Abstract
STING is a core adaptor in innate nucleic acid sensing in mammalian cells, on which different sensing pathways converge to induce type I interferon (IFN) production. Particularly, STING is activated by 2′3′-cGAMP, a cyclic dinucleotide containing mixed phosphodiester linkages and produced by cytoplasmic DNA sensor cGAS. Here, we reported on a novel transcript isoform of STING designated STING-β that dominantly inhibits innate nucleic acid sensing. STING-β without transmembrane domains was widely expressed at low levels in various human tissues and viral induction of STING-β correlated inversely with IFN-β production. The expression of STING-β declined in patients with lupus, in which type I IFNs are commonly overproduced. STING-β suppressed the induction of IFNs, IFN-stimulated genes and other cytokines by various immunostimulatory agents including cyclic dinucleotides, DNA, RNA and viruses, whereas depletion of STING-β showed the opposite effect. STING-β interacted with STING-α and antagonized its antiviral function. STING-β also interacted with TBK1 and prevented it from binding with STING-α, TRIF or other transducers. In addition, STING-β bound to 2′3′-cGAMP and impeded its binding with and activation of STING-α, leading to suppression of IFN-β production. Taken together, STING-β sequesters 2′3′-cGAMP second messenger and other transducer molecules to inhibit innate nucleic acid sensing dominantly.
INTRODUCTION
Sensing of foreign and intrinsic nucleic acids is an evolutionarily conserved component of host innate immunity. Nucleic acid sensors in mammalian cells include endosomal toll-like receptors (TLRs) such as TLR3, TLR7 and TLR9 as well as cytoplasmic RIG-I-like receptors (RLRs) such as RIG-I and MDA5 (1,2). Although multiple cytoplasmic DNA-sensing proteins such as DDX41, IFI16 and DNA-dependent protein kinase (DNA-PK) have been described (3–5), cyclic GMP-AMP (cGAMP) synthase (cGAS) is generally accepted as a primary DNA sensor (6,7). As a result of DNA sensing, type I interferons (IFNs), IFN-stimulated genes (ISGs) and other pro-inflammatory cytokines are induced through IRF3 and NF-κB pathways, the activation of which is critical for both viral clearance and pathogenesis (1,2).
The recognition of cytoplasmic DNA by cGAS produces a unique cyclic dinucleotide (CDN) c[G(2′,5′)pA(3′,5′)p], which is known to exist only in mammals. Distinct to its bacterial isomer that contains 3′-5′ phosphodiester bonds only, this mammalian CDN known as 2′3′-cGAMP contains 2′-5′ and 3′-5′ mixed phosphodiester linkages (8–10). For simplicity, hereafter we will use cGAMP to refer to mammalian 2′3′-cGAMP second messenger throughout our manuscript, whereas the bacterial isomer will be called 3′3′-cGAMP. cGAMP binds to and activates STING (7,11–13), known variously as MITA (14), MPYS (15), TMEM173 and ERIS (16), a central adaptor and converging point in DNA sensing.
STING is consisted of five transmembrane (TM) helical regions in the N-terminus and a large cytoplasmic domain in the C-terminus (11). Whereas the C-terminal domain (CTD) mediates protein-protein interaction, dimerization and ligand binding, the TM domains govern intracellular localisation (17,18). Bacterial CDN ligands that activate STING include cyclic diguanylate monophosphate (c-di-GMP), c-di-AMP and 3′3′-cGAMP (10,13,19,20). In addition, STING is also required for DNA sensing mediated by other cytoplasmic sensors including DDX41, IFI16 and DNA-PK (3–5). In resting cells, STING is localized to the endoplasmic reticulum (ER) with its C-terminal tail residing in the cytoplasm (11,14,15). Activated by ligand binding, STING translocates from the ER via the Golgi complex to perinuclear microsomes, where TBK1 phosphorylates STING and IRF3 (17,18). Additionally, STING recruits and activates STAT6 to induce the expression of pro-inflammatory cytokines (21). Notably, as part of the interconnected downstream signalling machinery activated by both DNA and RNA sensing (22), STING also directly transmits the activation signal of RIG-I and MAVS to TBK1 as demonstrated in human infection with Japanese encephalitis virus (23).
The essentiality of STING in innate nucleic acid sensing and antiviral response has been established in STING−/− cells and mice (12), which were unable to mobilize type I IFN response upon infection with herpes simplex virus 1 (HSV-1) or vesicular stomatitis virus (VSV). Conversely, various DNA and RNA viruses have developed distinct strategies, such as physical interaction, post-translational modification, mislocalisation and proteolysis, to circumvent STING function (1). Excessive activation of STING is detrimental not only to viruses but also to host cells (2,24–28). Autosomal dominant mutations in STING lead to constitutive activation of innate immune response, giving rise to a lupus-like infantile-onset autoinflammatory disease with elevated plasma levels of type I IFNs, ISGs and pro-inflammatory cytokines (29–31). To keep STING activity under control in healthy individuals, multiple negative regulatory mechanisms might be in place.
STING activation is regulated by post-translational modifications (1,32). STING is known to undergo K11-, K27-, K48- and K63-linked polyubiquitination catalyzed by different E3 ubiquitin ligases including TRIM32 (33), TRIM56 (34), TRIM30α (35), RNF5 (36), RNF26 (37) and AMFR (38). Whereas K48-linked ubiquitination targets STING to proteasomal degradation (35,36), the other three types of ubiquitination serve to bridge TBK1 and IRF3 (1,32). Other negative regulators of STING include autophagy-related multispanning transmembrane protein ATG9A (39), ULK1 kinase that phosphorylates STING to facilitate lysosomal degradation (40), NOD-like receptors NLRC3 (41) and NLRX1 (42), PPM1A phosphatase that dephosphorylates STING and TBK1 (43), as well as IFN-inducible protein ISG56 (44). On the other hand, optimal function of STING requires TRIF (45), S6K1 kinase (46) and ER-associated protein ZDHHC1 (47). Finally, MITA-related protein (MRP), an alternatively spliced isoform of STING lacking the conserved TBK1 interaction and CDN binding domain, was reported to inhibit STING-dependent IRF3 activation by disrupting the formation of STING-TBK1 complex (48). Thus, MRP acts as a dominant inactive regulator of STING-induced type I IFN production.
Surprisingly, some STING−/− mice were found to be more prone to autoimmune diseases and capable of mounting a more robust immune response (49). The identification of MRP indicates that negative regulators can be produced by alternative splicing from the same allele that expresses STING (48,50). It remains to be clarified whether the seemingly counterintuitive finding on hyperactivation of immune response in some STING−/− mice might be explained by concurrent loss of STING together with MRP and additional STING transcripts, which are also expressed from the deleted STING locus but exert a dominant negative effect. A thorough understanding of the negative regulatory mechanism of STING-dependent innate nucleic acid sensing will not only derive new knowledge but also instruct rational design of novel antiviral and anti-inflammatory agents. Here, we identified a novel transcript isoform of STING designated STING-β. Hereafter in our paper, the original STING isoform will be termed STING-α. STING-β contains the functional CTD but lacks the N-terminal TM domains. We performed both gain- and loss-of-function experiments to demonstrate the negative regulatory activity of STING-β on STING-α-mediated innate nucleic acid sensing. Our results support the model that STING-β preoccupies and sequesters STING-α, cGAMP second messenger and other transducer molecules to prevent them from binding with and activating their physiological effectors.
MATERIALS AND METHODS
Human blood samples
The human blood samples used in this study were collected from Queen Mary Hospital in Hong Kong. Blood samples were collected from a total of 24 Chinese individuals who fulfilled the American College of Rheumatology classification criteria for SLE as described in our previous papers (51–53). Control blood samples were collected from 24 blood donors who are healthy Chinese individuals. Total mRNA extracted from peripheral blood mononuclear cells (PBMCs) was reverse-transcribed into cDNA as described in our previous paper (53). The use of human blood samples was approved by the Joint Institutional Review Board of the University of Hong Kong and Hospital Authority Hong Kong West Cluster.
Real-time quantitative RT-PCR
Total RNA was extracted using TRIzol® reagent (Life Technologies). First-strand cDNA was synthesized with Transcriptor First Strand cDNA Synthesis Kit (Roche) and either stored at –20°C or used immediately. RT-PCR reactions were performed using Applied Biosystems® Veriti® thermal cycler. Quantitative RT-PCR (RT-qPCR) reactions and analysis were performed using Applied Biosystems StepOnePlus™ Real-Time PCR system. SYBR Premix Ex Taq kit (Takara Bio) was used for SYBR Green-based RT-qPCR according to the manufacturer's protocol. Comparative CT method was used for the calculation of fold changes in gene expression. Primers used in RT-PCR and RT-qPCR reactions were listed in Supplementary Table S1.
Plasmids and DNA oligonucleotides
pcDNA6B-STING-α-HA, pcDNA6B-STING-α-FLAG, pcDNA6B-STING-β-HA, pCAGEN-cGAS, pcDNA6B-STING-α-GFP, pcDNA6B-STING-β-GFP, pcDNA3.1-TRIF-V5/His and pcDNA6B-MyD88-FLAG were constructed using standard molecular cloning methods. Other expression plasmids for RIG-I, RIG-IN, MDA5, MAVS, TBK1, IKKϵ, TRIF and IRF3 (5D) have been described previously (54–56). IFNβ-luc and IRF3-luc reporter plasmids reflect the activity of IFN-β promoter and IRF3, respectively. The activation of type I IFN signalling and NF-κB is indicated by ISRE-luc (Clontech) and κB-luc (Clontech) reporter plasmids. Primers used in plasmid construction were listed in Supplementary Table S2. HSV-1-60mer was prepared as described (4).
Cell culture and transfection
HEK293, HEK293T, HeLa, MCR-5, IMR90, LX-2, MDA-MB-231, WI38, U2OS, HFF-1, HT-29, SF268 and Vero cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) with 10% heat-inactivated fetal bovine serum (FBS; Life Technologies). Caco-2 cells were cultured in Earle's Minimum Essential Medium with 10% heat inactivated FBS. hTERT-immortalized foreskin fibroblast cell line BJ-5ta (ATCC CRL-4001) was cultured in 4:1 ratio of DMEM:Medium 199 with 10% heat inactivated FBS. HEMa primary epidermal melanocytes (ATCC PCS-200-013) were cultured in dermal cell basal medium (ATCC PCS-200-030) supplemented with adult melanocyte growth kit (ATCC PCS-200-042). SK-MEL-31 (ATCC HTB-73) and HFL cells were cultured in minimum essential medium with 10% heat-inactivated FBS. Fetal normal colon FHC (ATCC CRL-1831) cells were cultured in DMEM:F12 medium supplied with extra 10 mM HEPES, 10 ng/mL cholera toxin, 0.005 mg/mL insulin, 0.005 mg/mL transferrin, 100 ng/mL hydrocortisone and 10% heat-inactivated FBS. CEM-T4, MT2, Jurkat, THP-1, U937 and HuT102 were cultured in RPMI 1640 medium (Life Technologies) with 10% heat-inactivated FBS. All cells were cultured at 37°C in a humidified incubator with 5% CO2. Plasmids were transfected into HEK293, HEK293T and HeLa cells using GeneJuice® transfection reagents (Novagen). THP-1 cells were transfected with GeneXPlus transfection reagents from ATCC. HSV-1-60mer, a dsDNA of 60 bp derived from HSV-1 genome, was prepared as previously described (4). poly (I:C) (Sigma P1530), HSV-1-60mer and 2′3′-cGAMP (InvivoGen tlrl-nacga23) were transfected into cells using Lipofectamine 2000 (Thermo Fisher).
Antibodies
Rabbit polyclonal anti-STING-β serum directed against the first 25 amino acids of STING-β was raised through Beijing Biodragon Immunotechnologies. These antibodies specifically react with STING-β with no cross-reactivity to STING-α. Rabbit anti-STING (Cell Signalling), mouse anti-HA (Santa Cruz), rabbit anti-HA (Santa Cruz), mouse anti-FLAG M2 (Sigma), rabbit anti-FLAG (Sigma), mouse anti-V5 (Invitrogen) and rabbit anti-V5 (Sigma) primary antibodies as well as sheep anti-mouse (GE Healthcare) and donkey anti-rabbit (GE Healthcare) secondary antibodies were purchased commercially. For western blotting and immunoprecipitation, rabbit polyclonal anti-STING and anti-STING-β antibodies were used at a dilution of 1:3000 and 1:200, respectively, in 5% bovine serum albumin (BSA).
Protein extraction and western blotting
Cells were lysed with RIPA-150 buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 1% NP-40, 0.5 mM EDTA) supplemented with protease inhibitor cocktail (Roche). Protein concentrations were determined by Bradford reagent (Bio-Rad). Cell lysate was mixed with SDS-PAGE sample buffer and boiled for 10 min prior to SDS-PAGE. Native PAGE was performed as described (43). In brief, cells were lysed with non-denaturing lysis buffer (20 mM Tris-Cl, pH 8.0, 137 mM NaCl, 1% NP-40, 2 mM EDTA) containing protease inhibitor cocktail (Roche). Cell lysate was mixed with 2 × non-denaturing loading buffer (125 mM Tris-Cl, pH 6.8, 30% glycerol, 0.01% bromophenol blue) and then resolved in 7.5% native PAGE gel. Proteins in the gel were transferred onto PVDF membranes (Millipore). After gel transfer and blocking, the membranes were incubated with primary antibody at 4°C overnight. After washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies diluted 1:5000 in 5% skim milk for 1–2 h at room temperature. Signals were visualized by WesternBright™ ECL (Advansta) and blots were exposed to X-ray film (Fuji-RX).
Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE)
SDD-AGE was carried out as described (43). Briefly, HEK293T cells were harvested 36 h after transfection and lysed with non-denaturing lysis buffer containing protease inhibitor cocktail (Roche). The cell lysate was mixed with 5 × sample loading buffer (2.5 × TBE, 2.5% SDS, 25% glycerol, 0.25% bromophenol blue) and then loaded onto a vertical 2% agarose gel. After electrophoresis in the running buffer (0.5 × TBE and 0.1% SDS) for 1 h with a constant voltage of 100 V at 4°C, the proteins were transferred to PVDF membranes (Millipore) for western blotting.
Co-immunoprecipitation
Immunoprecipitation was performed with protein G agarose beads (Invitrogen) using standard protocols as described (54–56). Briefly, plasmids that express the tagged protein were co-transfected into HEK293T cells. After 48 h, cells were harvested and lysed with RIPA-150 buffer. Cell lysates were incubated with primary antibody for 3 h at 4°C with low speed rocking. After binding, protein G agarose beads were added into the mixture and rocked overnight at 4°C. The beads were washed with RIPA-150 lysis buffer for three times. After washing, 50 μl of 1 × SDS-PAGE sample buffer was added into the tube containing the beads. The immunoprecipitated proteins were eluted from beads after 10 min of boiling. The samples were either stored at -20°C or analyzed by western blotting immediately.
Dual-luciferase reporter assay
Dual-luciferase reporter assays were performed by Dual-Luciferase® reporter assay system (Promega) according to the manufacturer's protocol and as described (54–56). Normalization of transfection efficiency was achieved by co-transfection of pRL-TK reporter (Promega). Relative luciferase activity was calculated by normalizing firefly luciferase activity to that of Renilla luciferase.
RNA interference
Small interfering RNAs (siRNAs) were designed using online siRNA tools of DsiRNA (https://sg.idtdna.com/site/order/designtool/index/DSIRNA_CUSTOM) and BLOCK-iT™ RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/). siRNAs were transfected into THP-1 cells using Lipofectamine 2000 (Invitrogen) as described previously (4,54,55). All siRNAs were synthesized by GenePharma. siRNA sequences were listed in Supplementary Table S3.
Confocal microscopy
HeLa cells were seeded on coverslips in 6-well plates the day before transfection and cultured for 24 h. Plasmids of interest were transfected for 36 h before cells were fixed with 4% paraformaldehyde. For immunostaining, cells were first blocked with 5% BSA for 1 h followed by 5-min permeabilization with 0.2% Triton X-100. Cells were then incubated with primary antibody (1:200; mouse anti-HA for STING-β and mouse anti-FLAG for TBK1) overnight at 4°C. After washing with 1× PBS for three times, goat anti-mouse IgG conjugated to TRITC (1:200; Zymed) was added and incubated for 1 h. After washing with 1× PBS for three times, the coverslips were mounted onto the slides for observation using Carl Zeiss LSM 510 META Multiphoton Confocal Microscope. DsRed2-ER and DsRed2-Mito (Clontech) served as ER and mitochondrial markers.
cGAMP pull-down assay
cGAMP pull-down assay was performed using cGAMP-agarose (Biolog). Briefly, control agarose and cGAMP-agarose were washed with 10 volume of non-denaturing lysis buffer for three times before use. Cell lysates containing proteins of interest were incubated with control agarose or cGAMP-agarose for 2 h at 4°C. After incubation, the agarose was washed by PBS containing 100 μM ATP for six times and was then mixed with 50 μl of 1 × SDS-PAGE sample buffer. Proteins were eluted by boiling for 10 min. Samples were either stored at –20°C or analyzed immediately by western blotting or silver staining.
Silver staining
After SDS-PAGE, protein gel was fixed in 50% methanol, 12% acetic acid for 1 h. The protein gel was washed with 35% ethanol three times for 20 min each and then sensitized in 0.02% Na2S2O3 solution for 2 min. After washing with distilled deionized water three times for 5 min each, the gel was stained with 0.2% AgNO3, 0.05% formalin for 20 min and then washed with distilled deionized water twice for 1 min each. The gel was developed with 6% Na2CO3, 0.05% formalin, 0.0004% Na2S2O3. When the gel was developed to the desired intensity, the development was terminated with 50% methanol, 12% acetic acid for 5 min. The gel was stored in distilled deionized water.
Viral infection
Sendai virus (SeV), VSV-GFP and HSV-1-GFP were described in our previous publications (54–56). ICP0-null HSV-1 virus (HSV-1-ΔICP0) was kindly provided by David Knipe from Harvard Medical School. ICP0 of HSV-1 strongly blocks IRF3-mediated activation of type I IFNs and ISGs. In contrast to wild-type HSV-1, HSV-1-ΔICP0 virus induces higher levels of type I IFNs and ISGs (57,58). Before virus infection, cells were washed with pre-warmed medium at 37°C twice. Virus was diluted at a certain ratio according to the desired multiplicity of infection (M.O.I.) in serum-free medium. Virus-medium mixtures were incubated with cells for 1 h in CO2 incubator. After infection, virus-medium mixtures were replaced with fresh culture medium with FBS.
Viral plaque assay
Viral plaque assay was performed on Vero or U2OS cells to measure the virus titre. Vero or U2OS cells were seeded on 6-well plates to achieve ∼90–100% confluency after 24 h. On the next day, cells were infected with serial dilutions of virus (i.e. 10−1, 10−2, 10−3, 10−4, 10−5 and 10−6 of original virus stock). After viral infection, the medium was removed and 0.5% agar in DMEM with 2% FBS was overlaid into the wells. After the agar overlay turned solid, cells were cultured for another 24 h for infection with VSV-GFP or 72 h for infection with HSV-1-GFP or HSV-1-ΔICP0. To fix the cells, 2 ml of 1:1 methanol-ethanol mixture was directly added into each well and incubated at 4°C for 30 min. The solid agarose-medium mix was removed carefully and the cells were stained with 0.05% crystal violet for 15 min. Then the plates were rinsed with water and plaques on the monolayer were counted to calculate the virus titre.
Statistical analysis
All results are representative of three independent experiments. Statistical analysis was performed by two-tailed unpaired Student's t test by GraphPad Prism 6.0. A difference was considered statistically significant when P < 0.05.
RESULTS
Expression and induction of STING-β as a novel transcript isoform of STING
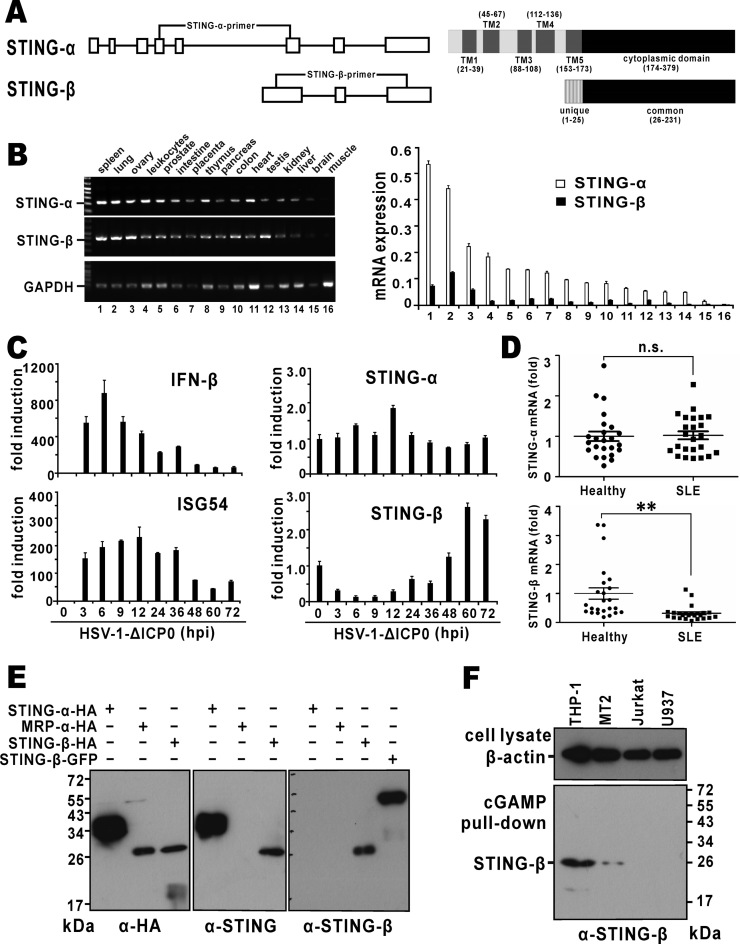
The human STING allele is composed of eight exons and seven introns (Figure 1A). In the expressed sequence tag (EST) database, multiple ESTs with a putative transcription start site (TSS) within intron 5 of STING allele were identified. Further analysis revealed that all ESTs in this group could be explained by an alternative STING-β transcript that starts from intron 5. This transcript was PCR-amplified and sequenced. The TSS was experimentally validated by rapid amplification of cDNA ends.
Figure 1.
Expression of STING-β mRNA and protein. (A) Genome and domain structure of STING-β. Boxes represent exons and the lines represent introns. Positions of isoform-specific primers used in RT-PCR are indicated. Also shown are transmembrane (TM) and cytoplasmic domains of STING-α and STING-β. Transcription of STING-β mRNA is driven by an alternative promoter located within intron 5 of STING-α. Compared to exon 6 of STING-α, exon 1 of STING-β contains an extra piece of sequence at the 5′ end, resulting in an additional 25 amino acids at the N-terminus. This unique N-terminal sequence is highlighted. (B) Expression of STING-α and STING-β transcripts in human tissues. Human MTC™ Panel I (Clontech, USA) containing the cDNA templates of various human tissues were used for RT-PCR (left panel) and RT-qPCR (right panel) analysis. The STING-β primers will amplify a fragment of the correct size from STING-β cDNA only, but neither STING-α cDNA nor genomic DNA. Likewise, the STING-α primers are specific to STING-α cDNA. mRNA level was obtained by the comparative Ct method. (C) Expression of STING-β transcript in virus-challenged THP-1 cells. A DNA virus HSV-1-ΔICP0 (5 M.O.I.), in which one major IFN antagonist named ICP0 is deleted, was used to stimulate type I IFN production in THP-1 cells. Samples were harvested at the indicated time points for RNA extraction. Temporal expression profile of STING-β was determined by RT-qPCR. (D) Expression of STING-β transcript in peripheral blood mononuclear cells of SLE patients. cDNA templates were prepared from peripheral blood mononuclear cells of SLE patients (n = 24) and healthy individuals (n = 24). Expression of STING-α and STING-β in theses samples was detected by RT-qPCR. Whereas there was no significant statistical difference (n.s.) in STING-α mRNA expression between the SLE and healthy groups, the levels of STING-β mRNA were significantly lower (∗∗P < 0.01) in SLE samples versus cells from healthy people. Statistical analysis was performed using Student's t test. (E) Detection of recombinant STING-β protein expressed in HEK293T cells by STING-β-specific antibodies. STING-α-HA, STING-β-HA, MRP-HA and STING-β-GFP were overexpressed in HEK293T cells. After 48 h, cells were lysed for western blotting using the indicated antibodies. Rabbit anti-STING-β antiserum specifically recognizes STING-β but not STING-α or MRP. (F) Detection of endogenous STING-β protein by STING-β-specific antibodies. THP-1, MT2, Jurkat and U937 cells were lysed and incubated with cGAMP agarose for 2 h at 4°C. After washing with PBS containing 100 μM ATP for six times, sample loading buffer was added. The samples were boiled for 10 min and then further analyzed by SDS-PAGE and western blotting with anti-STING-β antibodies. Results in each panel are representative of three independent experiments.
A cluster of transcription factor binding sites followed by TATA box sequences (Supplementary Figure S1A) were identified in the region upstream of the TSS by bioinformatic methods. ChIP-seq results from the ENCODE transcription factor database confirmed the binding of EGR1, STAT3, IRF1, NF-κB, MAX, MYC, CTCF and TATA box-binding protein (TBP) to this region. In addition, H3K27ac mark and DNase I hypersensitive sites, which are commonly associated with active enhancers, were also found in this region. Indeed, transcriptional activity of STING-β promoter as reflected in the activation of luciferase reporter expression was detected in THP-1 cells and it was weaker than that of STING-α or SV40 promoter (Supplementary Figure S1A). The binding of STAT3 and IRF1 prompted us to examine whether STING-β is an ISG. Because phorbol ester is a strong inducer of EGR1 (59), we also asked whether STING-β transcription might be affected by tetradecanoylphorbol acetate (TPA). STING-β promoter-driven luciferase reporter expression was induced by both IFN-β and TPA (Supplementary Figure S1B). Similar induction patterns were also observed when we measured the levels of STING-β mRNA (Supplementary Figure S1C). Although both STING-α and STING-β transcripts were induced by IFN-β, the induction kinetics was different (Supplementary Figure S1C). Furthermore, whereas the activity of STING-β promoter was boosted by TPA, it had an inhibitory effect on STING-α promoter (Supplementary Figure S1B and C). Thus, the expression of STING-β is controlled by its own promoter distinct to that of STING-α.
The cDNA sequence of STING-β is 1779 bp long and contains a 696-bp open reading frame encoding a protein of 231 amino acids (GenBank accession number: MF360993). STING-β transcript is expressed from an alternative promoter and its N-terminal 25 amino acids are translated from intron 5 of STING-α. No alternative start codon was found. STING-β not only shares the same CTD with STING-α but also has a unique N-terminus of 25 amino acids (Figure 1A). Interestingly, several proteins strikingly homologous to human STING-β were identified in Bactrian camel (Camelus bactrianus) (GenBank accession number: XP_010970027), Arabian camel (Camelus dromedaries) (GenBank accession number: NP_001306808), white-cheeked gibbon (Nomascus leucogenys) (GenBank accession number: XP_012360436) and chimpanzee (Pan troglodytes) (GenBank accession number: XP_016809410). The promoter regions and the sequences surrounding the start codon in these STING-β orthologs were highly conserved.
Both STING-α and STING-β were detectable in various human tissues and cell lines using RT-PCR and RT-qPCR (Figure 1B and Supplementary Figure S2A). STING-β mRNA could be detected at relatively low levels in most tissues including spleen, lung, ovary, peripheral leukocytes, prostate, small intestine, placenta, thymus, pancreas, colon (with mucosa), heart, testis, kidney and liver. However, it was barely expressed in brain and skeletal muscle. Although STING-α was more abundant than STING-β, both were expressed to reasonably higher levels in spleen, lung and ovary (Figure 1B). STING-α and STING-β were also detected in some human cell lines including THP-1, MT2, Jurkat and U937 (Supplementary Figure S2A). Notably, their basal levels in cell lines recently shown to have functional cGAS-STING signalling (60,61), including HFF-1, BJ-5ta, FHC, HEMa and HT-29, were also relatively high (Supplementary Figure S2A). Since STING-β was more abundant in THP-1 monocytic cells than most other cell lines, we chose this line for further analysis of STING-β expression. RT-qPCR results showed potent induction of both IFN-β and ISG54 by HSV-1-ΔICP0. This induction was attenuated gradually after 12 h, suggestive of a negative regulatory mechanism (Figure 1C). Notably, the HSV-1-ΔICP0-induced change in STING-α expression was not as dramatic and the range of fluctuation was within 2 fold. In stark contrast, the fall and rise in STING-β expression in infected cells were remarkably more substantial. It dropped in cells during the early phase of infection, but after 48 h its expression gradually increased to levels higher than the unstimulated sample. It is noteworthy that its expression was not only more inducible, but also correlated inversely with that of IFN-β and ISG54 (Figure 1C). Similar results were obtained when cells were transfected with poly(I:C) or HSV-1-60mer or infected with SeV or VSV (Supplementary Figure S2B). Whereas poly(I:C) is a synthetic analogue of dsRNA, HSV-1-60mer is a well-documented immunostimulatory dsDNA originally derived from HSV-1 genome (4). Thus, both RNA and DNA stimuli can induce rise and fall of STING-β expression, leading plausibly to the change in IFN and ISG production.
In further support of this notion, the mRNA levels of STING-β, but not of STING-α, decreased (Figure 1D) in peripheral blood samples of patients diagnosed with systemic lupus erythematosus (SLE), an interferonopathy characterized by excessive activation of type I IFN signaling (62). These results were compatible with the model that down regulation of STING-β expression might contribute to excessive activation of IFN signalling during the course of autoimmune disease. In addition, the underexpression of STING-β in SLE was also consistent with the underexpression of EGR1 (63,64), which might regulate STING-β promoter.
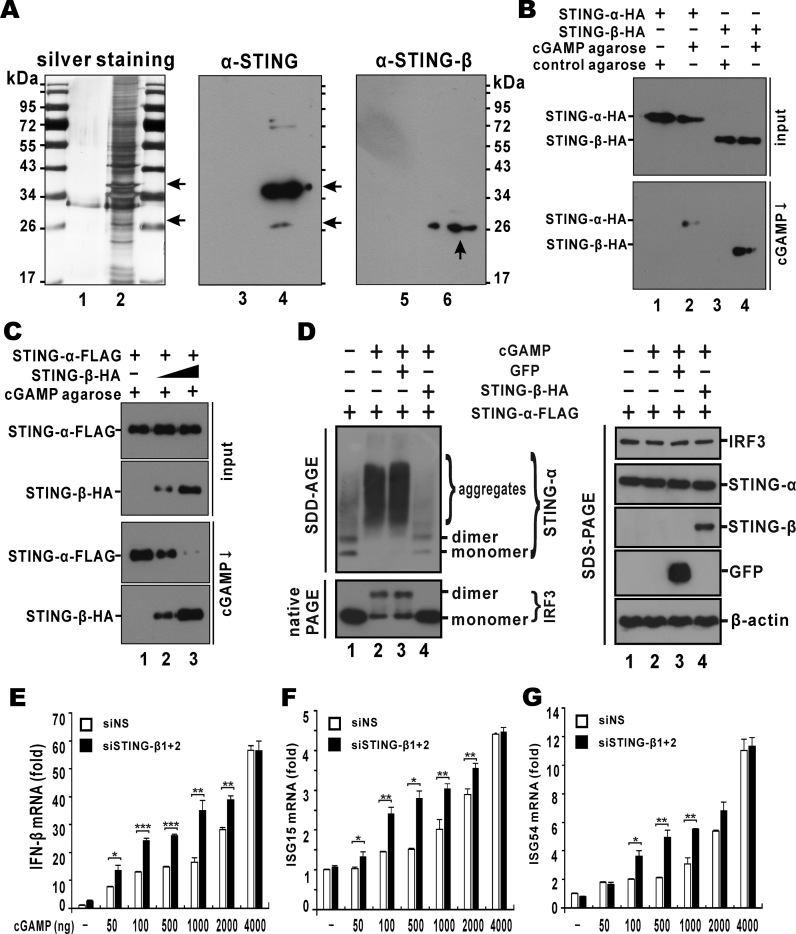
To detect the expression of STING-β protein, rabbit polyclonal antibodies were raised against a synthetic peptide corresponding to the unique N-terminus of STING-β (Figure 1A). We found that these antibodies could specifically recognize recombinant STING-β but not STING-α overexpressed in HEK293T cells. In contrast, the commercial anti-STING antibodies targeting the CTD recognized both STING-α and STING-β isoforms (Figure 1E). Because the expression levels of STING-β was very low and the avidity of the anti-STING-β antibodies was not sufficiently high, endogenous STING-β protein was not detected in the total cell lysates of THP-1, MT2, Jurkat or U937 cells. Given that STING-β contains the complete cGAMP-binding region of STING-α (8,20,65–67), we wondered whether cGAMP could be used to concentrate endogenous STING-β protein. Indeed, when we performed cGAMP pull-down assay to enrich STING-β protein from THP-1, MT2, Jurkat and U937 cells, a discrete STING-β protein band of about 26 kDa was detected in THP-1 and MT2 cells (Figure 1F). Hence, STING-β transcript is translated to a functional protein capable of cGAMP binding at least in some cultured human cells of monocytic and lymphocytic origin.
The TM domains help to anchor STING-α to the ER membrane (11,14,15). It remained to be seen whether the lack of TM domains could lead to dislocation of STING-β, together with its binding partners, from the ER. To address this issue, we compared the subcellular localisation of STING-α and STING-β by confocal microscopy. ER localisation of both STING-α and STING-β was observed in HeLa cells (Supplementary Figure S3). Some mitochondrial localisation was also found for STING-α, but not STING-β. How STING-β can be recruited to the ER in the absence of TM domains awaits further study.
STING-β antagonizes the antiviral activity of STING-α
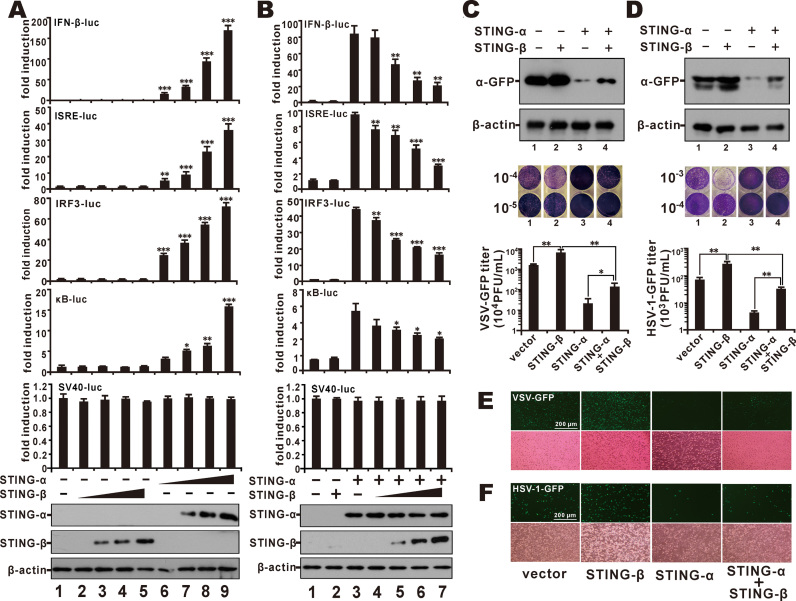
STING-β has the CTD of STING-α, which mediates ligand binding, protein interaction and signal transduction (1,11,17,18,66,68). With this in mind, we set out to compare the influence of STING-α and STING-β on type I IFN production, type I IFN signalling and NF-κB signalling as reflected by luciferase reporter expression under the control of IFN-β promoter (IFNβ-luc), IRF3-binding elements (IRF3-luc), ISRE (ISRE-luc) and κB elements (κB-luc). Consistent with previous findings (11,12,14,16), STING-α induced the activity of IFNβ-luc, IRF3-luc, ISRE-luc and κB-luc reporters in a dose-dependent manner when it was overexpressed in HEK293 cells. In contrast, overexpression of STING-β had no effect on these reporters (Figure 2A). Hence, STING-β did not share the same innate immunostimulatory properties with STING-α. Considering the inverse correlation of STING-β expression with IFN-β production (Figure 1C and D) as well as the domain structure (Figure 1A) and subcellular localisation (Supplementary Figure S3) of STING-β, we further asked whether and how STING-β might affect the activity of STING-α. When co-expressed with STING-α in HEK293 cells, STING-β suppressed the activity of STING-α on all four of the aforementioned reporters moderately but in a dose-dependent manner, and it had no influence on the activity of SV40 promoter (Figure 2B). Thus, STING-β antagonizes the immunostimulatory activity of STING-α.
Figure 2.
STING-β antagonizes the antiviral immunity of STING-α. (A) HEK293 cells were transfected with increasing doses (10, 50, 200, and 450 ng) of STING-β (lanes 2–5) or STING-α (lanes 6–9) plasmid together with IFNβ-luc (40 ng), IRF3-luc (40 ng), ISRE-luc (10 ng), κB-luc (10 ng) or SV40-luc (10 ng) reporter. Cells in lane 1 received pcDNA6 empty vector as control. The graphs show the means ± SD (n = 3). The differences between the indicated group and the control in lane 1 were statistically significant as judged by Student's t test (∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001). (B) STING-α (50 ng, lanes 3–7) were co-transfected into HEK293 cells together with increasing doses of STING-β plasmid (20, 100, 200 and 400 ng for lanes 4–7). Cells in lane 1 received pcDNA6 empty vector. Cells in lane 2 received STING-β plasmid alone. Additionally, 10 ng of pRL-TK reporter was added as an internal control. pcDNA6 empty vector was used to balance the total amount of transfected DNA. Dual luciferase assays were performed 36 h after transfection. The differences between the indicated group and the control in lane 1 were statistically significant as judged by Student's t test (∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001). (C, D) pcDNA6 empty vector (500 ng; lane 1), STING-β plasmid (500 ng; lane 2), STING-α plasmid (50 ng; lane 3) or STING-α (50 ng) + STING-β (450 ng) plasmids were transfected into HEK293T cells. After 36 h, cells were infected with VSV-GFP (0.1 M.O.I.) or HSV-1-GFP (1 M.O.I.) for another 12 h. The cells were then lysed for western blot analysis and the supernatant was collected for plaque assays. The differences between the indicated two groups were statistically significant as judged by Student's t test (∗P < 0.05 and ∗∗P < 0.01). (E, F) Fluorescent microscopic analysis of the proviral effect of STING-β in VSV-GFP- or HSV-1-GFP-infected HeLa cells. pcDNA6 empty vector (500 ng), STING-β plasmid (500 ng), STING-α plasmid (50 ng), STING-α (50 ng) + STING-β (450 ng) plasmids were transfected into HeLa cells. After 36 h, cells were infected with VSV-GFP (0.1 M.O.I.) (E) or HSV-1-GFP (1 M.O.I.) (F). Cells were analyzed by fluorescent microscopy 12 h after infection for VSV-GFP and 24 h after infection for HSV-1-GFP. Results in each panel are representative of three independent experiments.
To verify the STING-α antagonism of STING-β in antiviral response, we checked for the combined effect of STING-α and STING-β on the replication of two GFP-marked viruses, VSV-GFP and HSV-1-GFP. Notably, overexpression of STING-α and STING-β alone exhibited opposite effect on the replication of VSV-GFP and HSV-1-GFP as measured by GFP protein expression, plaque assays and GFP fluorescence. In addition, when STING-β was co-expressed with STING-α, the antiviral phenotype of STING-α was reversed (Figure 2C–F). Thus, our results from both luciferase assays and virus challenge experiments consistently demonstrated the proviral effect of STING-β.
STING-β suppresses IRF3 and NF-κB activation by cGAMP and other stimuli
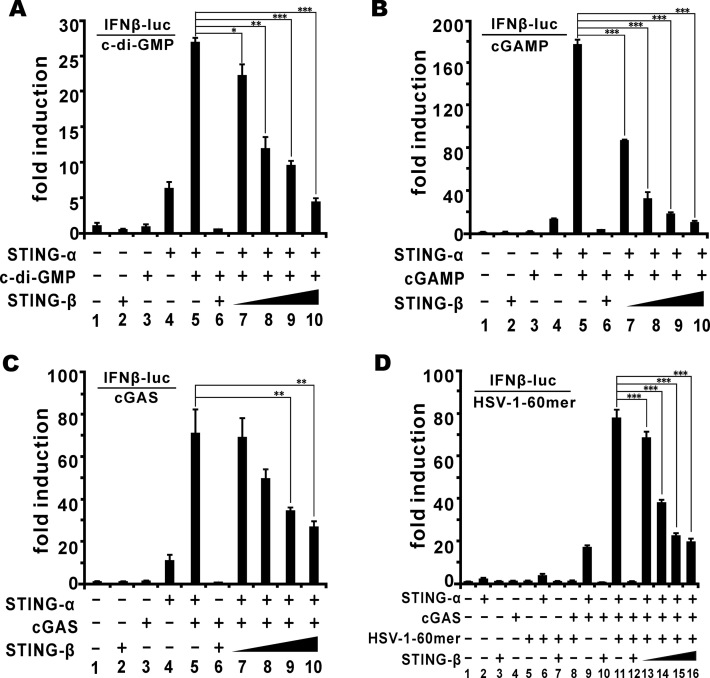
Both bacterial c-di-GMP and mammalian cGAMP bind to the CTD of STING-α to induce IRF3 phosphorylation by TBK1 and subsequent production of type I IFNs (6,8,13,20,65,68). Above we have also shown the concentration of STING-β from THP-1 and MT2 cell lysates by cGAMP pull-down (Figure 1F). We therefore sought to determine the impact of STING-β on CDN and cGAS signalling. Both c-di-GMP and cGAMP stimulated IFN-β promoter activity in HEK293T cells expressing STING-α (Figure 3A and B, lane 5) but not in cells expressing STING-β (Figure 3A and B, lane 6). Thus, although both STING-α and STING-β are capable of binding to cGAMP, STING-α but not STING-β served as the sensor of c-di-GMP and cGAMP. However, STING-β progressively mitigated c-di-GMP- and cGAMP-induced activation of STING-α and consequent induction of IFN-β promoter activity when STING-α was co-expressed with increasing doses of STING-β (Figure 3A and B, lanes 7–10). Because cGAMP is produced by cGAS upon binding with DNA (6,8,10), we repeated our experiments in the presence of cGAS or cGAS + HSV-1-60mer and obtained similar results (Figure 3C and D). Collectively, STING-β functions as a negative regulator of innate DNA sensing mediated by cGAS and CDNs.
Figure 3.
STING-β inhibits CDN- and DNA-induced activation of IFN-β promoter. (A, B) In HEK293T cells, STING-α plasmid (50 ng for lanes 4, 5 and 7–10) and increasing doses of STING-β plasmid (50, 50, 50, 200, 300 and 450 ng for lanes 2, 6 and 7–10) were co-transfected with pRL-TK (10 ng) and IFNβ-luc (40 ng) reporter plasmids as indicated. After 24 h, cells were stimulated by c-di-GMP (500 ng for lanes 3 and 5–10) (A) or cGAMP (500 ng for lanes 3 and 5–10) (B). The graphs show the means ± SD (n = 3). The differences between the indicated group and the control in lane 5 were statistically significant as judged by Student's t test (∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001). (C) STING-α plasmid (50 ng for lanes 4, 5 and 7–10), cGAS plasmid (50 ng for lanes 3, 5–10) and STING-β plasmid (50, 50, 50, 200, 300 and 450 ng for lanes 2, 6 and 7–10) were co-transfected into HEK293T cells with pRL-TK (10 ng) and IFNβ-luc (40 ng) reporter plasmids. The differences between the indicated group and the control in lane 5 were statistically significant as judged by Student's t test (∗∗P < 0.01). (D) STING-α plasmid (50 ng for lane 2, 6, 9, 11 and 13–16), cGAS plasmid (50 ng for lanes 4 and 8–16) plus STING-β plasmid (50 ng for lanes 3, 7, 10, 12 and 13 as well as 200, 300 and 350 ng for lanes 14–16) were co-transfected with pRL-TK (10 ng) and IFNβ-luc (40 ng) reporter plasmids. After 24 h, cells were stimulated with an immunostimulatory DNA known as HSV-1-60mer (500 ng for lanes 5–8 and 11–16) for another 12 h. Cells were then harvested for dual-luciferase assays. The differences between the indicated group and the control in lane 11 were statistically significant as judged by Student's t test (∗∗∗P < 0.001). Results in each panel are representative of three independent experiments.
Above we have shown that STING-β exerts a dominant inhibitory effect on the activation of IRF3 and NF-κB by STING-α (Figure 2B). Because STING-β has the CTD domain that also engages MAVS, TBK1 and other transducers, we extended our analysis to determine how STING-β might influence the activation of IFN-β promoter by additional stimuli including TBK1, IKKϵ, TRIF and IRF3 (5D). These stimuli are critical adaptors and transducers that might function upstream or downstream of STING-α in DNA and RNA sensing (1,2). STING-β exhibited an inhibitory activity on all stimuli tested except IRF3 (5D), a phosphomimetic and constitutively active mutant of IRF3 (Supplementary Figure S4A). The inability of STING-β to inhibit IRF3 (5D) activity indicated that STING-β functions upstream of IRF3, probably at the step of TBK1/IKKϵ. Consistent with these results, overexpression of STING-β inhibited the induction of IFN-β and ISG56 by TBK1, IKKϵ, TRIF but not IRF3 (5D) (Supplementary Figure S4B). When we repeated our experiments with two representative NF-κB-regulated genes encoding tumour necrosis factor α (TNF-α) and interleukin 8 (IL-8) (69), similar results were obtained. Particularly, overexpression of STING-β inhibited the induction of TNF-α and IL-8 transcripts by TBK1, IKKϵ, TRIF but not MyD88 (Supplementary Figure S4C). MyD88 is a transducer protein in TLR-induced activation of NF-κB (69). The lack of inhibition on MyD88 activity suggested that the action of STING-β is not promiscuous but has specificity. Thus, STING-β is capable of suppressing IRF3 and NF-κB activation by multiple stimuli. In keeping with this, induction of IFN-β, ISG56, TNF-α and IL-8 transcripts in HEK293 cells infected with SeV, VSV-GFP and HSV-1-ΔICP0 was augmented by STING-α but dampened by STING-β. In addition, co-expression of STING-α and STING-β resulted in the reversal of STING-α effect (Supplementary Figure S5). These results consistently demonstrated a dominant suppressive effect of STING-β in innate nucleic acid sensing.
Depletion of STING-β potentiates antiviral response
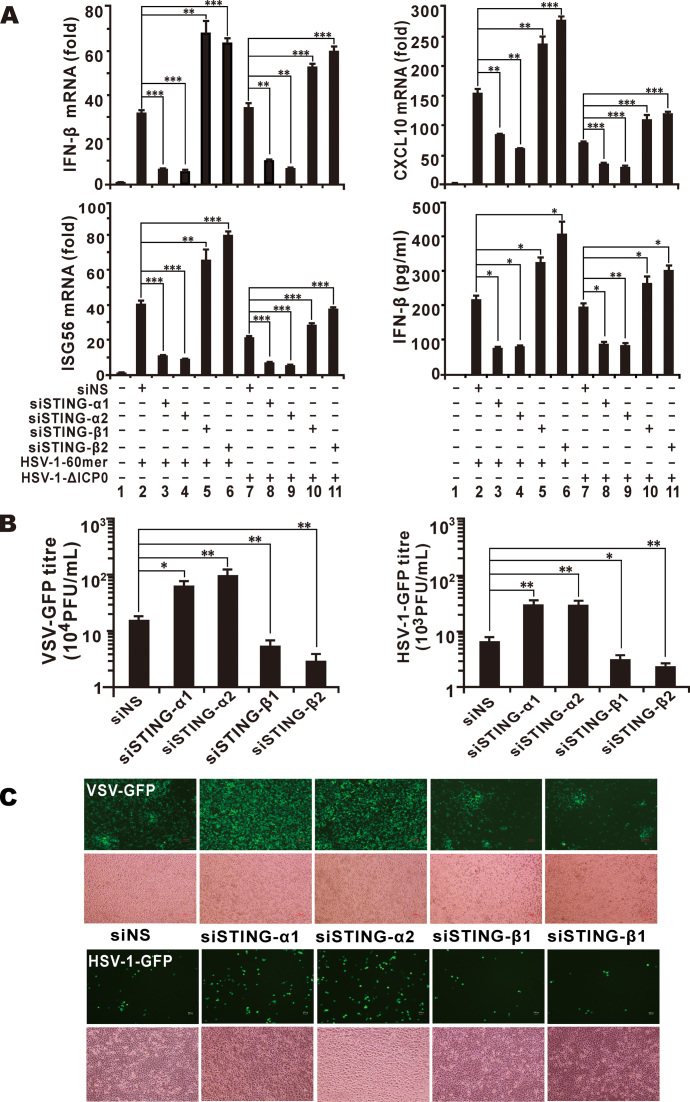
Unlike HEK293, HEK293T and HeLa cells, THP-1 cells possess an intact DNA sensing pathway (4). In addition, STING-β is also expressed relatively more abundantly in THP-1 cells (Supplementary Figure S2A). We therefore performed RNAi-based loss-of-function study of STING-β in these cells. Two pairs of siRNAs (siSTING-α1 and siSTING-α2) targeting the unique 5′ untranslated region of STING-α transcript were designed to specifically knockdown endogenous STING-α while sparing STING-β. On the other hand, two pairs of siRNAs (siSTING-β1 and siSTING-β2) targeting the N-terminal region of STING-β mRNA were designed to specifically knockdown endogenous STING-β without affecting STING-α. A pair of non-silencing siRNA (siNS) was used to control for any nonspecific effects induced by siRNA transfection. Indeed, siSTING-α specifically depleted endogenous STING-α but had no influence on STING-β. Likewise, siSTING-β selectively knocked down endogenous STING-β but did not affect STING-α (Supplementary Figure S6A and B). When endogenous STING-α was depleted in THP-1 cells, the induction of IFN-β, ISG56, CXCL10, ISG15 and ISG54 by HSV-1-60mer or HSV-1-ICP0 was blunted. On the contrary, the induction of these genes by HSV-1-60mer or HSV-1-ΔICP0 in STING-β-knockdown cells was further enhanced (Figure 4A plus Supplementary Figure S6C and D). Consistently, IFN-β detected in the conditioned media by ELISA declined in STING-α-knockdown cells but increased when STING-β expression was compromised (Figure 4A). These results revealed an inhibitory role of endogenous STING-β in DNA- and virus-induced expression of type I IFNs and ISGs.
Figure 4.
Depletion of STING-β by siRNA potentiates innate immune activation. (A) THP-1 cells cultured in six-well plates were transfected with 1 μl of siRNA (100 mM) targeting no human gene (siNS as a non-specific control), STING-α or STING-β. After 48 h, THP-1 cells were transfected with HSV-1-60mer (1000 ng/ml) or infected with 5 M.O.I. of HSV-1-ΔICP0 for another 9 h. THP-1 cells were then collected for RT-qPCR and the conditioned media of these samples were stored at –80°C for ELISA and subsequent analysis of antiviral activity. The differences between the indicated group and the siNS control group in lane 2 or 7 were statistically significant as judged by Student's t test (∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001). (B, C) Antiviral activity of conditioned media collected from siRNA-treated THP-1 cells. Conditioned media collected from THP-1 samples in (A) were used to stimulate HEK293T and HeLa cells by incubation for 3 h. After stimulation, HEK293T cells and HeLa cells were cultured for another 12 h and then infected with VSV-GFP (0.1 M.O.I.) or HSV-1-GFP (1 M.O.I.). The media from infected HEK293T cells were used for plaque assay (B) and GFP-positive HeLa cells can be observed under fluorescent microscope (C). The differences between the indicated group and the siNS control group in lane 2 in (A) were statistically significant as judged by Student's t test (∗P < 0.05 and ∗∗P < 0.01). Results in each panel are representative of three independent experiments.
To verify the role of STING-β in the context of viral infection, the conditioned media collected above for ELISA were further analyzed for antiviral activity. Both VSV-GFP and HSV-1-GFP replicated to higher titres in HEK293 cells incubated with conditioned media collected from siSTING-α-transfected THP-1 cells. However, viral replication was attenuated in HEK293 cells incubated with conditioned media from siSTING-β-transfected THP-1 cells (Figure 4B). When we repeated the same experiment in HeLa cells, similar results were obtained as reflected by the GFP fluorescence (Figure 4C). Hence, our results from RT-qPCR analysis of IFN-β and ISG transcripts, ELISA of IFN-β protein as well as virus challenge experiments were consistent and they corroborated with each other to support that STING-β acts as a negative regulator of type I IFN induction and antiviral response in THP-1 cells.
STING-β interacts with STING-α and TBK1 to prevent them from binding with each other
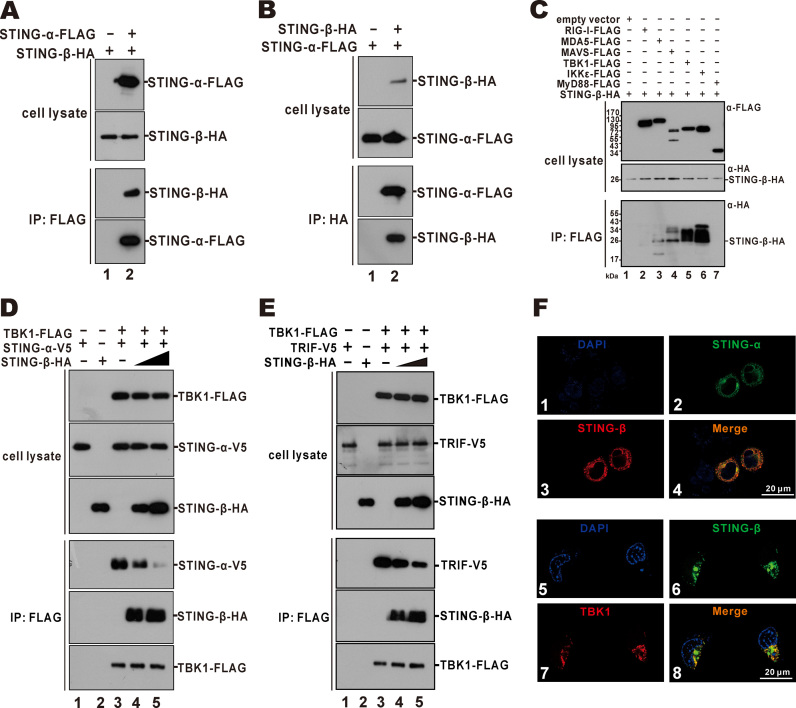
Because STING-α employs its CTD for dimer formation (66,68), it will be of interest to determine whether STING-β might interact with STING-α and perturb its activity using the same CTD. To investigate this, we used mouse anti-FLAG antibody to precipitate STING-α-containing protein complex from HEK293T cells and found STING-β-HA in this complex (Figure 5A). Reciprocally, when co-immunoprecipitation was carried out using mouse anti-HA antibody, STING-α-FLAG was detected in the STING-β-HA precipitate (Figure 5B). Thus, STING-β and STING-α interacted with each other when overexpressed in HEK293T cells.
Figure 5.
STING-β interacts with and inhibits STING-α and TBK1. (A, B) Interaction of STING-β with STING-α. Expression plasmids for STING-β-HA (9000 ng) and STING-α-FLAG (1000 ng) or STING-α-FLAG (1000 ng) and STING-β-HA (9000 ng) were co-transfected into HEK293T cells. After 48 h, cells were harvested and lysed. Immunoprecipitation (IP) was carried out with mouse anti-FLAG or anti-HA antibody. (C) Interaction of STING-β with MAVS, TBK1 and IKKϵ. Plasmids (1000 ng) expressing FLAG-tagged RIG-I, MDA5, MAVS, TBK1, IKKϵ and MyD88 were transfected individually into HEK293T cells together with STING-β-HA plasmid (9000 ng). Immunoprecipitation was performed with mouse anti-FLAG antibody. Input proteins were analyzed by western blotting with mouse anti-FLAG or anti-HA antibody. Immunoprecipitates were probed with rabbit anti-HA antibodies. (D) TBK1-FLAG plasmid (1000 ng) was co-transfected with STING-α-V5 plasmid (1000 ng) and increasing doses of STING-β-HA (4000 ng in lanes 2 and 4 as well as 8000 ng in lane 5) into HEK293T cells. Immunoprecipitation was carried out with mouse anti-FLAG antibody. (E) TBK1-FLAG plasmid (1000 ng) was co-transfected with TRIF-V5 plasmid (1000 ng) and increasing doses of STING-β-HA (4000 ng in lanes 2 and 4 as well as 8000 ng in lane 5) into HEK293T cells. Cells were harvested and lysed after 48 h. Immunoprecipitation was carried out with mouse anti-FLAG. Input proteins were probed with mouse anti-FLAG, anti-V5 or anti-HA antibody. Immunoprecipitates were probed with rabbit anti-V5, anti-HA or anti-FLAG antibodies. (F) Colocalisation of STING-β with STING-α and TBK1. STING-α-GFP plus STING-β-HA or STING-β-GFP plus TBK1-FLAG plasmids were co-transfected into HeLa cells. After 36 h, HeLa cells were fixed with 4% paraformaldehyde and then blocked with 5% bovine serum albumin. The cells were incubated with mouse anti-HA or anti-FLAG antibody and then goat anti-mouse IgG conjugated to TRITC (red) was used to stain for STING-β or TBK1. Nuclear morphology was revealed with DAPI (blue). Results in each panel are representative of three independent experiments.
The CTD is also known to interact with MAVS, TBK1 and IKKϵ (14,17,18). Therefore, it was not surprising that STING-β was also found to associate with MAVS, TBK1 and IKKϵ in the co-immunoprecipitation assay (Figure 5C). We next interrogated whether the interaction of STING-β with STING-α and TBK1 might impede the formation of STING-α-TBK1 complex, which is required for IRF3 phosphorylation and type I IFN induction (17). Detection of STING-α-V5 and STING-β-HA in the TBK1-FLAG precipitate (Figure 5D, lanes 3–5) raised the possibility that STING-β could also interact with TBK1 and compete with STING-α for binding with TBK1. Indeed, when STING-β-HA was more abundantly expressed, the level of STING-α-V5 in the precipitate was diminished (Figure 5D, lane 5). Results from a similar co-immunoprecipitation experiment also showed that STING-β was capable of impeding the formation of TBK1-TRIF complex (Figure 5E, lane 5). Plausibly, STING-β might act through two non-exclusive mechanisms to prevent the formation of STING-α-TBK1 and TBK1-TRIF complex. On one hand, STING-β interacts with STING-α and TBK1 to prevent them from contacting with each other or with other transducers such as TRIF. On the other hand, STING-β competes with STING-α and TBK1 for binding with their partners. In support of their interaction, STING-α and STING-β were found to co-localize in the cytoplasm of HeLa cells. Moreover, STING-β also co-localized with TBK1 (Figure 5F).
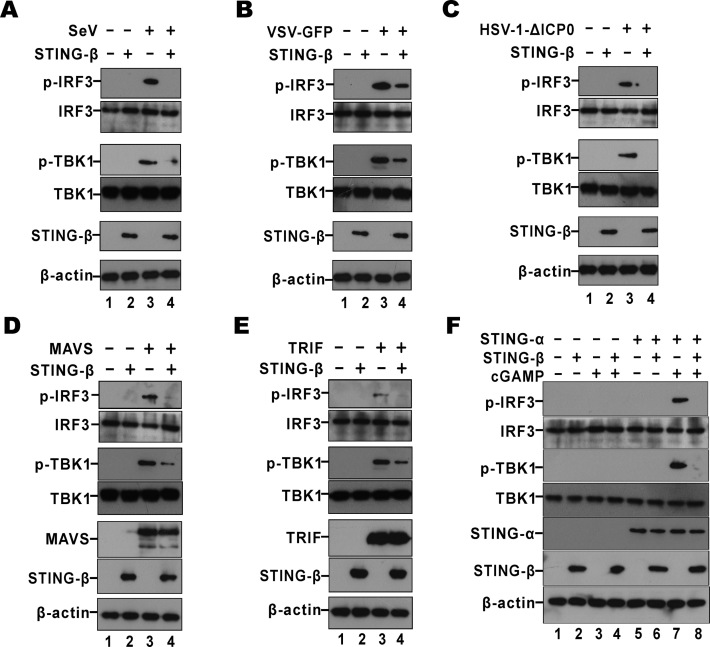
STING-β inhibits the phosphorylation of TBK1 and IRF3
The induction of type I IFNs requires the phosphorylation of TBK1 and IRF3 (17). Thus, the influence of STING-β on the phosphorylation of TBK1 and IRF3 was evaluated. First, the impact of STING-β on virus-induced phosphorylation of TBK1 and IRF3 was studied. In control cells receiving empty vector alone, SeV, VSV-GFP or HSV-1-ΔICP0 induced robust phosphorylation of TBK1 and IRF3 (Figure 6A–C). In contrast, the phosphorylated TBK1 and IRF3 species in STING-β-overexpressing cells were less pronounced or disappeared (Figure 6A–C). These results consistently suggested that STING-β negatively regulated the activation of IRF3 by SeV, VSV-GFP and HSV-1-ΔICP0 by preventing phosphorylation of TBK1 and IRF3. Our analysis was next extended to MAVS and TRIF. Whereas MAVS is a key adaptor between RIG-I and TBK1 (17), TRIF transduces the activation signal triggered by TLRs (2). However, as mentioned above, TRIF is also required for optimal STING-α function (45). Interestingly, the prominent phosphorylated TBK1 and IRF3 species seen in MAVS- or TRIF-overexpressing cells were diminished upon expression of STING-β (Figure 6D and E). Finally, we examined the impact of STING-β on cGAMP-induced phosphorylation of TBK1 and IRF3. The phosphorylation of TBK1 and IRF3 was robust when STING-α was simulated with cGAMP (Figure 6F, lane 7). However, this phenotype was abrogated by STING-β (Figure 6F, lane 8). Thus, STING-β dominantly inhibits the phosphorylation of TBK1 and IRF3 triggered by cGAMP binding to STING-α.
Figure 6.
STING-β inhibits phosphorylation of TBK1 and IRF3. (A–C) HEK293T cells were transfected with 450 ng of STING-β plasmid. After 24 h cells were infected with SeV (80 HA/ml), VSV-GFP (0.1 M.O.I.) or HSV-1-ΔICP0 (5 M.O.I.) for another 9 h. Cells were lysed for western blotting. (D, E) HEK293T cells were transfected with 450 ng of STING-β plasmid together with 50 ng of MAVS or TRIF plasmid. After 36 h, cells were lysed for western blotting. (F) STING-β plasmid (450 ng in lanes 2, 4, 6 and 8) was co-transfected with either empty vector or STING-α plasmid (50 ng in lanes 5–8) into HEK293T cells. After 24 h, 500 ng of cGAMP was transfected to stimulate the cells for another 12 h. Cells were then harvested and lysed for western blot analysis with anti-p-TBK1, anti-p-IRF3, anti-FLAG, anti-HA and anti-β-actin antibodies. Gel images in each panel are representative of three independent experiments.
STING-β binds to cGAMP and prevents it from binding with STING-α
Above we have experimentally validated the ability of STING-β to bind with cGAMP (Figure 1F), the inability of STING-β to activate IFN-β promoter in the presence of cGAMP, and the activity of STING-β to suppress IFN-β promoter in the presence of cGAMP and STING-α (Figure 3B). Plausibly, STING-β could interact with STING-α and cGAMP separately to prevent them from binding to each other. STING-β might also directly compete with STING-α for binding with cGAMP. To shed light on how STING-β exerts its suppressive effect on cGAMP signalling, the binding of endogenous STING-α and STING-β with cGAMP was studied using cGAMP pull-down assay. In THP-1 cells stimulated with DNA, endogenous cGAMP is produced by cGAS to induce type I IFNs by binding to STING-α and activating IRF3. THP-1 cell lysates were incubated with cGAMP-agarose and then washed with PBS containing 100 μM ATP. The collected cGAMP-bound proteins were analyzed by silver staining and western blotting. Compared to the control agarose, cGAMP-agarose was found to retain multiple proteins from THP-1 cell lysates (Figure 7A, lane 2). Among these, several protein bands with sizes compatible to those of STING-α (35–40 kDa) and STING-β (23–28 kDa) were observed (Figure 7A, lane 2). Next, western blotting was carried out to verify the pull down of STING-α and STING-β by cGAMP-agarose. Using STING antibodies, a strong band at the size of STING-α was found. Interestingly, another weaker band near the predicted size of STING-β was also visible (Figure 7A, lane 4). Using STING-β-specific antibodies, a specific band around the predicted size of STING-β was detected (Figure 7A, lane 6). Hence, both endogenous STING-α and STING-β were bound to cGAMP.
Figure 7.
STING-β impedes the binding of cGAMP to STING-α. (A) Endogenous STING-α and STING-β could be pulled down by cGAMP agarose. THP-1 cells were lysed and incubated with cGAMP agarose (lanes 2, 4 and 6) or control agarose (lanes 1, 3 and 5) for 2 h at 4°C. After washing with PBS containing 100 μM ATP for six times, sample loading buffer was added. The samples were boiled for 10 min and further analyzed by SDS-PAGE followed by silver staining (lanes 1–2) or western blotting with either anti-STING-α (lanes 3–4) or anti-STING-β (lanes 5–6). Arrows point to STING-α and STING-β bands of expected sizes. (B) Recombinant STING-α and STING-β could be pulled down by cGAMP agarose. STING-α-HA (1000 ng) and STING-β-HA (9000 ng) plasmids were transfected into HEK293T cells. After 48 h, cells were harvested and lysed. cGAMP pull-down (cGAMP ↓) assay was carried out with cGAMP agarose or control agarose. Input proteins and cGAMP pull-down products were analyzed by western blotting with mouse anti-HA or anti-FLAG antibody. (C) Overexpression of STING-β prevented STING-α from binding to cGAMP. STING-α-HA plasmid (1000 ng) and increasing doses of STING-β-HA plasmid (300 ng for lane 2 and 9000 ng for lane 3) were co-transfected into HEK293T cells. After 48 h, cells were lysed and cGAMP pull-down (cGAMP ↓) assay was carried out. (D) STING-β inhibits cGAMP-induced aggregation of STING-α. HEK293T cells were transfected with STING-α plasmid (50 ng for lanes 1–4), cGAMP (200 ng for lanes 2–4), GFP plasmid (200 ng for lane 3) and STING-β plasmid (200 ng for lane 4). The protein samples were analyzed with SDD-AGE, native PAGE and SDS-PAGE. (E-G) Knockdown of STING-β augments cGAMP-induced activation of IFN-β and ISG production. THP-1 cells cultured in six-well plates were transfected with either 1 μl of negative control siRNA (siNS; 100 mM) or 0.5 μl of siSTING-β1 (100 mM) plus 0.5 μl of siSTING-β2 (100 mM), denoted as siSTING-β1+2. After 48 h, THP-1 cells were transfected with cGAMP using Lipofectamine 2000 (Invitrogen). After another 12 h, THP-1 cells were collected for RT-qPCR analysis. The statistical differences between the indicated groups were judged by Student's t test (∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001). Results in each panel are representative of three independent experiments.
To further characterize cGAMP binding of STING-α and STING-β, STING-α-HA and STING-β-HA were expressed in HEK293T cells. The cell lysates containing recombinant STING-α-HA or STING-β-HA were used for cGAMP pull-down. The results showed that both STING-α-HA and STING-β-HA were pulled down by cGAMP-agarose but not by control agarose (Figure 7B). To further explore whether STING-β can affect the binding of cGAMP to STING-α, cGAMP pull-down was performed with HEK293T cells co-expressing STING-β and STING-α. Although both STING-α-FLAG and STING-β-HA could be pulled down by cGAMP-agarose, increasing doses of STING-β-HA resulted in diminution of cGAMP-bound STING-α-FLAG. That is to say, STING-β exerts a dominant inhibitory effect on the binding of STING-α to cGAMP (Figure 7C).
Activation of STING-α by TBK1 results in aggregation (12,43). Indeed, SDD-AGE analysis revealed the formation of STING-α aggregates in the lysate of cGAMP-stimulated HEK293T cells (Figure 7D, lane 2). Enforced expression of STING-β in these cells erased the stimulatory effect of cGAMP in the induction of STING-α aggregates (Figure 7D, lane 4), but the expression of an irrelevant GFP had no influence (lane 3). Consistent with this, the formation of IRF3 dimer in response to cGAMP stimulation was also seen on the native PAGE and it disappeared when STING-β was also expressed (Figure 7D). Thus, STING-β effectively suppressed cGAMP-induced aggregation of STING-α and activation of IRF3.
To determine how STING-β might affect the immunostimulatory activity of cGAMP under physiological condition, we activated the expression of IFN-β, ISG15 and ISG54 in THP-1 cells through cGAMP transfection. A dose-dependent induction of IFN-β, ISG15 and ISG54 expression by cGAMP was observed. Compromising STING-β with siSTING-β1+2 in cGAMP-transfected THP-1 cells resulted in further augmentation of IFN-β, ISG15 and ISG54 induction by cGAMP over a wide range of cGAMP concentrations. This effect of siSTING-β1+2 was not seen only when excess cGAMP was present (Figure 7E–G). These results indicated that depletion of STING-β in THP-1 cells sensitizes the cells to cGAMP stimulation. In other words, STING-β negatively regulates cGAMP-dependent activation of innate immune response.
DISCUSSION
In this study we identified and characterized STING-β as a novel transcript isoform that dominantly inhibits STING-α-mediated innate nucleic acid sensing. STING-β retains a CTD that interacts with cGAMP second messenger, STING-α, TBK1 and other transducers. However, its interaction with these molecules is counterproductive in innate immune signalling. STING-β preoccupies and sequesters these molecules to prevent them from engaging with their physiological effectors. STING-β also prevents aggregation and activation of STING-α. Our findings reveal new mechanistic insight on the regulation of innate immune response and have implications in the development of immunomodulatory agents.
Alternative splicing is an important regulatory mechanism in innate immunity and splice isoforms of various immunoregulatory proteins such as TLR3, TLR4, MyD88, IRAK2, TRAM, TRIF, TBK1, MAVS, IKKϵ, RIG-I and IRF3 have been described (50). STING isoforms generated by alternative splicing or single nucleotide polymorphisms have also been reported to influence CDN recognition, protein-protein interaction and signal transduction (9,27,48,70–72). Compared to other mechanisms for generation of RNA and protein isoforms, such as alternative exon inclusion, exon skipping, intron retention, alternative splice sites and alternative polyadenylation (50), STING-β is transcribed from an alternative promoter distinct from that of STING-α, resulting in the use of an alternative first exon and an alternative 5′ transcription initiation site. Intriguingly, STING-α and STING-β exhibit opposite expression patterns during virus infection. Whereas STING-α is an ISG in a positive feedback loop (73), STING-β expression correlates inversely with the induction of type I IFNs and might therefore constitute a negative feedback mechanism. The existence of STING-β-like proteins in other mammals provides crucial support to the biological importance of STING-β. In addition, a mouse isoform (GenBank accession number NP_001276521) that contains the CTD only might also serve the same function as human STING-β. Whether concurrent disruption of this isoform in the STING−/− mice might cause the previously described autoimmune-prone phenotype (49) remains to be clarified.
MRP is an alternatively spliced isoform of STING-α generated by skipping of exon 7. MRP shares the N-terminal portion with STING-α but possesses a unique stretch of 30 amino acids at the C-terminus, lacking the conserved CTD for TBK1 interaction and CDN binding. It is therefore not surprising that MRP exerts an inhibitory effect on IRF3 activation. Like STING-β, MRP is expressed in various tissues and cell lines. However, MRP expression was down-regulated by SeV but up-regulated by HSV-1. Opposite to the effect of STING-β, MRP is capable of activating NF-κB (48). MRP and STING-α share the same promoter, but STING-β has its own. It remains to be clarified how differential regulation of their expression can be achieved.
STING-β localizes to the ER although it does not have TM domains. Plausibly, STING-β can be recruited to the ER by STING-α or other ER-associated proteins such as ZDHHC1 (47) and AMFR (38). Overexpression of STING-β did not activate IRF3 or NF-κB, suggesting that the correct subcellular localisation mediated by the N-terminal TM domains might be required for STING-α activity. The role of TM domains in NF-κB activation has been shown in the MRP isoform (48). Additional roles of TM domains in signal transduction remain to be elucidated. Exactly how localisation of STING-β affects its suppressive function in nucleic acid sensing requires further study.
We have used several different methods including luciferase assays, RT-qPCR, ELISA, electrophoretic detection of STING-α aggregates and biological assays for antiviral activity to demonstrate the dominant inhibitory effect of STING-β in innate nucleic acid sensing. Results from these experiments were generally consistent and they corroborated with each other. Depletion of STING-β from THP-1 cells exhibited a potentiating effect on IFN-β and ISG induction by HSV-1, dsDNA and cGAMP, indicating its physiological role in nucleic acid sensing. Although enforced expression of STING-β alone had no influence on IRF3 and NF-κB activation in the luciferase reporter assays, it was sufficient to give a mild inhibitory effect in the biological assay for antiviral activity as shown consistently in all three readouts (i.e. GFP protein expression, virus titre and GFP fluorescence). This might be explained by the sensitivity of the different assays.
We proposed that STING-β suppresses innate nucleic acid sensing primarily through preoccupation and sequestration of its binding partners to prevent them from engaging physiological effectors. As such, STING-β antagonises STING-α function by not only inhibiting STING-α-TBK1 complex formation but also preventing the binding of cGAMP to STING-α. These two mechanisms are not mutually exclusive. On one hand, STING-β interacts with STING-α to prevent it from engaging cGAMP, TBK1 and other signal transducers. In other words, the formation of STING-α-STING-β heterodimer is unproductive or counterproductive in signalling. Several lines of our results are in favour of this model. First, STING-α interacted with STING-β. Second, the interaction between STING-α and TBK1 was compromised in the presence of STING-β. Third, the binding of STING-α with cGAMP was impeded when STING-β was expressed. Finally, the induction of TBK1 and IRF3 phosphorylation by cGAMP and STING-α was also suppressed by STING-β. Further validation of this model requires comparison of the structure and function of STING-α-STING-α and STING-α-STING-β complexes. On the other hand, STING-β also interacted with cGAMP, TBK1 and other transducers to prevent them from activating STING-α and downstream signalling. Comparing the affinity and dynamics of the interaction of STING-β versus STING-α with these ligands and signal transducers might shed light on exactly how STING-β perturbs their function in innate nucleic acid sensing. Further investigations will elucidate whether subcellular localisation, binding affinity, competition and other factors would be most critical in STING-β-mediated suppression of their activity. Our demonstration that STING-β can be enriched by cGAMP pull-down suggests that their binding affinity should be high, but in vitro studies with purified recombinant protein are still necessary and will clarify whether STING-β indeed binds more tightly to cGAMP and how it affects the binding of cGAMP to STING-α. Plausibly, the interaction of STING-β with other transducers such as TBK1, MAVS and TRIF could explain its suppressive effect on the activation of IRF3, NF-κB and other innate sensing pathways. STING-α is a shared downstream adaptor or transducer in both DNA and RNA sensing (22). Our findings on the inhibition of RNA sensing by STING-β are generally consistent with this model. The preoccupation and sequestration model described for STING-α might also operate in these scenarios. The suppression of TRIF activity by STING-β is particularly noteworthy. In addition to its function in adapting TLR signalling to STING-α (45), TRIF is also required for full activity of STING-α (45). Our finding on the suppression of TRIF activity by STING-β revealed a new facet of their interplay. Because STING-α interacts with TRIF through the CTD (45), STING-β should also be capable of binding with TRIF. Thus the action of STING-β might be mediated through preoccupation and sequestration of TRIF. It will be of interest to determine the mutual dependence of TRIF and STING-α as well as the influence of STING-β on STING-α-TRIF interaction.
STING-β is generally expressed at low levels. However, a functional STING-β protein capable of binding with cGAMP was detected in unstimulated THP-1 cells. In addition, both nucleic acid transfection and viral infection induced the fall and rise of STING-β transcript, which correlated inversely with IFN and ISG production. Furthermore, SLE is an interferonopathy associated with hyperactivation of IFN signalling and plasma levels of STING-β transcript in SLE patients remained low in our study. All the above three lines of evidence support the biological relevance of STING-β in viral infection and autoimmune response. In our new model, STING-β ambiently associates with STING-α and TBK1 to serve as a brake in innate IFN response. Upon immune challenge by DNA, RNA and pathogens, STING-β expression is down-regulated leading to the release of cGAMP, STING-α and TBK1 for activation of IFN production (Supplementary Figure S7). This model might also explain the relatively low expression of STING-β. If STING-β is much more abundant, the innate immune system cannot respond swiftly to invading nucleic acids and pathogens. STING-β suppresses innate nucleic acid sensing through a mechanism which is fundamentally different from those of known inhibitors such as E3 ubiquitin ligases (32), ATG9 (39), ISG56 (44), ULK1 (40), NLRC3 (41), NLRX1 (42) and MRP (48). Its probable role is to provide a negative regulatory mechanism that can be quickly brought into action at the right time and in the right subcellular location.
In this study, STING-β generated by alternative transcription initiation was identified as a novel negative regulator of DNA sensing. Further investigations in the following three areas may help to shed additional light on the regulation of STING-β expression, the physiological function of STING-β and the mechanism of STING-β-mediated suppression of nucleic acid sensing. First, the regulation of STING-β promoter should be characterized in more detail. Our findings suggest that the rise and fall of STING-β mRNA during the course of viral infection and autoimmune disease might be critical in the activation and termination of innate immune response. STING-α has recently been characterised as an ISG induced by type I IFNs (73). Although no ISRE was found in STING-β promoter, we demonstrated its induction by IFN-β and the mechanism merits investigations. Transcription factor EGR1 highly inducible by epidermal growth factor, phorbol ester and JAK-STAT signalling has been found to bind with STING-β promoter in the ENCODE database. STING-β promoter was also shown to be induced by TPA, which is known to activate EGR1 expression. Experimental validation of the role of EGR1 in the regulation of STING-β expression is warranted. Second, STING-β−/− THP-1 cells should be constructed and characterised. Knockdown of STING-β using siRNAs in THP-1 cells potentiated the induction of IFN-β and ISGs by cytosolic dsDNA, DNA virus and cGAMP. The moderate potentiating effect could be explained by incomplete depletion of STING-β. Because mouse STING-β has not be characterised, CRISPR/Cas9-mediated knockout of human STING-β is desirable. Third, the protein domain and residues that mediate the suppressive function of STING-β should be determined. This analysis may pave the way for design and development of peptide mimetics with immunosuppressive activity. In light of the relevance of STING-α to various diseases including cancer (1,27,70), the STING-α-inhibiting peptides might prove useful as versatile anti-inflammatory and anti-cancer agents.
DATA AVAILABILITY
The cDNA sequence of STING-β hs been deposited in GenBank under accession number MF360993.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues Mickey Wong, Roy Wong and Hinson Cheung for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Hong Kong Health and Medical Research Fund [15140682, HKM-15-M01]; Hong Kong Research Grants Council [HKU171091/14M, C7011-15R]. Funding for open access charge: Hong Kong Health and Medical Research Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen Q., Sun L., Chen Z.J.. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016; 17:1142–1149. [DOI] [PubMed] [Google Scholar]
- 2. Roers A., Hiller B., Hornung V.. Recognition of endogenous nucleic acids by the innate immune system. Immunity. 2016; 44:739–754. [DOI] [PubMed] [Google Scholar]
- 3. Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y.J.. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011; 12:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S. et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010; 11:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferguson B.J., Mansur D.S., Peters N.E., Ren H., Smith G.L.. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012; 1:e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun L., Wu J., Du F., Chen X., Chen Z.J.. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013; 339:786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu J., Sun L., Chen X., Du F., Shi H., Chen C., Chen Z.J.. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013; 339:826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Rohl I., Hopfner K.P., Ludwig J., Hornung V.. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013; 498:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diner E.J., Burdette D.L., Wilson S.C., Monroe K.M., Kellenberger C.A., Hyodo M., Hayakawa Y., Hammond M.C., Vance R.E.. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013; 3:1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao P., Ascano M., Wu Y., Barchet W., Gaffney B.L., Zillinger T., Serganov A.A., Liu Y., Jones R.A., Hartmann G. et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013; 153:1094–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishikawa H., Barber G.N.. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008; 455:674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ishikawa H., Ma Z., Barber G.N.. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009; 461:788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M., Hayakawa Y., Vance R.E.. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011; 478:515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P. et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008; 29:538–550. [DOI] [PubMed] [Google Scholar]
- 15. Jin L., Waterman P.M., Jonscher K.R., Short C.M., Reisdorph N.A., Cambier J.C.. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 2008; 28:5014–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z.. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:8653–8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015; 347:aaa2630. [DOI] [PubMed] [Google Scholar]
- 18. Tanaka Y., Chen Z.J.. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012; 5:ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davies B.W., Bogard R.W., Young T.S., Mekalanos J.J.. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012; 149:358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Danilchanka O., Mekalanos J.J.. Cyclic dinucleotides and the innate immune response. Cell. 2013; 154:962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen H., Sun H., You F., Sun W., Zhou X., Chen L., Yang J., Wang Y., Tang H., Guan Y. et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011; 147:436–446. [DOI] [PubMed] [Google Scholar]
- 22. Zevini A., Olagnier D., Hiscott J.. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol. 2017; 38:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nazmi A., Mukhopadhyay R., Dutta K., Basu A.. STING mediates neuronal innate immune response following Japanese encephalitis virus infection. Sci. Rep. 2012; 2:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahn J., Barber G.N.. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr. Opin. Immunol. 2014; 31:121–126. [DOI] [PubMed] [Google Scholar]
- 25. Klarquist J., Hennies C.M., Lehn M.A., Reboulet R.A., Feau S., Janssen E.M.. STING-mediated DNA sensing promotes antitumor and autoimmune responses to dying cells. J. Immunol. 2014; 193:6124–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao D., Li T., Li X.D., Chen X., Li Q.Z., Wight-Carter M., Chen Z.J.. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5699–E5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barber G.N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 2015; 15:760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crowl J.T., Gray E.E., Pestal K., Volkman H.E., Stetson D.B.. Intracellular nucleic acid detection in autoimmunity. Annu. Rev. Immunol. 2017; 35:313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Montealegre Sanchez G.A., Tenbrock K., Wittkowski H., Jones O.Y., Kuehn H.S. et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 2014; 371:507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jeremiah N., Neven B., Gentili M., Callebaut I., Maschalidi S., Stolzenberg M.C., Goudin N., Fremond M.L., Nitschke P., Molina T.J. et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest. 2014; 124:5516–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Konig N., Fiehn C., Wolf C., Schuster M., Cura Costa E., Tungler V., Alvarez H.A., Chara O., Engel K., Goldbach-Mansky R. et al. Familial chilblain lupus due to a gain-of-function mutation in STING. Ann. Rheum. Dis. 2017; 76:468–472. [DOI] [PubMed] [Google Scholar]
- 32. Davis M.E., Gack M.U.. Ubiquitination in the antiviral immune response. Virology. 2015; 479–480:52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J., Hu M.M., Wang Y.Y., Shu H.B.. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 2012; 287:28646–28655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y., Kawai T., Akira S.. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010; 33:765–776. [DOI] [PubMed] [Google Scholar]
- 35. Wang Y., Lian Q., Yang B., Yan S., Zhou H., He L., Lin G., Lian Z., Jiang Z., Sun B.. TRIM30α is a negative-feedback regulator of the intracellular DNA and DNA virus-triggered response by targeting STING. PLoS Pathog. 2015; 11:e1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhong B., Zhang L., Lei C., Li Y., Mao A.P., Yang Y., Wang Y.Y., Zhang X.L., Shu H.B.. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009; 30:397–407. [DOI] [PubMed] [Google Scholar]
- 37. Qin Y., Zhou M.T., Hu M.M., Hu Y.H., Zhang J., Guo L., Zhong B., Shu H.B.. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 2014; 10:e1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Q., Liu X., Cui Y., Tang Y., Chen W., Li S., Yu H., Pan Y., Wang C.. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 2014; 41:919–933. [DOI] [PubMed] [Google Scholar]
- 39. Saitoh T., Fujita N., Hayashi T., Takahara K., Satoh T., Lee H., Matsunaga K., Kageyama S., Omori H., Noda T. et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:20842–20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Konno H., Konno K., Barber G.N.. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013; 155:688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang L., Mo J., Swanson K.V., Wen H., Petrucelli A., Gregory S.M., Zhang Z., Schneider M., Jiang Y., Fitzgerald K.A. et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 2014; 40:329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo H., Konig R., Deng M., Riess M., Mo J., Zhang L., Petrucelli A., Yoh S.M., Barefoot B., Samo M. et al. NLRX1 sequesters STING to negatively regulate the interferon response, thereby facilitating the replication of HIV-1 and DNA viruses. Cell Host Microbe. 2016; 19:515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Z., Liu G., Sun L., Teng Y., Guo X., Jia J., Sha J., Yang X., Chen D., Sun Q.. PPM1A regulates antiviral signaling by antagonizing TBK1-mediated STING phosphorylation and aggregation. PLoS Pathog. 2015; 11:e1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Y., Li C., Xue P., Zhong B., Mao A.P., Ran Y., Chen H., Wang Y.Y., Yang F., Shu H.B.. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7945–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang X., Majumdar T., Kessler P., Ozhegov E., Zhang Y., Chattopadhyay S., Barik S., Sen G.C.. STING requires the adaptor TRIF to trigger innate immune responses to microbial infection. Cell Host Microbe. 2016; 20:329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang F., Alain T., Szretter K.J., Stephenson K., Pol J.G., Atherton M.J., Hoang H.D., Fonseca B.D., Zakaria C., Chen L. et al. S6K-STING interaction regulates cytosolic DNA-mediated activation of the transcription factor IRF3. Nat. Immunol. 2016; 17:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou Q., Lin H., Wang S., Wang S., Ran Y., Liu Y., Ye W., Xiong X., Zhong B., Shu H.B. et al. The ER-associated protein ZDHHC1 is a positive regulator of DNA virus-triggered, MITA/STING-dependent innate immune signaling. Cell Host Microbe. 2014; 16:450–461. [DOI] [PubMed] [Google Scholar]
- 48. Chen H., Pei R., Zhu W., Zeng R., Wang Y., Wang Y., Lu M., Chen X.. An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. J. Immunol. 2014; 192:1162–1170. [DOI] [PubMed] [Google Scholar]
- 49. Sharma S., Campbell A.M., Chan J., Schattgen S.A., Orlowski G.M., Nayar R., Huyler A.H., Nundel K., Mohan C., Berg L.J. et al. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E710–E717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carpenter S., Ricci E.P., Mercier B.C., Moore M.J., Fitzgerald K.A.. Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 2014; 14:361–376. [DOI] [PubMed] [Google Scholar]
- 51. Luo X., Yang W., Ye D.Q., Cui H., Zhang Y., Hirankarn N., Qian X., Tang Y., Lau Y.L., de Vries N. et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011; 7:e1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morris D.L., Sheng Y., Zhang Y., Wang Y.F., Zhu Z., Tombleson P., Chen L., Cunninghame Graham D.S., Bentham J., Roberts A.L. et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet. 2016; 48:940–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y., Yang J., Zhang J., Sun L., Hirankarn N., Pan H.F., Lau C.S., Chan T.M., Lee T.L., Leung A.M. et al. Genome-wide search followed by replication reveals genetic interaction of CD80 and ALOX5AP associated with systemic lupus erythematosus in Asian populations. Ann. Rheum. Dis. 2016; 75:891–898. [DOI] [PubMed] [Google Scholar]
- 54. Ng M.H., Ho T.H., Kok K.H., Siu K.L., Li J., Jin D.Y.. MIP-T3 is a negative regulator of innate type I IFN response. J. Immunol. 2011; 187:6473–6482. [DOI] [PubMed] [Google Scholar]
- 55. Kok K.H., Lui P.Y., Ng M.H., Siu K.L., Au S.W., Jin D.Y.. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe. 2011; 9:299–309. [DOI] [PubMed] [Google Scholar]
- 56. Kew C., Lui P.Y., Chan C.P., Liu X., Au S.W., Mohr I., Jin D.Y., Kok K.H.. Suppression of PACT-induced type I interferon production by herpes simplex virus 1 Us11 protein. J. Virol. 2013; 87:13141–13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Orzalli M.H., Conwell S.E., Berrios C., DeCaprio J.A., Knipe D.M.. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E4492–E4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kalamvoki M., Roizman B.. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E611–E617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bauer I., Hohl M., Al-Sarraj A., Vinson C., Thiel G.. Transcriptional activation of the Egr-1 gene mediated by tetradecanoylphorbol acetate and extracellular signal-regulated protein kinase. Arch. Biochem. Biophys. 2005; 438:36–52. [DOI] [PubMed] [Google Scholar]
- 60. Xia T., Konno H., Barber G.N.. Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res. 2016; 76:6747–6759. [DOI] [PubMed] [Google Scholar]
- 61. Xia T., Konno H., Ahn J., Barber G.N.. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 2016; 14:282–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eloranta M.L., Ronnblom L.. Cause and consequences of the activated type I interferon system in SLE. J. Mol. Med. (Berlin). 2016; 94:1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garaud J.C., Schickel J.N., Blaison G., Knapp A.M., Dembele D., Ruer-Laventie J., Korganow A.S., Martin T., Soulas-Sprauel P., Pasquali J.L.. B cell signature duringinactive systemic lupus is heterogeneous: toward a biological dissection of lupus. PLoS One. 2011; 6:e23900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ren L.-L., Li F.-R., Liu D.-Z., Qi H., Ouyang Z.-B.. Analysis of expression of early growth response 1 gene in patients with systemic lupus erythematosus by fluorescent quantitative PCR. Chin. J. Biolog. 2012; 25:102–110. [Google Scholar]
- 65. Zhang X., Shi H., Wu J., Zhang X., Sun L., Chen C., Chen Z.J.. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell. 2013; 51:226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shu C., Yi G., Watts T., Kao C.C., Li P.. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 2012; 19:722–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kranzusch P.J., Wilson S.C., Lee A.S., Berger J.M., Doudna J.A., Vance R.E.. Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell. 2015; 59:891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ouyang S., Song X., Wang Y., Ru H., Shaw N., Jiang Y., Niu F., Zhu Y., Qiu W., Parvatiyar K. et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012; 36:1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Q., Lenardo M.J., Baltimore D.. 30 years of NF-κB: A blossoming of relevance to human pathobiology. Cell. 2017; 168:37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Corrales L., McWhirter S.M., Dubensky T.W. Jr, Gajewski T.F.. The host STING pathway at the interface of cancer and immunity. J. Clin. Invest. 2016; 126:2404–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Patel S., Blaauboer S.M., Tucker H.R., Mansouri S., Ruiz-Moreno J.S., Hamann L., Schumann R.R., Opitz B., Jin L.. The common R71H-G230A-R293Q human TMEM173 is a null allele. J. Immunol. 2017; 198:776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gao P., Zillinger T., Wang W., Ascano M., Dai P., Hartmann G., Tuschl T., Deng L., Barchet W., Patel D.J.. Binding-pocket and lid-region substitutions render human STING sensitive to the species-specific drug DMXAA. Cell Rep. 2014; 8:1668–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ma F., Li B., Yu Y., Iyer S.S., Sun M., Cheng G.. Positive feedback regulation of type I interferon by the interferon-stimulated gene STING. EMBO Rep. 2015; 16:202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cDNA sequence of STING-β hs been deposited in GenBank under accession number MF360993.