Abstract
Background
Colostrum and milk are essential sources of antibodies and nutrients for the neonate, playing a key role in their survival and growth. Slight abnormalities in the timing of colostrogenesis/lactogenesis potentially threaten piglet survival. To further delineate the genes and transcription regulators implicated in the control of the transition from colostrogenesis to lactogenesis, we applied RNA-seq analysis of swine mammary gland tissue from late-gestation to farrowing. Three 2nd parity sows were used for mammary tissue biopsies on days 14, 10, 6 and 2 before (−) parturition and on day 1 after (+) parturition. A total of 15 mRNA libraries were sequenced on a HiSeq2500 (Illumina Inc.). The Dynamic Impact Approach and the Ingenuity Pathway Analysis were used for pathway analysis and gene network analysis, respectively.
Results
A large number of differentially expressed genes were detected very close to parturition (−2d) and at farrowing (+ 1d). The results reflect the extraordinary metabolic changes in the swine mammary gland once it enters into the crucial phases of lactogenesis and underscore a strong transcriptional component in the control of colostrogenesis. There was marked upregulation of genes involved in synthesis of colostrum and main milk components (i.e. proteins, fat, lactose and antimicrobial factors) with a pivotal role of CSN1S2, LALBA, WAP, SAA2, and BTN1A1. The sustained activation of transcription regulators such as SREBP1 and XBP1 suggested they help coordinate these adaptations.
Conclusions
Overall, the precise timing for the transition from colostrogenesis to lactogenesis in swine mammary gland remains uncharacterized. However, our transcriptomic data support the hypothesis that the transition occurs before parturition. This is likely attributable to upregulation of a wide array of genes including those involved in ‘Protein and Carbohydrate Metabolism’, ‘Immune System’, ‘Lipid Metabolism’, ‘PPAR signaling pathway’ and ‘Prolactin signaling pathway’ along with the activation of transcription regulators controlling lipid synthesis and endoplasmic reticulum biogenesis and stress response.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4719-5) contains supplementary material, which is available to authorized users.
Keywords: Colostrum, Mammary gland, Sow, Transcriptomics
Background
Colostrum and milk are essential sources of antibodies and nutrients for the neonate, playing a key role in their survival and growth [1, 2]. In particular, piglet mortality is a major problem especially during the first few days of life [2]. Development of mammary gland is particularly crucial during the final stages of gestation when alveoli begin to distend [3] and there is an abrupt increase in the concentration of colostrum and milk constituents in the swine mammary secretion just prior to parturition [4]. Due to all these rapid developments in such a small time it is clear that any slight abnormalities in colostrogenesis/lactogenesis potentially threaten piglet survival. Hence, characterizing the transcriptome profile and the metabolic and signaling pathways at that stage could provide a more detailed understanding of important molecular mechanisms occurring in the gland during this essential period of reproduction.
Longitudinal transcriptomic studies are ideally-suited for unravelling complex biological behavior at a genome-wide level and provide a more detailed view of the underlying physiological adaptations [5]. In this regard, the development of high-throughput technologies has revolutionized transcriptome analysis. In particular, RNA-Seq technology enables the generation of more extensive transcriptome information providing an advantage over microarray analyses, due to its capability to quantify all transcripts [6].
Recently, RNA-Seq technology has been used in several species to study the lactating mammary gland [7–9]. Although previous studies using microarrays have provided some preliminary insights into the differential expression of genes (DEG) in sow mammary glands around farrowing [10], our understanding of metabolic or signaling pathways in this species is still limited.
The aim of this study was to provide a comprehensive transcriptome profiling of the sow mammary gland from 14 days prior to parturition to day 1 in lactation using RNA Seq analysis and functional bioinformatics tools such as the Dynamic Impact Approach (DIA) [11] and Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA).
Methods
Animal sampling and RNA extraction
All procedures involving animals were in compliance with Danish laws and regulations for the humane care and use of animals in research [12]. Furthermore, the Danish Animal Experimentation Inspectorate approved the study protocols and supervised the experiment.
Sows used were a subset from an experimental cohort of 36, which involved stratifying animals for body weight at 105 days of gestation to receive one of nine diets (three fiber diets × three fat sources) until day 28 of lactation (weaning) [13]. The test sources of fiber were alfalfa meal or sugar beet pulp with wheat and barley as fiber sources in the control diet. The test fat sources (fed at 30 g/kg dry matter) were soybean oil or glycerol trioctanoate, with palm fatty acid distillate as the fat source in the control. Animals were housed individually in farrowing crates [13]. Mammary tissue collected on days 14, 10, 6 and 2 before (−) parturition and on day 1 after (+) parturition was from three 2nd parity crossbred sows (Danish Landrace × Yorkshire) with the highest colostrum yield (4.6 ± 1.3 kg vs. 2.9 ± 0.9 kg among 9 sows). One of the sows was fed alfalfa meal plus trioctanoate, one sugar beet pulp plus palm fatty acid distillate, and one alfalfa meal plus soybean oil. On the morning of biopsies, sows received only a portion of their meal and upon of milk letdown piglets were removed prior to general anesthesia by intramuscular injection of 1.0 mL/34 kg body weight of Telozol® (Fort Dodge Animal Health, Fort Dodge, IA) dissolved in 2.5 mL ketamine (VetaKet®; Lloyd Laboratories, Shenandoah, IA) and 2.5 mL xylazine-00 (AnaSed®; Lloyd Laboratories). After asceptic surgical prepartion a subcutaneous and intramammary injection of 1.0 mL of 2% lidocaine (Lidoject®; Butler Animal Health supply, Dublin, OH) was given prior to making a 2-cm incision vertical to the plica lateralis. A biopsy consisted of a maximum of three shots using a Manan ProMag 2.2 biopsy gun (Medical Device Technologies, Gainesville, FL) in the same intrusion site while the sow was under anesthesia and in lateral recumbency to expose one entire side of the udder. A total of 20 mg of mammary tissue was collected, after which the incision was closed with simple interrupted sutures [14]. Each sow received 1 mL/100 kg body weight of Banamine (Merck Animal Health, Summit, NJ) immediately after mammary biopsy and at 24 and 48 h post-biopsy. Upon recovery from anesthesia, sows were fed the remainder of their morning meal and piglets were returned to the sow and allowed to suckle normally. Extraction of RNA and quality evaluation was performed following protocols described previously [15]. The average yield of total RNA (from 20.3 ± 6.9 mg tissue) was 44 ± 19 μg, and the average RNA integrity number (Agilent Bioanalyzer) was 8.2 ± 0.8. An aggregate summary of RNA extraction and quality check for all the samples is reported in Additional file 1.
RNA-sequencing
Sequencing was performed by the High-Throughput Sequencing and Genotyping Unit of the W. M. Keck Biotechnology Center at the University of Illinois at Urbana Champaign (Urbana, IL, USA). A total of 15 mRNA libraries were quantified by qPCR and sequenced on two lanes for 101 cycles from one end of the fragments on a HiSeq2500 (Illumina Inc.), using v4 HiSeq SBS reagents. In total approximately 403 million single-read sequences of 100 nt in length were collected. Quality control metrics were performed on raw sequencing reads using the FASTQC v0.11.15 application. An index of the reference genome was built and single-end clean reads for each individual were aligned to the reference genome using STAR (v2.5.1b). Reads were mapped and annotated to the Sus scrofa genome (v10.2.86), downloaded from the EnsemblGenome website (Nov. 2016). Reads aligned were quantified with the Subread package (v1.5.0) based on the Refseq gene annotation.
Bioinformatics analysis
Identification of differentially expressed genes
Non-expressed and weakly expressed genes (i.e. without at least 1 read per million) were removed prior to differential expression (DE) analysis [16]. A TMM (trimmed mean of M-values) normalization was applied to all samples using edgeR [17]. After data were log2-transformed, limma-voom method (Bioconductor packages) was used to conduct DE analyses [18, 19]. The applied statistical model included time as fixed effect and animal as random effect. Differentially expressed genes (DEG) across different time points were defined as genes with a Benjamini–Hochberg multiple-testing adjusted p-value of ≤0.05. To identify the longitudinal transcriptional gene response close to parturition, the time point − 14 day was used as baseline for each time comparisons. In particular to highlight the metabolic processes underlying mammary changes associated with the colostrogenesis and the onset of lactogenesis in the last stages of gestation leading up to parturition, we relied on DEG between -10vs − 14, −6vs − 14, −2vs-14 and + 1vs-14 time comparisons.
Dynamic impact approach (DIA)
The DIA software described previously [11] was used for functional analyses. Briefly, DIA uses the systems information from the KEGG database and ranks pathways calculating the overall impact (importance of a given pathway) and flux (direction of impact; e.g., up-regulation, down-regulation, or no change). For this purpose, the whole dataset (minus weakly expressed genes) with Entrez gene IDs, FDR, FC, and p-values of each time comparison were uploaded in DIA and an overall cut-off (FDR and p-value ≤0.05) was applied as threshold.
Gene network analysis
Ingenuity Pathway Analysis (IPA) was performed to identify transcription regulators and their networks with other genes, within the list of significant DEG (similar cut-off as DIA analysis; FDR and p-value ≤0.05) at each time comparisons (https://www.qiagenbioinformatics.com).
Verification by real-time PCR
The expression of LALBA, CSN2, PAEP, and LTF was analyzed to verify the physiologic response of the mammary gland as farrowing approached. These genes are well-established markers of mammary-specific genes. Complete information about cDNA synthesis and qPCR performance are reported elsewhere [20]. After normalization with the geometric mean of three internal control genes (API5, VABP, and MRPL39), qPCR data were log2-transformed prior to statistical analysis to obtain a normal distribution. Statistical analysis was performed with SAS (v 9.4). Normalized, log2-transformed data were subjected to ANOVA with PROC MIXED. The statistical model included time (− 14, − 10, − 6, − 2, and + 1 day from farrowing) as fixed effect, and sow as the random effect. The Kenward-Roger statement was used for computing the denominator degrees of freedom. Fold change for the time comparison -10vs-14, −6vs-14, −2vs-14, and + 1vs-14 were then calculated from the estimates of the model. For each of the four genes and comparison FDR, fold change, and p-values are reported in Table 1, together with the respective results from the sequencing analysis.
Table 1.
Quantitative real time PCR (qPCR) validation of sequencing (Seq) results
| -14 vs −2 time comparison | -14 vs +1 time comparison | ||||
|---|---|---|---|---|---|
| Target | FDR | FC | P-value | FC | P-value |
| CSN2 | |||||
| qPCR | <.0001 | 51.9 | <.0001 | 254.8 | <.0001 |
| Seq | 0.001 | 31.0 | 0.003 | 33.5 | <.0001 |
| LALBA | |||||
| qPCR | <.0001 | 156.1 | <.0001 | 8321.9 | <.0001 |
| Seq | 0.003 | 128.5 | 0.03 | 1275.5 | 0.001 |
| LTF | |||||
| qPCR | <.0001 | 1.5 | 0.02 | 3.4 | <.0001 |
| Seq | 0.001 | 1.7 | 0.10 | 3.9 | <.0001 |
| PAEP | |||||
| qPCR | 0.0001 | 2.2 | 0.0005 | 3.4 | <.0001 |
| Seq | 0.004 | 1.8 | 0.01 | 1.9 | 0.001 |
The overall false discovery rate (FDR) together with the fold-change (FC) and P-value for the specific comparison is reported for each gene. The P-values were generated applying the same statistical model to either qPCR or Seq data
Results
RNAseq analysis and DEG
An aggregate summary of RNA sequencing and alignment for all the samples is reported in Additional file 2. Illumina sequencing was effective. A large numbers of high-quality reads were produced in all samples. On average, 92% of the total reads were successfully mapped. 91.8% of aligned reads were mapped to unique genomic regions. Results for total number of DEG due to time are presented in Fig. 1. Considering an FDR and p-value ≤0.05 among the 9393 genes (after annotation with the entrez genes ID) a total of 0, 17 (15 upregulated and 2 downregulated), 788 (451 upregulated and 337 downregulated), and 2884 (1508 upregulated and 1376 downregulated) were differentially expressed for -10vs-14, −6vs-14, −2vs-14, +1vs-14 time comparisons, respectively (Additional file 3).
Fig. 1.

Total number of differentially expressed genes (DEG) due to time resulting from differential expression (DE) analysis of RNAseq data of swine mammary gland tissue harvested in late-pregnancy through farrowing (FDR and p-value ≤0.05). The light blue bars indicate downregulation, while the yellow bars indicate upregulation
For further analysis with DIA [11] and IPA we focused on DEG between -2vs-14 and + 1vs-14 time comparisons, where the largest numbers of activated and inhibited genes were detected. The -2vs-14 comparison represents the difference in gene expression patterns between a gland with limited growth and a gland that is near full-term, i.e. genes that encompass the last stages of functional differentiation. In contrast, the +1vs-14 comparison represents the difference in mammary tissue between a stage with limited mammary growth and a functional mammary gland which is entered into the lactogenesis stage [4]. To highlight the overall weight of genes in each comparison, the top ten upregulated genes were underscored (Tables 2 and 3). CSN1S2 and LALBA were the most-expressed genes at 2d prepartum, whereas WAP, CSN1S2, SAA2 and LALBA had a marked upregulation at 1d postpartum. The overlap and specific upregulated genes between the last two time comparisons are reported in Table 4.
Table 2.
Top ten upregulated genes in the -2vs-14 time comparison (FDR and p-value ≤0.05)
| Gene symbol | Entrez gene ID | FDR | FC | P-value |
|---|---|---|---|---|
| CSN1S2 | 445,515 | ˂ .000 | 629.572 | 0.002 |
| LALBA | 397,647 | 0.003 | 128.511 | 0.032 |
| LOC100522145 | 100,522,145 | 0.002 | 95.634 | 0.019 |
| BTN1A1 | 100,153,328 | 0.007 | 90.488 | 0.023 |
| BTN1A1-like | 100,626,139 | 0.007 | 90.488 | 0.023 |
| CYP1A1 | 403,103 | 0.005 | 59.108 | 0.011 |
| LOC100524679 | 100,524,679 | 0.006 | 59.017 | 0.015 |
| CSN2 | 404,088 | 0.001 | 31.014 | 0.003 |
| HP | 397,061 | ˂ .000 | 29.272 | 0.001 |
| ACSL6 | 100,522,126 | 0.012 | 26.040 | 0.031 |
The overall false discovery rate (FDR) together with the fold-change (FC) and P-value for the specific comparison is reported for each gene
Table 3.
Top ten upregulated genes in the +1vs-14 time comparison (FDR and p-value ≤0.05)
| Gene symbol | Entrez gene ID | FDR | FC | P-value |
|---|---|---|---|---|
| WAP | 396,835 | 0.004 | 4205.803 | 0.001 |
| CSN1S2 | 445,515 | ˂ .000 | 3858.507 | ˂ .000 |
| SAA2 | 100,525,680 | ˂ .000 | 3039.234 | ˂ .000 |
| LALBA | 397,647 | 0.003 | 1275.540 | 0.001 |
| LOC100522145 | 100,522,145 | 0.002 | 568.374 | ˂ .000 |
| LBP | 397,303 | ˂ .000 | 372.186 | ˂ .000 |
| CP | 406,870 | 0.043 | 258.471 | 0.011 |
| HP | 397,061 | ˂ .000 | 204.864 | ˂ .000 |
| BTN1A1 | 100,153,328 | 0.007 | 202.517 | 0.002 |
| BTN1A1-like | 100,626,139 | 0.007 | 202.517 | 0.002 |
The overall false discovery rate (FDR) together with the fold-change (FC) and P-value for the specific comparison is reported for each gene
Table 4.
Summary of top 10 upregulated genes in both and specific time comparisons (FDR and p-value ≤0.05)
| Status | +1vs-14 time comparison | -2vs-14 time comparison | both time comparisons |
|---|---|---|---|
| upregulated | WAP, SAA2, LBP, CP | CYP1A1, LOC100524679, CSN2, ACSL6 | HP, LALBA, CSN1S2, BTN1A1, BTN1A1-like LOC100522145 |
Overall summary of KEGG categories
The DIA results are summarized in Fig. 2. They provide an overview of impact and flux for each KEGG category calculated following DIA procedures [11]. We clearly observed no significant changes in -10vs-14 and -6vs-14 time comparisons because of the lack of DEG associated with these comparisons (data not showed). Instead, closer to parturition (−2vs-14 comparison), we detected an evident activation of all main categories and in particular ‘Metabolism category’ and ‘Organismal Systems’ pathways, which became stronger considering the postpartum stage (+1vs-14). Focusing only on these main categories and considering the related subcategories with flux value at least 50% of impact value, clearly within ‘Metabolism’ the subcategory ‘Lipid Metabolism’ was the most-impacted and recurrent in both comparisons, followed by ‘Metabolism of Other Amino Acids’. It was not possible to highlight a recurrent subcategory within ‘Organismal Systems’ with flux value at least 50% of impact value in the last 2 comparisons, thus, we chose the recurrent subcategory with highest impact and upregulated flux: ‘Endocrine system’. Regarding the other KEGG pathway categories (i.e. ‘Genetic Information Processing’, ‘Environmental Information Processing’ and ‘Cellular Processes’), we observed a marked downregulation of all ‘Genetic Information Processing’ subcategories and a marked upregulation of ‘Environmental Information Processing’ at 1 day postpartum.
Fig. 2.

Summary of the main KEGG categories resulting from the Dynamic Impact Approach (DIA) analysis on differentially expressed genes (DEG) obtained by differential expression (DE) analysis of RNAseq data of swine mammary gland tissue harvested in late-pregnancy through farrowing (FDR and p-value ≤0.05). For each time comparison, the columns represent the effect (impact) and flux responses. The white bars represent the effect value (0 to 150), and the flux columns represent negative (−) and positive (+) flux (− 150 to + 150) based on the direction of the effect. The negative flux (light blue bars) indicates a downregulation, while the positive flux (yellow bars) indicates an upregulation
The above general results only provide information about the overall impact and the general direction of the impact (flux) of each significant category and subcategory in the dataset. With the aim to reach a better understanding of the biological relevance of each significant impacted category/subcategories, we focused on the single metabolic pathways falling within the main subcategories of interest particularly ‘Lipid Metabolism’ and ‘Endocrine system’ (Figs. 3 and 4). Except ‘Primary bile acid biosynthesis’, for the ‘Lipid Metabolism’ subcategory we uncovered a clear upregulation of all pathways. In particular, ‘Fatty acid biosynthesis’ was the most impacted and upregulated pathway in both comparisons followed by ‘Arachidonic acid metabolism’ and ‘Steroid hormone biosynthesis’ at 2d prepartum, and ‘Steroid biosynthesis’ and ‘Synthesis and degradation of ketone bodies’ at 1d postpartum. Within ‘Endocrine system’, we detected a marked upregulation of ‘Prolactin signaling pathway’ recurrent in the last 2 comparisons, followed by ‘Ovarian steroidogenesis’ and ‘PPAR signaling pathway’ at 2d prepartum and ‘PPAR signaling pathway’ and ‘Thyroid hormone synthesis’ at 1d postpartum. There was a marked downregulation of ‘Adipocytokine signaling pathway’ at 1d postpartum and ‘Insulin signaling pathway’ in the last 2 comparisons.
Fig. 3.

Summary of KEGG ‘Lipid Metabolism’ pathways resulting from the Dynamic Impact Approach (DIA) analysis on differentially expressed genes (DEG) obtained by differential expression (DE) analysis of RNAseq data on swine mammary gland from late pregnancy to farrowing (FDR and p-value ≤0.05). For each time comparison, the columns represent the effect (impact) and flux responses. The white bars represent the effect value (0 to 300), and the flux columns represent negative (−) and positive (+) flux (− 300 to + 300) based on the direction of the effect. The negative flux (light blue bars) indicates a downregulation, while the positive flux (yellow bars) indicates an upregulation
Fig. 4.
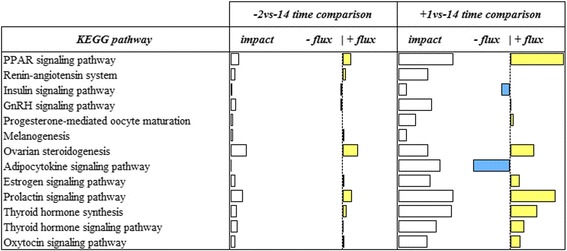
Summary of KEGG ‘Endocrine system’ pathways resulting from the Dynamic Impact Approach (DIA) analysis on differentially expressed genes (DEG) obtained by differential expression (DE) analysis of RNAseq data of swine mammary gland tissue harvested in late-pregnancy through farrowing (FDR and p-value ≤0.05). For each time comparison, the columns represent the effect (impact) and flux responses. The white bars represent the effect value (0 to 200), and the flux columns represent negative (−) and positive (+) flux (−200 to + 200) based on the direction of the effect. The negative flux (light blue bars) indicates a downregulation, while the positive flux (yellow bars) indicates an upregulation
Most impacted pathways
To highlight the overall most impacted and upregulated pathways, we considered all pathways without any category classification and with flux value at least 50% of impact value (Figs. 5 and 6). When the last stage (−2d vs -14d) of gestation was considered, ‘Fatty acid biosynthesis’, ‘Retinol metabolism’, ‘Drug metabolism – other enzymes’, ‘PPAR signaling pathway’, ‘Galactose metabolism’, ‘Steroid hormone biosynthesis’, ‘Metabolism of xenobiotics by cytochrome P450’, ‘Chemical carcinogenesis’, ‘Fatty acid degradation’, ‘Arachidonic acid metabolism’ were the most impacted and upregulated pathways. When the parturition stage (+1d vs -14d) was considered, ‘PPAR signaling pathway’, ‘Steroid biosynthesis’, ‘Fatty acid biosynthesis’, ‘Synthesis and degradation of ketone bodies’, ‘Mineral absorption’, ‘beta-Alanine metabolism’, ‘Galactose metabolism’, ‘Fatty acid degradation’, ‘Drug metabolism - other enzyme’ and ‘Histidine metabolism’ were the most impacted and upregulated pathways.
Fig. 5.

Top 10 upregulated KEGG pathways in the -2d vs −14 d comparison resulting from the Dynamic Impact Approach (DIA) analysis on differentially expressed genes (DEG) obtained by differential expression (DE) analysis of RNAseq data of swine mammary gland tissue harvested in late-pregnancy through farrowing (FDR and p-value ≤0.05). The columns represent the effect (impact) and flux responses. The white bars represent the effect value (0 to 150) and the yellow bars represent the flux (the direction of the effect)
Fig. 6.

Top 10 upregulated KEGG pathways in the + 1d vs − 14 d comparison resulting from the Dynamic Impact Approach (DIA) analysis on differentially expressed genes (DEG) obtained by differential expression (DE) analysis of RNAseq data of swine mammary gland tissue harvested in late-pregnancy through farrowing (FDR and p-value ≤0.05). The columns represent the effect (impact) and flux responses. The white bars represent the effect value (0 to 400) and the yellow bars represent the flux (the direction of the effect)
Enrichment analysis of genes in most recurrent pathway categories
As shown above, ‘Lipid metabolism’ and ‘Endocrine system’ were the most recurrent pathway subcategories. A KEGG enrichment analysis was performed to identify the most upregulated genes among these subcategories (FDR and p-value ≤0.05). To underscore the weight of a specific gene within the most recurrent KEGG pathway categories, the genes used by DIA for impact and flux calculations were extrapolated and divided into three groups: upregulated genes in ‘+1vs-14’, ‘-2vs-14’ and ‘both’ time comparisons. The summary of upregulated genes is reported in Table 5.
Table 5.
Summary of upregulated genes in the most-recurrent KEGG subcategories in both and specific comparisons (FDR and p-value ≤0.05)
| KEGG Category | Status | +1vs-14 time comparison | -2vs-14 time comparison | Both time comparisons |
|---|---|---|---|---|
| Lipid Metabolism | upregulated | ALDH2, HMGCS1, HSD11B1, SCD, ACSL3, FAXDC2, SGMS2, SC5D, 403,334, ACACB, SQLE, MSMO1, CYP2J2, CERS4, CYP2J34, LCLAT1, DHCR24, 100,233,182, PLPP3, FADS2, 100,170,845, 100,517,533, ACAT2, MGLL, CYP2D25, TM7SF2, ACADM, ARSA, PNPLA2, GLA, SMPD1, PTGS1, LPCAT3, GPCPD1, PAFAH2, ACADS, NSDHL, PLA2G12A, KDSR, ECI2, GBA2, COMT, 100515577 | CYP1A1, 100,157,065, GPAM, GPD2, SGMS1, CDIPT, DGAT1 | GGT1, 397,097, FADS1, AGPAT1, NEU1, GPAT4, CEPT1, HSD17B7, CDS2, PLA2G16, 100,522,126, 100,522,145, 100,522,692, GPAT3, 100,625,138, 100,625,332, 100,738,292 |
| Endocrine System | upregulated | CYP1A1, PYGB | 396,835, CD14, FXYD2, FOS, HK2, RCAN1, SH2B2, PFKFB2, STAT3, ACACB, PHKG1, ITGB3, HSPA5, OXTR, CYP2J2, PLN, PLCD3, SEC11C, CYP2J34, HSP90B1, MYL9, CPEB4, 100,514,493, ITGA11, RRAS, CREB3L2, CPEB3, CFL2, NFATC2, CTSB, SEC61G, SRP54, SPCS3, PRKCI, ITGA2, PLCD1, JUN, DIAPH1, SEC63, PRKAB2, SP1, BCAR1, B2M, 106,504,143, PRKAB1, PIKFYVE, KAT2B, CREB3, PRKCD, STAT1, RYR2, 100,522,756, 396,848, CASP9, PHKG2, JAK2, ITGAV, SRP68, SRP68, PPP1CB, ITPR3, SOCS6, IFI30, PAK1, PPP1R12C, ELK1, 100,517,270, RHOA | CTSV, KCNJ2, NOS3, PCK2, CSN2, FOXO3, MTOR, NCOA2, SPCS1, SRC, SEC61A1, HSD17B7, PDIA3, MAPK14, CALR, SEC61B, RPTOR, SRPRA, CREB3L1, CANX, VAV1, 100,522,176, PDIA4, GNA13, 100,523,015, 100,523,202, EEF2K, MRAS |
Gene network analysis
IPA allowed uncovering relationships between transcription factors and DEG. Considering a ± 2 ‘Activation z-score’ value and p-value cut-off of 0.01, we identified 6 and 55 upstream transcription regulators (TR) in -2vs-14 and + 1vs-14 time comparisons, respectively (Additional file 4). To highlight and summarize similarities and differences in the activation of TR, the overlap between the two comparisons was performed and the activated TR were extracted. The results are reported in Table 6.
Table 6.
Summary of the most-activated Transcription Regulators (TR) in both and specific comparisons (p-value ≤0.01; z-score ≥ ± 2)
| Status | +1vs-14 time comparison | -2vs-14 time comparison | Both time comparisons |
|---|---|---|---|
| Activated | ATF4, CDKN2A, CEBPA, CREB1, CREM, E2F6, ECSIT, EPAS1, FOXO3, GATA1, HIF1A, ID3, IRF1, IRF3, IRF5, IRF7, KDM5B, MEF2D, MXI1, NFATC2, NFKB1, NFKBIA, NUPR1, PDX1, PPARGC1B, RB1, RBL1, RELA, SMARCA4, SMARCB1, SREBP2, STAT1, STAT2, TCF3, TCF7L2, TOB1, TP53 | ATF6 | SREBP1 XBP1 |
Discussion
Although there is some discrepancy in the literature as to the specific timing of colostrogenesis and lactogenesis, the consensus is that swine lactogenesis is activated in late-pregnancy [4, 21]. Lactogenesis is further subdivided in two stages: lactogenesis I, which occurs in late-pregnancy and is linked to the synthesis of early milk components and to final structural mammary gland (MG) differentiation; and lactogenesis II, which is characterized by the onset of abundant milk secretion [21]. It is also accepted that colostrum production takes place during lactogenesis I and that the transition from colostrum to mature milk occurs within 48 h after parturition, influenced by suckling piglets whose effect enhances the rate of fat secretion and accelerates the increase in lactose concentrations [21]. Our results partly confirmed these timings and indirectly suggest that at 14 days prior to parturition (around day 100 of gestation) the MG already entered stage I lactogenesis, hence, the extremely low number of DEG when comparing both − 10, and -6d to -14d; alternatively, the very low number of DEG during those times could have been a reflection of the low activity of the MG. The large numbers (788) of DEG uncovered in the -2dvs-14d time comparison reflects a strong activation of many metabolic processes compatible with the shifting from stage I to stage II of lactogenesis occurring before parturition also in this species [22]. This is consistent with the consideration that MG reached the greatest degree of structural development at that time, and the preparation for copious milk synthesis and secretion had begun [4, 23].
The transition period from a non-lactating to lactating state requires important metabolic changes to enable the shift of nutrient prioritization from body reserves towards the mammary gland for milk production. It is clear that the marked number of DEG (2884) detected at day 1 postpartum vs 14 day prepartum reflects the extraordinary metabolic changes in the swine mammary gland once it fully entered into crucial phases of lactogenesis. A general overview of DIA results confirm this conclusion. In fact, the overall activation of all ‘Metabolic pathways’ and in particular of ‘Lipid Metabolism’, ‘Metabolism of Other Amino Acids’, ‘Carbohydrate Metabolism’ and ‘PPAR signaling pathways’ is compatible with the transition from a non-lactating to lactating state characterized by the expression of genes associated with synthesis of milk components (i.e. milk proteins, fat and lactose). At the same time, the general inhibition of ‘Genetic Information Processing’, the specific inhibition of ‘Cell Growth and Death’ in ‘Cellular Process’ and of ‘Development’ in ‘Organismal System’ is consistent with the fact that the mammary gland has already significantly grown in mass [3].
At farrowing, the MG is fully involved in the accumulation and secretion of colostrum and milk, with their nutritional and immunological proprieties confirmed by the activation of ‘Immune System’, ‘Endocrine System’ and ‘Excretory System’, as well as by the marked upregulation of OXTR (oxytocin receptor) [FC = 3.17] [24]. The role of prolactin secretion, which peaks around farrowing and drives the switch from formation and accumulation of colostrum to synthesis and secretion of milk components [25], is supported by the upregulation of ‘Prolactin signaling pathway’. In summary, although not directly compared, our results indicate that the transition from colostrogenesis to lactogenesis occurs between 6 and 2 days before expected parturition. This is likely attributable to upregulation of a wide array of genes including those involved in ‘Protein and Carbohydrate Metabolism’, ‘Immune System’, ‘Lipid Metabolism’, ‘PPAR signaling pathways’ and ‘Prolactin signaling pathway’.
Protein and carbohydrate metabolism
It is known that concentrations of total protein in sow mammary secretions are highest at parturition [26] The evident upregulation in the last comparison of ‘Amino Acid Metabolism’ and ‘Metabolism of Other Amino Acids’ pathways is consistent with the strong activation of synthesis of the major milk protein during the onset of lactation. These changes in protein concentrations mirror the changes in immunoglobulin content, highly abundant in colostrum with a gradual decrease in milk, and concurrently a lower casein content in colostrum followed by a high increase during the postpartum [26]. While immunoglobulin concentrations are significantly declining and β-lactoglobulin are relatively constant from the colostrum period through lactation, the proportion of casein and α-lactalbumin increases considerably in the postpartum period [26]. These previously reported responses are confirmed in our results by the strong upregulation of CSN2, CSN1S2, CSN1S1 and LALBA, which are among the overall top upregulated genes in the last 2 comparisons along with the marked upregulation of WAP at 1 day postpartum. The upregulation of LALBA deserves particular consideration because of its involvement in lactose biosynthesis [27]. Lactose is the major carbohydrate in sow milk and the major osmole in milk, responsible for drawing water into the secretory vesicles [26]. Lactose concentrations are low in colostrum then increase gradually over the first 2 to 3 days of lactation [26]. Our transcriptomic results confirmed this evidence, showing a marked upregulation of ‘Carbohydrate Metabolism’ during the transition from late pregnancy to parturition, driven by upregulation of many genes and particularly by LALBA, B4GALT1 and HK2 activity all involved in ‘Galactose Metabolism’ pathway.
Milk caseins
There was a marked upregulation of CSN1S2 (alpha-S2-casein) at 2d prepartum and 1d postpartum [FC = 629.57 and 3858.51]. Several studies reported CSN1S2 as one of the most up-regulated caseins increasing in expression during lactation in bovine [28], pig and mouse [29]. It was also found expressed in colostrum and mid lactation milk in goats [30]. The temporal expression pattern of α-casein genes is similar in many species with CSN1S2 as the most upregulated followed by CSN1S1. Our results confirmed this pattern also in pig with a lower expression of CSN1S1 [FC = 4.00 and 5.25] compared with CSN1S2.
An expected result was the marked upregulation of CSN2 [FC = 31.01 and 33.52], a member of the β-casein family. The principal protein in human milk is the β-casein, which represents the primary source of essential amino acids for a suckling infant. The increases in expression from 2d prepartum to 1d postpartum of CSN2 was not surprising because it is known that the proportion casein of total milk protein sharply increases by 24 h postpartum [26] and because caseins, together with the whey proteins, represent the highest percentage of milk protein fraction (~ 90%) [26]. In this regard, it is also known that the mRNA abundance of WAP in monogastric appears to be as high or higher than caseins [29]. This is in agreement with our results at 1d postpartum where we found the abrupt upregulation of WAP (whey acidic protein) [FC = 4205.80]. Whey acidic protein is the major whey protein in the milk of many species, including the pig where it is secreted at a consistent level throughout lactation [31]. The increase in expression of WAP in monogastrics was proportional to LALBA [29]. Our result confirmed this relationship [LALBA, FC = 128.51 and 1275.53].
It is well established that energy content of the diet and the simultaneous availability of amino acids (AA) affect milk protein content [29]. In this sense, one of the major limitations for milk protein synthesis is represented by transport of AA. A comprehensive literature review on AA transporters in the mammary gland was conducted by Shennan and Boyd [32]. The article describes a list of transporters including those presented in our results. In this regard, the marked upregulation of SLC7A4 in the last comparison [FC = 32.27] is noteworthy. SLC7A4 codes for the CAT-4 protein, related to other members of the SLC7 family of cationic amino acid transporters and found highly expressed in swine placental tissue [33]. Although the exact function of the SLC7A4 protein in the context of AA metabolism is not well-known, considering that SLC7A4 is an important paralog of SLC7A1, coding for the CAT-1 protein, which was identified in porcine MG where its abundance increases at early lactation compared with prepartum and it is positively correlated to β-CN and α-LA [34], we speculate that SLC7A4 in mammary epithelial cells (MEC) could participate in the uptake of leucine (Leu), hence, stimulating protein synthesis through activation of the mTOR cell signaling pathway [35, 36]. In fact, Krogh et al. [37] showed recently that Leu is the most extracted AA by the sow mammary gland in early lactation (d + 3) and in our study, we detected an upregulation [FC = 2.04] at 1d postpartum of another solute carrier family: SLC7A8, coding for LAT2, that, together with LAT1, have been proposed to be involved in Leu uptake in the mammary gland [38]. In the same way, insulin signaling, inducing translation via activation of the mTOR pathway, plays an important role in the control of milk protein synthesis [29]. In particular, the insulin effect prevents mTOR inhibition by blocking (via phosphorylation) the main inhibitors of mTOR: the tuberous sclerosis proteins (i.e., TSC1 and TSC2) [29]. In this regard, it was interesting to note the downregulation of TSC1 at postpartum stage [FC = − 1.24].
Lactose synthesis
Expression of LALBA, encoding α-lactalbumin, was strongly upregulated at 2d prepartum and 1d postpartum [FC = 128.51 and 1275.53]. This is one of the main milk proteins also involved in ‘Carbohydrate Metabolism’ via activation of ‘Galactose metabolism’. In fact, LALBA is part of the lactose synthetase complex that, inside the Golgi, uses glucose and UDP-galactose as substrates for the synthesis of lactose [39]. Our result is in agreement with findings of other studies, where LALBA upregulation was detected towards the end of gestation, just before parturition [40, 41]. This result is also consistent with the gradual increase in α-lactalbumin and lactose concentrations in colostrum compared with milk during the first days of lactation [26]. However, if glands are not suckled from 12 h after parturition (i.e. during the colostrum period), expression of LALBA is decreased 24 h after parturition in response to lack of colostrum removal [42].
Regarding the lactose synthase enzyme complex, it was noteworthy that in the last 2 comparisons B4GALT1 was upregulated [FC = 2.40 and 3.27]. The B4GALT1 gene encodes one of seven beta-1,4-galactosyltransferase (beta4GalT) proteins of the complex and is unique because it participates both in glycoconjugate and lactose biosynthesis [27].
The transport of UDP-galactose into the Golgi is regulated by SLC35A2, which is considered a rate-limiting process in lactose synthesis [43]. The expression of this gene was upregulated in the 2 last comparisons [FC = 2.05 and 2.01]. The marked upregulation of HK2 [FC = 5.02] at 1d postpartum is important in the context of lactose biosynthesis. In fact, hexokinase (HK) is considered to have a potential controlling step for glucose availability for lactose synthesis [43]. In rodents, HK2 is detected only after parturition and it was speculated that its presence may lead to an increase in free glucose for lactose synthesis and improved activity of the pentose phosphate shunt to generate reducing equivalents for lipogenesis [44].
Immune system
The concept that milk, mammary secretions, and the mammary gland have major roles in immune defense has long been proposed [45]. It is well-established that both colostrum and milk proteins have nutritive and immunological functions for the newborn [46]. This is crucial for pigs that have an epitheliochorial placenta impermeable to immunoglobulins (Ig) [1], thus, neonate survival depends upon the passive acquisition of maternal immunity [47]. Immunoglobulins are the primary protein components of colostrum with an immunological function [48]. Immunoglobulin G, in particular, is the major immunoglobulin in sow colostrum and its concentration remains elevated for the initial hours postpartum and then starts to decline consistently [26]. In bovine it is known that specific transport mechanism allows the transfer of a large amount of IgG immunoglobulins from the blood stream across the mammary barrier into colostrum and milk [49]. In pigs it would also appear that colostrum is not a true mammary secretion since 90% of its immunoglobulin content is of serum origin [50].
The transport of immunoglobulins from maternal plasma to colostrum is highly-selective [51] and it is known that FcRn plays an important role in the IgG transport during colostrum formation in several species [52, 53]. In this regard, our results showed no differential expression of FCGRT (Fc fragment of IgG receptor and transporter) among time comparisons. Considering the time-window of our experiment, we speculate that this result is consistent with the need for sustained expression of FCGRT as a way to support colostrum synthesis and also as a defense mechanism to protect mammary tissue. In fact, it is known that FcRn expression coincides with Stage 1 lactogenesis (the onset of colostrogenesis).
Antimicrobial components and chemoattractant activity
Antimicrobial proteins naturally present in colostrum and milk have the ability to kill and inhibit a broad spectrum of bacteria [54, 55]. In this regard, the marked upregulation of HP (haptoglobin) in the last 2 comparisons [FC = 29.27 and 204.86] was noteworthy. Haptoglobin is an acute-phase protein responsive to inflammation and infection [56] that has already been shown to exert immune modulating functions on the innate and adaptive immune system of the pig [57]. At 1d postpartum there was also a significant upregulation of LTF (lactotransferrin) [FC = 3.92]. This gene is a member of the transferrin gene family and is a major iron-binding protein in milk and body secretions with an antimicrobial activity and an important role in the non-specific immune system [45]. Our result is consistent with the fact that lactotransferrin concentrations in swine colostrum at parturition are high and remain elevated through the first days of lactation [26].
Milk is also known to exert a potent chemotactic activity on neutrophils [58]. The prompt recruitment of neutrophils is important for the containment of a number of pathogens at sites of infection, and represents an innate host defenses against microorganisms [59]. In this sense, the role of the chemokine superfamily that encodes secreted proteins involved in immunoregulatory and inflammatory processes must be underscored. Both CXCL2 and CXCL10 encode chemokine antimicrobial proteins with a marked upregulation at 1d postpartum [FC = 17.87 and 5.34, respectively]. Bovine colostrum contains all main chemokines (CXCL1, CXCL2 and CXCL3), but concentrations of CXCL2 are generally the lowest and decrease sharply such that it is undetectable in milk after the onset of lactation [58]. From that standpoint, the strong upregulation of growth-regulated protein homolog gamma (also known as CXCL3) at 1d postpartum [FC = 36.64] is noteworthy. CXCL3/GRO-gamma is involved in the chemokine signaling pathway and (in the absence of inflammation) is considered the major chemotactic factor for neutrophils secreted constitutively into milk [58]. Our results emphasized a major role of CXCL2 and CXCL3 in the transition from colostrum to mature milk in swine, probably to help in the prompt recruitment of neutrophils.
We also detected a marked upregulation of C7 [FC = 7.43] at 1d postpartum. This gene encodes a serum glycoprotein that forms a membrane attack complex together with complement components C5b, C6, C8, and C9 as part of the terminal complement pathway of the innate immune system. In bovine milk, these complement components are found in high concentration in the first 2 days after parturition and then decrease during the following days [60]. In the last 2 time comparisons we also detected the upregulation of C4A (Complement C4A) [FC = 2.05 and 5.73], which acts in concert with other complement components to hasten the destruction of pathogens by phagocytes [61]. It is known that milk and colostrum are rich in host-resistance factors, among the others C4 and C3 proactivators [62]. Even in the absence of cognate interactions, the complement system participates in innate immunity providing efficient and rapid protection [63]. The levels of complement fractions C3 and C4 have been studied in the human transition from colostrum to mature milk, where C3 and C4 decrease over lactation with a highest concentration of C3 in colostrum and a highest concentration of C4 in mature milk [63]. Thus, our results confirm a similar trend in the pig.
The marked upregulation of ceruloplasmin (CP) [FC = 258.47] was consistent with previous studies with pigs, where expression of CP increases in late pregnancy and especially upon lactation, with a correlation between the degree of mammary mRNA expression and the content of milk ceruloplasmin [64]. Although a specific function for CP in the mammary gland is unknown, it may participate in the metabolism of copper [64].
The antimicrobial protein encoded by LYZ (lysozyme) was downregulated in the last 2 comparisons [FC = − 3.22 and − 2.66]. Lysozyme has nonspecific antimicrobial activity that is present in many secretions, tissues, and phagocytic cells of mammals. Its role in swine mammary secretions is not yet well elucidated, even though it is thought to contribute to overall antibacterial activity [65]. Krakowski et al. [66] reported lysozyme activity in sow colostrum immediately after parturition, but Chandan et al. [67] did not find lysozyme activity in sow milk.
Proinflammatory cytokines mediate the early local and systemic responses to microbial challenges and may play a key role in development of the neonatal immune system [68]. In this regard, it was also interesting that IPA results showed a pattern of cytokines predicted to be activated: TNF, IFNG, OSM, IL6, IL1B, TNFSF11, IL5, IFNL1, CSF3, TNFSF13B, IL13, IFNB1, IL1A, IFNA2, IFNA1/IFNA13, IL15, IFNL4, THPO, and IFNK. We also detected the upregulation at 1d postpartum of IL13RA1 (Interleukin 13 Receptor Subunit Alpha 1) [FC = 1.95], IL15 (Interleukin 15) [FC = 1.57], IL17RB (Interleukin 17 Receptor B) [FC = 2.91], TNFSF13 (Tumor Necrosis Factor Superfamily Member 13) [FC = 2.31], TNFRSF12A (TNF Receptor Superfamily Member 12A) [FC = 1.76], and TNFRSF1A (TNF Receptor Superfamily Member 1A) [FC = 1.66]. The presence or transfer of these cytokines has not been clearly verified in porcine colostrum or milk. Furthermore, information about the persistence or function of these maternal cytokines in human suckling neonates, as well as in other species, are also limited [68]. The upregulation of F7 and F10 (Coagulation Factor VII and X) at 1d postpartum [FC = 6.44 and 5.01] also is noteworthy, because it is known that the coagulation system plays an important role in the innate immune system, particularly in the early host response to infection [69].
There was an abrupt upregulation of SAA2 (serum amyloid A-2 protein) [FC = 3039.23] at 1d postpartum. This isoform is considered the most predominant member of the SAA family expressed in the swine mammary gland [70]. Rodriguez et al. [70] reported that SAA mRNA production in swine increased during lactation and stimulates the neonatal immune response by enhancing the recruitment of mucosal gut B lymphoblasts and potentially influencing Ig concentrations. The authors speculated this confers active and passive protection on neonates and provides local protection for the mammary gland. The upregulation of CD14 [FC = 24.65] at 1d postpartum was noteworthy. This gene encodes for a surface antigen involved, with other proteins, in the innate immune response to bacterial lipopolysaccharide. Colostrum has high concentrations of soluble CD14 that decrease over time, with the highest concentration detected in “transitional” milk (0 to 4 d postpartum) [71]. Considering the enrichment of sCD14 in colostrum and milk, Filipp et al. [72] speculated it plays a role in actively stimulating the immune system and homeostasis of IgM of the suckling neonate.
The marked upregulation of SPP1 (also known as OST) at 1d postpartum [FC = 14.61] is in agreement with data from RNA isolated from colostrum and mid lactation milk from goats in which it was the most upregulated gene [30]. SPP1 encodes for the osteopontin protein. The precise role of osteopontin in the mammary gland is still unclear, but it seems to have a role in the modulation of milk protein gene expression, particularly by enhancing the expression of CSN2 [73, 74]. It has also been associated to mammary gland morphogenesis and newborn immunity [75, 76]. Our result seems to suggest a biologic role of this gene during swine lactation but further analyses are required.
Pathogen recognition
We detected the upregulation of TLR2 [FC = 4.45 and 7.32] at 2d prepartum and 1d postpartum. This gene encodes a protein member of the Toll-like receptor (TLR) family, playing a pivotal role in pathogen recognition and activation of innate immunity in human [77]. The main bacterial ligands for TLR2 are peptidoglycan and lipoteichoic acid (LTA) of Gram-positive bacteria [78]. TLR2 is known to be expressed in mammary epithelial cells in bovine, where, after the recognition of specific molecular motifs (i.e. PAMP), determines a rapid and complex innate cascade [79]. We also detected the upregulation at 1d postpartum of TLR4 [FC = 2.04], the main signaling receptor for most bacterial LPS, part of the outer membrane of Gram-negative bacteria. TLR4 acts as the signal-transducing receptor not only for whole Gram-negative bacteria but also for the fusion protein from respiratory syncytial virus [80].
The marked upregulation of LBP in the last 2 comparisons was surprising [FC = 372.18 and 20.89]. The lipopolysaccharide-binding protein (LBP) is one of the most-abundant proteins during infections with Gram-negative bacteria, and is involved in the acute-phase immunologic response. The main function of this protein is to bind bacterial lipopolysaccharides (LPS) expressed on the outer cell wall of bacteria, acting as a carrier for LPS and to help control LPS-dependent monocyte responses [81]. The expression of LBP was demonstrated also in mouse mammary gland early during involution, accompanied by a strong increase in the expression of CD14 protein [82]. Cow colostrum also contains LBP [83] and there is a marked upregulation of LBP in sow liver after parturition when animals experience a normal inflammatory state [84]. Our result seems to confirm an important role of LBP in swine mammary gland.
The upregulation of LY96 (Lymphocyte Antigen 96) [FC = 5.02] also appears biologically-relevant in the context of pathogen recognition. This gene encodes a protein associated with TLR4 on the cell surface and confers responsiveness to LPS, thus, providing a link between the receptor and LPS signaling. It is known that TLR4 cooperates with LY96 and CD14, both of which were upregulated at 1d postpartum and could indicate a response to mediate the innate immune response to bacterial LPS [85, 86]. The upregulation of ICAM1 at 1d postpartum was significant [FC = 4.42]. This gene encodes a cell surface glycoprotein, typically expressed on endothelial cells and cells of the immune system. The fact that human milk contains substantial amounts of slCAM-1 indicates that it could affect the immune system of the neonate [87]. This gene also could have a similar role in swine colostrum and milk.
Lipid metabolism
There was an evident activation of all lipid-related pathways very close to the parturition (2d prepartum). This is consistent with the consideration that mammary tissue is preparing to begin copious milk synthesis and secretion. The further upregulation of Lipid Metabolism pathways at 1d postpartum confirmed this and is consistent with the fact that the mammary gland retain fat in late gestation and synthesize great amounts of de novo fat in early lactation [26, 37]. Our results underscored that this transition is likely attributable to upregulation of many genes, including those involved in de novo fatty acid (FA) synthesis, FA activation and desaturation, cholesterol synthesis and ketone body utilization.
FA de novo synthesis
We observed at 1d postpartum the upregulation of ACACB (acetyl-CoA carboxylase-β) [FC = 3.65]. ACC is a complex multifunctional enzyme system, catalyzing the carboxylation of acetyl-CoA to malonyl-CoA, known as the rate-limiting step in fatty acid synthesis. This result is in agreement with expression profile of genes involved in de novo FA synthesis of human mammary gland during secretory activation, where a progressive increase of ACACB activity by day 4 postpartum was highlighted [88]. In contrast, in mouse mammary grand ACACA was the only isoform with significant upregulation of expression during lactation, while ACACB expression did not differ between pregnancy and lactation [89]. The ACACA and ACACB encode respectively the isoenzymic ACC proteins: ACCα and ACCβ. The ACACA is expressed in the lipogenic tissues and provides cytoplasmic malonyl-CoA for FA synthesis, whereas the ACACB is involved in the regulation of β-oxidation of FA in the mitochondria [90]. FASN encodes another rate-controlling enzyme in lipogenesis that works in concert with ACACA activity. Both genes play a key role in regulating de novo FA synthesis in bovine mammary gland [90]. However, we did not detect differential expression of FASN or ACACA between late-gestation and early lactation. Whether this represents a unique feature of the swine mammary gland will have to be established in future experiments.
FA desaturation genes
The pivotal enzyme implicated in monounsaturated FA synthesis is stearoyl-CoA desaturase (SCD), an important enzyme in the mammary gland, which introduces a double bond in the Δ-9 position of myristoyl-, palmitoyl-, and stearoyl-CoA, primarily [91]. The expression of SCD was upregulated at 1d postpartum [FC = 5.99], and appears to be central during milk fat synthesis at the onset of lactation in swine mammary gland. This result is in agreement with expression of desaturases in bovine during lactation [91, 92] but is opposite to data from human mammary epithelial cells where its expression decreased over the first 3 days and then gradually increased by day 21 of lactation [88].
Fatty acid desaturase 1 (FADS1) and 2 (FADS2) play an important role in the synthesis of very-long-chain FA, adding double bonds at the Δ-5 and Δ-6 position of PUFA [93]. FADS1 is involved in the synthesis of the long-chain PUFA arachidonic acid, eicosapentaenoic acid and docosahexaenoic acid. Stage of lactation alters mammary FADS1 and FADS2 expression in bovine [91], rat [94, 95], and mouse, with a marked upregulation after parturition. In the latter, FADS1 compared with FADS2 mRNA had a more pronounced and significant upregulation after parturition [89]. Yantao Lv et al. [96] suggested that from late-pregnancy and throughout lactation the swine mammary gland participates in LC-PUFA synthesis by altering the expression of FADS1 and FADS2. The authors speculated that FADS1 instead of FADS2–3 may play a major role in the biosynthesis of LC-PUFA in the lactating porcine mammary gland. Our results are in agreement with this consideration, since we found FADS1 gradually increased in the last 2 comparisons (2d prepartum and 1d postpartum) [FC = 2.59 and 4.35], while FADS2 was significantly activated only in the postpartum [FC = 2.32].
Glycerol backbone activation
To synthesize triacylglycerol (TAG), both fatty acyl-CoAs and glycerol 3-phosphate must be readily available [97]. The major steps in the pathway of TAG synthesis in mammary gland have been elucidated [98–100]. The activation of the glycerol carbon backbone, which is needed for further acylation, is the first and crucial step for further TAG assembly, and enzymes encoded by glycerol kinase (GK) (Ensembl:ENSSSCG00000012202; Additional file 3) and diacylglycerol kinase alpha (DGKA) (Ensembl:ENSSSCG00000000370; Additional file 3) play an important role. In particular, glycerol can enter the mammary epithelial cells from the plasma to be phosphorylated by GK [88], and in the last comparison we detected the upregulation of GK [FC = 2.41]. We also detected a moderate upregulation in expression of DGKA in the last 2 time comparisons [FC = 1.42 and 1.75]. DGKA plays an important role in the resynthesis of phosphatidylinositol and phosphorylation of diacylglycerol to phosphatidic acid. This indicates that the activation of the glycerol carbon backbone, which is needed for further acylation, is crucial during the onset of lactation in swine as in human [88].
FA internalization and activation
Protein-mediated FA uptake and the flip-flop mechanism play a major role compared with passive diffusion of FA across membranes [91]. The main proteins implicated in FA uptake in non-ruminant cells include fatty acid translocase FAT/CD36 (CD36) and fatty acid transport proteins (FATP or SLC27A) [101]. In bovine, CD36 was associated with mammary fatty acid uptake from the blood after parturition [91]. Our results underscored a strong upregulation of CD36 [FC = 7.47], hence, confirming its pivotal role in swine mammary gland. The strong upregulation of ACSL6 (acyl-CoA synthetase long-chain family member 6) in the last 2 comparisons [FC = 26.04 and 37.58] confirmed the importance of ACSL family member isoforms for the FA activation during the onset of lactation in swine as in human [88] and bovine [102]. In fact internalized FAs, prior to participating in further metabolism, must be esterified with CoA in the inner face of the plasma membrane via acyl-CoA (ASC) [91]. In this regard, the upregulation of ACSL3 at 1d postpartum [FC = 5.62] also is noteworthy given results reported by Lv et al. [96]. They reported that ACSL3 is the most abundant isoform in the porcine mammary gland, in contrast to ACSL1 which is the main isoform in lactating bovine [102] and human [88] mammary cells. The authors speculated that in swine mammary ACSL3 channels LCFA mainly towards TAG synthesis during lactation. This consideration was based on the fact that in the rat ACSL3 prefers C16-C20 unsaturated FA [103], which are major constituents of FA in sow milk [96]. The downregulation of ACSL1 and ACSL5 at 2 days prepartum [FC = − 1.44 and − 1.84] seems to support the idea of ACSL3 being more important during lactation.
Acyltransferases and TG assembly
From late-pregnancy to onset of lactation, we detected the upregulation in the last 2 comparisons of a cluster of genes involved in the first and rate-limiting step in the TAG biosynthesis pathway, i.e. GPAT3, GPAT4 and AGPAT1 [FC = 2.04, 2.09 and 1.44; 3.13, 4.46 and 1,47 respectively]. GPAT (glycerol-3-phosphate acyltransferase) enzymes, which reside in the endoplasmic reticulum (ER) and mitochondria, catalyze the first committed step in TAG synthesis via the glycerol phosphate pathway [104]. These enzymes add fatty-acyl groups to the sn-1 position of glycerol-3-phosphate, and lead to the production of monoacylglycerols (MAG) [97]. The conversion of lysophosphatidate to phosphatidate via AGPAT, which adds an acyl group to the sn-2 position of the glycerol backbone, represents the second acylation step in the glycerol phosphate pathway [104].
Regarding specific isoforms uncovered in our results, GPAT3 is a gene with a controversial identity. Current evidence suggests that it has both GPAT and AGPAT activities [104]. Based on its high amino acid similarity to AGPAT1 and AGPAT2, GPAT4 was initially classified as AGPAT6. Recently after the examination of its enzyme activity, it was considered a second ER-localized GPAT and renamed as GPAT4 [104]. Nagle et al. [105] revealed that GPAT4, expressed in cultured cells, can utilize a variety of substrates, including C12:0-, C16:0-, C18:0-, C18:1-, C18:2-, and C20:4-CoA substrates [105]. AGPAT1, which was upregulated in the last 2 time comparisons is a well-established AGPAT isoform, with a validated enzyme activity [106] and a preference for C12–16:0, C16:1, C18:2, and C18:3, followed by C18:0, C18:1, and C20:4, but with a poor activity for C20:0 and C24:0 [104]. Interestingly, AGPAT1 can also catalyze the reverse of the normal AGPAT reaction, that is the ATP-independent acyl-CoA and LPA (lysophosphatidic acid) synthesis from PA (phosphatidic acid) [107]. This activity suggested its possible implication in regulation of the levels of LPA and PA available to act as signaling molecules [104]. AGPAT1 (1-Acylglycerol-3-Phosphate O-Acyltransferase 1) was discovered to have a crucial role also during de novo synthesis of triacylglycerol in bovine mammary gland during lactation [102].
Once synthesized and activated, FAs are esterified to glycerol-3-phosphate to produce TAG [96]. Both GPAM and DGAT1 are responsible for the first and last step of esterification leading to TAG synthesis [91]. GPAM (glycerol-3-phosphate acyltransferase, mitochondrial) is a well-known gene, mostly expressed in tissues with high lipogenic activity and plays a key role in phospholipid and TAG biosynthesis [108]. DGAT1 (diacylglycerol acyltransferase 1) also is a well-characterized gene and catalyzes the esterification of the last FA to diacylglycerol leading to TAG synthesis. In the present study both GPAM and DGAT1 were upregulated only at 2d prepartum [FC = 1.91 and 1.31]. In the case of DGAT1, this result is not in agreement with studies in bovine and human, where its upregulation occurred postpartum [88, 91]. This may suggest that DGAT1 is of minor importance in the overall process of milk fat synthesis in the pig, compared with other genes involved in TAG synthesis. In a recent study, however, a western blot analysis of DGAT1 together with other proteins in porcine mammary tissue confirmed its increase during lactation compared with late-pregnancy [96]. Because our time frame of interest was around colostrogenesis, further protein expression and functional studies during these times would have to be conducted to clarify the importance of DGAT in colostrogenesis. The fact remains that DGAT1 is one of many proteins composing the TAG synthesis pathway [109, 110].
Because LPIN proteins act in an interdependent manner to optimize lipid homeostasis in various tissues, it is currently believed that their function and role in glycerolipid synthesis are influenced by intricate functional interactions among various LPIN family members [111, 112]. Lv et al. [96] argued for a major role of LPIN1 in TAG synthesis in the porcine mammary gland during lactation. In the present study, the upregulation of LPIN1 at 1d postpartum [FC = 2.26] seems to confirm this argument and is in agreement with other studies in human, mouse, and bovine mammary tissue, where a marked upregulation of LPIN1 during lactation was reported [88, 89, 102].
Lipid droplet formation in milk
After milk fat globule formation in the ER membrane via incorporation of newly-formed TAG, the globules are transported to the apical membrane and released during milk secretion [113]. Butyrophilin (BTN1A1) and xanthine dehydrogenase (XDH) are well-defined proteins involved in these processes in mammary [90], having a function as structural proteins in milk fat droplets in the lactating mammary gland [114]. Furthermore, the essential role of perilipins in droplet formation is well-known [115]. Our data are consistent with this evidence and support a significant role of all BTN1A1, XDH and PLIN5 genes in swine mammary lipid droplet formation. In fact, in the last 2 time comparisons we detected a strong upregulation of BTN1A1 [FC = 90.49 and 202.51] and at 1d postpartum we detected a marked upregulation of XDH [FC = 17.24] and PLIN5 [FC = 21.42]. This is in agreement with a recent study showing that fat is taken up in substantial amounts by the sow mammary glands in late gestation [37].
Cholesterol synthesis genes
The shift of nutrients from body stores towards the mammary gland for milk production requires not only the adaptation of glucose and lipid metabolism to the lactating state, but also cholesterol metabolism in particular during early lactation [116]. In our results the upregulation of HMGCS1, FAXDC2, NSDHL at 1d postpartum [FC = 7.63; 4.69 and; 1.50] seemed to confirm this evidence also in swine mammary gland. In particular HMGCS1, which is important for the regulation of cholesterol synthesis [117], was markedly upregulated during early lactation compared with late pregnancy in the bovine mammary gland [116].
Utilization of ketone bodies
On day 2 prepartum and day 1 postpartum, we detected moderate upregulation of BDH (3-hydroxybutyrate dehydrogenase), encoding a protein catalyzing the initial steps of BHBA utilization in mitochondria [118]. In human, cytosolic type BDH2 is involved in the utilization of ketone bodies, which can subsequently enter mitochondria and the tricarboxylic acid cycle [119]. In ruminants, previous studies showed that the mammary gland takes up large amounts of BHBA and that the use of BHBA (as 4-carbon units) by mammary cells is primarily for de novo FA synthesis [91, 120]. The moderate upregulation of BDH2 in the last 2 time comparisons [FC = 1.68; 1.93] suggested that ketone bodies likely are an energy source also for the sow mammary gland.
Ceramide-synthesis genes in mammary
There was moderate upregulation of SGMS1 (sphingomyelin synthase 1) in the prepartum [FC = 1.42] and higher upregulation of SGMS2 (sphingomyelin synthase 2) in the postpartum [FC = 4.37], and we found a concomitant downregulation of SGMS1 [FC = − 1.35]. Sphingomyelin synthases synthesize sphingomyelin through transfer of the phosphatidyl head group in phosphatidylcholine to the primary hydroxyl group of ceramide. Ceramide is one of the most-studied sphingolipids in nature, and is involved in cell signaling, cell cycle, and regulation of protein transport from ER to Golgi [121]. Sphingomyelin synthesis from ceramide is considered an important step because sphingomyelin constitutes about 25% of the total phospholipids in dairy products, having highly bioactive properties and considered to be functional in food [122]. The upregulation of SMPD1 (sphingomyelin phosphodiesterase 1) [FC = 1.67] and the simultaneously downregulation of CERS1 (Ceramide Synthase 1) [FC = − 2.12] at 1d postpartum appears to have a biologic role in the overall process of sphingomyelin metabolism. SMPD1 is involved in the conversion of sphingomyelin to ceramide, whereas CERS1 catalyzes the synthesis of ceramide. Further protein expression and functional studies during the entire lactation should be conducted to clarify the role of sphingolipids with signaling roles and the role of ceramide in swine mammary gland.
The marked upregulation of CYP4A21 [FC = 568.37], a member of the CYP4A subfamily discovered in pig [124], is noteworthy because the protein possesses taurochenodeoxycholic acid 6α-hydroxylase activity but does not metabolise lauric acid, a common substrate for other CYP4As [123]. The function of CYP4A in vivo is not well clarified but CYP4As are known for hydroxylating of a series of fatty acids, eicosanoids and prostaglandins (PG) [124–126]. The activity of CYP4A21 is still uncharacterized in mammary gland. CYP4A21 is believed to be responsible for formation of hyocholic acid, a bile acid typically found in porcine [127]. Further analysis is required to investigate the role of this gene during the onset of lactation in swine mammary gland.
Transcription factors
The first step of gene expression is transcription and represents the primary step at which gene expression is controlled. This is accomplished through the recruitment of several transcription factors, having the ability to bind to certain target-sequences of the genes, and promote or suppress transcription according to the stimuli [128]. Considering those transcription regulators in the IPA findings that overlap in the last 2 time comparisons (p-value cutoff ≤0.01 and activation z-score ≥ ± 2) the results supported the suggestion that SREBP1 and XBP1 are pivotal in the transition from colostrogenesis to lactogenesis in swine mammary gland. They likely act on regulation of lipid synthesis [129] and morphological mammary development [130], respectively.
Regulation of lipid biosynthesis
The function of SREBP1 (sterol regulatory element-binding protein 1) is well-established in rodents and it plays a crucial role in the regulation of hepatic cholesterol biosynthesis and FA metabolism, in particular the biosynthesis of fat [131, 132]. Our results show that SREBP1 is important also in the mammary gland for cholesterol biosynthesis and this is consistent with real-time PCR measurements that confirmed the upregulation of SREBP1 during the transition from pregnancy to lactation in murine mammary gland [133]. In non-ruminants, SREBP1 is transported from endoplasmic reticulum membrane, where it resides as an inactive precursor, to the Golgi for proteolytic cleavage (i.e., activation) prior to entering the nucleus where it causes the activation of sterol response element (SRE)-containing genes [90]. The transport step to the Golgi is regulated by sterols via the sterol-sensing protein SCAP (SREBP cleavage activating protein), which was modestly upregulated in the last comparison [FC = 1.26].
It is generally accepted that insulin induced gene (INSIG) 1 and 2 encode proteins which interact with SCAP in an oxysterol-dependent and independent fashion (in non-ruminants) and regulate the responsiveness of SREBP1 and 2 processing via SCAP. However, the precise role of INSIG1, strongly upregulated in our last comparison [FC = 13.22], is controversial. In fact, decreased SREBP activity as a consequence of increased INSIG1 has been observed in liver [134], but upregulation of INSIG1 was detected during lactation in bovine mammary gland, positively correlated with the ratio of synthesized/imported FA [91].
Our data support a need of INSIG1 in controlling the induction of gene expression by SREBP. Therefore, INSIG1 could play a central role in orchestrating lipid metabolism also in swine mammary tissue during lactation. In this regard, the predicted upregulation of PPARG expression at 1d postpartum (known to be involved in regulation of lipid synthesis in goat and bovine mammary cells [91, 135–137]) is noteworthy [PPARGC1B z-score = 2.06]. A potential role of this nuclear receptor in milk fat synthesis was already postulated in particular in bovine mammary gland, where INSIG1 was demonstrated to be a PPARG responsive gene [91]. PPARG could represent an important control point of milk fat synthesis, in particular in triacylglycerol synthesis and milk secretion in pig as well as in goat and bovine [136, 137], acting indirectly on SREBP1 protein activity through regulation of INSIG1 expression and directly on SREBP1.
Regulation of morphological mammary development
Colostrum and milk synthesis occur in alveolar structures composed of a single layer of MEC encircling a lumen where milk is secreted [129]. In order to become fully functional, MEC acquire a number of cellular characteristics during late pregnancy including the development of an elaborate endoplasmic reticulum (ER) system [138], which is required for the synthesis of secreted proteins but is also the site where fatty acids are assembled into TAG and phospholipids [139]. The transcription factor X-box binding protein 1 (XBP1) has multiple functions. Briefly, it promotes ER biogenesis [140] and is a component of a highly-conserved signaling cascade responsible for restoring homeostasis when the ER is confronted with various stresses, including increased protein synthesis and secretion [141, 142]. XBP1 is also implicated as a positive regulator of both lipogenesis and VLDL (very low density lipoprotein) secretion in hepatocytes [143, 144]. Recently, in murine, it was shown that XBP1 is required for MEC population expansion during lactation, enhancing the development of an elaborate endoplasmic reticulum compartment [130] and playing a central role in the coordination of synthesis and export of products in mammary epithelial cells [145].
All the above evidence is consistent with the suggestion that XBP1 may be dispensable for morphologically mammary development, colostrum and milk synthesis and secretion during late-pregnancy and the onset of lactation in pig. In particular, focusing on the upregulated genes involved in protein processing in endoplasmic reticulum that were detected in the last two comparisons, the significant upregulation of PDIA4 [FC = 2.42 and 3.11], PDIA3 [FC = 1.60 and 2.15], PDIA6 [FC = 1.49 and 2.05] and CALR (calreticulin) [FC = 1.63 and 2.11] is noteworthy. PDIA4, PDIA3 and PDIA6 are genes that encode for specific members of the disulfide isomerase (PDI) family of endoplasmic reticulum (ER) proteins catalyzing protein folding and thiol-disulfide interchange reactions. Calreticulin is a multifunctional protein acting in the lumen of the endoplasmic reticulum as a major Ca(2+)-binding (storage) protein. In MEC, it is not completely clear the role of ER-resident proteins on the folding and the retention of milk proteins. However, calreticulin and PDI have been detected in rat and goat lactating MEC, suggesting that these proteins could be involved in the formation of lipid droplets and raising questions about a possible link between the enzymes involved in protein and lipid synthesis [146].
Other upregulated transcription regulators
At 1d postpartum, we detected a marked upregulation of IRF7 [z-score = 4.69], TP53 [z-score = 4.25], NUPR1 [z-score = 4.10] and NFATC2 [z-score = 4.09] together with XBP1 and SREBP1 which had the highest z-score value. IRF7 encodes interferon regulatory factor 7, a member of the interferon regulatory transcription factor (IRF) family, it is a key transcriptional regulator of type I interferon (IFN)-dependent immune responses with an important role in the innate immune response against DNA and RNA viruses [147]. It regulates the transcription of type I IFN genes (IFN-α and IFN-β) and IFN-stimulated genes (ISG), which were markedly upregulated [ISG15, FC = 2.95], by binding in their promoters to an interferon-stimulated response element (ISRE). TP53 (Tumor Protein P53) is a tumor suppressor involved in several types of human tumors, acting both as a gene-specific transcription factor as well as a specific inhibitor of the transcription of certain genes [148]. Its tumor suppressor activity is implicated in the expression of genes involved in the control of cell cycle, cellular senescence, and apoptosis [149] but recently Munne et al. [150] suggested and demonstrated a role for TP53 in the epithelial-to-mesenchymal transition (EMT) and differentiation of mammary epithelia. NUPR1 is a nuclear protein transcriptional regulator involved in the negative regulation of the cell cycle [151]. Zhou et al. [152] reported a high expression of NUPR1, together with other TR, during lactation compared with pregnancy. This could explain why cell cycle-related genes are more active in pregnancy. NFATC2 (nuclear factor of activated t-cells 2) is a member of the nuclear factor of activated T cells (NFAT) family. Most of the work on NFAT proteins has been related to immune cell activation and its mediators, such as cytokines [153]. The product of this gene is a DNA-binding protein with a REL-homology region (RHR) and an NFAT-homology region (NHR) which is present in the cytosol and is only translocated to the nucleus upon T cell receptor (TCR) stimulation. Once in the nucleus it becomes a member of the nuclear factors of the activated T cells transcription complex with a central role in inducing gene transcription during the immune response [154].
Conclusions
The transcriptome changes greatly during the last week prepartum and these changes are highly likely involved in coordinating the synthesis of colostrum and main milk components (i.e. protein, fat, lactose and antimicrobial factors) as revealed by influenced pathways. The lipid metabolism pathway changes greatly and some of those adaptations are controlled at least in part via SREBP1 and XBP1, acting on regulation of lipid synthesis and morphological development of the mammary gland. Other transcription regulators including IRF7, TR53, NUPR1 and NFATC2 acting across a wide number of pathways become important at the onset of lactation. Further research will help confirm the functional relevance of the pathways uncovered, and how they influence the transition from colostrum to mature milk during a stage when slight abnormalities may potentially threaten piglet survival. Clearly, milk synthesis requires a myriad of factors beyond transcription of the major proteins involved in the synthesis and secretion of protein, fat, and lactose. Holistically, milk synthesis is the product of complex interactions among several tissues and organs that only an integrative systems-biology approach may help elucidate.
Additional files
Provides an aggregate summary of RNA extraction and quality check for all samples. Mammary tissue was collected on days 14, 10, 6 and 2 before (−) parturition and on day 1 after (+) parturition from three 2nd parity sows. Extraction of RNA and quality evaluation was performed following protocols described by Tramontana et al. (2008) [15]. (XLSX 10 kb)
Provides an aggregate summary of RNA sequencing and alignment for all the samples. mRNA libraries were sequenced on a HiSeq2500 (Illumina Inc.). Quality control metrics were performed on raw sequencing reads using the FASTQC v0.11.15 application. An index of the reference genome was built and single-end clean reads for each individual were aligned to the reference genome by STAR (v2.5.1b). Reads were mapped and annotated to the Sus scrofa genome (v10.2.86), downloaded from EnsemblGenome website (Nov. 2016). Reads aligned were quantified with Subread package (v1.5.0) based on the Refseq gene annotation. (XLSX 10 kb)
Provides the complete list of differentially expressed genes (DEG) resulting from differential expression (DE) analysis of RNAseq data of swine mammary gland tissue harvested in late-pregnancy through farrowing and used for Dynamic Impact Approach (DIA) and Ingenuity Pathway Analysis (IPA) analyses. Moreover, it provides additional discussion points, omitted from the main body of the manuscript. (XLSX 1239 kb)
Provides complete list of upstream regulator resulting from Ingenuity Pathway Analysis (IPA) analysis. Moreover, it provides additional discussion points, omitted from the main body of the manuscript. (XLSX 20 kb)
Acknowledgments
Funding
This research was supported in part by funds from the Danish Ministry for Food, Agriculture and Fishery (grant no. 3405–11-0342; to PKT) and Hatch funds (to JJL) under project ILLU-538–914, National Institute of Food and Agriculture (Washington, DC). The funders had no role in the design of the study, the collection, analysis, and interpretation of data, or in the writing of the manuscript.
Availability of data and materials
The data have been submitted to Gene Expression Omnibus database (BioProject ID: GSE101983).
Abbreviations
- AA
Amino acids
- ACACA
Acetyl-CoA carboxylase alpha
- ACACB
Acetyl-CoA carboxylase beta
- ACC
Acetyl-CoA carboxylase
- ACSL
Acyl-CoA synthetase long-chain
- ACSL1
Acyl-CoA synthetase long-chain family member 1
- ACSL3
Acyl-CoA synthetase long-chain family member 3
- ACSL5
Acyl-CoA synthetase long-chain family member 5
- ACSL6
Acyl-CoA synthetase long-chain family member 6
- AGPAT
1-Acylglycerol-3-phosphate O-acyltransferase
- AGPAT1
1-Acylglycerol-3-phosphate O-acyltransferase 1
- AGPAT2
1-Acylglycerol-3-phosphate O-acyltransferase 2
- ASC
Amino acid transport system ASC
- ATP
Adenosine triphosphate
- B4GALT1
Beta-1,4-galactosyltransferase 1
- BDH
3-Hydroxybutyrate dehydrogenase
- BDH2
3-Hydroxybutyrate dehydrogenase 2
- BHBA
Beta-hydroxybutyric acid
- BTN1A1
Butyrophilin subfamily 1 member A1
- C3
Complement component 3
- C4a
Complement component 4a
- C5b
Complement component 5b
- C6
Complement component 6
- C7
Complement component 7
- C8
Complement component 8
- C9
Complement component 9
- CAT-4
Cationic amino acid transporter 4
- CD14
CD14 molecule
- CD36
CD36 molecule
- CERS1
Ceramide synthase 1
- CP
Ceruloplasmin
- CSF3
Colony stimulating factor 3
- CSN1S2
Casein alpha-S2
- CSN2
Casein Beta
- CTGF
Connective Tissue Growth Factor
- CXCL1
C-X-C Motif chemokine ligand 1
- CXCL10
C-X-C Motif chemokine ligand 10
- CXCL2
C-X-C Motif chemokine ligand 2
- CXCL3
C-X-C Motif chemokine ligand 3
- CYP4A
Cytochrome P450 family 4
- CYP4A21
Cytochrome P450 family 8 subfamily B member 1
- DE
Differential expression
- DEG
Differentially expressed gene
- DGAT1
Diacylglycerol O-acyltransferase 1
- DGKA
Diacylglycerol kinase alpha
- DIA
Dynamic impact approach
- EMT
Epithelial-to-mesenchymal transition
- ER
Endoplasmic reticulum
- F10
Coagulation factor X
- F7
Coagulation factor VII
- FA
Fatty acid
- FADS1
Fatty acid desaturase 1
- FADS2
Fatty acid desaturase 2
- FASN
Fatty acid synthase
- FASTQC
Quality control application for FastQ files
- FAT
Fatty acid translocase
- FATP
Fatty acid transport protein
- FAXDC2
Fatty acid hydroxylase domain containing 2
- FC
Fold change
- FCGRT
Fc fragment of IgG receptor and transporter
- FcRn
Neonatal Fc receptor
- FDR
False discovery rate
- GK
Glycerol kinase
- GPAM
Glycerol-3-phosphate acyltransferase, mitochondrial
- GPAT
Glycerol-3-phosphate acyltransferase
- GPAT3
Glycerol-3-phosphate acyltransferase 3
- GPAT4
Glycerol-3-phosphate acyltransferase 4
- GRO
Growth-regulated protein homolog
- HK2
Hexokinase 2
- HMGCS1
3-Hydroxy-3-methylglutaryl-CoA synthase 1
- HP
Haptoglobin
- ICAM1
Intercellular adhesion molecule 1
- ID
Identification
- IFN
Interferon
- IFNA1
Interferon alpha 1
- IFNA13
Interferon alpha 13
- IFNA2
Interferon alpha 2
- IFNB1
Interferon beta 1
- IFNG
Interferon gamma
- IFNK
Interferon kappa
- IFNL1
Interferon lambda 1
- IFNL4
Interferon lambda 4
- Ig
Immunoglobulin
- IL13
Interleukin 13
- IL13RA1
Interleukin 13 receptor subunit alpha 1
- IL15
Interleukin 15
- IL17RB
Interleukin 17 receptor B
- IL1A
Interleukin 1 alpha
- IL1B
Interleukin 1 beta
- IL5
Interleukin 5
- IL6
Interleukin 6
- INSIG
Insulin induced gene
- INSIG1
Insulin induced gene 1
- IPA
Ingenuity pathway analysis
- IRF7
Interferon regulatory factor 7
- ISG15
Interferon-stimulated protein 15 KDa
- ISRE
Interferon-stimulated response element
- KEGG
Kyoto encyclopedia of genes and genomes
- LALBA
Lactalbumin alpha
- LAT1
Linker for activation of T-cells family member 1
- LAT2
Linker for activation of T-cells family member 2
- LBP
Lipopolysaccharide-binding protein
- LCFA
Long-chain fatty acid
- LC-PUFA
Long-chain polyunsaturated fatty acid
- Leu
Leucine
- LPA
Lysophosphatidic acid
- LPIN
Lipin
- LPIN1
Lipin 1
- LPS
Lipopolysaccharides
- LTA
Lipoteichoic acid
- LTF
Lactotransferrin
- LY96
Lymphocyte antigen 96
- LYZ
Lysozyme
- MAG
Monoacylglycerols
- MEC
Mammary epithelial cells
- MG
Mammary gland
- mTOR
Mechanistic target of rapamycin
- NFAT
Nuclear factor of activated T-cells
- NFATC2
Nuclear factor of activated T-cells 2
- NHR
NFAT-Homology Region
- NSDHL
NAD(P) Dependent steroid dehydrogenase-like
- NUPR1
Nuclear protein 1, transcriptional regulator
- OSM
Oncostatin M
- OST
Osteopontin
- OXTR
Oxytocin receptor
- PA
Phosphatidic acid
- PAEP
Progestagen associated endometrial protein
- PAMP
Pathogen associated molecular pattern
- PCR
Polymerase chain reaction
- PDI
Protein disulfide isomerase
- PDIA3
Protein disulfide isomerase family A member 3
- PDIA4
Protein disulfide isomerase family A member 4
- PDIA6
Protein disulfide isomerase family A member 6
- PG
Prostaglandin
- PLIN5
Perilipin 5
- PPAR
Peroxisome proliferator-activated receptor
- PPARG
Peroxisome proliferator activated receptor gamma
- PPARGC1B
PPARG coactivator 1 beta
- PUFA
Polyunsaturated fatty acid
- RHR
REL-Homology Region
- SAA2
Serum amyloid A2
- SCAP
SREBP Cleavage activating protein
- SCD
Stearoyl-CoA desaturase
- SCFA
Short-chain fatty acid
- SGMS1
Sphingomyelin synthase 1
- SGMS2
Sphingomyelin synthase 2
- SLC35A2
Solute carrier family 35 member A2
- SLC7A4
Solute carrier family 7 member 4
- SLC7A8
Solute carrier family 7 member 8
- SMPD1
Sphingomyelin phosphodiesterase 1
- SPP1
Secreted phosphoprotein 1
- SRE
Sterol response element
- SREBP1
Sterol regulatory element binding protein 1
- STAR
Spliced transcripts alignment to a reference
- TAG
Triacylglycerides
- TCR
T Cell receptor
- TF
Transcription factor
- THPO
Thrombopoietin
- TLR
Toll like receptor
- TLR2
Toll like receptor 2
- TLR4
Toll like receptor 4
- TMM
Trimmed mean of M-values
- TNF
Tumor necrosis factor
- TNFRSF12A
TNF Receptor superfamily member 12A
- TNFRSF1A
TNF Receptor superfamily member 1A
- TNFSF11
Tumor necrosis factor superfamily member 11
- TNFSF13
Tumor necrosis factor superfamily member 13
- TNFSF13B
Tumor necrosis factor superfamily member 13b
- TP53
Tumor protein P53
- TR
Transcription regulator
- TSC1
Tuberous sclerosis 1
- TSC2
Tuberous dclerosis 2
- UDP
Uridine diphosphate
- VLDL
Very-low-density lipoprotein
- WAP
Whey acidic protein
- XBP1
X-box binding protein 1
- XDH
Xanthine dehydrogenase
- α-LA
Lactalbumin alpha
- β-CN
Casein beta
Authors’ contributions
VP performed the statistical analysis of the RNAseq results, ran the DIA and IPA analysis, and wrote the main draft of the manuscript. UK and PKT designed and coordinated the animal study and harvested the mammary biopsies. JJL, MVR and MD interpreted data and contributed to writing the manuscript. KS extracted, assessed the RNA quality from the biopsy samples, and qPCR. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Sampling, housing and rearing were in compliance with Danish laws and regulations for the humane care and use of animals in research [The Danish Ministry of Justice, Animal Testing Act (Consolidation Act No. 726 of September 9, 1993, as amended by Act No. 1081 of December 20, 1995)]. Furthermore, the Danish Animal Experimentation Inspectorate approved the study protocols and supervised the experiment.
The animals were purchased from Ørgelgaard, DK 7840 Højslev, Denmark.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4719-5) contains supplementary material, which is available to authorized users.
Contributor Information
V. Palombo, Email: valentino.palombo@studenti.unimol.it
J. J. Loor, Email: jloor@illinois.edu
M. D’Andrea, Email: dandrea@unimol.it
M. Vailati-Riboni, Email: vailati2@illinois.edu
K. Shahzad, Email: khuramsb@gmail.com
U. Krogh, Email: uffekrogh@anis.au.dk
P. K. Theil, Email: peter.theil@anis.au.dk
References
- 1.Salmon H. Mammary gland immunology and neonate protection in pigs. Homing of lymphocytes into the MG. Adv Exp Med Biol. 2000;480:279–286. doi: 10.1007/0-306-46832-8_32. [DOI] [PubMed] [Google Scholar]
- 2.Theil PK, Lauridsen C, Quesnel H. Neonatal piglet survival: impact of sow nutrition around parturition on fetal glycogen deposition and production and composition of colostrum and transient milk. Animal. 2014;8:1021–1030. doi: 10.1017/S1751731114000950. [DOI] [PubMed] [Google Scholar]
- 3.Ji F, Hurley WL, Kim SW. Characterization of mammary gland development in pregnant gilts. J Anim Sci. 2006;84:579–587. doi: 10.2527/2006.843579x. [DOI] [PubMed] [Google Scholar]
- 4.Kensinger R, Collier R, Bazer F, Ducsay C, Becker H. Nucleic acid, metabolic and histological changes in gilt mammary tissue during pregnancy and lactogenesis. J Anim Sci. 1982;54:1297–1308. doi: 10.2527/jas1982.5461297x. [DOI] [PubMed] [Google Scholar]
- 5.Bionaz M, Loor JJ. Ruminant metabolic systems biology: reconstruction and integration of transcriptome dynamics underlying functional responses of tissues to nutrition and physiological state. Gene Regul Syst Biol. 2012;6:109–125. doi: 10.4137/GRSB.S9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kukurba KR, Montgomery SB. RNA sequencing and analysis. Cold Spring Harb Protoc. 2015;2015:951–969. doi: 10.1101/pdb.top084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wickramasinghe S, Rincon G, Islas-Trejo A, Medrano JF. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genomics. 2012;13:45. doi: 10.1186/1471-2164-13-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J, Bao ZK, Zhang Q, Hu WW, Yu QH, Yang Q. Transcriptome analysis of the mammary gland from GH transgenic goats during involution. Gene. 2015;565:228–234. doi: 10.1016/j.gene.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Suárez-Vega A, Gutiérrez-Gil B, Klopp C, Tosser-Klopp G, Arranz J-J. Comprehensive RNA-Seq profiling to evaluate lactating sheep mammary gland transcriptome. Sci Data. 2016;3:160051. doi: 10.1038/sdata.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao W, Shahzad K, Jiang M, Graugnard DE, Rodriguez-Zas SL, Luo J, et al. Bioinformatics and gene network analyses of the swine mammary gland transcriptome during late gestation. Bioinform Biol Insights. 2013;7:193–216. doi: 10.4137/BBI.S12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bionaz M, Periasamy K, Rodriguez-Zas SL, Hurley WL, Loor JJ. A novel dynamic impact approach (DIA) for functional analysis of time-course omics studies: validation using the bovine mammary transcriptome. PLoS One. 2012;7:e32455. doi: 10.1371/journal.pone.0032455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Danish Ministry of Justice. 1995. Animal testing act, consolidation act no. 726 of September 9, 1993 (as amended by act no. 1081 of December 20, 1995). The Danish Ministry of Justice, Copenhagen, Denmark. 1995.
- 13.Krogh U, Bruun TS, Poulsen J, Theil PK. Impact of fat source and dietary fibers on feed intake, plasma metabolites, litter gain and the yield and composition of milk in sows. Animal. 2017;11:975–983. doi: 10.1017/S1751731116002585. [DOI] [PubMed] [Google Scholar]
- 14.Theil PK, Flummer C, Hurley WL, Kristensen NB, Labouriau RL, Sørensen MT. Mechanistic model to predict colostrum intake based on deuterium oxide dilution technique data and impact of gestation and prefarrowing diets on piglet intake and sow yield of colostrum. J Anim Sci. 2014;92:5507–5519. doi: 10.2527/jas.2014-7841. [DOI] [PubMed] [Google Scholar]
- 15.Tramontana S, Bionaz M, Sharma A, Graugnard DE, Cutler EA, Ajmone-Marsan P, et al. Internal controls for quantitative polymerase chain reaction of swine mammary glands during pregnancy and lactation. J Dairy Sci. 2008;91:3057–3066. doi: 10.3168/jds.2008-1164. [DOI] [PubMed] [Google Scholar]
- 16.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 17.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vailati-Riboni M, Kanwal M, Bulgari O, Meier S, Priest NV, Burke CR, et al. Body condition score and plane of nutrition prepartum affect adipose tissue transcriptome regulators of metabolism and inflammation in grazing dairy cows during the transition period. J Dairy Sci. 2016;99:758–770. doi: 10.3168/jds.2015-10046. [DOI] [PubMed] [Google Scholar]
- 21.Farmer C, Devillers N, Rooke JN, Le Dividich J. Colostrum production in swine: from the mammary glands to the piglets. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour. 2006;1:16. [Google Scholar]
- 22.Hartmann PE, Smith NA, Thompson MJ, Wakeford CM, Arthur PG. The lactation cycle in the sow: physiological and management contradictions. Livest Prod Sci. 1997;50:75–87. [Google Scholar]
- 23.Farmer C. Review: mammary development in swine: effects of hormonal status, nutrition and management. Can J Anim Sci. 2012;93:1–7. [Google Scholar]
- 24.Kimura T, Tanizawa O, Mori K, Brownstein MJ, Okayama H. Structure and expression of a human oxytocin receptor. Nature. 1992;356:526–529. doi: 10.1038/356526a0. [DOI] [PubMed] [Google Scholar]
- 25.Devillers N, Farmer C, Mounier A-M, Le Dividich J, Prunier A. Hormones, IgG and lactose changes around parturition in plasma, and colostrum or saliva of multiparous sows. Reprod Nutr Dev. 2004;44:381–396. doi: 10.1051/rnd:2004043. [DOI] [PubMed] [Google Scholar]
- 26.Hurley WL. Composition of sow colostrum and milk. Chapter 9. In: Farmer, C. (ed.) The gestating and lactating sow. Wageningen: Wageningen Academic Publishers; 2015. pp. 193-229.
- 27.Ramakrishnan B, Shah PS, Qasba PK. Alpha-Lactalbumin (LA) stimulates milk beta-1,4-galactosyltransferase I (beta 4Gal-T1) to transfer glucose from UDP-glucose to N-acetylglucosamine. Crystal structure of beta 4Gal-T1 x LA complex with UDP-Glc. J Biol Chem. 2001;276:37665–37671. doi: 10.1074/jbc.M102458200. [DOI] [PubMed] [Google Scholar]
- 28.Gao Y, Lin X, Shi K, Yan Z, Wang Z. Bovine mammary gene expression profiling during the onset of lactation. PLoS One. 2013;8:e70393. doi: 10.1371/journal.pone.0070393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bionaz M, Hurley W, Loor J. (2012) Milk protein synthesis in the lactating mammary gland: Insights from transcriptomics analyses. In: Hurley WH, editor. Milk Protein, Chapter 11. InTech North America, New York, USA. 285–324. Available: 10.5772/46054. Accessed 12 Sept 2012.
- 30.Crisà A, Ferrè F, Chillemi G, Moioli B. RNA-sequencing for profiling goat milk transcriptome in colostrum and mature milk. BMC Vet Res. 2016;12:264. doi: 10.1186/s12917-016-0881-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson KJ, Bird P, Shaw D, Nicholas K. Molecular characterisation and hormone-dependent expression of the porcine whey acidic protein gene. J Mol Endocrinol. 1998;20:27–35. doi: 10.1677/jme.0.0200027. [DOI] [PubMed] [Google Scholar]
- 32.Shennan DB, Boyd CA. The functional and molecular entities underlying amino acid and peptide transport by the mammary gland under different physiological and pathological conditions. J Mammary Gland Biol Neoplasia. 2014;19:19–33. doi: 10.1007/s10911-013-9305-5. [DOI] [PubMed] [Google Scholar]
- 33.Vallet JL, McNeel AK, Miles JR, Freking BA. Placental accommodations for transport and metabolism during intra-uterine crowding in pigs. J. Anim. Sci. Biotechnol. 2014;5:55. doi: 10.1186/2049-1891-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manjarin R, Steibel JP, Zamora V, Am-in N, Kirkwood RN, Ernst CW, et al. Transcript abundance of amino acid transporters, β-casein, and α-lactalbumin in mammary tissue of periparturient, lactating, and postweaned sows. J Dairy Sci. 2011;94:3467–3476. doi: 10.3168/jds.2011-4163. [DOI] [PubMed] [Google Scholar]
- 35.Rezaei R, Wu Z, Hou Y, Bazer FW, Wu G. Amino acids and mammary gland development: nutritional implications for milk production and neonatal growth. J Anim Sci Biotechnol. 2016;7:20. doi: 10.1186/s40104-016-0078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao H, Hu H, Zheng N, Wang J. Leucine and histidine independently regulate milk protein synthesis in bovine mammary epithelial cells via mTOR signaling pathway. J Zhejiang Univ Sci B. 2015;16:560–572. doi: 10.1631/jzus.B1400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krogh U, Oksbjerg N, Storm AC, Feyera T, Theil PK. Mammary nutrient uptake in multiparous sows fed supplementary arginine during gestation and lactation. J Anim Sci. 2017;95:2517–2532. doi: 10.2527/jas.2016.1291. [DOI] [PubMed] [Google Scholar]
- 38.Shennan DB, Calvert DT, Travers MT, Kudo Y, Boyd CA. A study of L-leucine, L-phenylalanine and L-alanine transport in the perfused rat mammary gland: possible involvement of LAT1 and LAT2. Biochim Biophys Acta. 2002;1564:133–139. doi: 10.1016/s0005-2736(02)00410-8. [DOI] [PubMed] [Google Scholar]
- 39.Messer M, Elliott C. Changes in alpha-lactalbumin, total lactose, UDP-galactose hydrolase and other factors in tammar wallaby (Macropus eugenii) milk during lactation. Aust J Biol Sci. 1987;40:37–46. [PubMed] [Google Scholar]
- 40.Robinson GW, McKnight RA, Smith GH, Hennighausen L. Mammary epithelial cells undergo secretory differentiation in cycling virgins but require pregnancy for the establishment of terminal differentiation. Development. 1995;121:2079–2090. doi: 10.1242/dev.121.7.2079. [DOI] [PubMed] [Google Scholar]
- 41.Theil PK, Labouriau R, Sejrsen K, Thomsen B, Sørensen MT. Expression of genes involved in regulation of cell turnover during milk stasis and lactation rescue in sow mammary glands. J Anim Sci. 2005;83:2349–2356. doi: 10.2527/2005.83102349x. [DOI] [PubMed] [Google Scholar]
- 42.Theil PK, Sejrsen K, Hurley WL, Labouriau R, Thomsen B, Sørensen MT. Role of suckling in regulating cell turnover and onset and maintenance of lactation in individual mammary glands of sows. J Anim Sci. 2006;84:1691–1698. doi: 10.2527/jas.2005-518. [DOI] [PubMed] [Google Scholar]
- 43.Mohammad MA, Hadsell DL, Haymond MW. Gene regulation of UDP-galactose synthesis and transport: potential rate-limiting processes in initiation of milk production in humans. Am J Physiol Endocrinol Metab. 2012;303:E365–E376. doi: 10.1152/ajpendo.00175.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaselonis GL, McCabe ER, Gray SM. Expression of hexokinase 1 and hexokinase 2 in mammary tissue of nonlactating and lactating rats: evaluation by RT-PCR. Mol Genet Metab. 1999;68:371–374. doi: 10.1006/mgme.1999.2923. [DOI] [PubMed] [Google Scholar]
- 45.Wheeler TT, Hodgkinson AJ, Prosser CG, Davis SR. Immune components of colostrum and milk--a historical perspective. J Mammary Gland Biol Neoplasia. 2007;12:237–247. doi: 10.1007/s10911-007-9051-7. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez L, Lujan L, Oria R, Castillo H, Perez D, Ena JM, et al. Synthesis of lactoferrin and transport of transferrin in the lactating mammary gland of sheep. J Dairy Sci. 1992;75:1257–1262. doi: 10.3168/jds.S0022-0302(92)77875-8. [DOI] [PubMed] [Google Scholar]
- 47.Kruse PE. The importance of colostral immunoglobulins and their absorption from the intestine of the newborn animals. Ann Rech Veterinaires Ann Vet Res. 1983;14:349–353. [PubMed] [Google Scholar]
- 48.Hurley WL, Theil PK. Immunoglobulins in mammary secretions. Adv Dairy Chem. 2013;1:275–294. [Google Scholar]
- 49.Larson BL, Heary HL, Devery JE. Immunoglobulin production and transport by the mammary gland. J Dairy Sci. 1980;63:665–671. doi: 10.3168/jds.S0022-0302(80)82988-2. [DOI] [PubMed] [Google Scholar]
- 50.Bourne FJ, Curtis J. The transfer of immunoglobulins IgG, IgA and IgM from serum to colostrum and milk in the sow. Immunology. 1973;24:157. [PMC free article] [PubMed] [Google Scholar]
- 51.Mayer B, Doleschall M, Bender B, Bartyik J, Bosze Z, Frenyó LV, et al. Expression of the neonatal fc receptor (FcRn) in the bovine mammary gland. J Dairy Res. 2005;72 Spec No:107–112. [DOI] [PubMed]
- 52.Mayer B, Zolnai A, Frenyó LV, Jancsik V, Szentirmay Z, Hammarström L, et al. Localization of the sheep FcRn in the mammary gland. Vet Immunol Immunopathol. 2002;87:327–330. doi: 10.1016/s0165-2427(02)00059-4. [DOI] [PubMed] [Google Scholar]
- 53.Lu W, Zhao Z, Zhao Y, Yu S, Zhao Y, Fan B, et al. Over-expression of the bovine FcRn in the mammary gland results in increased IgG levels in both milk and serum of transgenic mice. Immunology. 2007;122:401–408. doi: 10.1111/j.1365-2567.2007.02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrington GM, Besser TE, Gay CC, Davis WC, Reeves JJ, McFadden TB, et al. Regulation of the immunoglobulin G1 receptor: effect of prolactin on in vivo expression of the bovine mammary immunoglobulin G1 receptor. J Endocrinol. 1999;163:25–31. doi: 10.1677/joe.0.1630025. [DOI] [PubMed] [Google Scholar]
- 55.Hurley WL, Theil PK. Perspectives on immunoglobulins in colostrum and milk. Nutrients. 2011;3:442–474. doi: 10.3390/nu3040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lai I-H, Tsao JH, Lu YP, Lee JW, Zhao X, Chien FL, et al. Neutrophils as one of the major haptoglobin sources in mastitis affected milk. Vet Res. 2009;40:17. doi: 10.1051/vetres:2008055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hiss-Pesch S, Daniel F, Dunkelberg-Denk S, Mielenz M, Sauerwein H. Transfer of maternal haptoglobin to suckling piglets. Vet Immunol Immunopathol. 2011;144:104–110. doi: 10.1016/j.vetimm.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 58.Rainard P, Riollet C, Berthon P, Cunha P, Fromageau A, Rossignol C, et al. The chemokine CXCL3 is responsible for the constitutive chemotactic activity of bovine milk for neutrophils. Mol Immunol. 2008;45:4020–4027. doi: 10.1016/j.molimm.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 59.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 60.Zhang L, Boeren S, Hageman JA, van Hooijdonk T, Vervoort J, Hettinga K. Bovine milk proteome in the first 9 days: protein interactions in maturation of the immune and digestive system of the newborn. PLoS One. 2015;10:e0116710. doi: 10.1371/journal.pone.0116710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janeway CA Jr, Travers P, Walport M, Shlomchik MJ. The complement system and innate immunity. N. Y. Garland Sci. 2001;Available from: https://www.ncbi.nlm.nih.gov/books/NBK27100/.
- 62.Goldman AS. Human milk, leukocytes, and immunity. J Pediatr. 1977;90:167–168. doi: 10.1016/s0022-3476(77)80805-6. [DOI] [PubMed] [Google Scholar]
- 63.Trégoat V, Montagne P, Cuillière M-L, Béné M-C, Faure G. C3/C4 concentration ratio reverses between colostrum and mature milk in human lactation. J Clin Immunol. 1999;19:300–304. doi: 10.1023/a:1020591508444. [DOI] [PubMed] [Google Scholar]
- 64.Cerveza PJ, Mehrbod F, Cotton SJ, Lomeli N, Linder MC, Fonda EG, et al. Milk ceruloplasmin and its expression by mammary gland and liver in pigs. Arch Biochem Biophys. 2000;373:451–461. doi: 10.1006/abbi.1999.1572. [DOI] [PubMed] [Google Scholar]
- 65.Wagstrom EA, Yoon KJ, Zimmerman JJ. Immune components in porcine mammary secretions. Viral Immunol. 2000;13:383–397. doi: 10.1089/08828240050144699. [DOI] [PubMed] [Google Scholar]
- 66.Krakowski L, Krzyzanowski J, Wrona Z, Kostro K, Siwicki AK. The influence of nonspecific immunostimulation of pregnant sows on the immunological value of colostrum. Vet Immunol Immunopathol. 2002;87:89–95. doi: 10.1016/s0165-2427(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 67.Chandan RC, Parry RM, Shahani KM. Lysozyme, lipase, and ribonuclease in milk of various species. J Dairy Sci. 1968;51:606–607. [Google Scholar]
- 68.Nguyen TV, Yuan L, Azevedo MSP, Jeong K, Gonzalez A-M, Saif LJ. Transfer of maternal cytokines to suckling piglets: in vivo and in vitro models with implications for immunomodulation of neonatal immunity. Vet Immunol Immunopathol. 2007;117:236. doi: 10.1016/j.vetimm.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Poll T, Herwald H. The coagulation system and its function in early immune defense. Thromb Haemost. 2014;112:640–648. doi: 10.1160/TH14-01-0053. [DOI] [PubMed] [Google Scholar]
- 70.de Jesus Rodriguez B, Chevaleyre C, Henry G, Mollé D, Virlogeux-Payant I, Berri M, et al. Identification in milk of a serum amyloid a peptide chemoattractant for B lymphoblasts. BMC Immunol. 2009;10:4. doi: 10.1186/1471-2172-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J-W, Paape MJ, Elsasser TH, Zhao X. Elevated milk soluble CD14 in bovine mammary glands challenged with Escherichia coli lipopolysaccharide. J Dairy Sci. 2003;86:2382–2389. doi: 10.3168/jds.S0022-0302(03)73832-6. [DOI] [PubMed] [Google Scholar]
- 72.Filipp D, Alizadeh-Khiavi K, Richardson C, Palma A, Paredes N, Takeuchi O, et al. Soluble CD14 enriched in colostrum and milk induces B cell growth and differentiation. Proc Natl Acad Sci U S A. 2001;98:603. doi: 10.1073/pnas.98.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dudemaine PL, Thibault C, Alain K, Bissonnette N. Genetic variations in the SPP1 promoter affect gene expression and the level of osteopontin secretion into bovine milk. Anim Genet. 2014;45:629–640. doi: 10.1111/age.12176. [DOI] [PubMed] [Google Scholar]
- 74.Sheehy PA, Riley LG, Raadsma HW, Williamson P, Wynn PC. A functional genomics approach to evaluate candidate genes located in a QTL interval for milk production traits on BTA6. Anim Genet. 2009;40:492–498. doi: 10.1111/j.1365-2052.2009.01862.x. [DOI] [PubMed] [Google Scholar]
- 75.Nemir M, Bhattacharyya D, Li X, Singh K, Mukherjee AB, Mukherjee BB. Targeted inhibition of osteopontin expression in the mammary gland causes abnormal morphogenesis and lactation deficiency. J Biol Chem. 2000;275:969–976. doi: 10.1074/jbc.275.2.969. [DOI] [PubMed] [Google Scholar]
- 76.Alain K, Karrow NA, Thibault C, St-Pierre J, Lessard M, Bissonnette N. Osteopontin: an early innate immune marker of Escherichia coli mastitis harbors genetic polymorphisms with possible links with resistance to mastitis. BMC Genomics. 2009;10:444. doi: 10.1186/1471-2164-10-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He Y, Lawlor NT, Newburg DS. Human milk components modulate toll-like receptor–mediated inflammation. Adv Nutr. 2016;7:102. doi: 10.3945/an.115.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rainard P, Riollet C. Innate immunity of the bovine mammary gland. Vet Res. 2006;37:369–400. doi: 10.1051/vetres:2006007. [DOI] [PubMed] [Google Scholar]
- 79.Stelwagen K, Carpenter E, Haigh B, Hodgkinson A, Wheeler TT. Immune components of bovine colostrum and milk. J Anim Sci. 2009;87:3–9. doi: 10.2527/jas.2008-1377. [DOI] [PubMed] [Google Scholar]
- 80.LeBouder E, Rey-Nores JE, Rushmere NK, Grigorov M, Lawn SD, Affolter M, et al. Soluble forms of toll-like receptor (TLR)2 capable of modulating TLR2 signaling are present in human plasma and breast milk. J Immunol. 2003;171:6680–6689. doi: 10.4049/jimmunol.171.12.6680. [DOI] [PubMed] [Google Scholar]
- 81.Vesy CJ, Kitchens RL, Wolfbauer G, Albers JJ, Munford RS. Lipopolysaccharide-binding protein and phospholipid transfer protein release lipopolysaccharides from gram-negative bacterial membranes. Infect Immun. 2000;68:2410–2417. doi: 10.1128/iai.68.5.2410-2417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy M-A, Heath VJ, et al. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004;6:R75–R91. doi: 10.1186/bcr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nissen A, Bendixen E, Ingvartsen KL, Røntved CM. In-depth analysis of low abundant proteins in bovine colostrum using different fractionation techniques. Proteomics. 2012;12:2866–2878. doi: 10.1002/pmic.201200231. [DOI] [PubMed] [Google Scholar]
- 84.Rosenbaum S, Ringseis R, Hillen S, Becker S, Erhardt G, Reiner G, Eder K. The stress signalling pathway nuclear factor E2-related factor 2 is activated in the liver of sows during lactation. Acta Vet Scand. 2012;54:59. doi: 10.1186/1751-0147-54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 86.Tsukamoto H, Fukudome K, Takao S, Tsuneyoshi N, Kimoto M. Lipopolysaccharide-binding protein-mediated toll-like receptor 4 dimerization enables rapid signal transduction against lipopolysaccharide stimulation on membrane-associated CD14-expressing cells. Int Immunol. 2010;22:271–280. doi: 10.1093/intimm/dxq005. [DOI] [PubMed] [Google Scholar]
- 87.Xyni K, Rizos D, Giannaki G, Sarandakou A, Phocas I, Creatsas G. Soluble form of ICAM-1, VCAM-1, E- and L-selectin in human milk. Mediat Inflamm. 2000;9:133–140. doi: 10.1080/09629350020003858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mohammad MA, Haymond MW. Regulation of lipid synthesis genes and milk fat production in human mammary epithelial cells during secretory activation. Am J Physiol Endocrinol Metab. 2013;305:E700–E716. doi: 10.1152/ajpendo.00052.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han LQ, Li HJ, Wang YY, Zhu HS, Wang LF, Guo YJ, et al. mRNA abundance and expression of SLC27A, ACC, SCD, FADS, LPIN, INSIG, and PPARGC1 gene isoforms in mouse mammary glands during the lactation cycle. Genet Mol Res GMR. 2010;9:1250–1257. doi: 10.4238/vol9-2gmr814. [DOI] [PubMed] [Google Scholar]
- 90.Abu-Elheiga L, Brinkley WR, Zhong L, Chirala SS, Woldegiorgis G, Wakil SJ. The subcellular localization of acetyl-CoA carboxylase 2. Proc Natl Acad Sci U S A. 2000;97:1444–1449. doi: 10.1073/pnas.97.4.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics. 2008;9:366. doi: 10.1186/1471-2164-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kinsella JE. Stearyl CoA as a precursor of oleic acid and glycerolipids in mammary microsomes from lactating bovine: possible regulatory step in milk triglyceride synthesis. Lipids. 1972;7:349–355. doi: 10.1007/BF02532654. [DOI] [PubMed] [Google Scholar]
- 93.Xie L, Innis SM. Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n-6) and (n-3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. J Nutr. 2008;138:2222–2228. doi: 10.3945/jn.108.096156. [DOI] [PubMed] [Google Scholar]
- 94.Rodriguez-Cruz M, Tovar AR, Palacios-González B, Del Prado M, Torres N. Synthesis of long-chain polyunsaturated fatty acids in lactating mammary gland: role of Delta5 and Delta6 desaturases, SREBP-1, PPARalpha, and PGC-1. J Lipid Res. 2006;47:553–560. doi: 10.1194/jlr.M500407-JLR200. [DOI] [PubMed] [Google Scholar]
- 95.Rodriguez-Cruz M, Sánchez R, Sánchez AM, Kelleher SL, Sánchez-Muñoz F, Maldonado J, et al. Participation of mammary gland in long-chain polyunsaturated fatty acid synthesis during pregnancy and lactation in rats. Biochim Biophys Acta. 1811;2011:284–293. doi: 10.1016/j.bbalip.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 96.Lv Y, Guan W, Qiao H, Wang C, Chen F, Zhang Y, et al. Veterinary medicine and omics (Veterinomics): metabolic transition of milk triacylglycerol synthesis in sows from late pregnancy to lactation. OMICS. 2015;19:602–616. doi: 10.1089/omi.2015.0102. [DOI] [PubMed] [Google Scholar]
- 97.Gonzalez-Baró MR, Lewin TM, Coleman RA. Regulation of triglyceride metabolism II. Function of mitochondrial GPAT1 in the regulation of triacylglycerol biosynthesis and insulin action. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1195. doi: 10.1152/ajpgi.00553.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.West CE, Bickerstaffe R, Annison EF, Linzell JL. Studies on the mode of uptake of blood triglycerides by the mammary gland of the lactating goat. The uptake and incorporation into milk fat and mammary lymph of labelled glycerol, fatty acids and triglycerides. Biochem J. 1972;126:477–490. doi: 10.1042/bj1260477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin CY, Abraham S, Smith S. Acyl specificity in triglyceride synthesis by lactating rat mammary gland. J Lipid Res. 1976;17:647–656. [PubMed] [Google Scholar]
- 100.Osorio JS, Lohakare J, Bionaz M. Biosynthesis of milk fat, protein, and lactose: roles of transcriptional and posttranscriptional regulation. Physiol Genomics. 2016;48:231–256. doi: 10.1152/physiolgenomics.00016.2015. [DOI] [PubMed] [Google Scholar]
- 101.Doege H, Stahl A. Protein-mediated fatty acid uptake: novel insights from in vivo models. Physiol Bethesda. 2006;21:259–268. doi: 10.1152/physiol.00014.2006. [DOI] [PubMed] [Google Scholar]
- 102.Bionaz M, Loor JJ. ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J Nutr. 2008;138:1019–1024. doi: 10.1093/jn/138.6.1019. [DOI] [PubMed] [Google Scholar]
- 103.Fujino T, Kang MJ, Suzuki H, Iijima H, Yamamoto T. Molecular characterization and expression of rat acyl-CoA synthetase 3. J Biol Chem. 1996;271:16748–16752. doi: 10.1074/jbc.271.28.16748. [DOI] [PubMed] [Google Scholar]
- 104.Takeuchi K, Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am J Physiol Endocrinol Metab. 2009;296:E1195–E1209. doi: 10.1152/ajpendo.90958.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nagle CA, Vergnes L, DeJong H, Wang S, Lewin TM, Reue K, et al. Identification of a novel sn-glycerol-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6−/− mice. J Lipid Res. 2008;49:823. doi: 10.1194/jlr.M700592-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leung DW. The structure and functions of human lysophosphatidic acid acyltransferases. Front Biosci. 2001;6:D944–D953. doi: 10.2741/leung. [DOI] [PubMed] [Google Scholar]
- 107.Yamashita A, Kawagishi N, Miyashita T, Nagatsuka T, Sugiura T, Kume K, et al. ATP-independent fatty acyl-coenzyme a synthesis from phospholipid: coenzyme A-dependent transacylation activity toward lysophosphatidic acid catalyzed by acyl-coenzyme a:lysophosphatidic acid acyltransferase. J Biol Chem. 2001;276:26745–26752. doi: 10.1074/jbc.M101795200. [DOI] [PubMed] [Google Scholar]
- 108.Tomàs A, Estellé J, Clop A, Gómez-Raya L, Noguera JL, Sànchez A, et al. Assignment of the mitochondrial glycerol-3-phosphate acyltransferase (GPAT) gene to porcine chromosome 14. Anim Genet. 2003;34:387. doi: 10.1046/j.1365-2052.2003.01041.x. [DOI] [PubMed] [Google Scholar]
- 109.Grisart B, Farnir F, Karim L, Cambisano N, Kim J-J, Kvasz A, et al. Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition. Proc Natl Acad Sci U S A. 2004;101:2398–2403. doi: 10.1073/pnas.0308518100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 111.Csaki LS, Dwyer JR, Li X, Nguyen MHK, Dewald J, Brindley DN, et al. Lipin-1 and lipin-3 together determine adiposity in vivo. Mol Metab. 2014;3:145–154. doi: 10.1016/j.molmet.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dwyer JR, Donkor J, Zhang P, Csaki LS, Vergnes L, Lee JM, et al. Mouse lipin-1 and lipin-2 cooperate to maintain glycerolipid homeostasis in liver and aging cerebellum. Proc Natl Acad Sci U S A. 2012;109:E2486–E2495. doi: 10.1073/pnas.1205221109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Keenan TW, Mather IH. Intracellular origin of milk fat globules and the nature of the milk fat globule membrane. Adv. Dairy Chem. 2006;2 Lipids:137–171.
- 114.Murakami N, Ohtsubo T, Kansui Y, Goto K, Noguchi H, Haga Y, et al. Mice heterozygous for the xanthine oxidoreductase gene facilitate lipid accumulation in adipocytes. Arterioscler Thromb Vasc Biol. 2014;34:44–51. doi: 10.1161/ATVBAHA.113.302214. [DOI] [PubMed] [Google Scholar]
- 115.Tansey JT, Sztalryd C, Hlavin EM, Kimmel AR, Londos C. The central role of perilipin a in lipid metabolism and adipocyte lipolysis. IUBMB Life. 2004;56:379–385. doi: 10.1080/15216540400009968. [DOI] [PubMed] [Google Scholar]
- 116.Kessler EC, Gross JJ, Bruckmaier RM, Albrecht C. Cholesterol metabolism, transport, and hepatic regulation in dairy cows during transition and early lactation. J Dairy Sci. 2014;97:5481–5490. doi: 10.3168/jds.2014-7926. [DOI] [PubMed] [Google Scholar]
- 117.Rikitake Y, Kawashima S, Takeshita S, Yamashita T, Azumi H, Yasuhara M, et al. Anti-oxidative properties of fluvastatin, an HMG-CoA reductase inhibitor, contribute to prevention of atherosclerosis in cholesterol-fed rabbits. Atherosclerosis. 2001;154:87–96. doi: 10.1016/s0021-9150(00)00468-8. [DOI] [PubMed] [Google Scholar]
- 118.Robinson AM, Williamson DH. Physiological roles of ketone bodies as substrates and signals in mammalian tissues. Physiol Rev. 1980;60:143–187. doi: 10.1152/physrev.1980.60.1.143. [DOI] [PubMed] [Google Scholar]
- 119.Yang W-C, Tsai W-C, Lin P-M, Yang M-Y, Liu Y-C, Chang C-S, et al. Human BDH2, an anti-apoptosis factor, is a novel poor prognostic factor for de novo cytogenetically normal acute myeloid leukemia. J Biomed Sci. 2013;20:58. doi: 10.1186/1423-0127-20-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Palmquist DL, Davis CL, Brown RE, Sachan DS. Availability and metabolism of various substrates in ruminants. V. Entry rate into the body and incorporation into milk fat of d(−)β-hydroxybutyrate1. J Dairy Sci. 1969;52:633–638. [Google Scholar]
- 121.Jazwinski SM, Conzelmann A. LAG1 puts the focus on ceramide signaling. Int J Biochem Cell Biol. 2002;34:1491–1495. doi: 10.1016/s1357-2725(02)00044-4. [DOI] [PubMed] [Google Scholar]
- 122.Palmquist DL. Milk Ffat: origin of fatty acids and influence of nutritional factors thereon. Adv Dairy Chem. 2006;2 Lipids:43–92.
- 123.Lundell K, Hansson R, Wikvall K. Cloning and expression of a pig liver taurochenodeoxycholic acid 6alpha-hydroxylase (CYP4A21): a novel member of the CYP4A subfamily. J Biol Chem. 2001;276:9606–9612. doi: 10.1074/jbc.M006584200. [DOI] [PubMed] [Google Scholar]
- 124.Simpson AE. The cytochrome P450 4 (CYP4) family. Gen Pharmacol. 1997;28:351–359. doi: 10.1016/s0306-3623(96)00246-7. [DOI] [PubMed] [Google Scholar]
- 125.Capdevila JH, Holla V, Helvig C, Falck JR. Microsomal cytochrome P450 and eicosanoid metabolism. Mol Asp Med. 1999;20:42–55. [PubMed] [Google Scholar]
- 126.Omura T. Forty years of cytochrome P450. Biochem Biophys Res Commun. 1999;266:690–698. doi: 10.1006/bbrc.1999.1887. [DOI] [PubMed] [Google Scholar]
- 127.Lundell K. The porcine taurochenodeoxycholic acid 6alpha-hydroxylase (CYP4A21) gene: evolution by gene duplication and gene conversion. Biochem J. 2004;378:1053. doi: 10.1042/BJ20031657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Laliotis GP, Bizelis I, Rogdakis E. Comparative approach of the de novo fatty acid synthesis (lipogenesis) between ruminant and non ruminant mammalian species: from biochemical level to the main regulatory lipogenic genes. Curr Genomics. 2010;11:168. doi: 10.2174/138920210791110960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Anderson SM, Rudolph MC, McManaman JL, Neville MC. Key stages in mammary gland development. Secretory activation in the mammary gland: it’s not just about milk protein synthesis! Breast Cancer Res. 2007;9:204. doi: 10.1186/bcr1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Davis KR, Giesy SL, Long Q, Krumm CS, Harvatine KJ, Boisclair YR. XBP1 regulates the biosynthetic capacity of the mammary gland during lactation by controlling epithelial expansion and endoplasmic reticulum formation. Endocrinology. 2016;157:417–428. doi: 10.1210/en.2015-1676. [DOI] [PubMed] [Google Scholar]
- 131.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 132.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 133.Rudolph MC, McManaman JL, Phang T, Russell T, Kominsky DJ, Serkova NJ, et al. Metabolic regulation in the lactating mammary gland: a lipid synthesizing machine. Physiol Genomics. 2007;28:323–336. doi: 10.1152/physiolgenomics.00020.2006. [DOI] [PubMed] [Google Scholar]
- 134.Engelking LJ, Kuriyama H, Hammer RE, Horton JD, Brown MS, Goldstein JL, et al. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J Clin Invest. 2004;113:1168–1175. doi: 10.1172/JCI20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shi H, Zhao W, Zhang C, Shahzad K, Luo J, Loor JJ. Transcriptome-wide analysis reveals the role of PPARγ controlling the lipid metabolism in goat mammary epithelial cells. PPAR Res. 2016;2016:9195680. doi: 10.1155/2016/9195680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shi H, Luo J, Zhu J, Li J, Sun Y, Lin X, et al. PPARγ regulates genes involved in triacylglycerol synthesis and secretion in mammary gland epithelial cells of dairy goats. PPAR Res. 2013;2013:310948. doi: 10.1155/2013/310948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kadegowda AKG, Bionaz M, Piperova LS, Erdman RA, Loor JJ. Peroxisome proliferator-activated receptor-γ activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J Dairy Sci. 2009;92:4276–4289. doi: 10.3168/jds.2008-1932. [DOI] [PubMed] [Google Scholar]
- 138.Akers RM, Bauman DE, Goodman GT, Capuco AV, Tucker HA. Prolactin regulation of cytological differentiation of mammary epithelial cells in periparturient cows. Endocrinology. 1981;109:31–40. doi: 10.1210/endo-109-1-31. [DOI] [PubMed] [Google Scholar]
- 139.Fagone P, Jackowski S. Membrane phospholipid synthesis and endoplasmic reticulum function. J Lipid Res. 2009;50 Suppl:S311–S316. [DOI] [PMC free article] [PubMed]
- 140.Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, et al. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem. 2007;282:7024–7034. doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- 141.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 142.Moore KA, Hollien J. The unfolded protein response in secretory cell function. Annu Rev Genet. 2012;46:165–183. doi: 10.1146/annurev-genet-110711-155644. [DOI] [PubMed] [Google Scholar]
- 143.Lee A-H, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang S, Chen Z, Lam V, Han J, Hassler J, Finck BN, et al. IRE1α-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab. 2012;16:473–486. doi: 10.1016/j.cmet.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim RS, Hasegawa D, Goossens N, Tsuchida T, Athwal V, Sun X, et al. The XBP1 arm of the unfolded protein response induces fibrogenic activity in hepatic stellate cells through autophagy. Sci Rep. 2016;6:39342. doi: 10.1038/srep39342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ghosal D, Shappell NW, Keenan TW. Endoplasmic reticulum lumenal proteins of rat mammary gland. Potential involvement in lipid droplet assembly during lactation. Biochim Biophys Acta. 1994;1200:175–181. doi: 10.1016/0304-4165(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 147.Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shaw PH. The role of p53 in cell cycle regulation. Pathol Res Pract. 1996;192:669–675. doi: 10.1016/S0344-0338(96)80088-4. [DOI] [PubMed] [Google Scholar]
- 149.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 150.Munne PM, Gu Y, Tumiati M, Gao P, Koopal S, Uusivirta S, et al. TP53 supports basal-like differentiation of mammary epithelial cells by preventing translocation of deltaNp63 into nucleoli. Sci Rep. 2014;4:4663. doi: 10.1038/srep04663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sambasivan R, Cheedipudi S, Pasupuleti N, Saleh A, Pavlath GK, Dhawan J. The small chromatin-binding protein p8 coordinates the association of anti-proliferative and pro-myogenic proteins at the myogenin promoter. J Cell Sci. 2009;122:3481–3491. doi: 10.1242/jcs.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhou Y, Gong W, Xiao J, Wu J, Pan L, Li X, et al. Transcriptomic analysis reveals key regulators of mammogenesis and the pregnancy-lactation cycle. Sci China Life Sci. 2014;57:340–355. doi: 10.1007/s11427-013-4579-9. [DOI] [PubMed] [Google Scholar]
- 153.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 154.Kuklina EM, Shirshev SV. Role of transcription factor NFAT in the immune response. Biochem Biokhimiia. 2001;66:467–475. doi: 10.1023/a:1010238931555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Provides an aggregate summary of RNA extraction and quality check for all samples. Mammary tissue was collected on days 14, 10, 6 and 2 before (−) parturition and on day 1 after (+) parturition from three 2nd parity sows. Extraction of RNA and quality evaluation was performed following protocols described by Tramontana et al. (2008) [15]. (XLSX 10 kb)
Provides an aggregate summary of RNA sequencing and alignment for all the samples. mRNA libraries were sequenced on a HiSeq2500 (Illumina Inc.). Quality control metrics were performed on raw sequencing reads using the FASTQC v0.11.15 application. An index of the reference genome was built and single-end clean reads for each individual were aligned to the reference genome by STAR (v2.5.1b). Reads were mapped and annotated to the Sus scrofa genome (v10.2.86), downloaded from EnsemblGenome website (Nov. 2016). Reads aligned were quantified with Subread package (v1.5.0) based on the Refseq gene annotation. (XLSX 10 kb)
Provides the complete list of differentially expressed genes (DEG) resulting from differential expression (DE) analysis of RNAseq data of swine mammary gland tissue harvested in late-pregnancy through farrowing and used for Dynamic Impact Approach (DIA) and Ingenuity Pathway Analysis (IPA) analyses. Moreover, it provides additional discussion points, omitted from the main body of the manuscript. (XLSX 1239 kb)
Provides complete list of upstream regulator resulting from Ingenuity Pathway Analysis (IPA) analysis. Moreover, it provides additional discussion points, omitted from the main body of the manuscript. (XLSX 20 kb)
Data Availability Statement
The data have been submitted to Gene Expression Omnibus database (BioProject ID: GSE101983).


