Abstract
Background
The nexus between resistance determinants, plasmid type, and clonality appears to play a crucial role in the dissemination and survival of carbapenem-resistant Klebsiella pneumoniae (CRKP). The incidence of infections involving CRKP in Saudi Arabia is increasing and there is a need for detailed molecular profiling of this pathogen for CRKP surveillance and control.
Methods
The resistance determinants of 71 non-redundant CRKP isolates were investigated by polymerase chain reaction (PCR) and sequencing. Plasmid typing was performed using PCR-based replicon typing and the clonality of isolates was determined by multilocus sequence typing. Capsular polysaccharide synthesis genes and other virulence factors were examined using multiplex PCR. Diversity was calculated using DIVEIN, clonal relationship was determined using eBURST, and phylogenetic analysis was performed using SplitsTree4.
Results
A polyclonal OXA-48 gene alone was the most common carbapenemase detected in 48/71 (67.6%) isolates followed by NDM-1 alone in 9/71 (12.7%) isolates. Coproduction of OXA-48 and NDM-1 was observed in 6/71 (8.5%) isolates. Both carbapenemase genes could be transferred into an Escherichia coli recipient. CTX-M-15 was the most abundant extended-spectrum β-lactamase gene detected in 47/71 (66.2%) isolates, whereas clone-specific CTX-M-14 (ST-199 and -709) was found in 15/71 (21%) isolates. Sixty-seven of 71 isolates were positive for one or more plasmid replicons. The replicons detected were: IncFII; IncFIIK; IncFIA; IncFIB; L/M; IncI1; and IncN. FIIK and L/M were predominant, with 69 and 67% positivity, respectively. All isolates were negative for the magA (K1), rmpA, and K2 genes and presented a non-hypermucoviscous phenotype.
Conclusion
A polyclonal CRKP reservoir of sequence types (STs)-37, − 199, and − 152 was observed and ST-152 appeared to be a “frequent carrier” of the NDM-1 gene. ST-199, a singleton not previously reported, showed a sequence diversity suggestive of positive selection. A significant association was evident between resistance determinants and the clonal types of K. pneumoniae: all ST-152 isolates were positive for NDM-1 but negative for OXA-48; ST-199 isolates were positive for OXA-48 but negative for NDM-1; and ST-709 and -199 isolates were positive for CTX-M-14. The incidence of certain clonal types in large numbers predicts an outbreak-like situation and warrants stringent surveillance and infection control.
Electronic supplementary material
The online version of this article (10.1186/s12879-018-3114-9) contains supplementary material, which is available to authorized users.
Keywords: Carbapenem-resistant Klebsiella pneumoniae (CRKP), Clonal diversity, Resistance determinants, Molecular epidemiology, Plasmid profile, Saudi Arabia
Background
Klebsiella pneumoniae (KP), a member of the Enterobacteriaceae family of Gram-negative bacteria, is an important human and animal pathogen [1]. High mortality rates are associated with infection with carbapenemase-producing KP [2]. Various carbapenemase genes, such as KP carbapenemase (KPC), NDM-1, VIM, IMP, and OXA-48, have been identified in KP [3, 4], some of which appear to be clone-specific and such clones form a reservoir for infection [5]. Although reports are available on the emergence of drug-resistant KP in Saudi Arabia [4, 6], few studies have examined the molecular basis of antibiotic resistance, plasmid profiles, virulence factors, and clonal diversity of drug-resistant KP in this country. In order to perform adequate infection control interventions and solicit support for effective stewardship programs, local data on antimicrobial resistance mechanisms and the clonal spread are important. Hence, baseline information on resistant clones assists in assessing the trends of resistance as well as their local epidemiology. Therefore, in this study, we investigated the clonal diversity and genetic profile of a population of multidrug-resistant, carbapenem-resistant KP (CRKP) isolates from patients admitted to our hospital in Riyadh, Saudi Arabia during years 2011 and 2012.
Methods
Bacterial isolates and testing of antibiotic resistance
Seventy-one non-redundant multidrug-resistant CRKP isolates were obtained from patients attending the Microbiology Section of the King Abdulaziz Medical City, Riyadh, Saudi Arabia, between January 2011 and December 2012. The isolates were identified to the species level using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (bioMérieux, Marcy-l’Étoile, France). The frozen isolates were sub-cultured on blood agar at 37 °C overnight. Single colonies were picked up and grown in liquid tryptic soy broth(TSB) medium 37 °C in a shaking incubator at 250 rpm overnight. Minimum inhibitory concentrations (MICs) were determined using a Micro VITEK® 2 microbial identification instrument (bioMérieux). The MIC breakpoints for carbapenem were defined according to the modified 2010 Clinical and Laboratory Standards Institute guidelines [7]. Isolates found to have elevated MICs for carbapenem by Micro VITEK® 2 analysis were confirmed by the manual ETEST® (bioMérieux) to have reduced susceptibility to either imipenem or/and meropenem.
Characterization of resistance genes
Bacterial DNA was isolated using a MagNA Pure kit (Roche Diagnostics, Risch-Rotkreuz, Switzerland). Uniplex polymerase chain reaction (PCR) was performed for bla-SHV, bla-TEM-1, bla-CTX-M, bla-KPC, bla-OXA-48, bla-VIM, and bla-IMP using primers reported previously [8]. Appropriate positive and negative controls were run simultaneously. Each PCR reaction was conducted at least twice. The primers 5’-GGTTTGGCGATCTGGTTTTC-3′ (F) and 5’-CGGAATGGCTCATCACGATC-3′ (R) were used for the amplification of bla-NDM-1, and the primers 5‘-CACCTCATGTTTGAATTCGCC-3′ (F) and 5‘-CTCTCTCACATCGAAATCGC-3′ (R) were used to determine its genetic environment [9]. The PCR products were sequenced on an ABI 3100 DNA analyzer (Life Technologies, Carlsbad, CA, USA). The DNA sequences were analyzed using SeqMan (DNASTAR, Inc., Madison, WI, USA) and MEGA4[10] and blasted against GenBank (National Center for Biotechnology Information, Bethesda, MD, USA). PCR-based replicon typing (PBRT) was performed on these isolates to identify the plasmids as per the protocol described in the PubMLST database (https://pubmlst.org/plasmid/).
Conjugation experiments
Conjugation experiments were performed on selected isolates (Table 2) using J53Azr Escherichia coli, a strain resistant to sodium azide, as a recipient (a kind gift from Dr. Timothy Walsh, University of Cardiff, Wales, UK). Cultures from both donor and recipient bacteria were grown in lysogeny broth (LB) to the logarithmic phase, mixed at a donor:recipient ratio of 1:3, and incubated at 37 °C overnight without shaking. Transconjugants were selected on LB agar plates supplemented with 100 μg/ml of sodium azide and 100 μg/ml of ampicillin. Positive clones were screened by colony PCR for the target resistance genes.
Table 2.
Results of conjugation experiments on selected isolates of KP
| Isolate | Resistance gene(s)/ Plasmid replicon(s) | |
|---|---|---|
| Clone | Transconjugant | |
| RD-99 | CTX-M-15, OXA-48/ L/M, FIIK | CTX-M-15, OXA-48/ L/M, FIIK |
| RD-76 | CTX-M-15, OXA-48/ L/M, FIIK | CTX-M-15, OXA-48/ L/M, FIIK |
| RD-75 | CTX-M-14, OXA-48/ L/M, FIIK, IncN1 | OXA-48/ L/M, FIIK |
| RD-62 | CTX-M-14, OXA-48/ L/M | OXA-48/ L/M |
| RD-123 | CTX-M-15, OXA-48/ L/M, FIIK | CTX-M-15, OXA-48/ FIIK |
| RD-122 | CTX-M-15, OXA-48/ L/M, FIIK, IncN1 | CTX-M-15, − / FIIK |
| RD-114 | CTX-M-15, OXA-48/ FIIK, FII | CTX-M-15, OXA-48/ FIIK, FII |
| RD-112 | CTX-M-15, OXA-48/ L/M, FIIK | CTX-M-15, OXA-48/ L/M, FIIK |
| RD-118 | CTX-M-15, OXA-48, NDM-1/ L/M, FIB | CTX-M-15, OXA-48, NDM-1/ L/M |
| RD-127 | CTX-M-15, NDM-1/ - | CTX-M-15, NDM-1/ - |
| RD-129 | CTX-M-15, NDM-1/ FIB | CTX-M-15, NDM-1/ - |
| RD-64 | CTX-M-15, OXA-48/ L/M, FII, IncN1 | CTX-M-15 / L/M, FII |
Detection of hypermucoviscocity phenotyping and virulence associated genes
Bacterial isolates grown on blood agar plates were checked for hypermucoviscocity. Hypermucoviscocity was considered present when an inoculation loop formed a viscous string of 5 mm or more in length from a bacterial colony [11]. Capsular polysaccharide synthesis (CPS) and virulence-associated genes were detected by multiplex PCR as described by Compain et al. [12]. The presence of CPS genes was reconfirmed by uniplex PCR.
Multilocus sequence typing, allelic diversity, and population structure analysis
Multilocus sequence typing (MLST) was conducted according to the method of Diancourt et al. [13] and sequence types (STs) were assigned through the Institut Pasteur (Paris, France) database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). Individual locus sequences, as well as the concatenated sequences of all seven MLST loci for each isolate, were analyzed for their diversity using DIVEIN [14]. Clonal groups were defined based on STs sharing six loci (single-locus variants) using eBURST [15]. A population snapshot was also drawn for the clonal relationship of these STs with those in the database of the Institut Pasteur using eBURST. Using the alignments of the concatenated sequences from the seven MLST loci for all isolates, a phylogenetic tree was constructed by the neighbor-net method using SplitsTree4 [16].
Results
Here, we present our findings on multidrug/carbapenem-resistant KP isolates obtained at a tertiary care hospital in Riyadh between January 2011 and December 2012. The demographic data on these isolates is given in Table 1.
Table 1.
Showing the distribution of resistance determinants, plasmid replicons and virulence factors in 71 KP isolates
| Isolate | ST | Sampling Date | Speciman | Carbapenemase gene | ESBLs | Other β-Lactamase | Plasmid Replicon |
|---|---|---|---|---|---|---|---|
| RD- 51 | 37 | 27-Oct-11 | Blood | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 52 | 974 | 22-Oct-11 | wound | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | FIIK, FII, L/M |
| RD- 53 | 29 | 8-Aug-11 | Rectal | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | FIIK, L/M |
| RD- 54 | 37 | 9-May-11 | Abd. Drain | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 56 | 37 | 31-Jul-11 | Sputum | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 57 | 694 | 4-Sep-11 | Urine | Negative | CTX-M 15 | SHV-1 | FIIK, L.M |
| RD- 58 | 199 | 9-Sep-11 | Wound | Negative | CTX-M 14 | TEM-1, SHV-1 | L/M |
| RD- 59 | 709 | 5-Sep-11 | Blood | OXA-48 | ND | SHV-11 | L/M |
| RD- 60 | 340 | 28-May-11 | Trachea | Negative | CTX-M 15 | SHV-11 | FII, FIIK |
| RD- 61 | 37 | 24-Sep-11 | Wound | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 62 | 709 | 9-Jan-11 | Blood | OXA-48 | CTX-M 14 | SHV-11 | L/M |
| RD- 63 | 37 | 28-Jan-11 | Sputum | Negative | CTX-M 15 | TEM-1, SHV-11 | FIIK |
| RD- 64 | 15 | 9-Jan-11 | Trachea | Negative | CTX-M 15 | SHV-1 | FII, L/M, IncN |
| RD- 66 | 199 | 13-Oct-11 | Wound | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | L/M |
| RD- 67 | 348 | 7-Feb-11 | Rectal | OXA-48 | ND | TEM-1, SHV-11 | FIIK, L/M |
| RD- 68 | 199 | 27-Jul-11 | Rectal | OXA-48 | ND | TEM-1, SHV-1 | L/M |
| RD- 69 | 974 | 8-Jan-11 | Urine | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | FIIK, L/M |
| RD- 70 | 48 | 26-Mar-11 | Trachea | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | FIIK |
| RD- 71 | 29 | 10-Jun-11 | Wound | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | L/M |
| RD- 72 | 37 | 2-Feb-11 | Blood | OXA-48 | ND | TEM-1, SHV-11 | L/M, FIIK |
| RD- 73 | 199 | 21-Mar-11 | Wound | OXA-48 | CTX-M 14 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 74 | 111 | 24-Mar-11 | Wound | OXA-48 | CTX-M 14 | TEM-1, SHV-11 | IncN |
| RD- 75 | 199 | 30-Mar-11 | Trachea | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | L/M, FIIK, IncN |
| RD- 76 | 37 | 8-Feb-11 | Trachea | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 78 | 152 | 14-Oct-11 | Rectal | OXA-48 + NDM-1 | CTX-M 15 | TEM-1, SHV-1 | Negative |
| RD- 79 | 37 | 31-Oct-11 | Trachea | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 80 | 199 | 13-Nov-11 | Wound | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | L/M, FIIK |
| RD- 82 | 37 | 24-Nov-11 | Trachea | OXA-48 | ND | TEM-1, SHV-11 | L/M, FIIK |
| RD- 83 | 348 | 28-Nov-11 | Rectal | OXA-48 + NDM-1 | CTX-M 15 | TEM-1, SHV-1 | FIIK, L/M |
| RD- 84 | 340 | 3-May-12 | Trachea | Negative | CTX-M 15 | SHV-11 | FII, FIB, FIA, FIIK |
| RD- 85 | 340 | 9-Nov-12 | Urine | OXA-48 | CTX-M 15 | SHV-11 | FIIK |
| RD- 86 | 152 | 16-Feb-12 | Urine | NDM-1 | CTX-M 15 | TEM-1, SHV-1 | FIA |
| RD- 87 | 17 | 6-Nov-12 | Trachea | OXA-48 | CTX-M 15 | SHV-11 | FIIK, L/M |
| RD- 88 | 152 | 3-Jan-12 | Wound | NDM-1 | CTX-M 15 | TEM-1, SHV-1 | FIB |
| RD- 89 | 298 | 6-Oct-12 | Urine | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | FIIK, L/M, IncN |
| RD- 90 | 37 | 6-May-12 | Blood | OXA-48 | ND | TEM-1, SHV-11 | L/M, FIIK |
| RD- 92 | 199 | 22-Jul-12 | Rectal | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | L/M, FIIK |
| RD- 93 | 199 | 22-Jan-12 | Trachea | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | L/M, FIIK, IncN |
| RD- 94 | 152 | 15-Apr-12 | Trachea | NDM-1 | CTX-M 15 | TEM-1, SHV-1 | FIB, FIA |
| RD- 95 | 340 | 16-Mar-12 | Blood | Negative | CTX-M 15 | SHV 11 | FIIK |
| RD- 96 | 16 | 16-Jul-12 | Trachea | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | FII, FIIK, FIA |
| RD- 97 | 48 | 1-Dec-12 | Trachea | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | FIIK |
| RD- 99 | 48 | 12-Jan-12 | Rectal | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | FIB, FIIK, L/M |
| RD- 100 | 37 | 2-Jul-12 | Urine | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 102 | 37 | 1-Sep-12 | TIP | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 103 | 199 | 7-May-12 | Rectal | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | L/M, FIIK |
| RD- 104 | 37 | 25-Jul-12 | Wound | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 105 | 152 | 29-Jan-12 | Blood | OXA-48 + NDM-1 | CTX-M 15 | TEM-1, SHV-1 | L/M, IncN |
| RD- 106 | 152 | 27-Jul-12 | Blood | NDM-1 | CTX-M 15 | TEM-1, SHV-1 | IncN |
| RD- 107 | 29 | 22-Oct-12 | Trachea | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | FIIK, L/M, IncN |
| RD- 108 | 199 | 14-Aug-12 | Wound | OXA-48 + NDM-1 | CTX-M 15 | TEM-1, SHV-1 | L/M, FIIK |
| RD- 109 | 199 | 16-Feb-12 | Urine | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | L/M, FIIK |
| RD- 111 | 152 | 26-Mar-12 | Urine | NDM-1 | CTX-M 15 | TEM-1, SHV-1 | Negative |
| RD- 112 | 15 | 28-Oct-12 | Wound | OXA-48 | CTX-M 15 | SHV-1 | FIIK, L/M |
| RD- 113 | 199 | 4-Oct-12 | Trachea | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | L/M, FIIK |
| RD- 114 | 16 | 18-Nov-12 | Trachea | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | FII, FIIK, FIA |
| RD- 116 | 48 | 7-Jun-12 | Rectal | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | FIIK |
| RD- 117 | 16 | 8-Jun-12 | Urine | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | FII, FIIK, L/M, FIA |
| RD- 118 | 152 | 7-Aug-12 | Trachea | OXA-48 + NDM-1 | CTX-M 15 | TEM-1, SHV-1 | L/M, FIB |
| RD- 119 | 709 | 3-Dec-12 | Wound | OXA-48 | CTX-M 14 | SHV-11 | L/M |
| RD- 120 | 48 | 11-Mar-12 | Rectal | OXA-48 | CTX-M 15 | TEM-1, SHV-1 | FIB, FIA |
| RD- 121 | 15 | 6-Nov-12 | Rectal | Negative | CTX-M 15 | TEM-1, SHV-1 | FIIK |
| RD- 122 | 11 | 17-Nov-12 | Urine | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | FIIK, IncI1, L/M |
| RD- 123 | 353 | 17-Nov-12 | Rectal | OXA-48 | CTX-M 15 | TEM-1, SHV-11 | L/M, FIIK |
| RD- 124 | 152 | 17-Sep-12 | Urine | OXA-48 + NDM-1 | CTX-M 15 | TEM-1, SHV-1 | Negative |
| RD- 125 | 199 | 11-Aug-12 | Wound | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | L/M, FIIK, IncN |
| RD- 126 | 199 | 25-Jun-12 | Trachea | OXA-48 | CTX-M 14 | TEM-1, SHV-1 | L/M, FIIK |
| RD- 127 | 152 | 12-May-12 | Trachea | NDM-1 | ND | TEM-1, SHV-1 | Negative |
| RD- 128 | 1045 | 12-Nov-12 | Urine | NDM-1 | ND | TEM-1, SHV-11 | FIIK |
| RD- 129 | 152 | 14-Dec-12 | Urine | NDM-1 | CTX-M 15 | TEM-1, SHV-1 | FIB |
| RD- 130 | 152 | 25-Nov-12 | Rectal | NDM-1 | ND | TEM-1, SHV-1 | FIB, FIA, IncN |
Isolates and their minimum inhibitory concentrations
Results on resistance determinants, virulence factors, plasmid types, and clonality are presented in Table 1. The clinical sources of the specimen were as follows; respiratory (n = 24); surgical wound (n = 14); rectal swabs (n = 14); urine (n = 13); blood (n = 9); and abdominal drainage (n = 2). Isolates RD-121 and RD-122, which were retrieved from different sites (rectal swab and urine sample) of a single patient 6 months apart, belonged to different STs (STs-15 and -11, respectively) and were considered independent isolates. All isolates were resistant to more than three types of antibiotic. Isolates with ST-199 were all resistant to amikacin (15/15), ‘intermediate’ to tobramycin (9/11), and susceptible to gentamicin (14/15). In contrast, isolates with ST-16 tended to be more susceptible to amikacin and gentamicin than tobramycin. ST-340 isolates were susceptible to amikacin alone. MIC levels are shown in Additional file 1.
Genes of resistance and their clonal distributions
The OXA-48 gene was the most common carbapenemase gene, found in 54/71 isolates, followed by the metallo-β-lactamase gene NDM-1. Of the 54 isolates positive for OXA-48, 48 exhibited OXA-48 alone and six exhibited OXA-48 in combination with NDM-1. Sequence analysis of the ~ 740-base pair PCR product revealed an OXA-48 gene allele sensu stricto in all the positive isolates. OXA-48 was found in the majority of the clones in this study; however, it was less prevalent in STs-152, − 340, and − 15. Among them, four of 12 isolates of ST-152, none of five isolates of ST-340, and one of three isolates of ST-15 were positive for this OXA-48 gene. The gene NDM-1 was less common, existing as the lone carbapenemase in nine isolates and in combination with OXA-48 in six isolates. The main clone carrying this gene was ST-152, with all of its 12 isolates positive for NDM-1. No other allelic form of this gene was detected in any isolate. The ~ 1-k base amplicon generated to study the genetic environment surrounding NDM-1 revealed the presence of ISAba125 and bleMBL flanking this gene. The co-existence of two carbapenemase genes (NDM-1 + OXA-48) was seen in six of 71 (8.5%) isolates. Eight isolates were negative for both the OXA-48 and NDM-1 genes. This included three isolates from ST-340, two from ST-15, and one each from STs-37, − 199, and − 694. None of the isolates, including those eight that were negative for the OXA-48 and NDM-1 genes, were positive for other carbapenemase genes such as KPC, IMP, and VIM. The CTX-M-15 gene allele was the major extended-spectrum β-lactamase (ESBL), with gene positivity in 47 isolates (69%), whereas CTX-M-14 was found in 16 isolates (21.1%). CTX-M-15 was carried by isolates of all STs except STs-199 and -709, whose main CTX-M phenotype was CTX-M-14. Eleven of 15 isolates of ST-199 and two of three isolates of ST-709 were carriers of the CTX-M-14 gene. The other β-lactamase genes identified in this collection of isolates were TEM-1 in 60/71 (84.5%) and SHV (SHV-1 and SHV-11 together) in 71/71 (100%) isolates. Isolates belonging to STs-340 and -709 were all negative for the TEM-1 gene. Two of three isolates of ST-15 were also negative for the TEM-1 gene. Isolates belonging to STs-37, − 340, − 709, and − 974 carried the SHV-11 phenotype, whereas SHV-1 was the main allele in the remaining STs (Table 1).
Plasmid profile
PBRT detected seven plasmid replicon types in the whole population of 71 isolates. The replicons identified were: L/M; IncFII; IncFIIK; IncFIA; IncFIB; IncI1; and IncN. Sixty-seven isolates carried one or more plasmid replicons, whereas four isolates were negative for any plasmid type studied. IncFIIK was the predominant plasmid, found in 49/71 (69%) isolates, followed by L/M, found in 47/71 (66.2%). Similarly, the incidence of IncN was 10/71 (14.1%), that of FIA was eight of 71 (11.3%), that of FIB was eight of 71 (11.3%), that of FII was five of 71 (7%), and that of IncI1 was one of 71 (1.4%). The four isolates negative for any plasmid type belonged to ST-152. RD-122 was the only isolate carrying the IncI1 plasmid replicon (Table 1).
Virulence genes and hypermucoviscocity phenotyping
The status of different virulence genes is shown in Table 1. All isolates were positive for entB and 68/71 (95.7%) isolates were positive for mrkD. Isolates belonging to ST-709 alone showed the plasmid borne iutA virulence factor. All were negative for rmpA and the allS, magA (K1), and K2 CPS genes. Similarly, none of the isolates showed the hypermucoviscocity phenotype (Table 1).
Conjugation experiments
The OXA-48 and NDM-1 genes from these isolates could be transferred into the recipient J53 Azr E. coli strain by the process of conjugation. The transconjugants were screened by colony PCR using gene-specific primers (Table 2). These experiments showed that all NDM-1-positive isolates tested (n = 3) could transfer the gene to their transconjugants. Similarly, the OXA-48 gene could be transferred from all positive isolates tested (n = 10) except for RD-122, the only isolate belonging to ST-11, which failed to transfer its OXA-48 gene. In contrast, only the CTX-M-15 gene could be transferred to the recipients, whereas the CTX-M-14 gene could not. These experiments also showed that the L/M, FIIK, and FII plasmids are conjugative plasmids and can be transferred into recipient bacteria, whereas the IncN1, IncI1, FIA, and FIB plasmids cannot. The results of these experiments are presented in Table 2.
Multilocus sequence typing, clonal distribution, and genetic diversity
The 71 isolates sequenced for the seven loci of the MLST scheme of Diancourt et al. [13] exhibited a total of 37 alleles and were distributed into 18 haplotypes/STs. The clonal distribution of the isolates was as follows: ST-199, n = 15; ST-152, n = 12; ST-37, n = 14; ST-48, n = 5; ST-340, n = 5; ST-29, n = 4; ST-16, n = 4; ST-709, n = 3; ST-15, n = 4; ST-348, n = 2; ST-974, n = 2; and seven other STs with single isolates each. Thirty-four isolates (48%) were distributed in four clonal complexes/groups (CGs) and remained as singletons. A SplitsTree4 diagram was drawn to show their phylogenetic relationship based on the concatenated sequences of all seven loci (Fig. 1). A population snapshot showed their relationship with the STs in the database of the Institut Pasteur (Fig. 2). The 37 alleles ranged from two (rpoB)–10 (tonB). There were no indels or tri-allelic single-nucleotide polymorphisms (SNPs). Of the 436 SNPs observed, 326 were transitional changes, whereas the remaining 110 were transvertional. The diversity index (π) ranged from 0.00055 (pgi)–0.0089 (tonB). Of the 39 mutations observed, 31 were synonymous, whereas eight were non-synonymous. Of the eight non-synonymous mutations, six were found at the tonB locus. The non-synonymous:synonymous mutations ratio (Ka:Ks) was 0.11 for the concatenated sequences of the seven loci (Table 3). The diversities of the concatenated sequences of the seven loci and individual locus sequences are given in Additional files 2 and 3.
Fig. 1.

Clonal diversity and phylogenetic relationship among 76 Klebsiella pneumoniae isolates. SplitTree4 analysis of the concatenated sequences of the seven loci. The numbers at the tips of the edges represent the isolate identity. Sequence type numbers are indicated beside the isolates/clusters and highlighted
Fig. 2.
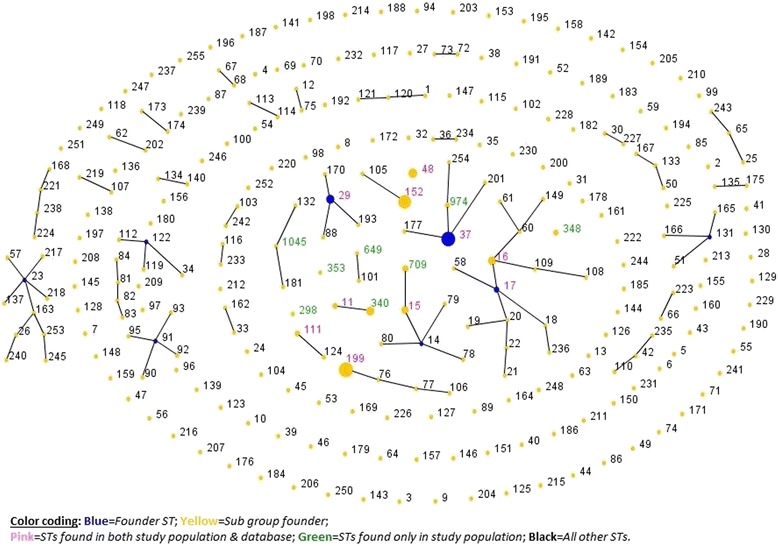
A population snapshot showing the sequence types of the study population of Klebsiella pneumoniae isolates compared with the entire multilocus sequence typing database of 3442 isolates of the Institute Pasteur. The size of the circles is proportionate to the number of isolates. For convenience, the study population is placed in the center of the diagram
Table 3.
Nucleotide polymorphism and allelic diversity among the loci of MLST
| Locus | Length (bp) | No. alleles present | No. Polymorphic sites (%) | No. Synonymous mutations | No. Non-synonymous mutations | # SNPs | Ts | Tv | Ks | Ka | Ka/Ks | Diversity Index (π) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gapA | 450 | 4 | 4 (0.89) | 4 | 0 | 32 | 26 | 6 | 0.0060 | NANa | – | 0.00161126 |
| InfB | 318 | 5 | 4 (1.25) | 4 | 0 | 69 | 69 | 0 | 0.0160 | NAN | – | 0.00393538 |
| mdh | 477 | 5 | 7 (1.47) | 6 | 1 | 35 | 33 | 2 | 0.0050 | 0.0000 | 0.0000 | 0.0014328 |
| pgi | 432 | 3 | 2 (0.46) | 2 | 0 | 9 | 0 | 9 | 0.0020 | NAN | – | 0.00055167 |
| phoE | 420 | 8 | 8 (1.90) | 8 | 0 | 77 | 73 | 4 | 0.0230 | NAN | – | 0.00533477 |
| rpoB | 501 | 2 | 1 (0.20) | 0 | 1 | 22 | 22 | 0 | NAN | 0.0010 | – | 0.00083368 |
| tonB | 414 | 10 | 13 (3.14) | 7 | 6 | 192 | 103 | 89 | 0.0160 | 0.0060 | 0.3750 | 0.0089006 |
| Concatenated | 3012 | 0 | 39 (1.29) | 31 | 8 | 436 | 326 | 110 | 0.0090 | 0.0010 | 0.1100 | 0.00304244 |
aNot a number; SNP single nucleotide polymorphism, Ts Transitions, Tv Transversions, Ks Average number of nucleotide substitution per aynonymous site, Ka Average number of nucleotide substitution per non-synonymous site
Discussion
Although a few reports are available on the antibiotic resistance profiles of CRKP and other members of the Enterobacteriaceae family in Saudi Arabia [4, 6, 8, 17], there is a paucity of studies on other characteristics such as the plasmid profiles, virulence factors, and clonality of CRKP. To the best of our knowledge, this is the first comprehensive report on these characteristics in multidrug-resistant CRKP isolates from a single hospital setting in this country.
Clonality and resistance profile
Multidrug resistance in members of the Enterobacteriaceae family has become a global phenomenon and for several reasons Saudi Arabia is viewed as a growing pool of these pathogens [4, 18–20]. The presence of large numbers of resistance determinants such as CTX-M, OXA-48, NDM-1, SHV, and TEM-1 in our KP isolates is reasonable, because all of them are resistant to several classes of antibiotic (Table 1, Additional file 1). A polyclonal OXA-48 gene was the most common carbapenemase found in our isolates and was distributed among all the STs. The OXA-48 gene has been reported to be endemic in Saudi Arabia [4, 8]. Although variants of OXA-48 such as OXA-204, − 232, − 181, − 162, and − 163 have also been reported in this region [4, 17, 21], none of our isolates showed any other OXA-48 variant. Similarly, the incidence of infections involving the NDM-1 gene is also being regularly reported in Asian countries, but limited data are available on the characteristics of this newly described gene in this region [4]. More recently, Mantilla-Calderon et al. [22] found the NDM-1 gene on an IncF plasmid in E. coli present in a non-nosocomial sample (sewage water) collected in Jeddah, Saudi Arabia, in 2013. The gene appeared to be on a conjugative plasmid because all of their E. coli transconjugants were positive for NDM-1. However, the genetic environment of NDM-1 on plasmids carrying the gene in our isolates appeared similar to that reported previously in KP and most other NDM-1 producers of the Enterobacteriaceae family: surrounded upstream by an ISAba125 insertion element and downstream by the bleomycin resistance gene bleMBL [23, 24]. Regarding the eight isolates that did not present any of the carbapenemase genes and yet were resistant to carbapenem, an alternative mechanism such as a defective or non-functional outer membrane porin, frequently reported in CRKP, may be present [25–27]. The CTX-M gene is associated with antibiotic resistance worldwide and its positivity rate of 93% (66/71) in our isolates indicates its endemicity in this region, in agreement with previous reports [8, 28–30]. The presence of CTX-M-15 (77%) and CTX-M-14 (23%) variants is also in agreement with previous reports from Saudi Arabia [31]. The conjugation experiment showed that CTX-M-15, OXA-48, and NDM-1 genes can be transferred to recipient E. coli, whereas CTX-M-14 appears to be on a non-conjugative plasmid, also evident in its limited dissemination.
Plasmid typing
Limited data are available on plasmid replicon typing in multidrug-resistant KP in Saudi Arabia. The incidence of seven types of plasmid replicon, ranging from one to four in a single isolate, is reasonable in the context of the large number of antibiotic-resistance determinants found in these isolates. The high incidence of conjugative FIIK (69%) and L/M plasmids (66%) found in our isolates is in agreement with reports from European and Asian countries [29, 32–34]. IncI1 is carried only by an isolate with ST-11, a member of clonal group 258, which is famous for carrying the KPC gene worldwide. The reason that four isolates lack any plasmid type may be due to differences in their geographic distribution or limitations in the PBRT protocol for their detection [35]. Except for IncI1, FIA, and FIB, all other plasmids could be transferred to recipient E. coli during conjugation, indicating that these plasmids contribute to the multidrug-resistant phenotype of these isolates. Because the conjugation experiments were carried out on a limited number of isolates in this study (Table 2), it is not possible to determine the relationship between the replicons identified and resistance determinants. However, these data may provide the international community with information on local plasmid profiles and their evolutionary origins [35].
Virulence factors
Hypervirulence is usually linked to the presence of the rmpA, magA (K1), and K2 genes and the hypermucoviscous phenotype is regarded as critical for an isolate to be virulent, whereas other virulence factors appear to be only contributory to this property [36]. Because all our isolates were negative for the rmpA, magA (K1), and K2 genes and did not present a hypermucoviscous phenotype (Table 1), they may have not been hypervirulent-KP. These results also support the notion that virulence and antibiotic resistance are independent and not overlapping traits of KP [37]. Although none of our isolates showed a hypervirulent phenotype, the presence of siderophores (such as ybtS, entB, and iutA) may contribute to the severity of infections in patients, as reported previously [38]. The virulence factor iutA and hypermucoviscous phenotype gene rmpA are usually carried together and co-transferred simultaneously by the plasmid pLVPK and its homologs; thus, the presence of iutA alone in ST-709 isolates warrants further investigation.
Clonality and genetic diversity
MLST is a valuable tool for drawing inferences on genetic diversity and population structure in an epidemiologic setting (https://pubmlst.org/references.shtml) [39]. A significant observation of this study is the presence of a polyclonal pool of several genetically unrelated and non-popular antibiotic-resistant KP STs at our hospital. Using the Breurec et al.[40]. Approach of grouping of single-and double-locus variants into a clonal group and applying e-BURST analysis, 35/71 (49%) isolates (belonging to 10 STs) were grouped in three clonal groups (CG-37, CG258, and CG17), whereas 51% remained singletons belonging to eight diverse and unrelated non-epidemic genetic lineage STs (Fig. 1). A population snapshot drawn against the KP MLST database of the Institut Pasteur also showed that only four of 18 (22%) STs aligned with any of known clonal group (Fig. 2). These findings are consistent with those of previous reports that described the spread of ESBL-producing KP largely as multi-clonal, in contrast to the spread of KPC-producing KP, which is limited to specific clones (CG258) [41]. None of our isolates, including those from CG258, carried a KPC gene. Several clones in this study, such as STs-29, − 37, − 709, and − 111, have been previously reported at this hospital as well as elsewhere [8, 42, 43]. These results also support previous observations that this region is becoming a pool for new antibiotic-resistant KP, but is still mostly free of KPC [4]. The most interesting finding is the presence in large numbers of the non-popular, singleton clone ST-199, which requires further investigation.
Nucleotide diversity in the seven loci sequences remained much lower and the Ka:Ks ratio of the concatenated sequences of these loci was 0.11, indicating a purifying selection against deleterious mutations in these isolates (Table 3). The diversity analysis of individual locus sequences showed a significant diversity in three loci: viz., infB, rpoB, and tonB. Of these, the tonB gene appears to be under more selection pressure because six of its 13 polymorphic sites showed non-synonymous mutations (Table 3). A clone-wise analysis of these sequences showed significant diversity between the tonB and infB genes of the ST-199 and -48 isolates (Fig. 1, Additional file 2). The maximum number of non-synonymous mutations in tonB in a single ST (three of six) belonged to ST-199, indicating strong positive selection of this clone.
A significant observation of our study is the association of certain resistance genes with certain clonal types/genotypes of KP. For example, the majority of isolates belonging to ST-152 were positive for the NDM-1 gene and negative for the OXA-48 gene, whereas isolates of STs-199 and -37 were positive for OXA-48 and negative for NDM-1. Although the NDM-1 gene is regularly reported [4, 17], OXA-48 remains the main carbapenemase in this country. We also found this gene in only 21% of the isolates. The interesting feature of this gene was that ST-152 was its main carrier and that all 12 isolates belonging to ST-152 were positive for it. Both this finding and a previous observation from the United Arab Emirates [18] indicate that clone ST-152 of KP is a “frequent carrier” of the NDM-1 gene exclusive to Saudi Arabia. What makes this more interesting is that all of our isolates belonging to clonal group ST-152 were of the KP subspecies ozaenae, a species not represented in large numbers among KP members [44]. This is the first report to describe the involvement of the ozaenae subspecies of KP in the carriage of the NDM-1 gene. The production of the OXA-48 and NDM-1 carbapenemases alone or in combination with other resistance genes, in isolates of genetic lineages that have not previously been described as high-risk clones, is alarming. It suggests that these clones in an outbreak-like situation may limit the spread of other high-risk clones and/or add to their increasing numbers. The CTX-M-15 phenotype has previously been associated with STs-11, − 15, − 23, and − 37 [45, 46]. In our study population, it was uniformly distributed among all STs except for STs-199 and -709. Isolates from these two clonal types exclusively exhibited CTX-M-14. To the best of our knowledge, this association of the CTX-M-14 phenotype with any ST has not been previously reported. Similarly, the majority of isolates of STs-199 and -709 were CTX-M-14 carriers. Another example of such an association is that of the SHV gene: most of the isolates belonging to STs-199 and -48 were SHV-11 carriers, whereas SHV-1 was the primary SHV phenotype in the other clonal groups. Similar selective resistance was observed in STs-199, − 340, and − 16 for different types of aminoglycoside, such as amikacin, gentamycin, and tobramycin.
The incidences of resistance gene profiles among the isolates of different STs indicate that there is a correlation between antibiotic-resistance pattern and clonal group. This is not due to mere clonal expansion of successful clones with certain resistance determinants (Table 4 and Additional files 2 and 3). A correlation between resistance phenotype and the genotypes of bacterial isolates has been reported previously [47]. Similarly, an association between Class-1 integrons, the carriers of resistance genes in bacteria, and certain STs of KP has also been reported [48, 49]. However, further studies are necessary to shed light on the mechanism underlying and factors associated with such resistance development in clones that are genetically fit in a particular clinical environment [47]. Our results also suggest a need to explore the mechanism underlying the mutual exclusion of genetic determinants in these phylogenetic groups of KP [50].
Table 4.
eBURST analysis of the MLST data showing clonal groups based on STs and their allelic profiles
| Sequence Type | n (% of total) | Clonal Group | Allelic Profile |
|---|---|---|---|
| 37 | 14 (18.4%) | CG37 | 2–9–2-1-13–1-16 |
| 974 | 2 (2.6%) | 4–9–2-1-13–1-16 | |
| 353 | 1(1.3%) | 3–9–47-1-13–1-16 | |
| 340 | 5 (6.6%) | CG258 | 3–3–1-1-1-1-18 |
| 11 | 1(1.3%) | 3–3–1-1-1-1-4 | |
| 16 | 4 (5.3%) | CG17 | 2–1–2-1-4-4-4 |
| 17 | 1(1.3%) | 2–1–1-1-4-4-4 | |
| 15 | 3 (3.9%) | CG14 | 1–1–1-1-1-1-1 |
| 709 | 3 (3.9%) | 1–1–1-1-1-1-4 | |
| 348 | 2 (2.6%) | Singleton | 2–1–20-1-12-1-16 |
| 298 | 1(1.3%) | Singleton | 4–1–2-1-1-1-7 |
| 111 | 1(1.3%) | Singleton | 4–1–5-1-17-1-42 |
| 1045 | 1(1.3%) | Singleton | 2–1–1-1-1-1-42 |
| 649 | 1(1.3%) | Singleton | 2–1–1-5-4-1-1 |
| 199 | 15 (19.5%) | Singleton | 4–34–1-1-21–1-35 |
| 152 | 12 (15.8%) | Singleton | 2–3–2-1-1-4-56 |
| 48 | 5 (6.6%) | Singleton | 2–5–2-2-7-1-10 |
| 29 | 4 (5.3%) | Singleton | 2–3–2-2-6-4-4 |
Additional files
Minimum inhibitory concentrations table. The minimum inhibitory concentrations for various antibiotics seen in 71 Klebsiella pneumoniae isolates. (DOCX 22 kb)
Clonal diversity analysis. Genetic diversity analysis at the nucleotide level across the seven multilocus sequence typing loci concatenated sequences, showing significant diversity for sequence types-48 and -199. (PDF 311 kb)
Individual diversity graphs. Diversity graphs for each of the seven multilocus sequence typing loci sequences i.e., gapA, mdh, pgi, phoE, rpoB, infB, and tonB. (PNG 92 kb)
Acknowledgements
We are thankful to Ms. Laila Qasem and Ms. Mashail Al-Ahmadi for their technical help.
Funding
This work was partially funded by a grant (No. ARP-28-112) from King Abdulaziz City of Science and Technology, Saudi Arabia, to Hanan Balkhy.
Availability of data and materials
All data generated or analyzed during this study are included in this manuscript and its supporting/additional attached information files.
Abbreviations
- CGs
Clonal complexes/groups
- CPS
capsular polysaccharide synthesis
- CRKP
Carbapenem-resistant Klebsiella pneumoniae
- ESBL
Extended-spectrum β-Lactamase
- Ka:Ks
Asynonymous:synonymous mutations
- KAIMRC
King Abdullah International Medical Research Center
- KP
Klebsiella pneumoniae
- KPC
Klebsiella pneumoniae carbapenemase
- LB
Luria bertani
- MICs
Minimum inhibitory concentrations
- MLST
Multilocus sequence typing
- PBRT
PCR-based replicon typing
- PCR
Polymerase chain reaction
- SNPs
Single-nucleotide polymorphisms
- STs
Sequence types
Authors’ contributions
Conception of idea: TU and HB. Experimental design and conduction: TU, MA, MA, MA and SA Analysis of data: TU, MIS and MA Manuscript writing: TU, MIS, and HB. All authors read and approved the final manuscript.
Ethics approval and consent to participate
There are no human participants involved, so ethical approval and consent to participate are not needed. Aditionally, the analyzed bacterial isolates were obtained as part of routine investigations. The information presented for these isolates does not compromise the confidentiality of the patients.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12879-018-3114-9) contains supplementary material, which is available to authorized users.
Contributor Information
Taher uz Zaman, Phone: +966-11-4294551, Email: uzzamanta@ngha.med.sa.
Maha Alrodayyan, Email: alrodayyanma@ngha.med.sa.
Maha Albladi, Email: albladima@ngha.med.sa.
Mohammed Aldrees, Email: aldreesmo@ngha.med.sa.
Mohammed Ismail Siddique, Email: muhammed.sid@gmail.com.
Sameera Aljohani, Email: johani@ngha.med.sa.
Hanan H. Balkhy, Phone: +966 11 8043718, Email: balkhyh@hotmail.com
References
- 1.Hasan B, Olsen B, Alam A, Akter L, Melhus A. Dissemination of the multidrug-resistant extended-spectrum beta-lactamase-producing Escherichia coli O25b-ST131 clone and the role of house crow (Corvus splendens) foraging on hospital waste in Bangladesh. Clin Microbiol Infect. 2015;21(11):1000 e1001–1000 e1004. doi: 10.1016/j.cmi.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 2.Endimiani A, Depasquale JM, Forero S, Perez F, Hujer AM, Roberts-Pollack D, Fiorella PD, Pickens N, Kitchel B, Casiano-Colon AE, et al. Emergence of blaKPC-containing Klebsiella pneumoniae in a long-term acute care hospital: a new challenge to our healthcare system. J Antimicrob Chemother. 2009;64(5):1102–1110. doi: 10.1093/jac/dkp327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammerum AM, Toleman MA, Hansen F, Kristensen B, Lester CH, Walsh TR, Fuursted K. Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect Dis. 2010;10(12):829–830. doi: 10.1016/S1473-3099(10)70276-0. [DOI] [PubMed] [Google Scholar]
- 4.Shibl A, Al-Agamy M, Memish Z, Senok A, Khader SA, Assiri A. The emergence of OXA-48- and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int J Infect Dis. 2013;17(12):e1130–e1133. doi: 10.1016/j.ijid.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 5.Woodford N, Turton JF, Livermore DM. Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 6.Tawfik AF, Alswailem AM, Shibl AM, Al-Agamy MH. Prevalence and genetic characteristics of TEM, SHV, and CTX-M in clinical Klebsiella pneumoniae isolates from Saudi Arabia. Microb Drug Resist. 2011;17(3):383–388. doi: 10.1089/mdr.2011.0011. [DOI] [PubMed] [Google Scholar]
- 7.CLSI: Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing: Twentieth Informational Supplement. 2010. pp. M100–MS20. [Google Scholar]
- 8.Uz Zaman T, Aldrees M, Al Johani SM, Alrodayyan M, Aldughashem FA, Balkhy HH. Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int J Infect Dis. 2014;28:186–192. doi: 10.1016/j.ijid.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 9.Nordmann P, Boulanger AE, Poirel L. NDM-4 metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother. 2012;56(4):2184–2186. doi: 10.1128/AAC.05961-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 11.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compain F, Babosan A, Brisse S, Genel N, Audo J, Ailloud F, Kassis-Chikhani N, Arlet G, Decre D. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol. 2014;52(12):4377–4380. doi: 10.1128/JCM.02316-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond SL, Mullins JI. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. BioTechniques. 2010;48(5):405–408. doi: 10.2144/000113370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186(5):1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 17.Zowawi HM, Sartor AL, Balkhy HH, Walsh TR, Al Johani SM, AlJindan RY, Alfaresi M, Ibrahim E, Al-Jardani A, Al-Abri S, et al. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother. 2014;58(6):3085–3090. doi: 10.1128/AAC.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnevend A, Ghazawi AA, Hashmey R, Jamal W, Rotimi VO, Shibl AM, Al-Jardani A, Al-Abri SS, Tariq WU, Weber S, et al. Characterization of Carbapenem-resistant Enterobacteriaceae with high rate of autochthonous transmission in the Arabian peninsula. PLoS One. 2015;10(6):e0131372. doi: 10.1371/journal.pone.0131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barguigua A, Zerouali K, Katfy K, El Otmani F, Timinouni M, Elmdaghri N. Occurrence of OXA-48 and NDM-1 carbapenemase-producing Klebsiella pneumoniae in a Moroccan university hospital in Casablanca, Morocco. Infect Genet Evol. 2015;31:142–148. doi: 10.1016/j.meegid.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Poirel L, Potron A, Nordmann P. OXA-48 carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67(7):1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 22.Mantilla-Calderon D, Jumat MR, Wang T, Ganesan P, Al-Jassim N, Hong PY. Isolation and characterization of NDM-positive Escherichia coli from municipal wastewater in Jeddah Saudi Arabia. Antimicrob Agents Chemother. 2016;60(9):5223–5231. doi: 10.1128/AAC.00236-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bastian S, Nordmann P, Creton E, Malpote E, Thiery G, Martino F, Breurec S, Dortet L. First case of NDM-1 producing Klebsiella pneumoniae in Caribbean islands. Int J Infect Dis. 2015;34:53–54. doi: 10.1016/j.ijid.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother. 2011;55(11):5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang XD, Cai JC, Zhou HW, Zhang R, Chen GX. Reduced susceptibility to carbapenems in Klebsiella pneumoniae clinical isolates associated with plasmid-mediated beta-lactamase production and OmpK36 porin deficiency. J Med Microbiol. 2009;58(Pt 9):1196–1202. doi: 10.1099/jmm.0.008094-0. [DOI] [PubMed] [Google Scholar]
- 26.Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2011;55(4):1485–1493. doi: 10.1128/AAC.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Fernandez A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, Carattoli A. An ertapenem-resistant extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother. 2010;54(10):4178–4184. doi: 10.1128/AAC.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, Song JH. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents. 2011;38(2):160–163. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Al-Marzooq F, Mohd Yusof MY, Tay ST. Molecular analysis of antibiotic resistance determinants and plasmids in Malaysian isolates of multidrug resistant Klebsiella pneumoniae. PLoS One. 2015;10(7):e0133654. doi: 10.1371/journal.pone.0133654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Agamy MH, Shibl AM, Tawfik AF. Prevalence and molecular characterization of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in Riyadh Saudi Arabia. Ann Saudi Med. 2009;29(4):253–257. doi: 10.4103/0256-4947.55306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamal WY, Albert MJ, Khodakhast F, Poirel L, Rotimi VO. Emergence of new sequence type OXA-48 Carbapenemase-producing Enterobacteriaceae in Kuwait. Microb Drug Resist. 2015;21(3):329–334. doi: 10.1089/mdr.2014.0123. [DOI] [PubMed] [Google Scholar]
- 32.Zhuo C, Li XQ, Zong ZY, Zhong NS. Epidemic plasmid carrying Bla(CTX-M-15) in Klebsiella penumoniae in China. PLoS One. 2013;8(1):e52222. doi: 10.1371/journal.pone.0052222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coelho A, Gonzalez-Lopez JJ, Miro E, Alonso-Tarres C, Mirelis B, Larrosa MN, Bartolome RM, Andreu A, Navarro F, Johnson JR, et al. Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int J Antimicrob Agents. 2010;36(1):73–78. doi: 10.1016/j.ijantimicag.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Huang XZ, Frye JG, Chahine MA, Glenn LM, Ake JA, Su W, Nikolich MP, Lesho EP. Characteristics of plasmids in multi-drug-resistant Enterobacteriaceae isolated during prospective surveillance of a newly opened hospital in Iraq. PLoS One. 2012;7(7):e40360. doi: 10.1371/journal.pone.0040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53(6):2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, Wang Q, Wang X, Chen H, Li H, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Inf Secur. 2015;71(5):553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20(11):1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo C, Yang X, Wu Y, Yang H, Han Y, Yang R, Hu L, Cui Y, Zhou D. MLST-based inference of genetic diversity and population structure of clinical Klebsiella pneumoniae China. Sci Rep. 2015;5:7612. doi: 10.1038/srep07612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breurec S, Guessennd N, Timinouni M, Le TA, Cao V, Ngandjio A, Randrianirina F, Thiberge JM, Kinana A, Dufougeray A, et al. Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin Microbiol Infect. 2013;19(4):349–355. doi: 10.1111/j.1469-0691.2012.03805.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinones D, Valverde A, Rodriguez-Banos M, Kobayashi N, Zayaz A, Abreu M, Canton R, del Campo R. High clonal diversity in a non-outbreak situation of clinical ESBL-producing Klebsiella pneumoniae isolates in the first national surveillance program in Cuba. Microb Drug Resist. 2014;20(1):45–51. doi: 10.1089/mdr.2013.0021. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Ye L, Guo L, Zhao Q, Chen R, Luo Y, Chen Y, Tian S, Zhao J, Shen D, et al. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect. 2013;19(11):E509–E515. doi: 10.1111/1469-0691.12275. [DOI] [PubMed] [Google Scholar]
- 44.Brisse S, Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J Syst Evol Microbiol. 2001;51(Pt 3):915–924. doi: 10.1099/00207713-51-3-915. [DOI] [PubMed] [Google Scholar]
- 45.Ewers C, Stamm I, Pfeifer Y, Wieler LH, Kopp PA, Schonning K, Prenger-Berninghoff E, Scheufen S, Stolle I, Gunther S, et al. Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J Antimicrob Chemother. 2014;69(10):2676–2680. doi: 10.1093/jac/dku217. [DOI] [PubMed] [Google Scholar]
- 46.Shin J, Soo Ko K. Single origin of three plasmids bearing blaCTX-M-15 from different Klebsiella pneumoniae clones. J Antimicrob Chemother. 2014;69(4):969–972. doi: 10.1093/jac/dkt464. [DOI] [PubMed] [Google Scholar]
- 47.Nazir H, Cao S, Hasan F, Hughes D. Can phylogenetic type predict resistance development? J Antimicrob Chemother. 2011;66(4):778–787. doi: 10.1093/jac/dkq505. [DOI] [PubMed] [Google Scholar]
- 48.Li B, Hu Y, Wang Q, Yi Y, Woo PC, Jing H, Zhu B, Liu CH. Structural diversity of class 1 integrons and their associated gene cassettes in Klebsiella pneumoniae isolates from a hospital in China. PLoS One. 2013;8(9):e75805. doi: 10.1371/journal.pone.0075805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maatallah M, Vading M, Kabir MH, Bakhrouf A, Kalin M, Naucler P, Brisse S, Giske CG. Klebsiella variicola is a frequent cause of bloodstream infection in the Stockholm area, and associated with higher mortality compared to K. Pneumoniae. PLoS One. 2014;9(11):e113539. doi: 10.1371/journal.pone.0113539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skippington E, Ragan MA. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol Rev. 2011;35(5):707–735. doi: 10.1111/j.1574-6976.2010.00261.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Minimum inhibitory concentrations table. The minimum inhibitory concentrations for various antibiotics seen in 71 Klebsiella pneumoniae isolates. (DOCX 22 kb)
Clonal diversity analysis. Genetic diversity analysis at the nucleotide level across the seven multilocus sequence typing loci concatenated sequences, showing significant diversity for sequence types-48 and -199. (PDF 311 kb)
Individual diversity graphs. Diversity graphs for each of the seven multilocus sequence typing loci sequences i.e., gapA, mdh, pgi, phoE, rpoB, infB, and tonB. (PNG 92 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript and its supporting/additional attached information files.


