Abstract
Background
The modification of histone acetylation and deacetylation is the most important mechanism of chromatin remodeling. These modifications are a subset of epigenetic alterations which affect tumorigenesis and progression through changes in gene expression and cell growth. Results of histone modification studies prompted us to explore the therapeutic and prognostic significance of histone deacetylase 3 (HDAC3) expression in patients with breast cancer.
Material/Methods
Immunohistochemical (IHC) staining was used to detect HDAC3 expression in a tissue microarray (TMA) that included 145 patients diagnosed with invasive ductal breast carcinoma. IHC scoring was used to evaluate the staining intensity and the proportion of positive cells.
Results
HDAC3 expression was significantly correlated with estrogen receptor (ER)-negative expression (P=0.036) and progesterone receptor (PR)-negative expression (P=0.024). Additionally, HDAC3 expression was significantly positively correlated with human epidermal growth factor 2 (HER2) overexpression (P=0.037). Our study also indicated that high expression of HDAC3 was more frequently observed in breast tumors with PT2 classification (74%) versus PT1 (50.0%) and PT3 (71.4%) (P=0.040). Furthermore, HDAC3 was correlated with clinical stage II (P=0.046). Univariate and multivariate survival analyses showed that high expression of HDAC3 was correlated with poor overall survival (OS) (P=0.029 and P=0.033, respectively) in patients without lymph node involvement.
Conclusions
High HDAC3 expression is closely correlated with ER-negative expression, PR-negative expression, HER2 overexpression, PT stage, and clinical stage of breast tumors. HDAC3 may be an appropriate prognostic indicator in patients with invasive ductal breast cancer.
MeSH Keywords: Breast Neoplasms, Histone Deacetylases, Prognosis
Background
With the rapid development of medical technology and individualized therapies in the present era, numerous breakthroughs have been made in the treatment of many illnesses. However, breast cancer is still the most common malignancy and the main cause of cancer-related death among women in less developed countries [1].
The origin of mammary cancer is associated with epigenetic alterations, including histone acetylation and DNA methylation, both of which are important mechanisms in the development of cancer [2,3] and can affect the expression of various oncogenes and tumor-suppressor factors. The expression of tumor-suppressor genes can be restricted when epigenetic controls are damaged. In particular, decreased expression of tumor-suppressor genes is significantly associated with the upregulation of histone deacetylases (HDACs) in patients diagnosed with breast cancer [4,5]. The upregulation of HDACs can cause cell proliferation, lack of differentiation, migration, cell invasion, and inhibition of cell apoptosis through the downregulation or inactivation of tumor-suppressor genes [6,7].
In vivo, histone acetylation and deacetylation require a dynamic balance. HDACs are categorized into 4 classes according to their distinct structure and biological functions [8] and each class plays a crucial role in the maintenance of chromatin structure during transcription, replication, recombination, and gene repair [9,10]. HDAC3 is a class I catalytic enzyme and is associated with nuclear hormone co-repressors [11]; it represses gene expression by directly binding to the nuclear hormone co-repressors (N-CoR or SMRT) [12]. Previous studies showed that HDAC3 plays a key role in the protection of genome stability [13].
To date, several studies have demonstrated the impact of HDAC overexpression as well as the anti-tumor effects of histone deacetylase inhibitors (HDACIs) and explored the relationship between the occurrence of tumor and HDACs from the perspective of epigenetics. This has put particular focus on HDACs in cancer research, especially in breast cancer research [3,4,14]. Approximately 70% of breast carcinomas express estrogen receptor (ER) and some of these will eventually develop into tamoxifen-resistant advanced breast cancer [15]. HDAC inhibitors can restore the efficacy of tamoxifen in the treatment of ER-negative breast cancer [16]. Previous in vitro studies have examined the efficacy of HDAC inhibitors in breast cancer overexpressing human epidermal growth factor [17,18].
In the present study, we examined the expression of HDAC3 in 145 patients with invasive ductal breast cancer by immunohistochemical staining on a tissue microarray (TMA). We also analyzed the role of HDAC3 in breast tumorigenesis and the association between HDAC3 expression and clinicopathological factors and prognostic significance, which may provide a theoretical basis for the treatment of breast cancer and prognosis evaluation.
Material and Methods
Patients and tissues
The tissue microarray was composed of a collection of paraffin specimens from the First People’s Hospital of Yibin affiliated with Southwest Medical University. This cohort included 145 female patients histologically diagnosed with invasive ductal breast cancer, and the age of patients at the time of diagnosis ranged from 29 to 83 years. A typical representative tumor region was selected from each of the 145 paraffin specimens. Cylindrical core tissue specimens (diameter 0.6 mm) were acquired from the obvious regions of each paraffin block and then precisely arrayed into a new recipient paraffin block (20×35 mm) using a precision instrument [4]. All patients underwent surgical treatment between 2001 and 2004, including modified radical mastectomy or lumpectomy with axillary lymphonodectomy. Follow-up time ranged from 7 to 150 months. Patients who lacked clear dates of histopathological diagnosis or whose samples did not present with enough cancer cells on the dot of the tissue chip were excluded. The relevant dates of clinicopathological parameters and long-term follow-up for the patients in this study were obtained from the hospital. The detailed clinicopathological factors are summarized in Table 1, including age at diagnosis, histological grade, tumor size, lymph node involvement status, and hormone receptor status. The 17 cases of adjacent-carcinoma tissues were collected from the First People’s Hospital of Yibin affiliated with Southwest Medical University. This study was performed in accordance with the Helsinki Declaration and the guidelines of the Ethics Review Committee of the First People’s Hospital of Yibin affiliated with Southwest Medical University.
Table 1.
Clinicopathological factors of patients with breast cancer.
| Clinicopathologic factor | No. of patients | % |
|---|---|---|
| ALL cases | 143 | 100.0 |
| Age | ||
| <50 | 62 | 43.4 |
| ≥50 | 81 | 56.6 |
| PT status | ||
| PT1 | 32 | 22.5 |
| PT2 | 96 | 67.6 |
| PT3 | 14 | 9.9 |
| Lymph nodes | ||
| Negative | 44 | 33.3 |
| Positive | 88 | 66.7 |
| Histologic grade | ||
| G1 | 5 | 3.5 |
| G2 | 111 | 77.6 |
| G3 | 27 | 18.9 |
| Clinical stages | ||
| I | 10 | 7.1 |
| II | 83 | 59.3 |
| III | 47 | 33.6 |
| ER | ||
| Negative | 54 | 40.6 |
| Positive | 79 | 59.4 |
| PR | ||
| Negative | 51 | 38.1 |
| Positive | 83 | 61.9 |
| HER2 status | ||
| Negative | 92 | 64.8 |
| Positive | 50 | 35.2 |
Immunohistochemical staining
Immunohistochemical staining was used to evaluate the expression of HDAC3. Samples were dewaxed in xylene twice for 15 minutes each time and rehydrated in a series of alcohol solutions with a decreasing concentration gradient. Antigen retrieval was performed in 10 mmol/L citrate buffer (pH 6.0) in a microwave oven (800 W) for 16 minutes, and 3% H2O2 was used at room temperature as a blocking agent to prevent nonspecific staining. Specimens were incubated with anti-human HDAC3 rabbit monoclonal antibody (1: 100, Abcam, Ab63353) at 4°C overnight. The slide was then washed with PBS for 5 minutes and incubated with the secondary antibody at 37°C for 1 hour. Color development was performed by DAB, and the dyeing time was monitored by microscopic visualization. Specimens were counterstained with hematoxylin to ensure clear visualization of the nucleus and cytoplasm of the breast cancer cells.
Western blotting
Breast cancer tissues and adjacent-carcinoma tissues were homogenized using protein lysis buffer. Protein lysates were separated via SDS-PAGE and transferred to PVDF membranes. Membranes were blocked with 5% milk and incubated with primary antibodies against βactin (1: 5000, Cell Signaling #8457) and anti-HDAC3 (1: 5000, Cell Signaling #3949) at 4°C overnight. Membranes were washed with TBST and incubated with a horseradish peroxidase-conjugated anti-rabbit anti-mouse secondary antibody. Proteins of interest were detected with an enhanced chemiluminescent detection substrate. The process was performed according to a previously described protocol.
Scoring of the staining results
The expression of HDAC3 was observed and analyzed according to staining intensity (0, negative; 1, weak; 2, moderate; 3, strong) and the percentage of positive cells (0, negative expression; 1, ≤10%; 2, >10% and ≤50%; 3, >50%). The final scores were obtained by multiplying the staining intensity by the proportion of positive cells as described previously [3]. Samples with IHC scores <3 were considered to have low HDAC3 expression and samples with IHC scores ≥3 were considered to have high HDAC3 expression. The stained tissue microarray was evaluated by 2 pathologists who had no prior clinicopathological information about the samples.
Statistical analysis
The SPSS 17.0 statistical software package was used to analyze the association between HDAC3 expression and clinicopathological parameters as well as overall survival. Linear correlations were analyzed using the chi-square test and Fisher exact test. The Kaplan-Meier method was used to evaluate prognostic significance and the log-rank test was used to evaluate the survival curves. Multivariate analysis of survival data was conducted using the Cox proportional hazard model method. P-value <0.05 was considered statistically significant.
Results
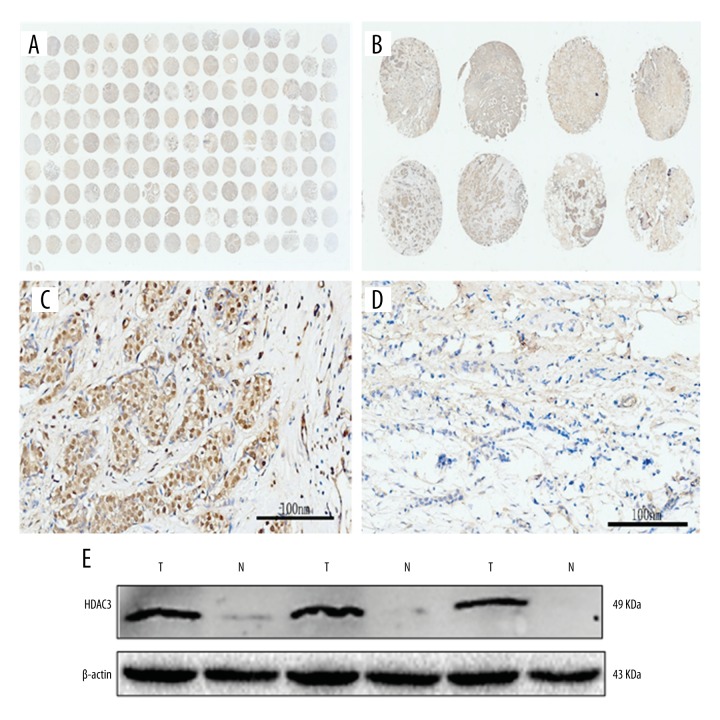
In this study, all samples in the tissue microarray came from patients who were diagnosed with invasive ductal mammary carcinoma. HDAC3 expression was higher in malignant breast cancer cells (67.8%) compared with normal mammary gland cells (23.5%) (P=0.001). HDAC3 was expressed in the nucleus and cytoplasm of breast cancer cells (Figure 1). Western blotting demonstrated that HDAC3 expression was significantly higher in breast cancer tissue than in the adjacent normal tissue (Figure 1E).
Figure 1.
The expression results of HDAC3. The tissue microarray (A, B); (C) was high expression of HDAC3; (D) represented for low expression of HDAC3; (E) the level of HDAC3 expression was significantly higher in breast cancer tissue than that of the adjacent normal tissue. T – tumor; N – normal tissue.
High HDAC3 expression was significantly correlated with negative estrogen receptor status (P=0.036) and negative progesterone receptor status (P=0.024). In addition, HDAC3 expression showed an apparent association with overexpression of human epidermal growth factor (HER2) (P=0.037). We also observed a close correlation between HDAC3 expression and PT stage (P=0.040), as well as clinical stage (P=0.046). We determined that 74% of PT2 tumors showed high HDAC3 expression and 74.7% of breast tumors in clinical stage II exhibited high HDAC3 expression. The correlation between clinicopathological factors and HDAC3 expression is demonstrated in Table 2. Associations between HDAC3 and age (P=0.475), histological grade (P=0.103), and lymph node involvement (P=0.413) were not statistically significant.
Table 2.
The correlation between HDAC3 expression and clinicopathological parameters.
| Clinicopathological parameter | Cases | HDAC3 high No. (%) |
HDAC3 low No. (%) |
P-value |
|---|---|---|---|---|
| ALL cases | 143 | 97 (67.8) | 46 (32.2) | |
| Age | 0.475 | |||
| <50 | 62 | 40 (64.5) | 22 (35.5) | |
| ≥50 | 81 | 57 (70.4) | 24 (29.6) | |
| PT stage | 0.040* | |||
| PT1 | 32 | 16 (50.0) | 16 (50.0) | |
| PT2 | 96 | 71 (74.0) | 25 (26.0) | |
| PT3 | 14 | 10 (71.4) | 4 (28.6) | |
| Lymph nodes | 0.413 | |||
| Negative | 44 | 34 (77.3) | 10 (22.7) | |
| Positive | 88 | 61 (69.3) | 27 (30.7) | |
| ER | 0.036** | |||
| Negative | 54 | 43 (79.6) | 11 (20.4) | |
| Positive | 79 | 49 (62.0) | 30 (38.0) | |
| PR | 0.024** | |||
| Negative | 51 | 40 (78.4) | 11 (21.6) | |
| Positive | 83 | 49 (59.0) | 34 (41.0) | |
| HER2 status | 0.037** | |||
| Negative | 92 | 57 (62.0) | 35 (38.0) | |
| Positive | 50 | 40 (80.0) | 10 (20.0) | |
| Clinical stage | 0.046* | |||
| I | 10 | 4 (40.0) | 6 (60.0) | |
| II | 83 | 62 (74.7) | 21 (25.3) | |
| III | 47 | 29 (61.7) | 18 (38.3) | |
| Histological grade | 0.103 | |||
| G1 | 5 | 2 (40.0) | 3 (60.0) | |
| G2 | 111 | 80 (73.1) | 31 (27.9) | |
| G3 | 27 | 15 (55.6) | 12 (44.4) |
Chi-square test;
Fisher exact test.
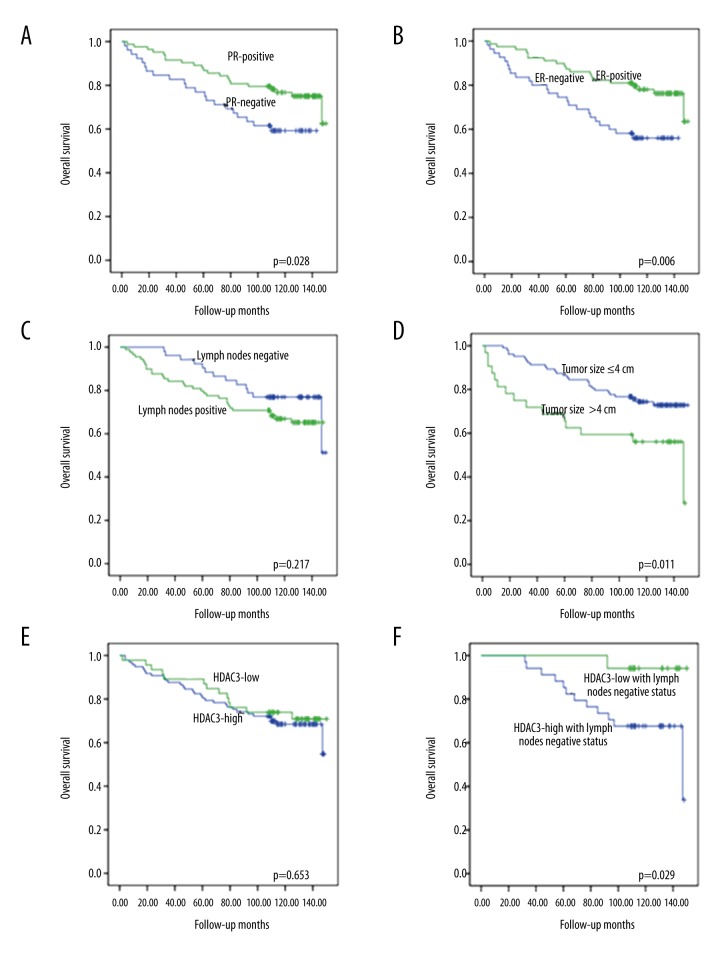
In univariate survival analyses using the Kaplan-Meier method and the log-rank test, overall survival (OS) was defined as the period of time from diagnosis to disease-related death. The results showed that high HDAC3 expression was not correlated with OS (P=0.653). In addition, high HDAC3 expression was significantly correlated with a short OS in patients negative for lymph node involvement (P=0.029). We also observed that ER (P=0.006), PR (P=0.028), and tumor size (P=0.011) were significantly correlated with OS in patients with breast cancer (Figure 2).
Figure 2.
Univariate survival analysis. (A) PR positive expression in patients with breast cancer was significantly associated with improved OS (P=0.028); (B) ER positive expression predicted prolonged OS in patients with breast cancer (P=0.006); (C) lymph nodes status had no statistical significance with the OS (P=0.217); (D) tumor size was significantly correlated with OS in our results (P=0.011); (E) there was no significance associated between HDAC3 expression and OS (P=0.653); (F) HDAC3 high expression predicted poor OS in patients with lymph nodes negative status (P=0.029).
Multivariate analysis in lymph nodes with negative status revealed that high HDAC3 expression and younger age at diagnosis were correlated with poor OS (P=0.033 and P=0.028, respectively). Tumor size ≤4 cm was correlated with an increased OS in patients without lymph node involvement (P=0.049) (Table 3). Multivariate analysis also showed that ER status (P<0.001), tumor size (P=0.041), lymph node involvement (P=0.001), and histological grade (P=0.036) were significantly correlated with OS (Table 4).
Table 3.
Multivariate analysis of overall survival in patients without lymph node involvement.
| Factor | Relevant-factor | OS HR (95%CI) |
P-Value |
|---|---|---|---|
| HDAC3 | High | 10.752 (1.211–95.500) | 0.033 |
| Tumor size | ≤4 cm | 0.163 (0.027–0.995) | 0.049 |
| HER2 | Negative | 2.251 (0.561–9.027) | 0.252 |
| Age | 1.068 (1.007–1.134) | 0.028 |
HR – hazard ratio.
Table 4.
Multivariate analysis of overall survival.
| Factor | Relevant-factor | OS HR (95%CI) |
P-Value |
|---|---|---|---|
| ER status | Negative | 8.538 (2.580–28.251) | <0.001 |
| Tumor size | ≤4 cm | 0.239 (0.061–0.942) | 0.041 |
| Histological grade | 0.036 | ||
| Grade1 | 4.489 (0.346–58.237) | 0.251 | |
| Grade2 | 0.292 (0.075–1.135) | 0.076 | |
| Lymph nodes | Negative | 0.097 (0.023–0.403) | 0.001 |
| Age | 0.994 |
HR – hazard ratio; CI – confidence interval.
Discussion
In recent years, HDACs have become an important topic in the study of the genesis and development of tumors, especially breast carcinomas. Histone deacetylases encourage tight bonding of DNA with histones through removal of the acetyl group. Additionally, they repress gene transcription by hindering the binding of the transcriptional unit to the promoter. In our study, we observed that HDAC3 expression was significantly higher in breast cancer tissue than in normal breast tissue, and we found that HDAC3 is similar to the partial anisotropy parameter of diffusion tensor imaging as a new malignant marker of breast tumor [19]. We also explored the correlation between the expression of HDAC3 and clinical parameters in breast cancer because this is important for the development of therapeutics and prognostic techniques. Tumors with an intermediate clinical stage and PT2 classification exhibited high expression of HDAC3 in our study.
Berit et al. [13] performed HDAC1, 2, and 3 expression analyses via immunohistochemical staining. Their study showed that HDAC3 expression was significantly associated with negative hormone receptor status, which is consistent with our findings. Additionally, they found that the expression of HDAC3 was correlated with poor differentiation in tumors. However, Krusche et al. [4] demonstrated that high HDAC3 expression was significantly correlated with ER and PR positivity. Their research also indicated that the expression of HDAC3 was associated with low rates of proliferation in breast cancer cells. In contrast to our study, neither of these studies reported a positive correlation between HDAC3 expression and HER2.
To date, several studies have demonstrated that HDAC inhibitors can improve interactions between ER-negative malignant breast tumors and the anti-estrogen tamoxifen [20,21]. Our findings are in accordance with the concept that HDAC3 overexpression can repress hormone receptor expression in breast cancer. Suppressing the activity or expression of HDAC3 results in re-expression of hormone receptors in aggressive ER- and PR-negative breast tumors. In contrast, another study has demonstrated that knockdown of HDAC3 can reduce ERa expression, and that HDAC3 plays a crucial role in maintaining the stability of ERa mRNA in breast cancer [22]. In vitro studies have shown that trastuzumab can cause apoptosis of HER2-overexpressing breast cancer cells when used in combination with HDACIs [23]. In our study, we observed a significant association between high HDAC3 expression and HER2 amplification, indicating that breast cancer patients with high HDAC3 expression experience a significant therapeutic effect from HDACIs.
In our study, HDAC3 expression was closely correlated with ER-negativity, PR-negativity, and HER2 over-expression in malignant breast tumors. This is the worst hormone receptor status according to our long-term clinical observations. This result suggests that patients with high HDAC3 expression may benefit more from treatment with specific HDAC3 inhibitors in combination with other therapies (including chemotherapy, trastuzumab-targeted therapy, and surgical treatment) compared to patients with low HDAC3 expression, HER2-amplification, and negative hormone receptor status. HDAC3 inhibitors such as capecitabine [24] may inhibit the growth of tumor cells and angiogenesis, but the specific therapeutic mechanism needs further study. Our experimental results also indicate that the inhibitors of HDAC3 are the same as those of magnetic multi-walled carbon nanotube-doxorubicin conjugate, which provides an experimental basis for novel targeted drugs [25].
Interestingly, our study demonstrated that, in patients with larger primary tumors and without lymph node involvement, high HDAC3 expression was correlated with a shorter OS. This result indicates that high HDAC3 expression is an early sign of malignancy and can predict a worse prognosis. Conversely, Berit et al. and Krusche et al. did not obtain prognostic value in their examination of HDAC3 expression in breast cancer. Therefore, our study is the first to propose that high HDAC3 expression is a sign of poor prognosis in breast cancer.
In this study, we observed that HDAC3 expression was significantly correlated with traditional clinicopathological parameters, and that HDAC3 can be considered a prognostic factor. This finding prompted us to further study the significance of HDAC3 expression in breast cancer as well as the correlation of HDAC3 expression with breast cancer treatment efficacy and prognosis.
Conclusions
Based on the results of our study, we propose that HDAC3 is an independent prognostic factor in breast cancer; however, further studies are needed to confirm the role of HDAC3 in breast cancer.
This manuscript has not been published and is not under consideration for publication elsewhere. We have no conflicts of interest to disclose. All authors have read and approved the final version of the manuscript. The authors declare no competing financial interests.
Footnotes
Source of support: Departmental sources
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Kristensen LS, Nielsen HM, Hansen LL. Epigenetics and cancer treatment. Eur J Pharmacol. 2009;625:131–42. doi: 10.1016/j.ejphar.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Seo J, Min SK, Park HR, et al. Expression of histone deacetylases HDAC1, HDAC2, HDAC3, and HDAC6 in invasive ductal carcinomas of the breast. J Breast Cancer. 2014;17:323–31. doi: 10.4048/jbc.2014.17.4.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krusche CA, Wulfing P, Kersting C, et al. Histone deacetylase-1 and -3 protein expression in human breast cancer: A tissue microarray analysis. Breast Cancer Res Treat. 2005;90:15–23. doi: 10.1007/s10549-004-1668-2. [DOI] [PubMed] [Google Scholar]
- 5.Thiagalingam S, Cheng KH, Lee HJ, et al. Histone deacetylases: Unique players in shaping the epigenetic histone code. Ann NY Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 6.Parbin S, Kar S, Shilpi A, et al. Histone deacetylases: A saga of perturbed acetylation homeostasis in cancer. J Histochem Cytochem. 2014;62:11–33. doi: 10.1369/0022155413506582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;265:420–32. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 8.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallinari P, Di Marco S, Jones P, et al. HDACs, histone deacetylation and gene transcription: From molecular biology to cancer therapeutics. Cell Res. 2007;17:195–211. doi: 10.1038/sj.cr.7310149. [DOI] [PubMed] [Google Scholar]
- 10.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37:569–76. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 11.Codina A, Love JD, Li Y, et al. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proc Natl Acad Sci USA. 2005;102:6009–14. doi: 10.1073/pnas.0500299102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhaskara S, Hiebert SW. Role for histone deacetylase 3 in maintenance of genome stability. Cell Cycle. 2011;10:727–28. doi: 10.4161/cc.10.5.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller BM, Jana L, Kasajima A, et al. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer – overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer. 2013;13:215. doi: 10.1186/1471-2407-13-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legare S, Basik M. Minireview: The link between ERalpha corepressors and histone deacetylases in tamoxifen resistance in breast cancer. Mol Endocrinol. 2016;30:965–76. doi: 10.1210/me.2016-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giacinti L, Claudio PP, Lopez M, Giordano A. Epigenetic information and estrogen receptor alpha expression in breast cancer. Oncologist. 2006;11:1–8. doi: 10.1634/theoncologist.11-1-1. [DOI] [PubMed] [Google Scholar]
- 17.Kim YJ, Greer CB, Cecchini KR, et al. HDAC inhibitors induce transcriptional repression of high copy number genes in breast cancer through elongation blockade. Oncogene. 2013;32:2828–35. doi: 10.1038/onc.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bali P, Pranpat M, Swaby R, et al. Activity of suberoylanilide hydroxamic Acid against human breast cancer cells with amplification of her-2. Clin Cancer Res. 2005;11:6382–89. doi: 10.1158/1078-0432.CCR-05-0344. [DOI] [PubMed] [Google Scholar]
- 19.Jiang R, Zeng X, Sun S, et al. Assessing detection, discrimination, and risk of breast cancer according to anisotropy parameters of diffusion tensor imaging. Med Sci Monit. 2016;22:1318–28. doi: 10.12659/MSM.895755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang ER, Lim SJ, Lee ES, et al. The histone deacetylase inhibitor trichostatin A sensitizes estrogen receptor alpha-negative breast cancer cells to tamoxifen. Oncogene. 2004;23:1724–36. doi: 10.1038/sj.onc.1207315. [DOI] [PubMed] [Google Scholar]
- 21.Fortunati N, Bertino S, Costantino L, et al. Valproic acid restores ER alpha and antiestrogen sensitivity to ER alpha-negative breast cancer cells. Mol Cell Endocrinol. 2010;314:17–22. doi: 10.1016/j.mce.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Oie S, Matsuzaki K, Yokoyama W, et al. HDAC3 regulates stability of estrogen receptor alpha mRNA. Biochem Biophys Res Commun. 2013;432:236–41. doi: 10.1016/j.bbrc.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Huang X, Gao L, Wang S, et al. HDAC inhibitor SNDX-275 induces apoptosis in erbB2-overexpressing breast cancer cells via down-regulation of erbB3 expression. Cancer Res. 2009;69:8403–11. doi: 10.1158/0008-5472.CAN-09-2146. [DOI] [PubMed] [Google Scholar]
- 24.Maur M, Omarini C, Piacentini F, et al. Metronomic capecitabine effectively blocks leptomeningeal carcinomatosis from breast cancer: A case report and literature review. Am J Case Rep. 2017;18:208–11. doi: 10.12659/AJCR.901812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji J, Liu M, Meng Y, et al. Experimental study of magnetic multi-walled carbon nanotube-doxorubicin conjugate in a lymph node metastatic model of breast cancer. Med Sci Monit. 2016;22:2363–73. doi: 10.12659/MSM.898597. [DOI] [PMC free article] [PubMed] [Google Scholar]