Abstract
Molecular crowding conditions provided by high concentration of cosolutes are utilized for characterization of biomolecules in cell-mimicking environment and development of drug-delivery systems. In this context, (poly)ethylene glycols are often used for studying non-canonical DNA structures termed G-quadruplexes, which came into focus by emerging structural biology findings and new therapeutic drug design approaches. Recently, several reports were made arguing against using (poly)ethylene glycols in role of molecular crowding agents due to their direct impact on DNA G-quadruplex stability and topology. However, the available data on structural details underlying DNA interaction is very scarce and thus limits in-depth comprehension. Herein, structural and thermodynamic analyses were strategically combined to assess G-quadruplex-cosolute interactions and address previously reported variances regarding the driving forces of G-rich DNA structural transformations under molecular crowding conditions. With the use of complementary (CD, NMR and UV) spectroscopic methods and model approach we characterized DNA G-quadruplex in the presence of the smallest and one of the largest typically used (poly)ethylene glycols. Dehydration effect is the key contributor to ethylene-glycol-induced increased stability of the G-quadruplex, which is in the case of the large cosolute mainly guided by the subtle direct interactions between PEG 8000 and the outer G-quartet regions.
INTRODUCTION
In cells, the genome is encoded in DNA, which canonically forms duplex through Watson-Crick base pairing. When the genetic information is replicated or expressed into functional proteins, the DNA duplex is temporarily dissociated by proteins such as DNA polymerase, RNA polymerase and helicase allowing the dissociated single stranded DNA to form non-canonical structures (1). One of the stable non-canonical structures that have been demonstrated to exist in living cells recently are G-quadruplexes (2). They are formed by G-rich sequences of both DNA and RNA. Various biological functions of the DNA and RNA G-quadruplexes that regulate gene expression processes such as replication, transcription, post-transcriptional RNA processing and translation have been demonstrated by biological and biochemical functional studies (3–6).
The basic building block of a G-quadruplex is G-quartet, which consists of four guanine nucleobases circularly interacting through Hoogsteen-type hydrogen-bonds (7). Pile of stacked G-quartets represents the core of a G-quadruplex structure. Although with some exceptions in intramolecular G-quadruplexes, the guanines located in the same corner of the stacked G-quartets are generally consistent with respect to the primary sequence and make backbone pillars like corner posts. While hitherto all the known RNA G-quadruplexes are monomorphic in that the strands in the core of the structure are oriented uniformly in the same direction and linked by three propeller-type loops, extreme polymorphism is the hallmark of G-rich DNA. Moreover, the variety of folding topologies accounts for several possible loop orientations, which are related to different orientations of the four stems in the G-quadruplex core and to alternative glycosidic bond arrangements, i.e. syn or anti conformation of 2′-deoxyguanosines comprising G-quartets (8). Besides propeller type loops found in parallel topology (9), the diagonal and lateral loop orientations are characteristic for anti-parallel and hybrid topologies (10–15). Notorious polymorphism of DNA G-quadruplex topologies is expanded further by peculiar loop orientations such as V-loop, D-loop and bulges (16–18). Additionally, DNA G-quadruplex topologies depend not only on the length and sequence of the loops, but also on the molecular environment such as nature and concentration of cations in solution (19,20).
Intracellular molecular environment is characterized as highly concentrated solution of various cosolutes, most notably proteins, nucleic acids and metabolites, which make up the so called molecular crowding conditions (21). The molecular crowding condition has been demonstrated to stabilize the G-quadruplex structures, while destabilize canonical duplexes (22). Additionally, it was shown that the equilibrium of different G-quadruplex structures adopted by G-rich DNA from human telomeric region was severely perturbed by supplementing aqueous solution with polyethylene glycol (PEG), which triggered conversion of hybrid and antiparallel topologies into parallel-stranded counterparts (23–25). These observations suggested that the parallel G-quadruplexes are most relevant in biological context, thereby establishing the need for further detailed investigations of their structures and physicochemical properties under conditions mimicking cellular environment. However, the polymorphic nature of DNA G-quadruplex structures and conformational transition depending on solution conditions and even during thermodynamic analyses make it difficult to perform such analyses in a quantitative manner. Interpretation of the experimental results is further complicated by the intricate mechanisms by which the commonly used crowding agents such as PEG influence DNA structural properties. In this regard, DNA features in the presence of PEG that had been discussed in terms of excluded volume and lowered water activity effects were challenged recently by ascribing PEG-induced DNA structural transformations to preferential and direct binding of the PEG to G-quartets of parallel-stranded DNA G-quadruplex (26). Additionally, insights into peculiarity of PEG and G-quadruplex interactions were expanded by exploring the interplay of metal ion- and cosolute-depended structural features (27), which further substantiated direct binding of PEG to parallel-stranded G-quadruplexes. Evidently, the interaction modes and moreover structural impacts of PEG on DNA G-quadruplexes need to be investigated further.
High-resolution NMR spectroscopy is a powerful technique for investigating molecular structure and dynamics with atomic resolution. On this account we assessed solution-state NMR data on naturally occurring G-rich DNA oligonucleotide and its derivatives, including artificially designed sequences in search for the model amenable for high-resolution studies. Among the screened DNA oligonucleotides, M2, 5′-d[TAGGGACGGGCGGGCAGGGT]-3′, was particularly interesting due to its distinguished characteristic to adopt a well-defined G-quadruplex structure in dilute solution and at different molecular crowding conditions. In addition, the primary sequence of M2 is consistent with the vastly accepted G-quadruplex consensus motif of four GGG tracts intervened by non-G residues. In this study, structural and thermodynamic analyses of M2 were strategically combined to assess G-quadruplex-cosolute interactions. Two grossly different cosolutes, ethylene glycol (EG) and its polymeric derivative polyethylene glycol 8000 (PEG 8000) were selected to address previously reported variances of the crowding agents’ effects. EG corresponds to the basic building block of PEGs, such as PEG 8000, which is one of the largest typically used crowding agents.
MATERIALS AND METHODS
Oligonucleotides—sample preparation
DNA oligonucleotides were synthesized on H-8 synthesizer (K&A LaborGeräte) with the use of standard phosphoramidite chemistry and deprotected with aqueous ammonia. Purification and desalting of DNA oligonucleotide was done with the use of RP-HPLC and Amicon-15 centrifuge filter with the cutoff 3000. Samples for NMR measurements were prepared in 100 mM aqueous KCl and 20 mM potassium phosphate, while the concentrations of EG and PEG 8000 were in the range between 10 and 20 wt%. The temperature treatment of samples included 5 min incubation at 90–95°C immediately followed by cooling in an ice-water bath for 5 min. For CD and UV analyses, DNA oligonucleotide was purchased from Japan Bio Services Co., Ltd.
NMR spectroscopy
NMR data were collected on Varian NMR Systems operating at 600 and 800 MHz in temperature range between 5 and 35°C with most of the experiments being performed at 25°C. 1H NMR samples were prepared in 90%/10% H2O/2H2O or 100% 2H2O at oligonucleotide concentrations between 0.1 and 1.0 mM per strand. NOESY spectra were acquired at mixing times between 80 and 300 ms with the use of DPFGSE solvent suppression method. DQF-COSY spectra and TOCSY spectra with mixing time of 80 ms were acquired in 100% 2H2O. Twenty different gradient strengths (2.4–59.6 G cm−1) were used in diffusion NMR experiments (DOSY). NMR spectra were processed and analysed with the use of VNMRJ (Varian Inc.) software and Sparky (UCSF) software (28).
Restraints and structure calculations
NOESY spectra with mixing times of 80, 150 and 200 ms were recorded to collect distance restraints for exchangeable and non-exchangeable protons. The NOE cross-peaks were classified as strong (1.8–3.6 Å), medium (2.6–5.0 Å) and weak (3.5–6.5 Å) relative to the distance-restraint reference of cytosine H5-H6 protons corresponding to 2.5 Å. Glycosidic torsion angle χ for guanine, thymine and cytosine residues were restrained to 240 ± 70°. South-type sugar puckering for all residues was inferred from large 3JH1′-H2′ coupling constants measured in DQF-COSY spectra. Correspondingly, conformational space related to pseudorotation phase angles was constraint in the range from C3′-exo to C1′-exo canonical forms by defining the endocyclic torsion angles ν1 (30 ± 7.5°), ν2 (-30 ± 10°), ν3 (10 ± 20°) and ν4 (5 ± 25°). AMBER 14 software (29,30) and parmbsc0 force field with parmχ OL4 (31) and parm ϵ/ζ OL1 (32) modifications and generalized Born implicit solvation model were used in calculations. For each simulated annealing calculation random velocity, collision frequency γ of 5 ps−1 and the cutoff for non-bonded interactions of 5000 Å were used. Twenty structures out of the total 100 were selected based on the smallest energy and amongst them 10 with the lowest NOE violations were subjected to a maximum of 10 000 steps of steepest descent energy minimization. The details of the SA protocols are given in the Supplementary Data. The resulting families of 10 structures at the three different conditions were used for structural statistics, which are for the M2 G-quadruplexes formed in the absence of cosolutes, in the presence of 20 wt% EG and 10 wt% PEG 8000 reported in Tables 2, 3 and 4, respectively. UCSF Chimera software (33) was used for visualization of the calculated structures and preparation of Figures 5, 8 and Supplementary Figures S6-S9. Coordinates of the M2 G-quadruplex structures in the absence of cosolutes, in the presence of 20 wt% EG and 10 wt% PEG 8000 have been deposited in the Protein Data Bank with accession codes 5NYS, 5NYT and 5NYU, respectively.
Table 2. NMR restraints and structural statistics for the M2 G-quadruplex in the absence of cosolutes.
| NOE-derived distance restraints | Non-exchangeable | Exchangeable |
|---|---|---|
| Intranucleotide NOEs | 323 | 0 |
| Sequential (i, i+1) | 64 | 8 |
| Long-range (i, > i+1) | 10 | 22 |
| Torsion angle restraints | 97 | |
| Hydrogen-bond restraints | 24 | |
| Structural statistics | ||
| NOE violations >0.2Å | 0 | |
| Deviations from idealized covalent geometry | ||
| Bonds (Å) | 0.011 ± 0.000 | |
| Angles (°) | 2.246 ± 0.064 | |
| Pairwise heavy atom RMSD (Å) | ||
| Overall | 0.890 | |
| Without 5′ TA overhang | 0.894 | |
| Without 3′ T overhang | 0.902 | |
| Without 5′ TA and 3′ T overhangs | 0.907 | |
| Without A6, C11 and C15, A16 loop residues | 0.929 | |
| Only core of the structure (only guanine residues) | 0.510 | |
Table 3. NMR restraints and structural statistics for the M2 G-quadruplex in the presence of 20 wt% EG.
| NOE-derived distance restraints | Non-exchangeable | Exchangeable |
|---|---|---|
| Intranucleotide NOEs | 335 | 0 |
| Sequential (i, i+1) | 70 | 9 |
| Long-range (i, > i+1) | 11 | 22 |
| Torsion angle restraints | 97 | |
| Hydrogen-bond restraints | 24 | |
| Structural statistics | ||
| NOE violations >0.2Å | 0 | |
| Deviations from idealized covalent geometry | ||
| Bonds (Å) | 0.011 ± 0.000 | |
| Angles (°) | 2.291 ± 0.068 | |
| Pairwise heavy atom RMSD (Å) | ||
| Overall | 1.270 | |
| Without 5′ TA overhang | 1.310 | |
| Without 3′ T overhang | 1.297 | |
| Without 5′ TA and 3′ T overhangs | 1.342 | |
| Without A6, C11 and C15, A16 loop residues | 1.316 | |
| Only core of the structure (only guanine residues) | 0.535 | |
Table 4. NMR restraints and structural statistics for the M2 G-quadruplex in the presence of 10 wt% PEG 8000.
| NOE-derived distance restraints | Non-exchangeable | Exchangeable |
|---|---|---|
| Intranucleotide NOEs | 345 | 0 |
| Sequential (i, i+1) | 68 | 10 |
| Long-range (i, > i+1) | 11 | 25 |
| Torsion angle restraints | 97 | |
| Hydrogen-bond restraints | 24 | |
| Structural statistics | ||
| NOE violations >0.2Å | 0 | |
| Deviations from idealized covalent geometry | ||
| Bonds (Å) | 0.011 ± 0.000 | |
| Angles (°) | 2.253 ± 0.034 | |
| Pairwise heavy atom RMSD (Å) | ||
| Overall | 1.055 | |
| Without 5′ TA overhang | 1.092 | |
| Without 3′ T overhang | 1.078 | |
| Without 5′ TA and 3′ T overhangs | 1.119 | |
| Without A6, C11 and C15, A16 loop residues | 1.089 | |
| Only core of the structure (only guanine residues) | 0.492 | |
Figure 5.
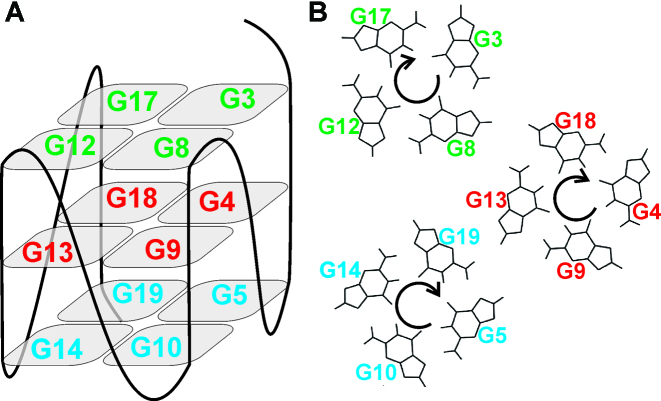
(A) Schematic presentation of G-quadruplex adopted by M2 with residues constituting G3–G8–G12–G17, G4–G9–G13–G18 and G5–G10–G14–G19 quartets designated in green, red and blue, respectively. (B) The directionalities of hydrogen-bonding within each of the three quartets.
Figure 8.
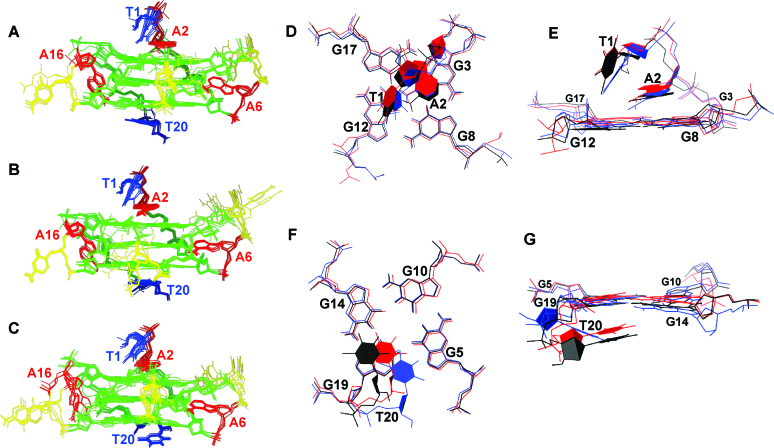
Superposition of ten NMR-based high-resolution refined structures of M2 G-quadruplex (A) in the absence of cosolutes (PDB ID: 5NYS), (B) in the presence of 20 wt% EG (PDB ID: 5NYT) and (C) in the presence of 10 wt% PEG 8000 (PDB ID: 5NYU). Bird's-eye and side views on the 5′-end overhang stacked over G3-G8-G12-G17 quartet are shown in (D) and (E), respectively. Bird's-eye and side views on the 3′-end overhang stacked over G5–G10–G14–G19 quartet are shown in (F) and (G), respectively. In panels A–C adenine, cytosine, guanine and thymine residues are colored in red, yellow, green and blue, respectively. In panels D–G, the structures of M2 G-quadruplex in the absence of cosolutes and in the presence of 20 wt% EG and 10 wt% PEG 8000 are coloured black, red and blue, respectively.
CD spectroscopy
DNA oligonucleotide (10 μM) was dissolved in buffer containing 20 mM potassium phosphate (pH 7.0), 100 mM KCl in the absence of cosolutes and in the presence of 20 wt% EG and PEG 8000, and refolded from 95 to 25°C at 0.5°C min−1. CD spectra of the samples from 200 to 350 nm were collected on a JASCO J-1500 spectropolarimeter at 25°C in 1.0-mm path length cuvettes. To measure temperature dependent CD spectra, DNA oligonucleotide (10 μM) was refolded from 95 to 10°C at 0.2°C min−1 in a buffer containing 20 mM potassium phosphate (pH 7.0) in the absence of cosolutes and in the presence of 20 wt% EG, 10 wt% PEG 8000 and 20 wt% PEG 8000. CD spectra of the samples from 200 to 350 nm were collected from 10 to 95°C at every 5°C interval in 1.0-mm path length cuvettes.
UV spectroscopy
DNA oligonucleotide (10 μM) was refolded from 95 to 10°C at 0.5°C min−1 in buffer containing 20 mM potassium phosphate (pH 7.0) in the absence of cosolutes and in the presence of 20 wt% EG, 10 wt% PEG 8000 and 20 wt% PEG 8000. UV absorbance at 295 nm was measured from 25 to 95°C at 0.2°C min−1. To evaluate stability of the G-quadruplex depending on potassium ion concentration, UV absorbance of DNA oligonucleotide (5 μM) was measured at 295 nm in a buffer containing 50 mM MES–LiOH (pH 7.0) and in the absence of cosolutes and in the presence of 20 wt% EG and PEG 8000. The thermodynamic stabilities at 25°C (ΔG°25) were calculated from the fit of the melting curves to a theoretical equation for an intramolecular association as described previously (22,34).
Measurements of physical properties of solutions
The water activity was determined by the osmotic stressing method via vapor phase osmometry using a model 5520XR pressure osmometer (Wescor, Logan, UT, USA) at 25°C. The dielectric constants were obtained from the maximum emission wavelength (λmax) of the fluorescence of 8-anilino-1-naphthalenesulfonic acid (ANS) at 25°C excited by 380 nm (35). The standard curve was determined by λmax of ANS with different contents of ethanol. The dielectric constants of ethanol standard solutions were measured by an M-870 liquid dielectric constant meter (Scientifica).
RESULTS
M2 adopts the same G-quadruplex topology in the absence and in the presence of EG and PEG 8000
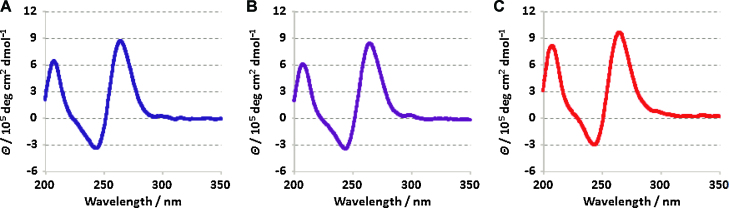
CD spectra of M2 were analyzed under diluted solution conditions and compared with data obtained under molecular crowding conditions induced by 20 wt% EG and PEG 8000 as cosolutes (Figure 1). All CD spectra show clear positive peaks around 208 and 265 nm and a negative peak around 243 nm, which are consistent with M2 adopting a parallel G-quadruplex irrespective of the additional cosolutes.
Figure 1.
CD spectra of M2 (A) in the absence of cosolute and in the presence of (B) 20 wt% EG or (C) 20 wt% PEG 8000. The spectra were recorded in a buffer containing 20 mM potassium phosphate (pH 7.0) and 100 mM KCl, at 25°C and 10 μM oligonucleotide concentration.
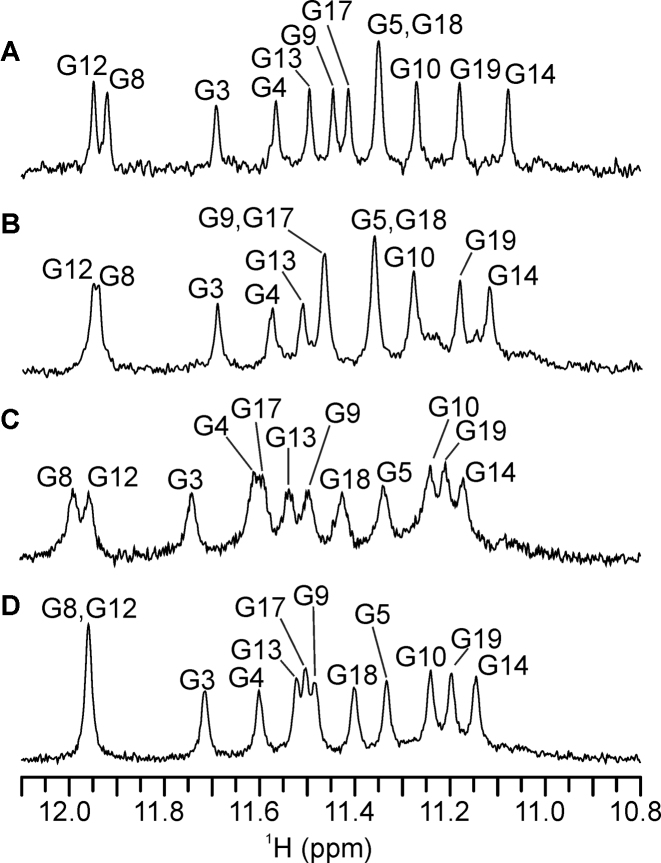
Imino proton signals are not observable in 1H NMR spectrum of the desalted sample of M2 which suggests absence of a well-defined high order structure (data not shown). Upon addition of potassium ions twelve imino signals are observed in the range between δ 11.07 and 11.95 ppm corresponding to twelve guanine residues assembled via Hoogsteen hydrogen-bonding within one predominant G-quadruplex structure (Figure 2A). Assignment of the imino as well as the aromatic, methyl and most anomeric protons was accomplished with the use of NOESY (Figure 3A), JRHMBC (Supplementary Figure S1) and TOCSY 2D NMR experiments (chemical shifts are reported in Supplementary Table S1). The NOE-derived correlations between the imino H1 and aromatic H8 protons (Figure 4A) are consistent with the following hydrogen-bond directionalities within the three G-quartets: G3→G8→G12→G17, G4→G9→G13→G18 and G5→G10→G14→G19 (Figure 5B). In addition, intra- and inter-G-quartet imino-imino NOE interactions are consistent with M2 adopting a unimolecular G-quadruplex structure exhibiting three consecutively stacked G-quartets with all four strands in parallel orientation (Supplementary Figure S2A). Furthermore, the characteristic interresidual H8-H1′, H8-H2′/H2″ as well as sequential guanine(i)-guanine(i+1) NOE cross-peaks reveal that all the guanine residues are in anti glycosidic conformation. Notably, the intramolecular M2 G-quadruplex fold is predominant species at up to 1 mM DNA oligonucleotide concentration (Supplementary Figure S3).
Figure 2.
1H NMR imino region of M2 (A) without cosolute and at (B) 20 wt% EG, (C) 20 wt% PEG 8000 and (D) 10 wt% PEG 8000. The spectra were recorded in 90% H2O/10% 2H2O at 25°C, 0.2 mM oligonucleotide concentration per strand, 100 mM KCl and 20 mM potassium phosphate (pH 7.0).
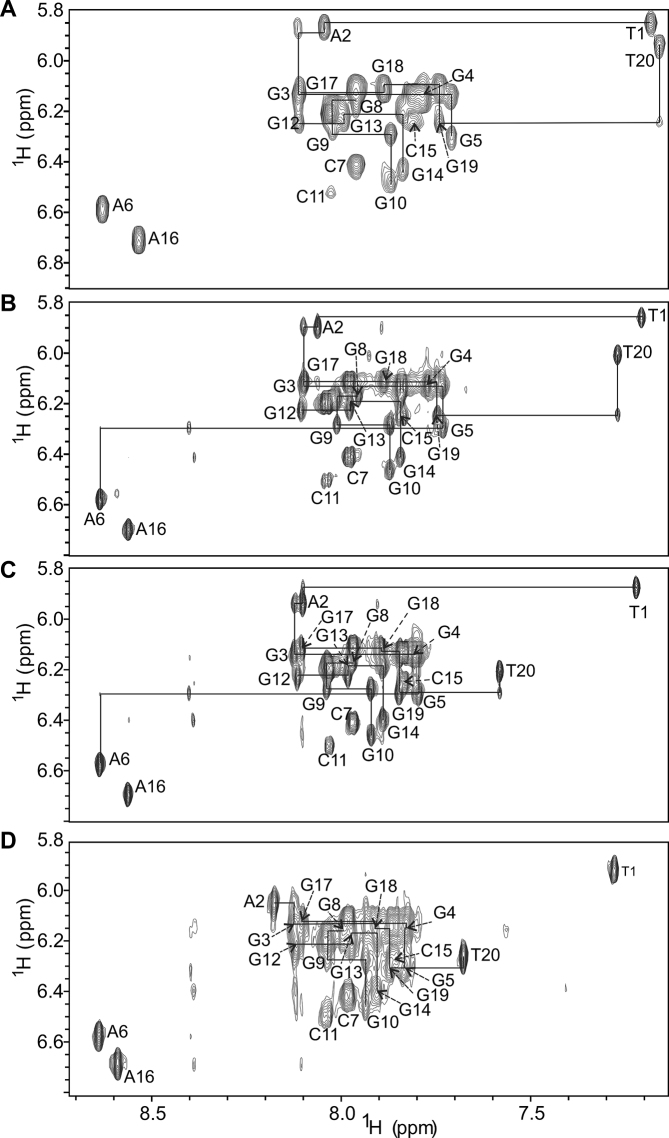
Figure 3.
Anomeric–aromatic region of NOESY spectrum of M2 (A) without cosolute, at (B) 20 wt% EG, (C) 10 wt% PEG 8000 and (D) 20 wt% PEG 8000. The spectra were recorded at mixing time of 300 ms in 90% H2O/10% 2H2O at 25°C, 0.2 mM oligonucleotide concentration per strand, 100 mM KCl and 20 mM potassium phosphate (pH 7.0). Residue numbers denote intraresidual H1′–H6/H8 NOE interactions and lines denote sequential correlations.
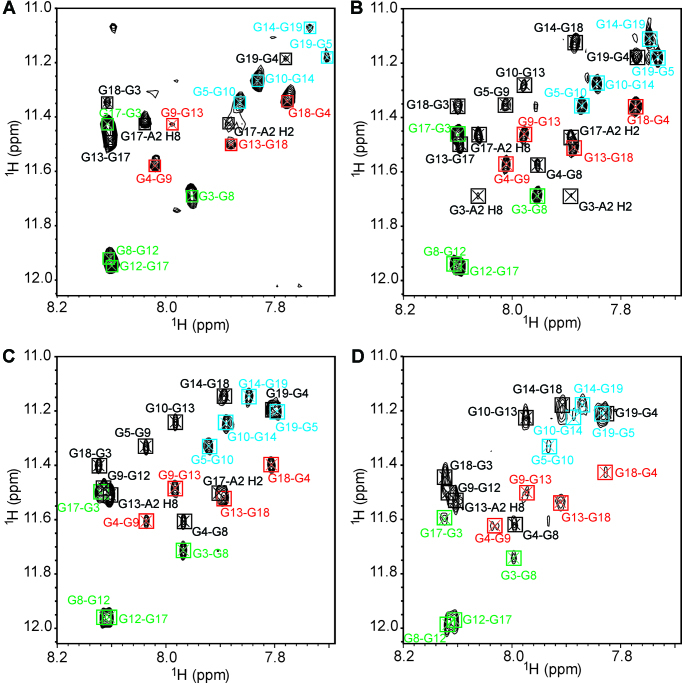
Figure 4.
Imino–aromatic region of NOESY spectrum of M2 (A) without cosolute and at (B) 20 wt% EG, (C) 10 wt% PEG 8000 and (D) 20 wt% PEG 8000. The spectra were recorded at mixing time of 300 ms in 90% H2O/10% 2H2O at 25°C, 0.2 mM oligonucleotide concentration per strand, 100 mM KCl, 20 mM potassium phosphate (pH 7.0). H1–H8 cross-peaks within G3–G8–G12–G17, G4–G9–G13–G18 and G5–G10–G14–G19 quartets are designated in green, red and blue, respectively. Cross-peaks corresponding to residues that are not part of the same G-quartet are designated in black.
Well-resolved resonances are observed in 1H NMR spectra of M2 in buffer solution containing potassium ions as well as in the presence of 20 wt% EG and 20 wt% PEG 8000 (Figure 2B and C). Most important regarding the focus of this study is observation of twelve major 1H NMR signals in the imino region characteristic of Hoogsteen hydrogen-bonded guanine residues, which is consistent with formation of a single G-quadruplex species exhibiting three G-quartets under all three conditions. Noteworthy, 1H NMR spectra of M2 without added cosolutes and at 20 wt% EG are characterized by narrow signals, while broadening of the 1H NMR signals in the presence of 20 wt% PEG 8000 implies equilibrium of different species or slowed down molecular tumbling. Moreover, DNA concentration of 0.2 mM, which is much higher in comparison to the one used in CD spectrum analysis (0.01 mM), might in addition to formation of predominant G-quadruplex lead to high-order species and aggregation. In this regard, further NMR-based characterization of M2 at 10 wt% PEG 8000 gave rise to narrow and well-resolved 1H NMR signals (Figure 2D).
2D NMR experiments were utilized in assigning of all imino, aromatic, methyl and most anomeric 1H NMR chemical shifts corresponding to M2 at 20 wt% EG and 10 wt% PEG 8000 (chemical shifts are reported in Supplementary Tables S2 and S3, respectively). Detailed examination of intra- and interresidual NOE cross-peaks demonstrated that M2 adopts a unimolecular G-quadruplex comprising three successively stacked G-quartets and exhibiting all strands in a parallel orientation in the presence of either of the two cosolutes (Figures 3B–D, 4B–D and 5). Further analysis of NOE interactions, in particular intraresidual aromatic-anomeric contacts showed that all guanines exhibit anti conformation of glycosidic torsion angle. Inter-G-quartet imino-imino as well as imino-H8 proton NOE interactions established formation of G3–G8–G12–G17, G4–G9–G13–G18 and G5–G10–G14–G19 quartets (Supplementary Figure S2b-d). Altogether, NMR data collected for M2 at 20 wt% EG, 10 wt% PEG 8000 and 20 wt% PEG 8000 demonstrate prevalence of a parallel G-quadruplex topology under molecular crowding conditions as well as in potassium ion containing solution.
Increase in thermodynamic stability of the M2 G-quadruplex induced by EG and PEG 8000 is coupled with higher potassium ion uptake
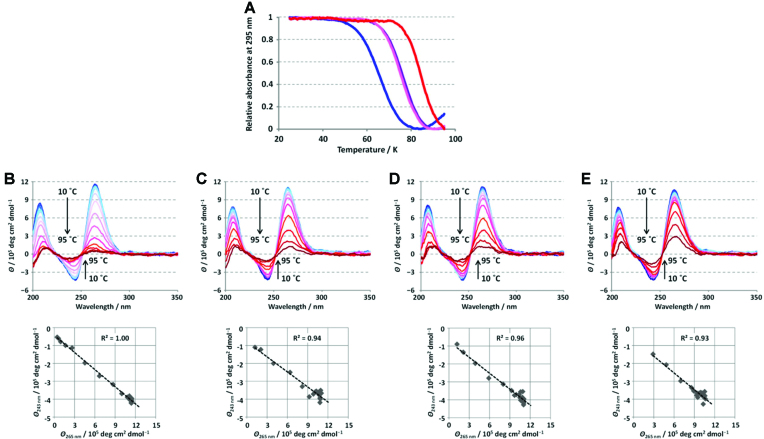
Thermodynamic stability of the M2 G-quadruplex was evaluated by using UV melting analysis. Since the M2 G-quadruplex does not melt even at 90°C in the presence of 100 mM KCl and cosolutes (data not shown), the melting properties were evaluated in a basal buffer containing 20 mM potassium phosphate in the absence and in the presence of 20 wt% EG, 20 wt% PEG 8000 and 10 wt% PEG 8000. At all studied conditions, the melting profiles show clear transition at 295 nm (Figure 6A). CD spectra at different temperatures in the same buffer show isosbestic points near 225 and 250 nm, and linear correlations between the signal changes at 265 and 243 nm during the melting (Figure 6). These results clearly indicate a two-state transition of the (un)folding equilibria of M2 G-quadruplex. Thermodynamic parameters, i.e. enthalpy (ΔH°) and entropy (ΔS°) changes and stability at 25°C (ΔG°25) for the formation of the G-quadruplex calculated by fitting the melting transitions are shown in Table 1. Addition of either EG or PEG 8000 stabilizes the M2 G-quadruplex due to favorable enthalpy contribution. Previous reports have demonstrated the dominant effect of dehydration for stabilization of not only G-quadruplexes, but also other nucleic acid structures (36–38). The stabilization of the M2 G-quadruplex by EG is likely caused by an osmolyte effect of EG involving dehydration upon formation of the G-quadruplex, because the water activity is clearly reduced by addition of EG. Interestingly, PEG 8000 stabilizes the M2 G-quadruplex to a greater extent than EG at 20 wt%, although PEG 8000 does not behave as an osmolyte and does not reduce the water activity comparing to EG (Supplementary Table S4). The energy difference in the presence and in the absence of cosolute (ΔΔG°25) is more than twofold higher in the presence of 20 wt% PEG 8000 (ΔΔG°25 = 7.37 kcal mol−1) comparing to the same weight percent of EG (ΔΔG°25 = 3.43 kcal mol−1). The stability of the M2 G-quadruplex in the presence of 20 wt% EG is comparable with that in the presence of 10 wt% PEG 8000. As dehydration is not the prime origin of PEG 8000-induced stabilization, the observed stabilization corresponds to other factors in solution or direct interaction between PEG 8000 and M2 G-quadruplex.
Figure 6.
Thermodynamic melting profile of M2 G-quadruplex. (A) UV melting transition of M2 G-quadruplex in the absence and in the presence of EG and PEG 8000. UV absorbance at 295 nm of M2 in the absence of cosolute (blue) and in the presence of 20 wt% EG (purple), 10 wt% PEG 8000 (pink) and 20 wt% PEG 8000 (red). The UV absorbance was measured at a rate of 0.2°C min−1. CD spectra of M2 in the range from 10 to 95°C (B) in the absence of cosolute and in the presence of (C) 20 wt% EG, (D) 10 wt% PEG 8000 or (E) 20 wt% PEG 8000. Plots of CD elipticity at 243 nm versus 265 nm are shown below. UV and CD spectra were recorded at 10 μM oligonucleotide concentration per strand and 20 mM potassium phosphate (pH 7.0).
Table 1. Thermodynamic parameters of the M2 G-quadruplex in a buffer containing 20 mM potassium phosphate (pH 7) in the absence and in the presence of cosolutes.
| Cosolute | ΔH° (kcal mol−1) | ΔS° (cal mol−1 K−1) | ΔG°25 (kcal mol−1) |
|---|---|---|---|
| without cosolute | −42.2 ± 0.9 | −124 ± 2.9 | −5.33 ± 0.04 |
| 20 wt% EG | −58.7 ± 0.4 | −168 ± 1.2 | −8.76 ± 0.10 |
| 20 wt% PEG 8000 | −76.6 ± 2.9 | −215 ± 8.3 | −12.7 ± 0.40 |
Values are average ± s.d. from at least triplicated experiments.
UV melting analyses were performed in a buffer containing 50 mM MES–LiOH (pH 7) in the absence and in the presence of 20 wt% cosolute with various concentrations of KCl to evaluate an effect of cation concentration on the G-quadruplex stability (Supplementary Figure S4a-c). Supplementary Table S5 shows thermodynamic parameters calculated from the UV meting curves. At a range of potassium ion concentrations, at which M2 G-quadruplex showed clear melting transition between 20 and 95°C, ΔG°25 of the M2 G-quadruplex and natural logarithm value of the potassium ion concentration (ln[KCl]) showed clear linear correlation (Supplementary Figure S4d). Slope value of the plots reflects a number of potassium ions involved in the G-quadruplex formation or folding cooperativity of the G-quadruplex in response to changing concentration of potassium ions (39,40). The absolute slope values increased in the presence of cosolutes, indicating that upon folding of M2 G-quadruplex under the molecular crowding conditions the uptake of potassium ions is higher than under the diluted condition. Thus, it is expected that in the presence of 100 mM potassium ions, at which we analysed NMR structure, the difference of G-quadruplex stability between the diluted and crowding conditions are larger than those reported in Table 1. UV melting profiles also indicate that the potassium ion concentration, at which a half of oligonucleotide forms G-quadruplex at 25°C (ΔG°25 = 0), is lower in the presence of cosolutes comparing to the diluted condition (Supplementary Figure S4D). These results suggest that not only cooperativity but also the observed binding affinity of potassium ions increases under the crowding condition. Importantly, interactions between potassium ions and M2 G-quadruplex seems more efficiently enhanced by PEG 8000, because the absolute slope value in ΔG°25vs. ln[KCl] plots is larger, while lower potassium ion concentration is required for the G-quadruplex folding in comparison to EG.
Overhanging residues of the M2 G-quadruplex in the role of antenna for cosolute-induced changes
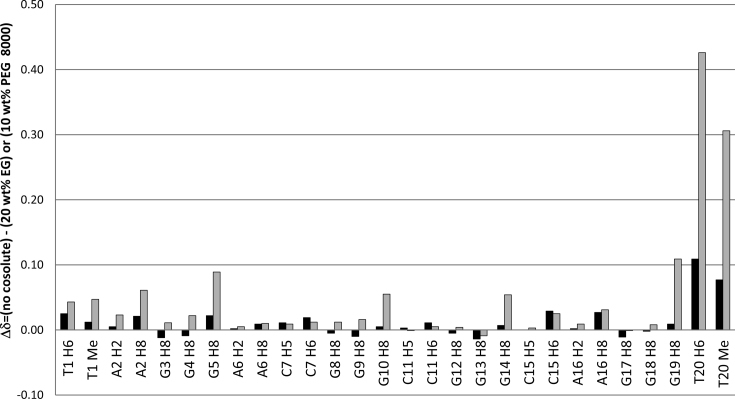
Upon addition of 20 wt% EG changes in 1H NMR chemical shifts of methyl and aromatic protons of T1-G19 residues of the M2 G-quadruplex are observed in the range between -0.01 and 0.03 ppm (Figure 7 and Supplementary Table S6). Noteworthy, these values are very close to the limits of the experimental error. On the other hand, there is a significant downfield shift of methyl as well as H6 protons of T20 by 0.08 and 0.11 ppm, respectively, indicating that their local environments are changed considerably upon addition of 20 wt% EG. These changes may result from decrease in water activity in the presence of 20 wt% EG and/or associated conformational change of residue T20. It is noteworthy that coupled alteration of M2 G-quadruplex structure and water network that are induced by the high concentrations of EG seem likely, especially as careful inspection of NOESY spectra recorded at mixing times in the range between 80 and 300 ms did not reveal any long-lived interactions between the M2 G-quadruplex and EG.
Figure 7.
1H NMR chemical shift differences of methyl and aromatic H8/H6/H5/H2 protons of the M2 G-quadruplex in the presence of 20 wt% EG and 10 wt% PEG 8000 with respect to the M2 G-quadruplex in the absence of cosolute. The bars in black and gray were derived by subtracting 1H NMR chemical shifts observed in the presence of 20 wt% EG and 10 wt% PEG 8000, respectively, from those observed in the absence of cosolute.
Perturbations in 1H NMR chemical shifts upon addition of 10 or 20 wt% PEG 8000 are evident for all residues corresponding to the M2 G-quadruplex (Figure 7 and Supplementary Table S6). Notably, the extent of 1H NMR chemical shift changes correlates with the increased concentration of PEG 8000 (Supplementary Figure S5). The effect is by far most pronounced for residue T20. Moreover, consequent with addition of 10 wt% and 20 wt% PEG 8000 the changes in 1H NMR chemical shift of T20 H6 aromatic proton are 0.43 and 0.52 ppm, respectively. The substantial PEG 8000-induced changes in the chemical environment at the 3′-end of the M2 G-quadruplex are further demonstrated by the large and gradual downfield shift of T20 methyl 1H NMR signal. Interestingly, at 20 wt% PEG 8000, the aromatic protons of G5, G10, G14 and G19 residues comprising the G-quartet positioned in vicinity of T20 are clearly shifted downfield by 0.13, 0.07, 0.07 and 0.13 ppm, respectively. In comparison, perturbations in 1H NMR chemical shifts for aromatic protons of guanine residues in the G3–G8–G12–G17 and G4–G9–G13–G18 quartets are in the range between –0.02 and 0.04 ppm and thus considerably smaller with respect to the G5–G10–G14–G19 quartet. Noteworthy, addition of 20 wt% EG (in contrast to PEG 8000) does not lead to notable deshielding of 1H NMR signals of guanine residues, which may reflect differences between EG- and PEG 8000-M2 G-quadruplex interactions. The addition of 10 wt% PEG 8000 gives rise to minor perturbations in methyl and aromatic 1H NMR chemical shifts (Δδ below 0.06 ppm) of the 5′-end overhanging residues of the M2 G-quadruplex. Interestingly, however, the increase of PEG 8000 to 20 wt% results in considerable downfield shifts of T1 methyl and H6 as well as of A2 H8 1H NMR signal, which are close or above 0.10 ppm with respect to cosolute-free condition. This indicates that PEG 8000, at least at 20 wt%, induces substantial changes in the chemical environment not only at the 3′, but also at the 5′-end overhanging region of the M2 G-quadruplex. Altogether, the analysis of 1H NMR chemical shifts perturbation for the M2 G-quadruplex upon addition of PEG 8000 indicates specificity of cosolute-induced effects, which are minor at the core of the structure, significant at the 5′-end and profound at the 3′-end of the structure. In this regard, it is noteworthy that perusal of NOESY spectra does not reveal any cross-peaks in support to direct binding of PEG 8000 to the M2 G-quadruplex.
EG and PEG 8000 modulate M2 G-quadruplex structure differently
We calculated the solution-state structure of the M2 G-quadruplex formed in the presence of potassium ions and absence of cosolutes with the use of 427 NOE-derived distance restraints together with 97 torsion angle and 24 hydrogen-bond restraints (Figure 8A). Well-converged ten representative structures exhibit a pairwise heavy atom RMSD of 0.89 Å (structural statistics are given in Table 2). All the guanine residues involved in G3–G8–G12–G17, G4–G9–G13–G18 and G5–G10–G14–G19 quartets exhibit anti glycosidic conformations. The three G-quartets are stacked on top of each other and represent the core of the structure. G-quartets are not perfectly planar as several guanines, in particular G4, G8, G9, G10, G14 and G17 exhibit orientation with O6 being slightly bent toward the 3′-end (Supplementary Figure S6). The 5′-end overhanging residues T1 and A2 are positioned on top of the central cation cavity (Figure 8D and E). Among the ensemble of the representative structures certain conformational freedom is observed for T1, whereas conformation of A2 is very well converged. The purine ring of A2 is parallel with respect to the plane of the nearby G3-G8-G12-G17 quartet. The 3′-end is well defined and exhibits T20 stacked over G19. A6 and C7 form propeller-type loop thereby linking G5 and G8 residues of the 5′- and 3′-end G-quartets, respectively. Orientation of A6 is well defined, with its purine ring inclined by ca. 45° with respect to the plane of the nearby central G4–G9–G13–G18 quartet (Supplementary Figure S7). C7 exhibits more conformational freedom in comparison to A6. The central loop comprises C11 and bridges the G8–G9–G10 and G12–G13–G14 stems of the M2 G-quadruplex via propeller-type arrangement. The loose conformation of C11, together with its orientation toward solvent is reminiscent of previous reports on single-nucleotide propeller-type loops in parallel stranded G-quadruplex structures (Supplementary Figure S8) (41–44). Structures of C15 and A16 are well defined and form the third propeller-type loop. Moreover, C15 is oriented away from the core of the structure toward the solvent, whereas A16 is inserted into the groove formed by G12–G14 and G17–G19 stems (Supplementary Figure S9). The M2 G-quadruplex is characterized by the average groove width of 11.1 (± 0.5) Å which is thus slightly broader with respect to the parallel-stranded N-myc (20) G-quadruplex (10.7 ± 0.5 Å) exhibiting three single-residue propeller-type loops and three G-quartets in the core of the structure.
High-resolution structure of M2 G-quadruplex in the presence of 20 wt% EG was calculated using 447 NOE-derived distance restraints. Ten representative structures of parallel stranded M2 G-quadruplex with the three stacked G-quartets in the core exhibit a pairwise heavy atom RMSD of 1.27 Å (Figure 8B; structural statistics are given in Table 3). This structure exhibits an average groove width of 11.0 (± 0.6) Å. All guanine residues adopt anti glycosidic conformation. T1 and more so A2 at the 5′-end overhang region exhibit well-defined conformations and together cap the nearby G3–G8–G12–G17 quartet (Figure 8D and E). T20 at the 3′-end exhibits a well-defined conformation and is stacked on the pyrimidine moiety of G19 (Figure 8F and G). In the first propeller-type loop of the M2 G-quadruplex in the presence of 20 wt% EG A6 exhibits a well-defined conformation, while conformation of C7 is poorly defined. C11 in the second propeller-type loop occupies the groove between G8–G10 and G12–G14 stems with the base moiety oriented inward the G5–G10–G14–G19 quartet (Supplementary Figure S8). Notably, eight among ensemble of ten representative structures exhibit C11 with a well-converged conformation. The two remaining structures are diverse, altogether indicating that conformation of C11 is not defined precisely. In contrary, the conformation of the third propeller-type loop is very well-defined with both residues, C15 and A16, being well converged among the ensemble of ten representative structures.
There is great resemblance between the calculated M2 G-quadruplex structures in the absence and in the presence of 20 wt% EG. Structures share orientations of residues at the 5′-end (Figure 8), albeit the ones corresponding to cosolute-free condition exhibit A2 seemingly slightly closer to the nearby G3–G8–G12–G17 quartet. Further, cores of the structures in the absence and in the presence of 20 wt% EG grossly match, with some deviations in conformations of guanine residues. In particular, in the presence of 20 wt% EG there is an out-of-plane bending of G4, G10, G14, G17, G18 and G19 toward the 5′-end and shifting of G4, G9, G12, G17 and G18 within the G-quartet planes in comparison to the structures in the absence of cosolutes (Supplementary Figure S6). These variances, however, could in principle be ascribed in the range of ‘breathing’ motions within G-quartets that are commonly observed in G-quadruplexes. The first and the third two-residue loops A6-C7 and C15-A16, respectively, appear very similar in the absence and in the presence of 20 wt% EG, with the exception that the cosolute imposes a slightly smaller inclination of A6 with respect to G4–G9–G13–G18 quartet plane, i.e. ca. 30° versus 45° (Supplementary Figure S7). On the other hand, a substantial structural change can be assigned to the presence of EG for the central one-residue propeller-type loop. Moreover, in eight out of ten representative structures of the M2 G-quadruplex in the presence of 20 wt% EG residue C11 is oriented toward the G5–G10–G14–G19 quartet and not aside the structural core as in the cosolute-free condition (Supplementary Figure S8). Coupled with the variance in C11 orientation the sugar-phosphate backbone in the G8–G13 segment is different between the structures in the absence and in the presence of 20 wt% EG. Notably, however, orientation of C11 is not firmly restrained by the experimental data in either of the conditions, thus suggesting that EG exhibits only a limited impact on orientation of loop residues. Yet, there exists a clear and consistent distinction between the M2 G-quadruplex structures in the presence and absence of EG and it is located at the 3′-ends of the structures (Figure 8F–G). In the absence of cosolutes T20 stacks over pyrimidine and imidazole moieties of G19, whereas in the presence of EG the interaction interface comprises almost exclusively pyrimidine moiety of G19. Additionally, the presence of EG is coupled with position of T20 closer to the top of the central cation channel of M2 G-quadruplex. Notably however, the overall distance between T20 and G19 appears similar in the absence and in the presence of EG, due to EG-induced change in their backbone conformations as well as out of plane bending of G19 toward the 5′-end. It is worth noting that for individual conditions, i.e. in the absence and in the presence of 20 wt% EG, the conformation of T20 is very well converged among the ensembles of the calculated structures. The prominent conformational changes of T20 in response to the presence of EG are consistent with the major perturbations of its 1H NMR chemical shifts (Figure 7). On the other hand, EG-induced changes in 1H NMR chemical shifts for residues in T1-A2 and G10–C11–G12 segments are only minor, although slight deviations in their conformations were observed with respect to the calculated structure in the absence of cosolutes. Overall, the NMR-based structural comparison suggests that the differences in biophysical properties of the M2 G-quadruplex in the absence and in the presence of EG (vide supra) are coupled to T20 conformational changes, while effect is considerably smaller for the three loops and the 5′-end overhang.
The high-resolution structure of parallel-stranded M2 G-quadruplex in the presence of 10 wt% PEG 8000 was calculated with the use of 459 NOE-derived distance restraints, 97 torsion angle restraints and 24 hydrogen-bond restraints. The ten representative structures exhibiting a pairwise heavy atom RMSD of 1.06 Å are shown in Figure 8C (structural statistics are given in Table 4). Twelve guanine residues, all characterized by anti glycosidic torsion angle values, form three stacked G-quartets at the M2 G-quadruplex core. The average groove width is 11.0 (±0.7) Å, matching well to the structures in cosolute-free and 20 wt% EG conditions. The G3–G8–G12–G17 quartet is capped with the 5′-overhang residues T1 and A2, which are successively stacked, whereby A2 is positioned partly above G3 and partly above the central cation channel. T1 and A2 are both well-converged among the ensemble of the ten representative structures. The conformation of T20 at the 3′-end is well-defined. Interestingly, T20 is oriented away from the G-quartet ‘platform’ and is positioned in extension of the groove defined by G3–G5 and G17–G19 stems, with its sugar moiety facing the solvent. A6, which is the first of the two residues in the first propeller-type loop, is well-defined and exhibits purine moiety inserted in the groove in the G3–G5 and G8–G10 region. Additionally, the purine base of A6 is slightly (ca. 30°) inclined with respect to the central G4–G9-G13–G18 quartet (Supplementary Figure S7). Conformation of C7 is rather flexible. The second propeller-type loop is well-defined and exhibits C11 residue oriented toward the solution. C15 in the third propeller-type loop is positioned away from the core of the structure enabling insertion of A16 into the groove defined by G12–G14 and G17–G19 stems. Among the ten representative structures of the M2 G-quadruplex in the presence of 10 wt% PEG 8000, orientation of A16 congregates into two clusters (Supplementary Figure S9D and E). In the cluster of seven structures the A16 purine ring caps the groove defined by G12–G14 and G17–G19 stems. The cluster of the remaining three structures is defined by A16 being closer to the central G4–G9–G13–G18 quartet and with plane of its base moiety appearing to intersect the nearby groove of G12–G14 and G17–G19 stems.
In the presence of PEG 8000 the conformation of T20 at the 3′-end is very different with respect to the structure in the absence of cosolutes (Figure 8F and G). In cosolute-free conditions the sugar-phosphate backbone progression in the G17-T20 segments of the M2 G-quadruplex is uniform, enabling T20 to stack on G19. In comparison, the addition of PEG 8000 results in T20 sugar moiety to orient closer to the groove defined by G12–G14 and G17–G19 stems, while its pyrimidine moiety is pushed aside of G5–G10–G14–G19 quartet. The conformational change of T20 corresponds to the largest difference between the structures in the absence and in the presence of 10 wt% PEG 8000 and thus corroborates the results of 1H NMR chemical shift perturbation analysis (Figure 7). In addition, the presence of 10 wt% PEG 8000 very modestly affects the conformation of the 5′-end of the M2 G-quadruplex in that T1 is slightly closer to A2. The conformation of T1 is more converged among the structures in the presence of 10 wt% PEG 8000, altogether suggesting more constrained disposition of the T1-A2 ‘cap’ over the nearby G-quartet in comparison to the structure in cosolute-free condition. Although, guanine residues involved in the core of the M2 G-quadruplex in the presence of 10 wt% PEG 8000 are not superimposable to the structures in the absence of cosolutes perfectly (Supplementary Figure S6), the differences are minor. In this regard, the distinction is even less prominent than between the structures in cosolute-free and EG conditions. PEG 8000-induced structural changes in the first propeller-type loop are confined to A6, whose purine moiety appears slightly less inclined with respect to the plane of G4–G9–G13–G18 quartet. Interestingly, the conformation of A6 is highly similar in the presence of 10 wt% PEG 8000 and 20 wt% EG (Supplementary Figure S7). The disposition of the second loop comprising C11 is not significantly affected by the presence of 10 wt% PEG 8000. However, PEG 8000 appears to promote moderate flexibility of the third propeller-type loop comprising C15 and A16. Furthermore, only three among the ensemble of the ten representative structures in the presence of 10 wt% PEG 8000 exhibit C15 and A16 in a disposition matching well to the conformation observed in the structures in the absence of cosolutes and in the presence of 20 wt% EG. However, the significance of the PEG 8000-induced variable disposition of C15 and A16 in the calculated structures is questionable, especially considering that only marginal 1H NMR chemical shifts changes are observed for the C15-A16 segment upon addition of PEG 8000. Overall, it seems that 10 wt% PEG 8000 only modestly effects disposition of loops and the 5′-end residues, while it has a significant impact on the conformation of T20 at the 3′-end of the M2 G-quadruplex.
In order to correlate the structural differences with intrinsic energy of the M2 G-quadruplexes under the three different conditions we performed electronic state calculations (Supplementary Data). Fragment molecular orbital (FMO) method in vacuum condition was utilized to carry out the interfragment interaction energy (IFIE) analysis to detect the direct molecular interactions between fragments (Supplementary Figure S10 and Supplementary Tables S7 to S10) (45–47). The orientation of C11 toward the G5–G10–G14–G19 quartet in the presence of 20 wt% EG contributes to lower IFIE of the M2 G-quadruplex, while the orientation toward the bulk solution observed in the presence of 10 wt% PEG 8000 and cosolute-free condition increases the IFIE. The position of A16 as a cap over the groove between G12–G14 and G17–G19 stems does not contribute to the IFIE as much as when it is located closer to the central G4–G9–G13–G18 quartet. Reduction of IFIE involving residue T20 follows the following order: 20 wt% EG > cosolute-free > 10 wt% PEG 8000. According to the above contributions, the overall IFIE of the M2 G-quadruplex is the highest in the presence of 20 wt% EG and is followed by cosolute-free and 10 wt% PEG 8000 conditions (Supplementary Table S10). However, due to considerably large standard deviations, the energy differences cannot be clearly distinguished between the structures under the three conditions.
DISCUSSION
Molecular crowding conditions with high concentration of cosolutes potentially alter the folding and thermodynamic stability of nucleic acids through interplay of various factors, such as reduced water activity, dielectric constant and also excluded volume effect that depend on chemical properties and molecular weight of the cosolutes (48). In the case of G-quadruplexes, stabilization and monomorphic property of their structures in the presence of the crowding cosolutes are caused likely by dehydration during their formation under the conditions of reduced water activity (22,49). Heddi et al. reported the first and one of the rare insights into high-resolution G-quadruplex structures in the presence of PEG (25). Moreover, these authors concluded that the conformational transition from polymorphic G-quadruplex mixture present under diluted solution to monomorphic parallel topology in the presence of PEG is predominantly caused by water depletion. However, apart from the indirect effects of the crowding cosolutes, also direct binding of PEG to DNA G-quadruplex should be noted. For example, not only the interactions between hydrophilic PEG moieties and DNA sugar-phosphate backbone, but also PEG amphiphilic nature and in this regard its organic-like properties were pointed out in deducing the driving force for PEG-DNA interactions (50). In fact, it is the hydrophobic counterparts that were perceived to note ‘non-inertness’ of PEG with respect to biomolecules (51). With regards to investigating structural properties of G-rich DNA, there are compelling arguments against the use of PEG as a molecular crowding agent due to its direct impact on G-quadruplex stability and topology (26,52). It is likely that there are different views on appropriateness of PEG as an inert crowding cosolute depending on its molecular size. Indeed, the solution properties very much depend on the length of PEG as evident from the water activities in buffers containing 20 wt% EG and PEG 8000 (Supplementary Table S4). Nevertheless, various polymeric ethylene glycol derivatives are still used in order to impose cell-like environment and thereupon perform physicochemical analyses of DNA G-quadruplexes. Notably, however, calculation and comparison of high-resolution structures in the presence of different cosolutes, which may behave as osmolyte and/or putative crowding agent were not reported. In this regard, generalization of PEG-DNA G-quadruplex interactions and the biological relevance of G-rich DNA features in solutions comprising PEGs remain unrewarding because very scarce structural data in the presence of these cosolutes limits in-depth comprehension. Thus, structural details underlying EG- and PEG 8000-M2 G-quadruplex interactions will be useful to constructively discuss the issue and further our understanding. This is especially relevant in light of the recent reports on nucleic acids-ligand interactions in different environments, which unrevealed the discrepancy of PEG-crowding systems and cell extracts, thereby further marking the importance of subtle hetero-molecular, i.e. DNA/RNA-protein interactions, which are generally absent in cell-mimicking systems (53,54).
At the herein used experimental setups M2 G-quadruplex exhibits preserved parallel topology both in the absence and in the presence of cosolutes as demonstrated by complementary spectroscopic methods. Since M2 may form high-order structures in the presence of 20 wt% PEG 8000, which mists and complicates NMR characterization, we analysed detailed structure of the M2 G-quadruplex in the absence of cosolute and in the presence of 20 wt% EG and 10 wt% PEG 8000. Importantly, the overall structure of the M2 G-quadruplex, especially the core G-quartets region, did not show considerable changes in the absence and in the presence of the cosolutes. In the case of loop nucleotides, certain differences were observed such as inward orientation of C11 in the presence of 20 wt% EG and capping-like orientation of A16 with respect to the groove defined by G12–G14 and G17–G19 segments in the presence of 10 wt% PEG 8000 (Supplementary Figures S8 and S9). In contrast, considerable and distinct structural changes were observed for residue T20 that are corroborated by perturbations in 1H NMR chemical shifts in the presence of both 20 wt% EG and 10 wt% PEG 8000 with respect to cosolute-free condition (Figure 8).
In the presence of 20 wt% EG, perturbations in the 1H NMR chemical shifts at the 3′-end region of the M2 G-quadruplex can be partly attributed to the fact that outer G-quartets are typically much more hydrophobic with respect to other G-quadruplex regions. In general, due to the hydrophobicity, the 5′- and 3′-flanking nucleobases in the G-quadruplexes tend to stack on the outer G-quartets. This kind of stacking interaction of flanking nucleobases that contributes to overall stability is also reported in single nucleotide overhanging regions of DNA and RNA duplexes (55,56). In the duplexes, the dangling end is hydrated more than the blunt end and destabilized by osmotic stress caused by a cosolute, which lowers water activity (57). In the herein studied system the addition of 20 wt% EG is accompanied by dehydration of residue T20 furthermore leading to its orientation toward the cation central-channel cavity. Thereby, the electrostatic repulsion between T20 and G5–G10–G14–G19 quartet is neutralized in favor of the stacking interactions. On the other hand, T1 and A2 at the 5′-end keep their conformation due to the stacking interactions impregnable to the osmotic stress. Moreover, the 5′-end adenine (purine) residue interacts with the outer G-quartet more tightly compared to the 3′-end thymine (pyrimidine) residue through stronger stacking interactions. The conformational change of C11 toward the groove in the region between G10 and G14 and closer to G5-G10-G14-G19 quartet is coupled with the dehydration effect of EG. Noteworthy, in the presence of PEG 8000 residue C11 retains its disposition toward the bulk solution.
The UV-melting experiments clearly demonstrate that the EG and PEG 8000 significantly stabilize the M2 G-quadruplex (Supplementary Figure S4 and Supplementary Table S5). These results together with IFIE analysis suggest that the intrinsic structural interaction energy itself does not correlate to the actual stability of the M2 G-quadruplex in the presence of cosolutes. Thus, the stabilization of the M2 G-quadruplex by EG and PEG 8000 is related to extrinsic factors. As mentioned above, EG behaves as osmolyte, which reduces water activity, rather than macromolecular crowding cosolute (Supplementary Table S4). The reduced water activity in the presence of 20 wt% EG is expected to be coupled with conformational changes of C11 and T20, and overall stabilization of the M2 G-quadruplex through intrinsic dehydration during formation of the core G-quartets. In addition, reduced dielectric constant caused by the EG might also contribute to the stabilization, although such effect is expected to be more prominent in the case of PEG 8000.
In the presence of PEG 8000, not only the 1H NMR chemical signals for residue T20, but also aromatic protons of guanines in G5–G10–G14–G19 quartet are significantly de-shielded, which is not the case in the presence of EG. These results suggest resolved stacking interactions between residue T20 and the outer G-quartet in the presence of PEG 8000, and different local environment of the 3′-end residue with respect to the presence of 20 wt% EG (and in the absence of cosolutes). Additionally, as depicted by the calculated structures in the presence of PEG 8000, T20 is oriented toward the groove region and is unstacked from G5–G10–G14–G19 guartet suggesting that the G-quartet platform is thereby available for interactions with the cosolute. Recently, Buscaglia et al. implied preferential binding of the high molecular-weight PEGs to external G-quartets. Additionally, we have also shown that oligoethylene glycols stabilize G-quadruplexes through CH-π and lone pair-π interactions (58). Thus, the structural change of T20 in the presence of PEG 8000 is potentially affected by direct interaction between PEG 8000 and M2 G-quadruplex. In this regard, scrutiny of cosolute-M2 G-quadruplex interactions and in particular the lack of NOE cross-peaks corresponding to close positioning of PEG 8000 to M2 G-quadruplex, i.e. within 5 Å, indicates the absence of long-lived intermolecular interactions. However, these observations do not disprove subsistence of direct DNA binding by PEG 8000. Previous reports estimated the dissociation constant KB of PEG to G-quadruplex forming DNA at molar to sub-molar range (23,26). By considering the molar concentrations of 10 wt% PEG 8000 and DNA at approximately 12 and 0.2 mM, respectively, the lack of NOE cross-peaks for their intermolecular interactions might be interpreted by fast exchange on NMR timescale of the PEG 8000 between bulk solution and DNA-binding platform(s) due to the weak interactions.
Notably, PEG 8000 is not an osmolyte and the water activity in the presence of 20 wt% PEG 8000 is considerably higher than in the presence of 20 wt% EG and is close to cosolute-free conditions (Supplementary Table S4). Thus, it is considered that the water activity cannot solely account for the stabilization of the M2 G-quadruplex. One of the potential factors for stabilization of the M2 G-quadruplex is excluded volume effect provided by PEG 8000. Excluded volume is a volume occupied by a molecule in a system and therefore inaccessible for other molecules. For considering the degree of the excluded volume effect it is important to evaluate hydrodynamic radius of the molecules in the system that defines their hydrodynamic volume in solution (59). Large excluded volume is provided to a solute molecule from co-existing crowding agent when they have similar hydrodynamic volumes (59–61). Large sized PEGs have previously been demonstrated to impose excluded volume effect against similar sized nucleic acids, which facilitates compaction of their structures including stabilization of canonical duplex and tertiary structures (62,63). In this regard, stabilization of the M2 G-quadruplex by PEG 8000 is expected. In addition to imposing the excluded volume, large sized PEGs such as PEG 8000 reduce dielectric constant of the solution more than EG (Supplementary Table S4), which could be also a reason for facilitation of ribozyme activity in the presence of PEG (64). Reduction of the dielectric constant of the solution should impact the G-quadruplex stability mediated by interactions between an ion and G-quadruplex. In fact, it has been shown that decrease in dielectric constant followed by addition of small organic solvents leads to increased stability of G-quadruplexes via favored electrostatic interactions (65). The plots of ΔG°25 versus ln[KCl] (Supplementary Figure S4) showed larger cooperativity and lower concentration of potassium ions required for the folding of M2 G-quadruplex in the presence of 20 wt% PEG 8000 compared to 20 wt% EG. EG also reduces dielectric constant and thus the cooperativity of potassium ion for the G-quadruplex folding in the presence of EG was higher than in the absence of cosolutes.
In conclusion, structural details on the M2 G-quadruplex in the presence and absence of crowding cosolutes reveal that the variances mainly concern T20 residue at the 3′-end. Furthermore, we show that PEG 8000 interacts with M2 G-quadruplex at its 3′-end, which is consistent with the suggested direct interaction between large size PEG and outer G-quartets. However, our complementary approach indicates that the interactions between PEG 8000 and the M2 G-quadruplex are weak and in this regard the stabilization contribution might be related also to the changes in solvation continuum and excluded volume by PEG 8000. On the other hand, the stability of M2 G-quadruplex is increased by EG mainly due to dehydration effect.
DATA AVAILABILITY
Data has been deposited in the Protein Data Bank (https://www.rcsb.org/) and Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu/) under the following accession numbers: PDB 5NYS, BMRB 34135, PDB 5NYT, BMRB 34136, PDB 5NYU, BMRB 34137.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Slovenian Research Agency (ARRS) [P1-0242, J1-6733]; CERIC-ERIC Consortium for the access to experimental facilities and financial support; Japan Society for the Promotion of Science (JSPS) and Slovenian Research Agency [BI-JP/15-17-002] under JSPS Bilateral Joint Research Program; Grant- in-Aid for Scientific Research on Innovative Areas ‘Chemistry for Multimolecular Crowding Biosystems’ (JSPS KAKENHI) [JP17H06351], for Scientific Research, Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; MEXT-Supported Program for the Strategic Research Foundation at Private Universities (2014–2019), Japan; Hirao Taro; Okazaki Kazuo Foundation of KONAN GAKUEN for Advanced Scientific Research; Chubei Itoh Foundation. Funding for open access charge: Slovenian Research Agency (ARRS) [P1-0242, J1-6733].
Conflict of interest statement. None declared.
REFERENCES
- 1. Paeschke K., McDonald K.R., Zakian V.A.. Telomeres: structures in need of unwinding. Febs Lett. 2010; 584:3760–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodrigez R., Miller K.M., Forment J.V., Bradshaw C.R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., Jackson S.P.. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012; 8:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simone R., Fratta P., Neidle S., Parkinson G.N., Isaacs A.M.. G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015; 589:1653–1668. [DOI] [PubMed] [Google Scholar]
- 4. Malgowska M., Czajczynska K., Gudanis D., Tworak A., Gdaniec Z.. Overview of the RNA G-quadruplex structures. Acta. Biochim. Pol. 2016; 63:609–621. [DOI] [PubMed] [Google Scholar]
- 5. David A.P., Margarit E., Domizi P., Banchio C., Armas P., Calcaterra N.B.. G-quadruplexes as novel cis-elements controlling transcription during embryonic development. Nucleic Acids Res. 2016; 44:4163–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng X.H., Nie X., Fang Y., Zhang Z., Xiao Y., Mao Z., Liu H., Ren J., Wang F., Xia L. et al. A cisplatin derivative tetra-Pt(bpy) as an oncotherapeutic agent for targeting ALT cancer. J. Natl. Cancer Inst. 2017; 109, doi:10.1093/jnci/djx061. [DOI] [PubMed] [Google Scholar]
- 7. Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. 2006; Cambridge: The Royal Society of Chemistry. [Google Scholar]
- 8. Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chem. Eur. J. 2007; 13:9738–9745. [DOI] [PubMed] [Google Scholar]
- 9. Parkinson G.N., Lee M.P.H., Neidle S.. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002; 417:876–880. [DOI] [PubMed] [Google Scholar]
- 10. Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J.. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:3745–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y., Patel D.J.. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993; 1:263–282. [DOI] [PubMed] [Google Scholar]
- 12. Patel D.J., Phan A.T., Kuryavyi V.. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007; 35:7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yue D.J., Lim K.W., Phan A.T.. Formation of (3+1) G-quadruplexes with a long loop by human telomeric DNA spanning five or more repeats. J. Am. Chem. Soc. 2011; 133:11462–11465. [DOI] [PubMed] [Google Scholar]
- 14. Ceru S., Sket P., Prislan I., Lah J., Plavec J.. A new pathway of DNA G-Quadruplex formation. Angew. Chem. Int. Ed. 2014; 53:4881–4884. [DOI] [PubMed] [Google Scholar]
- 15. Marusic M., Sket P., Bauer L., Viglasky V., Plavec J.. Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops. Nucleic Acids Res. 2012; 40:6946–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crnugelj M., Sket P., Plavec J.. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003; 125:7866–7871. [DOI] [PubMed] [Google Scholar]
- 17. Marusic M., Veedu R.N., Wengel J., Plavec J.. G-rich VEGF aptamer with locked and unlocked nucleic acid modifications exhibits a unique G-quadruplex fold. Nucleic Acids Res. 2013; 41:9524–9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mukundan V.T., Phan A.T.. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013; 135:5017–5028. [DOI] [PubMed] [Google Scholar]
- 19. Sket P., Crnugelj M., Plavec J.. Identification of mixed di-cation forms of G-quadruplex in solution. Nucleic Acids Res. 2005; 33:3691–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trajkovski M., da Silva M.W., Plavec J.. Unique structural features of interconverting monomeric and dimeric G-Quadruplexes adopted by a sequence from the intron of the N-myc gene. J. Am. Chem. Soc. 2012; 134:4132–4141. [DOI] [PubMed] [Google Scholar]
- 21. Miyoshi D., Sugimoto N.. Molecular crowding effects on structure and stability of DNA. Biochimie. 2008; 90:1040–1051. [DOI] [PubMed] [Google Scholar]
- 22. Miyoshi D., Karimata H., Sugimoto N.. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006; 128:7957–7963. [DOI] [PubMed] [Google Scholar]
- 23. Miyoshi D., Nakao A., Sugimoto N.. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry. 2002; 41:15017–15024. [DOI] [PubMed] [Google Scholar]
- 24. Xue Y., Kan Z.Y., Wang Q., Yao Y., Liu J., Hao Y.H., Tan Z.. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007; 129:11185–11191. [DOI] [PubMed] [Google Scholar]
- 25. Heddi B., Phan A.T.. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011; 133:9824–9833. [DOI] [PubMed] [Google Scholar]
- 26. Buscaglia R., Miller M.C., Dean W.L., Gray R.D., Lane A.N., Trent J.O., Chaires J.B.. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013; 41:7934–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fujii T., Podbevsek P., Plavec J., Sugimoto N.. Effects of metal ions and cosolutes on G-quadruplex topology. J. Inorg. Biochem. 2017; 166:190–198. [DOI] [PubMed] [Google Scholar]
- 28. Goddard T.D., Kneller D.G.. SPARKY 3. 2008; San Francisco: University of California. [Google Scholar]
- 29. Perez A., Marchan I., Svozil D., Sponer J., Cheatham T.E. III, Laughton C.A., Orozco M.. Refinenement of the AMBER force field for nucleic acids: Improving the description of alpha/gamma conformers. Biophys. J. 2007; 92:3817–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Case D.A., Babin V., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., Cheatham T.E. III, Darden T.A., Duke R.E., Gohlke H. et al. AMBER 14. 2014; San Francisco: University of California. [Google Scholar]
- 31. Krepl M., Zgarbova M., Stadlbauer P., Otyepka M., Banas P., Koca J., Cheatham T.E. III, Jurecka P., Sponer J.. Reference simulations of noncanonical nucleic acids with different chi variants of the AMBER force field: quadruplex DNA, quadruplex RNA and Z-DNA. J. Chem. Theory Comput. 2012; 8:2506–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zgarbova M., Luque F.J., Sponer J., Cheatham T.E. III, Otyepka M., Jurecka P.. Toward improved description of DNA Backbone: Revisiting Epsilon and Zeta torsion force field parameters. J. Chem. Theory Comput. 2013; 9:2339–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 34. Marky L.A., Breslauer K.J.. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987; 26:1601–1620. [DOI] [PubMed] [Google Scholar]
- 35. Tateishi-Karimata H., Nakano S., Sugimoto N.. Quantitative analyses of nucleic Acid stability under the molecular crowding condition induced by cosolutes. Curr. Protoc. Nucleic Acid Chem. 2013; 53:7.19.1–7.19.17. [DOI] [PubMed] [Google Scholar]
- 36. Miyoshi D., Nakamura K., Tateishi-Karimata H., Ohmichi T., Sugimoto N.. Hydration of Watson-Crick base pairs and dehydration of Hoogsteen base pairs inducing structural polymorphism under molecular crowding conditions. J. Am. Chem. Soc. 2009; 131:3522–3531. [DOI] [PubMed] [Google Scholar]
- 37. Lambert D., Draper D.E.. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ interactions. J. Mol. Biol. 2007; 370:993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lambert D., Leipply D., Draper D.E.. The osmolyte TMAO stabilizes native RNA tertiary structures in the absence of Mg2+: evidence for a large barrier to folding from phosphate dehydration. J. Mol. Biol. 2010; 404:138–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mullen M.A., Assmann S.M., Bevilacqua P.C.. Toward a digital gene response: RNA G-quadruplexes with fewer quartets fold with higher cooperativity. J. Am. Chem. Soc. 2012; 134:812–815. [DOI] [PubMed] [Google Scholar]
- 40. Risitano A., Fox K.R.. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004; 32:2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tong X., Lan W., Zhang X., Wu H., Liu M., Cao C.. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011; 39:6753–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai J.X., Chen D., Jones R.A., Hurley L.H., Yang D.Z.. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006; 34:5133–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuryavyi V., Phan A.T., Patel D.J.. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010; 38:6757–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Phan A.T., Kuryavyi V., Burge S., Neidle S., Patel D.J.. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007; 129:4386–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kitaura K., Ikeo E., Asada T., Nakano T., Uebayasi M.. Fragment molecular orbital method: an approximate computational method for large molecules. Chem. Phys. Lett. 1999; 313:701–706. [Google Scholar]
- 46. Pruitt S.R., Nakata H., Nagata T., Mayes M., Alexeev Y., Fletcher G., Fedorov D.G., Kitaura K., Gordon M.S.. Importance of Three-Body interactions in molecular dynamics simulations of water demonstrated with the fragment molecular orbital method. J. Chem. Theory Comput. 2016; 12:1423–1435. [DOI] [PubMed] [Google Scholar]
- 47. MIZUHO/BioStation Viewer, version 3.0 program. 2013; Tokyo: Mizuho Information and Research Institute Inc. [Google Scholar]
- 48. Nakano S.I., Sugimoto N.. The structural stability and catalytic activity of DNA and RNA oligonucleotides in the presence of organic solvents. Biophys. Rev. 2016; 8:11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang D.H., Fujimoto T., Saxena S., Yu H.Q., Miyoshi D., Sugimoto N.. Monomorphic RNA G-quadruplex and polymorphic DNA G-quadruplex structures responding to cellular environmental factors. Biochemistry. 2010; 49:4554–4563. [DOI] [PubMed] [Google Scholar]
- 50. Adali T., Bentaleb A., Elmarzugi N., Hamza A.M.. PEG-calf thymus DNA interactions: conformational, morphological and spectroscopic thermal studies. Int. J. Biol. Macromol. 2013; 61:373–378. [DOI] [PubMed] [Google Scholar]
- 51. Zhou H.X., Rivas G., Minton A.P.. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008; 37:375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hansel R., Lohr F., Foldynova-Trantirkova S., Bamberg E., Trantirek L., Dotsch V.. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011; 39:5768–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tyrrell J., Weeks K.M., Pielak G.J.. Challenge of mimicking the influences of the cellular environment on RNA structure by PEG-induced macromolecular crowding. Biochemistry. 2015; 54:6447–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang S.K., Su H.F., Gu Y.C., Lin S.L., Tan J.H., Huang Z.S., Ou T.M.. Complicated behavior of G-quadruplexes and evaluating G-quadruplexes' ligands in various systems mimicking cellular circumstance. Biochem. Biophys. Rep. 2016; 5:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Freier S.M., Sugimoto N., Sinclair A., Alkema D., Neilson T., Kierzek R., Caruthers M.H., Turner D.H.. Stability of XGCGCp, GCGCYp, and XGCGCYp helixes: an empirical estimate of the energetics of hydrogen bonds in nucleic acids. Biochemistry. 1986; 25:3214–3219. [DOI] [PubMed] [Google Scholar]
- 56. Sugimoto N., Kierzek R., Turner D.H.. Sequence dependence for the energetics of terminal mismatches in ribooligonucleotides. Biochemistry. 1987; 26:4559–4562. [DOI] [PubMed] [Google Scholar]
- 57. Tateishi-Karimata H., Pramanik S., Nakano S., Miyoshi D., Sugimoto N.. Dangling ends perturb the stability of RNA duplexes responsive to surrounding conditions. Chemmedchem. 2014; 9:2150–2155. [DOI] [PubMed] [Google Scholar]
- 58. Tateishi-Karimata H., Ohyama T., Muraoka T., Podbevsek P., Wawro A.M., Tanaka S., Nakano S.I., Kinbara K., Plavec J., Sugimoto N.. Newly characterized interaction stabilizes DNA structure: oligoethylene glycols stabilize G-quadruplexes CH-pi interactions. Nucleic Acids Res. 2017; 45:7021–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kuznetsova I.M., Zaslavsky B.Y., Breydo L., Turoverov K.K., Uversky V.N.. Beyond the excluded volume effects: mechanistic complexity of the crowded milieu. Molecules. 2015; 20:1377–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Minton A.P. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001; 276:10577–10580. [DOI] [PubMed] [Google Scholar]
- 61. Hall D., Minton A.P.. Macromolecular crowding: qualitative and semiquantitative successes, quantitative challenges. Biochim. Biophys. Acta. 2003; 1649:127–139. [DOI] [PubMed] [Google Scholar]
- 62. Knowles D.B., LaCroix A.S., Deines N.F., Shkel I., Record M.T. Jr. Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12699–12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kilburn D., Roh J.H., Guo L., Briber R.M., Woodson S.A.. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J. Am. Chem. Soc. 2010; 132:8690–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nakano S., Kitagawa Y., Miyoshi D., Sugimoto N.. Hammerhead ribozyme activity and oligonucleotide duplex stability in mixed solutions of water and organic compounds. FEBS Open Bio. 2014; 4:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smirnov I.V., Shafer R.H.. Electrostatics dominate quadruplex stability. Biopolymers. 2007; 85:91–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data has been deposited in the Protein Data Bank (https://www.rcsb.org/) and Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu/) under the following accession numbers: PDB 5NYS, BMRB 34135, PDB 5NYT, BMRB 34136, PDB 5NYU, BMRB 34137.