 mg/ml bovine serum albumin and 10% v/v glycerol. Free DNA and protein–DNA complexes were separated by electrophoresis through 8% polyacrylamide gels (29:1 acrylamide:bisacrylamide) in 30 mM MES/30 mM histidine (pH 6.0) for 45–60 min at 5 V/cm. Some experiments were also performed using a pH 8.3 binding and electrophoresis buffers (MES/histidine replaced with 40 mM Tris-acetate). Low power consumption, ∼2 W (110 V × ∼18 mA) per electrophoretic unit containing two gels (gel size—height:width:thickness 22:15:0.1 cm) and 1 l of electrophoretic buffer, ensured that the gels during electrophoresis remained at room temperature (∼22°C, well below the melting temperature of 12 bp duplexes used in the assay, >40°C). Radiolabeled DNA and protein–DNA complexes were detected using a ‘Cyclone’ phosphorimager and ‘OptiQuant’ software (Packard Instrument).
EMSA competition experiments
Samples contained 1 μM wt VAL1-B3, 1 μM of 33P-labeled cognate 12/12SP DNA and variable amounts (0.6–20 μM) of unlabeled competitor (12 bp oligoduplexes containing either the consensus Sph/RY motif 5′-TGCATG-3′ (5′-CATGCA-3′), or 18 variants containing single-base pair substitutions at each of the six positions) in the standard binding buffer described above (pH 6.0 MES/histidine). Radiolabeled free DNA and protein–DNA complexes were separated for 45 min at 3V/cm. Analysis of the dependence of the amount of the radiolabeled-specific complex on the concentration of the unlabeled competitor, performed as described in Supplementary Methods, allowed us to determine the relative affinities of VAL1-B3 to various recognition sequence variants. Relative affinities were converted into a sequence logo representation of the VAL1-B3 PWM as described in Supplementary Methods.
RESULTS
Overall structure of VAL1-B3 domain
We have solved the co-crystal structure of VAL1 B3 DNA recognition domain (VAL1-B3) with a 12 bp cognate oligoduplex DNA at 1.9 Å resolution (data collection and refinement statistics are summarized in Table 2). The asymmetric unit contains two almost identical DNA-bound protein molecules (RMSD 0.3 Å over 1100 protein and DNA atoms). In the crystal the two DNA-bound VAL1-B3 subunits form a back-to-back dimer with a surface area of ∼920 Å2 (determined using the PISA server (22), Supplementary Figure S1A). The predominantly polar intermolecular contact surface between VAL1-B3 molecules in the crystal, and the monomeric structure of VAL1-B3 in solution (Supplementary Figure S1B) imply that this interface plays no role in VAL1-B3 function. The 12 bp DNA oligoduplexes used for crystallization of VAL1-B3 form continuous double-stranded structures in the crystal (a common observation in protein–DNA complexes involving small DNA-binding domains), thereby defining the a = 40.1 Å (≈12 × 3.4 Å) edge of the unit cell (Supplementary Figure S1A).
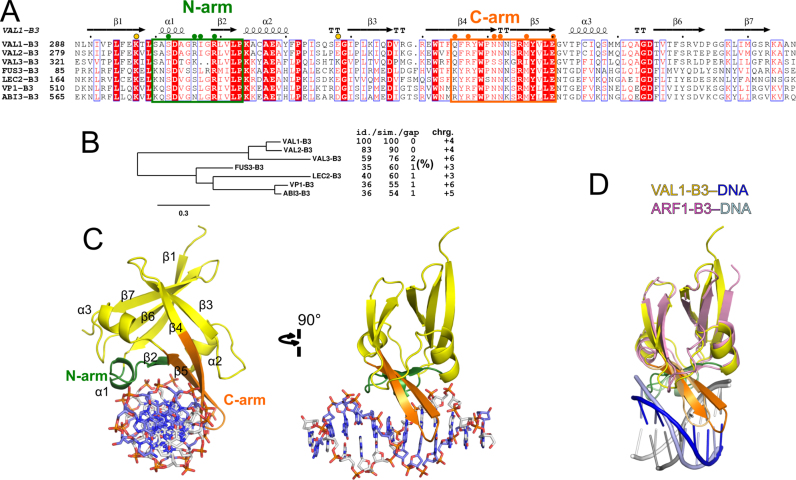
Like other plant B3 domains, VAL1-B3 is composed of seven β strands making a pseudo barrel decorated by short α helices (Figure 1A and B). VAL1-B3 is most similar to B3 DNA recognition domains of plant transcription factors ARF5 (PDB ID: 1ldu, DALI (23) Z-score 16.4), ARF1 (4ldx, 14.9) and RAV1 (1wid, 13.5). This reflects significant sequence similarity: VAL1-B3 shares ∼30% identical and ∼42% similar aa over a 104 aa region with ARF1/ARF5, and 38% identical 61% similar aa over a 116 aa region with RAV1.
Figure 1.
DNA recognition domain of Arabidopsis transcriptional repressor VAL1. (A) Sequence alignment of Arabidopsis thaliana LAV family B3 DNA binding domains and the B3 domain of maize ABI3 ortholog VP1. Residues forming the N-arm (green box) and C-arm (orange box) structural elements involved in DNA recognition are indicated. Secondary structure elements of VAL1-B3 are shown. Circles mark VAL1-B3 residues mutated in this work. (B) Phylogenetic tree of the aligned sequences. The theoretic charges (chrg.) at neutral pH of the aligned sequences, and % identity (id.)/similarity (sim.)/gaps of all family members relative to VAL1-B3 are given at the right-hand side of the alignment. (C) Overall structure of VAL1-B3–DNA complex. Secondary structure elements are numbered as in (A), N-arm is colored green, C-arm is orange, DNA strand containing the 5′-TGCATG-3′ sequence is colored blue, the complementary strand is colored white. (D) Superimposition of DNA-bound structures of VAL1-B3 (yellow) and ARF1-B3 (pink, PDB ID: 4ldx, residues 119–225). Overlay was generated with Multiprot (18). The N-arm residues of VAL1 and ARF1 are colored green and pale green, respectively; the C-arm residues of the corresponding B3 domains are colored orange and light orange. The DNA fragments of the cognate recognition sequences are shown in cartoon representation. The 5′-TGCATG-3′/5′-CATGCA-3′ fragments of VAL1-B3–DNA are blue/gray, the 5′-GAGACA-3′/5′-TGTCTC-3′ fragments of ARF1-B3–DNA are light blue/white.
Mode of DNA binding
In the crystal VAL1-B3 binds DNA as a monomer. The protein approaches DNA from the major groove side, and makes all of the base-specific and most DNA backbone contacts via structural elements termed N-arm (helix α1, strand β1 and the connecting loop), and C-arm (strands β4-β5, part of the pseudo-barrel), which form a wrench-like DNA-binding cleft (Figure 1A and B). Identical DNA binding orientation and DNA binding determinants are also employed by the B3 domain of plant transcription factor ARF1 (ARF1-B3, Figure 1C) (9) and all structurally characterized bacterial B3-like domains (DNA recognition domains of restriction endonucleases EcoRII, BfiI, NgoAVII and UbaLAI (10–13)). Like the above proteins, VAL1-B3 does not distort the DNA. The overall bend for the 12 bp oligoduplex, as determine by the Curves+ web server (24), is only ∼5° (Supplementary Figure S2); in comparison, the DNA fragment directly interacting with ARF1-B3 (PDB ID: 4ldx) is also bent by ∼3–5°, albeit the overall DNA axis direction relative to the protein is slightly different than in VAL1-B3–DNA complex (Supplementary Figure S2). The inter- and intra-base pair parameters determined by Curves+ for the VAL1-B3–DNA and ARF1-B3–DNA complexes are summarized in Supplementary Tables S1 and 2.
Sph/RY element recognition by VAL1-B3
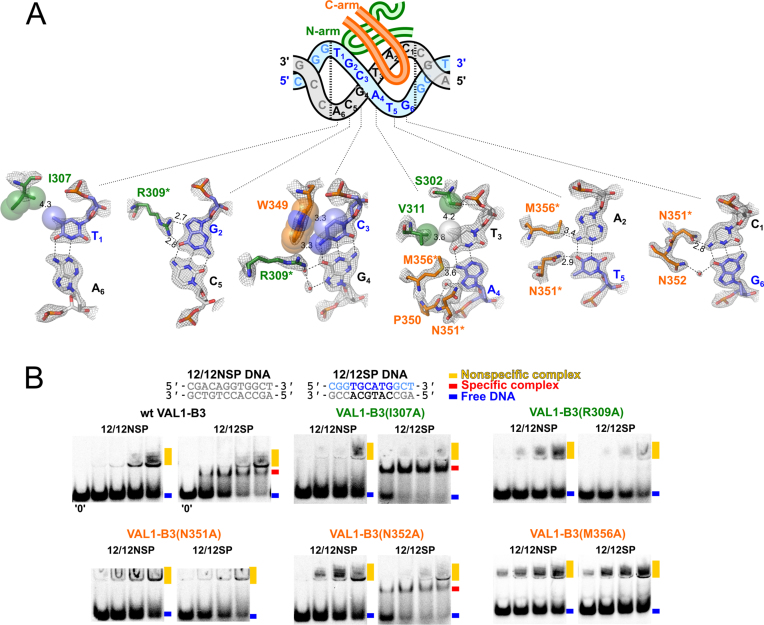
VAL1-B3 directly interacts with all 6 base pairs of the canonical Sph/RY DNA sequence 5′-CATGCA-3′/5′-TGCATG-3′ (Figure 2A). For convenience, we will henceforth number base pairs of the 5′-TGCATG-3′ Sph/RY DNA strand from 1 to 6 (this strand is colored blue/light blue in Figure 1 and all subsequent figures; the complementary 5′-CATGCA-3′ strand is colored gray).
Figure 2.
Recognition of the Sph/RY sequence by VAL1-B3. (A) The cartoon schematically depicts 12/12SP DNA used for VAL1-B3 crystallization and positions of the protein N- and C-arms. Numbering of the nucleotides forming the canonical Sph/RY sequence (5′-TGCATG-3′/5′-CATGCA-3′, blue/gray) is shown. Individual panels show contacts made by VAL1-B3 N-arm (green) and C-arm (orange) residues to 6 bp of the Sph/RY sequence. The 2mFo-DFc electron density map contoured at a 2.0 σ level for the corresponding residues, and distances of direct base-specific H-bond and van der Waals interactions (in Å) are also shown. Asterisks denote residues that make contacts to multiple base pairs. (B) DNA binding by wt VAL1-B3 and mutants. Sequences of 12/12NSP (non-cognate DNA) and 12/12SP (cognate) oligoduplexes used in EMSA are depicted. DNA concentration was 1 μM, protein concentrations were 1, 2, 4 and 10 μM, gel lanes marked ‘0’ contained no protein. EMSA experiments were performed in a pH 6.0 buffer as described in the ‘Materials and Methods’ section. Positions of the free DNA, specific complex and non-specific complex are marked by blue, red and yellow rectangles, respectively.
All base-specific contacts are made by the N-arm and C-arm residues, which primarily contact the 5′- and 3′ parts of the 5′-TGCATG-3′ sequence, respectively (Figure 2A). In particular, N-arm residue I307 makes a van der Waals (vdW) contact to the methyl group of T1 in the T1:A6 base pair, and N-arm arginine R309 makes direct H-bonds to O6 and N7 atoms of guanine G2 of the G2:C5 base pair (Figure 2A). R309 also makes a water-mediated contact to O6 of the complementary strand guanine G4 of the C3:G4 base pair, while N4 and C5 atoms of the ‘blue’ strand cytosine C3 make vdW contacts to the C-arm tryptophan W349 (Figure 2A).
N6 atoms of two adenines in the subsequent A4:T3 and T5:A2 base pairs are contacted by the C-arm methionine M356; the methyl group of the T3:A4 base pair thymine also makes vdW contacts to N-arm residues S302 and V311, while O4 atom of the T5:A4 base pair thymine makes a direct H-bond to C-arm asparagine N351 (Figure 2A). The same asparagine makes a direct H-bond to N4 group of cytosine C1 in the G6:C1 base pair, and an adjacent asparagine N352 makes a water-mediated contact to O6 atom of guanine G6.
Taken together, VAL1-B3 makes direct contacts to 8 out of 12 bases of the Sph/RY motif (6 H-bonds and 3 vdW interactions), and 3 water-mediated H-bonds to 2 more bases (Figure 2A); no contacts to DNA bases outside the canonical Sph/RY hexanucleotide are present.
DNA binding by VAL1-B3 in vitro
To date, specific binding of VAL1-B3 to Sph/RY sequence was detected in vivo (3,4), but experiments in vitro so far were inconclusive (7). We have failed to detect any protein–DNA complexes of VAL1-B3 in EMSA using a standard pH 8.3 Tris-acetate buffer, even with micromolar protein and DNA concentrations (Supplementary Figure S3A). Auspiciously, VAL1-B3–DNA complexes were detected in EMSA performed in a pH 6.0 Mes-histidine buffer (Figure 2B). Under these experimental conditions VAL1-B3 forms low-mobility DNA complexes with both cognate and non-cognate DNAs (Figure 2B, wt VAL1-B3), and an additional higher mobility band with cognate DNA. We presume that the latter band corresponds to the specific VAL1-B3–DNA complex (highlighted by red rectangles in Figure 2B), with a single protein bound to the Sph/RY sequence at the center of the 12/12SP oligoduplex. The low mobility band (yellow rectangles) most likely is a non-specific complex, formed by multiple protein copies bound to a single DNA. Due to positive charge (+4) of VAL1-B3 (Figure 1A) and the extra size, this complex fails to enter the gel. Noteworthy, the amount of specific complex with cognate DNA decreases at higher protein concentrations. Presumably, this happens due to non-specific binding of additional protein copies to the cognate complex.
We also find that formation of the specific complex in our experimental setup is observed only with the standard protein containing a C-terminal (His)6 tag, as protein variants with a C-terminal Strep-tag or lacking any tags failed to form a clearly defined specific band under identical conditions (Supplementary Figure S4). Presumably, the (partially) protonated C-terminal (His)6 tag at pH 6.0 provides just enough electrostatic attraction to the DNA to promote specific binding. Nevertheless, low stability of the cognate complex implies its formation should be highly sensitive to the loss of any DNA contacts. We have therefore mutated several residues implicated in base-specific contacts of VAL1, and tested the ability of VAL1 mutants, all containing an identical C-terminal (His)6 tag, to form the cognate complex (Figure 2B). We have found that alanine replacements of I307 (vdW contact to T1) and N352 (an indirect contact to G6) did not abolish formation of the high-mobility band, while mutations of R309 (H-bonds to G2), M356 (H-bonds/vdW contacts to A4:T3 and T5:A2 adenines) and N351 (2 H-bonds to T5 and C1 in the complementary strand, Figure 2A) compromised formation of the specific complex (Figure 2B).
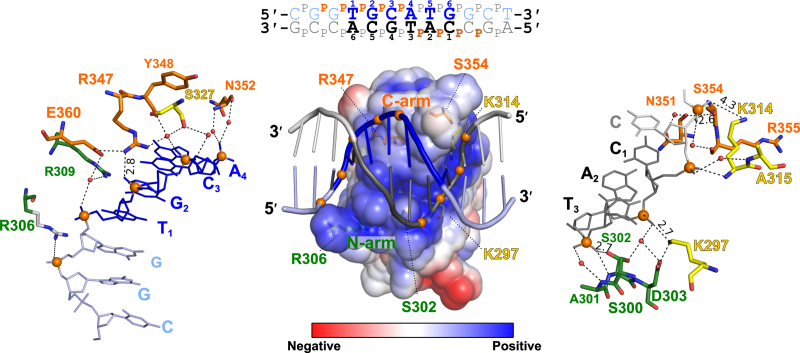
DNA backbone contacts
VAL1-B3 binds DNA in a positively charged cleft and makes multiple direct and water-mediated contacts to DNA phosphates both within and adjacent to the Sph/RY sequence (Figure 3). Backbone contacts are not limited to N-arm and C-arm residues, as several direct contacts are made by residues from other structural elements, e.g. K297 (β1 strand) and K314 (α2 helix, Figure 3). Alanine replacements of K297, S302 and R347 abolished formation of the specific complex or reduced its amount (Supplementary Figure S3B; as above, all mutants contained the C-terminal (His)6 tag). Though electron density of the N-arm R306 side chain was not resolved, we reasoned that it may form electrostatic or H-bond contacts to DNA backbone phosphate. In agreement with this model, the R306A mutant lost its ability to form a specific complex (Supplementary Figure S3B). Interestingly, VAL1-B3 residue Q345 occupies a position that in many other B3-domains contains an arginine; examples include ABI3-B3 (Figure 1A), RAV1 family domains (G. Sasnauskas et al., manuscript in preparation), EcoRII (R81) and BfiI (R272, the residue implicated in recognition of the ‘clamp’ phosphates (11)). Restoration of the positive charge at this position in the Q345R mutant enabled DNA binding under standard (pH 8.3) EMSA conditions (Supplementary Figure S3A).
Figure 3.
Contacts of VAL1-B3 to the DNA phosphates. Top: the 12/12SP oligoduplex used for VAL1-B3 crystallization. DNA phosphates making contacts to the protein are marked as orange ‘p’ letters. Center: an electrostatic potential surface of VAL1-B3 and a cartoon representation of the 12/12SP DNA (colored as in Figure 2). Phosphates that make contacts to VAL1-B3 are shown as orange spheres, VAL1-B3 residues making direct contacts to the phosphates are shown in stick representation. Left: direct and water-mediated contacts to the phosphates of the ‘blue’ strand. Right: direct and water-mediated contacts to the phosphates of the ‘gray’ strand. In all panels, N-arm residues are green, C-arm residues are orange, residues from other parts of the protein are yellow. The side chain of R306 (unresolved in the electron density map) is white.
Recognition stringency of Sph/RY sequence by VAL1-B3
To assess the stringency of Sph/RY sequence recognition by VAL1-B3, we have tested the effect of single base pair substitutions within the canonical Sph/RY hexanucleotide 5′-TGCATG-3′ on the ability of VAL1-B3 to form a specific complex. For that purpose we used a set of 12 bp oligonucleotides containing three possible replacements at each position of the Sph/RY element (3 × 6 = 18 variants, Table 1), and performed EMSA experiments using standard conditions (pH 6.0 buffer). We found that any single base pair replacement within the 5′-TGCATG-3′ sequence abolished specific complex formation by wt VAL1-B3, while replacements adjacent to the Sph/RY hexanucleotide had no effect (Supplementary Figure S5).
Based on the structure, T1:A6 base pair is recognized by a single vdW contact of I307 side chain to the methyl of T1. The I307A mutation did not abolish specific binding by VAL1-B3 (Figure 2A). Under our experimental conditions this mutant even formed a higher amount of the specific complex than wt VAL1-B3 (Figure 2B), suggesting that an alanine at this position may be sufficient for vdW interactions and residual specificity. Concomitantly, a truncated side chain could release some steric hindrance (not resolved in our structure), resulting in a more stable specific complex. But despite the non-critical nature of the I307 contact, wt VAL1-B3 did not form specific complexes with sequences containing replacements of the T1:A6 base pair. We reasoned that lack of binding in this case was due to low stability of VAL1-B3–DNA complexes under our EMSA conditions. To assess the relative preference of VAL1-B3 to all substituted variants of the Sph/RY element we have employed an EMSA-based competition assay (details described in Supplementary Methods) to determine the PWM (position weight matrix) for the VAL1-B3–DNA interaction. As expected, the experimentally determined PWM is consistent with the least stringent recognition of the T1-A6 base pair (WebLogo (25) representation in Figure 4). The FoldX algorithm (26) also predicts significant degeneracy in the T1:A6 recognition by VAL1-B3 (Figure 4).
Figure 4.
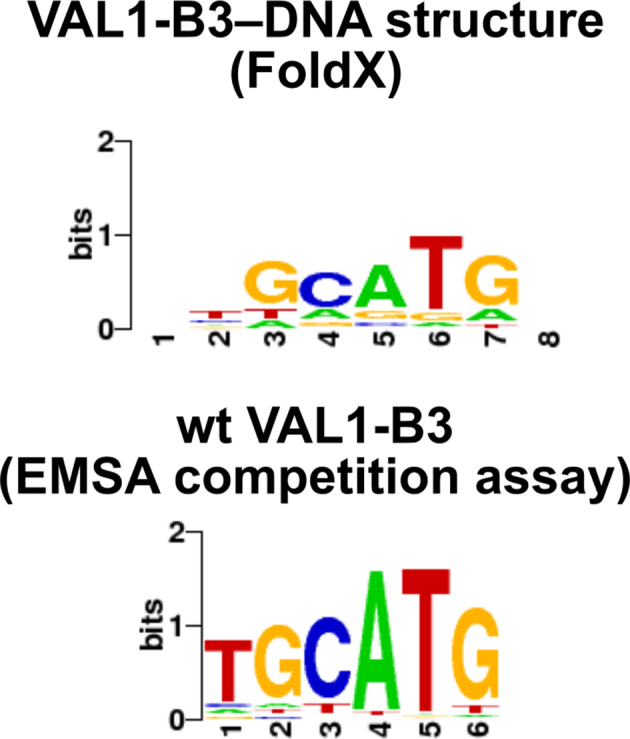
WebLogo representations of PWMs of wt VAL1-B3 domain. PWMs were either generated by analysis of the VAL1-B3–DNA structure using FoldX software, or determined experimentally using an EMSA-based competition assay as described in Supplementary Materials and Methods.
DISCUSSION
Base readout by VAL1-B3
The co-crystal structure of VAL1 uncovers the molecular basis of Sph/RY element recognition by plant B3 domains. VAL1-B3 makes direct hydrogen bonds, water-mediated hydrogen bonds or vdW contacts to all 6 base pairs of the canonical Sph/RY hexanucleotide (Figure 2). Of particular interest are base-specific contacts made by tryptophan W349 and methionine M356, residues that rarely participate in recognition of undistorted DNA.
The Sδ atom of M356 contacts the amino groups of adenines located on complementary strands in two subsequent base pairs (Figure 2A). The role of hydrogen bonds involving sulfur in protein structures, with cysteine thiol group as H-bond donor/acceptor and methionine sulfur as H-bond acceptor, was recognized decades ago (27). Due to the radius of sulfur atoms such H-bonds are significantly longer (3.3–3.7 Å between heavy atoms) than H-bonds involving N and O atoms (28). Interestingly, though sulfur was considered to be a very poor H-bond acceptor, and, consequently, methionine a poor candidate for making strong H-bonds, recent spectroscopic and computational studies revealed that amide-N–H···S H-bonds involving methionine Sδ are even stronger than amide-N–H···O = C H-bonds (29). Not surprisingly, interaction with main-chain amides is the major type of H-bonds involving methionine sulfur in proteins (28). In our structure, we find that M356 Sδ atom is located 3.52/3.33 Å (chains ACD) or 3.57/3.40 Å (chains BEF) away from the adenine amino group nitrogen atoms of the A4:T3 and T5:A2 base pairs (Figure 2A). These findings, together with the loss of specific DNA binding by the VAL1-B3 mutant M356A (Figure 2B), and impaired specific binding by the M356L and M356I mutants, (Supplementary Figure S6) indicate that M356 makes legitimate H-bonds to 6-amino groups of both adjacent adenines (Figure 2A).
The flat surface of W349 side chain contacts the N4-C4-C5-C6 edge of cytosine in the C3:G4 base pair (Figure 2A). Replacement of C3:G4 with a T:A base pair would be incompatible with specific interaction due to a steric clash between the methyl of T and W349 (a similar effect would be expected in the case of C3 methylation in the asymmetric 5′-CHH-3′ context); on the contrary, C3:G4 replacements with either G:C or A:T would considerably reduce the extent of vdW interactions. This tryptophan–cytosine contact is also conserved in RAV family B3 domains (G. Sasnauskas et al., manuscript in preparation). Interestingly, a similar contact is present in the B3-like domain of restriction endonuclease BfiI, where tryptophan W229 makes a vdW interaction to the 3rd cytosine in the sequence 5′-CCCAGT-3′ (11). However, the latter contact is not conserved: BfiI W229 is an N-arm arm residue (VAL1-B3 W349 is a C-arm residue), and the contacted cytosine is located on the opposite DNA strand.
Sph/RY recognition stringency
VAL1-B3 makes the smallest number of contacts to the T1:A6 base pair—a single vdW interaction by the I307 residue (Figure 2A). This agrees well with mutational analysis (I307A mutant retained the ability to form a cognate complex, Figure 2B), and the PWM of VAL1-B3–DNA interaction determined either in silico by the FoldX algorithm or using the competition-based EMSA assay (less stringent recognition of the T1:A6 base pair in comparison to other positions, Figure 4). Relaxed recognition of one terminus of the consensus sequence by the N-arm resembles ARF1-B3 and ARF5-B3, which both interact with a wide set of hexanucleotide sequences 5′-NNGACA-3′ as identified by saturating binding site selection, with the ‘canonical’ auxin response element sequence 5′-GAGACA-3′ found among the enriched motifs (9).
Stability of the cognate complex
VAL1-B3 makes 6/3 direct/water-mediated H-bonds and 4 vdW contacts to DNA bases of the hexanucleotide sequence (Figure 2A). This number is comparable to the number of interactions made by ARF1-B3 (5 H-bonds and 5 vdW contacts, PDB ID: 4ldx), but is considerably smaller than the number of interactions formed by the B3-like domains of bacterial restriction endonucleases known for their extraordinary DNA binding specificity. For example, BfiI makes 14 direct H-bonds and 3 vdW interactions to a hexanucleotide sequence, essentially saturating the major groove H-bonding potential of DNA bases (11). Relatively low number of direct VAL1-B3 interactions with the DNA might account for the low stability of cognate VAL1-B3–DNA complex. Indeed, under our experimental conditions cognate binding of VAL1-B3 was detectable only with micromolar protein and DNA concentrations in a pH 6.0 buffer, where protonation of the C-terminal HisTag provides additional attraction to the DNA (Supplementary Figure S4); no cognate complex was detected in a pH 8.3 buffer, or if lower protein and/or DNA concentrations were used. In contrast, B3-like domains of REases bind cognate DNA with nanomolar or lower KDs (10–13). We therefore believe that an important role in VAL1 binding to the Sph/RY elements in vivo is played by multivalent interactions involving B3 and other functional domains, in particular the PHD-L domain capable of H3K27me3 recognition (3). Interestingly, the wt FLC locus in A. thaliana contains two Sph/RY elements separated by 27 bp (3,4). The proposed dimerization of full-length VAL1 (3), or potentially formation of a heterodimer with VAL2 (4) may therefore result in cooperative DNA binding of a single VAL1/2 dimer to both Sph/RY sequences. Such mechanism would be reminiscent of homodimeric ARF1 and ARF5 proteins, which bind bipartite auxin response (AuxRE) sites as molecular calipers with ARF-specific spacing preferences (9). Enhanced binding of longer VAL1 (252 aa) constructs to DNA fragments containing two Sph/RY sequences was recently demonstrated on an oligoduplex mimicking DNA fragment upstream of the AGAMOUS-LIKE 15 (AGL15) coding region (30). However, an important difference between ARF1/5 and VAL1/2 recognition sites is that AuxREs recognized by the ARF1/5 B3 domains are in an inverted repeat orientation (9,31), whereas Sph/RY elements in the FLC locus are in direct repeat orientation. Inverted orientation of AuxREs enables formation of a symmetric complex with an ARF1/5 dimer (e.g. PDB ID: 4ldx (9)). In contrast, formation of a symmetric complex in the case of VAL1/2 would require bending of the intervening 27 bp DNA into an ‘U’-shaped loop; alternatively, VAL1/2 could form an asymmetric ‘head-to-tail’ heterodimer that could bind an undistorted FLC locus. Experimental validation of the FLC locus recognition mechanism and stoichiometry by VAL1/2, however, will require binding studies performed with longer protein constructs not analyzed in the current study.
The role of DNA backbone contacts in site-specific DNA interactions
As discussed above, the low stability of the specific VAL1-B3–DNA complex implies that any loss of positively charged DNA-contacting residues may abolish specific VAL1-B3 binding; in contrast, introduction of extra positively charged residues could stabilize specific interactions.
Indeed, mutations of several charged (K297, R306, R347) and even uncharged (S302) residues that make direct contacts to DNA phosphates abolished or severely impaired formation of the specific complex (Supplementary Figure S3B). In contrast, mutation Q345R, which restores an arginine found in many other B3 and B3-like domains, and increases the overall charge of VAL1-B3 to +5, enables DNA binding by VAL1-B3 at higher pH (Supplementary Figure S3A).
A similar effect was also observed upon mutation of negatively charged VAL1-B3 residues E328 and E360, which are both adjacent to the DNA phosphate 5′-TpGCATG-3′ (Supplementary Figure S3A). The fact that addition of a single positive charge/removal of a single negative charge in the vicinity of the DNA backbone enable specific complex formation at pH 8.3 (i.e. under conditions that preclude protonation of the hexahistidine tag) indicates that the key factor enabling formation of the cognate complex in our experimental system is sufficient electrostatic attraction, which may be achieved either via charge-increasing mutations or by protonation of protein residues. Interestingly, the VAL1-B3–DNA crystals were grown in a pH 5.6 buffer (‘Materials and Methods’ section), i.e. conditions that favor cognate complex formation by the standard VAL1-B3 protein containing the C-terminal (His)6 tag.
Base-specific contacts of other B3 domains
VAL1-B3 belongs to the LAV B3 family, which includes other transcriptional regulators capable of Sph/RY sequence binding (32). Despite of a limited sequence similarity to some family members (Figure 1A), all key residues involved in direct DNA contacts are conserved among VAL1, VAL2, FUS3, LEC2, ABI3 and maize ABI3 ortholog VP1, suggesting a conserved Sph/RY DNA recognition mechanism (Figure 1A). Modeling, mutational analysis and in vitro DNA binding studies of the most distant Arabidopsis LAV family member ABI3-B3 (36% identical, 54% similar aa residues to VAL1-B3, Figure 1A) confirm this conclusion (G. Sasnauskas et al., manuscript in preparation).
An obvious exception is VAL3-B3. Despite high overall sequence similarity to VAL1-B3 (59% identical, 76% similar residues), it has no counterparts for VAL1-B3 residues N351, N352 (both replaced by smaller serines), M356 (replaced by an isoleucine) and I307 (N-arm contains a deletion; this deletion may also perturb the position of adjacent R340, an equivalent of VAL1 R309). Given the detrimental effect of similar replacements on the cognate complex formation by VAL1-B3 (Figure 2B), we believe VAL3-B3 has no ability to specifically recognize the full Sph/RY sequence. This is in agreement with recent in vivo studies that failed to detect a role for VAL3 in vernalization (low expression level and no enrichment at Sph/RY elements (3,4)). Overall conservation of residues making DNA backbone contacts, and the highest positive charge (+6, Figure 1A) among Arabidopsis LAV B3 domains suggests it may be a DNA binding protein with reduced sequence specificity.
In conclusion, the co-crystal structure of VAL1-B3 provides molecular details for the Sph/RY sequence recognition by VAL1/2 transcriptional repressors and ABI3/FUS3/LEC2 transcriptional activators, key factors in vernalization, embryo maturation program and other phenomena central to plant life (7,8,33). This structure also helps rationalize target recognition by RAV family B3 domains that are specific for a different DNA sequence 5′-CACCTG-3′ (G. Sasnauskas et al., manuscript in preparation).
DATA AVAILABILITY
Coordinates and structure factors of VAL1-B3–DNA complex are deposited under PDB ID: 6fas.
Supplementary Material
ACKNOWLEDGEMENTS
Authors thank E. Rudokienė for sequencing services, K. Pranckevičiūtė for help with protein expression and EMBL DESY staff members Dr G. Bourenkov and Dr J. Kallio for help with beamline operation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Council of Lithuania [MIP-106/2015 to G.S.]. Funding for open access charge: Research Council of Lithuania.
Conflict of interest statement. None declared.
REFERENCES
- 1. Li C., Cui Y.. A DNA element that remembers winter. Nat. Genet. 2016; 48:1451–1452. [DOI] [PubMed] [Google Scholar]
- 2. Sheldon C.C., Rouse D.T., Finnegan E.J., Peacock W.J., Dennis E.S.. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. 2000; 97:3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yuan W., Luo X., Li Z., Yang W., Wang Y., Liu R., Du J., He Y.. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 2016; 48:1527–1534. [DOI] [PubMed] [Google Scholar]
- 4. Qüesta J.I., Song J., Geraldo N., An H., Dean C.. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science. 2016; 353:485–488. [DOI] [PubMed] [Google Scholar]
- 5. Tsukagoshi H., Saijo T., Shibata D., Morikami A., Nakamura K.. Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol. 2005; 138:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Veerappan V., Wang J., Kang M., Lee J., Tang Y., Jha A.K., Shi H., Palanivelu R., Allen R.D.. A novel HSI2 mutation in Arabidopsis affects the PHD-like domain and leads to derepression of seed-specific gene expression. Planta. 2012; 236:1–17. [DOI] [PubMed] [Google Scholar]
- 7. Suzuki M., Wang H.H.-Y., McCarty D.R.. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007; 143:902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suzuki M., Kao C.Y., McCarty D.R.. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 1997; 9:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boer D.R., Freire-Rios A., van den Berg W.A.M., Saaki T., Manfield I.W., Kepinski S., López-Vidrieo I., Franco-Zorrilla J.M., de Vries S.C., Solano R. et al. . Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014; 156:577–589. [DOI] [PubMed] [Google Scholar]
- 10. Golovenko D., Manakova E., Tamulaitiene G., Grazulis S., Siksnys V.. Structural mechanisms for the 5′-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII. Nucleic Acids Res. 2009; 37:6613–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Golovenko D., Manakova E., Zakrys L., Zaremba M., Sasnauskas G., Gražulis S., Siksnys V.. Structural insight into the specificity of the B3 DNA-binding domains provided by the co-crystal structure of the C-terminal fragment of BfiI restriction enzyme. Nucleic Acids Res. 2014; 42:4113–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tamulaitiene G., Silanskas A., Grazulis S., Zaremba M., Siksnys V.. Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII. Nucleic Acids Res. 2014; 42:14022–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sasnauskas G., Tamulaitienė G., Tamulaitis G., Čalyševa J., Laime M., Rimšelienė R., Lubys A., Siksnys V.. UbaLAI is a monomeric Type IIE restriction enzyme. Nucleic Acids Res. 2017; 44:W410–W415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kabsch W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 1994; 50:760–763. [DOI] [PubMed] [Google Scholar]
- 16. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. IUCr . Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Afonine P. V, Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shatsky M., Nussinov R., Wolfson H.J.. A method for simultaneous alignment of multiple protein structures. Proteins. 2004; 56:143–156. [DOI] [PubMed] [Google Scholar]
- 19. Shatsky M., Nussinov R., Wolfson H.J.. Optimization of multiple-sequence alignment based on multiple-structure alignment. Proteins. 2005; 62:209–217. [DOI] [PubMed] [Google Scholar]
- 20. Gouet P., Robert X., Courcelle E.. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003; 31:3320–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M. et al. . Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008; 36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu Q., Canutescu A.A., Wang G., Shapovalov M., Obradovic Z., Dunbrack R.L.. Statistical analysis of interface similarity in crystals of homologous proteins. J. Mol. Biol. 2008; 381:487–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holm L., Rosenstrom P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blanchet C., Pasi M., Zakrzewska K., Lavery R.. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011; 39:W68–W73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schneider T.D., Stephens R.M.. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990; 18:6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nadra A.D., Serrano L., Alibés A.. DNA-binding specificity prediction with FoldX. Methods Enzymol. 2011; 498:3–18. [DOI] [PubMed] [Google Scholar]
- 27. Gregoret L.M., Rader S.D., Fletterick R.J., Cohen F.E.. Hydrogen bonds involving sulfur atoms in proteins. Proteins Struct. Funct. Genet. 1991; 9:99–107. [DOI] [PubMed] [Google Scholar]
- 28. Zhou P., Tian F., Lv F., Shang Z.. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins. 2009; 76:151–163. [DOI] [PubMed] [Google Scholar]
- 29. Biswal H.S. Scheiner S. Hydrogen bonds involving Sulfur: new insights from ab initio calculations and gas phase laser spectroscopy. Noncovalent Forces. 2015; Cham: Springer International Publishing; 15–45. [Google Scholar]
- 30. Chen N., Veerappan V., Abdelmageed H., Kang M., Allen R.D.. HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in arabidopsis. Plant Cell. 2018; tpc.00655.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dinesh D.C., Calderón Villalobos L.I., Abel S.. Structural biology of nuclear auxin action. Trends Plant Sci. 2016; 21:302–316. [DOI] [PubMed] [Google Scholar]
- 32. Swaminathan K., Peterson K., Jack T.. The plant B3 superfamily. Trends Plant Sci. 2008; 13:647–655. [DOI] [PubMed] [Google Scholar]
- 33. Suzuki M., McCarty D.R.. Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 2008; 11:548–553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors of VAL1-B3–DNA complex are deposited under PDB ID: 6fas.