Abstract
The starch granules of hexaploid wheat (Triticum aestivum) contain a group of three proteins known as SGP-1 (starch granule protein-1) proteins, which have apparent molecular masses of 100, 108, and 115 kD. The nature and role of these proteins has not been defined previously. We demonstrate that these polypeptides are starch synthases that are present in both the starch granule and the soluble fraction at the early stages of wheat endosperm development, but that are exclusively granule bound at mid and late endosperm development. A partial cDNA clone encoding a fragment of the 100-kD protein was obtained by screening a wheat endosperm cDNA expression library using monoclonal antibodies. Three classes of cDNA were subsequently isolated from a wheat endosperm cDNA library by nucleic acid hybridization and were shown to encode the 100-, 108-, and 115-kD proteins. The cDNA sequences are highly homologous to class II starch synthases and have the highest homology with the maize SSIIa (starch synthase IIa) gene. mRNA for the SGP-1 proteins was detected in the leaf, pre-anthesis florets, and endosperm of wheat and is highly expressed in the leaf and in the grain during the early to mid stages of development. We discuss the roles of the SGP-1 proteins in starch biosynthesis in wheat.
The synthesis and assembly of the starch granule in the plastid is a complex process that involves the concerted action of a suite of biosynthetic enzymes. Enzymatic activities involved in the synthesis of granular starch include ADP-Glc pyrophosphorylase (EC 2.7.7.27), SS (EC 2.4.1.21), branching enzyme (EC 2.4.1.18), and debranching enzyme (EC 3.2.1.41 and EC 3.2.1.68) (Nelson and Pan, 1995; Ball et al., 1996). Bacterial glycogen biosynthesis requires ADP-Glc pyrophosphorylase, glycogen synthase and branching enzyme (Preiss and Romeo, 1994); however, mutagenesis studies demonstrate that the requirements for the synthesis of the starch granule are clearly more complex than for the synthesis of the glycogen molecule. To meet these requirements, plants contain multiple isozymes of starch biosynthetic enzymes. In maize, the most intensively studied cereal with respect to starch biosynthesis, three isoforms of branching enzyme (Boyer and Preiss, 1978), four isoforms of SS (Gao et al., 1998; Harn et al., 1998), and both isoamylase (James et al., 1995) and pullulanase (Beatty et al., 1999) types of debranching enzymes have been described from developing endosperm. The definition of how each of these isoforms contributes to the synthesis of granular starch remains a focus of research.
Isoforms of starch biosynthetic enzymes in plants are differentially located. At the subcellular level, ADP-Glc pyrophosphorylase is found in both the cytoplasm and the plastid (Denyer et al., 1996). Within the plastid, isoforms are found that are entirely soluble (branching enzyme I of wheat [Triticum aestivum], Rahman et al., 1995; the sugary-1 isoamylase of maize, Yu et al., 1998), entirely granule bound (GBSS [granule-bound SS], Mu-Forster et al., 1996), or partitioned between the granular and soluble phases (SSI, Yu et al., 1998).
The proteins within the matrix of the wheat starch granule have been extensively studied (Denyer et al., 1995; Rahman et al., 1995; Yamamori and Endo, 1996; Takaoka et al., 1997). The predominant 60-kD protein is exclusively granule bound and is analogous to the “waxy” GBSS gene in maize (Rahman et al., 1995). The 75-kD SSI is found in both the granule and the soluble fraction of wheat endosperm (Denyer et al., 1995; Li et al., 1999), and has been assigned to chromosomes 7A, 7B, and 7D (Yamamori and Endo, 1996; Li et al., 1999). The 85-kD band contains a class II branching enzyme and an unidentified polypeptide (Rahman et al., 1995).
Wheat starch granules contain a group of proteins known as the SGP-1 (starch granule protein-1) proteins (Yamamori and Endo, 1996), which have molecular masses of approximately 100, 108, and 115 kD. The precise molecular masses assigned to these proteins vary depending on the details of the SDS-PAGE separation system employed. The 100-, 108-, and 115-kD proteins have been designated SGP-B1, SGP-D1, and SGP-A1 (Yamamori and Endo, 1996; Takaoka et al., 1997), respectively, and have been shown to be encoded at loci located on the short arms of chromosome 7B, 7D, and 7A, respectively (Denyer et al., 1995; Yamamori and Endo, 1996). It has been suggested that the SGP-1 proteins are SSs, based on assays of SS activity following renaturation of SGPs separated by SDS-PAGE (Denyer et al., 1995) and on protein sequence data (Takaoka et al., 1997). In a previous report, we described the isolation of monoclonal antibodies to the SGP-1 proteins and N-terminal sequences determined from the 100-kD protein and a band containing both the 108- and 115-kD proteins (Rahman et al., 1995).
In this paper, we demonstrate that the SGP-1 proteins are SSs and that in early endosperm development they are partitioned between the granule and the soluble phase. However, the SGP-1 proteins become exclusively granule bound at mid to late stages of endosperm development. We report the cloning of cDNAs encoding for each of the SGP-1 polypeptides and demonstrate that they are members of class II of the SSs. The roles of SSII in starch biosynthesis are discussed.
MATERIALS AND METHODS
Plant Material
Genetic stocks of hexaploid bread wheat (Triticum aestivum cv Chinese Spring) with various chromosome additions and deletions were kindly supplied by Dr. E. Lagudah (Commonwealth Scientific and Industrial Research Organization, Plant Industry, Canberra, Australia) and derived from stocks described in Sears and Miller (1985). The hexaploid wheat cultivars Gabo and Wyuna were grown in controlled-environment (growth cabinet) conditions (cv Gabo: 18°C day/13°C night, with a photoperiod of 16 h; cv Wyuna; 24°C day and 16°C night, with a photoperiod of 16 h). Leaves, florets prior to anthesis, and endosperm were collected over the grain filling period, immediately frozen in liquid nitrogen, and stored at −80°C until use.
Gel Electrophoresis, Antibodies, and Immunoblotting
Three monoclonal antibodies (909/52, 911/17, and 910/35; Rahman et al., 1995) raised against the SGP-1 proteins were pooled with each antibody present at a 1:1,000 dilution. Preparation of starch granule and soluble extracts of wheat grain and immunoblotting of SDS-PAGE gels were as as described previously (Rahman et al., 1995) except that immunoreactive bands were revealed using enhanced chemiluminescent reagents (Amersham). SDS-PAGE was carried out according to the method of Takaoka et al. (1997), and zymogram analysis of SDS-denatured extracts was carried out according to the method of Buléon et al. (1997).
Construction and Screening of cDNA Libraries
An expression cDNA library of wheat endosperm was constructed in the vector λgt11 from mRNA isolated from cv Chinese Spring. RNA from 5, 7, 9, 11, and 13 DPA plants was pooled and random primers were used for the first strand of cDNA synthesis. A pool of three monoclonal antibodies (see above) was used for immunoscreening of the expression cDNA library.
A cDNA library was constructed in the vector pBluescript SK(+/−) (Stratagene) using cDNA synthesized from mRNA isolated from endosperm of the cv Wyuna 8 to 12 DPA endosperm, according to instructions provided by the supplier. The library was screened by DNA hybridization with the 85-bp insert from the clone pEL-1, which was obtained by immunoscreening of the expression cDNA library described above.
Amplification of Specific cDNA Regions of Wheat SSII Using PCR
Two PCR products, wSSIIp2 and wSSIIp3, were amplified from the cDNA clone wSSIIB and used for northern and Southern hybridization, respectively. wSSIIp2 was generated by PCR using the primers ssIIa (5′ TGTTGAGGTTCCATGGCACG TTC 3′) and ssIIb (5′ AGTCGTTCTGCCGTA TGATGTCG 3′), amplifying the region between nucleotide positions 1,435 and 1,835 of wCL-A1 (accession no. AF155217). The fragment wSSIIp3 was amplified using the primers ssIIc (5′ CCAAGTA CCAGTGGTGAACGC 3′) and ssIId (5′ CGGTGGGATCCAACGGCCC 3′), using wCL-B1 as template.
To establish the chromosome locations of the genes from which the wCL-A1, wCL-B1, and wCL-D1 cDNAs were transcribed, DNA was extracted from wild-type cv Chinese Spring wheat, from three nullisomic-tetrasomic lines of chromosome 7 of cv Chinese Spring wheat, and from Triticum tauschii var strangulata (accession no. CPI 100799), and analyzed by PCR using the primer combinations ssIIc and ssIId or ssIIc and ssIIe (5′ CATGTGAGCTAGCTTTCGCCC 3′).
PCR amplification was performed using a thermal sequencer (model FTS-1, Corbett Research, Sydney) for 1 cycle of 95°C for 2 min, 35 cycles of 95°C for 30 s, 60°C for 1 min, 72°C for 2 min, and 1 cycle of 25°C for 1 min.
DNA and RNA Analysis
Wheat genomic DNA was isolated and analyzed as previously described (Rahman et al., 1998). Approximately 20 μg of DNA was digested with restriction enzymes BamHI and EcoRI, separated on a 1% agarose gel, transferred to reinforced nitrocellulose membranes (Bio-Rad), and hybridized with the 32P-labeled cDNA probe wSSIIp3. The hybridization and wash conditions were performed as described in Rahman et al. (1997). For RNA analysis, 10 μg of total RNA was separated in a 1.4% agarose-formaldehyde gel, transferred to a Hybond N+ membrane (Amersham), and hybridized with the cDNA probe at 42°C as previously described (Rahman et al., 1998). A DNA fragment, wSSIIp2, was labeled with a DNA labeling kit (Rapid Multiprime, Promega) and used as probe. After washing for 30 min at 65°C with 2× SSC, 0.1% SDS, followed by three washes of 40 min at 65°C with 0.2× SSC, 1% SDS, the membrane was visualized by overnight exposure at −80°C with x-ray film (MR, Kodak).
DNA Sequencing and Analysis
DNA sequencing was performed using an automated system (ABI) with dye terminators as described by the manufacturer. DNA sequences were analyzed using the Genetics Computer Group suite of programs (Devereaux et al., 1984).
RESULTS
The SGP-1 Proteins Are SSs
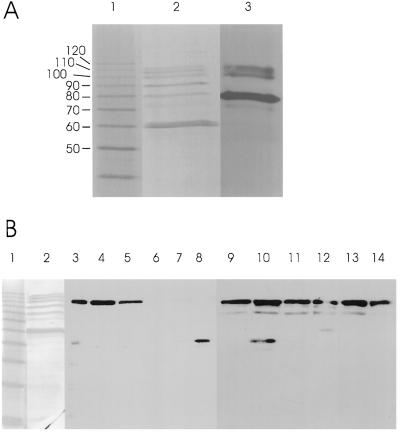
Figure 1 shows the analysis of SGPs using SDS-PAGE gels in which the acrylamide:bis-acrylamide ratio (30:0.135) was optimized for the separation of the SGP-1 proteins (Takaoka et al., 1997). Lane 2 shows the proteins revealed by silver staining, the 60-kD GBSS, an unidentified, weakly staining band of 72 kD, the 80-kD SSI, the 88-kD BEII band, and the 100-, 108-, and 115-kD SGP-1 proteins. Note that the apparent molecular mass of SSI is 75 kD using SDS-PAGE conditions with the commonly used acrylamide:bis-acrylamide ratio of (37.5:1) (Li et al., 1999). Lane 3 shows activity staining of a glycogen-containing SDS-PAGE gel following protein renaturation and incubation with a reaction mixture containing ADP-Glc. Alignment of the silver-stained and activity-stained gels showed that SSI is the predominant SS activity revealed by this assay procedure. The three SGP-1 proteins are also demonstrated to catalyze the formation of iodine-staining α-1,4-glucan. The weakly staining 72-kD band also catalyzes the synthesis of iodine-staining material. The presence of all bands following activity staining was dependent on incubation with ADP-Glc (data not shown).
Figure 1.
SDS-PAGE analysis of starch granule and soluble extracts from the endosperm of the wheat cv Wyuna. Each lane was loaded with the protein extracted from 5 mg of starch (starch granule extracts) or 20 μg of protein from soluble extracts. A, Lane 1, molecular mass markers; lane 2, silver-stained wheat SGPs; lane 3, activity stain of wheat SGPs. B, Lane 1, silver-stained protein standard ladder; lane 2, silver-stained wheat endosperm SGPs; lanes 3 to 14, immunoblots of SDS-PAGE gels loaded with either soluble extracts (lanes 3–8) or protease-treated starch granules (lanes 9–14). Samples were collected at different stages of endosperm development: 5 DPA (lanes 3 and 9), 7 DPA (lanes 4 and 10), 9 DPA (lanes 5 and 11), 13 DPA (lanes 6 and 12), 16 DPA (lanes 7 and 13), and 24 DPA (lanes 8 and 14). Occasional cross-reactivities of the monoclonal antibody with proteins of molecular mass lower than 100 kD were observed; however, these cross reactivities were not reproducible (unlike the consistent immunoreactivity of the 100-kD proteins) and are therefore considered to be nonspecific interactions.
Expression and Localization of Wheat SGP-1 Polypeptides in the Wheat Endosperm
Immunoblots of SDS-PAGE separations of wheat endosperm proteins are shown in Figure 1B, lanes 3 to 14. Proteins were extracted from the soluble phase (Fig. 1, lanes 3–8) and the starch granule (Fig. 1, lanes 9–14) during the development of the wheat endosperm. Monoclonal antibodies raised against the SGP-1 polypeptides were used (Rahman et al., 1995), which have been demonstrated by the analysis of nullisomic-tetrasomic lines of wheat (N7AT7B, N7BT7A, and N7DT7B; data not shown) to bind only to the 100-kD SGP-B1 protein (Fig. 1). SGP-B1 was present in the starch granule throughout endosperm development (Fig. 1, lanes 9–14). This result is consistent with Rahman et al. (1995), who used protein staining techniques to show that all three SGP-1 proteins are found in the starch granule throughout endosperm development. SGP-B1 is also present in the soluble fraction (Fig. 1, lanes 3–5); however, only during the early stages of endosperm development (5–9 DPA in the controlled-environment conditions used).
Isolation of cDNA Sequences for the SGP-1 Proteins
An expression cDNA library was constructed from mRNA from endosperm of the wheat cv Chinese Spring. Monoclonal antibodies to the SGP-B1 protein were used to probe the expression library. One immunoreactive clone was obtained, which was shown to contain an 85-bp cDNA insert (designated wEL-1).
The 85-bp wEL-1 sequence was used as a probe in the hybridization screening of a second wheat endosperm cDNA library that had been constructed from size-fractionated cDNAs greater than 1 kb in length. Ten hybridizing cDNA clones were selected and sequenced. On the basis of the DNA sequences obtained, the cDNA clones can be classified into three groups: group 1 contains seven cDNA clones, group 2 contains two cDNA clones, and group 3 contains one cDNA clone. The longest DNA clone from group 1 contains a 2,939-bp insert (designated wCL-B1) and encodes a 798-amino acid polypeptide starting at nucleotide 176 and terminating at nucleotide 2,572. The longest clone from group 2 contains a 2,807-bp insert (designated wCL-A1 [accession no. AF155217]) and encodes a 799-amino acid polypeptide starting at nucleotide 89 and terminating at nucleotide 2,488. The 2,107-bp insert from the group 3 clone (wCL-D1) encodes a 597-amino acid polypeptide starting at nucleotide 1 and terminating at nucleotide 1,794. The ORF of the truncated wCL-D1 clone encodes a protein approximately 200 amino acid residues shorter than that of polypeptides encoded by wCL-B1 or wCL-A1. For reasons discussed in detail below, the deduced amino acid sequences from wCL-A1, wCL-B1, and wCL-D1 are referred to as wSSII-A, wSSII-B, and wSSII-D, respectively. The deduced amino acid sequences are compared in Figure 2.
Figure 2.
Comparison of the deduced amino acid sequences of SSII from wheat (WSSIIA, WSSIIB, and WSSIID; this paper), maize (ZSSIIA and ZSSIIB; Harn et al., 1998), pea (PEASSII; Dry et al., 1992), and potato (POTSSII; van der Leij et al., 1991). Identical amino acid residues among each of these sequences are indicated below the sequences with an asterisk. The alignments of ZSSIIA with ZSSIIB, and PEASSII and POTSSII are essentially as described in Harn et al. (1998) and Edwards et al. (1995). All sequences are aligned to position the transit peptide cleavage site below the arrow (⇓) between residues 59 and 60 of the WSSIIA sequence. The deduced amino acid sequence from the wEL-1 insert, the sequence of SGP-B1 (peptide3), and the sequence of eight conserved regions are annotated and underlined.
Comparison of wSSII-B, wSSII-A, and wSSII-D shows that they share 95.7% to 96.6% identity (Table I), with variation at 44 positions among the three sequences (Fig. 2): 31 in the N-terminal region (residues 1–300), 10 in the central region (residues 301–729), and three in the C-terminal region (residues 730–799). wSSII-A and wSSII-B differ in length by a single amino acid residue, due to the deletion of an Asp residue at position 69 of the wSSII-B sequence.
Table I.
Homology between deduced amino acid sequences of SSIIs
| wSSII-A | wSSII-B | |
|---|---|---|
| % | % | |
| wSSII-A | 100 | – |
| wSSII-B | 95.9 | 100 |
| wSSII-Da | 96.3 | 96.7 |
| Maize SSIIa | 70.5 | 67.9 |
| Maize SSIIb | 61.9 | 61.7 |
| Pea SSII | 52.7 | 53.2 |
| Potato SSII | 49.7 | 49.7 |
The percentage of identical residues between pairs of SSII genes was calculated using the GAP program of the Genetics Computer Group (Devereaux et al., 1984).
Identities calculated over the region corresponding to the truncated wSSII-D deduced amino acid sequence.
The cDNA Clones Encode the SGP-1 Proteins
A comparison between the sequence of the wEL-1 cDNA obtained by immunoscreening and the three classes of wCL cDNAs obtained by hybridization showed that a sequence identical to the 85-bp wEL-1 sequence was found in wCL-B1, and highly homologous (83 of 85 bp identity) sequences are found in wCL-A1 and wCL-D1. The epitope that reacts with the anti-SGP-1 monoclonal antibodies can therefore be localized to between amino acid residues 272 and 298 in wSSII-B (Fig. 2). There is a single amino acid sequence difference between the epitope-containing regions of wSSII-B and wSSII-A. However, while the epitope-containing regions of wSSII-B and wSSII-D are identical, the monoclonal antibodies only react with SGP-B1 (Fig. 1B). This suggests that there may conformational or post-translational modification differences between wSSII-B and wSSII-D, which prevent the recognition of the wSSII-D product by the monoclonal antibodies.
AT/LGKKDAL, the sequence of the SGP-B1 protein (100 kD) given in Rahman et al. (1995), has been revised to AT/LGKKDAGID following further amino acid sequence analysis. wSSII-B contains a typical transit peptide sequence, followed by an amino acid sequence (AAGKKDAGID) that matches the N terminus of SGP-B1, with the exception of the second residue. wSSII-A contains an amino acid sequence (AAGKKDARVDDPA; see Fig. 2, wSSII-A, residues 60–73) that is identical at eight of 12 residues with an N-terminal sequence (ALGKKDAGIVDGA) determined by sequencing a band containing both the 108- and 115-kD (SGP-D1 and SGP-A1) proteins (Rahman et al., 1995).
Takaoka et al. (1997) reported sequences of three polypeptides obtained from sequencing SGPs derived from the SGP-1 proteins. Peptide 3 is found between residues 378 and 387 of wSSII-A cDNA (Fig. 2). Peptides 1 and 2 of SGP-B1 could not be detected in the amino acid sequences of these cDNA clones; however, peptide 1 can be found in the amino acid sequences of SSI from maize, rice, wheat, and potato (data not shown).
The Sequences of the SGP-1 Proteins Are Highly Homologous to SSs
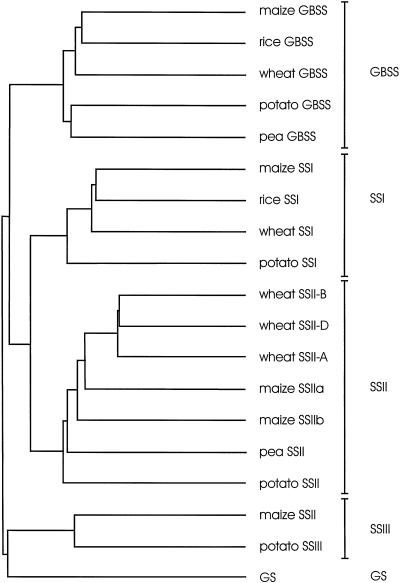
Comparison of the amino acid sequences of SSs from various sources with wSSII-A, wSSII-B and wSSII-D was conducted using the PileUp program from Genetics Computer Group (Devereaux et al., 1984), and the result is presented as a dendrogram (Fig. 3). The sequence of the glycogen synthase of Escherichia coli was also included. Four classes of plant SSs can be defined: GBSS, SSI, SSII, and SSIII. The deduced amino acid sequences encoded by the three wheat cDNA clones are members of the SSII group and are more similar in sequence to maize SSIIa than to maize SSIIb. Table I shows that levels of identity at the amino acid level between the wSSII sequences and other class II SSs range from 49% identity with potato SSII to 70% identity with maize SSIIa.
Figure 3.
Relationships between the primary amino acid sequences of SSs and glycogen synthase of E. coli. The dendrogram was generated using the PileUp program from Genetics Computer Group (Devereaux et al., 1984). The amino acid sequences used for the analysis are as follows (accession numbers given where reference not cited): wheat GBSS (Ainsworth et al., 1993), wheat SSI (Li et al., 1999), wheat SSII-A, wheat SSII-B, and wheat SSII-D (this paper), rice GBSS (X62134), rice SSI (D16202), maize GBSS (M24258), maize SSI (AF036891), maize SSIIa and maize SSIIb (Harn et al., 1998), maize SSII (Gao et al., 1998), pea GBSS (X88789), pea SSII (X88790), potato GBSS (X58453), potato SSI (Y10416), potato SSII (X87988), potato SSIII (X94400), and E. coli glycogen synthase (GS; J01616). Five groups of enzymes are labeled as GBSS, SSI, SSII, SSIII, and GS.
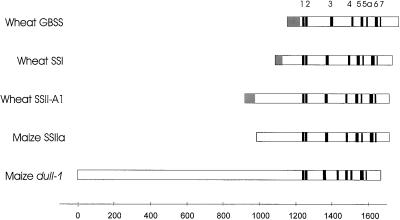
Comparison of the wSSII sequences with differing isoforms of SSs (GBSS, SSI, SSII, and SSIII) identified eight regions (defined in Fig. 2) that could be recognized in the deduced amino acid sequences of SSs from each class. Figure 4 shows an alignment of plant SS sequences, in which the position of the first homologous region is used as the basis of the alignment. This first homologous region contains the consensus motif KXGG that is believed to be present in the ADP-Glc binding site of starch and glycogen synthases (Furukawa et al., 1990).
Figure 4.
Alignment of conserved regions within cereal SS genes. Comparisons of cereal SSs were made based on their deduced amino acid sequences and eight conserved regions identified. Conserved regions are shown in bold and transit peptides (where defined) are shown in gray. Amino acid sequences were deduced from genes encoding; wheat GBSS (Ainsworth et al., 1993), wheat SSI (Li et al., 1999), wheat SSII-A1 (this report), maize SSIIa cDNA (Harn et al., 1998), and the maize dull-1 gene (Gao et al., 1998). The sequences of the conserved regions are defined in Figure 2.
Chromosomal Localization of the Wheat wSSII Genes
Total DNAs were purified from leaves of genetic stocks of wheat cv Chinese Spring with defined chromosome additions and deletions. Southern blot analysis of BamHI and EcoRI restricted DNA from each line was carried out using a fragment designated wSSIIp3 (corresponding to nucleotide positions 2,556–2,921 bp of wCL-B1) as a probe. The results (data not shown) showed that three hybridizing bands were obtained from cv Chinese Spring and that individual bands could be assigned to chromosomes 7A, 7B, and 7D, on the basis of their respective absences from the nullisomic-tetrasomic lines N7AT7B, N7BT7A, and N7DT7B. This analysis indicates that there is a single copy of the SSII gene in each genome in wheat, and is consistent with the findings of Yamamori and Endo (1996), who located the genes encoding SGP-A1, B1, and D1 proteins to the short arm of chromosome 7.
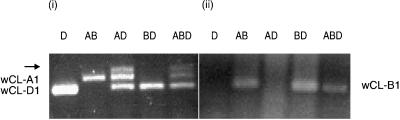
PCR analysis was used to assign each of the cDNA clones to the A, B, or D genomes. A single 365-bp PCR fragment was obtained from nullisomic-tetrasomic genomic DNA of Chinese Spring when primers ssIIc and ssIId were used for the PCR amplification (Fig. 5). This PCR product was obtained only from lines bearing the B genome. The fragment was cloned and sequenced and shown to be identical to a 365-bp region of the wCL-B1 cDNA. An identical fragment was obtained by PCR of the wCL-B1 cDNA clone (but not wCL-A1 or wCL-D1). The wCL-B1 cDNA is therefore the product of a gene located on chromosome 7 of the B genome. Two PCR products were amplified from nullisomic-tetrasomic genomic DNA of cv Chinese Spring when primers ssIIc and ssIIe were used for the PCR amplification (Fig. 5). One PCR fragment, approximately 350 bp long, was only amplified when the A genome was present, and a second 322-bp product was only amplified when the D-genome was present. The 350- and 322-bp PCR products were also cloned and sequenced and shown to be identical to the wCL-A1 and wCL-D1 cDNAs, respectively. The wCL-A1 and wCL-D1 cDNAs are therefore the product of genes located on chromosomes 7A and 7D, respectively. In this analysis, a third band (Fig. 5, arrow) appeared only when the A and D genes were amplified in the same mixture. This band is a heteroduplex, as confirmed by sequence analysis (data not shown).
Figure 5.
Localization of cDNAs on wheat genomes by PCR. (i), Amplification with primers ssIIc and ssIIe. (ii), Amplification with primers ssIIc and ssIId. Lanes D, T. tauschii; lanes AB, N7DT7B; lanes AD, N7BT7A; lanes BD, N7AT7B; and lanes ABD, cv Chinese Spring (wild type). PCR products related to each cDNA clone are labeled. The arrow indicates a heteroduplex band that appears only when the A and D genes are amplified in the same mixture (see text).
Northern Blot Hybridization Analysis of Wheat SSII mRNA Expression
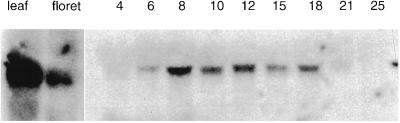
The mRNA for wheat SSII could be detected in leaves, pre-anthesis florets, and endosperm of wheat when total RNAs isolated from these tissues were probed with a PCR probe, wSSIIp2, corresponding to nucleotide positions 1,435 to 1,835 bp of the wCL-A1 cDNA (accession no. AF155217) (Fig. 6). Unlike wSSI, which could not be detected in the leaves under the conditions used (Li et al., 1999), mRNA for the wSSII genes is expressed in leaves and also at an intermediate level in pre-anthesis florets and during wheat endosperm development. However, SGP-B1 protein was not detected in leaf soluble or starch granule extracts using monoclonal antibodies (data not shown). Maize SSIIa mRNA is expressed predominantly in endosperm, while maize SSIIb mRNA is primarily detected in the leaf at low abundance (Harn et al., 1998). The SGP-1 mRNA was present 6 DPA and strongly expressed 8 to 18 DPA, after which time levels of mRNA decline. Southern blotting experiments in wheat using similar hybridization and wash conditions demonstrated that the wSSIIp2 probe used detected only a single copy of the SSII gene in each genome; therefore, it is unlikely that in these experiments this probe cross-hybridized with mRNAs from other SS genes.
Figure 6.
Northern blot analysis of wheat wSSII mRNA in wheat. Total RNA was isolated from leaves, pre-anthesis florets, and endosperm of the wheat cv Gabo grown under defined conditions (16-h day length, 18°C day/13°C night) and probed with the wSSIIp2 DNA fragment. RNAs from leaf and pre-anthesis florets were loaded in the two left lanes. The remaining lanes contained RNA from endosperm and the extraction times (in DPA) are given above the lanes. Equivalent amounts of RNA were loaded in each lane.
DISCUSSION
In this report the SGP-1 proteins have been unambiguously demonstrated to be SSs by the direct alignment of iodine staining bands in renatured SDS-PAGE gels with the SGP-1 proteins. These bands only appear if glycogen is contained in the SDS-PAGE gel and if the gel is incubated in a reaction mixture containing ADP-Glc. This result confirms two lines of evidence that previously suggested that these proteins were SSs. In the first, Denyer et al. (1995) excised sections of renatured SDS-PAGE gels containing the SGP-1 proteins and measured SS activity by direct enzyme assay. In the second, protein sequence data obtained by Takaoka et al. (1997) yielded sequences with homology to SSs. The result in Figure 1 shows a much greater intensity of staining by the SSI protein than the SGP-1 proteins; however, caution must be exercised in interpreting the in vivo significance of this result. Differences in efficiency of renaturation, activity with glycogen as the substrate, affinity for ADP-Glc, or activity in the reaction mixtures used could mean that the observed differences in activity may not accurately reflect the relative involvement of these enzymes in starch synthesis in vivo.
Previous work on the SGP-1 proteins (Denyer et al., 1995; Rahman et al., 1995) focused on the wheat starch granule, and no definitive conclusion concerning their presence or absence in soluble extracts of the wheat endosperm was presented. However, in this report, we demonstrate that a monoclonal antibody against SGP-B1 cross reacts strongly with a protein in the soluble fraction with identical mobility in SDS-PAGE to the granule-bound SGP-B1 protein. However, this protein was only present in the soluble fraction during the early stages of grain development, and no soluble protein could be detected during mid to late endosperm development. This observation is consistent with and extends the observation of Rahman et al. (1995), who suggested, based on studies of granules from endosperm in mid-late stages endosperm development, that the SGP-1 proteins were exclusively granule bound. Given the similarity in the expression of the SGP-A1, AGP-B1 and SGP-D1 proteins in the granule fraction (Rahman et al., 1995), it is reasonable to assume that SGP-A1 and SGP-D1 are also found in the soluble fraction during early endosperm development; however, the very tight specificity of the monoclonal antibody available precludes our addressing this hypothesis in the current set of experiments.
Comparison with the two maize SSII sequences obtained by Harn et al. (1998) demonstrates that the wheat clones are most highly homologous to the maize SSIIa gene. While the sequences of the maize SSIIa and SSIIb cDNAs have been published, the localization of the proteins they encode has not been defined (Imparl-Radosevich et al., 1999). Two isoforms of SS (originally designated SSI and SSII in order of elution from anion-exchange chromatography) have been described in soluble extracts of maize endosperm (Ozbun et al., 1973; Pollack and Preiss, 1980). It is now established that these isoforms are encoded by the SSI and dull1 genes, respectively (Harn et al., 1998; Imparl-Radosevich et al., 1999). Further analysis of the maize granule proteins is required to determine whether proteins that correspond to the SGP-1 proteins are present, and whether they are also found in the soluble fraction of maize endosperm in early endosperm development (Imparl-Radosevich et al., 1999). The expression of an analog to the maize SSIIb gene in wheat endosperm remains an open question that is currently being addressed.
Based on the nomenclature suggested by Harn et al. (1998), the SGP-1 proteins should be defined as SS rather than GBSS because they are not exclusively granule bound. The following nomenclature for SSs in wheat is suggested to provide consistency with the maize nomenclature (reference to the nomenclature of Yamamori and Endo [1996] is included in parentheses where appropriate): wGBSS for the 60-kD GBSS (Wx), wSSI for the 75-kD SSI (SGP-3), wSSII for the 100-, 108-, and 115-kD proteins (SGP-1), and wSSIII for the soluble, high-Mr SS (Li et al., 1999).
While the evidence is compelling that the wCL-A1, wCL-B1, and wCL-D1 cDNAs encode the SGP-A1, SGP-B1, and SGP-D1 proteins of the wheat starch granule, respectively, the calculated molecular masses of the protein products of the two full-length cDNAs, wSSII-A and wSSII-B, are considerably lower than estimates obtained from SDS-PAGE. The molecular mass calculated for the precursor protein represented by the wSSII-A deduced amino acid sequence is 87,229 D, and the mature protein sequence 81,164 D, yet the molecular mass of the mature protein determined by SDS-PAGE analysis is 115 kD. Similarly, the molecular mass calculated from the wSSII-B deduced amino acid sequence is 86,790 D and the mature protein 80,759 D, yet the molecular mass of the mature SGP-B1 protein is estimated to be 100 kD. The wSSII-A and wSSII-B amino acid sequences differ by just a single amino acid residue in length, yet there is an apparent difference of 15 kD in molecular mass when estimated by SDS-PAGE.
Several possibilities can be advanced to account for this apparent discrepancy in molecular mass. First, the wSSII proteins may not migrate in SDS-PAGE in accordance with expectations based on their molecular mass because they retain some conformation under the denaturing conditions used. Similar observations regarding the anomalous electrophoretic mobility of SSs in SDS-PAGE analysis have been made by Edwards et al. (1995), Knight et al. (1998), and Imparl-Radosevich et al. (1999). Second, the proteins may be subject to post-translational modification. While it is also possible that the proteins may be linked to starch through a high-affinity starch binding site or glucosylation reaction, the modified proteins would be unlikely to migrate as distinct and highly reproducible bands given the highly polydisperse nature of starch.
Figure 4 shows a schematic comparison of the structures of SS genes from the four classes of genes that have been sequenced to date from plants: GBSS, SSI, SSII, and SSIII. As has been noted previously (Gao et al., 1998; Harn et al., 1998), the catalytic domain of the SSs is found at the C-terminal end of the gene. Harn et al. (1998) identified seven conserved regions among the SSIIa, SSIIb, SSI, and GBSS sequences. In this report we include an additional region characteristic of SSs (designated region 5a in order to retain conservation of numbering of conserved regions) and extend this identification of regions characteristic of SSs to include the wheat SSII sequences and the dull1 SS sequences reported by Gao et al. (1998). The conservation of the eight conserved regions among the four classes of SSs is striking in terms of their sequence homologies and their alignment.
The major differences in structure between the classes of genes are found in the length of the N-terminal region between the transit peptide and the first conserved region. At one extreme, the GBSS genes have a very short N-terminal arm, whereas the du-1 SS contains a very long N-terminal extension containing several distinct regions (Gao et al., 1998). The wSSII genes contain an N-terminal extension that is longer than GBSS, SSI, or SSIIb, and slightly longer than the maize SSIIa gene. Analysis of the wheat SSII genes shows that the motif PVNGENK is repeated. The area surrounding the repeated PVNGENK motif is not homologous to maize SSIIa and the insertion of this region is responsible for the difference in length between the wheat SSII and maize SSIIa genes. Deletion of the N-terminal arms of the maize SSIIa and maize SSIIb genes expressed in E. coli yields truncated enzymes with altered kinetic properties, but the truncated enzymes retain substantial catalytic potency (Imparl-Radosevich et al., 1999). This is consistent with the working hypothesis that the N-terminal region may be more involved in the localization and substrate recognition of SSs than in catalysis.
Surveys of other species indicate that analogous proteins to the SGP-1 proteins are not present (Rahman et al., 1995; Yamamori and Endo, 1996). Two possibilities that might arise as consequences of the extended N-terminal region of the wheat SGP-1 proteins can be explored. The SGP-1 analogs are essentially soluble in other species; however, the wheat SGP-1 proteins are partitioned into the starch granule, as demonstrated in Figure 1. Also, the wheat SGP-1 proteins migrate in an anomalous manner in SDS-PAGE, and the analog proteins are to be found at a different apparent molecular mass in other species.
Mutations in SSs are known in three other species. In pea, a mutation in SSII gives rise to starch with altered granule morphology and an amylopectin that yields an oligosaccharide distribution with reduced chain length on debranching compared with the wild type (Craig et al., 1998). A similar mutation eliminating a soluble SS designated SSII is known in Chlamydomonas reinhardtii (the sta-3 mutation), and similar effects on granule morphology and amylopectin structure are observed (Fontaine et al., 1993). In maize, two mutations affecting SSs are known. First, the dull1 mutation has been shown to be caused by a lesion within the dull1 SSIII-type SS gene (Gao et al., 1998). A second mutation, sugary-2, yields a starch with reduced amylopectin chain lengths on debranching. This mutation co-segregates with the SSIIa locus (Harn et al., 1998), although direct evidence that the sugary-2 mutation is caused by a lesion in the SSIIa gene remains to be confirmed.
In the SSII mutants of each of these species, the capacity for amylose biosynthesis is retained. This demonstrates clearly different roles in amylose and amylopectin synthesis for the GBSS and SSII genes. Given the conservation in overall organization of the GBSS and SSII genes (Fig. 4) when an alignment is made based on the KTGGL motif of the first conserved region, this focuses attention on the role(s) of the N-terminal region in defining substrate specificity and the localization of the proteins, as the N-terminal region is the major area of divergence between the four classes of SSs. However, it is premature to exclude the influence of more subtle mutations in central and C-terminal regions of the gene.
The generation of a wheat line combining null alleles at each of the three SGP-1 loci, SGP-A1, SGP-B1, and SGP-D1, has been reported recently by Yamamori (1998). In this triple null line, the large starch granules were reported to be mostly deformed and a novel starch with elevated iodine binding capacity was observed. This result demonstrates that SSII is a key enzyme for the development of the starch granule in wheat. Analysis of the triple null lines is currently under way to understand the molecular basis of the mutations. This research will help to further define the effects of the mutation on starch structure and properties and on the expression and localization of other enzymes in the starch biosynthesis pathway.
NOTE ADDED IN PROOF
The 100-, 108-, and 115-kD bands were confirmed as products of the wSSII genes using peptide mass fingerprinting/MALDI-TOF analysis (Australian Proteome Analysis Facility, Macquarie University, Sydney).
ACKNOWLEDGMENTS
We thank Tanya Phongkham for assistance in the preparation of wheat mRNA blots and cDNA libraries, Dr. Mrinal Bhave for supervision of Liuling Yan, and Dr. W.C. Taylor and Dr. A. Pryor for constructive comments on the manuscript.
Abbreviations:
- GBSS
granule-bound starch synthase
- N7AT7B
wheat lacking chromosome 7A but containing two copies of chromosome 7B
- N7BT7A
wheat lacking chromosome 7B but containing two copies of chromosome 7A
- N7DT7B
wheat lacking chromosome 7D but containing two copies of chromosome 7B
- SGP
starch granule protein
- SS
starch synthase
Footnotes
This work was supported by Goodman Fielder (Sydney) and Groupe Limagrain (Paris). IACR receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
LITERATURE CITED
- Ainsworth C, Clark J, Balsdon J. Expression, organisation and structure of the genes encoding the waxy protein (granule-bound starch synthase) in wheat. Plant Mol Biol. 1993;22:67–82. doi: 10.1007/BF00038996. [DOI] [PubMed] [Google Scholar]
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J (1996) From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell 349–352 [DOI] [PubMed]
- Beatty MK, Rahman A, Cao HP, Woodman W, Lee M, Myers AM, James MG. Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize. Plant Physiol. 1999;119:255–266. doi: 10.1104/pp.119.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer CD, Preiss J. Multiple forms of starch branching enzyme of maize: evidence for independent genetic control. Biochem Biophys Res Comm. 1978;80:169–175. doi: 10.1016/0006-291x(78)91119-1. [DOI] [PubMed] [Google Scholar]
- Buléon A, Gallant DJ, Ball S. Starches from A to C: Chlamydomonas reinhardtii as a model microbial system to investigate the biosynthesis of the plant amylopectin crystal. Plant Physiol. 1997;115:949–957. doi: 10.1104/pp.115.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J, Lloyd JR, Tomlinson K, Barber L, Edwards A, Wang TL, Martin C, Hedley CL, Smith AM. Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. Plant Cell. 1998;10:413–426. doi: 10.1105/tpc.10.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer K, Dunlap F, Thorbjornsen T, Keeling P, Smith AM. The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant Physiol. 1996;112:779–785. doi: 10.1104/pp.112.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer K, Hylton CM, Jennet CF, Smith AM. Identification of multiple isoforms of soluble and granule-bound starch synthase in developing wheat endosperm. Planta. 1995;196:256–265. [Google Scholar]
- Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dry I, Smith A, Edwards A, Bhattacharyya M, Dunn P, Martin C. Characterisation of cDNAs encoding two isoforms of granule-bound starch synthase which show differential expression in developing storage organ of pea and potato. Plant J. 1992;2:193–202. [PubMed] [Google Scholar]
- Edwards A, Marshall J, Sidebottom C, Visser RGF, Smith AM, Martin C. Biochemical and molecular characterization of a novel starch synthase from potato tubers. Plant J. 1995;8:283–294. doi: 10.1046/j.1365-313x.1995.08020283.x. [DOI] [PubMed] [Google Scholar]
- Fontaine T, D'Hulst C, Maddelein M-L, Routier F, Pépin TM, Decq A, Wieruszeski J-M, Delrue B, Van den Koornhuyse N, Bossu J-P. Toward an understanding of the biogenesis of the starch granule: evidence that Chlamydomonas soluble starch synthase II controls the synthesis of intermediate size glucans of amylopectin. J Biol Chem. 1993;22:16223–16230. [PubMed] [Google Scholar]
- Furukawa K, Tagaya M, Inouye M, Preiss J, Fukui T. Identification of lysine 15 at the active site in Escherichia coli glycogen synthase. J Biol Chem. 1990;265:2086–2090. [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM. Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell. 1998;10:399–412. doi: 10.1105/tpc.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harn C, Knight M, Ramakrishnan A, Guan H, Keeling PL, Wasserman BP. Isolation and characterization of the zSSIIa and zSSIIb starch synthase cDNA clones from maize endosperm. Plant Mol Biol. 1998;37:639–649. doi: 10.1023/a:1006079009072. [DOI] [PubMed] [Google Scholar]
- Imparl-Radosevich JM, Nichols DJ, Li P, McKean AL, Keeling PL, Guan HP. Analysis of purified maize starch synthases IIa and IIb: SS isoforms can be distinguished based on their kinetic properties. Arch Biochem Biophys. 1999;362:131–138. doi: 10.1006/abbi.1998.1028. [DOI] [PubMed] [Google Scholar]
- James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ME, Harn C, Lilley CER, Guan H, Singletary GW, Mu-Forster C, Wasserman BP, Keeling PL. Plant J. 1998;14:613–622. doi: 10.1046/j.1365-313x.1998.00150.x. [DOI] [PubMed] [Google Scholar]
- Leij van der FR, Visser RGF, Ponstein AS, Jacobsen E, Feenstra WJ. Sequence of the structural gene for granule-bound starch synthase of potato (Solanum tuberosum L.) and evidence for a single point deletion in the amf allele. Mol Gen Genet. 1991;228:240–248. doi: 10.1007/BF00282472. [DOI] [PubMed] [Google Scholar]
- Li Z, Rahman S, Kosar-Hashemi B, Mouille G, Appels R, Morell MK. Cloning and characterisation of a gene encoding wheat starch synthase I. Theor Appl Genet. 1999;98:1208–1216. [Google Scholar]
- Mu-Forster C, Huang RM, Powers JR, Harriman RW, Knight M, Singletary GW, Keeling PL, Wasserman BP. Physical association of starch biosynthetic enzymes with starch granules of maize endosperm: granule-associated forms of starch synthase I and starch branching enzyme II. Plant Physiol. 1996;111:821–829. doi: 10.1104/pp.111.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson O, Pan D. Starch synthesis in maize endosperms. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:475–496. [Google Scholar]
- Ozbun JL, Hawker JS, Greenberg E, Lammel C, Preiss J, Lee EYC. Starch synthetase, phosphorylase, ADPglucose pyrophosphorylase, and UDPglucose pyrophosphorylase in developing maize kernels. Plant Physiol. 1973;51:1–5. doi: 10.1104/pp.51.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock C, Preiss J. The citrate-stimulated starch synthase of starchy maize kernels: purification and properties. Arch Biochem Biophys. 1980;204:578–588. doi: 10.1016/0003-9861(80)90070-3. [DOI] [PubMed] [Google Scholar]
- Preiss J, Romeo T. Molecular biology and regulatory aspects of glycogen biosynthesis in bacteria. Prog Nucleic Acids Res Mol Biol. 1994;47:299–329. doi: 10.1016/s0079-6603(08)60255-x. [DOI] [PubMed] [Google Scholar]
- Rahman S, Abrahams S, Abbott D, Mukai Y, Samuel M, Morell M, Appels R. A complex arrangement of genes at a starch branching enzyme I locus in D-genome donor of wheat. Genome. 1997;40:465–474. doi: 10.1139/g97-062. [DOI] [PubMed] [Google Scholar]
- Rahman S, Kosar-Hashemi B, Samuel M, Hill A, Abbott DC, Skerritt JH, Preiss J, Appels R, Morell M. The major proteins of wheat endosperm starch granules. Aust J Plant Physiol. 1995;22:793–803. [Google Scholar]
- Rahman S, Li Z, Abrahams S, Abbott D, Appels R, Morell M. Characterisation of a gene encoding wheat endosperm starch branching enzyme-I. Theor Appl Genet. 1998;98:156–163. [Google Scholar]
- Sears ER, Miller TG. The history of Chinese spring wheat. Cereal Res Commun. 1985;13:261–263. [Google Scholar]
- Takaoka M, Watanabe S, Sassa H, Yamamori M, Nakamura T, Sasakuma T, Hirano H. Structural characterisation of high molecular weight starch granule-bound proteins in wheat (Triticum aestivum L) J Agric Food Chem. 1997;45:2929–2934. [Google Scholar]
- Yamamori M (1998) Selection of a wheat lacking a putative enzyme for starch synthesis, SGP-1. In Proceedings of the Ninth International Wheat Genetics Symposium, Vol 4. University Extension Press, University of Saskatchewan, Saskatoon, Saskatchewan, Canada, pp 300–302
- Yamamori M, Endo TR. Variation of starch granule proteins and chromosome mapping of their coding genes in common wheat. Theor Appl Genet. 1996;93:275–281. doi: 10.1007/BF00225757. [DOI] [PubMed] [Google Scholar]
- Yu Y, Mu HH, Wasserman BP. Polypeptides of the maize amyloplast stroma: stromal localization of starch-biosynthetic enzymes and identification of an 81-kilodalton amyloplast stromal heat-shock cognate. Plant Physiol. 1998;116:1451–1460. doi: 10.1104/pp.116.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]