Abstract
The speed at which new genomes are being sequenced highlights the need for genome-wide methods capable of predicting protein–DNA interactions. Here, we present PADA1, a generic algorithm that accurately models structural complexes and predicts the DNA-binding regions of resolved protein structures. PADA1 relies on a library of protein and double-stranded DNA fragment pairs obtained from a training set of 2103 DNA–protein complexes. It includes a fast statistical force field computed from atom-atom distances, to evaluate and filter the 3D docking models. Using published benchmark validation sets and 212 DNA–protein structures published after 2016 we predicted the DNA-binding regions with an RMSD of <1.8 Å per residue in >95% of the cases. We show that the quality of the docked templates is compatible with FoldX protein design tool suite to identify the crystallized DNA molecule sequence as the most energetically favorable in 80% of the cases. We highlighted the biological potential of PADA1 by reconstituting DNA and protein conformational changes upon protein mutagenesis of a meganuclease and its variants, and by predicting DNA-binding regions and nucleotide sequences in proteins crystallized without DNA. These results opens up new perspectives for the engineering of DNA–protein interfaces.
INTRODUCTION
It is estimated that around 6% of the eukaryotic genome encodes for DNA-binding proteins (1,2). These proteins, which form DNA–protein interactions (DPIs) through different types of protein domains and domain architectures, are involved in numerous processes including DNA replication, DNA repair, gene regulation, recombination, DNA packing, etc. Currently, although there are >120 000 structures deposited in the PDB (3), only ∼5000 of them involve DNA–protein complexes. When considering the rate at which new genomes are being sequenced, the resolution of novel DNA–protein structures is relatively scarce. As such, we need to develop methods not only capable of predicting whether a protein can interact with DNA, but also capable of determining the protein's 3D binding region, and the DNA sequence to which it can bind (4). Several sequence-based (direct read-out) methods have already been developed for predicting whether a protein can bind to DNA, and by means of sequence homology, also determine which residues are involved in the interaction (5–9). However, although these methods are useful in many specific cases, they lack 3D information, such as atomic distances and dihedral restraints, regarding the interactions. As a consequence, these methods are unable to predict the DNA sequence that is recognized by a protein, the effect of mutations found in sequenced genomes and are not suitable for rational protein design, or to interpret the effect of these mutations. Structural-based (indirect read-out) methods on the other hand, have the potential to address these issues. In this case, both the 3D structure of a target protein (or a good homology model) and an algorithm that can dock DNA structures, identify the best docking, and search for the best DNA recognition sequence are required.
The development of a general method to blindly predict double-stranded DNA–protein (dsDP) binding sites still poses an important challenge (10). Historically, docking models have been designed in numerous ways, each with their own characteristics and limitations. For instance, approaches based on molecular dynamics (11,12) require long computations to simulate a small fraction of time, which in many cases is not enough to achieve the equilibrium. Other methods such as Monte-Carlo sampling (13–15) are based on non-observed structural configurations, and rigid solid approaches (16) are unable to deal with small, binding-induced backbone conformational changes. Furthermore, alternative methods are often biased towards specific families of proteins, and thereby fail to predict docking for proteins outside these targeted families (17–21). While some accurate methods use canonical DNA templates (22), they require both protein and DNA structures as input (23). Scoring functions for the previously cited methods use empirical knowledge-based or physical force fields. Other strategies use ab-initio methods (24), which although very accurate, can require huge computational resources or time. Some of the scoring functions include different energy terms, such as base coplanarity and electrostatics (25) as the main force driving binding, while others include H-bonding and Lennard–Jones (26) (to check for clashing elements) potential. These methods are computationally expensive since they calculate all pairwise atomic interactions, and therefore cannot be used with large protein datasets. Some of these cited methods were developed before 2006, and as such do not take into account the current completeness of the PDB. This makes it hard to measure their accuracy because deposited dsDP complexes were about a quarter of the current number (Supplementary Figure S5).
An alternative possibility, which is exploited by protein design algorithms like Rosetta (27) and FoldX (28), uses libraries made of protein fragments (29) and the spatial relationships (interactions) between these fragments (30) to model protein–protein and protein–peptide interactions. This strategy relies on the following assumptions: (i) the conformational space of single protein structures can be described by combining recurrent structural patterns (31); (ii) protein interactions can be captured in motifs of repetitive patterns (32–35); (iii) there are enough structures to cover the largest part of the possible conformational space of protein–protein interactions and (iv) exhaustive fragment libraries can be used to generate conformational backbone ensembles for predicting protein folding and/or protein complexes.
Here, based on the above assumptions, we have developed the protein-assisted DNA assembly version 1 (PADA1) algorithm to predict and model the binding of double-stranded DNA (dsDNA) to proteins. PADA1 includes an empirical interaction model generator in combination with an ultra-fast statistical knowledge-based force field, which together perform dsDP docking. This algorithm uses fragment pairs (peptide or pepX paired to short dsDNA or dnaX) that represent empirical, compatible backbone conformations found in nature. Our interaction database contains ∼18 million (17 920 450) atomic coordinates of pepX and dnaX pairs (intX), and is ready to be trained with more interactions as dsDP complexes continue to be deposited in the PDB. For a validation set of 212 structures deposited after 2016, our algorithm was able to predict the DNA-binding region with an RMSD per residue of <1.8 Å in 209 cases (>95%, see Validation section). DNA–protein structures modeled by PADA1 can be used in combination with protein design software like FoldX to predict DNA recognition sequences. This cooperativity between PADA1, used to predict backbone compatibility, and FoldX, for sidechain refinement and interface optimization, turns ModelX in a powerful modelling toolsuite with potential further applications in other kind of interactions like protein–RNA, protein–protein and protein–drug interactions. Using different examples, we not only highlight the potential biological application of PADA1, but also demonstrate how it can be used to discover DNA-binding regions, dock dsDNA molecules, generate conformational diversity, and in combination with protein design force fields, identify DNA recognition sequences.
MATERIALS AND METHODS
Algorithm and database remarks
PADA1 is a command-line tool included in a more complex object-oriented application named ModelX. It is written in C++ and stores all data in a relational MySQL database using the InnoDB engine. It is compiled with only C11++ support for the three main platforms (Linux-64bit, MacOS-64bit, Windows-32bit) and Raspberry PI. It uses Mysqlpp (https://tangentsoft.net/mysql++) as database connector and Boost (http://www.boost.org) for eventual standardization. The software is a portable executable with the only dependency of the MySQL database, and can be downloaded freely for academic users from http://modelx.crg.es. The application has a standard C-type layer of parameters that are set with default values to make things easier for the users. Parallelization is easy by overwriting these default parameters. The executable can be interactively queried for help about the program arguments and command mode. The algorithm has been developed using design patterns, inheritance, a data access object layer (DAO), and a complex class hierarchy that allows it, as ModelXDB, to be easily extended for other biomolecules such as RNA, ssDNA or small drugs. The database is optimized for speed, with keys and indexes for the fields that will be used for querying from inside the code. The database was digested for several combinations (intX) of pepX-dnaX fragment lengths (i.e. peptide fragments of 6–12 amino acids in length and dsDNA fragments of 4–8 bp in length) Moreover, it can be trained with new interacting structural motifs as they are deposited in the PDB, thereby increasing its prediction capabilities. In fact, the release of dsDP structures in the PDB has followed an exponential trend since the first structure was deposited in 1986 (36). We thus expect to further improve the prediction capabilities of our algorithm in the upcoming years by releasing newer and more complete versions of the database. The Mysqlpp connector allows data retrieval from either local or remote databases. The commands for building fragment libraries are hidden from the user. A FoldX force field connector for rapid evaluation within the ModelX toolsuite was developed in-house allowing to accurately compute free energies over structures taking sidechain atoms into account in docking refinement steps.
The PADA1 fragment retrieval and superimposition methods
The ModelXDB stores the CαNter-CαCter distance for every pepX fragment, and uses it to retrieve pepX fragment lengths within a given distance uncertainty (dubiety parameter, default = 0.1 Å). The PADA1 algorithm then scans along an input PDB protein, superimposing different pepX fragment lengths as anchor segments to retrieve compatible 3D interactions with dnaX. These pepX fragments have slightly different backbone conformations that mimic flexibility upon binding, and are used to place the corresponding interacting DNA fragment onto the input protein. In this manner, we are able to generate dsDNA clouds containing compatible interacting models. These clouds can be used to explore backbone flexibility upon binding and sidechain refinement using FoldX.
The superimposition method (Supplementary Figure S2) is used to scan an input protein with compatible peptide fragments. Using an overlapping sliding window on the input protein, we select (by default) peptides of six amino acids in length (pepx-length parameter). Scan peptide geometry is used to retrieve pepXs with similar CαNter-CαCter distances. For the scanned peptide, we convert CαNter and CαCter into a vector (Scan P) and by means of one translation and two rotations we reference it to the x-axis of a Cartesian coordinate system. The timeline for the required referencing movements is stored inside the peptide object. After retrieval of pepXs, using the same strategy the database pepX is referenced and rotated through the x-axis (Supplementary Figure S2a) in order to find the best overlap, also applying these movements to the dnaX that is stored with it in the interaction database. The timeline for the scanned pepX is also stored. We consider two peptides to be overlapping when the angles between CαNter–CβNter from the scan peptide and CαNter–CβNter from the pepX are smaller than 5° (default cb-angle parameter). The atoms consecutive to CαNter and CαCter in the pepX peptide are not allowed to deviate more than 0.5 Å (default fit-threshold parameter) from the scan peptide. Thus, for a fit level of 2 (default fit-level parameter), the best overlap is obtained considering the minimum distance between these atoms and their first neighbors. A fit level of 3, on the other hand, also takes into account the corresponding second neighbor distances, and so on for longer peptides (Supplementary Figure S2B). The distance distributions (Supplementary Figure S1) that give rise to the force field were computed for the backbone atomic pairs using only dsDNA fragments and dsDNA–dsRNA hybrids. As the differences among the means and the standard deviations were in the order of the mili-Armstrong for both cases, we decided to include the hybrid information when building the force field.
Dock positioning and optimization strategy
Once a good fitting peptide is found, we apply the timeline (movements) of the scanned pepX in reverse, to both the referenced database pepX and its corresponding dnaX in order to place them on the input protein. Afterwards, we delete the database pepX and evaluate the docking model using the developed force field. Also, is possible to substitute the WT pepX by the scanned pepX in order to generate flexibility on the DNA-binding protein. In order to increase the speed of this method, we first evaluate the energy of the atoms that are closer than 18 Å to the scanned pepX, and if this energy is favorable, we then evaluate it again for the full neighbourhood to accept or reject the docked fragments.
Statistical knowledge based force field
The energetic evaluation in the PADA1 force field is performed using the equation ΔG = –RTLn(Kp), where R is the Boltzmann constant and Kp is Pr/Po. These parameters are calculated with a statistical test. Pr is the probability of finding a pair of atomic contacts for a given amino acid, a correction based on the deviation from the extracted normal distribution for that pair of atoms is included. Po is the probability of finding a pair of atomic contacts for any amino acid, also a correction based on the deviation from the extracted normal distribution for that pair is included. In the case of glycine, which has dihedral angles compatible with other amino acids, a dummy Cβ was placed averaging the Cβ coordinates of two alanines included in the dihedral database: one with the smaller difference from φ, and the other with the smaller difference from ψ.
Sequence profiling pipeline
To test the ability of our method at predicting the binding nucleotide sequence, we developed a validation pipeline consisting of the following steps: (i) DNA molecules are removed from the target crystal structure; (ii) the FoldX software processes each unbounded structure in order to relax the sidechains and mimic an ‘apo’ configuration; (iii) PADA1 is used to dock dsDNA over the apo protein structure and (iv) FoldX is used again to repair the interface of the proposed DNA docks and ‘apo’ structures, and to mutate each DNA base pair to all four nucleotides to find the more energetically favorable sequence. All residues contacting the DNA are compared with the original crystallized version. To evaluate the accuracy of the predictions, only those nucleotides contacting the protein target are taken into account.
DNA–protein interface flexibility
For the meganuclease analysis discussed in the results section, the following strategy was applied: i) after removing all deposited structures of the protein from the database, we performed a docking over the constructed models with PADA1, relaxing the dubiety (0.5 Å) and cb-angle (8°) parameters to give flexibility to the binding site; ii) FoldX was used (DNAScan command) to measure the binding energy differences between both the WT and the constructed proteins in combination with the WT DNA, the crystallized XPC DNA and an XPC-built dock made by visually merging those dsDNA docked fragments with best overlapping. The resulting ΔΔG values reproduce the experimental relative affinity observations (see results and Figure 6D).
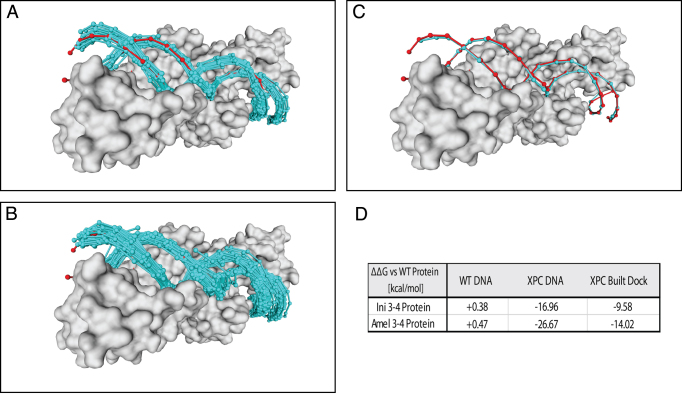
Figure 6.
I-Crel Amel3-4 (PDB: 4aqu) engineered protein: (A) DNA Docked molecules obtained using PADA1 default parameters in blue, XPC DNA in red. (B) Dockings obtained with relaxed parameters (dubiety = 0.5, cb-angle = 8°). (C) The merged DNA fragment using RMSD criteria against XPC DNA superimposed on the crystallographic DNA. (D) Energy variation for both Ini3-4 and Amel3-4 engineered proteins against the different DNAs. The built models show the same specificity tendencies experimentally reported for crystallographic DNAs (WT and XPC), and the PADA1 built dock shows that the full in-silico analysis reproduces the experimental affinity tendencies studied.
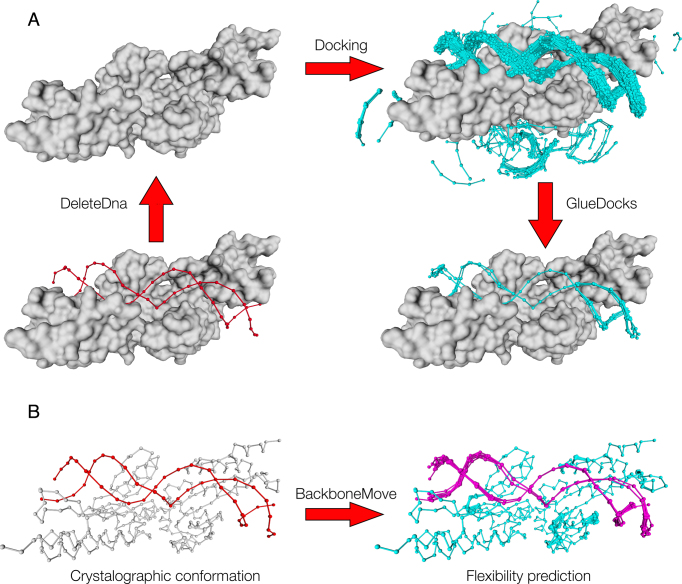
To generate backbone conformational variability on the DNA, we developed a branch and bound algorithm (GlueDocks command, Supplementary Figure S6) that automatically glues the compatible fragments of a cloud returned by a docking run resulting in an ensemble of extended fragments. The algorithm combines in a new extended fragment, fragments having overlapping residues below 0.4 RMSD threshold (rmsd-threshold parameter). The new fragment is recursively combined with the remaining fragments until reaching the maximum longitude for each possible extension. The fragments are connected through O3′–P5′ atoms and the bound distances of the recently formed phosphodiester bond are checked, if the bond distance violates the maximum O3′–P5′ distance found in our database we perform a phosphate repair. Phosphate repair is done by exchanging fragments (C3′–O3′)NUC1–(PO2–O5′)NUC2 (PhoX) from database dinucleotides having the same C3′–O5′ distances than the one containing the wrong distances for the O3′–P5′ bond. The strategy to graft the PhoX is the same that the one used to replace the peptides for the BackboneMove command: the wrong PhoX is reference to the x-axis by means of one translation and two rotations and so the database-fragments, the lasts are rotated around the axis till the best overlap (smallest RMSD) beetwen the firsts nucleotides of the dinucleotide is found, then we apply the timeline from the wrong bonded dinucleotide to the database PhoX fragment in order to place it back in the glued structure. This allows us to analyze the flexibility of the docked fragments and the maximal longitude of the dsDNA docked strands (Figure 7A). Once the dsDNA strands are computed, protein backbone flexibility upon binding can be studied using the command BackboneMove. The command first evaluates the protein regions with high free energy residues and replace them with geometrically compatible pepX fragments using the same strategy described in the fragment retrieval section above (allowing more or less pep-mismatches with its computational performance impact). Replacement is carried out from higher to lower protein energy regions in a step-descendant manner.
Figure 7.
Modeling of flexibility upon binding. (A) DNA flexibility prediction: We first remove the Crystallographic DNA, then we do DNA Docking and select the fragments that will be used for reconstructing the DNA molecule. Once the fragments are selected we join those that are compatible using (GlueDocks; see Materials and Methods). As can be seen the cleavage site is quite rigid, while the DNA backbone becomes more flexible farther away from it. This flexibility could be used to design protein mutants that will recognize other DNA sequences. (B) Protein flexibility prediction: the BackboneMove command can be used to model protein backbone variability over the high free energy regions generated upon DNA flexibility prediction.
RESULTS
ModelXDB: Database genesis
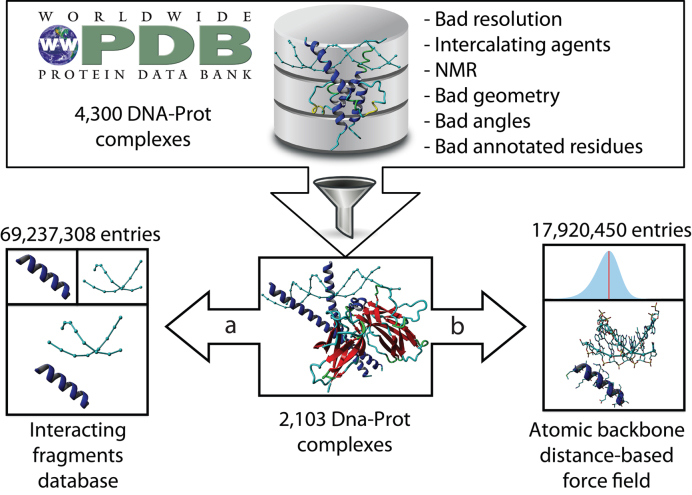
To generate the ModelXDB database (Figure 1), we started with 4300 DNA–protein X-ray complexes extracted from the PDB. We filtered out any complexes which had low resolution (worse than 3.4 Å), intercalating agents, structural defects, duplicated atoms, and/or only single-stranded DNA. After filtering, we ended up with 2103 high quality structures that were in silico digested to generate the ModelXDB in four steps: (i) all atoms and their coordinates were included in the database for further modeling purposes; (ii) dsDNA fragments were broken into smaller fragments of different lengths (4–8 bases, dnaXs), their coordinates stored in the database, and nucleotide hybridization partners (pair bases) found using the software x3DNA (37); (iii) for every dsDNA fragment, we mined all the contacting protein residues and retained all pepXs of 6–12 amino acids and (iv) all combinations of interacting pepX and dnaX fragments (in which at least one of the atoms was closer than the threshold distance of 4.5 Å to another atom of the second molecule) were stored in the database and linked through their database identifiers. After these four steps, we ended up with a database of peptides and short dsDNA fragments containing ∼70 million (69 237 308) spatial relationships that could be used for structural interaction modeling. We did not filter the training set by sequence similarity because redundant sequences can possess different configurations that can posteriorly be used to generate conformational flexibility. The database was optimized and indexed for high-throughput querying.
Figure 1.
Database and force field genesis: (A) Digestion of complexes into peptide–dsDNA (pepX–dnaX) fragment pairs as database records (intXs); Poor quality structures (ie NMR, bad resolution) are filtered in this step. (B) Atomic distance measurement and force field generation.
The PADA1 fragment retrieval
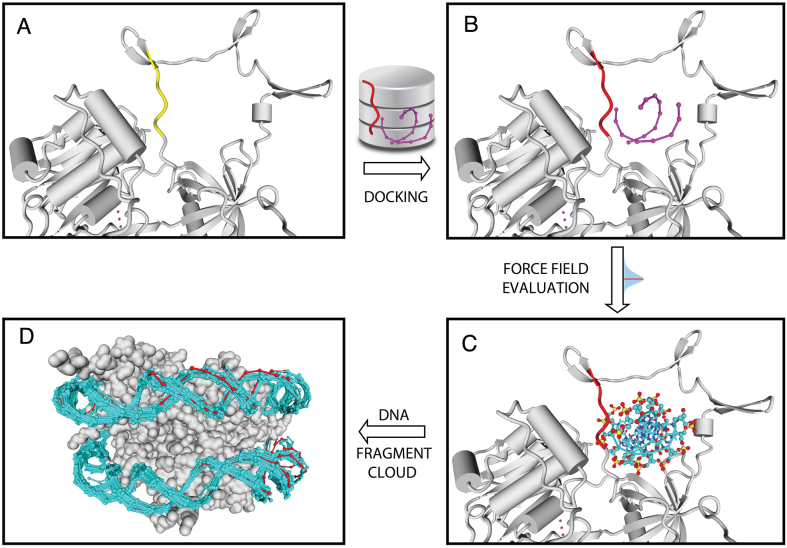
The ModelXDB stores CαNter–CαCter distances for every pepX fragment. This distance is used to retrieve pepX fragment lengths with a given distance uncertainty (dubiety parameter). The PADA1 algorithm scans along a PDB input protein and superimposes different pepX fragment lengths as anchor segments to retrieve compatible 3D interactions with dnaXs (see Materials and Methods). These pepX fragments have slightly different backbone conformations that can mimic flexibility upon binding, and are used to place the corresponding interacting DNA fragment onto the input protein (Figure 2A and B). In this manner, we are able to generate dsDNA clouds containing compatible interacting models (Figure 2D). These clouds can be used, as well, for sidechain refinement using FoldX (see below).
Figure 2.
Docking procedure: (A) a protein fragment (yellow) is used to query the pepX database for a compatible fragment; (B) the retrieved pepX fragment (red) is superimposed on the yellow one placing the associated DNA fragment (dnaX, purple); (C) backbone dnaX atoms are evaluated with the PADA1 force field; (D) an example with a histone octamer showing all dnaX docked models (cyan) fully covering the crystallographic DNA (red).
Statistical knowledge based force field and filtering of spurious docking
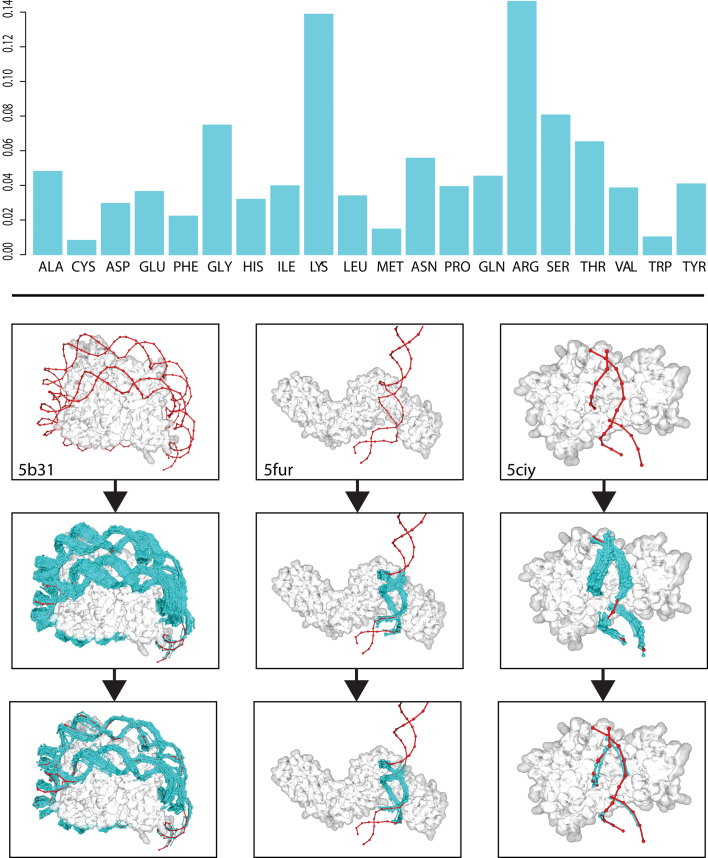
To distinguish between true binders and false positives, it is necessary to define a scoring function, or force field. For this purpose, we considered only those structures of the interacting fragment database which had a resolution equal to or lower than 2.5 Å (1295 structures). For every pepX–dnaX pair (intX), we measured the atomic all-to-all distances between the pepX protein fragment and the corresponding dnaX fragment. Then, using the atomic distance distributions (Supplementary Figure S1), we extracted the statistical parameters (mean and standard deviation of the distances) for all possible contacts between the protein and dsDNA fragments included in the interaction database. All-to-all distances between contacting nucleotide-amino acid pairs were measured (a contact is considered when at least one atom of the amino acid, including sidechains, is 4 Å or less from any atom in the nucleotide). Statistics were computed in two ways: (i) by not considering the nucleotide identity (i.e. unbiased force field); and (ii) by considering the identity of the nucleotide to which a DNA atom belongs (i.e., nucleotide-based force field) We used the unbiased force field for removing DNA fragments from the cloud, and the nucleotide-based one to search for the DNA sequence that is recognized by the target protein. Using a Boltzmann device (38) with the Kono (39) modification of the Sippl method, we calculated the force field free energies for every pair of amino acid and DNA base. This was computed using the protein (N, Cα, Cβ, C, O) and DNA (P, OP1, OP2, O5′, C5′, C4′, O4′, C3′, O3′, C2′, C1′) backbone atoms for residues that were closer than 4 Å. For both force fields the identity of the amino acid is considered on the probabilistic terms of the scoring function (see Materials and Methods). The global energy of a dsDNA–protein interaction was calculated by adding together all the atom-atom partial free energies. As expected, the propensity of a nucleotide to bind an amino acid is higher for lysine and arginine (40) (Figure 3, upper panel).
Figure 3.
(Upper) DNA-amino acid binding propensities for all residues against any given nucleotide on the ModelXDB. (Lower) Examples (PDBs: 5b31, 5fur, 5ciy, top panel) of PADA1 predictions: fragment clouds (cyan) are filtered using the PADA1 force field going from a disperse cloud (medium panel) to a refined cluster (bottom panel) containing the most energetically favorable docks. Crystallographic DNA in red.
Rather than evaluating all atoms in the input structure, only protein backbone atoms and Cβs that are up to 18 Å (the default pep-threshold parameter value) away from the scanning peptide were considered in a first round. Then a second round is performed with the selected fragments in which all the backbone atoms of the input protein are included. In both rounds, docking models are accepted if their energy values are lower than those established by the energy-threshold parameter (default −0.1 kcal/mol per nucleotide). The force field energy is used to reduce the conformational diversity and approach the crystallographic conformation by choosing the most favorable result for every scanned peptide′s cluster of solutions (clusters have <1.5 Å RMSD per residue). Since the forcefield propensities can be loaded into memory the statistical evaluation can be performed for large sets of docks in an ultra rapid manner. By combining binding energy with the number of contacting nucleotides, spurious dockings can be removed (Figure 3, and Supplementary Figures S4 and S5). Although our database includes all combinations of dnaX fragment lengths (4–8 amino acids) paired to pepX fragments lengths (6 to 12 bases), we only used those pepX fragments of 6 amino acids with dnaX fragments of 4–6 bases. Once spurious docks are filtered a knowledge based forcefield (like FoldX, integrated within ModelX to PADA1, or others like Rosseta) could be used in order to refine the dsDP interfaces at sidechine level.
Validation
We used three different datasets for validation purposes: (i) 212 dsDP complexes released after 2016 and not included in our database (Supplementary Table S2); (ii) a standard benchmark validation set (41) of 47 proteins crystallized with and without DNA (Supplementary Table S3) and (iii) all available validation sets (42–46) mentioned in a recent review (10) as a standard for benchmarking DNA docking algorithms. We also include a small set of negative controls, containing for example, YFP (PDB: 1kyp), a FAB region of an antibody (PDB: 1bog), and serum albumin (PDB: 1n5u). For all validations, we excluded the PDBs which belonged to the evaluated protein.
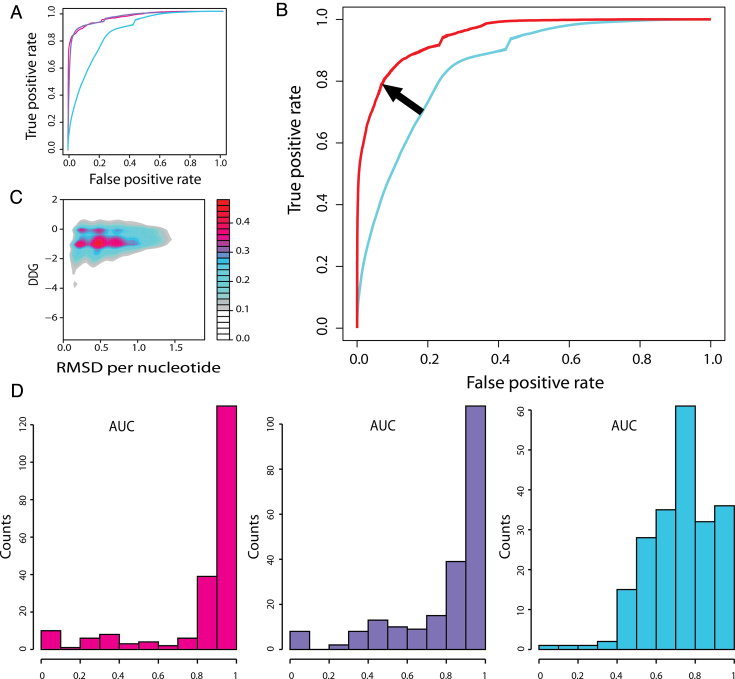
Given that the computational cost of allowing six mismatches for the scan with peptides of length six (exhaustive mode) is very expensive, we explored the validity of our predictions allowing for only 1–3 mismatches (pep-mismatches parameter, see Methods). We used the crystallographic dsDNA of the validation sets to calculate the RMSD of the predicted docking fragments for each nucleotide using the following atoms: C1′, C2′, C3′, O3′, C4′, O4′, C5′, O5′, P, OP1, OP2. When allowing one and two sequence mismatches with the first validation set, we obtained a ROC curve with an extremely low number of false positives (Figure 4A). However, upon using three mismatches, the number of false positives was increased (Figure 4A). Using the number of contacts between the protein and the DNA, as well as the interaction energy (Figure 5F), we were able to filter out the majority of the false positives (Figure 4B).
Figure 4.
Density maps and roc curves: (A) ROC curves for all predicted 4 base pair length dnaX fragments against the 212 validation complexes considering an RMSD threshold per residue <1.8 Å allowing 1 (pink), 2 (purple), 3 (cyan) sequence mismatches in the search; (B) ROC curves for three mismatches before (cyan) and after (red) filtering results by contacts and energy; (C) ROC space density map, X-axis represents RMSD per residue and Y-axis binding energy for 1 mismatch predictions; (D) histogram with the frequency of cases for a given area under the curve or TP/FN rate for 1 (pink), 2 (purple), and 3 (blue) mismatches.
Figure 5.
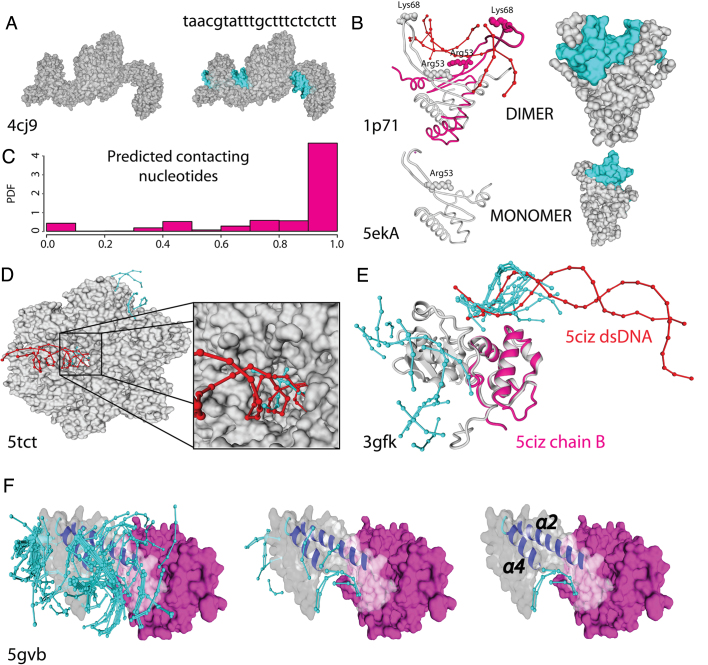
Accuracy on the docking predictions: In all cases cyan colour is used for docked DNA and red for the crystallographic one. (A) TAL effector (PDB: 4jc9), protein–DNA interface regions and nucleotide sequence specificity are correctly predicted. (B) Left and right: Cartoon and VdW surface style for a dimeric structure with DNA (PDB: 1p71; Upper) and the related protein crystallized as a monomer missing part of the structure (PDB: 5eka; lower). Predicted docked DNA in both structures in cyan. (C) histogram of sequence specificity accuracy over 13 proteins of different PFam family. (D) Humanized yeast ACC carboxyltransferase (PDB: 5tct) binding region (zoomed) was found within the five best energy docks. (E) the DNA-directed RNA polymerase subunit alpha of Bacillus subtilis (PDB: 3gfk, grey) superimposed to the overlapping domain of the E. coli ortholog with low sequence identity (PDB: 5ciz chainB, magenta). (F) The helical bundle of AND-1 human protein (PDB: 5gvb) present a dense cloud of docked fragments (left side) in the binding groove formed by the α2 and α4 helices. Within the six best energy docks for AND-1 human protein we found three docks (right side) placed within the predicted binding region.
For the 212 PDB validation set, only three crystal structures (PDBs: 5exh, 5j3e, and 5tct) did not yield docking results upon allowing 1–3 pep-mismatches. In the case of 5tct, we recovered the binding zone within the five best dockings after allowing for six mismatches and ranking the results by energy and contacts (Figure 5D). In contrast, the 5exh and 5j3e structures represent true, novel structural configurations not found in our database, the most probable reason is the PDB incompleteness of the interaction landscape containing no suitable IntXs to properly model the interface present on those two complexes. Further versions of DNAXB will be populated with more structures in order to overcome this limitation.
With respect to the standard benchmark validation set, our docking algorithm predicted the binding zone with an RMSD of <1.8 Å per residue in 96% of the cases. As expected, no dockings were found for the set of proteins that do not bind DNA when setting the pep-mismatches value to 1. However, upon enabling PADA1 to allow up to six mismatches, at least one dock was found for 141 of the 250 structures of the negative dataset. Nonetheless, these docked DNA molecules were filtered out in all cases using the contacting nucleotides and computed energy filters (Supplementary Figure S4).
PADA1 biological applications
In the previous section, we clearly demonstrate the high performance of PADA1 using different validation sets. However, in all cases, we used structures crystallized with bound DNA. For true biological applications, PADA1 should be able to predict the DNA-binding region, and if possible, the DNA sequences recognized by proteins crystallized without DNA. Furthermore, it should be capable of predicting the effect of protein point mutations on DNA structure and the sequence recognized by the mutant protein. In the following section, we will demonstrate the ability of PADA1 to tackle both these applications using different examples.
To demonstrate that we can predict the DNA recognition sequence of a protein that was crystallized without DNA, we selected the TAL-effector family. This protein family was chosen as a validation case because the DNA recognition sequence can easily be inferred from its protein sequence (47). As shown in Figure 5A, we were able to find not only the binding region but also the binding motif of a TAL-effector protein (PDB: 4cj9 (48), Uniprot accession E5AV36).
The UniProt entry P19436, is a non-specific HU histone-like DNA-binding protein (49) that was partially crystallized in its apo form (PDB: 5eka (50)). It is known that residues R53 and K68 of this protein are involved in DNA binding. In the crystal structure, only R53 is present as residue K68 belongs to an unresolved region. This protein binds DNA in a dimeric conformation but the 5eka structure presents only one monomer. Despite all these difficulties (incomplete coverage, incomplete quaternary structure, and non-specific binding nature of the protein), our algorithm accurately predicts the binding region around residue R53 (Figure 5B). This example highlights the benefits of a peptide/DNA fragment-based prediction strategy over typical protein/DNA-based methods.
The 3D crystal structure of the DNA-directed RNA polymerase subunit alpha of Bacillus subtilis (UniProt P20429) has been resolved (PDB: 3gfk (51)) without DNA. Similarly, the Escherichia coli ortholog, which has the highest sequence identity (only 42%, with a sequence coverage of 82%), has also had its crystal structure solved (PDB: 5ciz). By executing PADA1 with parameters that allowed any peptide sequence, we were able to identify the binding zone (Figure 5E).
As another example, the human AND-1 adaptor protein has recently been crystallized (PDB: 5gvb (52)), and based on its homology with yeast genes, it is thought that the multi-helical domain acts as a DNA-binding groove. Our predictions place several dnaX fragments in the multi-helical domain, with an especially dense cloud (containing the best docks according to our filtering criteria, see Supplementary Figure S4) in the zone between helices α2 and α4, exactly where the binding groove is reported to be (Figure 5F). Accordingly, the non-DNA-binding domain was predicted to have no docks. In addition, we tested the ability of PADA1 to correctly predict not only the DNA-binding region but also the DNA recognition sequence. For this purpose, we chose a set of 13 crystal structures from the validation set (Supplementary Tables S1 and S2) which belonged to different proteins (distinct UniProt and Pfam families), and subjected them to a sequence validation pipeline (See methods). This test computed a total of 2348 docks (each six nucleotides in length) over the analyzed proteins. The proportion of favorable and unfavorable contacts in the dsDNA–protein interface is variable, but the best predicted sequence conserves at least 80% of the contacting nucleotides of the crystal structure in most of the predictions (∼80%). Furthermore, accuracy is reduced (<50% of contacting nucleotides correctly predicted) in only less than 6% of the docks (Figure 5C). Supplementary Table S1 contains detailed information of these predictions.
When used in combination with FoldX, PADA1 has a great potential for protein engineering. As an illustrative example, we took a WT meganuclease (PDB: 4aqu) and engineered (See methods) two mutated protein structures in-silico: the Ini3-4 and Amel3-4 configurations (Figure 6). Is known that these mutant proteins alter the DNA binding conformation and bind to a DNA motif named XPC in a more specific manner compared to the WT (53). We show that by using PADA1, we can retrieve the conformation of the mutated DNA, and correctly distinguish between experimental specificities for the different DNA motifs against the different engineered mutants. Algorithms to automatize this process are included within the modelX toolsuite (GlueDocks and BackboneMove commands; Figure 7A and Materials and Methods) to model both the flexibility of the protein and the dsDNA backbone flexibility (see Figure 7B and Materials and Methods). These algorithms also analyse the stability and change in specificity for the different combinations in protein and DNA sequences by sidechain refinement. For the mentioned WT meganuclease, a set of strands with similar lengths than the crystallographic DNA remained after the GlueDocks execution over an exahustive-mode Docking (Figure 7A). The cleavage region for the «glued» strands presents strong rigidity, while some flexibility arises in the adjacent zones. This allows the user to analyze the plausibility of binding for different DNA motifs subsequently moving the protein backbones in order to induce a fit for the new complex using the BackboneMove command (Figure 7B).
DISCUSSION
The validations performed in this study emphasize the high accuracy of PADA1 in predicting DNA-binding regions. This method is based on a novel built database that has a wide coverage of backbone conformational spaces of dsDNA–protein interfaces. The peptide-DNA scan basis of this method enables the correct prediction of binding regions even when crystallographic information is incomplete, such as for structures with unresolved protein regions. From the statistics analysis of the validation sets, we find that our predicted binding regions are mostly correct. Our method yields a very low rate of false positives that only increases when the number of allowed peptide sequence mismatches is increased. Exhaustive-mode searches can be filtered to reduce noisy predictions by incorporating energy ranking, nucleotide contact number threshold, sequence similarity and/or protein family of the crystal structure where the docks come from.
The limitations of our method are, of course, related with the coverage of the PDB over the structural space of DNA–protein interactions. This coverage will continue to expand however, as new structures are released. Nonetheless, after applying exhaustive-mode search parameters, we still obtained good results for novel protein configurations, as well as for proteins with low sequence identity to the peptide sequences in ModelXDB. Unfortunately however, for such cases the docking quality is probably not good enough for predicting DNA recognition sequences. The prediction ability for structurally unexplored interfaces is hard to measure since this interaction space is sparse and depends on multiple factors (54).
Protein sequence redundancy stemming from the in-silico digestion of the complexes, is a valuable advantage that provides flexibility in the modelling of either the docked DNA or the input protein, and as we have shown using a meganuclease, allows to model structural changes induced by mutation.
The PADA1 force field, which was designed to permit rapid evaluation of the docked DNA, is accurate enough to achieve the desired goals. However, once the binding zone and the dnaX cloud are determined, the necessity of a finer force field emerges, depending on the specific case study. The FoldX tool allows both sidechain refinement upon docking, and greater precision with respect to other terms, such as hydrogen bonds, solvation, π interactions, Van der Waals forces, clashes, and electrostatics, which were not incorporated in the PADA1 force field. Through combining these tools, we were able to reproduce, using only the structure of the protein, the rational design of a meganuclease–dsDP complex and the experimental specificities over different DNA recognition sequences. We have also developed algorithms to automatize this process and generate conformational diversity for the protein and DNA backbone. In further versions of the software we plan to deepen this aspect (i.e. automatically modelling all the backbone moves energetically favorable for the docked dsDNA strands and computing its PSSM upon DNA mutagenesis) by repairing the sidechains (with FoldX RepairPDB command) and selecting those with best free energy values calculated with FoldX Pssm command (Figure 7B). It is expected that it can lead to engineer new interfaces and predict its specificities.
Last but not least, PADA1 was tested as an in silico predictor of DNA recognition sequences with accurate results. The main limitation with respect to this, is the structural coverage of DPIs. Currently, we can correctly predict the dsDNA-binding region in 98% of the 212 DNA structures of the validation dataset, and the sequence profile in 80% of the cases. It is expected that the continual increase in the number of available structures will enhance the accuracy of our force field, and at the same time remove the necessity of exhaustive searches. This in turn, will improve DNA positioning on the docking site, reduce the number of false positives, and decrease the computation time of our docking algorithm.
DATA AVAILABILITY
This software is freely available for academic users through the web site at http://modelx.crg.es/.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Professor Jesús Delgado Calvo for the inspiring mathematical discussions that helped us improving algorithm's computing efficiency, Tony Ferrar for manuscript revision and language editing (http://theeditorsite.com), the CRG TBDO for supporting with licensing information, the CRG TIC for helping with web hosting, and the CRG SIT for fruitful discussions about database performance and tuning. We appreciate all the feedback from the Serrano lab members.
Author Contributions: J.D. and L.S. conceived the idea. J.D. designed the software architecture. J.D. and L.R. developed the software. J.D., L.R. and H.C. built the database. J.D., L.R. and L.S. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013–2017′; CERCA Programme/Generalitat de Catalunya; Ministerio de Economia y Competitividad and FEDER funds (European Regional Development Fund) [BFU2015-63571-P]. Funding for open access charge: Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013–2017’; CERCA Programme/Generalitat de Catalunya; Ministerio de Economia y Competitividad and FEDER funds (European Regional Development Fund) [BFU2015-63571-P].
Conflict of interest statement. None declared.
REFERENCES
- 1. Walter M.C., Rattei T., Arnold R., Güldener U., Münsterkötter M., Nenova K., Kastenmüller G., Tischler P., Wölling A., Volz A. et al. . PEDANT covers all complete RefSeq genomes. Nucleic Acids Res. 2009; 37:D408–D411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Luscombe N.M., Austin S.E., Berman H.M., Thornton J.M.. An overview of the structures of protein–DNA complexes. Genome Biol. 2000; 1:REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000; 16:16–23. [DOI] [PubMed] [Google Scholar]
- 5. Hwang S., Gou Z., Kuznetsov I.B.. DP-Bind: a web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics. 2007; 23:634–636. [DOI] [PubMed] [Google Scholar]
- 6. Ofran Y., Mysore V., Rost B.. Prediction of DNA-binding residues from sequence. Bioinformatics. 2007; 23:i347–i353. [DOI] [PubMed] [Google Scholar]
- 7. Chen Y.C., Wu C.Y., Lim C.. Predicting DNA-binding amino acid residues from electrostatic stabilization upon mutation to Asp/Glu and evolutionary conservation. Proteins: Struct. Funct. Bioinf. 2007; 67:671–680. [DOI] [PubMed] [Google Scholar]
- 8. Wang L., Huang C., Yang M.Q., Yang J.Y.. BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC Syst. Biol. 2010; 4(Suppl. 1):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang L., Brown S.J.. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006; 34:W243–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Si J., Zhao R., Wu R.. An overview of the prediction of protein DNA-Binding sites. Int. J. Mol. Sci. 2015; 16:5194–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamasaki S., Terada T., Kono H., Shimizu K., Sarai A.. A new method for evaluating the specificity of indirect readout in protein–DNA recognition. Nucleic Acids Res. 2012; 40:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. MacKerell A.D., Banavali N.K.. All-atom empirical force field for nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem. 2000; 21:105–120. [Google Scholar]
- 13. Liu Z., Guo J.-T., Li T., Xu Y.. Structure-based prediction of transcription factor binding sites using a protein–DNA docking approach. Proteins. 2008; 72:1114–1124. [DOI] [PubMed] [Google Scholar]
- 14. Bastard K., Thureau A., Lavery R., Prévost C.. Docking macromolecules with flexible segments. J. Comput. Chem. 2003; 24:1910–1920. [DOI] [PubMed] [Google Scholar]
- 15. Knegtel R.M., Boelens R., Kaptein R.. Monte Carlo docking of protein–DNA complexes: incorporation of DNA flexibility and experimental data. Protein Eng. 1994; 7:761–767. [DOI] [PubMed] [Google Scholar]
- 16. Aloy P., Moont G., Gabb H.A., Querol E., Aviles F.X., Sternberg M.J.E.. Modelling repressor proteins docking to DNA. Proteins: Struct. Funct. Genet. 1998; 33:535–549. [PubMed] [Google Scholar]
- 17. Fanelli F., Ferrari S.. Prediction of MEF2A–DNA interface by rigid body docking: A tool for fast estimation of protein mutational effects on DNA binding. J. Struct. Biol. 2006; 153:278–283. [DOI] [PubMed] [Google Scholar]
- 18. Fan L., Roberts V.A.. Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:8384–8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roberts V.A., Case D.A., Tsui V.. Predicting interactions of winged-helix transcription factors with DNA. Proteins. 2004; 57:172–187. [DOI] [PubMed] [Google Scholar]
- 20. Adesokan A.A., Roberts V.A., Lee K.W., Lins R.D., Briggs J.M.. Prediction of HIV-1 Integrase/Viral DNA interactions in the catalytic domain by fast molecular docking. J. Med. Chem. 2004; 47:821–828. [DOI] [PubMed] [Google Scholar]
- 21. Sandmann C., Cordes F., Saenger W.. Structure model of a complex between the factor for inversion stimulation (FIS) and DNA: modeling protein–DNA complexes with dyad symmetry and known protein structures. Proteins. 1996; 25:486–500. [DOI] [PubMed] [Google Scholar]
- 22. Buchete N.-V., Straub J.E., Thirumalai D.. Orientational potentials extracted from protein structures improve native fold recognition. Protein Sci. 2004; 13:862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tuszynska I., Magnus M., Jonak K., Dawson W., Bujnicki J.M.. NPDock: a web server for protein-nucleic acid docking. Nucleic Acids Res. 2015; 43:W425–W430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaplan T., Friedman N., Margalit H.. Ab initio prediction of transcription factor targets using structural knowledge. PLoS Comput. Biol. 2005; 1:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banitt I., Wolfson H.J.. ParaDock: a flexible non-specific DNA–rigid protein docking algorithm. Nucleic Acids Res. 2011; 39:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morozov A.V. protein–DNA binding specificity predictions with structural models. Nucleic Acids Res. 2005; 33:5781–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Havranek J.J., Duarte C.M., Baker D.. A simple physical model for the prediction and design of Protein–DNA interactions. J. Mol. Biol. 2004; 344:59–70. [DOI] [PubMed] [Google Scholar]
- 28. Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L.. The FoldX web server: an online force field. Nucleic Acids Res. 2005; 33:W382–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verschueren E., Vanhee P., van der Sloot A.M., Serrano L., Rousseau F., Schymkowitz J.. Protein design with fragment databases. Curr. Opin. Struct. Biol. 2011; 21:452–459. [DOI] [PubMed] [Google Scholar]
- 30. Vanhee P., Reumers J., Stricher F., Baeten L., Serrano L., Schymkowitz J., Rousseau F.. PepX: a structural database of non-redundant protein–peptide complexes. Nucleic Acids Res. 2010; 38:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vanhee P., Verschueren E., Baeten L., Stricher F., Serrano L., Rousseau F., Schymkowitz J.. BriX: a database of protein building blocks for structural analysis, modeling and design. Nucleic Acids Res. 2010; 39:D435–D442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davey N.E., Van Roey K., Weatheritt R.J., Toedt G., Uyar B., Altenberg B., Budd A., Diella F., Dinkel H., Gibson T.J.. Attributes of short linear motifs. Mol. Biosyst. 2012; 8:268–281. [DOI] [PubMed] [Google Scholar]
- 33. Obenauer J.C., Cantley L.C., Yaffe M.B.. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003; 31:3635–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hofmann K., Bucher P., Falquet L., Bairoch A.. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999; 27:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Puntervoll P., Linding R., Gemünd C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D.M.A., Ausiello G., Brannetti B., Costantini A. et al. . ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003; 31:3625–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lahm A., Suck D.. DNase I-induced DNA conformation. 2 A structure of a DNase I-octamer complex. J. Mol. Biol. 1991; 222:645–667. [DOI] [PubMed] [Google Scholar]
- 37. Kumar R., Grubmüller H.. do_x3dna: a tool to analyze structural fluctuations of dsDNA or dsRNA from molecular dynamics simulations: Fig. 1. Bioinformatics. 2015; 31:2583–2585. [DOI] [PubMed] [Google Scholar]
- 38. Sippl M.J. Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J. Mol. Biol. 1990; 213:859–883. [DOI] [PubMed] [Google Scholar]
- 39. Kono H., Sarai A.. Structure‐based prediction of DNA target sites by regulatory proteins. Proteins: Struct. Funct. Genet. 1999; 35:114–131. [PubMed] [Google Scholar]
- 40. Luscombe N.M. Amino acid-base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 2001; 29:2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Dijk M., Bonvin A.M.J.J.. A protein–DNA docking benchmark. Nucleic Acids Res. 2008; 36:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao M., Skolnick J.. DBD-Hunter: a knowledge-based method for the prediction of DNA–protein interactions. Nucleic Acids Res. 2008; 36:3978–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szilágyi A., Skolnick J.. Efficient prediction of nucleic acid binding function from low-resolution protein structures. J. Mol. Biol. 2006; 358:922–933. [DOI] [PubMed] [Google Scholar]
- 44. Ahmad S., Gromiha M.M., Sarai A.. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004; 20:477–486. [DOI] [PubMed] [Google Scholar]
- 45. Stawiski E.W., Gregoret L.M., Mandel-Gutfreund Y.. Annotating nucleic acid-binding function based on protein structure. J. Mol. Biol. 2003; 326:1065–1079. [DOI] [PubMed] [Google Scholar]
- 46. Si J., Zhang Z., Lin B., Schroeder M., Huang B.. MetaDBSite: a meta approach to improve protein DNA-binding sites prediction. BMC Syst. Biol. 2011; 5(Suppl. 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moscou M.J., Bogdanove A.J.. A simple cipher governs DNA recognition by TAL effectors. Science. 2009; 326:1501. [DOI] [PubMed] [Google Scholar]
- 48. Stella S., Molina R., López-Méndez B., Juillerat A., Bertonati C., Daboussi F., Campos-Olivas R., Duchateau P., Montoya G.. BuD, a helix-loop-helix DNA-binding domain for genome modification. Acta Crystallogr. D Biol. Crystallogr. 2014; 70:2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Swinger K.K., Lemberg K.M., Zhang Y., Rice P.A.. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003; 22:3749–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Papageorgiou A.C., Adam P.S., Stavros P., Nounesis G., Meijers R., Petratos K., Vorgias C.E.. HU histone-like DNA-binding protein from Thermus thermophilus: structural and evolutionary analyses. Extremophiles. 2016; 20:695–709. [DOI] [PubMed] [Google Scholar]
- 51. Lamour V., Westblade L.F., Campbell E.A., Darst S.A.. Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase α subunit C-terminal domain complex. J. Struct. Biol. 2009; 168:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guan C., Li J., Sun D., Liu Y., Liang H.. The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome. J. Biol. Chem. 2017; 292:9627–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Redondo P., Prieto J., Muñoz I.G., Alibés A., Stricher F., Serrano L., Cabaniols J.-P., Daboussi F., Arnould S., Perez C. et al. . Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008; 456:107–111. [DOI] [PubMed] [Google Scholar]
- 54. Jayaram B., McConnell K., Dixit S.B., Das A., Beveridge D.L.. Free-energy component analysis of 40 protein–DNA complexes: a consensus view on the thermodynamics of binding at the molecular level. J. Comput. Chem. 2002; 23:1–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This software is freely available for academic users through the web site at http://modelx.crg.es/.