Abstract
The identification of regulatory targets of all TFs is critical for understanding the entire network of the genome regulation. The lac regulon of Escherichia coli K-12 W3110 is composed of the lacZYA operon and its repressor lacI gene, and has long been recognized as the seminal model of transcription regulation in bacteria with only one highly preferred target. After the Genomic SELEX screening in vitro of more than 200 transcription factors (TFs) from E. coli K-12, however, we found that most TFs regulate multiple target genes. With respect to the number of regulatory targets, a total of these 200 E. coli TFs form a hierarchy ranging from a single target to as many as 1000 targets. Here we focus a total of 13 single-target TFs, 9 known TFs (BetI, KdpE, LacI, MarR, NanR, RpiR, TorR, UlaR and UxuR) and 4 uncharacterized TFs (YagI, YbaO, YbiH and YeaM), altogether forming only a minor group of TFs in E. coli. These single-target TFs were classified into three groups based on their functional regulation.
INTRODUCTION
Bacteria constantly monitor physical, chemical and biological conditions in environment, and respond for adaptation and survival by modifying the expression pattern of their genomes. Transcription, the major step of regulation of the genome expression, is carried out by a single species of RNA polymerase (RNAP). The intracellular concentration of RNAP in growing Escherichia coli K-12 W3110 strain is about 2000 molecules per genome, which is less than a total of about 4500 genes on its genome (1,2). The pattern of genome transcription, however, changes through modulation of the selectivity of transcription targets of RNAP by two steps of protein–protein interaction, i.e. seven species of the RNAP sigma subunit with the promoter recognition activity in the first step (3–5) and about 300 species of the DNA-binding transcription factors (TFs) in the second step (6–9). Based on the protein structure of DNA-binding motifs, we classified these TFs into 63 families (6,10; also cited in TEC database [www.shigen.nig.ac.jp/ecoli/tec/]). When bound to the target DNA, the activator-type TFs interact directly with one of the RNAP subunits for function (1,9,11). A tight correlation exists between the mode of transcription regulation and the contact subunit (class-I, II, III and IV for contact with α, σ, β and β’ subunit, respectively) (6,12). In order to facilitate the frequent and quick replacement of RNAP-interacting TFs, the affinity of protein–protein interaction between RNAP and TFs must be weak enough, but the binding of TFs at specific target sites near promoter enables the effective protein–protein interaction by increasing the local concentration of pairing TFs at the promoter region.
In advance in the genome-wide research technologies such as transcriptomics (13–16), ChIP-chip (17–19) and ChIP-seq analyses (19,20), the regulatory role of each TF has been identified mainly based on the transcription patterns in vivo in the absence of test regulator or after its over-expression. A large amount of knowledge of TFs is assembled in databases such as EcoCyc (21,22) and RegulonDB (23,24). Up to the present time, about 70–80% of the estimated 300 TFs in E. coli have been linked to at least one regulatory target gene or operon in the genome, but with use of in vivo analyses, it is in principle difficult to get the complete set of regulatory target promoters, genes or operons because the binding in vivo of TFs to their DNA targets is interfered by ∼300 co-existing TFs (10,25) and more than 200 species of other DNA-binding proteins that are involved in DNA functions (26). The majority of regulatory targets of TFs so far detected in vivo represent those not under the direct control of TFs but instead indirectly regulated by TFs (1,6) because the regulatory targets of TFs often include the genes encoding other TFs, forming the hierarchy of TF network (10). In addition, another serious problem of using unselected data sets of in vivo transcription is related to the difference in genetic backgrounds of bacterial strains used in these experiments. Up to the present time, the whole sequence has been determined for more than 1000 E. coli strains and it turned clear that a high-level difference exists in the genome sequence between E. coli strains (27,28). The difference exists even in both sigma and TF sequences not only between different E. coli strains but also even between laboratory stock strains of the same E. coli strain. For instance, the sequence difference exists in the rpoS gene encoding the stationary-phase sigma factor even between stock strains of the same E. coli K-12 W3110 from different laboratories (29). In addition, some of the regulatory targets listed in databases were predicted in silico simply based on the presence of sequences similar to the TF recognition sequence of low-level accuracy but without experimental confirmations. Thus, even for the characterized TFs, we have only fragmentary knowledge even for the best characterized model prokaryote E. coli K-12.
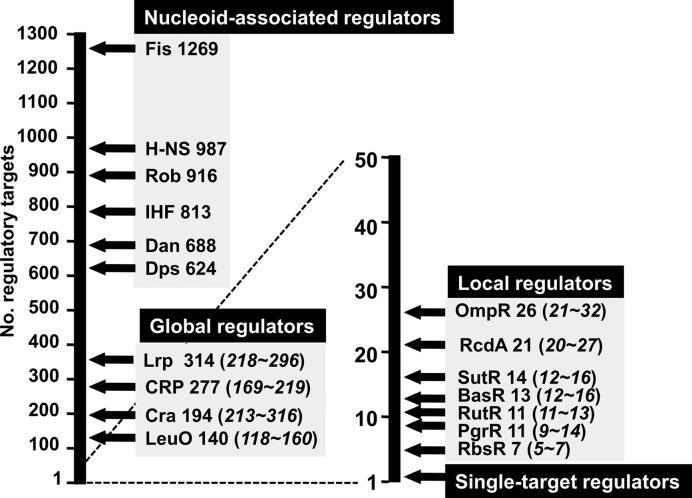
Identification of the connections between TFs and their direct targets represents a major bottleneck for modeling the transcriptional regulatory networks. The first step of a bottom-up approach to make a breakthrough toward understanding the regulatory network is to make the complete list of regulatory targets for all TFs. The knowledge of such a list of the targets under the direct control of TFs and the intracellular concentrations of these TFs under test environmental conditions are absolutely needed for detailed understanding the whole view of the genome regulation. To avoid the problems included in in vivo approaches as noted above, we then decided to employ several lines of in vitro approach. The Genomic SELEX (systematic evolution of ligands by exponential enrichment) is one short-cut approach for the identification of regulatory targets under the direct control of a test TF, because bacterial TFs generally bind to the recognition sequences located near the regulation target promoters (10,25,30). For identification of the regulatory targets of TF, the Genomic SELEX in vitro offers a number of advantages over in vivo analyses such as anti-TF antibody-based ChIP-chip. First, the binding site of TF can be identified in the absence of other interfering competitive DNA-binding proteins. Second, the TF-binding affinity to target can be monitored by using various concentrations of test TF or controlling the SELEX cycles. Third, possible influence of effectors on TF function can be examined in the presence and absence of effectors. Forth, only the direct targets under a test TF can be identified, excluding indirectly affected targets that are associated with in vivo data (see above). Using the newly developed Genomic SELEX system, we have so far identified the complete set of the constitutive promoters for five sigma factors (RpoD, RpoS, RpoH, RpoF and RpoE) (31,32). In parallel, we have performed a systematic Genomic SELEX screening for all 300 TFs from E. coli K-12 W3110, including both characterized and uncharacterized TFs. One unexpected finding of our ongoing research is that most of E. coli TFs regulate multiple target genes or operons. Based on the number of regulatory targets, we propose a novel classification system: single-target regulators (number of targets, 1 or a few); local TFs (targets ranging 10–50); global regulators (more than 100 targets); and nucleoid-associated regulators (as many as 1000 binding sites) (Figure 1). Since we performed the SELEX screening using the purified TF alone, TFs that require as yet unidentified effector for target recognition were not identified in this study. In this report, we describe a total of 13 single-target TFs, altogether forming a minor group among the total of as many as 200 E. coli K-12 TFs so far examined. With respect to the mode of modulation of TF activity, these TFs could be classified into three groups.
Figure 1.
Classification of TFs of Escherichia coli K-12 W3110. The Genetic SELEX screening of regulatory targets has been performed for more than 200 TFs from E. coli K-12 W3110 (reference 10; also see TEC database [www.shigen.nig.ac/ecoli/tec/]). Based on the number of regulatory targets, we propose to classify E. coli K-12 TFs into four groups: nucleoid-associated regulators; global regulators; local regulators; and single-target regulators. Some representative regulators are shown. The estimated number of the regulatory targets is shown after each TF (note that: the numbers in parenthesis shown the range of minimum and maximum number of regulatory targets). Some representative TFs analyzed by SELEX are shown in each group. In the case of nucleoid-associated regulators, the total number of regulatory targets has not been identified at this moment, and thus the Y-axis represents the total number of binding sites on the E. coli K-12 genome. Details of the single-target TFs are described in this report.
MATERIALS AND METHODS
Escherichia coli strains and culture conditions
The genome of E. coli K-12 W3110 type-A (29) was used as the source for construction of TF expression plasmids, and the DNA library for SELEX screening of regulatory targets of TFs. Escherichia coli DH5α was used for plasmid amplification. Escherichia coli BL21(DE3) was used for over-expression of all TFs. Cells were grown in LB medium with shaking at 37°C in the presence of 100 μg/ml ampicillin.
Expression and purification of TFs
Expression plasmid of all TFs was constructed according to the standard procedure (33). In brief, the TF-coding sequences were polymerase chain reaction (PCR)-purified using the E. coli K-12 W3110 type-A genome DNA as a template, and inserted into pET21α vector. The expression of His-tagged TFs was performed in E. coli BL21(DE3). His-tagged TFs were affinity-purified according to the standard procedure (33). The purity of all TFs used in this study was more than 95% as detected by protein-staining of polyacrylamide gel electrophoresis (PAGE) pattern. All the purified TFs used in this study were obtained from the E. coli TF collection in Ishihama laboratory (Hosei University, Japan).
Genomic SELEX screening of TF-binding sequences
The Genomic SELEX was performed according to the standard procedure (25,30). In each SELEX screening, the substrate DNA mixture was generated by PCR using the genome DNA library of E. coli K-12 W3110 type-A as template. For SELEX screening, the DNA mixture (5 pmol) and TF (10 pmol) were mixed in the binding buffer. This SELEX cycle was repeated several times depends on each TF. The original mixture of genomic DNA fragments formed smear bands on PAGE, but after several cycles of genomic SELEX, DNA fragments with high affinity to TF were enriched, forming sharper bands on PAGE gels. DNA was isolated from gel bands of DNA–TF complexes and PCR amplified. Mapping of SELEX fragments along the E. coli genome was performed by the SELEX-chip system by using a 43 450-feature DNA microarray (Oxford Gene Technology) (25). Approximately 300 bp long SELEX fragments should bind to two or more consecutive 60 bp long probes aligned at 105 bp intervals. The genomic SELEX sample obtained with use of TF was labeled with Cy3, while the reference SELEX sample obtained in the absence of TF addition was labeled with Cy5. After hybridization of both samples with the same DNA tilling array, the Cy5/Cy3 ratio was measured for each probe. The scanned pattern was plotted along the E. coli K-12 genome. Some of SELEX patterns shown in this report include minor non-specific peaks, but these peaks disappear after repeated cycles of SELEX [for the change of SELEX pattern after repetition of SELEX cycle see ref. 25]. All the SELEX-chip data described in this report were submitted to the ‘Transcription Factor Profiling of Escherichia coli‘ (TEC) database at the National Institute of Genetics (https://shigen.nig.ac.jp/ecoli/tec/) under the accession codes: BetI, KdpE, LacI, MarR, NanR, RpiR, TorR, UlaR, UxuR, CecR(YbiH), DecR(YbaO), NimR(YeaM) and XynR(YagI).
Gel shift assay
The gel shift assay was performed according to the standard procedure (34). Probes containing the recognition target sequences of test TFs were generated by PCR amplification using a pair of primers and Ex Taq DNA polymerase (Takara, Kusatsu, Japan). For gel shift assays, a mixture of each probe and each purified TF was incubated at 37°C for 30 min in the gel shift buffer. After addition of a DNA loading solution, the mixture was directly subjected to PAGE. Probe DNA in gels was stained with GelRed (Biotium, Fremont, CA, USA) and detected using LAS-4000 IR multi-colour (GE Healthcare, Little Chalfont, UK).
RESULTS AND DISCUSSION
Genomic SELEX search for regulatory targets of TFs
For quick search of DNA sequences that are recognized by DNA-binding proteins, we developed the Genomic SELEX system (25,30), in which TF-associated DNA segments can be isolated from mixtures of purified TF and a library of genome DNA fragments. Sequences of the protein-bound SELEX DNA fragments can be determined by either SELEX-clos (cloning and sequencing) and SELEX-chip (mapping by tilling array consisting of 43 450 species of 60 bp long oligonucleotide probe aligned at 105 bp intervals along the E. coli genome) (25,30). Combination of the SELEX-clos and SELEX-chip patterns provides not only a more reliable set of regulatory targets of the test TF but also the order of binding affinity between the predicted targets. The SELEX screening system is also useful for quick search of the influence of effector ligands and protein modification on the target selectivity of TFs. However, TFs that require as yet unidentified effectors for the target recognition were not analyzed in this study.
In the classic molecular genetic studies, E. coli promoters were considered to be regulated by a single specific regulatory protein, either a repressor or an activator, as originally identified in regulation of the lac operon by LacI repressor (35). Accordingly, most of TFs in E. coli have been believed to be the LacI-type TF, herein referred to ‘single-target TF’, that regulates transcription of one specific target gene (36). Until recently only a small number of TFs have been classified into the global regulators, which influence the expression of a large number of transcription units that belong to different metabolic pathways, thereby exhibiting pleiotropic phenotypes (8,37,38). After Genomic SELEX screening of TFs with known regulatory roles, however, we realized that the number of regulation targets of most E. coli TFs are more than those hitherto identified or predicted (1,6,10). Even though the regulation of multiple targets has been indicated in vivo by using modern technologies, it was not discriminated, in most cases, whether the targets were under direct or indirect control. Based on the results of the Genomic SELEX screening (see TEC database [www.shigen.nig.ac/ecoli/tec/]), we propose in this report an improved classification system of E. coli TFs (Figure 1). Among a total of more than 200 TFs so examined, the single-target regulators were found to be only 14, including 9 known TFs and 5 unknown TFs. From the ongoing SELEX screening of the remaining TFs, we estimate that the total number of single-target TFs are less than 20 at most, altogether forming only a minor group within a total of about 300 E. coli TFs. This report describes the detailed analysis of the single-target TFs so far identified.
LacI as the seminal model of single-target regulator
In the presence of glucose, E. coli utilizes this carbon source exclusively, even when other sugars are present. Based on the study on lactose utilization system, Jacob and Monod (39) proposed the epoch-making framework, the operon theory, which delineated how E. coli switches from glucose to lactose. When glucose is depleted, E. coli upregulates expression of the lac operon encoding proteins that transport and metabolize lactose. Transcription of the lac operon is regulated by LacI TF, which was the first regulatory protein isolated and tested for its function in vitro (40). In the absence of lactose, LacI binds with high affinity to the promoter of the lac operon at a specific operator DNA sequence, leading to repression of transcription (41). The regulation model of the lac operon has since served as a model system in the gene regulation in bacteria.
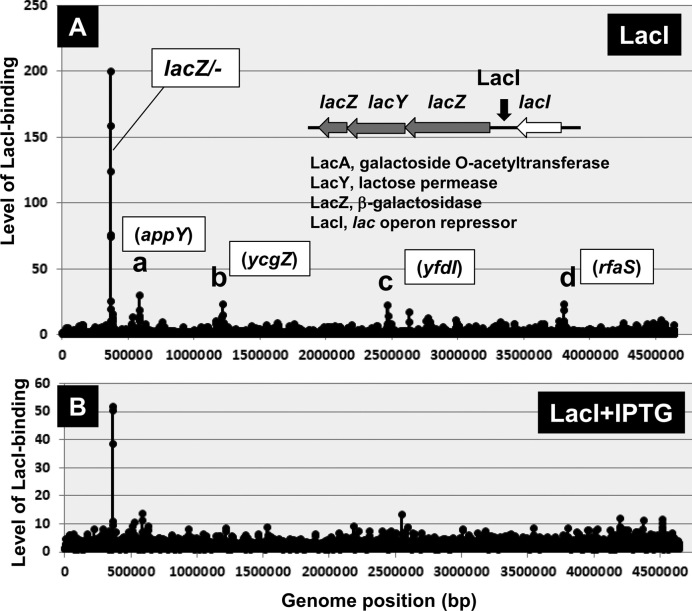
After the Genomic SELEX screening of more than 200 TFs from E. coli K-12, however, we found that most TFs regulate multiple target genes and operons whereas the single-target TFs are rare (reviewed in 6,10). We then examined the regulatory target of LacI by using the SELEX screening in vitro of LacI-binding sites along the E. coli K-12 genome. The SELEX pattern, however, indicated only a single binding site upstream of the lacZYA operon (Figure 2A). Low-level peaks were identified, but inside open reading frames (ORFs) of several genes. Furthermore the appearance of these peaks was not reproducible after repeated tilling array analysis. As expected from the regulation model of lac operon, the level of LacI binding at the lacZ promoter region decreased concomitant with the increased addition of IPTG (isopropyl-β-D-thiogalactoside), the synthetic inducer for lac operon expression (Figure 2B), indicating that LacI binding to the target is interfered by the inducer such as IPTG. Within the promoter region of the lac operon, three LacI-binding sites have been identified, all involved in the regulation of the lac operon (42). By SELEX screening, multiple TF-binding sites have been identified for some TFs near promoters, implying the regulation of a single and the same target. We then concluded that LacI is a typical single-target TF.
Figure 2.
Genomic SELEX pattern of LacI. The Genomic SELEX screening was performed for LacI in the absence (A) and presence (B) of the inducer IPTG. In the absence of IPTG, a single peak was detected within the spacer between the lacZYA and lacI operons. The appearance of low-level peaks was not reproducible, suggesting no specific binding to these sites. By adding increased concentrations of IPTG, the level of LacI binding to the single target decreased gradually. Minor peaks in (A) are located inside ORF of appY (a), ycgZ (b), yfdI (c) and rfaS (d). These one-point peaks might be non-specific background noise because ∼300 bp long SELEX fragments should bind to two or more consecutive 60 bp long probes aligned at 105 bp intervals on the tilling array used (25).
Identification of single-target TFs within the uncharacterized TF group
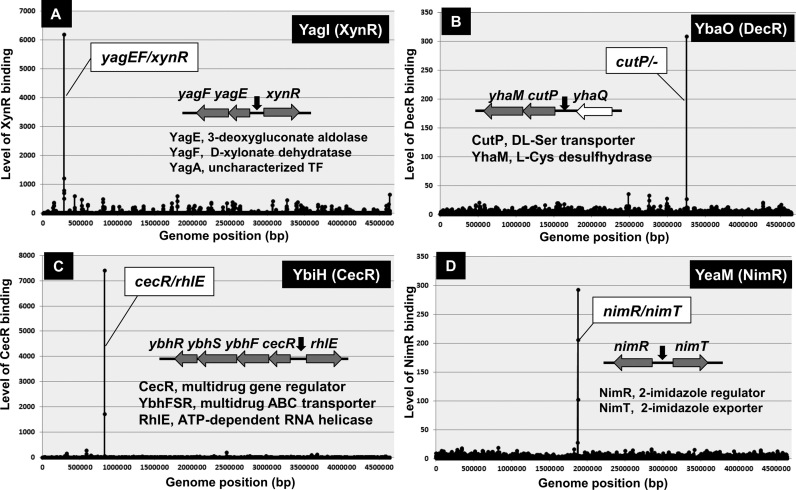
Bacterial DNA-binding TFs generally bind to DNA near promoters for effective interaction of RNAP (6,10). The Genetic SELEX screening was then developed for mapping the TF-binding sites along the E. coli genome (25,30). The SELEX screening is, in particular, a useful short-cut approach for identification of the regulatory targets of as yet uncharacterized TFs because the prediction of regulatory function of TFs with unknown functions is not easy with use of the phenotype screening. So far we performed SELEX screening for all 53 uncharacterized TFs, referred to Y-gene TFs (see TEC database [www.shigen.nig.ac.jp/ecoli/tec/]) for the complete list). After experimental confirmation of each of the predicted targets, we have so far published the regulatory functions for 15 TFs (Table 1). Among these characterized Y-TFs, five have been identified to belong to the group of single-target TFs, including YagI (renamed to XynR; regulator of xylonate catabolism) (43), YbiH (renamed to CecR; regulator of cefoperazone and chloramphenicol sensitivity) (44), YbaO (renamed to DecR; regulator of cysteine detoxification) (45) and YeaM (renamed to NimR; nitroimidazole regulator) (46) (Figure 3).
Table 1. Regulatory functions of Y-gene TFs.
| TF | Regulatory function | Family | Size (aa) | Data source | Target gene or operon | |
|---|---|---|---|---|---|---|
| Y-name | Renamed | |||||
| YagI | XynR | Regulator of xylonate catabolism | IclR | 252 | SELEX | yagE/yagA (single target)yagE/yagA (single target) |
| Regulon | None | |||||
| YbaO | DecR | Regulator of cysteine detoxification | AsnC | 152 | SELEX | yhaO (single target)yhaO (single target) |
| Regulon | None | |||||
| YbiH | CecR | Regulator of cefoperazone-chloramphenicol sensitivity | TetR | 223 | SELEX | cecR (single target)cecR (single target) |
| Regulon | cecR-ybhGFSR | |||||
| YbjK | RcdA | Regulator of csgD (master regulator of biofilm formation)Regulator of csgD (master regulator of biofilm formation) | TetR | 178 | SELEX | ybjJ/ybjK,sulA/sxy,ycdT,csgD,ycgF/ycgZ,ydeI,asr,yehA,rrlD |
| Regulon | asr,bluF,csgDEFG,sulA,yagK,ydeI | |||||
| YcdC | RutR | Regiator of pyrimidine metabolisma and purine degradation | TetR | 212 | SELEX | carA,gadW,gadC,glnE,glxR,deoL,ycdM/ycdC |
| Regulon | carAB,fepB,gadAXW,nemR,pdeR,rutA,rutA | |||||
| YcjZ | PgrR | Regulator of peptidoglycan (PG) recycling | LysR | 299 | SELEX | leuL/leuO,pheP,ycjY/ycjZ,ydjE,yedX/yedS,xapR,yfgF/yfgG,yhiI,rfaL,rfaS,yjgL |
| Regulon | ycjG,ycjXF-tyrR,ycjY-ymfDC-mpaA | |||||
| YdeO | PhhR | Regulator of intracellular pH homeostatis | AraC | 253 | SELEX | hyaA,appC,cadX,hdeD,nhaA,slp,yiiS |
| Regulon | appCBXA,gadAXW,gadEF-mdtEF,hyaABCDEF,safA-ydeO,slp-dctR | |||||
| YdcN | SutR | Regulator of sulfur utilization | Xre | 178 | SELEX | yagR,xseB,ydcO/ydcN,ynfG,pfkB,fliZ,ypfN,yfgM,yfiC,tdcF,yjcS |
| Regulon | cysE,fliAZ-tcyJ,sutR | |||||
| YdhM | NemR | Regulator of N-ethylmaleimide reductase | TetR | 199 | SELEX | nemR (single target) |
| Regulon | nemRA-gloA | |||||
| YeaM | NimR | Regulator of resistance to 2-nitroimidazole | AraC | 273 | SELEX | yeaM/yeaN (single target) |
| Regulon | nimR,nimT | |||||
| YedW | HprR | HprSR-TCS response regulator of H2O2 sensitivity | OmpR | 223 | SELEX | ydeS,ybaS/yegT,dctA |
| Regulon | cusCFBA,cusRS,cyoABCDE,yedX | |||||
| YgiP | Dan | Nucleoid-associated regulator for anaerobic growth | LysR | 310 | SELEX | (688 binding sites) |
| Regulon | ttdA,ttdR | |||||
Figure 3.
Genomic SELEX patterns of uncharacterized TFs. A total of about 50 uncharacterized TFs from Escherichia coli K-12 W3110 (for the list see TEC database) were subjected to the Genomic SELEX screening, of which four were identified to be the single-target TFs. The regulatory functions for these TFs have been described elsewhere: (A) YagI (renamed to XynR) (43); (B) YbaO (renamed to DecR) (45); (C) YbiH (renamed to CecR) (44); and (D) YeaM (renamed to NimR) (46).
Identification of single-target TFs within the characterized TF group
The regulatory functions of hitherto characterized TFs have mostly been analyzed in details with use of one or a few representative targets, and thus the whole set of regulatory targets have not been identified for most of E. coli TFs. We then extended the Genomic SELEX screening for all the known TFs from E. coli K-12 W3110. As a result, we found that most of TFs regulate multiple targets, and LacI-type single-target regulators are rather rare, forming only a small group of <20 TFs (Table 2). We summarize in this report the whole set of experimentally confirmed TFs including five uncharacterized single-target TFs (see above) and eight known single-target regulators: BetI (regulator of glycine betaine synthesis), KdpE (regulator of K+ uptake operon), MarR (regulator of multi-antibiotic resistance), NanR (regulator of sialic acid utilization), RpiA (regulator of D-ribose utilization), TorR (regulator of TMAO-based respiration), UlaR (regulator of L-ascorbate utilization) and UxuR (regulator of hexuronic acid utilization). Since the regulatory function of these TFs is modulated through chemical or physical modification of proteins, these single-target TFs are classified into three subgroups.
Table 2. Regulatory targets of single-target TFs with known functions.
| TF | Family | Size (aa) | Target operon | |
|---|---|---|---|---|
| Group-S1 | ||||
| LacI | GalR/LacI | 360 | SELEX | lacZ (single-target)lacZ (single-target) |
| Regulator of lactose utilization | Regulon | lacZYA | ||
| MarR | MarR | 144 | SELEX | nanC/marR (single-target)nanC/marR (single-target) |
| Multiple antibiotic resistance regulator | Regulon | marRAB | ||
| RpiR | RpiR | 296 | SELEX | rpiR/rpiB (single-target)rpiR/rpiB (single-target) |
| Regulator of D-ribose utilization | Regulon | rpiB,rpiRalsBACE | ||
| UlaR | DeoR | 251 | SELEX | ulaR,ulaG/ulaA (single-target)ulaR,ulaG/ulaA (single-target) |
| Regulator of ascorbate utilization | Regulon | ulaABCDEF,ulaG | ||
| UxuR | GntR | 257 | SELEX | gntP/uxuABR (single-target)gntP/uxuABR (single-target) |
| Regulator of hexuronic acid utilization | Regulon | exuR,gntR,lgoR,uidABC,uxuAB,uxuR | ||
| Group-S2 | ||||
| BetI | TetR/AcrR | 195 | SELEX | betI/betT (single-target)betI/betT (single-target) |
| Osmo regulator of blycine betaine synthesis | Regulon | betIBA,betT | ||
| NanR | GntR | 263 | SELEX | nanC/fimB (single-target) |
| Regulator for sialic acid utilization | Regulon | fimB,nanATEKyhcH,nanCMS,yjhBC | ||
| Group-S3 | ||||
| KdpE | OmpR | 255 | SELEX | kdpF/ybfA (single-target) |
| Tugor-keeping regulator for controlling K+ level | Regulon | kdpFABC | ||
| TorR | OmpR | 230 | SELEX | torR/torC (single-target) |
| Regulator for TMAO-based respiration | Regulon | gadAXW,hdeAByhiD,tnaCAB,torCAD,torR | ||
Group-S1 (LacI-type): recognition of the single-target by TF alone
MarR: multiple antibiotic resistance regulator
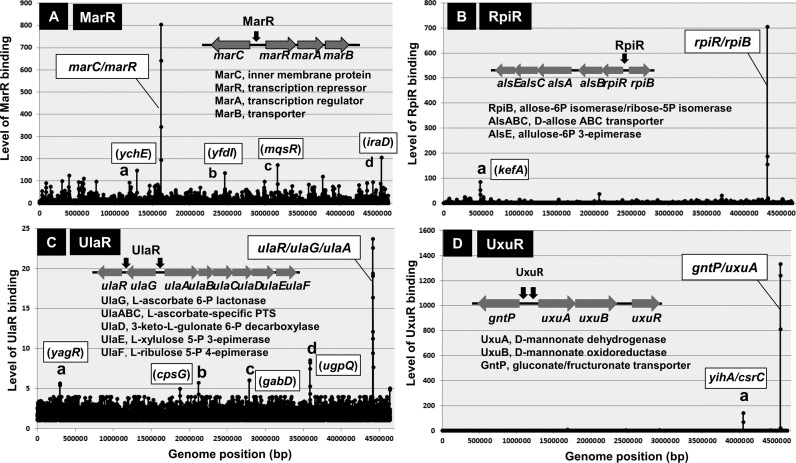
The multiple antibiotic resistance regulator (MarR) in E. coli was discovered based on genetic selections to identify mutants that conferred resistance to a range of antibiotics (47,48). MarR controls a wide variety of phenotype including not only multiple antibiotic resistance (MDP) but also the catabolism of aromatic compounds, stress responses and virulence in the case of pathogenic E. coli. The well-characterized target of MarR is located within the intergenic region between marC and marRAB, thereby repressing the bidirectional transcription of both operons (49,50). MarC is an inner membrane protein but is not essential for MDR while MarB is a periplasmic protein that is needed for expression of the full activity of MDR but MarR and MarA are both TFs. After the Genomic SELEX screening with use of purified MarR and a library of E. coli K-12 genome segments, we identified only a single target for MarR at the spacer between the marC and marRAB operons (Figure 4A), indicating that MarR is a single-target TF. However, more than 30 genes have been identified in vivo to be under the influence of MarR (51). These genes are, however, not under the direct control of MarR, but instead regulated by MarA, which is under the control of MarR. Taken together we concluded that the mar phenotype is determined mainly by MarA, which is under the control of single-target MarR. The activity of MarR is controlled by aromatic acids and antibiotics (52,53) (Table 3).
Figure 4.
Genomic SELEX patterns of group-S1 single-target TFs. After Genomix SELEX screening of more than 150 TFs with known regulatory functions, a total of 8 TFs have been identified to regulate only single targets, of which four showed specific binding to the respective targets in the presence and absence of effectors. This group-S1 of single-target TFs includes MarR (A); RpiR (B); UlaR (C); and UxuR (D). The binding site of each TF and the physiological function of each target gene are shown inside each SELEX pattern. In the case of UlaR and UxuR, two binding sites were identified within the same operons. The location of minor peaks is indicated in each panel. Each of these minor peaks represents TF-binding to a single probe, and the binding center is located inside ORF (except for peak of panel D). Thus, we predicted these peaks represent non-specific background noise.
Table 3. Modulation of target-selectivity of TFs.
| TF | Family | Size(aa) | Effector | Reference |
|---|---|---|---|---|
| Group-S1 | ||||
| LacI | GalR/LacI | 360 | Allolactose, IPTG | Jobe and Bourgeois (118) Barkley et al. (121) |
| MarR | MarR | 144 | Aromatic acids, Antibiotics | Alekshun and Levy (53) Chubiz and Rao (52) |
| RpiR | RpiR | 296 | D-Allose, D-Ribose | Poulsen et al. (58) |
| UlaR | DeoR | 251 | L-Ascorbate, L-Ascorbate-6-phosphate | Garces et al. (63) |
| UxuR | GntR | 257 | D-Glucuronate, D-Fructuronate | Bates Utz et al. (122) Tutukina et al. (68) |
| Group-S2 | ||||
| BetI | TetR/AcrR | 195 | NaCl, Choline, Glycine betaine | Esho et al. (123) Rokenes et al. (124) |
| NanR | GntR | 263 | N-Acetylneuramic acid | Kalivoda et al. (80) Kalivoda et al. (83) |
| Group-S3 | ||||
| KdpE | OmpR | 255 | Phosphorylation by KdpD, AcP | Walderhaug et al. (90) |
| TorR | OmpR | 230 | Phosphoorylation by TorS, AcP | Joutlin et al. (125) |
RpiR: regulator of D-ribose utilization
For growth on D-ribose as a carbon source, E. coli K-12 employs two-types of uptake system, the high-affinity ABC-type RbsACB transporter (54) and the low-affinity ABC-type AlsBAC transporter (55). Inside the cell, D-ribose is first converted by ribokinase (RbsK) to ribose-5-phosphate (56). The RbsACB transporter and RbsK ribokinase are encoded by the rbsDACBKR operon (54), which is regulated by RbsR. Previously we identified by Genomic SELEX screening that the LacI-family RbsR regulates not only the rbsDABCKR operon but also a set of genes involved in purine and pyrimidine nucleotide metabolism (34). On the other hand, the AlsBAC transporter is encoded by the rpiR-alsBACE operon (57) that is regulated by RpiR (or AlsR) (55). RpiR was identified as a negative regulator of the expression of genes involved in transport and catabolism of D-allose. The catabolism of ribose 5-phosphate requires the participation of enzymes of the pentose phosphate pathway. By this pathway, ribose 5-phosphate, ribulose 5-phosphate and xylulose 5-phosphate are inter-converted in reactions catalyzed by the action of RpiB and UlaE. These three species of the pentose phosphate are all converted into fructose 6-phosphate and glyceraldehyde 3-phosphate, the glycolytic pathway intermediates. Ribose-5-phosphate also serves as the substrate for the synthesis of nucleotides and amino acids, histidine and tryptophan (58).
The rpiR-alsBACE operon containing the genes for transport and metabolism of both D-allose and D-ribose is organized in a divergent transcription unit with the rpiB gene coding for a secondary D-ribose-5-phosphate isomerase (57,59). Here we attempted to identify the whole set of regulatory targets of RpiR. After SELEX-chip analysis using the purified RpiR alone, we identified a single high-level peak within the spacer of bidirectional rpiR-alsBACE operon and rpiB gene (Figure 4B), indicating that RpiR is a single-target TF. The rpiR gene is located at 5′-proximal end of the rpiR operon. Expression of the rpiR and rpiB genes is both induced when E. coli is grown on D-allose in the absence of glucose, suggesting that D-allose binds to RpiR as an inducer (57,59). Transport systems of D-allose and D-ribose are functionally redundant. The reporter assay of the rpiB promoter, however, indicated that D-ribose is more potent inducer than D-allose (see below).
UlaR: regulator of ascorbate utilization
Enteric bacteria are able to ferment and oxidize L-ascorbate. In E. coli, the divergent ulaG and ulaABCDEF operons are involved in transport and catabolism of L-ascorbate (60). The ulaG gene encodes L-ascorbate-6-P lactonase (61) while the ulaABCDEF operon encodes the three components of the L-ascorbate phosphotransferase transport system (UlaABC) for uptake and phosphorylation of L-ascorbate (62) and three catabolic enzymes (UlaDEF) (61). Intracellular L-ascorbate-6-P is transformed by L-ascorbate-6-phosphate lactonase (UlaG) to 3-keto-L-gulonate-6-phosphate, which is decarboxylated by UlaD to L-xylulose-5-phosphate, and then converted to D-xylulose-5-phosphate by the sequential action of UlaE (3-epimerase activity) and UlaF (4-epimerase activity) (61). Overall the function of the gene products of the ula system is the transport of L-ascorbate and its transformation into D-xylulose-5-phosphate (61), which is subsequently metabolized by the pentose phosphate pathway.
The ula regulon is under the control of the DeoR-family UlaR repressor (60). After SELEX-chip analysis, a single peak was identified within the ulaR-ulaG-ulaABCDEF region (Figure 4C), indicating that UlaR is a single-target TF. After high-resolution analysis of the SELEX pattern, however, two UlaR-binding sites were identified in this peak, one upstream of ulaR (but downstream of ulaG) and another between the ulaG and ulaABCEDEF operons. The binding of UlaR upstream of its own gene indicates the autogenous regulation of the ulaR gene. The activity of UlaR is controlled in vitro by L-ascorbate 6-phosphate while its activity is controlled in vivo by the addition of ascorbate (Table 3) (63).
UxuR: regulator of hexuronic acid utilization
Hexuronates are abundant in natural environments, including the bodies of warm-blooded animals, and thus the hexuronate catabolism is important for the colonization of E. coli in host animals (64,65). D-Galacturonate and D-glucuronate are transported into E. coli K-12 by the action of hexuronate transporter (ExuT) (66,67). By the action of the uronic isomerase (UxaC), D-galacturonate and D-glucuronate are converted into D-tagaturonate and D-fructuronate, respectively. D-Glucuronate is then degraded into 2-keto-3-deoxy-D-gluconate by the action of UxuB (D-mannonate oxidoreductase) plus UxuA (D-mannonate dehydratase) (66) while D-galacturonate is degraded into 2-keto-3-deoxy-D-gluconate by the action of UxaB (altronate oxidoreductase) (67). Finally 2-keto-3-deoxy-D-gluconate is converted to pyruvate via the Entner-Doudoroff pathway. These genes are organized in the uxaCA, uxuAB and uxaB operons. Three local regulators, UxuR, ExuR and UidR, are involved in the regulation of this hexuronate pathway (68–70).
In order to get insight into the cross-regulation between three local regulations, we first analyzed the set of regulatory targets of UxuR by using the SELEX screening system. The purified UxuR alone binds to only a single-specific site within the intergenic spacer between gntP and uxuA in the absence of glucuronate (Figure 4D). Thus, we concluded that UxuR is one of the single-target TFs. Upon addition of increasing concentrations of D-glucuronate, the level of UxuR binding to this probe decreased (data not shown), supporting the hypothesis that glucuronate is an inducer for inactivation of UxuR. In the presence of D-glucuronate, the divergent lgoRT and lgoD operons are induced, each encoding a uncharacterized TF (LgoR) and galactonate:H+ symporter (LgoT) and L-galactonate oxidoreductase (LdoD), respectively (70). This finding apparently disagrees with the single-target TF model for UxuR predicted based on the SELEX screening in the presence of UxuR alone. However, the lgoRT and lgoD operons might be regulated in vivo by ExuR-UidR heterodimer.
Group-2 (BetI-type): recognition of the single-target in the presence of effector
BetI: regulator of glycine betaine synthesis for osmoprotection
Escherichia coli is able to adapt to environments of high osmolarity by the intracellular accumulation of compatible solutes, so-called osmoprotectants, which accumulate in large amounts for protection against osmotic stress (71). The molecular species of the osmoprotectant varies depending on the cell growth conditions, including the presence of potassium glutamate, trehalose, proline or glycine betaine. The highest osmotolerance is achieved by the accumulation of glycine betaine (72,73). When added exogenously to E. coli at an inhibitory osmotic strength, glycine betaine can be taken up from the environment through the ProU and ProP transport systems (74,75). In the absence of glycine betaine in environments, it can be synthesized de novo from its precursor, choline, by the choline-to-glycine betaine pathway (71,72).
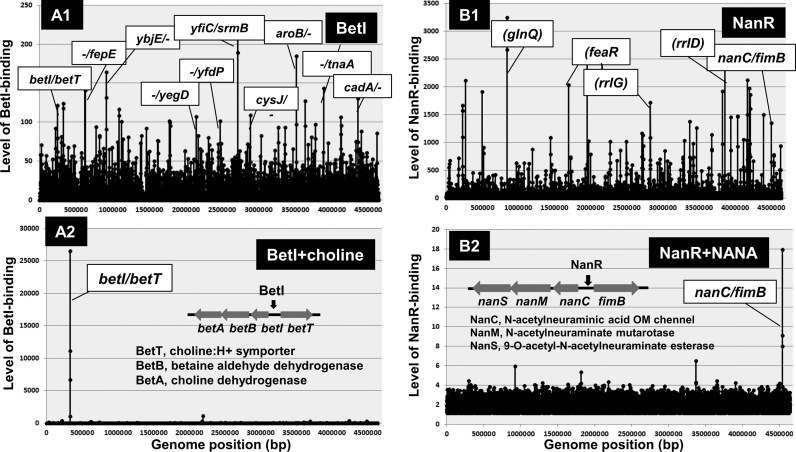
The bet regulon comprises a regulatory gene, betI and three structural genes: betT (choline porter), betA (choline dehydrogenase) and betB (betaine aldehyde dehydrogenase) (73), which are organized into the divergently transcribed betIBA and betT operons. Expression of the bet genes is induced by oxygen, choline and osmotic stress (71,76). Except for the regulation of the divergent betI and betTAB operons, possible participation of BetI in regulation of as yet unidentified operons has not been seriously examined. In this study, we attempted to identify a complete set of the regulatory targets of BetI using the Genomic SELEX screening system (25,30). After SELEX screening, BetI binds to a number of sites (Figure 5A1) but in the presence of choline, the inducer of bet regulon expression, BetI remained bound only at a single site inside the overlapping promoter region within the betI-betT spacer (Figure 5A2). This unique property can be used for discrimination between specific and non-specific binding (76). After addition of choline and KCl, BetI binding disappeared except the betI-betT spacer site, indicating that all other bindings are non-specific (Figure 5A2). The gel shift assay also supported the stable binding of BetI to a target irrespective of the presence and absence of effector choline (see below).
Figure 5.
Genomic SELEX patterns of group-S2 single-target TFs. In the absence of effectors, BetI and NanR bound to a number of non-specific sites (A1 and B1). In the presence of specific effectors (choline for BetI and NANA for NanR), both BetI and NanR bound only to the respective specific target site, but all the non-specific binding disappeared (A2 and B2). TFs showing this type of binding specificity are classified into group-S3 single-target TFs.
NanR: regulator for sialic acid utilization
The sialic acids are a unique family of about 40 natural derivatives of the nine-carbon monosaccharide N-acetylneuraminic acid (Neu5Ac or NANA). In eukaryotes, sialic acids play important roles and sialate biosynthesis is generally confined to higher eukaryotes. In the case of bacteria, however, sialic acids are used as sources of carbon, nitrogen and metabolic energy. Sialic acid metabolism is also central in host-microbe interactions (77,78). Utilization of sialic acid by E. coli K-12 requires the nanATEK operon (79). The first products of sialic acid catabolism by the N-acetyl-β-neuraminidase (the nanA gene product) are pyruvate and a six-carbon amino sugar, N-acetylmannosamine (ManNAc).
The nanATEK operon is negatively regulated by the upstream nanR-encoded NanR (80–82). When E. coli is growing in glucose minimal medium, nanA expression is barely detectable due to repression by NanR. The exogenous addition of sialic acid results in the marked induction of the nanATEK operon (82). The most common sialic acid, N-acetylneuraminate (Neu5Ac or NANA), has also been implicated as an additional inducer of the E. coli nan system (81,83–84). By the Genomic SELEX screening, the purified NanR alone was found to bind to a number of sites along the entire E. coli genome including the intergenic spacer between the nanCMS operon and the fimB gene (Figure 5B1). The binding of NanR to these non-specific sites disappeared in the presence of Neu5Ac but NanR remained bound at a single target for the divergent expression of nanCMS and fimB operons (Figure 5B2). NanR remains bound to the target site even in the presence of inducer Neu5Ac. This unique property is similar to that of BetI and thus we propose a subgroup of the single-target TFs, including both BetI and NanR that remain bound at the operator in the inactive form with respect to repression.
Group-3 (TCS response regulator): recognition of the single-target by phosphorylated TF
KdpE: turgor-keeping regulator for controlling K+ level
K+ is important for maintenance of turgor (85) and the intracellular pH (86). Escherichia coli K-12 has at least three K+ uptake systems, the constitutive TrkG/TrkH and Kup systems and the inducible high-affinity K+ uptake KdpFABC system, altogether contributing to adjust the intracellular K+ concentration at an optimum level under a given environmental condition (87), but only the Kdp system is controlled by medium K+ concentration and thus serves as an emergency system to scavenge K+ (85). The Kdp system is composed of four proteins, KdpFABC, altogether forming the K+-translocating Kdp-adenosine triphosphatase (ATPase) for high affinity K+ uptake (88,89). The genes encoding KdpFABC are organized in a single kdpFABC operon.
Transcription of the kdpFABC operon is under the control of a two-component system (TCS) consisting of the sensor kinase KdpD and the response regulator KdpE. The kdpED operon is located downstream of the kdpFABC operon. Under K+-limiting conditions, KdpD and KdpE together induce expression of the kdpED operon (90–92). Under K+ limitation or high osmolarity imposed by a salt, KdpD autophosphorylates and transfers the phosphoryl group to the response regulator KdpE (92). The response regulator KdpE receives the phosphoryl group from KdpD and induces kdpFABC transcription. Phosphorylated KdpE exhibits increased affinity for a KdpE-box located upstream of the kdpFABC promoter and thereby triggers kdpFABC transcription (93). Except for the kdpFABC operon, serious search has never been performed to identify additional regulatory targets.
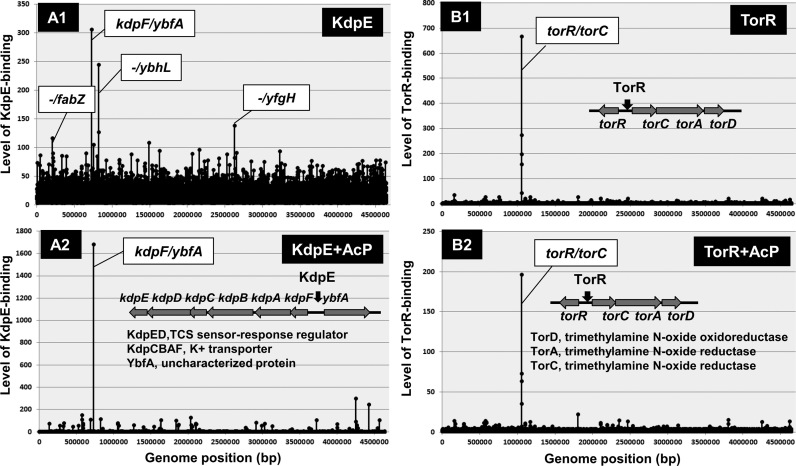
We then carried out the Genomic SELEX search for an additional target(s) of KdpE. After SELEX screening in the absence of AcP (acetyl phosphate), the binding of unphosphorylated KdpE was detected at the known target within the spacer between the kpdF and ybfA genes (Figure 6A1). Besides this peak, several low-level and supposedly non-specific peaks were detected in the absence of AcP. In the presence of AcP that is able to phosphorylate KdpE in the absence of KdpD sensor kinase, however, only a single and high-level peak was identified at the promoter region within the kdpFABC operon (Figure 6A2). The binding affinity to this single target increased in the presence of AcP, but the binding to non-specific sites almost disappeared. We then concluded that the activated form of KdpE is a single-target TF.
Figure 6.
Genomix SELEX patterns of group-S3 single-target TFs. Two TCA response regulators, KdpE (A) and TorR (B), were found to regulate only single targets, and thus are classified into group-S3 single-target TFs. In the absence of AcP for protein phosphorylation, KdpE bound to several sites (A1), of which the specific target within the spacer between the kdpABCDE and ybfA operons remained bound in the presence of ApC (A2). On the other hand, TorR bound only to the spacer between the torR and torcAD operons in the presence and absence of AcP (B1 and B2).
TorR: regulator for TMAO-based respiration
Under anaerobic conditions, E. coli is able to use alternative terminal electron acceptors such as nitrate, nitrite, fumarate, dimethyl sulfoxide (DMSO) or trimethylamine N-oxide (TMAO) for respiration (94). The TMAO-based respiration system is composed of two parts: a catalytic respiration TorCAD pathway and a regulatory TorSR two-component pathway (TCS) (95). The TorC protein is a c-type cytochrome anchored to the membrane and oriented toward the periplasmic compartment. The TorA protein, located in the periplasm, is the terminal reductase and receives the electrons from TorC (96). TorD is a cytoplasmic protein that acts as both a specific chaperone for maturation of TorA (97). In the absence of TMAO, the TorCAD system is not expressed.
TorR forms, together with TorS, an alkaline- and acid-stress response TCS system. TorS is a typical TCS histidine kinase receptor, but containing three phosphorylation sites while TorR is an OmpR-family response regulator (98,99). TMAO stimulates its His phosphorylation and the phosphotransfer cascade, leading to formation of functional phosphorylated TorR (100). In order to identify the set of regulatory targets of TorR, we carried out SELEX screening in the presence and absence of AcP. In the presence and absence of AcP, a single high-level peak of TorR binding was identified within the spacer between the divergent torR and torCAD operons although the level of TorR binding to this target was higher in the presence of AcP (Figure 6B). We then concluded that the phosphorylated form of TorR is a single-target TF. This finding agrees with the previous observations that TorR is a transcriptional DNA-binding positive regulator in transcription of the genes related to TMAO induction (101,102).
Functional modulation of single-target TFs
Single-target TFs were classified into three groups (Table 3). Group-S1 TFs such as LacI repressor recognize and bind to promoter regions of the respective single-target genes in the absence of effector ligands, but after interaction with effectors, the binding affinity to targets decreases, leading to derepression of the respective targets. The effector ligands are known for LacI, MarR, RpiR, UlaR and UxuR. LacI repressor is inactivated by allolactose, but IPTG, a non-hydrolyzable analog of lactate, is widely used for induction of the lac promoter. The activity of MarR, RpiR, UlaR and UxuR is all controlled by at least two different inducers (Table 3). Recently the increased number of TFs have been recognized to be under the control of more than two effector ligands such as allantoin and glyoxalate for AllR (103), arginine and lysine for ArgP (104), glyoxylate and pyruvate for IclR (105), ATP and maltoriose for MalT (106), and uracil and thymine for RutR (107). In the case of SdiA, at least three analogs of homoserine lactone (HSL) participate, beside the natural HSL, in the activity control of SdiA (108). The single-target TFs appear to monitor multiple effectors, thereby leading to control their targets at various levels in response to various external signals.
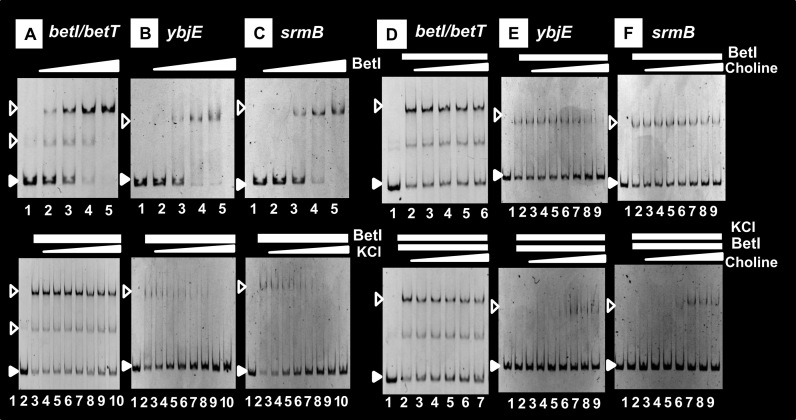
The regulatory mode of group-S2 TFs such as BetI is different remarkably from that of group-S1 TFs. Group-S2 TFs alone bind to not only specific targets but also a number of non-specific sites, but in the presence of specific effectors, the recognition specificity is defined so as to inhibit its binding to non-specific DNA sites (see Figure 3). For instance, BetI alone binds to a number of sites but in the presence of 0.5 M KCl, its binding was detected only at the specific betI-betT site (Figure 7A), but the non-specific binding to the non-specific ybjE and srmB probes disappeared (Figure 7B and C), indicating that the functional holo-conformation of BetI is induced in the presence of 0.5 M KCl. The effector K+ plays a role in transient (or Neu5Ac) increase of the intracellular turgor for survival upon exposure to elevated salt (109). For the permanent adaptation, glycine betaine is synthesized by the bet regulon, of which transcription is regulated by BetI. By adding another effector choline, BetI still remained bound at the specific probe (Figure 7D). This unexpected finding agrees with the prediction that BetI remains bound to the operator even in the presence of choline (76,110). One possibility of this unique characteristic is that the inducer binds to the group-S2 TFs, leading to inactivation of a site other than DNA-binding such as the site needed for interaction with RNAP. The binding of BetI to the non-specific probes, however, increased by the addition of choline even in the presence of KCl (Figure 7E and F). Recruiting of BetI to non-specific sites might lead to decrease in the intracellular level of available BetI, ultimately reducing the expression of targets. In addition to BetI, we found that NanR is another example of group-S2 TF, which remained bound to the operator even after addition of the inducer Neu5Ac (see Figure 3).
Figure 7.
Control of the activity of BetI by two effectors. Activity control of BetI by KCl and choline. Gel shift assay was performed to observe the influence of KCl and choline, the predicted effectors of BetI, on the BetI activity. In the presence of 0.5 M KCl, the binding of Bet was observed only at the spacer between betI and betT (A and D), the specific single-target of BetI, but non-specific binding inside ORFs of ybjE (B and E) and srmB (C and F) genes disappeared. Details of the gel shift assay are described in the ‘Materials and Methods’ section. The concentration of choline added was: 0 (lane 2), 10 nM (lane 3), 100 nM (lane 4); 1 μM (lane 5); 10 μM (lane 6); 100 μM (lane 7); 1 mM (lane 8); 10 mM (lane 9); 100 mM (lane 10).
Group-S3 TFs recognize and bind to the respective single-targets only when phosphorylated. Group-S3 KdpE is the response regulator of KdpDE TCS involved in osmoregulation and K+ sensing. Under K+ limitation or high osmolarity imposed by a salt, KdpD autophosphorylates and transfers the phosphoryl group to the response regulator KdpE (92). KdpE is also phosphorylated through TCS cross-talk by non-cognate sensor kinases UhpB, which is responsible for sensing and transport of the metabolic intermediate glucose-6-phosphate (33), and PhoB, which responds to phosphate limitation (111). Thus, the level of activated KdpE is controlled by three different sensor kinases, each monitoring a different environmental signal. We found that TorR, the response regulator of TorSR TCS, regulates only a single target, the torCAD operon for TMAO-based respiration system (see Figure 6). The activation process of TorR is unique, involving four-step of phosphorelay within TorR protein (96) by TorS that senses both alkaline- and acid-stresses. Again this single-target TF is able to response to multiple environmental signals.
The single-target TFs respond to multiple signals in environment through different mechanisms, and regulate the respective targets at different levels. Toward understanding the whole view of genome regulation by these single-target TFs; however, another important background knowledge is their intracellular concentrations. We have determined the level of all seven sigma factors (cited in 7) and of more than 70 TFs in a single and the same E. coli K-12 W3110 type-A strain (112). With respect to the intracellular level, the single-target TFs herein described belong to the group of low abundance, being <100 molecules per genome in growing cells of E. coli K-12 W3110.
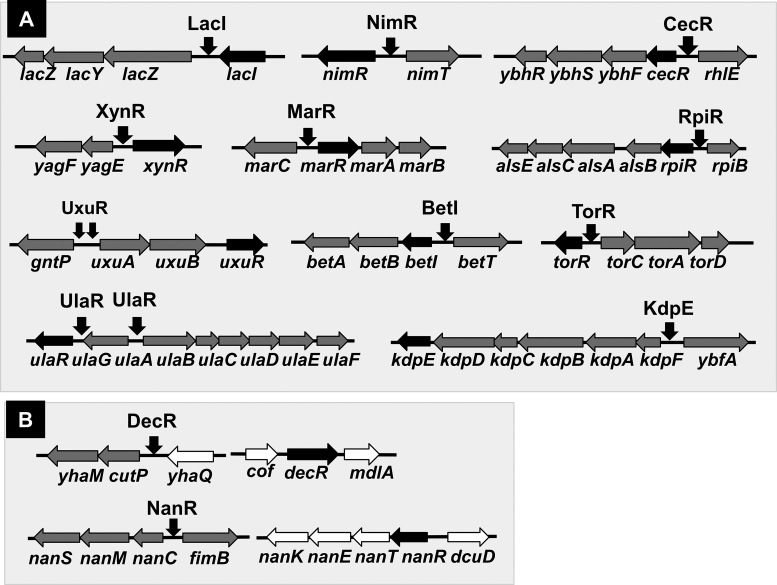
Gene organization of single-target TFs
Here we identified a total of 13 single-target TFs, 9 hitherto identified TFs and 4 uncharacterized TFs [the regulatory functions of these TFs have been identified by the Genomic SELEX screening as noted in this report]. Most of the genes encoding these single-target TFs are located close or adjacent to their regulatory target genes (Figure 8), because this type of the gene organization is easily transferred into E. coli from other bacteria existing in the same environment in nature, and moreover, the set of single-target TF and its targets can be easily retained in E. coli if the products of target genes confer a benefit to the host. One major pathway of the gene transmission is phage infection. In E. coli K-12, phage fossils exist as a total of ten prophages (113,114), which altogether comprise about 3.6% of the E. coli K12 genome and carry a total of 14 TF genes (43,115), of which the regulatory function remains unidentified for 10 TFs. Using the Genomic SELEX screening, we have identified the function of XynR (renamed YagI), which encodes the single-target regulator of xylonate catabolism (see Figure 2) (43,115). The set of the xynR gene and its regulatory target genes exists inside CP4-6 prophage in E. coli K-12 (see Figure 8). With use of the xynR regulon, E. coli is able to utilize xylonate, a degradation product of xylan, the representative hemicellulose in plants. The regulatory targets of XynR in E. coli K-12 are still fixed within the prophage. In agreement of the regulation of only one specific target, the single-target TFs recognize and bind to one unique sequences.
Figure 8.
Organization of the genes for single-target TFs and their regulatory targets. (A) Most of the TF genes are adjacently arranged on the Escherichia coli K-12 genome with the genes encoding their regulatory target genes. (B) The TF gene and its regulatory target genes are separated on the E. coli genome for two single-target TFs, DecR and NanR.
LacI regulates the operon consisting of a set of the genes for transport and catabolism of lactose. Likewise, the single-target TFs herein identified regulate the operons involved in transport and catabolism of carbon sources such as NanR for Neu5Ac, RpiR for D-allose and D-ribose, UlaR for L-ascorbate and UxuR for glucuronate and gluconate. The operons under the control of CecR and DecR are also composed of a set of the genes for transport and metabolism of transported chemicals. This type of the regulon could be transferred from donor microorganisms into E. coli and maintained in functional forms in the host.
After the Genomic SELEX screening of a total of 53 uncharacterized Y-gene TFs (see TEC database for the complete list), we realized that a majority of these TFs are involved in either regulation of the genes involved in utilization of natural materials in nature or for response to varieties of the environmental stress (reference 10; details will be described elsewhere). Thus, the functional characterization of these uncharacterized TFs could not be identified in the absence of stresses such as under the ordinary laboratory culture conditions.
DATA AVAILABILITY
All the SELEX-chip data described in this report were submitted to the ‘TEC‘ database at the National Institute of Genetics (https://shigen.nig.ac.jp/ecoli/tec/) under the accession codes: BetI, KdpE, LacI, MarR, NanR, RpiR, TorR, UlaR, UxuR, CecR(YbiH), DecR(YbaO), NimR(YeaM) and XynR(YagI).
ACKNOWLEDGEMENTS
We thank Ayako Kori (Hosei University) and Kayoko Yamada (Hosei Univ) for expression and purification of TFs, and Kaneyoshi Yamamoto (Hosei University) for support and discussion.
FUNDING
MEXT Grants-in-Aid for Scientific Research (C) [16K07195 to T.S.]; Grants-in-Aid for Young Scientist (B) [24780070]; Grants-in-Aid for Young Scientist (B) [15K18670 to H.O.]; MEXT Cooperative Research Program of Network Joint Research Center for Materials and Devices (to A.I., T.S.); MEXT-Supported Program for the Strategic Research Foundation at Private Universities (to A.I.). Funding for open access charge: MEXT Grants-in-Aid for Scientific Research (C) [16K07195 to T.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ishihama A. Prokaryotic genome regulation: multi-factor promoters, multi-target regulators and hierarchic networks. FEMS Microbiol. Rev. 2010; 34:628–645. [DOI] [PubMed] [Google Scholar]
- 2. Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 2000; 54:499–518. [DOI] [PubMed] [Google Scholar]
- 3. Ishihama A. Building the whole image of genome regulation in the model organism Escherichia coli K-12. J. Gen. Appl. Microbiol. 2017; 63:311–324. [DOI] [PubMed] [Google Scholar]
- 4. Gourse R.L., Ross W., Rutherford S.T.. General pathway for turning on promoters transcribed by RNA polymerase containing alternative sigma factors. J. Bacteriol. 2006; 188:4589–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gruber T.M., Gross C.A.. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003; 57:441–466. [DOI] [PubMed] [Google Scholar]
- 6. Ishihama A. Prokaryotic genome regulation: a revolutionary paradigm. Proc. Jpn. Acad. Ser B Phys. Biol. Sci. 2012; 88:485–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Babu M.M., Teichmann S.A.. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003; 31:1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perez-Rueda E., Collado-Vides J.. The repertoire of DNA-binding transcription regulators in Escherichia coli K-12. Nucleic Acids Res. 2000; 28:1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ishihama A. Role of the RNA polymerase alpha subunit in tanscription activation. Mol. Microbiol. 1992; 6:3283–3288. [DOI] [PubMed] [Google Scholar]
- 10. Ishihama A., Shimada T., Yamazaki Y.. Transcription profile of Escherichia coli: Genomic SELEX search for regulatory targets of transcription factors. Nucleic Acids Res. 2016; 44:2058–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Busby S., Ebright R.H.. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 1999; 293:199–213. [DOI] [PubMed] [Google Scholar]
- 12. Ishihama A. Protein-protein communication within the transcription apparatus. J. Bacteriol. 1993; 175:2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oshima T., Aiba H., Masuda Y., Kanaya S., Sugiura M., Wanner B.L., Mori H., Mizuno T.. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 2002; 46:281–291. [DOI] [PubMed] [Google Scholar]
- 14. Richmond C.S., Glasner J.D., Jin H.R., Blattner F.R.. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999; 27:3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tao H., Bausch C., Richmond C., Blattner F.R., Conway T.. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 1999; 181:6425–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bulyk M.L. DNA microarray technologies for measuring protein-DNA interactions. Curr. Opin. Biochem. 2006; 17:422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wade J.T., Struhl K., Busby S.J.W., Grainger D.C.. Genomic analysis of protein-DNA interactions in bacteria: insights into transcription and chromosome organization. Mol. Microbiol. 2007; 65:21–26. [DOI] [PubMed] [Google Scholar]
- 18. Grainger D.C., Lee D.J., Busby S.J.W.. Direct methods for studying transcription regulatory proteins and RNA polymerase in bacteria. Curr. Opin. Microbiol. 2009; 12:531–535. [DOI] [PubMed] [Google Scholar]
- 19. Kahramanoglou C., Seshasayee A.S.N., Prieto A.I., Ibberson D., Schimidt S., Zimmermann J., Benes V., Flaser G.M., Luscombe N.M.. Direct and indirect effects of H-NS and Fig on global gene expression control in Escherichia coli. Nucleic Acids Res. 2010; 39:2073–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peano C., Wolf J., Demol J., Rossi E., Petiti L., De Bellis G., Geiselmann J., Egli T., Lacour S., Landini P.. Characterization of the Escherichia coli sigma-S core region by chromatin immunoprecipitation-sequencing (ChIP-seq) analysis. Sci. Rep. 2015; 5:10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keseler I.M., Collado-Vides J., Santos-Zavaleta A., Peralta-Gil M., Gama-Castro S., Muniz-Rascado L., Bonavides-Martinez C., Paley S., Krummenacker M., Altman T. et al. . EcoCyc: a compehensive database of Escherichia coli biology. Nucleic Acids Res. 2011; 39:D583–D590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham S., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D.. EcoCyc: a compehensive database resource for Escherichia coli. Nucleic Acids Res. 2005; 33:D334–D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Salgado H., Peralta-Gil M., Gama-Castro S., Santos-Zavaleta A., Muniz-Rascado L., Garcia-Sotelo J.S., Weiss V., Solano-Lira H., Martinez-Flores I., Medina-Revera A. et al. . RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res. 2013; 41:D203–D213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salgado H., Gama-Castro S., Martinez-Antonio A., Diaz-Peredo E., Sanchez-Solano F., Peralta-Gil M., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C. et al. . RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions. Nucleic Acids Res. 2006; 32:D303–D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shimada T., Ogasawara H., Ishihama A.. Genomic SELEX screening of regulatory targets of Escherichia coli transcription factors. Meth. Mol. Biol. 2018; in press. [DOI] [PubMed] [Google Scholar]
- 26. Ishihama A. Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nystrom T, Slauch JM, Squires CL, Ussery D. The nucleoid: an overview. EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. 2009; Washington, DC: ASM Press. [Google Scholar]
- 27. Dunne K.A., Chaudhuri R.R., Rossiter A.E., Beriotto I., Browning D.F., Squire D., Cunningham A.F., Cole J.A., Loman N., Henderson I.R.. Sequencing a piece of history: complete genome sequence of the original Escherichia coli strain. Microbial. Genomics. 2017; 3, doi:10.1099/mgen.0.000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Land M., Hauser L., Jun S-R., Nookaew I., Leuze M.R., Ahn T-H., Karpinets T., Lund O., Kora G., Wassenaar T. et al. . Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics. 2015; 15:141–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jishage M., Ishihama A.. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 1997; 179:959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shimada T., Fujita N., Maeda M., Ishihama A.. Systematic search for the Cra-binding promoters using genomic SELEX systems. Genes Cells. 2005; 10:907–918. [DOI] [PubMed] [Google Scholar]
- 31. Shimada T., Tanaka K., Ishihama A.. The whole set of the constitutive promoters recognized by four minor sigma subunits of Escherichia coli RNA polymerase. PLoS One. 2017; 12:e0179181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shimada T., Yamazaki Y., Tanaka K., Ishihama A.. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PLoS One. 2014; 9:e90447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamamoto K., Hirao K., Oshima T., Aiba H., Utsumi R., Ishihama A.. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 2005; 280:1448–1456. [DOI] [PubMed] [Google Scholar]
- 34. Shimada T., Kori A., Ishihama A.. Involvement of the ribosome operon repressor RbsR in regulation of purine nucleotide synthesis in Escherichia coli. FEMS Microbiol. Lett. 2013; 344:159–165. [DOI] [PubMed] [Google Scholar]
- 35. Reznikoff W.S. Catabolite gene activator protein activation of lac transcription. J. Bacteriol. 1992; 174:655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martinez-Antonio A., Collado-Vides J.. Identifying global regulators in transcription regulatory networks in bacteria. Curr. Opin. Microbiol. 2003; 6:482–489. [DOI] [PubMed] [Google Scholar]
- 37. Balderas-Martinez Y.I., Savageau M., Salgado H., Perez-Rueda E., Morrett E., Collado-Vides J.. Transcription factors in Escherichia coli prefer the holo conformation. PLoS One. 2013; 8:e65723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gottesman S. Bacterial regulation: global regulatory neworks. Ann. Rev. Genet. 1984; 18:415–441. [DOI] [PubMed] [Google Scholar]
- 39. Jacob F., Monod J.. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961; 3:318–356. [DOI] [PubMed] [Google Scholar]
- 40. Gilbert W., Müller-Hill B.. Isolation of the Lac repressor. Proc. Natl. Acad. Sci. U.S.A. 1966; 56:1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson C.J., Zhan H., Swint-Kruse L., Matthews K.S.. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell. Mol. Life Sci. 2006; 64:3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oehler S., Eismann E.R., Kramer H., Muller-Hill B.. The three operators of the lac operon cooperate in repression. EMBO J. 1990; 9:973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shimada T., Momiyama E., Yamanaka Y., Watanabe H., Yamamoto K., Ishihama A.. Regulatory role of XynR (YagI) in catabolism of xylonate in Escherichia coli K-12. FEMS Microbiol. Lett. 2017; 364, doi:10.1093/femsle/fnx220. [DOI] [PubMed] [Google Scholar]
- 44. Yamanaka Y., Shimada T., Yamamoto K., Ishihama A.. Transcription factor CecR (YbiH) regulates a set of genes affecting the sensitivity of Escherichia coli against cefoperazone and chloramphenicol. Microbiology. 2016; 162:1253–1264. [DOI] [PubMed] [Google Scholar]
- 45. Shimada T., Tanaka K., Ishihama A.. Transcription factor DecR (YbaO) controls detoxification of L-cysteine in Escherichia coli. Microbiology. 2016; 162:1698–1707. [DOI] [PubMed] [Google Scholar]
- 46. Ogasawara H., Ohe S., Ishihama A.. Role of transcription factor NimR (YeaM) in sensitivity control of Escherichia coli to 2-nitroimidazole. FEMS Microbiol. Lett. 2015; 362:1–8. [DOI] [PubMed] [Google Scholar]
- 47. Cohen S.P., Hachler H., Levy S.B.. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 1993; 175:1484–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. George A.M., Levy S.B.. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 1983; 155:541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martin R.G., Rosner J.L.. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5456–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martin R.G., Nyantakyi P.S., Rosner J.L.. Regulation of the multiple antibiotic resistance (mar) regulon by marORA sequences in Escherichia coli. J. Bacteriol. 1995; 177:4176–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alekshun M.N., Levy S.B.. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 1999; 7:410–413. [DOI] [PubMed] [Google Scholar]
- 52. Chubiz L.M., Rao C.V.. Aromatic acid metabolites of Escherichia coli K-12 can induce the marRAB operon. J. Bacteriol. 2010; 192:4786–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alekshun M.N., Levy S.B.. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 1999; 181:4669–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Iida A., Harayama S., Iino T., Hazelbauer G.L.. Molecular cloning and characterization of genes required for ribose transport and utilization in Escherichia coli K-12. J. Bacteriol. 1984; 158:674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sorensen K.I., Hove-Jensen B.. Ribose catabolism of Escherichia coli; Characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J. Bacteriol. 1996; 178:1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hope J.N., Bell A.W., Hermodson M.R., Groarke J.M.. Ribokinase from Escherichia coli K12: Nucleotide sequence and overexpression of the rbsK gene and purification of ribokinase. J. Biol. Chem. 1986; 261:7663–7668. [PubMed] [Google Scholar]
- 57. Kim C., Song S., Park C.. The D-allose operon of Escherichia coli K-12. J. Bacteriol. 1997; 179:7631–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Poulsen T.S., Chang Y-Y., Hove-Jensen B.. D-Allose catabolism of Escherichia coli: Involvement of alsI and regulation of als regulon expression by allose and ribose. J. Bacteriol. 1999; 181:7126–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lopilato J.E., Garwin J.L., Emr S.D., Silhavy T.J., Beckwith J.R.. D-Ribose metabolism in Escherichia coli K-12: genetics, regulation, and transport. J. Bacteriol. 1984; 158:665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Campos E., Baldoma L., Aguilar J., Badia J.. Regulation of expression of the divergent ulaG and ulaABCDEF operons involved in L-ascorbate dissimilation in Escherichia coli. J. Bacteriol. 2004; 186:1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yew W.S., Gerlt J.A.. Utilization of L-ascorbate by Escherichia coli K-12: assignments of functions to products of the yif-sga and yia-sgb operons. J. Bacteriol. 2002; 184:302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Z., Aboulwafa M., Smith M.H., Saier M.H. Jr. The ascorbate transporter of Escherichia coli. J. Bacteriol. 2003; 185:2243–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garces F., Fernandez F.J., Gomez A.M., Perez-Luque R., Campos E., Prohens R., Aguilar J., Baldoma L., Coll M., Badia J. et al. . Quaternary structural transitions in the DeoR-type repressor UlaR control transcriptional readout from the L-ascorbate utilization regulon in Escherichia coli. Biochemistry. 2008; 47:11424–11433. [DOI] [PubMed] [Google Scholar]
- 64. Fabich A.J., Jones S.A., Chowdhury F.Z., Cernosek A., Anderson A., Smalley D., McHargue J.W., Hightower G.A., Smith J.T., Autieri S.M. et al. . Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 2008; 76:1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chang D.E., Smalley D.J., Tucker D.L., Leatham M.P., Norris W.E., Stevenson S.J., Anderson A.B., Grissom J.E., Laux D.C., Cohen P.S. et al. . Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Portalier R., Robert-Baudouy J., Stoeber F.. Regulation of Escherichia coli K-12 hexuronate system genes: exu regulon. J. Bacteriol. 1980; 143:1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nemoz G., Robert-Baudouy J., Stoeber F.. Physiological and genetic regulation of the aldohexuronate transport system in Escherichia coli. J. Bacteriol. 1976; 127:706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tutukina M.N., Potapova A.V., Cole J.A., Ozoline O.N.. Control of hexuronate metabolism in Escherichia coli by the two interdependent regulators, ExuR and UxuR: derepression by heterodimer formation. Microbiology. 2016; 162:1220–1231. [DOI] [PubMed] [Google Scholar]
- 69. Robert-Baudouy J., Portalier R., Stoeber F.. Regulation of hexuronate system genes in Escherichia coli K-12: multiple regulation of the uxu operon by exuR and uxuR gene products. J. Bacteriol. 1981; 145:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Suvorova J.A., Tutukina M.N., Ravcheev D.A., Rodionov D.A., Ozoline O.N., Gelfand M.S.. Comparative genomic analysis of the hexuronate metabolism genes and their regulation in Gammaproteobacteria. J. Bacteriol. 2011; 193:3956–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Le Rudulier D., Strom A.R., Dandekar A.M., Smith L.T., Valentine R.C.. Molecular biology of osmoregulation. Science. 1984; 224:1064–1068. [DOI] [PubMed] [Google Scholar]
- 72. Landfald B., Strøm A.R.. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J. Bacteriol. 1986; 165:849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Styrvold O.B., Falkenberg P., Landfald B., Eshoo M.W., Bjørnsen T., Strøm A.R.. Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-glycine betaine pathway. J. Bacteriol. 1986; 165:856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gowrishankar J., Jayashree P., Rajkumari K.. Molecular cloning of an osmorergulatory locus in Escherichia coli: Increased proU gene dosage results in enhanced osmotolerance. J. Bacteriol. 1986; 168:1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. May G., Faatz E., Villarejo M., Bremer E.. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K12. Mol. Gen. Genet. 1986; 205:225–233. [DOI] [PubMed] [Google Scholar]
- 76. Lamark T., Rokenes T.P., McDougall J., Strom A.R.. The complex bet promoters of Escherichia coli; Regulation by oxygen (ArcA), chloine (BetI) and osmotic stress. J. Bacteriol. 1996; 178:1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vimr E.R. Unified theory of bacterial sialometabolism: how and why bacteria metabolize host sialic acids. ISRN Microbiol. 2013; 2013:816713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vimr E.R., Kalivoda K.A., Deszo E.L., Steenbergen S.M.. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 2004; 68:132–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Plumbridge J., Vimr E.. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 1999; 181:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kalivoda K.A., Steenbergen S.M., Vimr E.R., Plumbridge J.. Regulation of sialic acid catabolism by the DNA binding protein NanR in Escherichia coli. J. Bacteriol. 2003; 185:4806–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ringenberg M., Lichtensteiger C., Vimr E.. Redirection of sialic acid metabolism in genetically engineered Escherichia coli. Glycobiology. 2001; 11:533–539. [DOI] [PubMed] [Google Scholar]
- 82. Sohanpal B.K., El-Labany S., Lahooti M., Plumbridge J.A., Blomfield I.C.. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:16322–16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kalivoda K.A., Steenbergen S.M., Vimr E.R.. Control of the Escherichia coli sailoregulon by transcription repressor NanR. J. Bacteriol. 2013; 195:4689–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Vimr E.R., Troy F.A.. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J. Bacteriol. 1985; 164:845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Epstein W. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 2003; 75:293–320. [DOI] [PubMed] [Google Scholar]
- 86. Booth I.R. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 1985; 49:359–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Buurman E.T., McLaggan D., Naprstek J., Epstein W.. Multiple paths for nonphysiological transport of K+ in Escherichia coli. J. Bacteriol. 2004; 186:4238–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Laimins L.A., Rhoads D.B., Epstein W.. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:464–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rhoads D.B., Laimins L., Epstein W.. Functional organization of the kdp genes of Escherichia coli K-12. J. Bacteriol. 1978; 135:445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Walderhaug M.O., Polarek J.W., Voelkner P., Daniel J.M., Hesse J.E., Altendorf K., Epstein W.. KdpD and KdpE proteins that control expression of the kdpABC operon are members of the two-component sensor-effector class of regulators. J. Bacteriol. 1992; 174:2152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Altendorf K., Voelkner P., Puppe W.. The sensor kinase KdpD and the response regulator KdpE control expression of the kdpFABC operon in Escherichia coli. Res. Microbiol. 1994; 145:374–381. [DOI] [PubMed] [Google Scholar]
- 92. Voelkner P., Puppe W., Altendorf K.. Characterization of the KdpD protein, the sensor kinase of the K+-translocating Kdp system of Escherichia coli. Eur. J. Biochem. 1993; 217:1019–1026. [DOI] [PubMed] [Google Scholar]
- 93. Sugiura A., Nakashima K., Tanaka K., Mizuno T.. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol. Microbiol. 1992; 6:1769–1776. [DOI] [PubMed] [Google Scholar]
- 94. Ansaldi M., Theraulaz L., Baraquet C., Panis G., Mejean V.. Aerobic TMAO respiration in Escherichia coli. Mol. Microbiol. 2007; 66:484–494. [DOI] [PubMed] [Google Scholar]
- 95. Barrett E.L., Kwan H.S.. Bacterial reduction of trimethylamine oxide. Annu. Rev. Microbiol. 1985; 39:131–149. [DOI] [PubMed] [Google Scholar]
- 96. Gon S., Giudici-Orticoni M.T., Mejean V., Iobbi-Nivol C.. Electron transfer and binding of the c-type cytochrome TorC to the trimethylamine N-oxide reductase in Escherichia coli. J. Biol. Chem. 2001; 276:11545–11551. [DOI] [PubMed] [Google Scholar]
- 97. Ilbert M., Mejean V., Iobbi-Nivol C.. Functional and structural analysis of members of the TorD family, a large chaperone family dedicated to molybdoproteins. Microbiology. 2004; 150:935–943. [DOI] [PubMed] [Google Scholar]
- 98. Simon G., Mejean V., Jourlin C., Chippaux M., Pascal M.C.. The torR gene of Escherichia coli encodes a response regulator protein involved in the expression of the trimethylamine N-oxide reductase genes. J. Bacteriol. 1994; 176:5601–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jourlin C., Ansaldi M., Mejean V.. Transphosphorylation of the TorR response regulator requires the three phosphorylation sites of the TorS unorthodox sensor in Escherichia coli. J. Mol. Biol. 1997; 267:770–777. [DOI] [PubMed] [Google Scholar]
- 100. Gao R., Stock A.M.. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009; 63:133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ansaldi M., Jourlin-Castelli C., Lepelletier M., Theraulaz L., Mejean V.. Rapid dephosphorylation of the TorR response regulator by the TorS unorthodox sensor in Escherichia coli. J. Bacteriol. 2001; 183:2691–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Simon G., Jourlin C., Ansaldi M., Pascal M.C., Chippaux M., Mejean V.. Binding of the TorR regulator to cis-acting direct repeats activates tor operon expression. Mol. Microbiol. 1995; 17:971–980. [DOI] [PubMed] [Google Scholar]
- 103. Hasegawa A., Ogasawara H., Kori A., Teramoto J., Ishihama A.. The transcription factor AllR senses both allantoin and glyoxylate and controls a set of genes for degradation and reutilization of purines. Microbiology. 2008; 154:3366–3378. [DOI] [PubMed] [Google Scholar]
- 104. Marbaniang C.S., Gowrishankar J.. Role of ArgP (IciA) in lysine-mediated repression in Escherichia coli. J. Bacteriol. 2011; 193:585–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lorca G.L., Ezersky A., Lunin V.V., Walker J.R., Altamentova S., Evdokimova E., Vedadi M., Bochkarev A., Savchenko A.. Glyoxylate and pyruvate are antagonistic effectors of the Escherichia coli IclR transcriptional regulator. J. Biol. Chem. 2007; 282:16476–16491. [DOI] [PubMed] [Google Scholar]
- 106. Schraiber V., Richet E.. Self-association of the Escherichia coli trascription activator MalT in the presence of a maltotriose and ATP. J. Biol. Chem. 1999; 274:33220–33226. [DOI] [PubMed] [Google Scholar]
- 107. Shimada T., Hirao K., Kori A., Yamamoto K., Ishihama A.. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimdines. Mol. Microbiol. 2007; 66:744–779. [DOI] [PubMed] [Google Scholar]
- 108. Shimada T., Shimada K., Matsui M., Kitai Y., Igarashi J., Suga H., Ishihama A.. Roles of cell division control factor SdiA: Recognition of quorum sensing signals and modulation of transcription regulation targets. Genes Cells. 2014; 19:405–418. [DOI] [PubMed] [Google Scholar]
- 109. Dinnbier U., Limpinsel E., Schmid R., Bakker E.P.. Transient accumulation of potassium glutamate and its replacement by trelalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 1988; 150:348–357. [DOI] [PubMed] [Google Scholar]
- 110. Røkenes T.P., Lamark T., Strom A.R.. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation. J. Bacteriol. 1996; 178:1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Schrame H., Laermann V., Tegetmeyer H.E., Brachmann A., Jung K., Altendorf K.. Revisiting regulatioin of potassium homeostatis in Escherichia coli: the connection to phosphate limitation. Microbiol. Open. 2017; 6:e438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ishihama A., Kori A., Koshio E., Yamada K., Maeda H., Shimada T., Makinoshima H., Iwata A., Fujita N.. Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J. Bacteriol. 2014; 196:2718–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang X., Kim Y., Ma Q., Hong S.H., Pokusaeva K., Sturino J.M., Wood T.K.. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010; 1, doi:10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Casjens S. Prophages and bacterilal genomics: what have we learned so far. Mol. Microbiol. 2003; 49:277–300. [DOI] [PubMed] [Google Scholar]
- 115. Yamamoto K., Yamanaka Y., Shimada T., Sarkar P., Yoshida M., Bharadwaj N., Watanabe H., Taira Y., Chatterji D., Ishihama A.. Altered distribution of RNA polymerase lacking the omega subunit within the prophages along the Escherichia coli K-12 genome. mSystems. 2018; e00172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Grainger D.C., Hurd D., Harrison M., Holdstock J., Busby S.J.W.. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17693–17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muniz-Rascado L., Garcia-Sotelo J.S., Alquicira-Hernandez K., Martinez-Flores I., Pannier L., Castro-Mondragon A. et al. . RegulonDB v9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016; 44:D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jobe A., Bourgeois S.. lac repressor-operator interaction. VI. The natural inducer of the lac operon. J. Mol. Biol. 1972; 69:397–408. [DOI] [PubMed] [Google Scholar]
- 119. Ashwell G. Enzymes of glucuronic and galacturonic acid metabolism in bacteria. Methods Enzymol. 1962; 5:190–208. [Google Scholar]
- 120. Polarek J.W., Williams G., Epstein W.. The products of the kdpDE operon are required for expression of the Kpd ATPase of Escherichia coli. J. Bacteriol. 1992; 174:2145–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Barkley M.D., Riggs A.D., Jobe A., Burgeois S.. Interaction of effecting ligands with lac repressor and repressor-operator complex. Biochemistry. 1975; 14:1700–1712. [DOI] [PubMed] [Google Scholar]
- 122. Bates Utz C., Nguyen A.B., Smalley D.J., Anderson A.B., Conway T.. GntP is the Escherichia coli Fructuronic acid transporter and belongs to the UxuR regulon. J. Bacteriol. 2004; 186:7690–7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Eshoo M.W. lac fusion analysis of the bet genes of Escherichia coli: regulation by osmolarity, temperature, oxygen, choline, and glycine betaine. J. Bacteriol. 1988; 170:5208–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Rkenes T.P., Lamark T., Strøm A.R.. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation. J. Bacteriol. 1996; 178:1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Jourlin C., Ansaldi M., Méjean V.. Transphosphorylation of the TorR response regulator requires the three phosphorylation sites of the TorS unorthodox sensor in Escherichia coli. J. Mol. Biol. 1997; 267:770–777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the SELEX-chip data described in this report were submitted to the ‘TEC‘ database at the National Institute of Genetics (https://shigen.nig.ac.jp/ecoli/tec/) under the accession codes: BetI, KdpE, LacI, MarR, NanR, RpiR, TorR, UlaR, UxuR, CecR(YbiH), DecR(YbaO), NimR(YeaM) and XynR(YagI).