Abstract
To screen for drug resistance and possible treatment with Dolutegravir (DTG) in treatment-naive patients and those experiencing virologic failure during first-, second-, and third-line combined antiretroviral therapy (cART) in Uganda. Samples from 417 patients in Uganda were analyzed for predicted drug resistance upon failing a first- (N = 158), second- (N = 121), or third-line [all 51 involving Raltegravir (RAL)] treatment regimen. HIV-1 pol gene was amplified and sequenced from plasma samples. Drug susceptibility was interpreted using the Stanford HIV database algorithm and SCUEAL was used for HIV-1 subtyping. Frequency of resistance to nucleoside reverse transcriptase inhibitors (NRTIs) (95%) and non-NRTI (NNRTI, 96%) was high in first-line treatment failures. Despite lack of NNRTI-based treatment for years, NNRTI resistance remained stable in 55% of patients failing second-line or third-line treatment, and was also at 10% in treatment-naive Ugandans. DTG resistance (n = 366) was not observed in treatment-naive individuals or individuals failing first- and second-line cART, and only found in two patients failing third-line cART, while 47% of the latter had RAL- and Elvitegravir-resistant HIV-1. Secondary mutations associated with DTG resistance were found in 2%–10% of patients failing third-line cART. Of 14 drugs currently available for cART in Uganda, resistance was readily observed to all antiretroviral drugs (except for DTG) in Ugandan patients failing first-, second-, or even third-line treatment regimens. The high NNRTI resistance in first-line treatment in Uganda even among treatment-naive patients calls for the use of DTG to reach the UNAIDS 90:90:90 goals.
Keywords: : HIV drug resistance, integrase inhibitors, dolutegravir, Uganda
Introduction
By 2016, there were 36.7 million people living with HIV-1 infection worldwide and 1.8 million new infections diagnosed in the same year.1 Uganda is among the countries with the highest burden of HIV-1 infections, with ∼1.5 million (7.1%) people living with HIV/AIDS and 57% of them receiving combined antiretroviral therapy (cART).2 Unfortunately, due to various sociological and economic factors, HIV-1 drug resistance has been rapidly emerging in patients receiving cART, such that HIV-1 drug-resistant variants are now responsible for at least 9% of the new infections.3
In Uganda, as most of sub-Saharan Africa, over 57% of patients have access to tenofovir- and efavirenz (EFV)-based first-line regimen recommended by the World Health Organization (WHO),1 while <50% of patients are receiving the dolutegravir (DTG)- and darunavir-based first-line regimens recommended by the International Antiviral Society—United States of America.4 However, rapid emergence of HIV-1 drug resistance to first-line B1a rated treatment regimens argues for increasing access to more effective cART, which may lead to better treatment outcomes, lower adverse events/drug toxicity, and higher barriers for drug resistance. DTG, a second-generation integrase strand transfer inhibitor (INSTI), Elvitegravir/cobicistat (EVG/cobi), or Raltegravir (RAL) are three “backbone” INSTIs recommended for first-line treatment regimens in combination with tenofovir disoproxil fumarate/emtricitabine (FTC) (or with tenofovir alafenamide/FTC when combined with EVG/cobi).5 DTG/abacavir/lamivudine is also employed if the patients are screened to exclude those with an HLA B57 allele. Rilpivirine along with the latter nucleoside reverse transcriptase inhibitors (NRTIs) are used for new first-line treatment regimens. Despite these preferences for treatment in high income countries (HICs), at the Joint Clinical Research Center (JCRC) in Uganda, currently <1% of the 60,000+ patients receive this first-line cART recommended by the Centers for Disease Control and Prevention (CDC) for HICs.
Equally important, in vivo DTG resistance has rarely been observed even in those few patients showing virological failure in NRTI-experienced, but INSTI-naive patients.6 In vitro studies characterizing the DTG-associated R263K mutation in subtype B, C, and CRF02_AG HIV-1 strains showed early emergence of R263K in subtypes B and CRF02_AG, whereas the G118R substitution was only observed in subtype C and CRF02_AG.7 It is possible that HIV-1 strains from different subtypes may explore distinct paths, selecting for different mutations, to escape DTG pressure. Moreover, while resistance to EVG and RAL comes at high fitness cost,8 the DTG-resistant R263K mutation is even slower to emerge and more debilitating to HIV-1 replication. The R263K mutation observed in clinical and in vitro studies, and secondary mutations such as H51Y, M50I, and E138K observed in cell culture studies do emerge during DTG treatment, but appear unable to restore replication capacity of the virus.9–11 DTG can also inhibit most HIV-1 isolates resistant to RAL and EVG, which relates to its success in RAL-experienced patients in the VIKING-3 study.12
Despite the strong safety profile and success of DTG in both first- and second-line treatments, its use in low and middle income countries (LMICs) has been minimal. DTG has been extensively tested in patients infected with subtype B, with no difference on DTG susceptibility been observed in non-B subtype HIV-1 strains in dose-escalating/selection experiments.13 However, little is known about DTG treatment outcomes in patients infected with subtype A and D HIV-1 primarily found in Uganda.14
In this study, we evaluated the possible treatment with DTG in cART to treat HIV-1-infected Ugandan individuals based on the current drug resistance profile in treatment-naive and highly treatment-experienced patients. Drug susceptibility was predicted in 417 plasma samples from treatment-naive individuals (N) (n = 87), first-line failures (FF) (n = 158), second-line failures (SF) (n = 121), or third-line/RAL-based antiretroviral failures (RF) (n = 51). Our results did not predict DTG resistance in HIV-1-infected treatment-naive patients, nor from individuals failing different cART regimens, highlighting the suitability of DTG-based therapies to treat HIV-1-infected patients in Uganda at any stage of the disease.
Materials and Methods
Samples for the study
This was a retrospective study that assessed the prevalence and impact of DTG-associated mutations in patients failing on different treatment regimens. Samples were collected from the WHO, College of American Pathologist (CAP), and National Institutes of Health-Virology Quality Assurance (NIH-VQA)-accredited Center for AIDS Research (CFAR) Laboratory of the JCRC in Kampala, Uganda. The JCRC is one of the first HIV treatment centers in the country to roll out cART and currently, the only site licensed to provide INSTIs in the country. The patient database in the CFAR laboratory was used to access the patient demographic, medical, and treatment history of the study samples. A total of 440 plasma samples were collected from patients receiving routine treatment care at the JCRC and with virological failure, defined by a viral load above 1,000 copies/ml and/or CD4+ T cell counts below 250 cells/mm3. These virological failures included plasma samples from 90 N, 165 FF, 125 SF, and 60 RF patients. Antiretroviral therapy (ART)-naive samples came from the Pan-African Studies to Evaluate Resistance network,15 the Monitoring Antiretroviral Resistance in Children observational cohort study,16 and the Hormonal Contraception and HIV-1 Genital Shedding (GS) and Disease Progression among Women with Primary HIV Infection study.17 Forty-two of one hundred sixty-five FF samples came from the Europe-Africa Research Network for Evaluation of Second-line Therapy trial.18 The rest of the samples were patient samples collected during routine Sanger genotyping testing at the JCRC. Ethical clearance was obtained from the IRBs at the JCRC and UHCMC/CWRU (EM-10-07 and 10-05-35).
RNA extraction and PCR amplification
Viral RNA was extracted from 440 plasma samples using a QIAamp viral RNA Mini Kit (Qiagen) according to the manufacturer's instructions. Reverse transcription of the full-length HIV-1 integrase (IN)-coding region from extracted viral RNA and amplification was done with the sense primer RTA9F (5′-TATGGGGAAAGACTCCTAAATTTA-3′) and antisense primer 3Vif (5′-AGCTAGTGTCCATTCATTG-3′) using a Superscript III single RT-PCR system with Platinum Taq DNA polymerase kit (Thermo Fisher Scientific) as per the manufacturer's instructions. Nested PCR was done using the sense primer INTFEXT1 (5′-AGAAGTAAACATAGTAACAGACTCACA-3′) and antisense Vif 3 reverse 1 primer (5′-GTCCTGCTTGATATTCACACC-3′) using a Platinum Taq kit (Thermo Fisher Scientific) as per the manufacturer's instructions to generate amplicon of 1,433 base pairs. Amplification of protease (PR) and reverse transcriptase (RT) regions was done as previously described.3 The amplicon was purified using ExoSAP-IT enzyme (Thermo Fisher Scientific) and quantified using a Qubit fluorometer (Thermo Fisher Scientific).
Cycle sequencing and sequence analysis
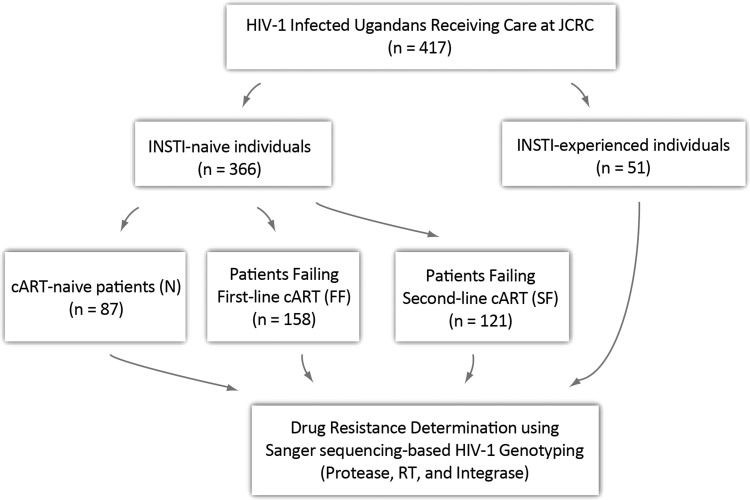
The HIV-1 IN as well as the PR and RT coding regions were amplified and analyzed on a Sanger sequencing platform. Briefly, a quantified and purified PCR product was sequenced with primers spanning the full length of the IN gene (1–288 amino acids): Vif 3 reverse1 (5′-GTCCTGCTTGATATTCACACC-3′), INTREXT (5′-AATCCTCATCCTGTCTAC-3′), and INTFEXT1 (5′-AGAAGTAAACATAGTAACAGACTCACA-3′). PCR product was sequenced with ABI Big dye terminator (v3.1) (Thermo Fisher Scientific) according to the manufacturer's instructions on ABI 3730xl sequencing platform (Life Technologies, Carlsbad, CA). A total of 417 samples (N = 87, FF = 158, SF = 121, and RF = 51) were amplified and sequenced successfully for the IN gene (Fig. 1; Table 1). Sequences were analyzed using RECall (beta v3.02) program as recommended by the WHO.19 Stanford resistance predictions were obtained using a Python client, SierraPy 0.1.2, to automate transactions with the Stanford HIVdb Sierra web service algorithm v8.3.20
FIG. 1.
The work flow chart of the patient numbers and their respective groups. FF consisted of patients who were on NNRTI-based combination therapy, SF had PI-based combination, RF had RAL backbone and N had not been exposed to ART. INSTI, integrase strand transfer inhibitor; RT, reverse transcriptase; PR, protease; INT, integrase; NNRTI, nonnucleoside reverse transcriptase inhibitor; FF, first-line failures; SF, second-line failures; PI, protease inhibitor; RAL, raltegravir; RF, antiretroviral failures; ART, antiretroviral therapy.
Table 1.
Clinical and Virological Characteristics of the Patients in the Study
| Subtype no. (%) | |||||||
|---|---|---|---|---|---|---|---|
| Group of patientsa | cART regimen (no. patients)b | Mean HIV-1 RNA log10 c/mlc | A | D | C | A/D | Otherd |
| cART naive (n = 87) | None | 4.64 | 42 (47.1) | 17 (19.5) | 3 (3.4) | 9 (10.3) | 17 (19.5) |
| Failing first-line cART (n = 158) | AZT, 3TC, NVP (38) | 0.54 | 14 (36.9) | 11 (28.9) | 4 (10.5) | 2 (5.3) | 7 (18.4) |
| TDF, 3TC, EFV (34) | 1.21 | 14 (41.2) | 10 (29.4) | 2 (5.9) | 1 (2.9) | 7 (20.6) | |
| AZT, 3TC, EFV (18) | 1.31 | 18 (44.4) | 6 (33.3) | 1 (5.6) | 1 (5.6) | 2 (11.1) | |
| TDF, 3TC, FTC (15) | 0.43 | 1 (20) | 1 (20) | 1 (20) | 0 (0.0) | 2 (40) | |
| Other (14) | 7.5 | 5 (35.7) | 5 (35.7) | 2 (14.3) | 2 (14.3) | 0 (0.0) | |
| 3TC, D4T, NVP (13) | 2.2 | 8 (61.5) | 1 (7.7) | 0 (0.0) | 1 (7.7) | 3 (23.1) | |
| ABC, 3TC, NVP (10) | 2.56 | 4 (40) | 1 (10) | 1 (10) | 1 (10) | 3 (30) | |
| ABC, 3TC, EFV (7) | 0.64 | 7 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| TDF, 3TC, NVP (6) | 0.97 | 0 (0.0) | 3 (50) | 0 (0.0) | 1 (16.7) | 2 (33.3) | |
| 3TC, AZT, NVP (4) | 2.22 | 0 (0.0) | 1 (25) | 1 (25) | 1 (25) | 1 (25) | |
| d4T, 3TC, NVP (3) | 0.97 | 0 (0.0) | 3 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| AZT, 3TC, ABC (2) | ND | 0 (0.0) | 1 (50) | 0 (0.0) | 0 (0.0) | 1 (50) | |
| FTC, TDF, EFV (2) | 0.7 | 1 (50) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50) | |
| TDF, ABC, AZT (2) | ND | 1 (50) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50) | |
| Failing second-line cART (n = 121) | TDF, 3TC, LPVr (30) | 1.22 | 13 (43.3) | 4 (13.3) | 2 (6.7) | 2 (6.7) | 9 (30) |
| TDF, 3TC, ATVr (30) | 2.2 | 14 (46.7) | 7 (23.3) | 1 (3.3) | 2 (6.7) | 6 (20) | |
| ABC, 3TC, LPVr (25) | 2.13 | 12 (48) | 8 (32) | 1 (4) | 0 (0.0) | 4 (16) | |
| ABC, 3TC, ATVr (25) | 3.64 | 3 (60) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (40) | |
| AZT, 3TC, LPVr (13) | 7.82 | 4 (30.7) | 3 (23.1) | 0 (0.0) | 3 (23.1) | 3 (23.1) | |
| LPVr (7) | 0.05 | 3 (42.9) | 4 (57.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| AZT, 3TC, ATVr (6) | 2.53 | 4 (66.7) | 2 (33.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Other (5) | 0.7 | 1 (20) | 1 (20) | 0 (0.0) | 1 (20) | 2 (40) | |
| Failing RAL-based cART (n = 51) | RAL, LPVr (28) | 3.1 | 15 (53.5) | 7 (25.0) | 1 (3.5) | 3 (10.7) | 2 (7.1) |
| Other (3) | 0.3 | 2 (67.0) | 1 (33.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| RAL, DRVr (6) | 6.4 | 5 (83.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (16.6) | |
| RAL, ATVr (2) | 8.5 | 1 (50) | 1 (50) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| RAL, TDF, 3TC, LPVr (3) | 2.7 | 3 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| TDF, 3TC, RAL, DRVr (7) | 1.6 | 5 (71.4) | 2 (28.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| RAL, ETR, DRVr (2) | 1.1 | 2 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
The HIV subtype was predicted using SCUEAL subtype classification algorithm. Viral loads were assayed using Abbott m2000sp/rt or Roche COBAS Amplicor Monitor ultrasensitive tests, v1.5.
The total number of patients tested for INSTI resistance in each patient group.
Description of the ART combinations in the study, and the number of patients taking that combination. Other, the number of ART combinations, which were less prescribed, that is, only one patient for each of these ART drug combinations: (3TC, TDF, NVP) (AZT, D4T, 3TC, NVP) (3TC, EFV, LPVr) (3TC, EFV, ATVr) (ABC, DDI, LPVr) (TDF, 3TC, ATVr) (EFV, DDI, LPVr) (EFV, RAL, DRVr) (TDF,3TC, RAL, ETR) (TDF, FTC, RAL, DRVr) (RAL, ETR, LPVr).
The average number of patient viral loads (copies/ml × 105).
The percentage of patients with HIV-1 unique circulating recombinant forms (CRFs); A1/C, A1/AE, D/U, J/A1,C/G, AE/D, A1/U, A3/U, and CRF35. Until the time ATVr was available and incorporated into national treatment guidelines, LPVr was infrequently provided as second-line monotherapy if resistance patient had drug resistance to 3TC and AZT.
cART, combined antiretroviral therapy; ND, not determined; INSTI, integrase strand transfer inhibitor; ART, antiretroviral therapy; ABC, abacavir; AZT, zidovudine; D4T, stavudine; DDI, didanosine; FTC, emtricitabine; 3TC, lamivudine; TDF, tenofovir; EFV, efavirenz; ETR, etravirine; NVP, nevirapine; RPV, rilpivirine; ATVr, atazanavir/r; DRVr, darunavir/r; FPVr, fosamprenavir/r; IDVr, indinavir/r; LPVr, lopinavir/r; NFV, nelfinavir; SQVr, saquinavir/r; TPVr, tipranavir /r; DTG, dolutegravir; EVG, elvitegravir; RAL, raltegravir.
Subtype classification
The resulting JSON files were converted into CSV files using an in-house R script. SCUEAL was used for HIV-1 subtype classification and recombination detection.21 This algorithm maps sequences to a phylogeny of subtype reference sequences by maximum likelihood to classify subtypes and detect recombination. Sequences with two or more recombination breakpoints with multiple subtype or sub-subtype (e.g., A1, A2) parents are labeled “complex” recombinants. Amino acid polymorphisms were extracted by pairwise alignment of the consensus sequence to the HXB2 reference integrase gene sequence using an in-house Python script. Insertions relative to this reference were discarded and the aligned sequences were translated into amino acids with an HXB2 coordinate system. We excluded circulating recombinant forms (CRFs) from the phylogenetic analysis and aligned all the data using MAFFT v7.305b,22 including the HIV-1 subtype reference sequences of HIV-1 subtypes A1, A2, C, and D. We manually adjusted the resulting alignment using AliView v1.19-beta-3.23 A phylogenetic tree was reconstructed by maximum likelihood using PHYML v20160207 with the default parametric bootstrap support analysis.24 The general time-reversible model incorporating invariant sites and a gamma distribution for rate variation across sites (GTR + I + G) was selected using the Akaike Information Criterion using jModeltest v2.1.10.25 The tree was visualized and manually annotated in FigTree (A. Rambaut, http://tree.bio.ed.ac.uk/software figtree) and Archaeopteryx v0.9920 beta.26
Statistical analyses
Statistical analyses were performed using GraphPad Prism 7.03 (GraphPad Software, La Jolla, CA) using one-way analysis of variance unless otherwise specified.
Results
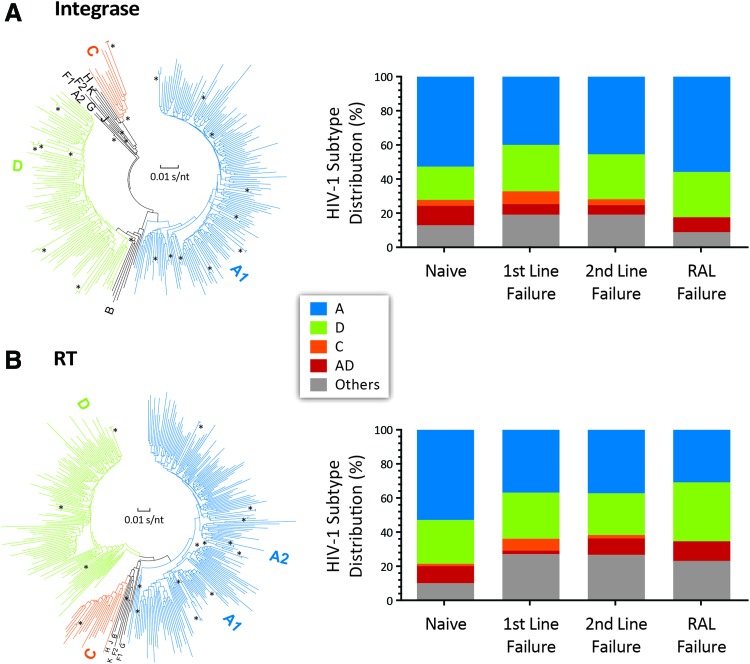
Based on sequencing of the PR, RT, and IN-coding regions, HIV-1 subtype distributions did not significantly differ among the four groups (N, FF, SF, and RF). Subtype A virus was the predominant subtype found in nearly half of patients in each group followed by subtype D and C. Our observations are consistent with our previous study on Uganda HIV-1 subtype distribution in the last 10 years3 (Fig. 2). As previously described,3 a higher proportion of patients failing treatment appeared to be infected with subtype D, but this was not statistically significant (Fig. 2). No subtype C infections were identified in the RAL-treated group, most likely due to the smaller sample size. AD recombinants were observed at higher frequency in the naive than treatment-failure populations, whereas complex recombinants/CRFs comprised the remaining 13%–19% (Fig. 2).
FIG. 2.
HIV-1 subtype classification of RT, protease, and IN regions. (A) The bar graph (right) and phylogenetic tree (left) describe the HIV subtype classification of the IN gene (percentages) of ART-naive patients, FF, SF and RF. (B) The bar graph (right) and phylogenetic tree (left) of HIV subtype classification of RT, and PR regions of N, FF, SF, and RF. Subtype descriptions are embedded in the figure. An in-house Python script was used to label tips with subtype classifications from SCUEAL. A maximum likelihood phylogenetic tree was reconstructed from the alignment using PHYML v20160207 with the default parametric bootstrap estimation of branch supports given the data. The tree was visualized and manually annotated in FigTree (available at: http://tree.bio.ed.ac.uk/software/figtree) and Archaeopteryx v0.9920 beta. IN, integrase. Color images available online at www.liebertpub.com/aid
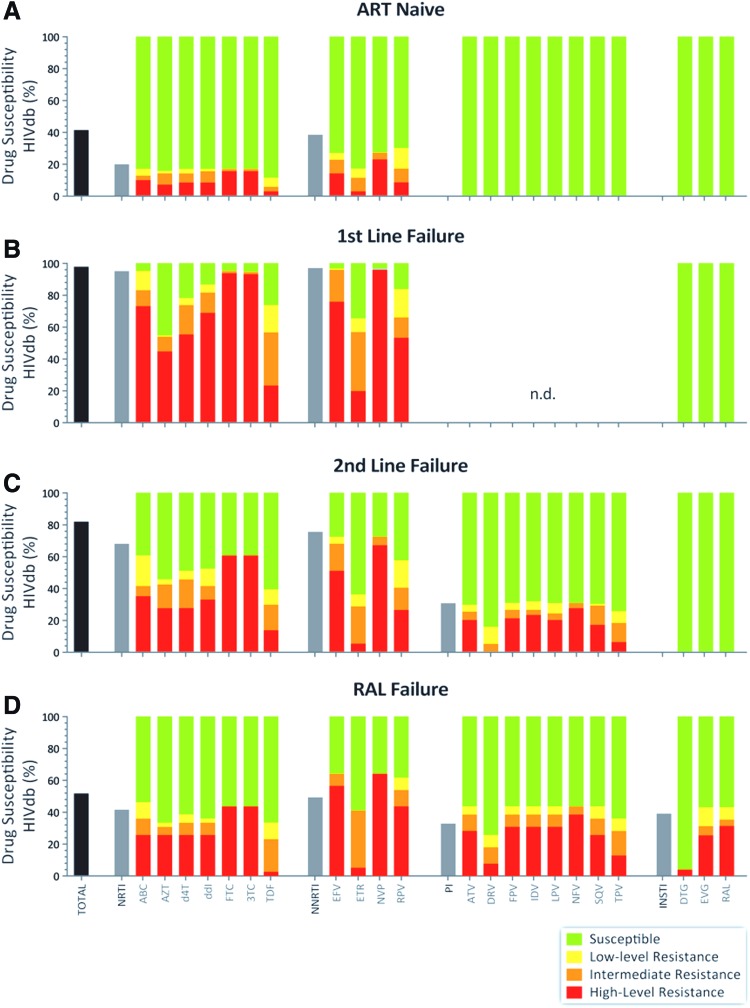
On average, 10% of the treatment-naive patients were infected with HIV-1 variants harboring primary resistance mutations to at least one NRTI or non-NRTI (NNRTI) (Figs. 3 and 4). This treatment-naive group was recruited from 2007 to 2011 during chronic disease and tested for HIV-1 drug resistance genotype.15,16,18 A 10% incidence of drug resistance in chronically infected patients from 2007 to 2011 is likely an underestimate of current rates of transmitted drug- resistant HIV-1 in the treatment-naive population. Recruitment of these patients during chronic disease likely resulted in a loss of drug resistance and reversion to wild-type HIV-1 following initial infections with a drug-resistant HIV-1 variant. A recent WHO report 2017 shows pretreatment HIV-1 drug resistance to EFV/nevirapine (NVP) in 15.4% of the HIV-positive population in Uganda.27 A slightly higher frequency of NNRTI over NRTI resistance was observed in our treatment-naive population, which may be due to higher fitness costs of NRTI-resistant over NNRTI-resistant mutations and faster reversion to wild-type HIV-1.28,29 Finally, no protease inhibitor (PI) or INSTI resistance was observed in the naive patients from 2007 to 2011, which reflects very limited use of these treatments in Uganda during this time period.
FIG. 3.
Drug resistance predictions based on pol sequences of treatment-naive patients, first- and second-line treatment failures, or in patients receiving RAL. Genotypic resistance/drug susceptibility prediction was performed on 417 HIV pol sequences from Uganda using the HIVdb genotypic resistance interpretation algorithm from Stanford University. (A–D) Show the level of drug resistance in N, FF, SF, and RF, respectively. The first bar in each patient group represents total drug resistance in that group, and the first bar in each drug class represent, total drug resistance in that respective drug class. The rest of description of level of drug resistance is embedded in the figure. Color images available online at www.liebertpub.com/aid
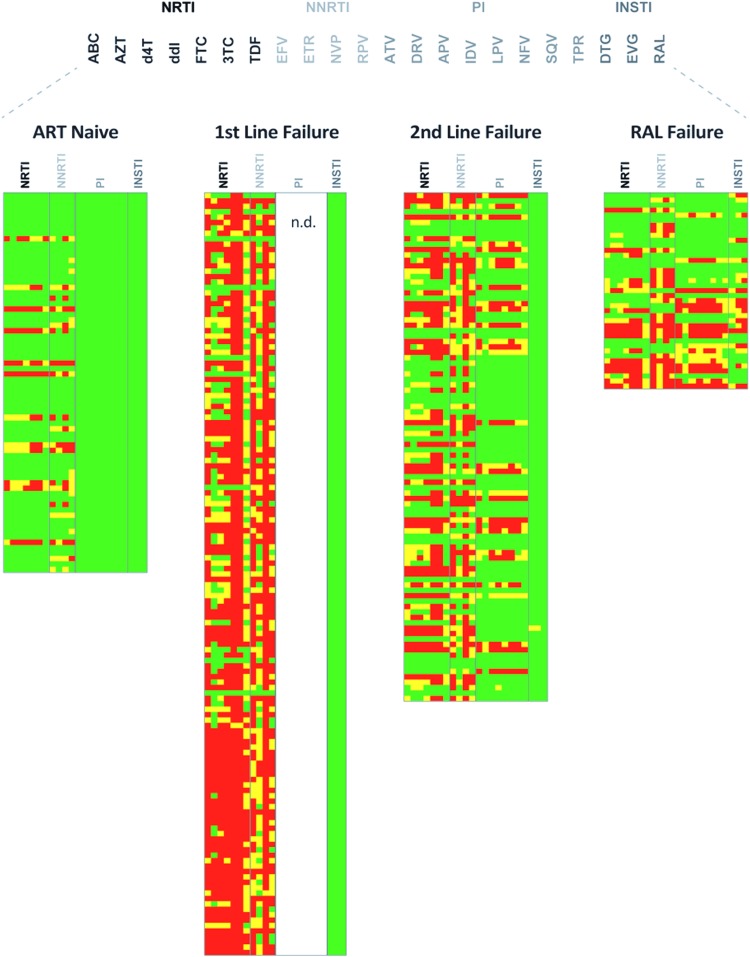
FIG. 4.
HIV-1 genotypic resistance interpretation based on Sanger sequencing. Amino acid substitutions was used with HIVdb program Genotypic resistance interpretation algorithm from Stanford university HIV drug resistance database (https://hivdb.stanford.edu) to predict the levels of susceptibility to PR, RT, and INSTIs. A susceptible genotype is shown in green, intermediate- and high-level resistance is shown in yellow and red, respectively. ABC, abacavir; AZT, zidovudine; D4T, stavudine; DDI, didanosine; FTC, emtricitabine; 3TC, lamivudine; TDF, tenofovir; EFV, efavirenz; ETR, etravirine; NVP, nevirapine; RPV, rilpivirine; ATVr, atazanavir/r; DRVr, darunavir/r; FPVr, fosamprenavir/r; IDVr, indinavir/r; LPVr, lopinavir/r; NFV, nelfinavir; SQVr, saquinavir/r TPVr, tipranavir/r; DTG, dolutegravir; EVG, elvitegravir. Color images available online at www.liebertpub.com/aid
Following first-and second-line treatment failures, 97.8% and 81.9% of the HIV-1 sequences showed resistance to at least one ART drug, respectively (Fig. 3). Resistance and presence of drug-resistant mutations were mainly observed in the NRTI and NNRTI drug classes following first-line treatment failures due to exclusive use of these drug classes on cART initiation (Fig. 3; Table 2). Due to complete absence of PI treatment in the first-line treatment regimens, the PR coding region was not sequenced upon treatment failure. However, previous studies in Uganda have confirmed the near absence of PR-resistant mutations in first-line treatment regimens involving two NRTIs and an NNRTI.30 PIs, typically lopinavir/ritonavir or atazanavir/ritonavir, were prescribed in nearly all second-line treatments, hence the appearance of PI resistance upon failure. Despite the absence of NNRTIs in second- or third-line treatment with RAL, NNRTI resistance remained and was still the most common in these patients, found in 75.5% and 49.0% patients, respectively. Once PIs are administered, PIs are typically retained in the regimen even upon treatment failure and emergence of PI-resistant mutations. As a consequence, we have little to no data on the potential loss or reversion of PI-resistant mutations, except that PI resistance is still less frequent than NNRTI or NRTI resistance in newly infected or treatment-naive patients.
Table 2.
Frequency of Mutations Associated with Reduced Susceptibility to Protease, Reverse Transcriptase, and Integrase Strand Transfer Inhibitors
| PI (n)a | NRTIs (n)b | NNRTIs (n)c | INSTIs (n)d | |
|---|---|---|---|---|
| ART naive | None | E44D (4) | G190A (3) | M50I/L (29) |
| T69D (5) | E138A (7) | L74I/M (5) | ||
| K70R (7) | Y181C (6) | T97A/T (7) | ||
| M184V (11) | K101E (3) | Others (1) | ||
| M41L (3) | K238T (2) | |||
| L210W (2) | K103N (4) | |||
| D67N/G (6) | A98G (5) | |||
| K219Q/E (5) | V108I (2) | |||
| T215Y/I/F (7) | Others (7) | |||
| Others (5) | ||||
| First-line failures | n.d. | E44D/E (16) | Y181V (5) | M50I/L (46) |
| T69D/N (10) | E138A (3) | L74I/M (6) | ||
| K70R (32) | K238T (6) | T97A/T (8) | ||
| M41L (47) | H221Y (18) | E157Q (2) | ||
| L210W (31) | V108I/V (40) | Others (1) | ||
| K70KR (4) | H221HY (11) | |||
| V75V/I/M (45) | V179D/T (6) | |||
| A62AV (6) | P225H/P (8) | |||
| F77FL (2) | A98G/A (26) | |||
| K70KE (3) | M230L (6) | |||
| L210LW (4) | F227L (4) | |||
| M184V/M/I (131) | L100I (7) | |||
| D67/N/G (46) | Y188L (6) | |||
| L74V/L/I (18) | Y181C/V (49) | |||
| Y115F/Y (15) | E138E/A/G (5) | |||
| K65R/K (27) | K101P/H/E (37) | |||
| T215I/F/Y/T (71) | G190A/G/Q/S (52) | |||
| K219N/K/E/Q (42) | K103N/K/S (58) | |||
| Others (19) | Others (10) | |||
| Second-line failures | M46M/K/L (4) | K65R (5) | E138A (7) | M50I/L (34) |
| I47A (5) | Y115F/Y (12) | Y181C (17) | L74I/M (9) | |
| V82A/F (19) | E44D (12) | K103N/K (28) | T97A/T (9) | |
| I84V (6) | T69D (6) | K101E (9) | ||
| L76V (5) | K70R (12) | K238T (4) | ||
| I54V (12) | M184V/M/I (57) | P225H (6) | ||
| L90M (2) | M41L/M (20) | A98G/A (14) | ||
| N88S 92) | L210W (10) | V108I/V (23) | ||
| V82S/C (2) | T215F/Y/I/T/S (62) | H221H/Y (7) | ||
| Others (1) | D67N/D (18) | M230L (3) | ||
| K219Q (9) | G190S/A/G (23) | |||
| T215I (3) | Y188L (5) | |||
| K219E (9) | H221HY (3) | |||
| Y115Y/F (11) | V179I/T/A (5) | |||
| F116Y (3) | K101E/H (16) | |||
| Q151M (4) | Others (9) | |||
| V75M/I (11) | ||||
| K70K/R/E (8) | ||||
| F77L (3) | ||||
| L210LW (3) | ||||
| L74I (7) | ||||
| Others (11) | ||||
| RAL failures | I54V/L/I (12) | M184V/M (18) | Y181C (9) | M50I/L/M (10) |
| M46M/I/L (11) | K219E/Q (6) | G190A (4) | L74I/M (8) | |
| V82F/A/V (8) | M41L/M (4) | K103N/S/K/D (13) | T97A/T (16) | |
| L76V/L (4) | E44D (4) | E138Q/EA (4) | E138K/A (3) | |
| I84V (5) | T69G/D (2) | Y188L (4) | E157Q (3) | |
| I47A/I/V (2) | K70R (4) | K101H/P/Q/E (3) | G163R (7) | |
| V82A/V (6) | T215I/V/TFS (5) | M230M/I (2) | Y143R/S (4) | |
| L90M/L (2) | A62A/V (2) | L100LI (3) | N155H (9) | |
| V32I/V (2) | L74V/I (3) | Others (14) | G140A (2) | |
| I50IV (2) | V75I/M/V (5) | Q148K/R (2) | ||
| Others (6) | Y155F/Y (2) | T66A/IV (2) | ||
| Q151M/Q (2) | Others (1) | |||
| D67N (4) | ||||
| Others (16) |
The number of primary resistance mutations in protease region of HIV found in the study patients.
Primary resistant mutations, which confer resistance to NRTIs.
Primary resistant mutations, which result in resistance to NNRTIs.
Primary resistant mutations, which confer resistance to INSTIs. n.d., drug-resistant mutations not determined. Others, drug-resistant mutations with one frequency in each patient group.
PI, protease inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor.
Unlike NNRTI, NRTI, and PI resistance, major drug resistance mutations (DRMs) to INSTIs were noticeably absent in N, FF, and SF, that is, Y143R/C/H, Q148R/K/H/N, N155H, E92Q, E138A/K/T, G140S/A/C, S147G, T66A/I/K, and R263K, conferring resistance to RAL, EVG, and/or DTG. Nevertheless, we detected some minor IN mutations described as secondary or compensatory mutations: T97A/T (9.6%), M50I (6.7%), L74M/I (3.1%), E157Q (1.43%), V151I/A (2.0%), and G163R (2.0%) (Fig. 3; Table 3). L74M was observed in 0.8% of INSTI-naive patients (N, FF, and SF) and 9.8% of RF patients. It is a polymorphic accessory mutation selected by RAL and EVG,31 and selected by DTG in previously RAL-treated patients with DTG primary mutations.32 The M50I polymorphism was observed in 6.2% of INSTI-naive patients and 9.8% of RF patients, and increases resistance to DTG in combination with R263K to 5.6-fold in cell culture, although it does not increase the replication capacity of the virus.10 E157Q was found in 0.8% of INSTI-naive and 5.8% of RF patients. It has been identified as a compensatory mutation for the N115H mutation,33 but also tends to increase susceptibility to DTG.34 T97A was found in 6.5% of INSTI-naive and 29.4% of RF patients. T97A has been shown to preexist in 5%–10% of INSTI-naive patients infected with subtype A virus.35 In addition, T97A was previously coselected in the presence of primary INSTI DRMs by RAL in many clinical studies,36,37 and by DTG in treatment-experienced patients with preexisting RAL-associated resistance mutations.32 As expected, the lack of INSTI resistance is attributable to the absence of INSTI treatment in Uganda and most of sub-Saharan Africa. However, previous reports and data presented herein also indicate that natural polymorphisms conferring resistance to INSTIs are extremely rare in these subtype A and D isolates, also reported as rare in subtype B.38 Predicted DTG resistance was also absent in HIV-1 variants from 366 treatment-naive or treatment-experienced patients (FF and SF treatment failures) (Figs. 3 and 4).
Table 3.
HIV-1-Infected Patients Failing on Raltegravir-Based Regimen with Primary and/or Secondary (Compensatory) Integrase Strand Transfer Inhibitor Mutations
| INSTI susceptibility | ||||
|---|---|---|---|---|
| Primary/secondary mutations | n (%) | DTG | RAL | EVG |
| M50I | 1 (2.0) | S | S | S |
| M50IM | 1 (2.0) | S | S | S |
| M50I, L74I | 1 (2.0) | S | S | S |
| T97A | 1 (2.0) | S | P | P |
| T97A, G163R, L74M | 3 (5.8) | S | L | L |
| N155H | 2 (4.0) | P | H | H |
| N155H, M50I | 1 (2.0) | P | H | H |
| N155H, T97A | 1 (2.0) | P | H | H |
| N155H, T97AT | 1 (2.0) | P | H | H |
| N155NH, T97AT, M50L | 1 (2.0) | P | H | H |
| N155H, T97A, E157Q, L74I | 1 (2.0) | P | H | H |
| N155H, E157Q, G163R, M50L, L74I | 1 (2.0) | P | H | H |
| Y143S, T97A | 1 (2.0) | S | H | H |
| Y143R, T97A | 2 (4.0) | S | H | L |
| Y143R, T97AT, G163R | 1 (2.0) | P | H | I |
| Y143R, T97A, M50I, L74LM | 1 (2.0) | P | H | I |
| E138A, T97A, V151A | 1 (2.0) | P | I | I |
| E138A, G140A, Q148R, G163R | 1 (2.0) | H | H | H |
| E138K, G140A, S147G, Q148K | 1 (2.0) | H | H | H |
| T66AIV, T97A | 1 (2.0) | S | L | H |
| T66A, T97A, G163R, L74M | 1 (2.0) | S | I | H |
Secondary mutations (not included in the table) found in N, FF, and SF, were classified as follows; M50I was found in 12 (7.5%) of FF, 4 (3.3%) of SF, and 7 (8.0%) of N. L74M found in 1 (0.8%) of SF and 1 (1.1%) of N. T97A was found in 8 (5.0%) of FF, 9 (7.4%) of SF, and 7 (8.0%) of N. E157Q was found in 2(1.2%) of FF and 1 (1.1%) of N. In bold, the major INSTI primary resistance mutations, which confer resistance to INSTIs.
FF, first-line failures; SF, second-line failures; H, high-level resistance; I, intermediate-level resistance; L, low-level resistance; S, susceptible genotype.
INSTI resistance major mutations Y143R/S (0.9%), Q148K/R (0.47%), N155H (2.1%), E138A/K (0.7%), G140A (0.47%), T66A/TAIV (0.47%), and S147G (0.25%) (Fig. 3; Table 3) were only observed in RF. However, only two patients (DR-206-12, DR-1059-17) in RF had genotypes with potential resistance to DTG (i.e., G140A, S147G, Q148K, and E138K, and G140A, Q148R, E138A, and G163R, respectively). In addition, R263K, the mutation most commonly associated with DTG resistance, was not observed. Over 47% RF had RAL and EVG resistance, but only 23.5% were predicted to have weak and moderate resistance to DTG.
Discussion
The Ministry of Health in Uganda has implemented the WHO “Treat All” recommendation, which states that every person tested HIV-1 positive be started on treatment irrespective of his/her virological and immunological status. With increasing emergence of drug resistance in treatment-naive population in lower income countries (LICs)27,29,39,40 and the number of patients on cART increasing, HIV-1 drug resistance prevalence will inevitably also rise. More potent antiretroviral drugs, such as DTG, have shown to be active in treatment-experienced patients. More importantly, resistance to DTG seems to be infrequent in cART-naive individuals treated with this integrase inhibitor.41 The purpose of the study was to screen for drug resistance and possible treatment with DTG in treatment-naive patients, and those experiencing virologic failure during first-, second-, and third-line cART in Uganda. To our knowledge, this is the first study to look at INSTI-associated drug resistance in both cART-naive and cART-experienced patients in Uganda and most other countries in sub-Saharan Africa.
Among minor INSTI resistance mutations, T97A mutation was observed in both INSTIs-naive and RF patients and has been shown to reduce sensitivity of virus to INSTIs and/or rescue viral fitness in combination with Y143C/R, Q148+G140S, or N155H. However, a previous study shows that T97A does not significantly reduce susceptibility to INSTI with up to 94% and 97% viral suppression achievable in HIV-1 patients with preexisting and emerging T97A, respectively.35 The prevalence of the M50I polymorphism observed in INSTI-naive patients is lower compared to the 10% found in patients infected with HIV-1 subtype B virus, which shows variation in the evolution of INSTI-associated mutations in different viral subtype populations. A relatively small number of patients were infected with viruses carrying the E157Q mutation, which has shown to increase susceptibility to DTG.34 However, the combination of E157Q and R263K increases DTG resistance by 10-fold.34
We found that neither of the two rare mutations associated with DTG resistance, R263K and G118R. R263K has been identified in both clinical samples and cell culture assays6,7 and G118R in cell culture tests.7 The major mutation pathways for RAL, Y143R/C, Q148R/K/H, and N155H42 were not found in INSTI-naive patients in agreement with previous study done in treatment-naive patients in South Africa.43 In RF, 18 (35%) of patients had Y143R/S, Q148R/K, G140A, E138A, S147G, T66A/TAIV, and N155H INSTI major mutations, which could explain the virological failure observed in these patients.
This study shows that the accumulation of INSTI DRMs at positions G140, S147, Q148, and E138 after RAL failure can potentially predict the potential loss of susceptibility of the virus to DTG as seen previously.44 For example, we found two individuals infected with a virus carrying the Q148K/R resistant mutation, which when present alone, moderately reduces RAL and EVG susceptibility, while having a minimal effect on DTG susceptibility; however, in combination with G140S/A/C and/or E138K/A, it may reduce DTG susceptibility up to 10-fold.20,45 In RF, N155H had highest frequency 9 (17.6%) compared to other major INSTI DRMs, which may be due to early selection of this resistant mutation under RAL pressure as observed previously in a phase II study looking at long-term efficacy and safety of RAL in patients with limited treatment options.46 Among the six major mutations, which reduce susceptibility to EVG, T66I, E92Q, T97A, S147G, Q148K, and N155H,47 only substitution E92Q was not selected by the virus in the study patients, which could be due to cross-resistance as EVG is currently unavailable in the country.
Based on our analyses of 417 Ugandan HIV-1 pol sequences, DTG would possibly be effective at any stage of cART treatment in sub-Saharan Africa. DTG has become the preferred drug for the majority of new FL or salvage treatments in HICs. In our Ugandan cohort, we did not observe a higher frequency of mutations conferring DTG or any other primary INSTI resistance mutations. We observed a high frequency of NNRTI resistance in FF (96.4%), and in patients who remain on treatment, but who have not received NNRTI for years, SF (75.5%) and RF (49.0%). This observation complements the fact that over 50% of treatment failures in Uganda retain NNRTI-resistant virus, and often for years following the last dose of NVP or EFV. In contrast, the frequency of NRTI and PI resistance is lower in all of these HIV-1-infected groups in Uganda. With the UNAIDS/WHO 90:90:90 goals, continued use of NNRTIs (EFV or NVP) may help increase access to treatment since these drugs are readily available in this setting, but may not impact treatment outcomes due to associated high drug resistance, high pill burden, and poor tolerability profiles. However, to achieve continued viral suppression in 90% of treated individuals, we should abandon the continued use of NNRTIs (NVP or EFV) in first-line treatment and strongly advocate for the use of second-generation INSTIs, such as DTG or even Bictegravir, in first-line treatment regimens in LICs, such as Uganda and other East African countries.
Acknowledgment
This work was supported by grants from National Institutes of Health (AI-49170 and AI-71747 to E.J.A. and M.E.Q.-M., respectively). M.E.Q.-M. was also partially supported by grants, the CWRU/UH Center for AIDS Research (P30 AI036219). A.F.Y.P. and M.A. are supported by grants from the Canadian Institutes for Health Research (CIHR PJT-153391 and BOP-149562) and by the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-131). E.nd was also supported by funding from Queen Elizabeth II Diamond Jubilee scholarship through Western University DLI O19375892122. The authors thank JCRC HIV Drug resistance working team, which includes Leonard Bagenda (Africa biosystems, Kampala, Uganda), Regina Namusisi (Joint Clinical Research Center, Kampala, Uganda), Francis Ssali (Joint Clinical Research Center, Kampala, Uganda), Helen Musana (Joint Clinical Research Center, Kampala, Uganda), Joselyne Nansimbe (Joint Clinical Research Center, Kampala, Uganda), Aggrey Bukuru (Joint Clinical Research Center, Kampala, Uganda), William Tamale (Joint Clinical Research Center, Kampala, Uganda), Victor Musiime (Joint Clinical Research Center, Kampala, Uganda; Makerere University, Kampala, Uganda) and all the staff of Joint Clinical Research Center, Kampala, Uganda.
Feedback from other members of the Arts laboratory and colleagues at the Department of Microbiology and Immunology at Western University, London Ontario, are highly appreciated.
The Sequence Data
Pol sequences have been submitted to GenBank (Sequin v15.50; available at: www.ncbi.nlm.nih.gov/Sequin), with accession numbers MF138165-MF138546, MF138547-MF138684, and MF138685-MF138863.
Author's Contributions
E.nd, F.K., I.N., E.na, performed all genotyping drug resistance assays and experimentation in this article and E.nd, M.A., A.F.Y.P., R.M.G., and M.E.Q.-M. performed data analyses. C.K. and P.M. recruited all the patients for this study. E.J.A. and M.E.Q-M. procured the funding and E.J.A. had the concept, and was the principal investigator who guided the overall direction of the study.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.UNAIDS: UNAIDS data 2017. Available at http://unaids.org (2017), accessed July22, 2017
- 2.WHO: Global AIDS update 2016-enormous gains,persistent challenges. Available at http:who.int (2016), accessed July2, 2017
- 3.Kyeyune F, Nankya I, Metha S, et al. : Treatment failure and drug resistance is more frequent in HIV-1 subtype D versus subtype A-infected Ugandans over a 10-year study period. AIDS 2013;27:1899–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunthard HF, Aberg JA, Eron JJ, et al. : Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA 2014;312:410–425 [DOI] [PubMed] [Google Scholar]
- 5.CDC: CDC HIV/AIDS guidelines and recommendations. Available at www.cdc.gov/hiv/guidelines/index.html (2017), accessed August30, 2017
- 6.Cahn P, Pozniak AL, Mingrone H, et al. : Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: Week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 2013;382:700–708 [DOI] [PubMed] [Google Scholar]
- 7.Quashie PK, Mesplede T, Han YS, et al. : Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir. J Virol 2012;86:2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson RM, Weber J, Winner D, Miller MD, Quiñones-Mateu ME: Contribution of human immunodeficiency virus type 1 minority variants to reduced drug susceptibility in patients on an integrase strand transfer inhibitor-based therapy. PLOS ONE 2014;9(8):e104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesplede T, Quashie PK, Osman N, et al. : Viral fitness cost prevents HIV-1 from evading dolutegravir drug pressure. Retrovirology 2013;10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wares M, Mesplede T, Quashie PK, Osman N, Han Y, Wainberg MA: The M50I polymorphic substitution in association with the R263K mutation in HIV-1 subtype B integrase increases drug resistance but does not restore viral replicative fitness. Retrovirology 2014;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mesplede T, Osman N, Wares M, et al. : Addition of E138K to R263K in HIV integrase increases resistance to dolutegravir, but fails to restore activity of the HIV integrase enzyme and viral replication capacity. J Antimicrob Chemother 2014;69:2733–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castagna A, Maggiolo F, Penco G, et al. : Dolutegravir in antiretroviral-experienced patients with raltegravir- and/or elvitegravir-resistant HIV-1: 24-Week results of the phase III VIKING-3 study. J Infect Dis 2014;210:354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charpentier C, Bertine M, Visseaux B, et al. : In-vitro phenotypic susceptibility of HIV-1 ‘non-B’ integrase inhibitors naive clinical isolates to dolutegravir and raltegravir. AIDS 2013;27:2959–2961 [DOI] [PubMed] [Google Scholar]
- 14.Ssemwanga D, Ndembi N, Lyagoba F, et al. : Transmitted antiretroviral drug resistance among drug-naive female sex workers with recent infection in Kampala, Uganda. Clin Infect Dis 2012;54(Suppl 4):S339–S342 [DOI] [PubMed] [Google Scholar]
- 15.Hamers RL, Straatsma E, Kityo C, et al. : Building capacity for the assessment of HIV drug resistance: Experiences from the pharmaccess African studies to evaluate resistance network. Clin Infect Dis 2012;54(Suppl 4):S261–S265 [DOI] [PubMed] [Google Scholar]
- 16.Boender TS, Sigaloff KC, Kayiwa J, et al. : Barriers to initiation of pediatric HIV treatment in Uganda: A mixed-method study. AIDS Res Treat 2012;2012:817506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison CS, Demers K, Kwok C, et al. : Plasma and cervical viral loads among Ugandan and Zimbabwean women during acute and early HIV-1 infection. AIDS 2010;24:573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paton NI, Kityo C, Hoppe A, et al. : Assessment of second-line antiretroviral regimens for HIV therapy in Africa. N Engl J Med 2014;371:234–247 [DOI] [PubMed] [Google Scholar]
- 19.Woods CK, Brumme CJ, Liu TF, et al. : Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol 2012;50:1936–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang MW, Liu TF, Shafer RW: The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology 2012;55:98–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosakovsky Pond SL, Posada D, Stawiski E, et al. : An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol 2009;5:e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K, Standley DM: MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 2013;30:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson A: AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014;30:3276–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guindon S, Gascuel O: A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003;52:696–704 [DOI] [PubMed] [Google Scholar]
- 25.Darriba D, Taboada GL, Doallo R, Posada D: jModelTest 2: More models, new heuristics and parallel computing. Nat Methods 2012;9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zmasek CM, Eddy SR: ATV: Display and manipulation of annotated phylogenetic trees. Bioinformatics 2001;17:383–384 [DOI] [PubMed] [Google Scholar]
- 27.WHO: HIV drug resistance report 2017. Available at http://who.int/hiv/topics/drugresistance/en (2017), accessed August14, 2017
- 28.Garcia-Lerma JG: Diversity of thymidine analogue resistance genotypes among newly diagnosed HIV-1-infected persons. J Antimicrob Chemother 2005;56:265–269 [DOI] [PubMed] [Google Scholar]
- 29.Boerma RS, Sigaloff KC, Akanmu AS, et al. : Alarming increase in pretreatment HIV drug resistance in children living in sub-Saharan Africa: A systematic review and meta-analysis. J Antimicrob Chemother 2017;72:365–371 [DOI] [PubMed] [Google Scholar]
- 30.Kyeyune F, Gibson RM, Nankya I, et al. : Low-frequency drug resistance in HIV-infected Ugandans on antiretroviral treatment is associated with regimen failure. Antimicrob Agents Chemother 2016;60:3380–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW: HIV-1 integrase inhibitor resistance and its clinical implications. J Infect Dis 2011;203:1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eron JJ, Clotet B, Durant J, et al. : Safety and efficacy of dolutegravir in treatment-experienced subjects with raltegravir-resistant HIV type 1 infection: 24-week results of the VIKING Study. J Infect Dis 2013;207:740–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anstett K, Mesplede T, Oliveira M, Cutillas V, Wainberg MA: Dolutegravir resistance mutation R263K cannot coexist in combination with many classical integrase inhibitor resistance substitutions. J Virol 2015;89:4681–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anstett K, Cutillas V, Fusco R, Mesplede T, Wainberg MA: Polymorphic substitution E157Q in HIV-1 integrase increases R263K-mediated dolutegravir resistance and decreases DNA binding activity. J Antimicrob Chemother 2016;71:2083–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abram ME, Ram RR, Margot NA, et al. : Lack of impact of pre-existing T97A HIV-1 integrase mutation on integrase strand transfer inhibitor resistance and treatment outcome. PLoS One 2017;12:e0172206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molina JM, Lamarca A, Andrade-Villanueva J, et al. : Efficacy and safety of once daily elvitegravir versus twice daily raltegravir in treatment-experienced patients with HIV-1 receiving a ritonavir-boosted protease inhibitor: Randomised, double-blind, phase 3, non-inferiority study. Lancet Infect Dis 2012;12:27–35 [DOI] [PubMed] [Google Scholar]
- 37.Malet I, Delelis O, Valantin MA, et al. : Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob Agents Chemother 2008;52:1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Low A, Prada N, Topper M, et al. : Natural polymorphisms of human immunodeficiency virus type 1 integrase and inherent susceptibilities to a panel of integrase inhibitors. Antimicrob Agents Chemother 2009;53:4275–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamers RL, Wallis CL, Kityo C, et al. : HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: A multicentre observational study. Lancet Infect Dis 2011;11:750–759 [DOI] [PubMed] [Google Scholar]
- 40.Gupta RK, Jordan MR, Sultan BJ, et al. : Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: A global collaborative study and meta-regression analysis. Lancet 2012;380:1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raffi F, Rachlis A, Stellbrink HJ, et al. : Once-daily dolutegravir versus raltegravir in antiretroviral-naive adults with HIV-1 infection: 48 week results from the randomised, double-blind, non-inferiority SPRING-2 study. Lancet 2013;381:735–743 [DOI] [PubMed] [Google Scholar]
- 42.Cooper DA, Steigbigel RT, Gatell JM, et al. : Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med 2008;359:355–365 [DOI] [PubMed] [Google Scholar]
- 43.Fish MQ, Hewer R, Wallis CL, Venter WD, Stevens WS, Papathanasopoulos MA: Natural polymorphisms of integrase among HIV type 1-infected South African patients. AIDS Res Hum Retroviruses 2010;26:489–493 [DOI] [PubMed] [Google Scholar]
- 44.Hurt CB, Sebastian J, Hicks CB, Eron JJ: Resistance to HIV integrase strand transfer inhibitors among clinical specimens in the United States, 2009–2012. Clin Infect Dis 2014;58:423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danion F, Belissa E, Peytavin G, et al. : Non-virological response to a dolutegravir-containing regimen in a patient harbouring a E157Q-mutated virus in the integrase region. J Antimicrob Chemother 2015;70:1921–1923 [DOI] [PubMed] [Google Scholar]
- 46.Gatell JM, Katlama C, Grinsztejn B, et al. : Long-term efficacy and safety of the HIV integrase inhibitor raltegravir in patients with limited treatment options in a Phase II study. J Acquir Immune Defic Syndr 2010;53:456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeJesus E, Rockstroh JK, Henry K, et al. : Co-formulated elvitegravir, cobicistat, emtricitabine, and tenofovir disoproxil fumarate versus ritonavir-boosted atazanavir plus co-formulated emtricitabine and tenofovir disoproxil fumarate for initial treatment of HIV-1 infection: A randomised, double-blind, phase 3, non-inferiority trial. Lancet 2012;379:2429–2438 [DOI] [PubMed] [Google Scholar]