Abstract
CHD1 is a conserved chromatin remodeling enzyme required for development and linked to prostate cancer in adults, yet its role in human cells is poorly understood. Here, we show that targeted disruption of the CHD1 gene in human cells leads to a defect in early double-strand break (DSB) repair via homologous recombination (HR), resulting in hypersensitivity to ionizing radiation as well as PARP and PTEN inhibition. CHD1 knockout cells show reduced H2AX phosphorylation (γH2AX) and foci formation as well as impairments in CtIP recruitment to the damaged sites. Chromatin immunoprecipitation following a single DSB shows that the reduced levels of γH2AX accumulation at DSBs in CHD1-KO cells are due to both a global reduction in H2AX incorporation and poor retention of H2AX at the DSBs. We also identified a unique N-terminal region of CHD1 that inhibits the DNA binding, ATPase, and chromatin assembly and remodeling activities of CHD1. CHD1 lacking the N terminus was more active in rescuing the defects in γH2AX formation and CtIP recruitment in CHD1-KO cells than full-length CHD1, suggesting the N terminus is a negative regulator in cells. Our data point to a role for CHD1 in the DSB repair process and identify a novel regulatory region of the protein.
INTRODUCTION
Chromodomain-helicase-DNA-binding protein 1 (CHD1) is a member of the SNF2-like family of helicase-related enzymes, and part of a subgroup of CHD proteins that contain tandem chromodomains (CDs) (5–8). Only one CHD protein is expressed in yeast (Chd1p), whereas nine CHD proteins are expressed in vertebrates (i.e. CHD1 through CHD9). Members of the vertebrate CHD family are further divided into subgroups, based on the presence of additional accessory domains. For example, CHD1 and CHD2 are grouped together based on the presence of a putative SANT-SLIDE-like DNA-binding domain (DBD) near their C terminus (6).
Several studies have shown that CHD1 associates with active genes and is involved in regulating transcription. For example, CHD1 from yeast and flies can assemble periodic nucleosomal arrays and is required for establishing or maintaining the proper chromatin structure at promoters or across gene bodies (9–14). One role for the assembly activity of CHD1 may be to help suppress cryptic transcription initiation, which is supported by the finding that deletion of CHD1 in yeast gives rise to an increase in antisense transcripts (15).
The co-expression of CHD1 and CHD2 implies these two proteins have more specialized roles than their yeast CHD1 counterpart. This is supported by the fact that CHD1 and CHD2 have different patterns of distribution around transcriptional start sites (TSSs); CHD1 is only present near TSSs whereas CHD2 is present across gene bodies (16). Although CHD1 and CHD2 share ∼67% identity, the N terminus of human CHD1 contains a unique serine-rich region that is not present in CHD2. These findings imply that the N-terminal region of CHD1 may play a regulatory role that separates CHD1 from CHD2. In fact, it was reported that this region is phosphorylated in mouse cell extracts, and deletion of more than half of those sites in ES cells disrupts the differentiation of those cells into specific cell lineages (17).
While the role of CHD1 in gene transcription is well-studied, the role of CHD1 in other cellular processes is less understood. CHD1 has been identified as a key suppressor of prostate cancer, and loss of CHD1 sensitizes prostate cancer cells to chemotherapeutic DNA-damaging agents, suggesting CHD1 may be involved in the repair of double-strand DNA breaks (18,19). In both breast and prostate cancer cells, loss of CHD1 is synthetically lethal with loss of PTEN (20), and PTEN-deficient tumors may benefit from selective inhibition of CHD1. While the mechanisms underlying the tumor suppressor role of CHD1 are unclear, the ability of CHD1 to remodel chromatin during transcription and DNA repair is likely a key aspect of its tumor suppressor role. Despite the importance of CHD1 in preventing tumorigenesis, our knowledge of how CHD1 functions in cells is just emerging (19), and the molecular mechanism of how the activity of CHD1 is regulated and linked to double-strand break repair is largely unclear. A recent report shows that the related protein CHD2 is involved in a distinct DSB repair process, the non-homologous end joining (NHEJ) pathway (21). Understanding how the remodeling and assembling activities of CHD1 are regulated and identifying the differences between CHD1 and other remodeling proteins (i.e. CHD2) will further our understanding of its cellular function.
In this study, we found that human CHD1 is critical for the efficient repair of DSBs through homologous recombination (HR). CHD1 is required for the early steps in the HR pathway, including efficient γH2AX signaling and recruitment of CtIP. Thus, loss of CHD1 leads to an increased sensitivity to ionizing radiation. In addition, we discovered a new autoinhibitory region of CHD1 at the N terminus. The N terminus inhibits the DNA-binding and ATP-dependent activities of CHD1 in vitro as well as the DNA repair activities in cells. The repressive region of CHD1 is largely outside of the serine-rich region and is not conserved in CHD2 or in yeast and Drosophila CHD1 proteins. Together, our data demonstrate that human CHD1 is regulated by its unique N terminus and suggest at least one mechanism by which CHD1 and CHD2 are differentially regulated, specifically in their functions in DNA double-strand break repair.
MATERIALS AND METHODS
Purification of recombinant CHD1
The human CHD1 proteins were cloned and expressed as N-terminal FLAG-tagged fusion proteins in Sf9 cells using the Bac-to-Bac baculovirus expression system (Invitrogen). Infected Sf9 cells were suspended in cold Lysis Buffer (20 mM HEPES–KOH-pH 7.6, 500 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 20% glycerol, 0.01% NP-40, 1 mM DTT and the following protease inhibitors: 0.5 mM benzamidine, 1 μg/ml of leupeptin, aprotinin, pepstatin A, and 0.2 mM PMSF). The cells were lysed using a Dounce homogenizer on ice for 30 min and the lysate was centrifuged at 21 000 × g for 15 min at 4°C followed by a second centrifugation for 10 min. The cleared cell lysate was then incubated with anti-FLAG resin (Sigma) for 4 h at 4°C on a rotator. The resin was pelleted by centrifugation and washed three times with Lysis Buffer. The CHD1 proteins were eluted with two sequential incubations of the resin with an equal volume of Elution Buffer (Lysis Buffer containing 0.2 mg/ml FLAG peptide, Sigma). The concentration and quality of the purified CHD1 proteins were assessed by Bradford assay and SDS-PAGE/Coomassie staining. The eluted protein was then flash-frozen with liquid nitrogen and stored at −80°C in small aliquots for subsequent biochemical experiments.
Electrophoretic mobility gel shift assay (EMSA)
The dsDNA probes were generated by annealing a 5′-IRDye-700 labeled 40 or 60 bp oligonucleotide (IDT) with its complementary strand. CHD1 or other remodeling enzymes (0, 10, 20, 50, 100, 200 or 400 nM) and a dsDNA probe (5 nM) were mixed in the binding buffer (45 mM Tris-borate, 50 mM KCl, 5 mM magnesium acetate, 0.1 μg/μl BSA, 5% glycerol, 1 mM DTT, 1.2 mM EDTA and 0.01% NP-40) to a final volume of 10 μl. The samples were incubated for 1 h on ice and resolved on a 5% native polyacrylamide gel made with 0.5× TBE and 5% glycerol, running in 0.5× TBE at 4°C. The gel was then scanned on a LI-COR Odyssey Imaging System.
Chromatin remodeling-restriction endonuclease accessibility (REA) assay
A standard 25 μl REA reaction contained 1 μg of plasmid DNA (pGIE-0, 3.2 kb) or plasmid pre-assembled into chromatin by salt dialysis, 25 units of HaeIII (NEB) in a buffer consisting of 20 mM Tris acetate, 10 mM magnesium acetate, 50 mM potassium acetate, pH 7.9, and either 5 mM AMP-PNP (as a minus ATP control) or ATP. 1 μg of WT CHD1 or CHD1 mutants were added to each reaction where indicated. After 2 h in a 30°C water bath, the reactions were stopped by the addition of 125 μl of Stop Buffer (1% [w/v] SDS, 200 mM NaCl, 250 μg/ml glycogen, 20 mM EDTA pH 8.0) supplemented with 50 μg/ml proteinase K (Worthington). The DNA was then phenol:chloroform extracted, ethanol precipitated, resolved by native gel electrophoresis and visualized by SYBR Safe or ethidium bromide staining. A Digestion Index (DI) was calculated for each lane, using a method as described previously (1).
Nucleosome assembly—supercoiling analysis
The nucleosome assembly assay was performed as previously described (2). A standard 70 μl assembly reaction contained core histones (0.4 μg), plasmid DNA (0.4 μg; pGIE-0; 3.2 kb;), Topo I, NAP-1 (2 μg), purified ACF or CHD1 (1 μg), and an ATP regeneration system (3 mM ATP or AMP-PNP, 5 mM MgCl2, 30 mM phosphocreatine, and 5 ng/μl creatine phosphokinase). Briefly, the histones were incubated first with the histone chaperone NAP-1 for 30 min on ice. In parallel, the plasmid DNA/Topo I reaction was set up for 10 min at 30°C and kept at room temperature until use. After 30 min, the ATP regeneration system, relaxed plasmid DNA (with Topo I still present), and CHD1 were added to the histone/chaperone mixture. For minus ATP reactions, AMP-PNP was used instead of ATP. The reactions were stopped after 1 h incubation at room temperature with the addition of 125 μl of Stop Buffer and proteinase K at a final concentration of 50 μg/ml. The DNA was then extracted with phenol:chloroform, ethanol precipitated, resolved by agarose gel electrophoresis, and visualized by SYBR Safe staining. Plasmid DNA or DNA extracted from the plasmid DNA/Topo I mixture was used to assess where supercoiled (sc) and relaxed (rel) DNA run for comparison in agarose gel electrophoresis.
ATPase activity assays
The ATPase activity assay reactions (20 μl) contained the reaction buffer (20 mM HEPES-KOH (pH 7.6), 100 mM KCl, 5 mM MgCl2, 0.25 μg/μl BSA, 0.05 mM EDTA, 0.5 mM DTT, 3% glycerol, and 0.01% NP-40), the substrate (100 μM ATP, and 1 μCi of [γ-32P]-ATP as a tracer) and the enzyme (CHD1-WT and truncations). Where indicated, reactions also contained 25 ng/μl double-stranded plasmid DNA (pGIE-0, 3.2 kb), or plasmid that had been pre-assembled into chromatin by dialysis (containing 25 ng/μl of DNA). ATP and the tracer [γ-32P]-ATP were added to the reaction buffer first and 1 μg of CHD1 protein was added to start the ATPase reactions. Aliquots (2 μl) of the reaction were removed and immediately dotted to a dry PEI-cellulose plate (Sigma) at indicated time points. The samples were resolved by thin-layer chromatography (TLC) in 4.5% formic acid supplied with 0.5 M lithium chloride. The TLC plate was then air-dried and exposed to a phosphor screen for visualization of radioactivity by a Personal Molecular Imager (PMI) System (Bio-Rad). Quantification of the [γ-32P]-ATP and the released inorganic phosphate (32Pi) was performed using Quantity One software. After background subtraction, the fraction of hydrolyzed ATP was calculated and plotted versus time. Mean and standard deviation (SD) from at least three experiments were shown. The data points were fitted to a curve by Prism GraphPad 7 using the one-phase association model.
Plasmids and siRNA
For transient transfection, CHD1-WT coding sequence and CHD1-ΔN were cloned into the pOZ-FH-N plasmid between XhoI and NotI sites, in frame with the N-term Flag and HA tags. The constructs were also used for retroviral infection to deliver CHD1. Two days after viral infection, cells were sorted by the co-expressing IL-2α receptor for CHD1 expression. siRNA against CHD1 and CtIP were designed and synthesized by IDT. siRNA was transfected into cells by HiPerfect (Qiagen) at a final concentration of 20 nM. Experiments with siRNA were performed 48–72 h after transfection to allow effective knockdown.
Cell lines and CHD1 knock out
U2OS and RPE1 cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. The CHD1 knock out cell lines were generated using CRISPR-Cas9 system following the protocol from Zhang lab at MIT (3). RPE1 cells stably expressing Cas9 is a gift from Chowdhury lab at Dana-Farber Cancer Institute. The designed oligos (5′-CACCGACCCAGAATCATCATCCGAC-3′ and 5′-AAACGTCGGATGATGATTCTGGTC-3′, targeting exon 1 of the CHD1 gene) were annealed and inserted into lentiGuide-Puro plasmid (Addgene #52963). The plasmid expressing gRNA was then transfected to RPE1-Cas9 cells by Lipofectamine 2000 and the transfected cells were selected by 10 μg/mL puromycin for 2 weeks. One thousand of selected cells were seeded onto a 175 mm dish to form colonies. Individual colonies were picked and CHD1-KO clones were verified by Western blotting. For rescue experiments, CHD1-WT coding sequence or CHD1-ΔN in pOZ-FH-N was delivered to cells via retroviral infection, and cells were sorted using IL-2α receptor for CHD1 expressing cells.
Clonogenic survival assay
RPE1 control cells or CHD1-KO cells were trypsinized and counted, and 1000 cells were seeded onto a 10 cm dish. The cells were treated with 0, 0.5, 1 or 2 Gy of irradiation. Irradiated cells were allowed to grow for 11–14 days to form colonies. The dishes were stained with 0.5% of crystal violet in methanol for 10 min and rinsed with H2O four times. The stained dishes were dried and the number of colonies was counted.
Immunofluorescence microscopy
Cells were seeded onto 35mm glass bottom dishes (MatTek), and were fixed with 3.2% PFA for 10 min at the time of experiments. Cells were washed with PBS, permeabilized with PBS supplemented with 0.5% Triton X-100 for 10 min and blocked with blocking reagent (Licor, diluted 1:1 in PBS with 0.2% Triton X-100) for 1 h. Cells in the dish were incubated with primary antibody at 4°C overnight. The dishes were washed with PBS-T (0.2% Tween-20) three times and incubated with fluorophore-conjugated secondary antibodies (and DAPI) for 1 h. The dishes were then washed with PBS-T three times and mounted in Anti-Fade mounting solution (Life Technology). Images were taken using a Zeiss Observer.Z1 fluorescence microscope under a Plan-Apochromat 63x/1.4Oil object.
Laser striping
U2OS and RPE1 cells were seeded onto 35mm glass bottom dishes (MatTek). siRNA and plasmid transfection were performed within the same dish following the protocol above. Micro-irradiation was performed on a Zeiss PALM MicroBeam system equipped with a 355 nm UV laser source (E < 60 uJ, f = 1000 Hz, t < 2 ns). The laser micro-irradiation was focused and delivered through a 63× object (LD Plan-Neofluar 63×/0.75Corr) of a Zeiss Observer.Z1 microscope. The power of output (cut) was set at 28% and the speed of cutting was set at 50%. The laser striping was done at room temperature and the cells were allowed to recover at 37°C incubator for indicated time. Cells were fixed with 3.2% PFA after recovery and regular immunofluorescence procedures were followed as described above. γH2AX signal was used as a marker for the DNA damage created by the laser.
Irradiation-induced foci (IRIF) assay
Cells were seeded onto a 35mm glass bottom dish (MatTek) the day before irradiation. For γH2AX focus formation, cells were generally treated with 2 Gy of irradiation and allowed to recover at 37°C. Cells were fixed by 3.2% PFA at indicated time and immunofluorescence microscopy procedures were carried out as described above. Acquired images were imported to Zen software (Zeiss) and applied with the same display settings of the channels. γH2AX foci in the nuclei were counted for each cell and documented. The average number of foci per cell was shown. The number of CtIP foci was counted by a custom-designed macro that runs in ImageJ. The macro defines nuclei by DAPI then finds maxima of pixels in the nuclei and defines them as CtIP foci (details available upon request).
GFP reporter assays
The DR-GFP and EJ-5 repair substrates were gifts from Dr Jeremy Stark at Beckman Research Institute of the City of Hope (4). To measure the repair efficiency in the cell lines, 5 × 104 cells were plated in each well of a 12-well plate. Cells were transfected with siRNA (20 nM final) and HiPerfect (3 ul, Qiagen) in 1 ml of serum free medium per well. 48–72 h after siRNA transfection, cells were infected with Adenoviruses that express the I-SceI enzyme. Two days after infection, cells were trypsinized and GFP positive cells were analyzed by flow cytometry (Beckman Coulter). Analysis of the GFP positive cells were performed using the CytExpert software (Beckman Coulter), following the gating scheme shown in Supplementary Figure S2A. Mean and standard deviation of at least three tests were shown.
Chromatin immunoprecipitation (ChIP) and quantitative PCR (qPCR)
T98G cells were transfected with p84-ZFN vector 48–72 h after CHD1 knockdown. 18 h after transfection of p84-ZFN, the cells were cross-linked by 1% paraformaldehyde for 10 min at room temperature and the reaction was stopped by glycine solution. ChIP was done by using the SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling Technology, #9003). Nuclei preparation, chromatin digestion, chromatin immunoprecipitation, reversal of cross-linking and DNA purification were performed following the manufacturer's protocol. In each ChIP, 4 × 106 cells were used to prepare the chromatin extract, and 5 μg of phospho-Ser139-H2AX antibody (Millipore 05–636) or H2AX antibody (ABCAM ab11175) was used.
Quantitative PCR was used to measure the γH2AX or H2AX bound DNA around the DSB site, operated on a StepOnePlus qPCR machine (Applied Biosystems). 2× SYBR Green qPCR Master Mix was purchased from Applied Biosystems (4367659) and used following the manufacturer's instructions. Sequences of the primers used for the qPCR can be found in Supplementary Table S1. The primers were used at the final concentration of 500 nM. Primer annealing and DNA extension was set for 45 s at 60°C in each cycle. The percentage of DNA captured by ChIP was normalized to 2% input DNA using the cycle threshold (Ct) values.
RESULTS
CHD1 is critical for the efficient repair of DSBs via HR
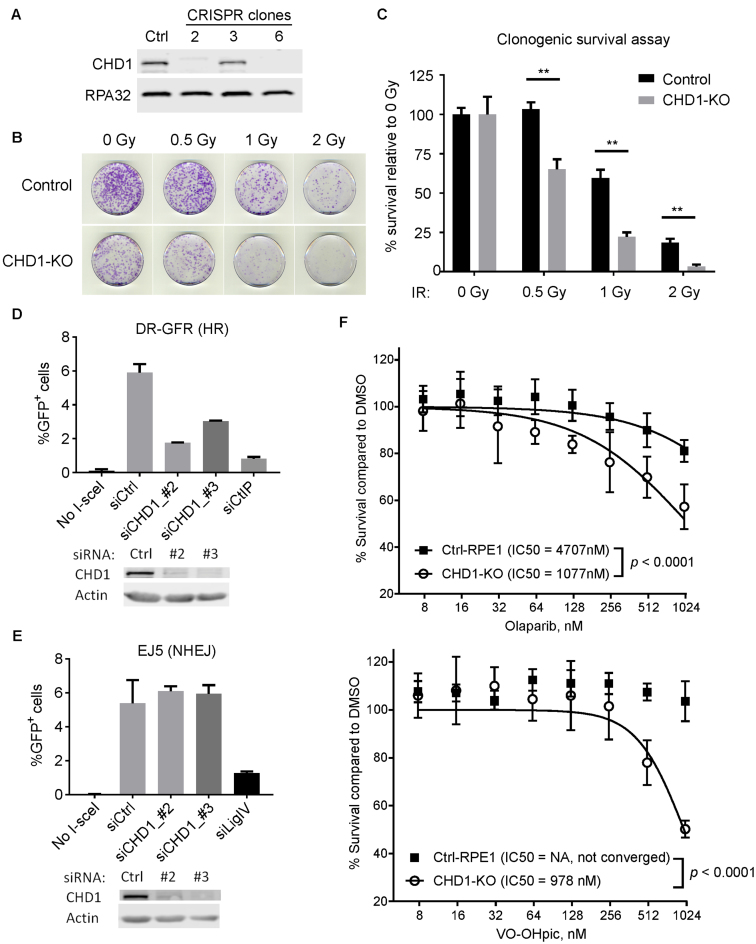
Mutations in CHD1 have been linked to tumorigenesis in prostate cancer (22), and CHD1-deficient cells were reported to be sensitive to DNA damaging agents (23). We started our investigation by asking if cells lacking CHD1 are hypersensitive to ionizing irradiation (IR). We knocked out the CHD1 gene in immortalized human RPE1 cells using the CRISPR-Cas9 system and isolated clones from the cell pools. Clones with CHD1 knocked out (CHD1-KO) were identified by Western blotting (Figure 1A). The CHD1-KO cells were viable and continued to proliferate without impaired growth rates (Supplementary Figure S1A). We then performed clonogenic survival assays on a CHD1-KO line (clone #6) and found that the CHD1-KO cells were more sensitive to IR than the WT parental control cells (i.e. Cas9 expressing; Figure 1B and C). We observed sensitivity of CHD1-KO cells to doses as low as 0.5 Gy while the WT control cells were not significantly sensitive at this dose. At 2 Gy, the CHD1-KO cells were almost completely lost, whereas a significant fraction of the WT control cells survived this higher dose (Figure 1B and C). Similar results have been recently reported in CHD1 knockdown cells by shRNA, with milder sensitivity (19).
Figure 1.
Human CHD1 is required for repair of DSBs through HR. (A) A Western blot shows that CHD1 was successfully knocked out in RPE1 cells by CRISPR-Cas9. Clone 6 was used for subsequent assays. (B) Clonogenic survival assays with WT or CHD1-KO RPE1 cells. The dishes were stained 12 days after IR. (C) Quantification of the clonogenic survival assays shown in (B). Mean and SD of three experiments were shown. **, p < 0.01. (D) Quantification of the DR-GFP (HR) assay in control or CHD1 depleted cells. CtIP knockdown (siCtIP) was used as a positive control. (E) Quantification of the EJ5 (NHEJ) assay. Ligase-IV knockdown (siLigIV) was used as a positive control. Western blots shown below each graph indicate successful knockdown. (F) Sensitivity of CHD1-KO cells and control cells to PARP inhibitor Olaparib (top) and PTEN inhibitor VO-OHpic. Cells were treated with the drugs for 6 days, and cell viability was measured by the CellTiter-Glo Assay.
To better understand the link between CHD1 and DNA repair, we measured the efficiencies of HR and NHEJ, the two major DSB repair pathways, in the presence or absence of CHD1. For this analysis, we took advantage of a well-established GFP reporter system established in the human U2OS cell line; the DR-GFP reporter line detects HR events while the EJ5 reporter line detects NHEJ events (4,24). In these reporter lines, a single DSB is introduced by the I-SceI restriction enzyme, and successful repair of the break leads to the creation of a functional GFP reporter. The efficiency of repair can then be monitored by FACS analysis (see Supplementary Figure S2 for schematics of the repair assays, gating methods, and representative raw FACS data). To assess which repair pathway requires CHD1, we depleted CHD1 by two different siRNAs (or non-targeting siRNAs as controls) in the U2OS cells containing one of the two GFP reporter substrates (Figure 1D and E). The knockdown of CHD1 in these cell lines was confirmed by Western blotting. We also depleted CtIP and Ligase-IV as positive controls. The knockdown of CtIP and Ligase-IV resulted in greatly reduced HR and NHEJ efficiency, respectively, which confirms the identities of the repair substrates in these U2OS cell lines. Upon CHD1 knockdown by both siRNAs, we observed a decrease of more than 50% in HR efficiency when compared to siControl (Figure 1D). However, we did not find a significant difference in NHEJ efficiency between the siControl and siCHD1 (Figure 1E). One possible explanation for the reduced HR efficiency is that CHD1 depletion alters the cell cycle and reduces the S/G2 cell population. To exclude this possibility, we analyzed the cell cycle profiles of CHD1-KO and WT control RPE1 cells by flow cytometry and did not see a significant difference in S/G2 population between the two cell lines (Supplementary Figure S1C). After IR, an accumulation of the G2/M population and a decrease of the S population were observed in both WT and CHD1-KO RPE1 cells (Supplementary Figure S1B and C), indicating CHD1-KO cells were proficient in G2/M and G1/S checkpoints. In addition, the levels of phosphorylation of the G2/M checkpoint protein, CHK2, were similar between WT and CHD1-KO cells (Supplementary Figure S1D). These data indicate that CHD1 is required for efficient DSB repair by HR, and loss of CHD1 results in IR sensitivity.
Because CHD1 is frequently mutated or deleted in prostate cancers, inhibiting key pathways that are synthetically-lethal with CHD1 could provide a therapeutic benefit. For example, HR-deficient cancer cells are generally sensitive to PARP inhibitors (25). Since the CHD1-KO cells are defective in HR, we measured the sensitivity of CHD1-KO cells to the FDA-approved PARP inhibitor drug, Olaparib. We found that CHD1 knockout RPE1 cells were significantly more sensitive to Olaparib, when compared to the CHD1-WT control RPE1 cells (Figure 1F, top). In addition, a recent study identified CHD1 as an essential gene in PTEN-deficient cancers (20), indicating a synthetic lethality between PTEN and CHD1. We measured the sensitivity of the CHD1-KO RPE1 cells to the commercially-available PTEN inhibitor VO-OHpic and found the CHD1-KO cells were highly sensitive to the PTEN inhibitor, when compared to the control REP1 cells (Figure 1F, bottom).
CHD1 is required for the recruitment of CtIP to damage sites
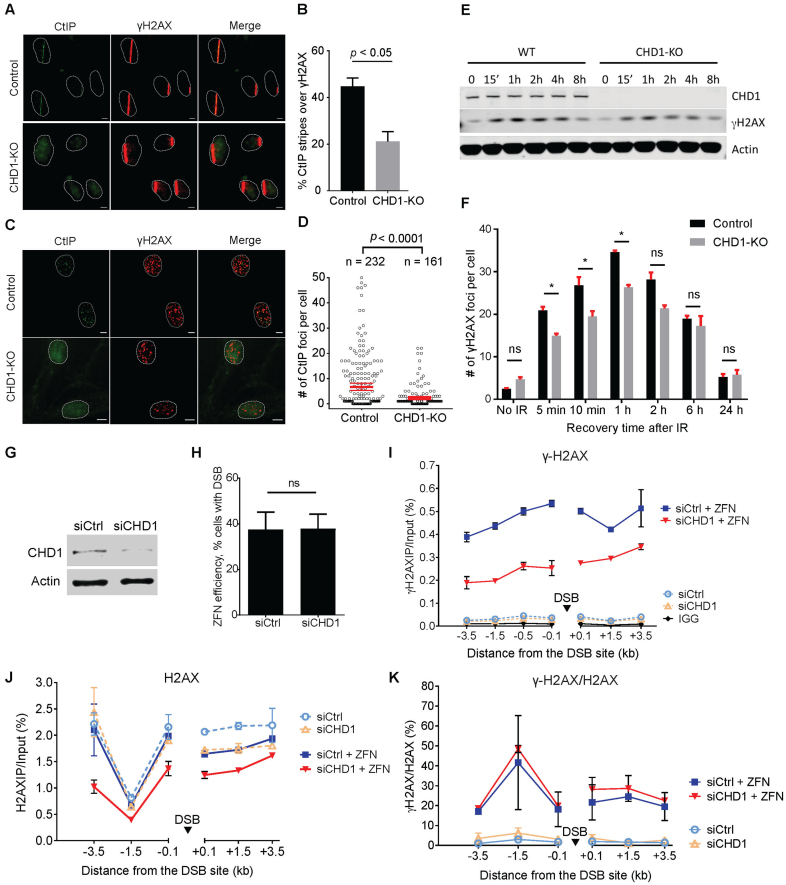
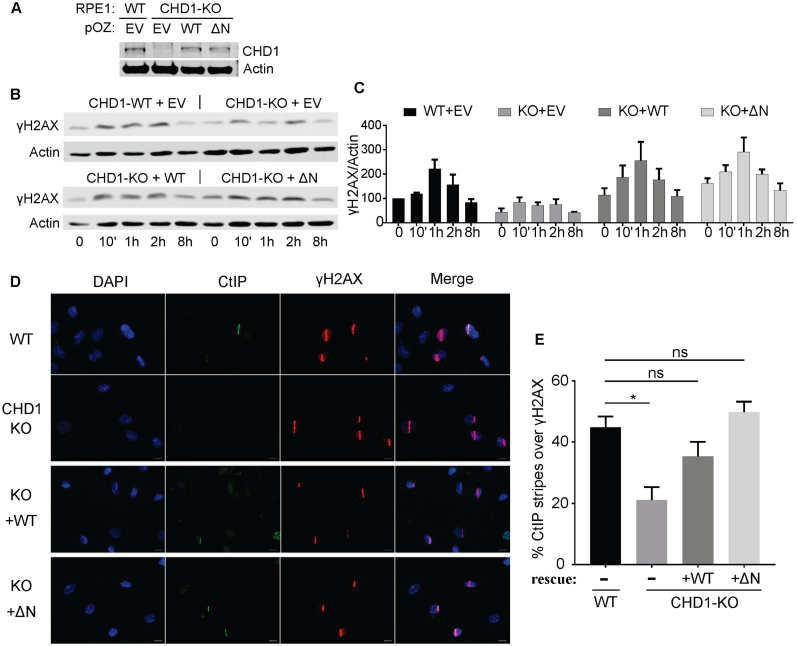
End resection is a critical step for the successful repair of DSBs through HR. To understand how CHD1 facilitates HR, we first asked if CHD1 is important for the recruitment of CtIP, a nuclease that is recruited to DSBs and is involved in resection of the DNA ends (26). We first performed laser striping (micro-irradiation) experiments followed by immunofluorescence staining (Figure 2A–B). CtIP can be readily detected in the laser-micro-irradiated tracks in WT control RPE1 cells. However, we found that the recruitment of CtIP to the laser tracks was significantly reduced in CHD1-KO cells when compared to the WT control cells. Approximately 45% of the laser-striped WT cells showed positive staining for CtIP in the laser track whereas fewer than 20% of the CHD1-KO cells showed positive CtIP recruitment (Figure 2A and B). In these experiments, γH2AX was stained as a positive control marker for successful micro-irradiation. To confirm the laser striping observations, we performed IR-induced foci formation (IR-IF) assays, in which WT and CHD1-KO cells were treated with 2 Gy of IR, and the recruitment of CtIP to repair foci was observed by immunofluorescence. The number of CtIP foci were then automatically counted by a custom-designed algorithm running in ImageJ (see Methods). We found that the CHD1-KO cells showed much fewer CtIP foci after IR as compared to WT control cells (Figure 2C and D), in agreement with the results from the laser striping experiments. These data are consistent with a previous report showing that knockdown of CHD1 decreases the recruitment of CtIP to I-SceI-induced DSB in U2OS19 cells (19). These data demonstrate that CHD1 is required for the proper recruitment of CtIP and suggest that CHD1 is important for early events during HR.
Figure 2.
CHD1 is critical for formation of γH2AX and recruitment of CtIP at DSBs. (A) Representative images of laser striping experiments showing CtIP recruitment to laser tracts. Cells were fixed 10 min after laser striping. γH2AX was used as a marker for DSBs. (B) Quantification of CtIP recruitment in the laser tracts. The number of cells with CtIP and γH2AX staining were counted, and the percentage of γH2AX tracts with CtIP recruitment is shown. The average number of foci per cell and the 95% confidence interval of two experiments are shown for control [n = 232] and CHD1-KO [n = 161] cells. (C) Representative images of immunofluorescence staining for CtIP and γH2AX after 2 Gy of IR. Cells were fixed 10 min after IR. (D) Quantification of the CtIP foci after IR. Foci were automatically identified and counted by a macro running in Fiji (ImageJ). (E) Western blotting of γH2AX after IR. WT and CHD1-KO cells were exposed to 5 Gy of IR. (F) γH2AX foci were analyzed and counted following IR by Immunofluorescence staining. Mean and SD from three separate experiments are shown. (G) Western blotting showing that CHD1 was successfully knocked down in T98G used in ChIP assays. (H) T98G cells were transfected with mock plasmid or p84-ZFN 72 h after siRNA treatment. Cells were fixed and chromatin samples were purified 18 h post-transfection. The percent of cells with DSBs was measured by qPCR. (I) γH2AX formation at a single DSB site created by ZFN nuclease in control siRNA- or siCHD1-treated T98G cells. The percentage of DNA immunoprecipitated by ChIP was normalized to input. (J) H2AX levels before and after ZFN cleavage at the DSB site. (K) The ratio of γH2AX levels were normalized to total H2AX levels from two ChIP experiments. All samples in the ChIP experiments were taken 18 h after p84-ZFN transfection.
CHD1 is essential for γH2AX formation early in the DNA-damage response
Our data so far show that CHD1 is critical for CtIP recruitment, and thus HR, which agree with a recent study by Kari et al., who also show reduced RPA and Rad51 foci because of impaired CtIP in CHD1 depleted cells(19). Yet, the CHD1-dependent events required for CtIP recruitment are unknown. We therefore focused on determining how CHD1 is linked to CtIP recruitment. One of the earliest events in the response to DNA damage is phosphorylation of the histone variant H2AX by PI3-kinases, such as the ATM kinase (27). Using our CRISPR-generated knockout RPE1 cells, we measured the formation and resolution of phosphorylated H2AX (γH2AX) following IR in WT and CHD1 KO cells. We first measured the levels of γH2AX by Western blotting of extracts from irradiated cells. We found that the levels of γH2AX in CHD1-KO cells were lower than in WT cells by Western blot (Figure 2E and also Supplementary Figure S4A). We then examined γH2AX foci formation following IR and found that the CHD1-KO cells exhibit delayed γH2AX foci formation following IR, starting as early as 5 min (Figure 2F). The CHD1-KO cells also exhibit a lower plateau in the number of γH2AX foci 1 h after IR. In addition, the CHD1-KO cells appear to have a slower resolution of γH2AX foci, since 45% (or average 16 foci) of γH2AX foci were resolved in WT after 6 h (compared to 1 h) while only 34% of foci (or average 9 foci) were resolved in CHD1-KO cells (Figure 2F). The slower kinetics of γH2AX formation and resolution in CHD1-KO cells compared to WT cells are consistent with the reduced levels of HR in these cells (Figure 1D). Although the CHD1-KO cells appear have slightly increased number of γH2AX endogenous foci, presumably due to the impaired repair of the spontaneous DSBs in the CHD1-KO cells, statistical analysis revealed no significant difference. These findings suggest that CHD1 plays a key role in the early DNA damage response by modulating γH2AX levels. In contrast, Kari et al. did not observe a difference in γH2AX formation (1 h after IR) (19). The discrepancy may be explained by the lower efficiency of shRNA compared to CRISPR-Cas9 knock out, or the fact that earlier time points are needed to determine γH2AX formation.
To confirm CHD1 is critical for the formation of γH2AX at DSB sites, we introduced a single DSB in T98G cells using a zinc finger nuclease (ZFN) and then performed chromatin immunoprecipitation (ChIP) experiments using a γH2AX specific antibody, as previously described (28). Following immunoprecipitation, the purified DNA was analyzed by qPCR using multiple primers flanking both sides of the break. For this analysis, we used siRNA-depleted CHD1 cells or control siRNA-treated cells to compare the levels of γH2AX flanking the break site in the presence or absence of ZFN. The efficiency of knockdown of CHD1 was confirmed by Western blotting (Figure 2G), and qPCR revealed that the ZFN nuclease created similar amount of DSBs in siControl- and siCHD1-treated cells (Figure 2H). In the ChIP experiments, we could clearly detect accumulated γH2AX around the break (0.1–50 kb) after introducing ZFN. Moreover, we found that the levels of γH2AX on both sides of the break were much lower in the siCHD1-treated cells compared to the siControl-treated cells (Figure 2I). These results were consistent across different data analysis methods (Supplementary Figure S3A, fold change of γH2AX) and experimental replicates (Supplementary Figure S3B). These data are also consistent with the γH2AX foci formation assays, and together, confirming that CHD1 is required for the robust accumulation of γH2AX at DSBs.
The decrease in the levels of break-induced γH2AX in CHD1-depleted cells could be explained by a decrease in the accessibility or phosphorylation of H2AX, or by a decrease in the overall levels of H2AX on the chromatin. To investigate these possibilities, we performed the ChIP experiments with antibodies against total H2AX (i.e. non-phosphorylated and phosphorylated) and compared the levels of H2AX in siCHD1 and siControl-treated cells. In the absence of the ZFN, we observed moderately lower levels of H2AX in the CHD1-depleted cells (Figure 2J and Supplementary Figure S3C and D), suggesting CHD1 is required for the efficient incorporation of H2AX. The difference in H2AX levels between the CHD1-depleted and control cells was greater after transfection of the ZFN, suggesting that CHD1 promotes stability of H2AX at DSBs (Figure 2J and Supplementary Figure S3C-D). When the γH2AX levels were normalized to H2AX in chromatin, we did not observe a difference in the ratio between siCHD1-treated and siControl-treated cells (Figure 2K), suggesting that CHD1 promotes incorporation and stability of H2AX in chromatin at DSBs but not the rate of H2AX phosphorylation. In agreement with these data, MRE11 recruitment to micro-irradiated DNA was not affected by CHD1 knockout (Supplementary Figure S4B and C), suggesting CHD1 functions after the MRN complex during DSB repair. These results suggest that CHD1 plays a role in H2AX incorporation and retention at the DSB, to ensure sufficient H2AX is present for phosphorylation and signaling.
The N terminus of CHD1 suppresses DNA binding
Our observation that loss of CHD1 leads to a DNA repair defect suggests other chromatin remodeling factors are not functionally redundant with CHD1 during HR. In particular, CHD2 shares ∼67% identity with CHD1 but appears non-redundant with CHD1 in our DNA repair experiments and, as reported, during development (29). To identify potentially unique regions of CHD1 that could distinguish it from CHD2 and other remodelers, we aligned the sequences of CHD1 and CHD2 and found that the N terminus of the two proteins are not well conserved (Supplementary Figure S5). Thus, we decided to investigate the role of the N terminus in regulating the activity of CHD1.
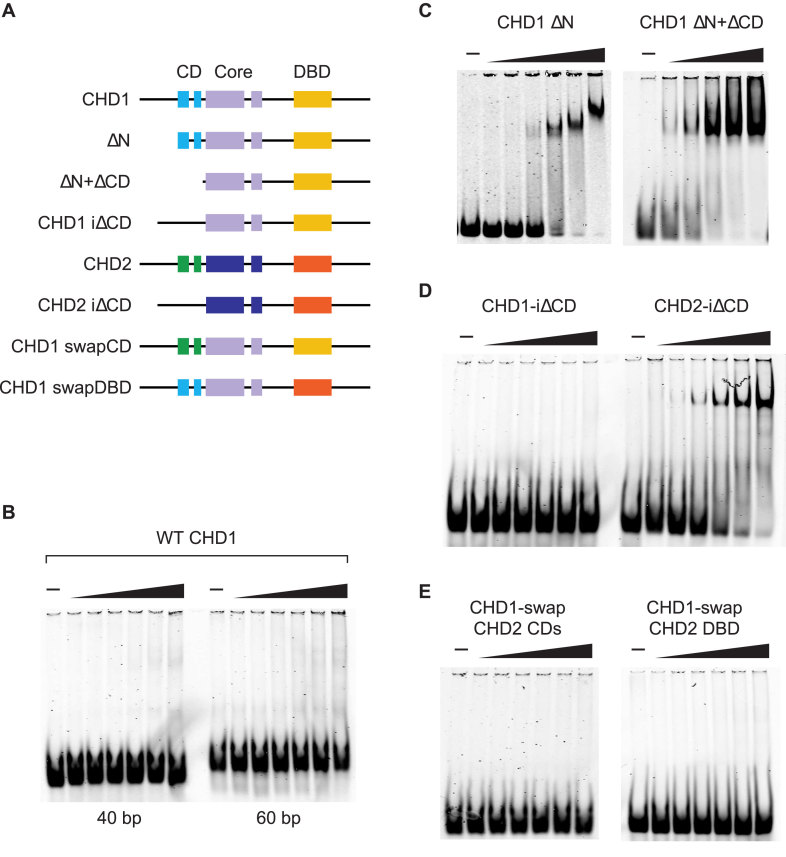
CHD1 and CHD2 belong to a subgroup of the CHD family largely due to the presence of a conserved SANT-SLIDE-like DBD near their C termini. The presence of this DBD implies that the interaction of CHD1 and CHD2 with naked DNA, such as at sites of under-assembled chromatin or long linker regions, is a core feature of their activities. We previously reported that CHD2 uses its DBD to bind DNA molecules that are greater than 30 bp in length (1). To investigate the binding of CHD1 to DNA, we cloned, expressed, and purified FLAG-tagged wild-type (WT) human CHD1 using a baculovirus expression system. In parallel, we expressed and purified a series of deletion and domain-swap constructs of CHD1 (Figure 3A). We first measured the binding of WT CHD1 to DNA by incubating purified CHD1 with IRDYE-labeled dsDNA that was either 40 or 60 bp in length and resolving the reactions by native PAGE. Unlike the strong binding to dsDNA that we previously observed for WT CHD2 (1), we were unable to observe binding by WT CHD1 to either the 40 or 60 bp probe (Figure 3B).
Figure 3.
The N terminus of human CHD1 suppresses binding to DNA. (A) A series of deletion and domain-swap proteins were purified and used in electrophoretic mobility shift assays. (B) Full-length WT CHD1 has low affinity for 40 or 60 bp dsDNA substrates. (C) Deletion of the N terminus (271 amino acids; ΔN) or the N terminus plus tandem chromodomains (ΔN+ ΔCD) leads to an increase in affinity to a 40 bp dsDNA probe. (D) An internal deletion of the tandem chromodomains of CHD1 (left; CHD1-i ΔCD) does not increase binding to the 40 bp probe. For comparison, internal deletions of the chromodomains of human CHD2 (right; CHD2-i ΔCD) does bind the 40 bp probe. (E) Domain-swap experiments in which the chromodomains (left; CHD1-swap CHD2 CD) or the DNA-binding domain (right; CHD1-swap CHD2 DBD) of CHD1 were swapped for those of CHD2 did not increase the affinity of CHD1 to the 40 bp probe, suggesting the suppression of DNA binding requires the N terminus of CHD1.
We predicted that the lack of apparent binding of the WT CHD1 protein to DNA was either due to the lack of strong DNA binding activity from the DBD or from repressive sequences in the protein that were inhibiting the DBD. We previously showed that deletion of the chromodomains and N terminus of CHD2 increases its affinity for DNA, and thus we constructed a series of deletion proteins of CHD1 to investigate whether the N terminus of CHD1 is repressing its binding to DNA. Moreover, because WT CHD2 binds DNA and CHD1 and CHD2 share homology within their chromodomains, we considered the possibility that sequences outside of the chromodomains were inhibiting the DNA binding activity of CHD1. An alignment of human CHD1 and CHD2 proteins show a high degree of identity across the two proteins except near their N and C termini (Supplementary Figure S5). We analyzed the binding of purified CHD1 proteins lacking either the N terminus (up to the chromodomains; ΔN) or lacking both the N terminus and chromodomains (ΔN + ΔCD) to the 40 bp DNA probe. We found that deletion of the N terminus alone leads to a substantial increase in DNA binding when compared with the binding by WT CHD1, while additional deletion of the chromodomains leads to a further increase in DNA binding (Figure 3C).
We next decided to examine how loss of just the chromodomains affects DNA binding and expressed CHD1 or CHD2 proteins which had an internal deletion of their chromodomains (iΔCD). CHD1-iΔCD did not show significant binding to the DNA and appeared similar to WT CHD1 (Figure 3D). In contrast, CHD2-iΔCD did show DNA binding activity that was more robust than previously reported for WT CHD2 (1). These results suggest that, unlike CHD2, the N terminus is the dominant domain that represses DNA binding activity of CHD1, as deletion of the N terminus alone stimulates DNA binding whereas deletion of the chromodomains alone does not. We next constructed domain-swap versions of CHD1 in which we replaced the CHD1 chromodomains or the DBD with those of CHD2 (CHD1-swap CHD2 CDs or CHD1-swap CHD2 DBD, respectively) and analyzed their binding to the 40 bp DNA probe (Figure 3E). In both cases, we were unable to observe DNA binding greater than WT CHD1, indicating that the reason WT CHD1 exhibits low affinity for DNA as compared to WT CHD2 is mostly due to the N terminus and not the chromodomains nor DBD.
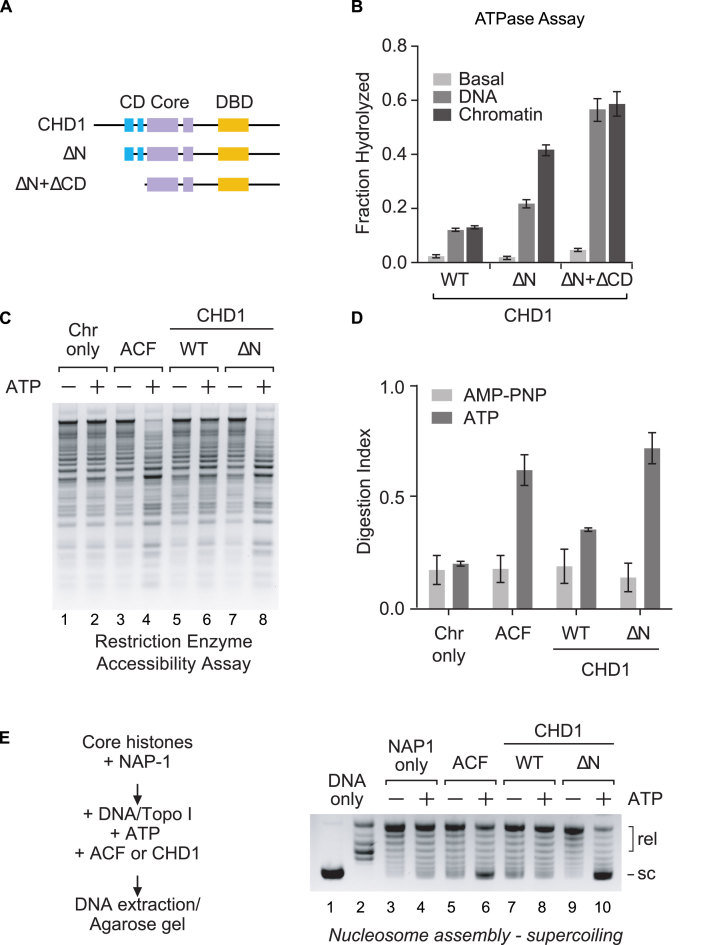
The N terminus represses CHD1 ATPase and remodeling activities
After determining that the N terminus of CHD1 represses its DNA binding activity, we next investigated whether the N terminus inhibits the ATPase activity of CHD1. We incubated purified WT CHD1, ΔN, and ΔN + ΔCD proteins (Figure 4A) with ATP and either plasmid DNA, plasmid chromatin, or without any cofactor and then measured the amount of ATP hydrolyzed (Figure 4B). We found that WT CHD1 shows little ATPase activity in the presence of plasmid DNA or chromatin and its activity is only modestly better than the basal ATPase activity (i.e. without DNA or chromatin cofactors). In contrast, deletion of the N terminus results in a robust increase in the ATPase activity in the presence of DNA and chromatin, with chromatin showing the most stimulation. Deletion of both the N terminus and chromodomains resulted in an additional stimulation by both DNA and chromatin, with DNA and chromatin stimulating the ATPase activity of CHD1 at almost equal levels. The jump in ATPase activity by plasmid DNA following the loss of the chromodomains suggests the chromodomains are repressing the ability of the DBD to bind DNA. We previously reported a similar finding for CHD2 containing a deletion of its N terminus and chromodomains (1).
Figure 4.
The N terminus represses the ATP-dependent activity of CHD1. (A and B) Deletion of the N terminus (ΔN) or N terminus and chromodomains (ΔN + ΔCD) leads to an increase in ATP hydrolysis. CHD1 proteins were incubated with plasmid DNA (DNA), plasmid DNA assembled into chromatin (Chromatin), or without any cofactor (Basal) in the presence of ATP (with 32-P-ATP). The ATPase assays were stopped and the hydrolyzed ATP resolved by thin-layer chromatography. The fraction of hydrolyzed ATP was quantified. (C) Deletion of the N terminus (ΔN) leads to an increase in chromatin remodeling activity in a restriction enzyme accessibility assay. CHD1 or ACF (as a positive control) were incubated with pre-assembled chromatin which contains 15 restriction sites for the enzyme HaeIII. The reactions also included ATP or AMP-PNP (as a minus ATP control). Following digestion with HaeIII, the DNA was deproteinized, purified, and resolved by agarose gel electrophoresis. (D) Quantification of the digestion for the REA assays, as shown in C (mean ± SD; n = 3). (E) Human CHD1 exhibits ATP-dependent chromatin assembly activity that is repressed by the N terminus. Core histones (0.4 μg) were pre-incubated with the histone chaperone NAP-1 (2 μg). Naked plasmid DNA (0.4 μg), Topo I, and WT or ΔN CHD1 (1 μg) was then added. Finally, an ATP regeneration system containing either ATP or AMP-PNP (as a minus ATP control) was included. The chromatin assembly factor ACF (1 μg) was used as a positive control. The plasmid DNA was then purified and the degree of chromatin assembly was monitored by DNA supercoiling and agarose gel electrophoresis.
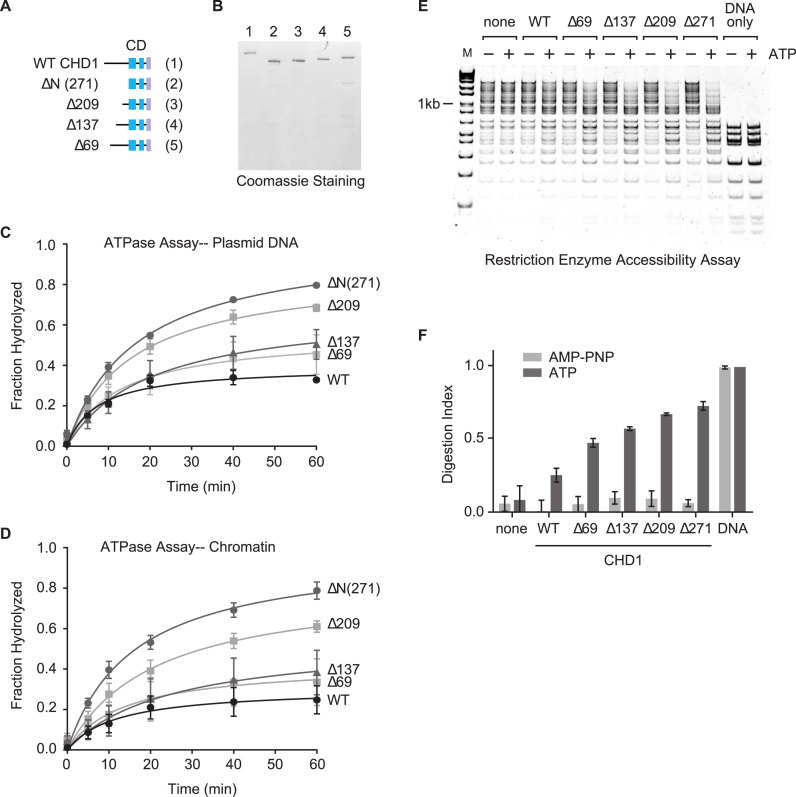
To investigate the remodeling activities of human CHD1, we first performed a general chromatin remodeling assay to determine whether CHD1 is an active remodeling protein. We performed restriction enzyme accessibility (REA) assays using plasmid-assembled chromatin that contains 15 HaeIII restriction sites, as previously described (1). The presence of nucleosomes occludes some of the restriction sites and, in the absence of any remodeling activity, the plasmid chromatin is only partially digested by the restriction enzyme (Figure 4C, lanes 1–2). More restriction sites are accessible in the presence of a remodeling factor (e.g. ACF) and ATP, leading to an increase in the digestion of the chromatin (Figure 4C, lanes 3–4). Using this remodeling assay, we found that WT CHD1 exhibits low remodeling activity, as indicated by the minimal increase in digestion in the presence of ATP (Figure 4C, lanes 5–6). Deletion of the N terminus leads to a noticeable increase in the remodeling activity (Figure 4C, lanes 7–8), and the amount of chromatin digestion in the presence of ΔN was similar to that observed with ACF (Figure 4D).
CHD1 has chromatin assembling activity that is restricted by the N terminus
Several studies have examined the remodeling activities of yeast and Drosophila CHD1, but little is known about the remodeling activity of human CHD1. Drosophila and yeast CHD1 proteins are capable of assembling periodic arrays of nucleosomes and along with ISWI and human CHD2 are the only known remodeling factors that can assemble chromatin (1,2,30). We next determined whether purified human CHD1 can assemble chromatin using a chromatin assembly assay. We incubated core histones with the histone chaperone NAP1, and then added plasmid DNA, topoisomerase I (TopoI), ATP, and either ACF or CHD1. We then extracted the DNA and analyzed the amount of supercoiling by agarose gel electrophoresis; plasmid DNA that is assembled into chromatin will be supercoiled and migrate faster in the gel (e.g. Figure 4E, lane 1) while unassembled DNA will be relaxed (e.g. Figure 4E, lane 2). In the absence of a chromatin assembly factor, only minimal supercoiling of the DNA is observed (Figure 4E, lanes 3–4). In the presence of the assembly factor ACF, which contains the ISWI ATPase, we observed an increase in DNA supercoiling in reactions that also contain ATP (Figure 4E, lanes 5–6). We then examined WT CHD1 and found that reactions containing WT CHD1 do not generate supercoiled DNA, indicating WT CHD1 is a poor chromatin assembly factor under the conditions we tested (Figure 4E, lanes 7–8). In contrast, the ΔN deletion shows significant ATP-dependent chromatin assembly activity, as indicated by the majority of the template DNA becoming supercoiled (Figure 4E, lanes 9–10). The results from the chromatin assembly assay mirrored those of the REA chromatin remodeling assay and suggest that the ATP-dependent activities of WT CHD1 are strongly repressed by its N terminus.
The CHD1 N terminus suppresses the activity of CHD1 in a step-wise manner
We predict that unique sequences within the N terminus of CHD1 but not CHD2 act to repress the activities of CHD1, and an alignment of the N termini of CHD1 and CHD2 show only partial conservation between the two factors (Supplementary Figure S5). The N termini of both CHD1 and CHD2 are serine-rich; the N terminus of CHD1 contains 26% serine while the N terminus of CHD2 contains 20% serine. The highest numbers of serines are present in the extreme N terminus with 33 of the first 69 (48%) amino acids of CHD1, and 29 of the first 73 (40%) amino acids of CHD2 being serine. In addition to the serine-rich regions, CHD1 and CHD2 both contain patches of basic and acidic residues. Despite the lack of a clearly distinguishable region in the N terminus that could repress the activity of CHD1, we decided to better map the repressive regions by generating a series of deletion proteins in which we removed the first 69 (Δ69), 137(Δ137), 209 (Δ209) amino acids, or the whole N terminus before the chromatin domains (ΔN, 271 aa) (Figure 5A and B). ATPase assays with these truncated or WT proteins were then performed in the presence of DNA or chromatin (Figure 5C and D). We found that deletion of amino acids up to 137 (i.e. the Δ69 and Δ137 proteins) showed only a minimal increase in ATPase activation when compared to WT CHD1. Deletion of the first 209 amino acids (i.e. Δ209) yielded a more significant increase in ATPase activity, and further deletion up to the chromodomains (i.e. ΔN) showed maximum stimulation. We also measured the ability of the deletion proteins to remodel chromatin. Using the REA chromatin remodeling assay, we observed a stepwise increase in remodeling activity that was consistent with the increase in ATPase activity (Figure 5E and F).
Figure 5.
A step-wise loss of the N terminus of CHD1 leads to a length-dependent increase in enzymatic activity. (A and B) A series of deletions from the N terminus was expressed and purified, similar to the WT CHD1 protein. The schematic only highlights the N terminus although the full-length proteins minus the deletion were expressed. Time-course ATPase assays using either plasmid DNA (C) or a chromatin substrate (D) as a cofactor were performed using the N terminal deletion proteins. (E) Successive deletions from the N terminus of CHD1 lead to a step-wise increase in the remodeling activity of CHD1 as measured by REA. (F) Quantification of the REA assays shown in (E). Mean and SD of the digestion index (DI) from three experiments are shown.
Deletion of the N terminus stimulates the DNA repair activity of CHD1 in cells
To determine whether the N terminus of CHD1 plays a suppressive role in cells, we compared the ability of WT and ΔN CHD1 to rescue the repair defects observed in the CHD1-KO cells. We first measured the kinetics of γH2AX formation following IR by Western blotting in our CHD1-KO cells ectopically expressing WT or ΔN CHD1 delivered by retroviral infection (Figure 6A). We found that both WT and ΔN CHD1 can restore the slow kinetics of γH2AX in CHD1-KO cells (Figure 6B and C). Reintroducing WT and ΔN can also restore the recruitment of CtIP to laser-induced DNA damage (Figure 6D and E). For both γH2AX kinetics and CtIP recruitment, we consistently observed an increased rescue response with the ΔN CHD1 when compared to WT CHD1. These findings demonstrate that the N terminus of CHD1 is not required for the DNA repair activities of CHD1 and likely play a suppressive role in regulating CHD1 in human cells.
Figure 6.
CHD1-ΔN showed hyperactivity compared to the WT CHD1 in cells. (A) Western blotting showing that expression of CHD1-WT and CHD1-ΔN in the CHD1-KO cells is close to the endogenous CHD1 levels. (B) Representative Western blotting for γH2AX following IR in CHD1-KO cells rescued by expression of CHD1-WT and CHD1-ΔN. (C) Quantification of the Western blots, such as in (B), across replicate experiments. Normalized γH2AX signal was shown. (D) CtIP recruitment to laser stipes was measured in the CHD1-WT or CHD1-ΔN rescued cells. Cells were fixed 10 min after laser striping and stained for CtIP and γH2AX. (E) Quantification of the rescue experiments in (D). Mean and SD from three experiments were shown. ns, not significantly different; *,p < 0.05.
DISCUSSION
In this study, we discovered that CHD1 is required for the efficient repair of DSBs through HR. We show that loss of CHD1 results in decreased global incorporation of H2AX and decreased retention of H2AX at DSBs, which accounts for the reduced phosphorylation of H2AX and impaired CtIP recruitment to the DSBs. We also identified the N-terminal region of CHD1 as a novel autoinhibitory domain that regulates its biochemical activities and repair functions in cells.
Although chromatin remodelers have been implicated in the repair of DSBs, the mechanisms through which they regulate repair remain to be understood. The related factor, CHD2, has been reported to play a key role in the NHEJ pathway (21), and knockdown of the SWI/SNF BRG1 and BRM remodeling complexes sensitizes cells to DSB-inducing agents (31). Likewise, the results from our clonogenic survival assays demonstrate that knockout of CHD1 sensitizes cells to IR. Our GFP reporter assays show CHD1 is critical for efficient HR but not for NHEJ (Figure 1), suggesting distinct, non-redundant roles of CHD1 and CHD2 in DSB repair. We further show that CHD1 is required for full CtIP recruitment (Figure 2) to facilitate HR. Our data are consistent with a recent study using prostate cancer cells that showed CtIP recruitment involves CHD1 (19); however, the underlying mechanism linking CHD1 to CtIP was not known, nor whether CHD1 is involved in γH2AX foci formation (i.e. earlier than 1 h after IR). In our study, we have extended our understanding of the roles of CHD1 in DSB repair by showing that CHD1 functions in HR prior to CtIP recruitment, as early as the formation of γH2AX foci (Figure 2). Since γH2AX signaling is an initiating event in the HR pathway, our data place CHD1 as one of the earliest enzymes involved in the HR pathway.
CHD1 is one of the most frequently deleted genes in prostate cancer (22,32). PARP inhibitors have been used successfully in treating HR-deficient cancers, such as BRCA1 mutated ovarian tumors. Our data show that CHD1-null cells are hypersensitive to the PARP inhibitor Olaparib (Figure 1F), suggesting prostate cancer patients with alteration in CHD1 gene may respond to PARP inhibition. In addition, we demonstrated that CHD1-null cells are hypersensitive to PTEN inhibition (Figure 1F). These results are in agreement with a recent report that show PTEN mutant cells are hyper-dependent on CHD1 (20), and indicate that a potent PTEN inhibitor may be useful in the treatment of CHD1-deficient tumors. Alternatively, small molecule inhibitors against CHD1 may be an effective way for treating PTEN-mutated cancers.
We also provide mechanistic data connecting CHD1 with γH2AX signaling. We showed that CHD1 promotes stabilization of H2AX at DSBs to ensure sufficient accumulation of γH2AX for HR signaling. Knockdown of CHD1 reduces H2AX accumulation on chromatin even in the absence of a DSB, suggesting CHD1 promotes global H2AX incorporation to chromatin. We also found that CHD1 stabilizes H2AX near DSBs, thereby facilitating H2AX phosphorylation.
It has been reported that the chromatin around DSBs is decondensed to facilitate processing of the break ends, a process involving eviction of histones (33,34). In contrast, H2AX is transiently yet rapidly stabilized at DSBs, through a mechanism that involves chromatin incorporation and stabilization of H2AX at the breaks, mediated by the ATM kinase, the sirtuin protein SIRT6, and the chromatin remodeler ISWI (35). Our data show a reduction of H2AX on chromatin at DSBs after CHD1 knockdown (Figure 2J), suggesting CHD1 may have a similar role as ISWI in the establishment of specific chromatin states.
We also performed in-depth analysis of the biochemical activities of CHD1. In contrast to CHD2, which readily hydrolyzes ATP to ADP in the presence of DNA or chromatin (1), WT CHD1 exhibits little to no ATPase activity by itself (Figure 4B). In addition, WT CHD1 has no detectable chromatin assembly capability (Figure 4E) and only weak remodeling activity (Figure 4C–D). This may be due to the low affinity that WT CHD1 has for dsDNA substrates or because of distinct auto-repressive features of the proteins (Figure 3). Since CHD1 and CHD2 have high sequence homology except at their N termini, the N terminus of CHD1 is likely to be a key inhibitory region of the protein. Indeed, removal of the N terminus drastically enhances the DNA binding affinity, ATPase activity, and chromatin assembly and remodeling activities of CHD1 (Figures 3–5).
Our biochemical assays show that the N terminus domain of CHD1 inhibits the activities of the enzyme. The N terminus is separate from the chromodomains, and its inhibitory nature in the human CHD1 protein contrasts that of the yeast protein, for which the chromodomains have been reported to regulate its ATPase activity (36,37). Similar inhibitory domains in the N terminus of ISWI have been reported (36,38).
In contrast to our biochemical assays, which suggest that WT CHD1 does not have much activity by itself, we were able to rescue the repair defects of CHD1-KO cells with both WT and ΔN CHD1. WT CHD1 and ΔN both restored the capability of γH2AX formation and CtIP recruitment of CHD1-KO cells (Figure 6). Reintroducing ΔN rescued the impairment of CtIP recruitment of CHD-KO cells better than re-expressing CHD1-WT. These findings suggest that a mechanism exists in vivo to relieve the inhibitory effects of the N terminus. This leads us to speculate that in cancer cells, such as in prostate tumors, mutations in the regulatory N terminus may cause deregulation of CHD1.
In summary, we show that the DNA binding, ATPase, chromatin assembly, and remodeling activities of CHD1 are largely restrained by the N terminus. Our study also revealed a new role of CHD1 in DNA double-strand break repair, which is to ensure the sufficient H2AX incorporation in chromatin and stabilization of H2AX at DSBs and thus proper formation of γH2AX. The fact that homozygous deletion of CHD1 is associated with increased rates of homozygous deletions (32) is consistent with our finding that CHD1 is important for accurate repair of DSBs by HR. While CHD1 is one of the most frequently deleted genes in prostate cancer, it has also been shown that knockdown of CHD1 is synthetically lethal in PTEN-deficient tumors (20). Therefore, the development of drugs that inhibit CHD1 via stabilization of its N terminus may provide therapeutic benefit to patients with PTEN-deficient tumors while minimizing the chance of targeting other chromatin remodelers such as CHD2.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Chowdhury lab for providing us with the Cas9-expressing RPE1 cells. We thank Dr Benjamin Manning for critical reading of the manuscript.
Author Contributions: J.Z., B.D.P. and T.Y. designed the experiments. J.Z., J.L. and T.Y. wrote and edited the manuscript. All authors commented on the manuscript. All authors were involved in conducting the experiments and acquiring and analyzing the data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [GM114304 to T.Y., CA64585, CA177884 to B.D.P.]. Funding for open access charge: TBD.
Conflict of interest statement. None declared.
REFERENCES
- 1. Liu J.C., Ferreira C.G., Yusufzai T.. Human CHD2 is a chromatin assembly ATPase regulated by its chromo- and DNA-binding domains. J. Biol. Chem. 2015; 290:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ito T., Bulger M., Pazin M.J., Kobayashi R., Kadonaga J.T.. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997; 90:145–155. [DOI] [PubMed] [Google Scholar]
- 3. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bennardo N., Cheng A., Huang N., Stark J.M.. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008; 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eisen J.A., Sweder K.S., Hanawalt P.C.. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995; 23:2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Flaus A., Martin D.M.A., Barton G.J., Owen-Hughes T.. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006; 34:2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gorbalenya A.E., Koonin E.V.. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993; 3:419–429. [Google Scholar]
- 8. Marfella C.G., Imbalzano A.N.. The Chd family of chromatin remodelers. Mutat. Res. 2007; 618:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F.. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007; 446:572–576. [DOI] [PubMed] [Google Scholar]
- 10. Cheung V., Chua G., Batada N.N., Landry C.R., Michnick S.W., Hughes T.R., Winston F.. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008; 6:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mavrich T.N., Jiang C., Ioshikhes I.P., Li X., Venters B.J., Zanton S.J., Tomsho L.P., Qi J., Glaser R., Schuster S.C. et al. Nucleosome organization in the Drosophila genome. Nature. 2008; 453:358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pointner J., Persson J., Prasad P., Norman-Axelsson U., Strålfors A., Khorosjutina O., Krietenstein N., Peter Svensson J., Ekwall K., Korber P.. CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe. EMBO J. 2012; 31:4388–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smolle M., Venkatesh S., Gogol M.M., Li H., Zhang Y., Florens L., Washburn M.P., Workman J.L.. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012; 19:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valouev A., Johnson S.M., Boyd S.D., Smith C.L., Fire A.Z., Sidow A.. Determinants of nucleosome organization in primary human cells. Nature. 2011; 474:516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hennig B.P., Bendrin K., Zhou Y., Fischer T.. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep. 2012; 13:997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Dieuleveult M., Yen K., Hmitou I., Depaux A., Boussouar F., Bou Dargham D., Jounier S., Humbertclaude H., Ribierre F., Baulard C. et al. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016; 530:113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Piatti P., Lim C.Y., Nat R., Villunger A., Geley S., Shue Y.T., Soratroi C., Moser M., Lusser A.. Embryonic stem cell differentiation requires full length Chd1. Sci. Rep. 2015; 5:8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burkhardt L., Fuchs S., Krohn A., Masser S., Mader M., Kluth M., Bachmann F., Huland H., Steuber T., Graefen M. et al. CHD1 Is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 2013; 73:2795–2805. [DOI] [PubMed] [Google Scholar]
- 19. Kari V., Mansour W.Y., Raul S.K., Baumgart S.J., Mund A., Grade M., Sirma H., Simon R., Will H., Dobbelstein M. et al. Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep. 2016; 17:1609–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao D., Lu X., Wang G., Lan Z., Liao W., Li J., Liang X., Chen J.R., Shah S., Shang X. et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature. 2017; 542:484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luijsterburg M.S., de Krijger I., Wiegant W.W., Shah R.G., Smeenk G., de Groot A.J.L., Pines A., Vertegaal A.C.O., Jacobs J.J.L., Shah G.M. et al. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell. 2016; 61:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang S., Gulzar Z.G., Salari K., Lapointe J., Brooks J.D., Pollack J.R.. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene. 2012; 31:4164–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shenoy T.R., Boysen G., Wang M.Y., Xu Q.Z., Guo W., Koh F.M., Wang C., Zhang L.Z., Wang Y., Gil V. et al. CHD1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann. Oncol. 2017; 28:1495–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Konstantinopoulos P.A., Ceccaldi R., Shapiro G.I., D’Andrea A.D.. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015; 5:1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P.. Human CtIP promotes DNA end resection. Nature. 2007; 450:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J.. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001; 276:42462–42467. [DOI] [PubMed] [Google Scholar]
- 28. Xu Y., Ayrapetov M.K., Xu C., Gursoy-Yuzugullu O., Hu Y., Price B.D.. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell. 2012; 48:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E. et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009; 460:863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robinson K.M., Schultz M.C.. Replication-independent assembly of nucleosome arrays in a novel yeast chromatin reconstitution system involves antisilencing factor Asf1p and chromodomain protein Chd1p. Mol. Cell. Biol. 2003; 23:7937–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park J.-H., Park E.-J., Lee H.-S., Kim S.J., Hur S.-K., Imbalzano A.N., Kwon J.. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J. 2006; 25:3986–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu W., Lindberg J., Sui G., Luo J., Egevad L., Li T., Xie C., Wan M., Kim S.T., Wang Z. et al. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene. 2012; 31:3939–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lans H., Marteijn J.A., Vermeulen W.. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet. Chromatin. 2012; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Price B.D., D’Andrea A.D.. Chromatin remodeling at DNA double-strand breaks. Cell. 2013; 152:1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Atsumi Y., Minakawa Y., Ono M., Dobashi S., Shinohe K., Shinohara A., Takeda S., Takagi M., Takamatsu N., Nakagama H. et al. ATM and SIRT6/SNF2H mediate transient H2AX stabilization when DSBs form by blocking HUWE1 to allow efficient gammaH2AX foci formation. Cell Rep. 2015; 13:2728–2740. [DOI] [PubMed] [Google Scholar]
- 36. Clapier C.R., Cairns B.R.. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature. 2012; 492:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hauk G., McKnight J.N., Nodelman I.M., Bowman G.D.. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol. Cell. 2010; 39:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ludwigsen J., Pfennig S., Singh A.K., Schindler C., Harrer N., Forne I., Zacharias M., Mueller-Planitz F.. Concerted regulation of ISWI by an autoinhibitory domain and the H4 N-terminal tail. Elife. 2017; 6:e21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.