Abstract
Background
Glycerol, an inevitable byproduct of biodiesel, has become an attractive feedstock for the production of value-added chemicals due to its availability and low price. Pseudomonas chlororaphis HT66 can use glycerol to synthesize phenazine-1-carboxamide (PCN), a phenazine derivative, which is strongly antagonistic to fungal phytopathogens. A systematic understanding of underlying mechanisms for the PCN overproduction will be important for the further improvement and industrialization.
Results
We constructed a PCN-overproducing strain (HT66LSP) through knocking out three negative regulatory genes, lon, parS, and prsA in HT66. The strain HT66LSP produced 4.10 g/L of PCN with a yield of 0.23 (g/g) from glycerol, which was of the highest titer and the yield obtained among PCN-producing strains. We studied gene expression, metabolomics, and dynamic 13C tracer in HT66 and HT66LSP. In response to the phenotype changes, the transcript levels of phz biosynthetic genes, which are responsible for PCN biosynthesis, were all upregulated in HT66LSP. Central carbon was rerouted to the shikimate pathway, which was shown by the modulation of specific genes involved in the lower glycolysis, the TCA cycle, and the shikimate pathway, as well as changes in abundances of intracellular metabolites and flux distribution to increase the precursor availability for PCN biosynthesis. Moreover, dynamic 13C-labeling experiments revealed that the presence of metabolite channeling of 3-phosphoglyceric acid to phosphoenolpyruvate and shikimate to trans-2,3-dihydro-3-hydroxyanthranilic acid in HT66LSP could enable high-yielding synthesis of PCN.
Conclusions
The integrated analysis of gene expression, metabolomics, and dynamic 13C tracer enabled us to gain a more in-depth insight into complex mechanisms for the PCN overproduction. This study provides important basis for further engineering P. chlororaphis for high PCN production and efficient glycerol conversion.
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1123-y) contains supplementary material, which is available to authorized users.
Keywords: Pseudomonas chlororaphis, Phenazine-1-carboxamide, Glycerol, Metabolomics, 13C, Channeling
Background
In recent years, the fast development of biofuel and bioethanol industries has led to the production of large quantities of waste glycerol [1]. Significant amounts of glycerol surplus have given rise to a sharp drop in the glycerol price, making it a promising industrial carbon source for the production of value-added chemicals [2, 3]. Glycerol can be used to synthesize phenazine-1-carboxamide (PCN), a phenazine derivative, by Pseudomonas strains [4–6]. Phenazines are heterocyclic, nitrogen-containing compounds, which have attracted attention due to their broad spectrum antibiotic properties and roles in virulence, and been widely used in the biological control of various fungal phytopathogens [7]. PCN is one of the important molecules among phenazine compounds [8]. Figure 1 shows the metabolic pathways for PCN biosynthesis in Pseudomonas chlororaphis grown on glycerol. The gene cluster phzABCDEFGH is responsible for PCN biosynthesis [8, 9]. The production of PCN is regulated by different groups of genes: the phzI/phzR quorum-sensing system [10], the gacS/gacA regulatory genes [11], the parS/parR regulatory genes [12], and the psrA gene [13]. PhzI produces the autoinducer N-hexanoyl-l-homoserine lactone (C6-HSL), which binds to PhzR. Subsequently, the PhzR-C6-HSL complex probably binds to the upstream of phz operon, resulting in the initiation of this operon [10, 14]. The GacS/GacA two-component system is a master regulator of secondary metabolism, stimulates the production of phenazines in Pseudomonas chlororaphis and other Pseudomonas species [15, 16]. In contrast to PhzI/PhzR and GacS/GacA, ParS/ParR and PsrA act as repressors for the production of PCN [13]. Modifying these regulatory genes led to the improvements of the PCN titer and yield [10, 11, 13]. However, until now, underlying mechanisms for the PCN overproduction still remain obscure, which have been the bottleneck for the further improvement and industrialization. To fully understand the mechanisms, it requires further quantifying and integrating the metabolic function, regulation, and the physiological parameters at a systematic level.
Fig. 1.
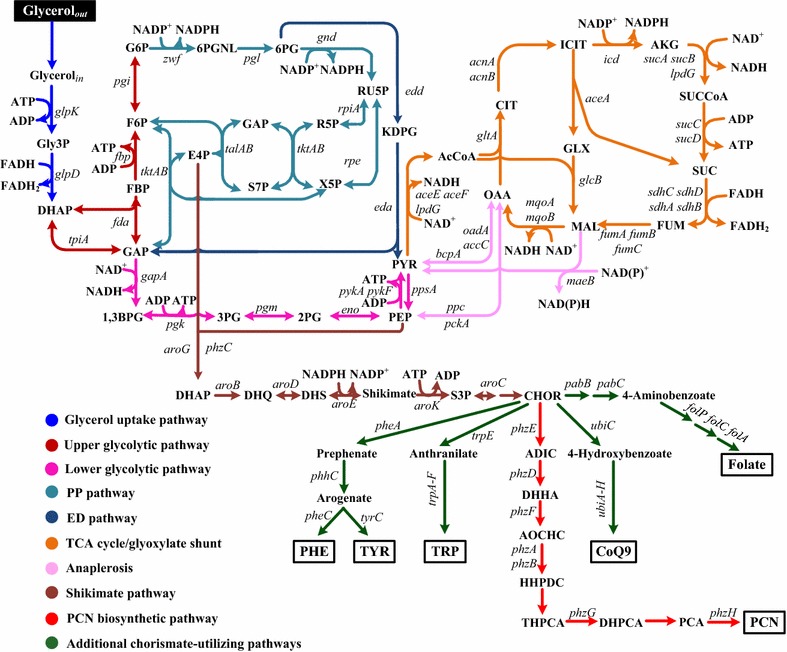
Metabolic pathways for PCN biosynthesis in P. chlororaphis. Broken lines illustrate multiple steps
Metabolomics, comprehensive analysis of a wide range of metabolites, is a broad and sensitive method to detect differences in metabolic states between conditions [17]. Combining metabolite levels with dynamic 13C-labeling is a powerful tool to peak inside the black box of the cellular metabolism, even in the absence of quantitative flux analysis [18]. It can provide useful information on the relative pathway activities and qualitative changes in pathway contributions, identify bottlenecks for the target product synthesis within the linear pathways, discover the latent pathway interactions, and help infer fluxes [19–21].
In our previous work, Pseudomonas chlororaphis HT66 isolated from the rice rhizosphere produced 0.42 g/L of PCN in King’s B medium [6]. In this study, we generated PCN-overproducing P. chlororaphis starting from HT66 by deleting regulatory genes. To interrogate the metabolism for the PCN overproduction and enable metabolic engineering approach, we studied gene expression, metabolomics, and dynamic 13C tracer in the base strain and the engineered strain. To the best of our knowledge, this is the first study to evaluate metabolic responses to the PCN overproduction on different organizational levels.
Results
Construction of PCN-overproducing strains
The GacS/GacA two-component system is a key element in the Gac/Rsm cascade [22]. This cascade activates the production of phenazine [23], which is negatively regulated by Lon protease [24]. It was reported that the Δlon mutant showed elevated antibiotic production in P. protegens [24]. Thus, the strain HT66L was constructed by the disruption of lon in HT66. Disruption of lon diminished the maximum biomass (Xmax) produced by 19.0% and the specific growth rate (μ) by 27.2%, but did not cause significant changes to the specific glycerol consumption rate (Table 1). As expected, HT66L produced 2.05 g/L of PCN (Fig. 2b), which was 4.9 times of that produced in the base strain (Fig. 2a).
Table 1.
Growth kinetic parameters of PCN-producing P. chlororaphis strains
| Strain | Specific growth rate (h−1) | Specific glycerol consumption rate (mmol/g/h) | Specific PCN production rate (mmol/g/h) | PCN titer (g/L) | YP/G (mol/mol) |
|---|---|---|---|---|---|
| HT66 | 0.11 ± 0.003 | 2.51 ± 0.07 | 0.01 ± 0.0006 | 0.42 ± 0.02 | 0.01 ± 0.001 |
| HT66L | 0.08 ± 0.003 | 2.51 ± 0.07 | 0.10 ± 0.002 | 2.05 ± 0.10 | 0.05 ± 0.002 |
| HT66LS | 0.08 ± 0.003 | 2.52 ± 0.07 | 0.13 ± 0.003 | 2.43 ± 0.12 | 0.05 ± 0.002 |
| HT66LR | 0.08 ± 0.003 | 2.51 ± 0.06 | 0.11 ± 0.002 | 2.22 ± 0.11 | 0.05 ± 0.002 |
| HT66LSP | 0.08 ± 0.003 | 2.51 ± 0.07 | 0.23 ± 0.005 | 4.10 ± 0.21 | 0.09 ± 0.005 |
YP/G PCN yield on glycerol
Fig. 2.
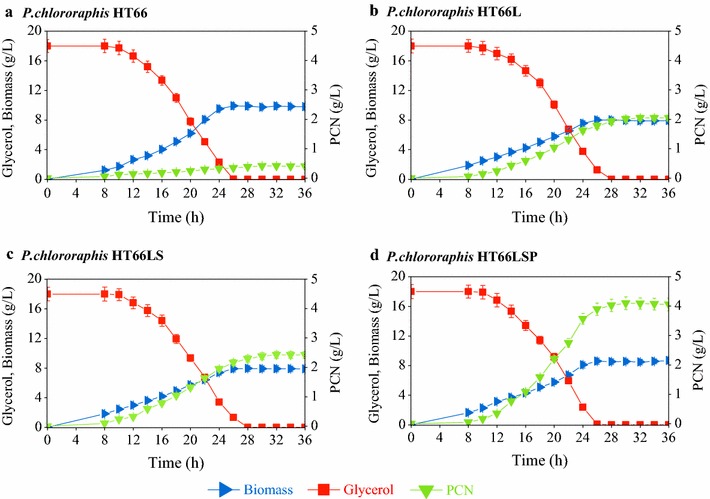
Culture profiles of PCN-producing P. chlororaphis strains HT66 (a), HT66L (b), HT66LS (c), and HT66LSP (d). Data represent the mean ± SD from three independent cultures
The ParS/ParR two-component system, which consists of a membrane-bound histidine sensor kinase (ParS) and a cytoplasmic response regulator (ParR), plays a negative regulatory role in phenazine biosynthesis [12, 25]. Deletion of parS or parR led to increased pyocyanin production [12]. In order to gain more PCN, parS was deleted from HT66L to construct HT66LS, and parR was deleted from HT66L to construct HT66LR. The specific growth rates and the specific glycerol consumption rates in HT66LS and HT66LR were comparable with HT66L (Table 1). The PCN productions in HT66LS and HT66LR were increased to 2.43 and 2.22 g/L (Fig. 2c; Additional file 1), 5.9 and 5.3 times higher than that in HT66 (Fig. 2a). Thus, HT66LS was used for the further studies.
PsrA belongs to the TetR/AcrR family of transcriptional regulators, which inhibits the PCN production in P. chlororaphis PCL1391 in rich growth medium [13]. In order to further enhance the PCN production, the prsA gene was knocked out in HT66LS, and the resulting strain was named HT66LSP. HT66LSP exhibited a 13.1% decrease in the Xmax and a 27.2% decrease in μ compared with HT66 (Table 1). The glycerol consumption rate kept constant. The strain HT66LSP produced 4.10 g/L of PCN at a yield of 0.23 (g/g) from glycerol after 30 h (Fig. 2d; Table 1), which represents, to our knowledge, the highest PCN titer and the yield ever obtained among engineered PCN-producing strains. Meanwhile, the specific PCN production rate was also improved in HT66LSP (Table 1). To determine the metabolic responses to the PCN overproduction, HT66 and HT66LSP were selected for a comparative analysis on different organizational levels.
Relative gene transcription levels
To investigate the transcriptional responses to the PCN overproduction, we analyzed the transcription levels of the HT66LSP genes by qRT-PCR, and compared the data with those of HT66 (Fig. 3). Compared with HT66, the expression levels of glpF, glpK, and glpD in the glycerol utilization pathway, tpiA in glycolysis, and zwf and gnd in the oxidative pentose phosphate (PP) pathway were not significantly altered in HT66LSP. In addition, the transcription levels of pykF and aceE in the lower glycolytic pathway, and gltA, icd, sucA, and sucC in the TCA cycle were downregulated in HT66LSP. Due to the deletion of lon, the transcript levels of gacA and gacS were upregulated in HT66LSP compared with the wild-type strain, confirming its key role for PCN synthesis. In addition, the transcription of autoinducer synthase and transcriptional activator genes phzI and phzR increased in HT66LSP. In accordance with the activation of phzI/phzR, the transcript levels of phz biosynthetic genes were all upregulated in HT66LSP, which could explain the observed high PCN production. The overexpression of phzI/phzR and the phz biosynthetic operon in a psrA mutant of P. chlororaphis PCL1391, and the upregulation of phzA, phzB, and phzH in a parS mutant of P. aeruginosa have been previously reported [12, 13]. These results were consistent with our findings.
Fig. 3.
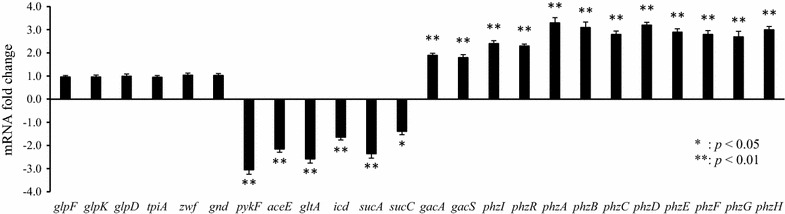
Fold changes of transcription levels of selected genes in P. chlororaphis HT66LSP compared with HT66
Metabolome analysis
To gain a further insight into mechanisms for the PCN overproduction, comparative metabolomics approach was employed to analyze metabolite concentrations difference of HT66 and HT66LSP (Fig. 4). A total of 38 metabolites were determined by isotope-assisted LC–MS, including central carbon metabolites, shikimate pathway metabolites, PCN biosynthetic pathway metabolites, chorismate-utilizing pathways metabolites, amino acids, and cofactors.
Fig. 4.
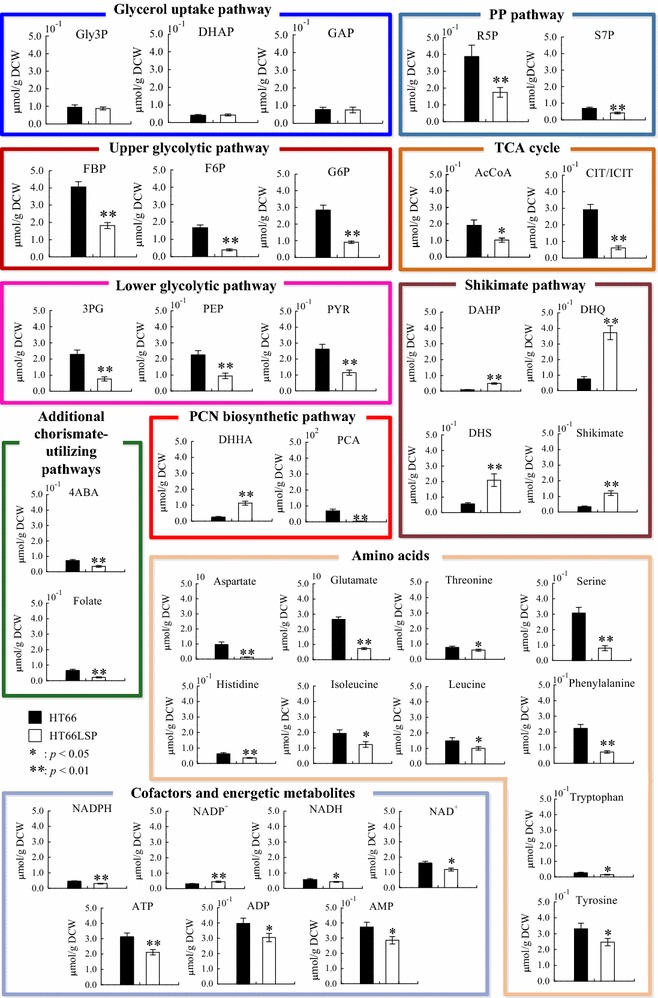
Comparison of intracellular metabolite concentrations of P. chlororaphis strains HT66 and HT66LSP. Data represent the mean ± SD from five independent cultures
In the glycerol uptake pathway, the pool sizes of Gly3P, DHAP, and GAP were nearly constant for the two strains, indicating their pool sizes were strictly controlled. Central carbon metabolites in glycolysis (FBP, F6P, G6P, 3PG, PEP, and PYR), the PP pathway (R5P and S7P), and the TCA cycle (AcCoA and CIT + ICIT) were present in significantly lower concentrations in HT66LSP than in HT66. In addition, the intracellular concentrations of AKG, SUC, and FUM downstream of ICIT were below detection levels. It was reported that a low activity of the enzymes within the reductive TCA cycle and a high activity of the glyoxylate shunt were observed in P. putida KT2440 when grown on glycerol [26], which may explain the poor pool sizes of these three metabolites. The drops in levels of precursors for biosynthesis might be linked to decreased amino acid pools in the central metabolic pathway, leading to the reduced biomass production and μ in HT66LSP.
Shikimate pathway converts the primary carbon metabolites via shikimate to chorismate. DAHP, DHQ, DHS, and shikimate were more abundant in HT66LSP. Chorismate serves mainly as a common precursor for the syntheses of PCN, aromatic amino acids, folate, and co-enzyme Q. DHHA, the last stable intermediate in the pathway leading to PCA, was more abundant in HT66LSP. However, the level of PCA was lower. One of the possible explanations could be that PCA was more rapidly catabolized by PhzH which was transcriptionally upregulated than biosynthesized from DHPCA. The conversion of PCA to PCN was also shown to be essential for the biocontrol activity of strain P. chlororaphis PCL1391 [27]. The levels of other competing pathways metabolites derived from chorismate were lower in HT66LSP, including 4-aminobenzoate, folate, phenylalanine, tryptophan, and tyrosine. In addition, 4-hydroxybenzoate was below the detection level in both strains. These results indicated that manipulations of regulatory genes reduced competing drains on additional chorismate-utilizing pathways and promoted PCN biosynthesis.
NADPH provides the reducing power that drives numerous anabolic reactions, including those responsible for the biosynthesis of all major cell components in the central carbon metabolic pathway and the shikimate pathway. NADH is involved in ATP generation of oxidative phosphorylation. Compared with HT66, NADPH, NADH, NAD+, ATP, ADP, and AMP were higher, while NADP+ was lower in HT66LSP. The energy charges in HT66 and HT66LSP were 0.85 ± 0.05 and 0.84 ± 0.01, respectively, consistent with the previous reported values (0.80–0.95) [28].
Qualitative assessment of dynamic labeling data with metabolomics data
To determine the relative pathway activities and help infer metabolic fluxes in response to the PCN overproduction, dynamic 13C-labeling experiments were performed, and integrated with metabolomics data to perform crossover analyses in HT66 and HT66LSP (Figs. 5, 6). Immediately after the introduction of [1,3-13C]glycerol or [U-13C]glycerol to the culture, the labeling dynamics of key metabolites were measured over a 1-h time window (Fig. 5; Additional file 2a). The average 13C-enrichment of each metabolite increased monotonically over time (Fig. 6; Additional file 2b), with the MDV shifting gradually toward heavier mass isotopomers. In addition to determining the level of enrichment, LC–MS analysis also provides information on the distribution of mass isotopomers of metabolites. The labeling dynamics were similar in two glycerol tracers.
Fig. 5.
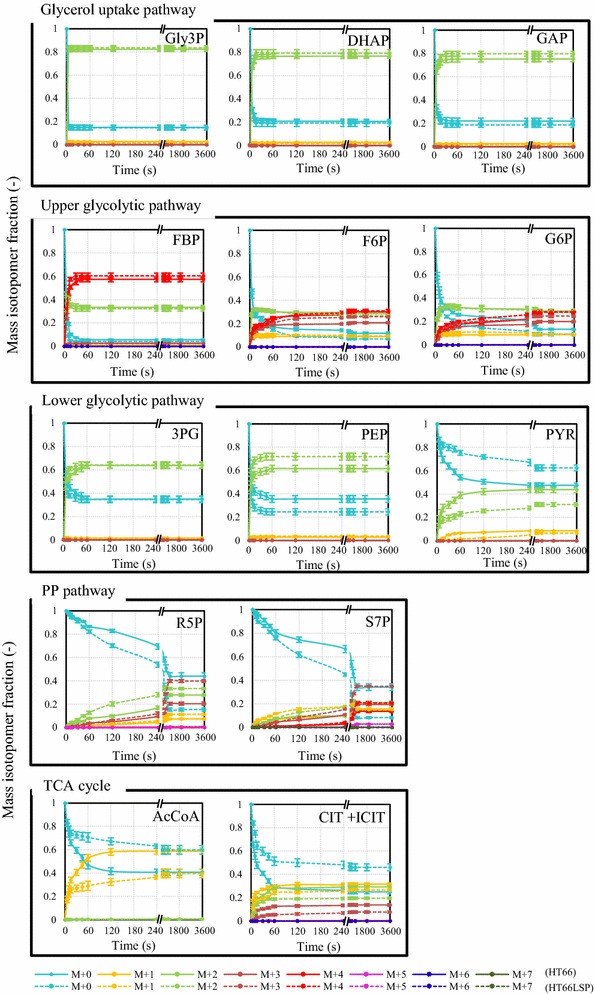
13C-labeling trajectories of selected intracellular metabolites after the introduction of [1,3-13C]glycerol at the exponential phase. Mass isotopomer data corrected for natural isotopic abundances are shown. The solid lines (HT66) and dashed lines (HT66LSP) illustrate the measured labeling trends
Fig. 6.
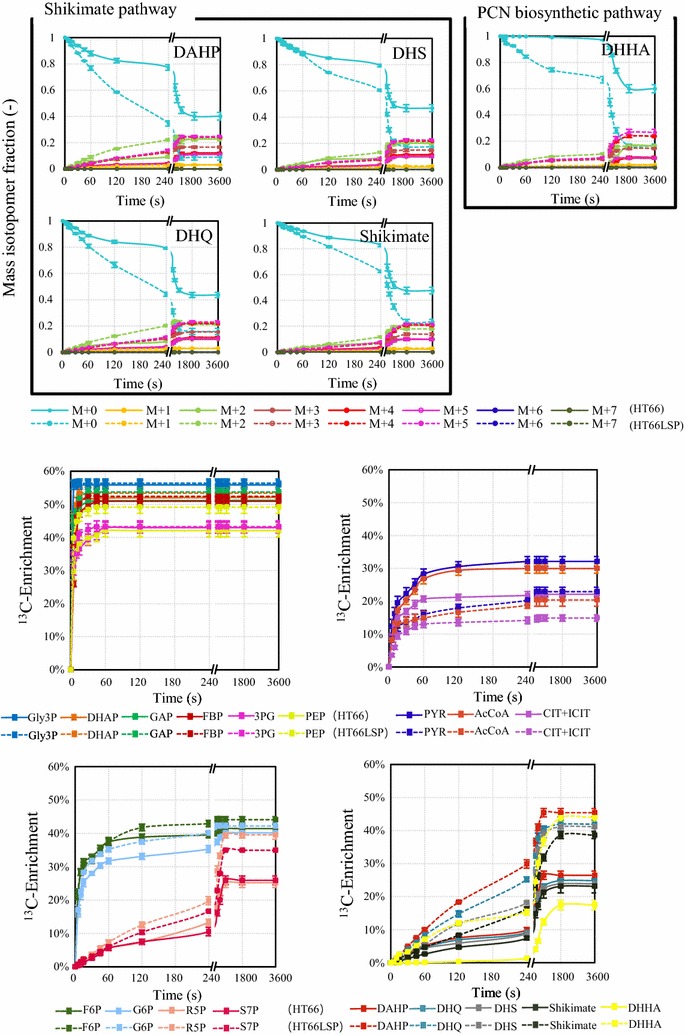
Average 13C-enrichments calculated using the formula , where N is the number of carbon atoms in the metabolite and Mi is the fractional abundance of the ith mass isotopomer. The solid lines (HT66) and dashed lines (HT66LSP) illustrate the measured labeling trends
The glycerol utilization pathway and glycolysis intermediates Gly3P, DHAP, GAP, 3PG, PEP, and FBP, distances of which from the entry point of the tracer is near, had the short response times, and a saturation in the labeling state was already reached within 60 s in both strains. The labeling dynamics of these metabolites were comparable with the exception of PEP in two strains. The identical labeling dynamics and intracellular concentrations of Gly3P, DHAP, and GAP indicated the similar flux through the glycerol utilization pathway in the two strains. In HT66LSP, the enrichment of PEP, which is the downstream metabolite of 3PG, was higher compared with 3PG throughout the labeling period, revealing unusual labeling patterns caused by metabolite channeling. A slight delay was found in the enrichment of metabolites in the other parts of central carbon metabolism, whereby isotopic stationary was reached within the time frame of 240 ≤ t ≤ 900 s. The two strains had the similar rates of labeling incorporation of F6P and G6P, together with intracellular concentrations, suggesting a lower flux through the upper glycolytic pathway or faster exchange with the nonoxidative PP pathway in HT66LSP. S7P and R5P which connect to the glycolytic pathway reached higher isotopic ratios more quickly in HT66LSP than in HT66. Slower decreases in M + 0 of PYR, AcCoA, and CIT + ICIT downstream of PEP were observed, together with their lower concentrations in HT66LSP than in HT66, signifying a lower flux through the lower glycolysis and the TCA cycle in the engineered strain.
Compared with glycolysis, the labeling dynamics in the shikimate pathway and PCN biosynthetic pathway were orders of magnitude slower, whereby isotopic stationary was reached within 30 min. A time lag of at least 30 s was observed before the labeling reached metabolites involved in these pathways followed by a further decrease in the labeling dynamics in both strains. HT66LSP had higher rates of labeling incorporation of DAHP, DHQ, DHS, shikimate and DHHA than HT66. Interestingly, shikimate was less labeled than its downstream product DHHA in HT66LSP, which implied the presence of metabolite channeling. In addition, distributions of labeled mass isotopomers of DAHP, DHQ, DHS, shikimate and DHHA were also different: predominantly either M + 2 ([1,3-13C]glycerol) or M + 3 ([U-13C]glycerol) in HT66 and either M + 5 ([1,3-13C]glycerol) or M + 7 ([U-13C]glycerol) in HT66LSP. The higher mass isotopomer of these metabolites formed in the condensation of a 13C-labeled PEP and a 13C-labeled E4P, while the lower mass isotopomer came from either 13C-labeled PEP or E4P. The faster labeling dynamics and heavier mass isotopomers integrating with higher levels of shikimate pathway intermediates in HT66LSP inferred a higher flux through this pathway.
Discussion
Glycerol utilization
In the glycerol uptake pathway, the gene transcript levels, pool sizes of metabolites, and the flux were not substantially affected by the PCN overproduction (Figs. 3, 4, 5 and 6; Additional file 2), which led to the constant glycerol consumption rate in HT66LSP (Table 1). This indicated the structural robustness of the metabolic network itself.
Regulatory mechanisms involved in rerouting of central carbon to shikimate pathway
Shikimate pathway, a bridge between primary and secondary metabolism, links the carbohydrate metabolism to the biosynthesis of phenazines [29, 30]. The shikimate pathway starts with DAHP-synthase-mediated condensation of PEP and E4P into DAHP. This step is one of the primary limiting reactions for the whole pathway [31]. In the starting strain HT66, the labeling dynamics of DAHP were much slower than PEP (Figs. 5, 6; Additional file 2), suggesting a potential bottleneck at this step. DAHP synthase is a key enzyme controlling the shikimate pathway, which is supported by several genome-scale and enzymatic analyses of mutant strains with improved shikimate production, in which this enzyme has been found to be overexpressed [32, 33]. By modifying regulatory genes, we found that phzC encoding DAHP synthase was upregulated, accompanied with more abundances of intracellular metabolites and the increased flux through the shikimate pathway, finally translating into PCN overproduction in the engineered strain (Figs. 2d, 3, 4, 5, 6, and Additional file 2). Our research confirmed the biological function of DAHP synthase to regulate the flux from the central metabolic pathway toward the shikimate pathway.
PEP, the precursor of the shikimate pathway, is a key intermediate in the glycolytic pathway. The carbon flux of PEP to its derived compounds usually competes with that of the lower glycolysis and the TCA cycle [34]. Compared with HT66, the carbon partitioning at PEP node directed relatively less carbon into the lower glycolysis and the TCA cycle (Figs. 5, 6; Additional file 2), associated with the downregulation of the genes (pykF, aceE, gltA, icd, sucA, and sucC) involved in this pathway (Fig. 3) in HT66LSP. The results demonstrated that reducing the carbon flux below the PYR node had a clear benefit to the PCN production. Beckers et al. [35] reported that the reductive TCA cycle influenced the production of poly(3-hydroxyalkanoate) which was synthesized from PYR in P. putida KT2440 grown on glycerol adversely.
Metabolomics analysis revealed that a handful of metabolites in the central carbon metabolism were less abundant, together with increases in the shikimate pathway intermediates in HT66LSP (Fig. 4), indicating significant drains on central carbon to provide precursors for the shikimate pathway and again confirming effective manipulations.
PCN production starts from chorismate [29], and the shikimate pathway converts central carbon metabolites via shikimate to chorismate [31], further away from glycerol consumption. Thus, metabolic engineering of the shikimate pathway and the PCN biosynthetic pathway alone maybe insufficient for increasing the yield and productivity from glycerol. Thus, genetic modifications to alter central carbon metabolism to supply the needed precursors are required. To improve E4P availability, the overexpressions of tktA and talA in the PP pathway are a successful strategy for the optimization of shikimate-producing strains [36], which can be applied for the efficient production of PCN. For the PEP supply, since metabolic flux through the TCA cycle is theoretically not needed for shikimate production [37], inactivation of pyruvate kinase represents a potential target to further enhance PCN production, which has been successively applied in its upstream metabolites chorsimate and PCA [15, 38].
Cofactors and energy metabolism
Compared with HT66, the NADPH, NADH, and ATP were all present in lower concentrations in HT66LSP (Fig. 4), which can be explained by the following reasons: (1) the lower flux from PYR toward the TCA cycle yielded less cofactors and ATP; (2) the conversion from DHS to shikimate consumed more NADPH; (3) S3P formation from shikimate needed more ATP; (4) the expression levels of zwf and gnd were not substantially modified, which may not generate more NADPH. The results indicated that the PCN production is constrained to a greater extent by precursor availability than by cofactors and ATP-deficiency.
Effective synthesis of PCN through metabolite channeling
Metabolite channeling, also known as substrate channeling, is a process of direct transfer of the product of an enzyme to another proximate enzyme or cell as its substrate without equilibration with the bulk phase [39]. Zhao et al. [40] showed that the chemotactic assembly of enzymes occurs even under cytosolic crowding conditions, which indicates that all native enzymes can naturally form a channel. Isotope dilution and enrichment has proved useful in measuring metabolite channeling [41–43]. In this study, we noted the metabolite channeling of 3PG into PEP and shikimate into DHHA in HT66LSP by this method (Figs. 5, 6; Additional file 2). In previous studies, metabolite channeling has been reported in the glycolytic pathway [43, 44], the Calvin cycle [45], the oxidative PP pathway [46], the Calvin–Benson–Bassham cycle [41], and so on. Our results provide the first evidence of metabolic channeling in the PCN biosynthetic pathway.
The effect of channeling could significantly improve the reaction thermodynamics and accelerated reaction rates by enzyme complexes [47]. In this research, metabolite channeling could enable high-yielding synthesis of PCN. First, PEP and DHHA would be synthesized more efficiently with high catalytic rates. Moreover, lower levels of metabolites in additional chorismate-utilizing pathways in HT66LSP indicated the forestallment of the chorismate competition among different pathways through channeling of shikimate into DHHA. In addition, when shikimate was directed toward DHHA, the unstable chorismate might be protected. By mimicking natural enzyme complexes for metabolite channeling, the strategy of structuring nonnatural cascade reactions has also made considerable success in the fields of metabolic engineering and synthetic biology, such as co-localization of cascade enzymes by protein, nucleic acid, and polymer scaffolds [48–50]. The downstream metabolites of DHHA in the phenazine biosynthetic pathway are not stable [51]. In addition, AOCHC is likely toxic to the cell so that its accumulation must be prevented [51]. To overcome these obstacles, co-localization of the core phenazine biosynthetic enzymes (PhzA, PhzB, PhzF, and PhzG) via protein scaffold would increase the robustness of the PCN biosynthetic pathway and prevent this pathway from releasing intermediates. The channeling strategy would have great biotechnological potentials for the overproduction of PCN.
Conclusions
In this work, we obtained a PCN-overproducing strain through knocking out three regulatory genes, lon, pars, and psrA in P. chlororaphis HT66. The engineered strain produced 4.10 g/L of PCN with a yield of 0.23 (g/g) from glycerol, which was the highest PCN titer and the yield ever obtained so far among PCN-producing strains. The integrated analysis of gene expression, metabolomics, and dynamic 13C tracer enabled us to gain a more in-depth insight into complex mechanisms for the PCN overproduction. Rerouting of central carbon to the shikimate pathway and metabolite channeling facilitated the PCN production. This study demonstrated that integrating systems biology analysis could be an efficient way to identify metabolic responses to the PCN overproduction, forming the basis for the rational design and engineering of the high efficiency strains using glycerol as the cost-effective carbon source. This approach would have a great potential for the future industrial production and agricultural application of biopesticide phenazines.
Methods
Strains, culture medium, and growth conditions
The strains, plasmids, and primers used in this study are listed in Table 2. The wild-type strain P. chlororaphis HT66 (CCTCC, M2013467) was obtained from China Center for Type Culture Collection. For genetic manipulations, E. coli strains were grown at 37 °C in LB medium. P. chlororaphis HT66 and its derivative strains stored in a − 80 °C freezer were activated at 28 °C for 12-24 h in King’s B agar media (glycerol 18 g/L, tryptone 20 g/L, MgSO4 0.732 g/L, K2HPO4 0.514 g/L). Selection of single colonies from Petri plates was performed, which were then used to inoculate 10 mL of King’s B broth in 50 mL flasks. Cultures were then grown at 28 °C in a flask kept under stirring at a speed of 180 rpm overnight. Portions of these cultures were then inoculated into 500-mL baffled flasks containing 120 mL King’s B broth to achieve an initial OD600 = 0.02. If necessary, ampicillin (100 μg/mL) and kanamycin (50 μg/mL) were added into the culture medium of E. coli and P. chlororaphis. After 8–36-h growth at 28 °C and 180 rpm, cultures were collected for the measurements of cell growth, glycerol, and PCN.
Table 2.
Strains and plasmids used in this study
| Strains and plasmids | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains E. coli DH5α | E. coli F−Ф80lacZΔM15 Δ(lacZYA-argF) M15 U169 recA1 endA1 hsdR17 (rk− mk−) phoA supE44 thi−1 gyrA96 relA1 | Invitrogen |
| E. coli S17-1 (λpir) | res− pro mod+ integrated copy of RP4, mob+, used for incorporating constructs into P. chlororaphis | [58] |
| P. chlororaphis HT66 | P. chlororaphis HT66 Wild-type, Apr Spr | China Center for type culture collection |
| HT66L | lon in-frame deletion mutant of HT66 | This study |
| HT66LS | parS in-frame deletion mutant of HT66L | This study |
| HT66LR | parR in-frame deletion mutant of HT66L | This study |
| HT66LSP | psrA in-frame deletion mutant of HT66LS | This study |
| Plasmids pK18mobsacB | Broad-host-range gene replacement vector; sacB, Kanr | [59] |
| pK18-lon | pK18mobsacB containing lon flanking region | This study |
| pK18-parS | pK18mobsacB containing parS flanking region | This study |
| pK18-parR | pK18mobsacB containing parR flanking region | This study |
| pK18-psrA | pK18mobsacB containing psrA flanking region | This study |
Construction of nonscar deletion mutant strains
The lon gene involved in the biosynthesis of PCN in P. chlororaphis HT66 was disrupted using the nonscar deletion method as described by Du et al. [52]. Briefly, two DNA fragments flanking the upstream and downstream area were amplified by PCR from the genome and combined using overlap PCR. The nonscar-modified DNA fragment was cloned into pK18mobsacB, and the resulting plasmid was then transferred into E. coli S17-1 (λpir) to be mobilized into HT66 by biparental mating. The colones occurring as a single-crossover event were selected from plates containing 100 μg/mL of ampicillin and 50 μg/mL of kanamycin, and then clones occurring as a double-crossover events were selected from plates containing 15% (w/v) sucrose and 50 μg/mL of kanamycin [6]. The mutant strain HT66L was further verified by PCR and DNA sequencing. In similar ways, the parS, parR, and prsA nonscar-deleted mutants were constructed in their corresponding strains. All the primers used to delete genes are shown in Additional file 3.
Analytical methods
Dry cell weight (DCW) in King’s B medium was calculated from the optical density at 600 nm (1 OD600 = 0.4114 g DCW L−1). Concentrations of glycerol were measured by high-performance liquid chromatography (HPLC) (model 1260, Agilent, Santa Clara, USA) using a cation-exchange column (HPX-87H, Bio-Rad, Hercules, CA) and a differential refractive index (RI) detector. A mobile phase of 5 mM H2SO4 at 0.5 mL/min flow rate was used, and the column was operated at 60 °C. In order to extract PCN, fermentation broth (400 μL) was acidified to pH 2.0 with 6 M HCl, and then 3.6 mL of ethyl acetate was added. The sample was vigorously agitated and centrifuged at 13,000×g for 5 min. A 400-μL portion of the organic layer was collected and evaporated. The residue was dissolved in 1 mL of acetonitrile. Concentrations of PCN were measured by HPLC using a C18 reverse-phase column (Agilent Eclipse XDB-C18, 4.6 mm × 250 mm, 5μΜ, Santa Clara, USA) by means of a UV light detector [6]. A mobile phase of 8% acetonitrile and 92% 5 mM NH4Ac at 1 mL/min flow rate was used, and the column was operated at 30 °C.
Quantitative real-time PCR (qRT-PCR) analysis
Culture samples at the mid-logarithmic growth phase were collected for RNA extraction. Total RNA was isolated using an RNA Extraction Kit (ABigen Corporation, China). Contaminating DNA was removed with RNase-free DNase I (ABigen Corporation, China). The first-strand cDNA was synthesized using PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara Co. Ltd., China). QRT-PCR was performed with the SYBR® Premix Ex Taq™ Kit (Takara Co. Ltd., China) on an ABI Stepone Real-Time PCR System (Applied Biosystems, USA). The primers used for qRT-PCR are listed in Additional file 1. The PCR conditions were 95 °C for 4 min, followed by 35 cycles of denaturation at 95 °C for 15 s, annealing at 57 °C for 15 s, and extension at 72 °C for 20 s. Three biological samples were analyzed, and each sample was analyzed three times. The housekeeping gene 16S rRNA was used as an internal standard. Fold changes of genes of interest were calculated as 2−ΔΔCT according to Schmittgen and Livak [53]. The data were averaged and presented as the mean ± standard deviation. Significant differences were determined by one-way analysis of variance (ANOVA). Statistical significance was defined as p < 0.05.
Sampling, quenching, and extraction of intracellular metabolites
Samples for intracellular metabolome analysis were withdrawn from the mid-logarithmic growth phase. For quenching and extraction, the protocol was adapted from the works of Millard et al. [54] and Toya et al. [55] with slight modifications. In brief, 10 mL of culture was poured into 50-mL falcon-tube containing 5 mL of 0.9% NaCl solution precooled at 0 °C, and the tube was immediately immersed in liquid N2. The sample solutions were manually agitated using a digital thermometer, and then it could be cooled to 0 °C within 10 s. Extracellular medium was removed by centrifugation at 8000 rpm at 0 °C for 3 min. To accurately determine the metabolites’ concentrations, isotope dilution mass spectrometry (IDMS) [56] was adopted in this study. After quenching, the cell pellet was suspended in 5 mL of cooled methanol containing U-13C-labeled cell extract as internal standards. After 30 s of sonication, 4 mL of chloroform and 1.6 mL of Milli-Q water were added to the solution and mixed, and the mixture was centrifuged at 4600×g at 4 °C for 5 min. The methanol layer was filtered by centrifugation through a 5-kDa cutoff filter (Merck Millipore Ltd., Darmstadt, Germany) to extract most of the intracellular metabolites, with the exception of 4-aminobenzoate, 4-hydroxybenzoate, folate, and PCA, which were extracted from the chloroform layer. The filtrate was thoroughly lyophilized (FreeZone 6 Liter, Labconco, USA) and reconstituted in 50 μL of methanol–water (1:1, v/v) just prior to measurement.
13C-pulse experiments
A sample of corresponding to time zero (unlabeled) was withdrawn just before the 13C-pulse experiments. After 23.6 h, labeling experiments were started by the rapid addition of 165.1 mM of either 100% [1,3-13C]glycerol (99% purity; Cambridge Isotope Laboratories Inc., Andover, MA) or 100% [U-13C]glycerol (99% purity; Cambridge Isotope Laboratories Inc., Andover, MA) to 30.4 mM unlabeled glycerol present in the flask, leading to a final glycerol concentration of 195.5 mM. To measure the 13C-incorporation into intracellular metabolites, 10 mL samples were withdrawn and rapidly quenched at time points of 5, 10, 15, 30, 45, 60, 120, 240, 450, 600, 900, 1800, and 3600 s, each in three biological replicates.
LC–MS measurement of metabolite labeling and concentration
Metabolite samples were analyzed via high resolution mass spectrometry analysis, carried on a Waters I-Class Acquity UPLC (Waters, UK) coupled with a Vion IMS QToF (Waters, UK) using a SeQuant ZIC-HILIC column (100 mm × 2.1 mm i.d., 3.5 µm) (Merck, Germany). The mobile phase A was 50 mM ammonium formate in water, and mobile phase B was acetonitrile. Metabolites were separated via gradient elution under the following conditions: 0–10 min, 90–50% B; 10–12 min, 50–90% B; 12–15 min, 90% B; and the column was maintained at 45 °C. The flow rate was 0.4 mL/min. The parameters of high-resolution mass spectrometry analysis on full scan mass spectrometry were as follows: MS range, m/z 50–1000; scan time, 0.3 s; CE 6 eV; desolvation temperature, 500 °C; source temperature, 120 °C; desolvation gas, 1000 L/h; cone gas, 50 L/h; capillary voltage, 2000 V. The lock correction (lock sprayer reference: mass, 556.2766 m/z; interval, 0.5 min; sample time, 0.5 min; CE, 6 eV; flow rate 10 μL/min) enabled isotopic m/z screen, with tolerance of 3 mDa mass error, the expected adduct –H. Data were acquired and processed using UNIFI 1.8.1. All purified metabolite standards were purchased from Sigma-Aldrich (St. Louis, MO, USA), with the exception of DHHA, which was produced in our own lab [57]. Quantitation of the peaks was achieved by comparing peak areas with the standards.
Significant differences were determined by a two-tailed Student’s t test using Microsoft Excel 2013 program. Statistical significance was defined as p < 0.05. Metabolite mass isotopomer distribution was determined based on the ratio of the integrated peak areas of the chosen isotopomer to the sum of all the integrated peak areas of the possible isotopomers for the given metabolites.
Abbreviations
Metabolites
AcCoA: acetyl-CoA; 4ABA: 4-aminobenzoate; ADIC: 2-amino-4-deoxychorismic acid; AKG: α-ketoglutarate; AOCHC: 6-amino-5-oxocyclohex-2-ene-1-carboxylic acid; 1,3 BPG: 1,3-bisphosphoglyceric acid; CHOR: chorismate; CIT: citrate; DAHP: 3-deoxy-D-arabino-heptulosonate 7-phosphate; DHAP: dihydroxyacetone phosphate; DHHA: trans-2,3-dihydro-3-hydroxyanthranilic acid; DHPCA: 5,10-dihydrophenazine-1-carboxylic acid; DHQ: 3-dehydroquinate; DHS: 3-dehydro-shikimate; E4P: erythrose 4-phosphate; F6P: fructose 6-phosphate; FBP: fructose 1,6-bisphosphate; FUM: fumarate; G6P: glucose-6-phosphate; GAP: glyceraldehyde 3-phosphate; GLX: glyoxylate; Gly3P: glycerol-3-phosphate; HHPDC: hexahydrophenazine-1,6-dicarboxylic acid; ICIT: isocitrate; KDPG: 2-keto-3-deoxy-6-phosphogluconate; MAL: malate; OAA: oxaloacetate; 2PG: 2-phosphoglyceric acid; 3PG: 3-phosphoglyceric acid; 6PG: 6-phosphogluconate; 6PGNL: 6-phosphogluconolactone; PCA: phenazine-1-carboxylic acid; PEP: phosphoenolpyruvate; Phe, phenylalanine; PYR: pyruvate; R5P: ribose 5-phosphate; RU5P: ribulose 5-phosphate; S3P: shikimate 3-phosphate; S7P: sedoheptulose 7-phosphate; SUCCoA: succinyl-CoA; SUC: succinate; THPCA: tetrahydrophenazine-1-carboxylic acid; Trp, tryptophan; Tyr, tyrosine; X5P: xylulose 5-phosphate.
Proteins (enzymes)
PhzA: phenazine biosynthesis protein PhzA; PhzB: phenazine biosynthesis protein PhzB; PhzF: phenazine biosynthesis protein PhzF family; PhzG: pyridoxamine 5′-phosphate oxidase.
Additional files
Additional file 1. Culture profiles of HT66LR.
Additional file 2. 13C-labeling trajectories of selected intracellular metabolites after the introduction of [U-13C]glycerol at the exponential phase. a Mass isotopomer abundances after correction for natural isotopic abundances. b Average 13C-enrichments calculated using the formula , where N is the number of carbon atoms in the metabolite and Mi is the fractional abundance of the ith mass isotopomer. The solid lines (HT66) and dashed lines (HT66LSP) illustrate the measured labeling trends.
Additional file 3. Primers used in this study.
Authors’ contributions
RY designed and performed most of the experiments, analyzed the result, and wrote the manuscript. KP assisted with experiments. LF carried out LC–MS analysis. HP, HH, and XZ considered the experimental design, analyzed the result, and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (31400086 and 31670033) and 973 Programs of China (2012CB721005).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1123-y) contains supplementary material, which is available to authorized users.
Contributor Information
Ruilian Yao, Email: yaoruilian@sjtu.edu.cn.
Keli Pan, Email: pankeli@sjtu.edu.cn.
Huasong Peng, Email: hspeng@sjtu.edu.cn.
Lei Feng, Email: fiona.fenglei@sjtu.edu.cn.
Hongbo Hu, Email: hbhu@sjtu.edu.cn.
Xuehong Zhang, Email: xuehzhang@sjtu.edu.cn.
References
- 1.Chen Z, Liu D. Toward glycerol biorefinery: metabolic engineering for the production of biofuels and chemicals from glycerol. Biotechnol Biofuels. 2016;9:205. doi: 10.1186/s13068-016-0625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clomburg JM, Gonzalez R. Anaerobic fermentation of glycerol: a platform for renewable fuels and chemicals. Trends Biotechnol. 2013;31:20–28. doi: 10.1016/j.tibtech.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Vivek N, Sindhu R, Madhavan A, Anju AJ, Castro E, Faraco V, Pandey A, Binod P. Recent advances in the production of value added chemicals and lipids utilizing biodiesel industry generated crude glycerol as a substrate—metabolic aspects, challenges and possibilities: an overview. Bioresour Technol. 2017;239:507–517. doi: 10.1016/j.biortech.2017.05.056. [DOI] [PubMed] [Google Scholar]
- 4.van Rij ET, Wesselink M, Chin-A-Woeng TF, Bloemberg GV, Lugtenberg BJ. Influence of environmental conditions on the production of phenazine-1-carboxamide by Pseudomonas chlororaphis PCL1391. Mol Plant Microbe Interact. 2004;17:557–566. doi: 10.1094/MPMI.2004.17.5.557. [DOI] [PubMed] [Google Scholar]
- 5.Shanmugaiah V, Mathivanan N, Varghese B. Purification, crystal structure and antimicrobial activity of phenazine-1-carboxamide produced by a growth-promoting biocontrol bacterium, Pseudomonas aeruginosa MML2212. J Appl Microbiol. 2010;108:703–711. doi: 10.1111/j.1365-2672.2009.04466.x. [DOI] [PubMed] [Google Scholar]
- 6.Jin XJ, Peng HS, Hu HB, Huang XQ, Wang W, Zhang XH. iTRAQ-based quantitative proteomic analysis reveals potential factors associated with the enhancement of phenazine-1-carboxamide production in Pseudomonas chlororaphis P3. Sci Rep. 2016;6:27393. doi: 10.1038/srep27393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavrodi DV, Parejko JA, Mavrodi OV, Kwak YS, Weller DM, Blankenfeldt W, Thomashow LS. Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ Microbiol. 2013;15:675–686. doi: 10.1111/j.1462-2920.2012.02846.x. [DOI] [PubMed] [Google Scholar]
- 8.Chin-A-Woeng TF, Bloemberg GV, van der Bij AJ, van der Drift KM, Schripsema J, Kroon B, et al. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant Microbe Interact. 1998;11:1069–1077. doi: 10.1094/MPMI.1998.11.11.1069. [DOI] [Google Scholar]
- 9.Chen Y, Shen X, Peng H, Hu H, Wang W, Zhang X. Comparative genomic analysis and phenazine production of Pseudomonas chlororaphis, a plant growth-promoting rhizobacterium. Genom Data. 2015;4:33–42. doi: 10.1016/j.gdata.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin-A-Woeng TF, van den Broek D, de Voer G, van der Drift KM, Tuinman S, Thomas-Oates JE, Lugtenberg BJ, Bloemberg GV. Phenazine-1-carboxamide production in the biocontrol strain Pseudomonas chlororaphis PCL1391 is regulated by multiple factors secreted into the growth medium. Mol Plant Microbe Interact. 2001;14:969–979. doi: 10.1094/MPMI.2001.14.8.969. [DOI] [PubMed] [Google Scholar]
- 11.Girard G, van Rij ET, Lugtenberg BJ, Bloemberg GV. Regulatory roles of psrA and rpoS in phenazine-1-carboxamide synthesis by Pseudomonas chlororaphis PCL1391. Microbiology. 2006;152:43–58. doi: 10.1099/mic.0.28284-0. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Seeve C, Pierson LS, 3rd, Pierson EA. Transcriptome profiling reveals links between ParS/ParR, MexEF-OprN, and quorum sensing in the regulation of adaptation and virulence in Pseudomonas aeruginosa. BMC Genom. 2013;14:618. doi: 10.1186/1471-2164-14-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin-A-Woeng TF, van den Broek D, Lugtenberg BJ, Bloemberg GV. The Pseudomonas chlororaphis PCL1391 sigma regulator psrA represses the production of the antifungal metabolite phenazine-1-carboxamide. Mol Plant Microbe Interact. 2005;18:244–253. doi: 10.1094/MPMI-18-0244. [DOI] [PubMed] [Google Scholar]
- 14.Khan SR, Mavrodi DV, Jog GJ, Suga H, Thomashow LS, Farrand SK. Activation of the phz operon of Pseudomonas fluorescens 2-79 requires the LuxR homolog PhzR, N-(3-OH-Hexanoyl)-L-homoserine lactone produced by the LuxI homolog PhzI, and a cis-acting phz box. J Bacteriol. 2005;187:6517–6527. doi: 10.1128/JB.187.18.6517-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu K, Hu H, Wang W, Zhang X. Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-hydroxyphenazine. Microb Cell Fact. 2016;15:131. doi: 10.1186/s12934-016-0529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas luorescens CHA0. J Bacteriol. 2002;184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sévin DC, Kuehne A, Zamboni N, Sauer U. Biological insights through nontargeted metabolomics. Curr Opin Biotechnol. 2015;34:1–8. doi: 10.1016/j.copbio.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Buescher JM, Antoniewicz MR, Boros LG, Burgess SC, Brunengraber H, Clish CB, DeBerardinis RJ, Feron O, et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jazmin LJ, Xu Y, Cheah YE, Adebiyi AO, Johnson CH, Young JD. Isotopically nonstationary 13C flux analysis of cyanobacterial isobutyraldehyde production. Metab Eng. 2017;42:9–18. doi: 10.1016/j.ymben.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAtee AG, Jazmin LJ, Young JD. Application of isotope labeling experiments and 13C flux analysis to enable rational pathway engineering. Curr Opin Biotechnol. 2015;36:50–56. doi: 10.1016/j.copbio.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Nöh K, Grönke K, Luo B, Takors R, Oldiges M, Wiechert W. Metabolic flux analysis at ultra short time scale: isotopically non-stationary 13C labeling experiments. J Biotechnol. 2007;129:249–267. doi: 10.1016/j.jbiotec.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang D, Lee SH, Seeve C, Yu JM, Pierson LS, Pierson EA. Roles of the Gac-Rsm pathway in the regulation of phenzaine biosynthesis in Pesudomonas chlororaphis 30-84. MicrobiologyOpen. 2013;2:505–524. doi: 10.1002/mbo3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi K, Tsuchiya W, Noda N, Suzuki R, Yamazaki T, Haas D. Lon protease negatively affects GacA protein stability and expression of the Gac/Rsm signal transduction pathway in Pseudomonas protegens. Environ Microbiol. 2014;16:2538–2549. doi: 10.1111/1462-2920.12394. [DOI] [PubMed] [Google Scholar]
- 25.Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother. 2010;54(8):3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikel PI, Kim J, de Lorenzo V. Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT2440. Environ Microbiol. 2014;16:239–254. doi: 10.1111/1462-2920.12224. [DOI] [PubMed] [Google Scholar]
- 27.Chin-A-Woeng TF, Thomas-Oates JE, Lugtenberg BJ, Bloemberg GV. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol Plant Microbe Interact. 2001;14:1006–1015. doi: 10.1094/MPMI.2001.14.8.1006. [DOI] [PubMed] [Google Scholar]
- 28.Atkinson DE. The energy charge of the adenylate pool as a regulatory parameter. Interact Feedback modifiers. Biochem. 1968;7:4030–4034. doi: 10.1021/bi00851a033. [DOI] [PubMed] [Google Scholar]
- 29.Mentel M, Ahuja EG, Mavrodi DV, Breinbauer R, Thomashow LS, Blankenfeldt W. Of two make one: the biosynthesis of phenazines. ChemBioChem. 2009;10:2295–2304. doi: 10.1002/cbic.200900323. [DOI] [PubMed] [Google Scholar]
- 30.Jin K, Zhou L, Jiang H, Sun S, Fang Y, Liu J, Zhang X, He YW. Engineering the central biosynthetic and secondary metabolic pathways of Pseudomonas aeruginosa strain PA1201 to improve phenazine-1-carboxylic acid production. Metab Eng. 2015;32:30–38. doi: 10.1016/j.ymben.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Herrmann KM, Weaver LM. The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- 32.Kogure T, Kubota T, Suda M, Hiraga K, Inui M. Metabolic engineering of Corynebacterium glutamicum for shikimate overproduction by growth-arrested cell reaction. Metab Eng. 2016;38:204–216. doi: 10.1016/j.ymben.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Suástegui M, Guo W, Feng X, Shao Z. Investigating strain dependency in the production of aromatic compounds in Saccharomyces cerevisiae. Biotechnol Bioeng. 2016;113:2676–2685. doi: 10.1002/bit.26037. [DOI] [PubMed] [Google Scholar]
- 34.Noda S, Kondo A. Recent advances in microbial production of aromatic chemicals and derivatives. Trends Biotechnol. 2017;35:785–796. doi: 10.1016/j.tibtech.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Beckers V, Poblete-Castro I, Tomasch J, Wittmann C. Integrated analysis of gene expression and metabolic fluxes in PHA-producing Pseudomonas putida grown on glycerol. Microb Cell Fact. 2016;15:73. doi: 10.1186/s12934-016-0470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez A, Martínez JA, Báez-Viveros JL, Flores N, Hernández-Chávez G, Ramírez OT, Gosset G, Bolivar F. Constitutive expression of selected genes from the pentose phosphate and aromatic pathways increases the shikimic acid yield in high-glucose batch cultures of an Escherichia coli strain lacking PTS and pykF. Microb Cell Fact. 2013;12:86. doi: 10.1186/1475-2859-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patnaik R, Liao JC. Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield. Appl Environ Microbiol. 1994;60:3903–3908. doi: 10.1128/aem.60.11.3903-3908.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noda S, Shirai T, Oyama S, Kondo A. Metabolic design of a platform Escherichia coli strain producing various chorismate derivatives. Metab Eng. 2016;33:119–129. doi: 10.1016/j.ymben.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Geck MK, Kirsch JF. A novel, definitive test for substrate channeling illustrated with the aspartate aminotransferase/malate dehydrogenase system. Biochemistry. 1999;38:8032–8037. doi: 10.1021/bi983029c. [DOI] [PubMed] [Google Scholar]
- 40.Zhao X, Palacci H, Yadav V, Spiering MM, Gilson MK, Butler PJ, Hess H, Benkovic SJ, Sen A. Substrate-driven chemotactic assembly in an enzyme cascade. Nat Chem. 2018;10(3):311–317. doi: 10.1038/nchem.2905. [DOI] [PubMed] [Google Scholar]
- 41.Young JD, Shastri AA, Stephanopoulos G, Morgan JA. Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab Eng. 2011;13:656–665. doi: 10.1016/j.ymben.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wheeldon I, Minteer SD, Banta S, Barton SC, Atanassov P, Sigman M. Substrate channelling as an approach to cascade reactions. Nat Chem. 2016;8:299–309. doi: 10.1038/nchem.2459. [DOI] [PubMed] [Google Scholar]
- 43.Hollinshead WD, Rodriguez S, Martin HG, Wang G, Baidoo EE, Sale KL, Keasling JD, Mukhopadhyay A, Tang YJ. Examining Escherichia coli glycolytic pathways, catabolite repression, and metabolite channeling using Δpfk mutants. Biotechnol Biofuels. 2016;9:212. doi: 10.1186/s13068-016-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shearer G, Lee JC, Koo JA, Kohl DH. Quantitative estimation of channeling from early glycolytic intermediates to CO2 in intact Escherichia coli. FEBS J. 2005;272:3260–3269. doi: 10.1111/j.1742-4658.2005.04712.x. [DOI] [PubMed] [Google Scholar]
- 45.Suss KH, Arkona C, Manteuffel R, Adler K. Calvin cycle multienzyme complexes are bound to chloroplast thylakoid membranes of higher plants in situ. Proc Natl Acad Sci USA. 1993;90:5514–5518. doi: 10.1073/pnas.90.12.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Debnam PM, Shearer G, Blackwood L, Kohl DH. Evidence for channeling of intermediates in the oxidative pentose phosphate pathway by soybean and pea nodule extracts, yeast extracts, and purified yeast enzymes. Eur J Biochem. 1997;246:283–290. doi: 10.1111/j.1432-1033.1997.00283.x. [DOI] [PubMed] [Google Scholar]
- 47.Abernathy MH, He L, Tang YJ. Channeling in native microbial pathways: implications and challenges for metabolic engineering. Biotechnol Adv. 2017;35(35):805–814. doi: 10.1016/j.biotechadv.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 49.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 50.Wang XL, Li Z, Shi JF, et al. Bioinspired approach to multienzyme cascade system construction for efficient carbon dioxide reduction. ACS Catal. 2014;4:962–972. doi: 10.1021/cs401096c. [DOI] [Google Scholar]
- 51.Blankenfeldt W, Parsons JF. The structural biology of phenazine biosynthesis. Curr Opin Struct Biol. 2014;29:26–33. doi: 10.1016/j.sbi.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du X, Li Y, Zhou W, Zhou Q, Liu H, Xu Y. Phenazine-1-carboxylic acid production in a chromosomally non-scar triple-deleted mutant Pseudomonas aeruginosa using statistical experimental designs to optimize yield. Appl Microbiol Biotechnol. 2013;97:7767–7778. doi: 10.1007/s00253-013-4921-y. [DOI] [PubMed] [Google Scholar]
- 53.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 54.Millard P, Massou S, Wittmann C, Portais JC, Létisse F. Sampling of intracellular metabolites for stationary and non-stationary 13C metabolic flux analysis in Escherichia coli. Anal Biochem. 2014;465:38–49. doi: 10.1016/j.ab.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 55.Toya Y, Nakahigashi K, Tomita M, Shimizu K. Metabolic regulation analysis of wild-type and arcA mutant Escherichia coli under nitrate conditions using different levels of omics data. Mol BioSyst. 2012;8:2593–2604. doi: 10.1039/c2mb25069a. [DOI] [PubMed] [Google Scholar]
- 56.Wu L, Mashego MR, van Dam JC, Proell AM, Vinke JL, Ras C, van Winden WA, van Gulik WM, Heijnen JJ. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal Biochem. 2005;336:164–171. doi: 10.1016/j.ab.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 57.Hu H, Li Y, Liu K, Zhao J, Wang W, Zhang X. Production of trans-2,3-dihydro-3-hydroxyanthranilic acid by engineered Pseudomonas chlororaphis GP72. Appl Microbiol Biotechnol. 2017;101(17):6607–6613. doi: 10.1007/s00253-017-8408-0. [DOI] [PubMed] [Google Scholar]
- 58.Hofmann A, Thimm T, Droge M, Moore ER, Munch JC, Tebbe CC. Inter-generic transfer of conjugative and mobilizable plasmids harbored by Escherichia coli in the gut of the soil microarthropod Folsomia candida (Collembola) Appl Environ Microbiol. 1998;64:2652–2659. doi: 10.1128/aem.64.7.2652-2659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Culture profiles of HT66LR.
Additional file 2. 13C-labeling trajectories of selected intracellular metabolites after the introduction of [U-13C]glycerol at the exponential phase. a Mass isotopomer abundances after correction for natural isotopic abundances. b Average 13C-enrichments calculated using the formula , where N is the number of carbon atoms in the metabolite and Mi is the fractional abundance of the ith mass isotopomer. The solid lines (HT66) and dashed lines (HT66LSP) illustrate the measured labeling trends.
Additional file 3. Primers used in this study.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.


