Abstract
Initiation is the rate-limiting step in translation. It is well-known that stable structure at a ribosome binding site (RBS) impedes initiation. The ribosome standby model of de Smit and van Duin, based on studies of the MS2 phage coat cistron, proposed how high translation rates can be reconciled with stable, inhibitory structures at an RBS. Here, we revisited the coat protein system and assessed the translation efficiency from its sequestered RBS by introducing standby mutations. Further experiments with gfp reporter constructs assessed the effects of 5′-tails—as standby sites—with respect to length and sequence contributions. In particular, combining in vivo and in vitro assays, we can show that tails of CA-dinucleotide repeats—and to a lesser extent, AU-repeats—dramatically increase translation rates. Tails of increasing length reach maximal rate-enhancing effects at 16–18 nucleotides. These standby tails are single-stranded and do not exert their effect by structure changes in the neighboring RBS stem–loop. In vitro translation and toeprinting assays furthermore demonstrate that standby effects are exerted at the level of translation initiation. Finally, as expected, destabilizing mutations within the coat RBS indicate an interplay with the effects of standby tails.
INTRODUCTION
Bacterial protein synthesis has been studied extensively for decades, mostly in enterobacterial systems like Escherichia coli. Generally speaking, all steps in translation can contribute to the output of polypeptides. However, even though elongation rates, e.g. via codon bias effects, contribute to overall protein synthesis [e.g. (1,2)], it is clear that translation initiation is rate-limiting for protein synthesis (3–6).
In the canonical mode of enterobacterial translation initiation, the 30S ribosomal subunit binds to the translation initiation region (TIR) of an mRNA which comprises all elements required for initiation to take place (6,7). Proper start site selection usually involves a base-pairing interaction between the Shine-Dalgarno (SD) sequence in the mRNA and the antiSD sequence in the 16S rRNA (8,9). An appropriately spaced downstream AUG (or GUG/UUG) start codon binds fMet-tRNAfmet and GTP-bound initiation factor IF2 to set the reading frame. This complex is now ready for 50S subunit joining, recruitment of the ternary complex EF-Tu/ aa-tRNA, followed by the elongation phase. Sometimes, sequences called translational enhancers (often A/C-, A/U-, or U-rich, e.g. (10–16), and protein factors such as ribosomal protein S1 (13,17,18) modulate the efficiency of initiation. Alternative modes distinct from canonical initiation are known. E.g., leaderless mRNAs, lacking SD sequences, can be efficiently translated (19–21) in an S1-independent manner, and a 70S-scanning initiation mechanism has been reported (22).
The ribosome binding site (RBS), defined as the region within the mRNA that is protected by the bound 30S subunit in the initiation complex, was reported to extend from positions –18 to +10 relative to the start codon (9). Hydroxyl radical footprinting indicated protection from –35 to +19 (23), reflecting RNA backbone contacts and the two base-pairing interactions (SD-antiSD; tRNA-start codon). This extensive interaction area suggests that stable structures within an RBS should impair binding and interfere with initiation. Mutational as well as global analyses indeed support this and, moreover, show that local structure rather than the stability of an SD-antiSD interaction predicts output. For example, extensive mutagenesis of GFP reporter constructs indicated that low local structure from position +4 to +38 after the AUG correlates with high GFP synthesis. Instead, rare codons, the strength of an SD, or the frequency of optimal codons failed to explain the results (24). Similarly, structure-destabilizing mutations from the start codon to +25 increase in vitro translation (25), and stable structures within the first five codons impair translation (26,27). A bias towards low local structure within the RBSs of most genes was also observed in global studies, and high translation rates—assessed by ribosome profiling—countercorrelate with RBS structure (28–30). A ‘ramp model’ suggested that rare codons near the start of a coding sequence (CDS) promote high expression. However, since rare codons are biased towards A or U in their third positions, they decrease the propensity of forming stable structures (31,32).
Since the accessibility of an RBS affects protein output, translational regulation often involves induced RNA structure change (33). For example, proteins, small ‘antisense’-type RNAs, metabolites, pH, or temperature [e.g. (34–41)] can change mRNA structure to modulate translation initiation rates. Other cases involve translational coupling in which upstream translation irons out a stably folded downstream RBS (42,43).
Work by de Smit and van Duin (44) on an RNA bacteriophage showed an inverse relationship between the efficiency of translation of the MS2 coat protein and the thermodynamic stability of an RNA stem–loop that encompasses its RBS. Each mutation that changes the ΔG°-value by –1.4 kcal/mol gives a tenfold change in expression (45). That is, ribosomes compete with folding and need to capture an RBS in its unfolded state. However, the calculated ΔG°-value of the wild-type RNA structure [–11 kcal/mol; (46)] raised a problem: the fraction of time the stem–loop would spend in an unfolded state, exposing the RBS, is in the microsecond range. This is incompatible with significant coat protein translation, considering known 30S association rates and the time the RBS is in its unfolded state; calculations predicted >10 000-fold lower coat protein translation (47,48). The authors solved this apparent paradox by suggesting the ‘standby’ model. Sequence-nonspecific binding of 30S subunits to accessible single-stranded regions, in the vicinity of a stably structured RBS stem–loop, kinetically overcomes the effect of the inhibitory structure. 30S subunits pre-bound to the mRNA can access the RBS as soon as it is transiently unfolded (‘breathing’). This turns a second-order into a first-order reaction, simply requiring the relocation of the ribosome from the standby site to the proper start site (48,49). Replacing a thermodynamically based model by a kinetically driven mechanism resolved the coat protein paradox.
Based on structure predictions of the coat protein RBS, de Smit and van Duin (49) suggested that several upstream and downstream unstructured regions, adding up to 45–50 nucleotides altogether [i.e. roughly the size of a 30S footprint (23)], might act as standby sites. Some studies suggested that 30S ribosomes are able to bind single-stranded surface areas even when interrupted by structural elements (50,51). More recent work has provided biochemical and genetic support for the standby model. Studer and Joseph (52) used a fluorescence-based assay to study binding of the 30S subunit to short model mRNAs. 30S initiation complexes failed to form on an RNA that consisted of a stable stem–loop comprising both the SD and the start codon. However, the addition of a single-stranded 5′-oligo-U-tail (12 nt) promoted initiation complex formation, even though the stability of the stem–loop was unchanged (52). In a second study, our lab addressed the function and regulation of the bacterial toxin-antitoxin system tisB/istR-1 (53,54). In short, the toxin RBS is embedded in a very stable structure, and a single-stranded sequence ≈100 nt upstream serves as a standby site which is required to permit translation. Sequestration of this standby site, either by formation of a structure element in the 5′-UTR of tisB mRNA, or by binding of the antisense antitoxin RNA IstR-1, prevents translation (53,55,56). As in the Studer and Joseph study (52), the standby site promotes translation, even though the inhibitory structure of the RBS is unchanged.
In this study, we revisited the MS2 standby model, and performed proof-of-principle studies on a simplified standby reporter based on the coat RBS structure and a GFP readout system. In vivo and in vitro results confirm the de Smit and van Duin conclusions concerning the complexity of the natural standby site (48,49). In our test system, translation of GFP initiating from only the coat RBS stem–loop was at background level, but the addition of unstructured 5′-tails conferred dramatic enhancement of GFP translation. A systematic assessment of an AU- and CA-series of tails shows that both sequence and length of a tail determines expression levels.
MATERIALS AND METHODS
Plasmid constructions
All primers are listed in Supplementary Tables S1 and S2. Constructs were derived from the pSC101* plasmid pXG-10 carrying a PLTetO-driven lacZ-gfp translational fusion (57). The lacZ-segment is bordered by NsiI (immediately downstream of the promoter) and NheI sites (at the start of gfp). Plasmid pXG-10 DNA was digested with both enzymes and dephosphorylated. For pMS2 and the truncated series (pMS2–0 to -11; Figure 2), PCR-fragments were amplified from plasmid pK2, containing the MS2 coat gene (44), using various primers (Supplementary Tables S1 and S3), generating flanking NsiI and NheI sites. After digestion with both enzymes, each fragment was inserted into the vector; the coat start codon is in frame with gfp (Figure 1A).
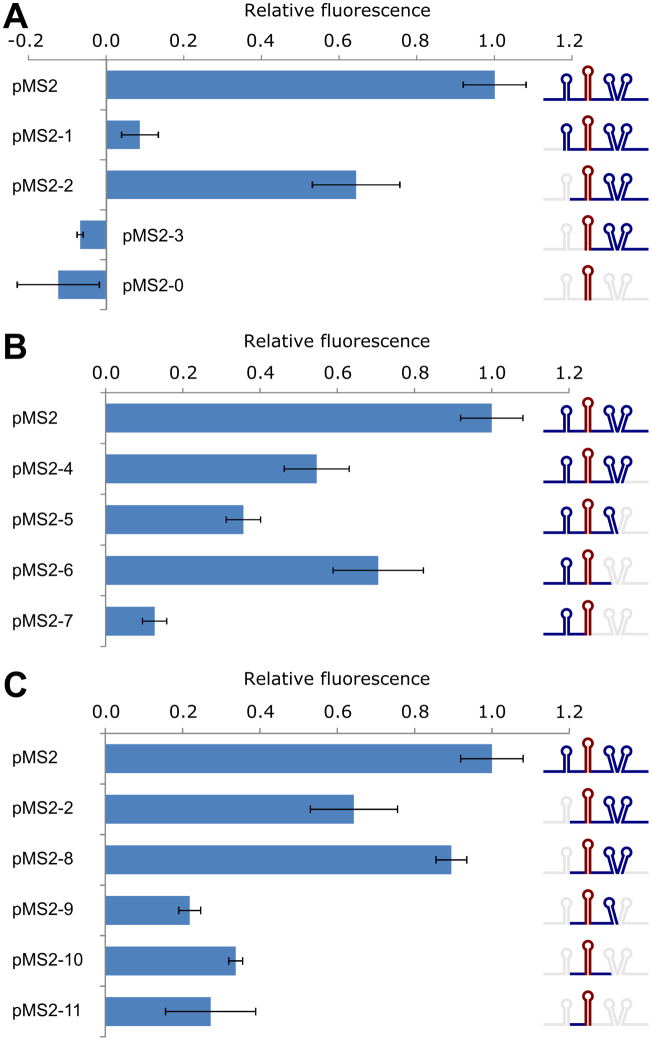
Figure 2.
RNA segments flanking the coat RBS stem–loop contribute to standby activity. Expression of gfp was assessed by fluorescence measurements in cells carrying various deletion plasmids (Materials and Methods). Inserts had flanking segments deleted as shown schematically, either upstream (A), downstream (B), or on either side of the RBS stem–loop (C). Experiments were done in biological triplicates, and error bars show the standard error.
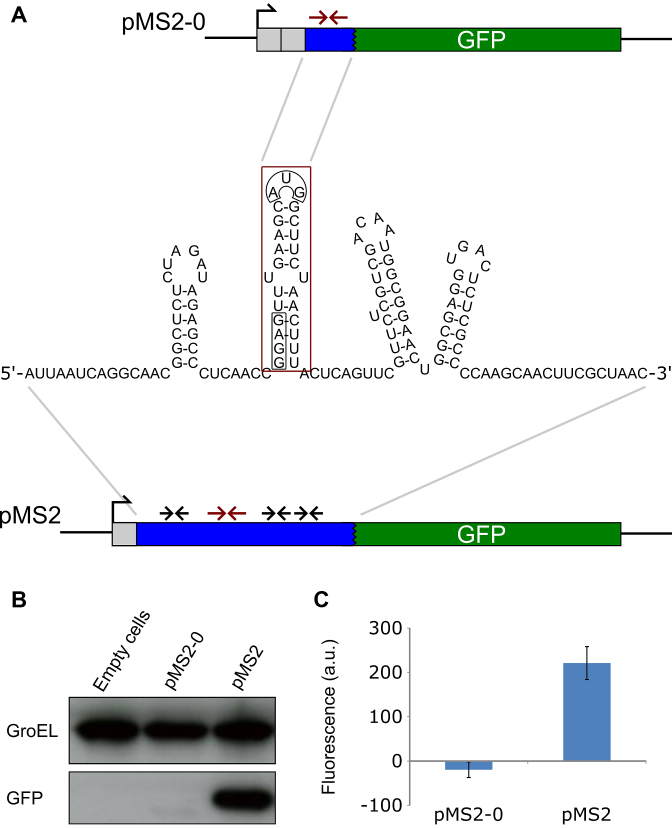
Figure 1.
Experimental design and test of reporter system. (A) The DNA sequence encoding the sequestered RBS of the MS2 coat protein (red box) was translationally fused to gfp (green) either including (pMS2) or lacking (pMS2–0) the relatively unstructured flanking regions. The coat SD sequence and start codon are boxed (black). Grey squares indicate restriction sites. Red and black arrows indicate inverted repeat sequences. (B) Expression levels of gfp as detected by Western blot. Detection of GroEL was used as a loading control. This gel is representative of five replicates. (C) Expression levels of gfp as detected by plate reader. Fluorescence was measured as in Materials and Methods. Growth curves were similar between strains. The data are averages of three biological replicates. Error bars indicate the standard error.
Plasmids with 5′-tails immediately preceding the MS2 RBS stem–loop (Figure 3 and further), were created as follows. First, a PCR fragment generated from plasmid pEH87 (58) using primers EHO-444 and EHO-445 was circularized by ligation to create plasmid pEH108. The pEH108 plasmid was PCR amplified with Phusion polymerase (NEB) using phosphorylated outward primers (Supplementary Table S4), and the PCR product was circularized by ligation. Plasmids generated by this strategy all contain: promoter::AA::standby-tail::MS2-RBS::site-NheI::GFP.
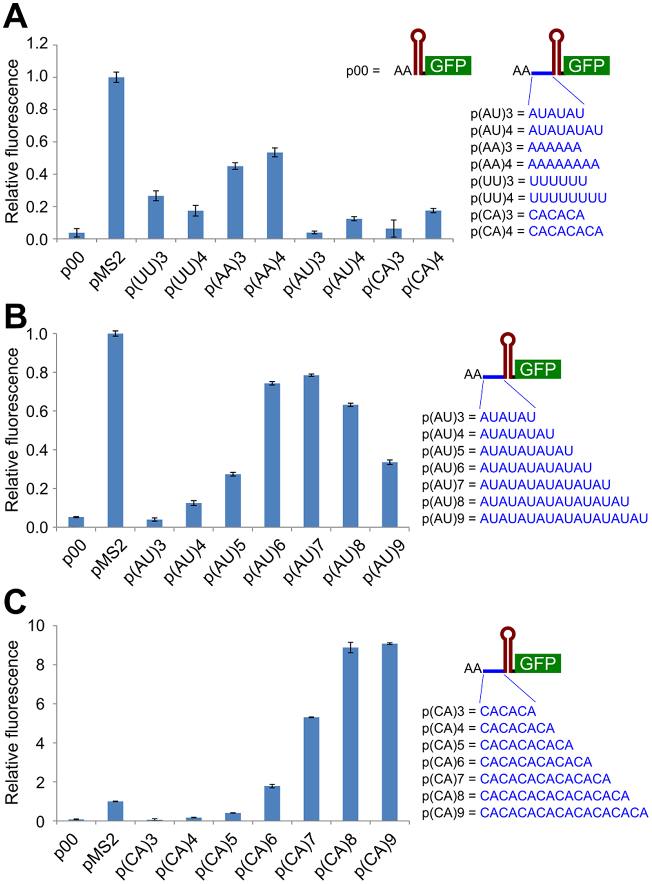
Figure 3.
Short sequences in front of the inhibitory RBS affect translation rates. The DNA sequence of the coat RBS stem–loop (in red) was translationally fused to gfp (green). Short sequences were inserted upstream of the stem–loop, immediately downstream of two A residues (positions +1 and +2 in mRNA). These were mono- or dinucleotide repeats of six or eight nucleotides (A), increasing numbers of (AU)-repeats (B) or of (CA)-repeats (C). Fluorescence levels are shown relative to that of the reference strain carrying pMS2 (see Figure 1), with error bars for three biological replicates.
Microplate reader experiments
Cell cultures (biological triplicates) were grown in LB medium containing chloramphenicol (15 μg/ml) for 2–4 h at 37°C, and then diluted 50×. Technical triplicates of each sample were used in 96-well plates. Control wells contained only LB medium ± chloramphenicol, or plasmid-free cells. Plates were incubated for 20 h at 37°C in a microplate reader (Infinite M200 Pro; Tecan). Cell density (OD600) and fluorescence (excitation at 480 nm, emission at 520 nm) were measured at 5 min intervals, after 1 min of shaking.
Data analysis was done by subtracting the average background cell density from all cell density measurements, and subtracting the background fluorescence from all samples separately for each time point. The fluorescence values at time points at which OD600 was ≈0.4 (late exponential phase; this OD value measured in the plate reader corresponds to a proper OD value of 1.4) were used for further calculations: the fluorescence/cell density ratio was calculated, and the corresponding values from empty cells was subtracted from all other samples. Averages were normalized against the positive control (pMS2). Standard errors were calculated as the standard deviation divided by the square root of the number of biological replicates.
Western analysis of in vivo translation
For in vivo analysis of GFP-fusion protein levels, cells were grown in LB containing chloramphenicol (15 μg/ml) until an OD600 of 0.5. One ml of cells was centrifuged, and pellets resuspended in 50 μl protein loading buffer (Fermentas #R0891). Samples were denatured for 3 min at 90°C and separated on 10% denaturing PAGE in running buffer (25 mM Tris, pH 8.8; 190 mM glycine; 0.1% SDS). Proteins were transferred to Immobilion-P PVDF membranes (Millipore) o/n at 35 mA at 4°C in transfer buffer (20% methanol; 25 mM Tris; 300 mM glycine; 0.01% SDS) using an electroblotting system (Bio-Rad Trans Blot). Membranes were blocked for 1 h in 20 ml of 3% BSA in PBST (PBS with 0.1% Tween20) and then incubated in 10 ml of 3% BSA in PBST, containing conjugated anti-GFP-HRP (Horseradish Peroxidase; 1/5,000; Miltenyi Biotec) or anti-GroEL-HRP (1/50,000; Sigma) for 1 h. Membranes were washed 3 × 5 min in PBST and 2 × 5 min in PBS. Blots were developed for 1–5 min in ECL Prime (GE Healthcare) and exposed in a PMI™ system (Bio-Rad).
In vitro transcription
For use in translation experiments, DNA sequences were PCR-amplified from gfp-fusion plasmid templates. Forward primers contained the T7-promoter sequence, one G for transcriptional efficiency, and the 5′-part of the gene. The reverse primer EHO-715 binds 30 nucleotides downstream of the gfp stop codon. (For template and primers used, see Supplementary Table S5.) For structure probing, reverse primer EHO-828, positioned ≈80 nucleotides into gfp, was used to create a shorter mRNA variant (Supplementary Table S5). PCR products were transcribed at 37°C by 400 U of T7 RNA polymerase (Ambion) in the presence of 7 mM of each NTP in transcription buffer (40 mM Tris–HCl [pH 7.5], 30 mM MgCl2, 11 mM DTT, 2 mM spermidine, 0.01% Triton X-100) in a total volume of 200 μl. DNA templates were degraded by 2 μl of DNase I (1 U/μl; Thermo Scientific) for 15 min at 37°C. In vitro transcribed RNA was extracted twice with acidic phenol/chloroform/isoamylalcohol (25/24/1) and once with chloroform/isoamylalcohol (24/1), followed by ethanol precipitation. Pellets were washed with 70% ethanol and resuspended in water. Nucleotides were removed using Illustra MicroSpin G-50 columns (GE Healthcare). RNA concentrations were measured by Nanodrop, and RNA quality was validated by agarose gel electrophoresis.
In vitro translation assay
Full-length in vitro transcribed mRNA (gfp-fusions; 7.5 pmol) in a total volume of 6 μl was pre-incubated for 3 min at 90°C and 2 min on ice, and then diluted in TMN buffer (20 mM Tris, 5 mM Mg-acetate, 100 mM NaCl [pH 7.5]) to a total volume of 7.5 μl. For each reaction, 3 μl of the pre-incubation mix was added to 7 μl of translation mix (PURExpress In Vitro Protein Synthesis kit; NEB #E6800). For the experiments in Figure 6, pre-incubation mixes contained 50 pmol of either an antisense (MS213) or control (MS073) oligo. Translation was performed for 30 min at 37°C, and reactions were stopped by adding protein loading buffer (Fermentas #R0891) on ice. Samples were run on a 10% PAGE, followed by Western blot as described.
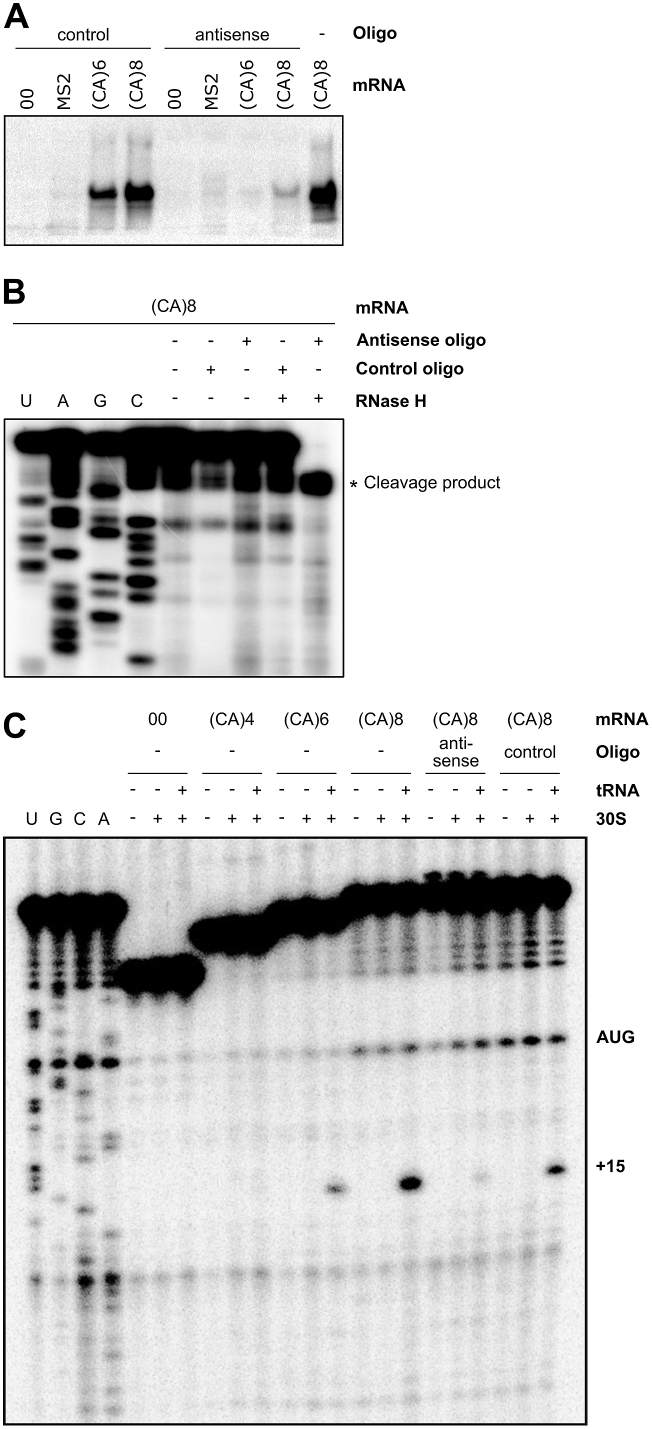
Figure 6.
The 5′ standby tails need to be single-stranded to promote translation and initiation complex formation. (A) The effect of blocking of the (CA)6 and (CA)8 standby tail by an antisense oligo (at 6.7 times molar excess), or a control oligo, was assayed by in vitro translation. The mRNAs used are indicated, and GFP was detected by Western blot. (B) Formation of a heteroduplex between the antisense oligo and the standby tail of (CA)8 mRNA was tested by RNase H cleavage (Materials and Methods). Cleavage was observed after reverse transcription using a 5′-labeled oligo annealed downstream. Cleavage near the base of the stem is observed only in the presence of the antisense oligo and RNase H (far right lane). (C) The toeprint experiment was conducted on several mRNA variants, with 30S and tRNAfMet, with or without antisense or control oligo (Materials and Methods), as indicated. The position of the characteristic toeprint at +15 is shown. UAGC shows a sequencing ladder for reference. The gel is representative of three technical replicates.
Radioactive labeling of oligodeoxyribonucleotides
20 pmol of an oligo was incubated for 30 min at 37°C with γ-[32P]-ATP and 10 U of T4 polynucleotide kinase (Thermo Scientific) in (50 mM Tris–HCl [pH 7.6], 10 mM MgCl2, 5 mM DTT, 0.1 mM spermidine) in a total volume of 20 μl. After incubation for 10 min at 80°C, free γ-[32P]-ATP was removed using an Illustra MicroSpin G-50 column (GE Healthcare).
RNase H cleavage assay
7.5 pmol of in vitro transcribed RNA and 50 pmol of antisense (MS213) or control (MS073) oligo were incubated for 10 min at 37°C in TMN buffer in a total volume of 15 μl. RNase H (5 U; Thermo Scientific) and RNase H buffer (200 mM Tris–HCl [pH 7.8], 400 mM KCl, 80 mM MgCl2, 10 mM DTT) were added to a total volume of 20 μl, and incubated for 15 min at 37°C. Samples were phenol-extracted and precipitated with ethanol. Redissolved RNA was denatured in the presence of 5′-end-labeled oligo MS294, and reverse transcribed in the presence of dNTPs (0.5 mM each) and SSII RT (200 U/μl; Thermo Scientific) in SB-Mg10 buffer (10 mM Tris-acetate [pH 7.6], 1 mM DTT, 100 mM K-acetate, 10 mM Mg-acetate) for 30 min at 37°C. The cDNA was phenol-extracted and precipitated with ethanol. Sequencing reactions were performed as for the toeprinting experiment, and samples were separated by 7.5% denaturing PAGE. Detection was done using a Phosphoimager screen and a PMI™ system (Biorad).
Toeprinting analysis
For each 4 μl reaction, an annealing mixture contained 0.2 pmol of in vitro transcribed RNA and 0.4 pmol of [32P]-end-labeled oligo MS294 (complementary to +41 to +57 in mRNA) in SB1x buffer (10 mM Tris-acetate [pH 7.6], 1 mM DTT, 100 mM K-acetate). When specified, 10 pmol of an antisense (MS213) or control (MS073) oligo were included. Annealing mixtures were incubated for 1 min at 90°C and then placed on ice. Initiation complexes in a final volume of 8 μl were formed by adding 3.2 μl of the annealing mixtures to dNTPs (final concentration 625 μM each) and 30S subunits (250 nM) in SB-Mg10 buffer (10 mM Tris-acetate [pH 7.6], 1 mM DTT, 100 mM K-acetate, 10 mM Mg-acetate). After pre-incubation for 10 min at 37°C, 2 μl of tRNAfMet was added to a final concentration of 0.4 μM and the mixture incubated continued for 15 min at 37°C. Reactions without 30S or tRNA received equivalent volumes of SB1x-Mg10 buffer. For reverse transcription, 200 U of SuperScript III RT (Thermo Scientific) were added for 15 min at 37°C. To stop the reactions, 110 μl of 300 mM Na-acetate was added on ice. After phenol- extraction and ethanol precipitation, pellets were dissolved in loading buffer (0.02% xylene cyanol, 0.02% bromophenol blue in formamide). Corresponding sequencing ladders were generated using mRNA, with the same RT primer as above, and SuperScript RT, but in reactions containing 25 μM of one dNTP and 1.7 μM of one ddNTP for chain termination (4 different reactions) and 100 μM of each other dNTP. The cDNA products were run on 7.5% sequencing gels which were fixed in 10% ethanol and 6% acetic acid for 5 min, transferred onto Whatmann paper 3MM, and dried at 80°C. Detection of bands was by Phosphoimager as above.
RNA structure probing
2 pmol of gel-purified mRNA was denatured for 1 min at 95°C and chilled on ice for 2 min. Renaturation was done in native buffer (10 mM Hepes [pH 7.5], 1 mM Mg acetate, 100 mM K acetate) at 37°C for 10 min, followed by addition of 2 μg of carrier tRNA (ThermoFisher Scientific, #AM7119). RNase V1 probing used a final concentration of 0.01 U/μl (ThermoFisher Scientific, #AM2275) for 5 min at 37°C. Reactions were stopped by addition of ice-cold sodium acetate to 0.3 M. RNAs were phenol-extracted and precipitated with ethanol. Lead(II) acetate (Sigma, #316512) was used at 6.25 mM final concentration for 2.5 min at 37°C. Reactions were stopped with EDTA, final concentration 35 mM. RNAs were directly precipitated with 3 vol of ethanol. RNA pellets were washed twice with 70% ethanol and redissolved in water, with 5′-end-labeled oligo EHO-828 and dNTPs (0.5 mM each). Samples were heated at 65°C for 3 min and cooled to 4°C. Reverse transcription was done at 48°C for 25 min with 100 U of SuperScript® IV RT and corresponding buffer (ThermoFisher Scientific, #18090010). The cDNAs were phenol-extracted, precipitated, and washed with ethanol. Sequencing reactions were performed as for toeprinting. The cDNA products were run on a 7.5% sequencing gel, and processed for analysis as above.
RESULTS
The standby site of the coat protein gene of phage MS2 is required for efficient translation in vivo
First, we revisited the MS2 model system to systematically assess the contribution of sequence and structure elements to standby. Earlier work had shown that deletions upstream of the coat cistron decreased translation (47,59), and the relatively unstructured regions surrounding the coat RBS were proposed as a standby site (49). The predicted structures around the coat protein start site are shown in Figure 1A. The corresponding DNA segment, including the RBS stem–loop, was translationally fused to GFP (Figure 1A; pMS2) on a low-copy pSC101* plasmid (for details, see Materials and Methods). A second fusion only contained the coat RBS stem–loop, lacking the unstructured flanking regions (Figure 1A; pMS2–0). The reporter protein GFP was detected by Western blot, and GFP fluorescence in cells was in parallel monitored in a microplate reader. Cells carrying plasmid pMS2 expressed high levels of GFP, shown both by Western blot and plate reader, whereas cells carrying pMS2–0 supported insignificant GFP synthesis (Figure 1B and C). Thus, the absence of the flanking regions caused translation to drop to background levels.
Subsequently, elements of the flanking regions around the coat RBS in pMS2 were systematically deleted, and gfp expression levels examined in the microplate reader. When parts of the region upstream of the sequestered start site were removed, expression levels decreased, and complete removal of upstream sequences completely abolished expression (Figure 2A). For deletions of the downstream elements, we ensured that all fusions remained in frame. Also here, gfp expression decreased as more segments were deleted (Figure 2B). Generally, downstream deletions mostly gave smaller effects than upstream deletions. Additionally, deletions of both upstream and downstream segments (Figure 2C) indicate that unstructured regions on both sides contribute to wild-type, high translation rates, congruent with the model of de Smit and van Duin (48). Overall, it appears that removal of 5′-tails contributes more to decreased output than most other deletions (cf. Figure 2A pMS2 versus pMS2–1, pMS2–2 versus pMS2–3). It cannot be excluded that differences in mRNA stability in part contribute to these results (but see below).
Short sequences in front of the coat RBS stem–loop enhance translation in vivo
The experiments above are consistent with the non-sequence-specific binding sites surrounding the translation initiation site being essential for translation (48). However, the complexity of this system makes is difficult to pinpoint the contributions of particular sequences or structural motifs to standby and, consequently, translation rates. We designed a simpler, more easily manipulated system, to assess features of putative standby sites that affect translational efficiency. Six or eight nucleotides were inserted immediately upstream of the inhibitory coat RBS in plasmid pMS2–0 (Figure 3A). As constructed, all encoded mRNAs carried two A’s at their 5′-ends (Materials and Methods). For simplicity, and to minimize the putative effects of alternative secondary structures, we inserted homopolymers or dinucleotide repeats. These included an oligo-U insert similar to the one in (52) that stimulated initiation complex formation on a stable RBS.
The in vivo levels of GFP were assessed using a plate reader. Figure 3 shows the expression values of cells carrying each of the fusion plasmids in late exponential phase (at the same cell density). The pMS2 construct with the ‘wild-type standby site’ (49) was chosen as an arbitrary reference for levels of expression and normalization. The results indicate that some 5′-tails, but not others, increase gfp expression over the background obtained with a tail-less construct (p00, Figure 3A), yet lower than the reference fusion pMS2. Most 10 nt tails (AA plus an inserted 8-mer) enhanced translation slightly more than those of 8 nt, suggesting a length-dependence. At this point, these results could not distinguish between effects due to sequence motifs, secondary structure, or both.
Increasing numbers of AU- and CA-repeats strongly enhance expression levels in vivo and in vitro
Since Figure 3A suggested a length-dependence of putative standby sites, a more systematic set of constructs was tested. AU- and CA-repeats have a low propensity for forming stable secondary structure. Moreover, ACA-motifs adjacent to the RBSs of yifK and dppA stimulate translation initiation (60), and the sRNA GcvB represses its target mRNAs by binding to upstream C/A-rich regions, which may serve as translational enhancers (15). AU-rich sequences in 5′-UTRs also sometimes enhance translation (13,14). The series of plasmids created had dinucleotide repeats, AU or CA (ranging from 6–18 nt), inserted between the AA 5′-end of the mRNA and the sequence of the inhibitory RBS stem–loop. Fluorescence levels are shown relative to the reference strain, carrying pMS2 (Figure 3B and C). For both AU- and CA- repeats, translation levels show length-dependent translation-enhancing activity. For AU-repeats, translation levels increased, reached a maximum from AU(5–8) (12–18 nt tails), and then decreased slightly. With the CA-series, dramatically higher expression was obtained, with top levels for (CA)8 being nine-fold higher than for the reference (pMS2); note the difference in scale between Figure 3B and C. Again, a length-dependence was observed, with a maximum at 18 nt tails. Clearly, the two dinucleotide repeat tails promote strikingly different gfp expression levels.
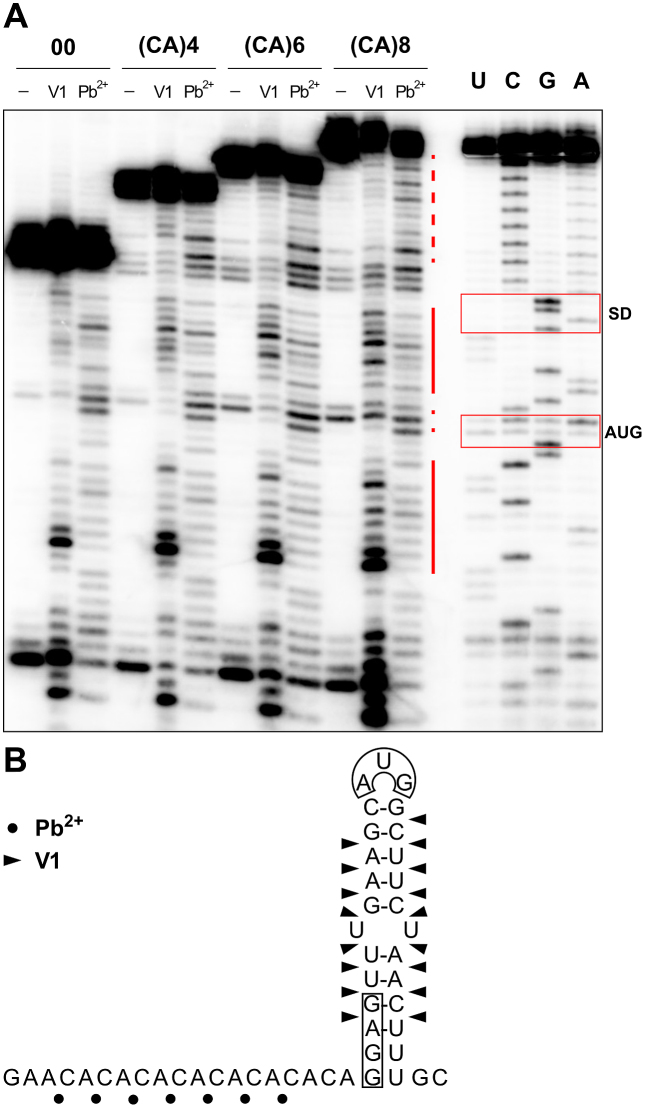
In order to test whether the tails may enhance translation by changing the structure of the inhibitory stem–loop, structural probing was conducted on the 5′-segments of in vitro transcribed mRNAs derived from plasmids p00, p(CA)4, p(CA)6 and p(CA)8. The cleavage patterns observed with RNase V1 (specific for double-stranded RNA) and lead(II), [specific for single-stranded RNA (61)], indicated no change within the RBS stem–loop in all four tested RNAs (Figure 4). RNase V1 cleavages mapped to the stem, whereas lead(II) cleavages were strongest in the loops and the CA-tails. Thus, the structure of the RBS stem–loop is maintained upon addition of these 5′-tails, suggesting that the observed effects (Figure 3) are not due to structural rearrangements. Structure predictions of these and other mRNAs furthermore suggest that the gfp sequences downstream do not affect the structures of the 5′ regions (Supplementary Figure S1).
Figure 4.
Addition of CA-repeats does not change the secondary structure of the coat RBS stem–loop. (A) Enzymatic and chemical structure probing was conducted on control mRNA (00) or mRNAs with tails of 4, 6 or 8 (CA)-repeats, (see Materials and Methods), as indicated. The mRNAs were mock-treated (lanes ‘–’), partially digested with double-strand-specific RNase V1 (V1), or treated with lead(II) acetate (Pb2+). UCGA: sequencing reactions on (CA)8 mRNA. The position of the SD and AUG start codon are indicated by red boxes. Regions of reactivity toward RNase V1 (red solid line) and lead(II) acetate (red dashed line) are indicated on the autoradiogram. (B) The localization of RNase V1 (filled triangles) and lead(II) acetate (black dots) cuts are shown on the secondary structure of the 5′-segment of (CA)8 mRNA. Black boxes: SD and AUG.
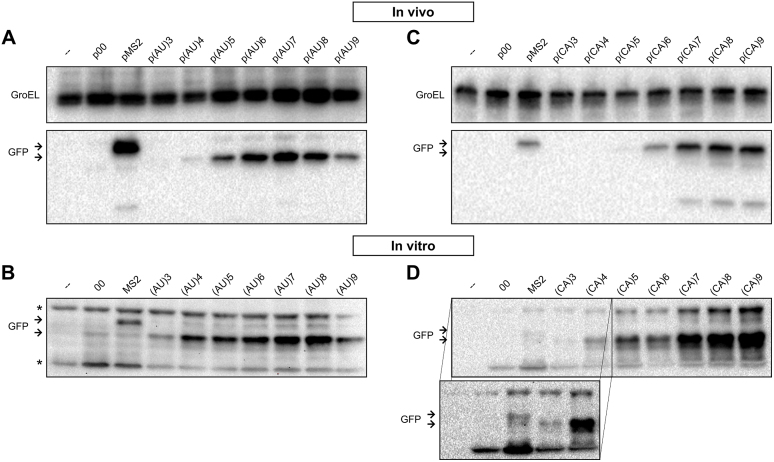
The stimulatory effect on GFP expression in vivo by addition of single-stranded tails in front of the structured MS2 RBS could be either due to enhanced translation rates, effects on mRNA turnover, or a combination of both. To disentangle these possibilities, we tested if translation-enhancing effects of 5′-tails on GFP expression in vivo were consistently observed in an in vitro translation system. In vitro translation assays contained purified ribosomes and all other components needed, and added mRNAs. GFP production was measured after 30 min; pilot experiments showed that translation rates were linear past the sampling time. The mRNAs were derived from the same series of plasmids used for in vivo experiments, and translation rates were assessed by Western blot. These experiments are shown in Figure 5B and D for the AU- and CA-series, respectively. The in vitro results largely recapitulated the pattern of fluorescence readouts for strains with the corresponding fusion plasmids in vivo (compare to Figure 3B and C), and a Western analysis of GFP protein produced in vivo (Figure 5A and C). Together, these experiments indicate that the effects observed in vivo are not due to differential mRNA degradation, since mRNAs are stable in the in vitro assay (PURExpress, NEB; Materials and Methods). Both in vitro and in vivo, CA-tails were far superior over AU-tails, and similar length maxima for an enhancement effect were supported. In quantitative terms, in vitro effects were somewhat more pronounced for the CA-series (Figure 5D). The reason for this is unclear, but it may be related to the absence of other mRNAs to compete for ribosomes.
Figure 5.
Western analysis of translation rates on mRNAs with AU- and CA-tails in vivo and in vitro. GFP levels were monitored by Western analysis either of total protein (in vivo) extracted from cells carrying plasmids of the AU-series (A) and CA-series (C), or after in vitro translation of purified mRNAs from the AU-series (B) and CA-series (D). Detection used an antibody against GFP, and subsequently an anti-GroEL antibody (loading control). Arrows show the position of the longer GFP fusion protein (from pMS2), and all other GFP products (same size). The expanded box in (D) shows a longer exposure of this part of the gel, since the output from the CA-series was much stronger (see Results). Asterisks indicate unspecific antibody binding to proteins in the assay mix.
Blocking of the standby site inhibits translation in vitro
The experiments so far suggested that the translation-enhancing effects of 5′-tails reflected their accessibility as standby sites by their single-stranded character, modulated by sequence-dependent effects. If so, rendering these tails double-stranded should prevent standby binding and block translation. This was tested in the in vitro translation system by pre-incubation with either an oligodeoxyribonucleotide (hereafter ‘oligo’), 5′-TGTGTGTGTG (antisense), complementary to the CA-region of the 5′-tail, or a control oligo, 5′-TATTGATCGC, lacking complementarity anywhere in the mRNA. Figure 6A shows that the matching antisense DNA oligo almost completely abolished translation, whereas the control oligo had no effect. Thus, accessibility of the single-stranded CA-tail is required for standby and efficient translation. An RNase H cleavage assay verified that the antisense oligo, but not the control oligo, had base-paired as expected (Figure 6B).
Formation of translation initiation complexes requires a single-stranded standby site
The above results suggest that translation initiation on the stem–loop-only mRNA is ineffective due to impeding structure, but that a CA-tail of sufficient length can overcome inhibition. To directly monitor the effect of CA-tails on initiation complex formation, a toeprint analysis was conducted on several mRNAs with increasing number of CA repeats as standby sites. Figure 6C shows that the initiation complex formed readily when 8 CA-repeats preceded the sequestered start site, whereas it fails to do so on the mRNA lacking 5′-tails (Figure 6C, cf. CA(8) and 00). The toeprint signal was weaker on (CA)6 RNA, consistent with lower activity than (CA)8 in all previous experiments (Figures 3C, 5A and C). Congruent with the in vitro translation results (Figure 6A), annealing of the matching antisense DNA oligo, but not the control oligo, significantly blocked initiation complex formation (Figure 6C). Together, these experiments show that single-stranded 5′-tails act as standby sites to enable initiation complex formation on an otherwise inaccessible RBS stem–loop.
The presence of standby sites and structural destabilization contribute to translation efficiency
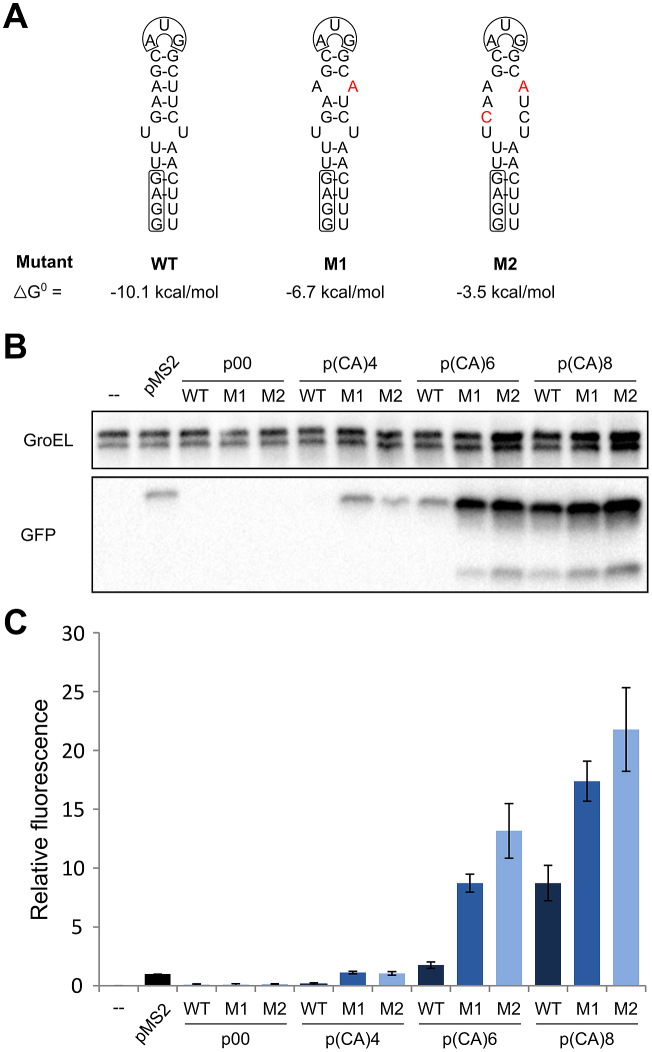
The above results showed the contribution of CA- and AU-tails to translation efficiency relative to each other, and relative to the ‘natural’ pMS2 standby construct. This did not address how standby enhancement of translation rates relates to gfp expression when the stem–loop structure is destabilized. Therefore, point mutations were introduced to destabilize the sequestered stem–loop (Figure 7A). The predicted ΔG0 value for the stem–loop was –10.1 kcal/mol [according to (62) version 3.5 at 37°C], and –6.7 and –3.5 kcal/mol for mutants M1 and M2, respectively.
Figure 7.
Effects of destabilizing the inhibitory RBS stem–loop. (A) Structure predictions for the coat RBS stem–loop and the two mutant variants. Calculated ΔG° values are shown (acc. to reference (62) version 3.5 at 37°C). (B) Western blot showing the effects of stem–loop destabilization in the absence or presence of standby tails in vivo. GroEL served as loading control. (C) Relative gfp expression obtained with the same strains as in (B) measured in the microplate reader. Error bars are given as the standard error for three biological replicates.
Interestingly, the constructs with a destabilized stem–loop, but without standby tail (p00-M1 and p00-M2; Figure 7B), failed to confer higher levels of gfp translation compared to a construct with the sequestered start site (p00; Figure 7B). As these constructs carry the SD sequence almost immediately downstream of the 5′-end of the mRNA, this might impede translation initiation. We therefore introduced the destabilizing mutations in several constructs that encode 5′-tails. Upon destabilization of stem–loops preceded by short tails, translation levels increased dramatically over that of the tail-less mRNA (Figure 7B; (CA)4/6-M1/2). In all cases, destabilizing the stem–loop (from WT to M1) had quantitatively similar effects as adding four additional nt of standby tail (Figure 7B), as can be seen from the relative expression based on microplate reader measurements (Figure 7C). Together, these results show that the effects of both standby tails and accessibility of the RBS together contribute to translation efficiency.
DISCUSSION
Formation of an initiation complex is the rate-limiting step in translation, and structural accessibility at/around RBS sequences is the major determinant for protein output (1,24,25,28,29,32,33,63). The pioneering work by de Smit and van Duin (44–49) proposed a solution for the paradoxical finding that high translation rates can sometimes be achieved even from a very stable RBS stem–loop. Here, we revisited the standby model in its original MS2 coat sequence context, and—subsequently using a simplified experimental set-up—addressed some of its mechanistic aspects.
First, GFP reporter constructs were employed to measure translation in the context of the original coat standby site (Figure 1A), and truncated variants thereof. As predicted (49), a complex set of single-stranded sequences surrounding the coat RBS stem–loop is required to overcome inhibition and to obtain high translation rates (Figure 2). Systematic deletion of sequence/ structure elements gradually decreased expression to background levels, in line with an on-site standby model (49). Similar contributions of several single-stranded sequences (‘surface area’) in a different gene context also suggested them to be standby sites contacted by initiating ribosomes (64). In several papers, the Salis lab has developed a biophysical model to predict translation initiation rates (27,64–66). It is worth noting that our operational definition of standby is slightly different. Here, as in the de Smit and van Duin interpretation, a standby site permits substantial translation from a stably sequestered RBS which, on its own, is inactive. In the Salis papers, binding free energy components of standby, SD-antiSD, AUG-tRNAfMet, as well as the SD-AUG spacing, each contribute to a total ΔG0-value that predicts translation rate. The Salis lab initially defined standby sequences as the four nt preceding the SD (66), and more recently the combined surface area contacted by ribosomes (27,64).
For mechanistically more tractable in vitro and in vivo experiments, we used simpler standby sites consisting of 5′-tails inserted upstream of the stable coat RBS. The rationale for using upstream sequences stems from several observations. Firstly, a 5′-U-tail works as a standby site in vitro (52). Secondly, a genome-wide RNA structure study suggested a single-stranded ‘docking site’; a stretch of sequences ≈20 nt upstream of the AUG start codon tends to be accessible (28). Thirdly, this region coincides with the binding site of ribosomal protein S1, about 11 nt upstream of the SD according to cryo-EM (67). S1 is implicated in binding to single-stranded RNA on and off the ribosome (51,68).
Using selected series of homopolymeric and dinucleotide repeat sequences as tails, the contributions of tail sequence and length were analyzed. Strikingly, increasing lengths of AU- and CA-repeat tails strongly enhance translation in vivo and in vitro (Figures 3B, C and 5A–D). For both CA- and AU-repeats, 16–18 nt tails give maximum activity, with ≈11-fold higher values obtained for the CA-series than for the AU-series (Figure 3B and C). Moreover, (CA)8 even promoted activity exceeding that of the natural standby site in pMS2 by 9-fold (Figure 3C). Thus, sequence matters, as does length, and a low structure content can be inferred. Incidentally, an oligo-U tail of 12 nt, like the one in (50), enhanced expression to the level obtained with the reference pMS2 (data not shown).
Structure mapping experiments (Figure 4) suggest that addition of single-stranded tails does not affect expression through RBS structure change. It is worth noting that simply adding any 5′-tail does not increase translation. Out of 100 random 12 nt insertion tails [length as (CA)6], only few enhanced expression, whereas most gave background levels (Supplementary Figure S2).
The good agreement between fold-effects of the CA-series in in vitro- and in vivo-assays (Figures 3B and C; 5A–D) argues that mRNA decay rates do not contribute much to the results in cells. This is because the in vitro assay (PureSystem) monitors translation-only effects; added mRNAs are not degraded throughout the incubation time. Thus, protein output primarily or exclusively reflects differences in translation rates. The validity of translation-only assays can be limited if coupled transcription-translation effects are expected. However, translation of coat protein occurs on MS2 RNA after infection, and thus is well-mimicked in our experimental setting.
The in vitro experiments corroborate that standby activity depends on single-strandedness. An oligo that base-paired to a CA-standby tail, but not a control oligo, strongly and specifically decreased translation (Figure 6A) as well as initiation complex formation (toeprint; Figure 6C). Furthermore, since quantitatively similar rate-increasing effects of CA-tails were observed in vivo (Figures 3C and 5C), in cell-free translation assays (Figure 5D), and in toeprinting experiments (Figure 6C), this confirms that standby acts on initiation rates.
In a final set of experiments, the effect of tails was tested in conjunction with a destabilized coat RBS (Figure 7A). An interplay is suggested by the results in Figure 7, shown as Western (Figure 7B) and plate reader results (Figure 7C): short standby tails in (CA)4 failed to significantly activate translation in the absence of destabilizing mutations. Upon tail extension to a moderately standby-active (CA)6, destabilization gave an additional 5- to 8-fold increase. With a highly active (CA)8 tail, only a 2- to 2.5-fold activation was due to destabilization. Overall, the result that both structure, and a standby site, contribute to total translation rate is unsurprising. Why the M2 mutation, predicted to give >100-fold lower folding than M1, does not increase translation further is unclear. In any case, the data are congruent with the de Smit and van Duin model (48) in that standby sites can compensate for inhibitory structure at the RBS.
What is the nature of a standby site? As our results show, in agreement with available literature (44–48,52,53), a single-stranded segment of RNA in the vicinity of a structured RBS fits the bill. Two issues are of concern. Firstly, in spite of strong circumstantial evidence for standby, no studies have so far biochemically detected a 30S ribosome transiently bound to a standby site. We are currently addressing this by mapping of RNA-segments crosslinked to standby ribosomes on the tisB mRNA (53). Secondly, several studies reported generally increased translation rates—not specifically in the context of structured RBSs—that were attributed to so-called translational enhancers. For example, U-rich regions bind 30S ribosomes in vitro (10,52) and act as translational enhancers of rnd mRNA in vivo (10). Similarly, AU-rich 5′-regions were shown to affect ribosome binding and enhance translation (11,69). Also, several CA-rich sequences in front of mRNAs increase translation (15), and an ACA-motif near the yifK RBS has been proposed as an enhancer (60). In a different study, multimers of CA-repeats, 3′ of the start codon of a leaderless lacZ gene, increased expression in a length-dependent manner, reminiscent of our results (Figure 3C); these CA-repeats bound ribosomes efficiently (12). We suspect that these and other reported enhancer elements may be similar or identical to standby sites as analyzed here. Whether they are responsible for overcoming structural constraints according to the original standby concept, or are one of several elements contributing to a total expression value (27,64,66), likely differs in a case-by-case manner. At this point, the apparent strong translation-stimulating power of CA-rich sequences is puzzling and deserves further studies.
A further question concerns requirements for ribosomal components in standby mechanisms. Clearly, ribosomal protein S1 is a main suspect, since AU-rich regions (upstream of SD sequences) are binding targets for this protein (13,14). As a single-strand RNA-binding protein—on and off the ribosome—it is a prime candidate for transiently anchoring the 30S subunit to a single-stranded stretch of RNA. Whether this indeed occurs is currently under investigation in our lab. Given that ∼10 nt of single-stranded RNA can be bound by one molecule of S1, and that optical tweezer experiments suggest unwinding activity on neighboring structures (70), this protein might act as an anchor and/or a structure-changing factor. Another paper indicates that S1 binding to upstream enhancers may promote translation by faster dissociation of strong SD-antiSD interactions (71). Notably, a recent cryo-EM study suggests that S1, at least within 70S ribosomes, may contact mRNA segments near both the mRNA entry and exit tunnels (72). In particular, S1 has also been analyzed with respect to the contributions of its six RNA-binding domains to translation of mRNAs (18). We presume that, whether unwinding plays a role or not, S1 may be the dominant RNA-binding protein that provides a sufficiently long bound state to make standby work and to overcome inhibitory structure. Another important aspect is that the 30S platform can accommodate structured mRNAs which, in some cases, can become unfolded for entry into an initiation-competent state (51).
This study has purposely ignored aspects of initiation that may or may not be relevant to standby mechanisms. For instance, little is known about determinants important to leaderless or 70S-only initiation (20–22). Also, a recent study indicated that RNA structures may affect initiation efficiency by restricting sideway diffusion of ribosomes, affecting appropriate positioning for initiation (73). Finally, in a kinetic ‘ribosome drafting’ model, it was suggested that the rates of local folding/unfolding at an RBS—rather than RNA stability as such—and the rates of ribosome binding, together can cause widely different expression levels (74).
In conclusion, this paper provides a proof-of-principle study in which we demonstrate that single-stranded tails in front of a structurally sequestered RBS can promote dramatically increased translation rates. The differences between presumably unstructured tails of different sequences (CA- or AU-series) indicate that sequence matters as well, though low structure is a prerequisite. We are in the process of following up on this by using a random mutagenesis approach as in (75) to identify positive and negative determinants for standby.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Erik Holmqvist and Mikael Sellin for critical reading of the manuscript. We are grateful to Maarten de Smit for providing plasmid pK2 for plasmid constructions, and thank Måns Ehrenberg's group for providing 30S ribosomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council [VR 621-2010-5233 to E.G.H.W.]. Funding for open access charge: Swedish Research Council [to E.G.H.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Quax T.E.F., Wolf Y.I., Koehorst J.J., Wurtzel O., van der Oost R., Ran W., Blombach F., Makarova K.S., Brouns S.J.J., Forster A.C. et al. Differential translation tunes uneven production of operon-encoded proteins. Cell Rep. 2013; 4:938–944. [DOI] [PubMed] [Google Scholar]
- 2. Li G.-W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schlax P.J., Worhunsky D.J.. Translational repression mechanisms in prokaryotes. Mol. Microbiol. 2003; 48:1157–1169. [DOI] [PubMed] [Google Scholar]
- 5. Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005; 361:13–37. [DOI] [PubMed] [Google Scholar]
- 6. Milón P., Rodnina M.V.. Kinetic control of translation initiation in bacteria. Crit. Rev. Biochem. Mol. Biol. 2012; 47:334–348. [DOI] [PubMed] [Google Scholar]
- 7. Simonetti A., Marzi S., Jenner L., Myasnikov A., Romby P., Yusupova G., Klaholz B.P., Yusupov M.. A structural view of translation initiation in bacteria. Cell. Mol. Life Sci. CMLS. 2009; 66:423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shine J., Dalgarno L.. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steitz J.A., Jakes K.. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:4734–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J., Deutscher M.P.. A uridine-rich sequence required for translation of prokaryotic mRNA. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:2605–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hook-Barnard I.G., Brickman T.J., McIntosh M.A.. Identification of an AU-rich translational enhancer within the Escherichia coli fepB leader RNA. J. Bacteriol. 2007; 189:4028–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin-Farmer J., Janssen G.R.. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol. Microbiol. 1999; 31:1025–1038. [DOI] [PubMed] [Google Scholar]
- 13. Komarova A.V., Tchufistova L.S., Supina E.V., Boni I.V.. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA N. Y. 2002; 8:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komarova A.V., Tchufistova L.S., Dreyfus M., Boni I.V.. AU-rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 2005; 187:1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma C.M., Darfeuille F., Plantinga T.H., Vogel J.. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007; 21:2804–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vimberg V., Tats A., Remm M., Tenson T.. Translation initiation region sequence preferences in Escherichia coli. BMC Mol. Biol. 2007; 8:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boni I.V., Isaeva D.M., Musychenko M.L., Tzareva N.V.. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991; 19:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duval M., Korepanov A., Fuchsbauer O., Fechter P., Haller A., Fabbretti A., Choulier L., Micura R., Klaholz B.P., Romby P. et al. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol. 2013; 11:e1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Udagawa T., Shimizu Y., Ueda T.. Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria. J. Biol. Chem. 2004; 279:8539–8546. [DOI] [PubMed] [Google Scholar]
- 20. Moll I., Hirokawa G., Kiel M.C., Kaji A., Bläsi U.. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004; 32:3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A.C., Engelberg-Kulka H., Moll I.. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011; 147:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamamoto H., Wittek D., Gupta R., Qin B., Ueda T., Krause R., Yamamoto K., Albrecht R., Pech M., Nierhaus K.H.. 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1180–E1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hüttenhofer A., Noller H.F.. Footprinting mRNA-ribosome complexes with chemical probes. EMBO J. 1994; 13:3892–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kudla G., Murray A.W., Tollervey D., Plotkin J.B.. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009; 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Voges D., Watzele M., Nemetz C., Wizemann S., Buchberger B.. Analyzing and enhancing mRNA translational efficiency in an Escherichia coli in vitro expression system. Biochem. Biophys. Res. Commun. 2004; 318:601–614. [DOI] [PubMed] [Google Scholar]
- 26. Bouvier M., Sharma C.M., Mika F., Nierhaus K.H., Vogel J.. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol. Cell. 2008; 32:827–837. [DOI] [PubMed] [Google Scholar]
- 27. Espah Borujeni A., Cetnar D., Farasat I., Smith A., Lundgren N., Salis H.M.. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Res. 2017; 45:5437–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Del Campo C., Bartholomäus A., Fedyunin I., Ignatova Z.. Secondary structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet. 2015; 11:e1005613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burkhardt D.H., Rouskin S., Zhang Y., Li G.-W., Weissman J.S., Gross C.A.. Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. eLife. 2017; 6:e22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evfratov S.A., Osterman I.A., Komarova E.S., Pogorelskaya A.M., Rubtsova M.P., Zatsepin T.S., Semashko T.A., Kostryukova E.S., Mironov A.A., Burnaev E. et al. Application of sorting and next generation sequencing to study 5′-UTR influence on translation efficiency in Escherichia coli. Nucleic Acids Res. 2017; 45:3487–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bentele K., Saffert P., Rauscher R., Ignatova Z., Blüthgen N.. Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol. 2013; 9:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goodman D.B., Church G.M., Kosuri S.. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013; 342:475–479. [DOI] [PubMed] [Google Scholar]
- 33. Meyer M.M. The role of mRNA structure in bacterial translational regulation. Wiley Interdiscip. Rev. RNA. 2017; 8, doi:10.1002/wrna.1370. [DOI] [PubMed] [Google Scholar]
- 34. Moine H., Romby P., Springer M., Grunberg-Manago M., Ebel J.P., Ehresmann B., Ehresmann C.. Escherichia coli threonyl-tRNA synthetase and tRNA(Thr) modulate the binding of the ribosome to the translational initiation site of the thrS mRNA. J. Mol. Biol. 1990; 216:299–310. [DOI] [PubMed] [Google Scholar]
- 35. Du H., Babitzke P.. trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis. J. Biol. Chem. 1998; 273:20494–20503. [DOI] [PubMed] [Google Scholar]
- 36. Majdalani N., Cunning C., Sledjeski D., Elliott T., Gottesman S.. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12462–12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tucker B.J., Breaker R.R.. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005; 15:342–348. [DOI] [PubMed] [Google Scholar]
- 38. Johansson J., Mandin P., Renzoni A., Chiaruttini C., Springer M., Cossart P.. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002; 110:551–561. [DOI] [PubMed] [Google Scholar]
- 39. Krajewski S.S., Narberhaus F.. Temperature-driven differential gene expression by RNA thermosensors. Biochim. Biophys. Acta. 2014; 1839:978–988. [DOI] [PubMed] [Google Scholar]
- 40. Nechooshtan G., Elgrably-Weiss M., Sheaffer A., Westhof E., Altuvia S.. A pH-responsive riboregulator. Genes Dev. 2009; 23:2650–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Giuliodori A.M., Di Pietro F., Marzi S., Masquida B., Wagner R., Romby P., Gualerzi C.O., Pon C.L.. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell. 2010; 37:21–33. [DOI] [PubMed] [Google Scholar]
- 42. Schmidt B.F., Berkhout B., Overbeek G.P., van Strien A., van Duin J.. Determination of the RNA secondary structure that regulates lysis gene expression in bacteriophage MS2. J. Mol. Biol. 1987; 195:505–516. [DOI] [PubMed] [Google Scholar]
- 43. Blomberg P., Nordström K., Wagner E.G.. Replication control of plasmid R1: RepA synthesis is regulated by CopA RNA through inhibition of leader peptide translation. EMBO J. 1992; 11:2675–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. de Smit M.H., van Duin J.. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Smit M.H., van Duin J.. Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J. Mol. Biol. 1994; 244:144–150. [DOI] [PubMed] [Google Scholar]
- 46. de Smit M.H., van Duin J.. Translational initiation on structured messengers. Another role for the Shine-Dalgarno interaction. J. Mol. Biol. 1994; 235:173–184. [DOI] [PubMed] [Google Scholar]
- 47. de Smit M.H., van Duin J.. Translational initiation at the coat-protein gene of phage MS2: native upstream RNA relieves inhibition by local secondary structure. Mol. Microbiol. 1993; 9:1079–1088. [DOI] [PubMed] [Google Scholar]
- 48. de Smit M.H., van Duin J.. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 2003; 331:737–743. [DOI] [PubMed] [Google Scholar]
- 49. de Smit M.H., van Duin J.. A Prelude to Translational (Re)Initiation. Translation Mechanisms. 2003; Georgetown: Landes Bioscience; 298–321. [Google Scholar]
- 50. Sacerdot C., Caillet J., Graffe M., Eyermann F., Ehresmann B., Ehresmann C., Springer M., Romby P.. The Escherichia coli threonyl-tRNA synthetase gene contains a split ribosomal binding site interrupted by a hairpin structure that is essential for autoregulation. Mol. Microbiol. 1998; 29:1077–1090. [DOI] [PubMed] [Google Scholar]
- 51. Marzi S., Myasnikov A.G., Serganov A., Ehresmann C., Romby P., Yusupov M., Klaholz B.P.. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell. 2007; 130:1019–1031. [DOI] [PubMed] [Google Scholar]
- 52. Studer S.M., Joseph S.. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell. 2006; 22:105–115. [DOI] [PubMed] [Google Scholar]
- 53. Darfeuille F., Unoson C., Vogel J., Wagner E.G.H.. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007; 26:381–392. [DOI] [PubMed] [Google Scholar]
- 54. Vogel J., Argaman L., Wagner E.G.H., Altuvia S.. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. CB. 2004; 14:2271–2276. [DOI] [PubMed] [Google Scholar]
- 55. Berghoff B.A., Hoekzema M., Aulbach L., Wagner E.G.H.. Two regulatory RNA elements affect TisB-dependent depolarization and persister formation. Mol. Microbiol. 2017; 103:1020–1033. [DOI] [PubMed] [Google Scholar]
- 56. Berghoff B.A., Wagner E.G.H.. RNA-based regulation in type I toxin-antitoxin systems and its implication for bacterial persistence. Curr. Genet. 2017; 63:1011–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Urban J.H., Vogel J.. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007; 35:1018–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Holmqvist E., Reimegård J., Sterk M., Grantcharova N., Römling U., Wagner E.G.H.. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010; 29:1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kastelein R.A., Berkhout B., Overbeek G.P., van Duin J.. Effect of the sequences upstream from the ribosome-binding site on the yield of protein from the cloned gene for phage MS2 coat protein. Gene. 1983; 23:245–254. [DOI] [PubMed] [Google Scholar]
- 60. Yang Q., Figueroa-Bossi N., Bossi L.. Translation enhancing ACA motifs and their silencing by a bacterial small regulatory RNA. PLoS Genet. 2014; 10:e1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lindell M., Romby P., Wagner E.G.H.. Lead(II) as a probe for investigating RNA structure in vivo. RNA N. Y. N. 2002; 8:534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scharff L.B., Childs L., Walther D., Bock R.. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet. 2011; 7:e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Espah Borujeni A., Channarasappa A.S., Salis H.M.. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014; 42:2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Salis H.M. The ribosome binding site calculator. Methods Enzymol. 2011; 498:19–42. [DOI] [PubMed] [Google Scholar]
- 66. Salis H.M., Mirsky E.A., Voigt C.A.. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009; 27:946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sengupta J., Agrawal R.K., Frank J.. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:11991–11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Draper D.E., von Hippel P.H.. Nucleic acid binding properties of Escherichia coli ribosomal protein S1. I. Structure and interactions of binding site I. J. Mol. Biol. 1978; 122:321–338. [DOI] [PubMed] [Google Scholar]
- 69. Osterman I.A., Evfratov S.A., Sergiev P.V., Dontsova O.A.. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res. 2013; 41:474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Qu X., Lancaster L., Noller H.F., Bustamante C., Tinoco I.. Ribosomal protein S1 unwinds double-stranded RNA in multiple steps. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14458–14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Takahashi S., Furusawa H., Ueda T., Okahata Y.. Translation enhancer improves the ribosome liberation from translation initiation. J. Am. Chem. Soc. 2013; 135:13096–13106. [DOI] [PubMed] [Google Scholar]
- 72. Loveland A.B., Korostelev A.A.. Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM. Methods San Diego Calif. 2017; 10.1016/j.ymeth.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jagodnik J., Chiaruttini C., Guillier M.. Stem–loop structures within mRNA coding sequences activate translation initiation and mediate control by small regulatory RNAs. Mol. Cell. 2017; 68:158–170. [DOI] [PubMed] [Google Scholar]
- 74. Espah Borujeni A., Salis H.M.. Translation Initiation is Controlled by RNA Folding Kinetics via a Ribosome Drafting Mechanism. J. Am. Chem. Soc. 2016; 138:7016–7023. [DOI] [PubMed] [Google Scholar]
- 75. Holmqvist E., Reimegård J., Wagner E.G.H.. Massive functional mapping of a 5′-UTR by saturation mutagenesis, phenotypic sorting and deep sequencing. Nucleic Acids Res. 2013; 41:e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.