 We have developed a novel AND logic based fluorescence probe for the simultaneous detection of ONOO– and GSH (GSH-PF3).
We have developed a novel AND logic based fluorescence probe for the simultaneous detection of ONOO– and GSH (GSH-PF3).
Abstract
We have developed a novel AND logic based fluorescence probe for the simultaneous detection of ONOO– and GSH (GSH-PF3). The GSH-PF3 probe was synthesised over three steps starting from commercially available fluorescein. The probe was constructed by attaching the GSH reactive motif, 2,4-dinitrobenzenesulfonyl, to the previously reported boronate fluorescence based probe, PF3. GSH-PF3 produced only a small fluorescence response towards the addition of GSH or ONOO– separately. However, when the probe was exposed to both analytes, there was a significant (40-fold) fluorescence enhancement. GSH-PF3 demonstrated an excellent selectivity towards both GSH and ONOO–. In cellular imaging experiments the probe was shown to be cell permeable with no ‘turn-on’ response observed for the addition of either GSH or ONOO– separately. However, in the presence of both analytes, a clear fluorescence response was observed in live cells. GSH-PF3 was further able to monitor the co-existence of metabolically produced ONOO– and GSH by exogenous stimulation.
Peroxynitrite (ONOO–), is a highly reactive nitrogen species that is formed via the diffusion controlled reaction between superoxide anion (O2˙–) and nitric oxide (NO˙).1,2 ONOO– acts as a signalling molecule in vivo for a number of pathways.1,3 However, ONOO– is more commonly known for its deleterious properties, causing irreversible damage to a range of biological targets such as lipids, proteins and DNA.4 Therefore, ONOO– has been implicated as a key pathogenic factor for a number of diseases, which include inflammation, cancer, ischemia-reperfusion and neurodegenerative diseases.5–7 Glutathione (GSH) is a natural tripeptide (γ-l-glutamyl-l-cysteinyl-glycine), that exists in the thiol reduced form (GSH) and disulphide-oxidised (GSSG) form. GSH is the predominant form, existing at millimolar concentrations in most cells.8 However, GSH can be directly oxidised by ONOO– therefore acting as a cellular defence by serving as an ONOO– scavenger. Elevated levels of GSH are common in cells under oxidative stress and the susceptibility of a cell towards ONOO– largely depends on the concentration of intracellular GSH.1,9,10
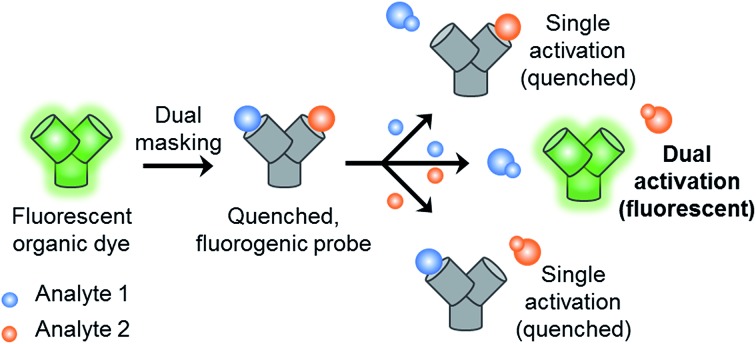
Therefore, with this work we set out to develop a fluorescence-based probe capable of monitoring the close relationship between ONOO– and GSH. Traditionally, most fluorescence probes require a single analyte to produce a fluorescence response.11–19 However, in recent years a number of fluorescence based probes for dual or multi-analyte detection have been developed.20–27 These types of fluorescence based probes have been used for the construction of molecular logic gates or for medical diagnostics.28 AND logic based fluorescence probes require both analytes to be simultaneously present or work in tandem in order to elicit a fluorescence response. This method has a number of advantages including: being faster than serial measurements for different analytes within the same biological sample and can provide a method for monitoring bimolecular events, which may contribute to a specific disease.20
Currently, only a few reversible fluorescence based probes for the detection of ONOO– and GSH have been developed to monitor the relationship between these analytes.29,30 These include a selenium based fluorescence probe, which is oxidised by ONOO– (turn “on”) and reduced by GSH (turn “off”). The fluorescence of the CyPSe probe is initially quenched by a photoinduced electron transfer (PET) process. The presence of ONOO– results in the oxidation of selenium to CyPSe O causing the fluorescence emission to be “turned on”. Then in the presence of biological thiols such as cysteine and GSH, the CyPSe O probe is reduced back to its non-fluorescent selenide form. A similar reversible NIR Tellurium-based fluorescence probe was later developed for monitoring the redox cycles between ONOO– and GSH. This probe was successfully applied for the visualisation of the redox cycles of ONOO– and GSH in live cells and animals.29 The probes developed by Han et al. are turn “on” with ONOO– and “off” with GSH. As far as we are aware, there are currently no AND logic based fluorescence probes for GSH “and” ONOO–.
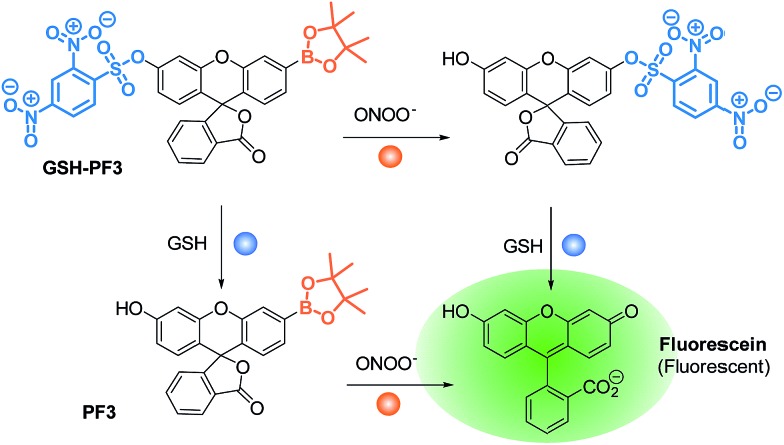
With this research, we aimed to develop a GSH “and” ONOO– logic based fluorescence probe (Scheme 1). We identified a suitable mono-boronate fluorescein based fluorescence probe, PF3 that had been previously developed by Chang et. al., shown in Scheme 2. As previously reported the reactivity of boronate based fluorescence probes with ONOO–,12,14,15 are significantly greater than hypochlorite (ClO–) and H2O2.31 Therefore, we anticipated that the attachment of a GSH reactive motif to PF3 would produce a selective GSH-ONOO– AND logic based fluorescence probe, GSH-PF3 (Scheme 2). GSH-PF3 was readily synthesised in three steps. Fluorescein was triflated using N-phenyl bis(trifluoromethanesulfonamide) to afford fluorescein mono-triflate in good yield. Suzuki–Miyaura conditions were then carried out to provide fluorescein mono-boronate, PF3. The 2,4-dinitrobenzenesulfonyl unit was then attached to PF3 using 2,4-dinitrobenzenesulfonyl chloride, CH2Cl2 and NEt3 at 0 °C. Using these conditions GSH-PF3 was prepared in a reasonable yield of 52%.
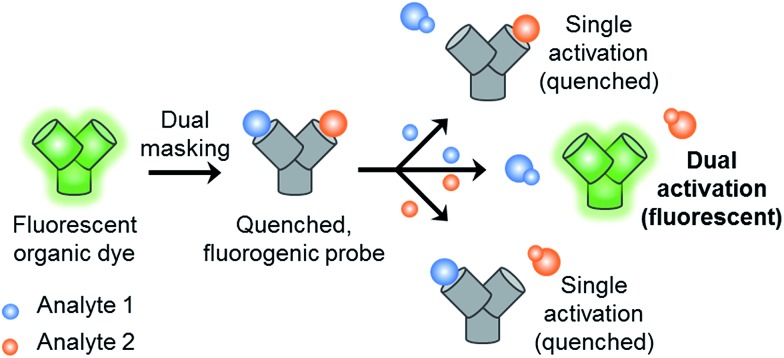
Scheme 1. Design concept for AND logic based fluorogenic probe. A fluorescence dye is masked by two functional groups, which respond to two different analytes. The fluorogenic probe requires both analytes to be present or to work in tandem in order to produce a response.
Scheme 2. Structure of the GSH-PF3 probe and proposed sensing mechanism for the simultaneous detection of ONOO– and GSH.
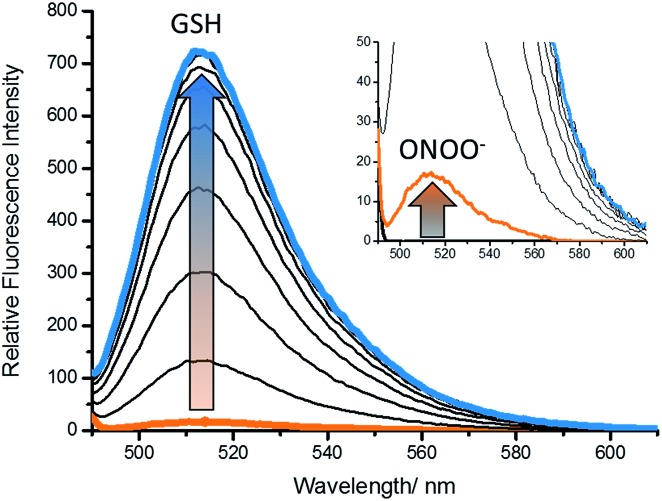
With GSH-PF3 in hand, fluorescence experiments for the detection of GSH and ONOO– were performed. As shown in Fig. 1, GSH-PF3 was initially non-fluorescent and with the addition of ONOO– (10 μM), a small fluorescence increase was observed. However, incremental additions of GSH resulted in a much larger increase in fluorescence intensity (>30-fold see ESI – Fig. S1†), clearly demonstrating the need for the addition of both GSH and ONOO– in order to achieve a full ‘turn-on’ response.
Fig. 1. Fluorescence spectra of GSH-PF3 (0.5 μM) with addition of ONOO– (10 μM) (inset) followed by the addition of GSH (0–80 μM), 5 min wait between addition in buffer solution [52 wt% methanol] (pH = 8.21 at 25 °C). Fluorescence intensities were measured with λex = 488 nm with slit widths ex slit: 5 nm and em slit: 2.5 nm.
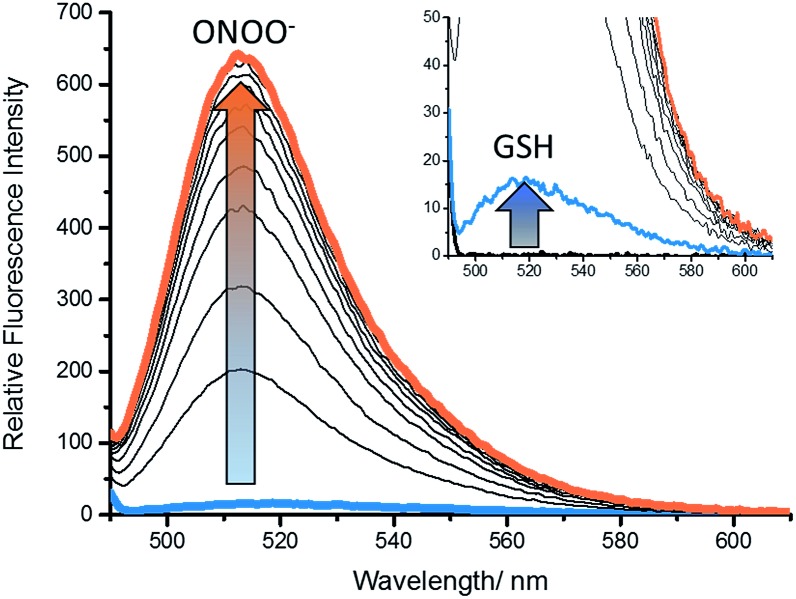
In order to demonstrate that GSH-PF3 required both GSH and ONOO– for a complete ‘turn-on’ response, the fluorescence experiments were performed the other way around. Therefore, an excess of GSH (200 μM) was added to GSH-PF3, and the probe was incubated for 10 min. Remarkably, this only led to a small increase in fluorescence intensity and the subsequent additions of ONOO– resulted in a large fluorescence increase (Fig. 2 and ESI – Fig S2†). These results confirm that the probe requires both GSH and ONOO– for a full fluorescence ‘turn-on’ response.
Fig. 2. Fluorescence spectra of GSH-PF3 (0.5 μM) with addition of GSH (200 μM), 10 min wait (inset), then addition of ONOO– (0–10 μM) in buffer solution [52 wt% methanol] (pH = 8.21 at 25 °C). Fluorescence intensities were measured with λex = 488 nm with slit widths ex slit: 5 nm and em slit: 2.5 nm.
The selectivity of GSH-PF3 was then evaluated against a series of amino acids (l-cysteine, l-methionine, l-tryptophan, l-serine, l-lysine, l-leucine, l-glutamic acid, l-valine, l-arginine, l-histidine and l-aspartic acid) – see ESI Fig. S3.† Unsurprisingly, the probe responded to the other sulfhydryl containing amino acid cysteine. However, the biological concentrations of cysteine are low in cells.32 More importantly, GSH-PF3 displayed an excellent selectivity against other amino acids including serine, methionine and lysine. The probe demonstrated excellent selectivity for ONOO– over many other ROS including H2O2 (see ESI – Fig. S4†). The excellent selectivity for both GSH and ONOO– allowed us to evaluate GSH-PF3 in cell imaging experiments.
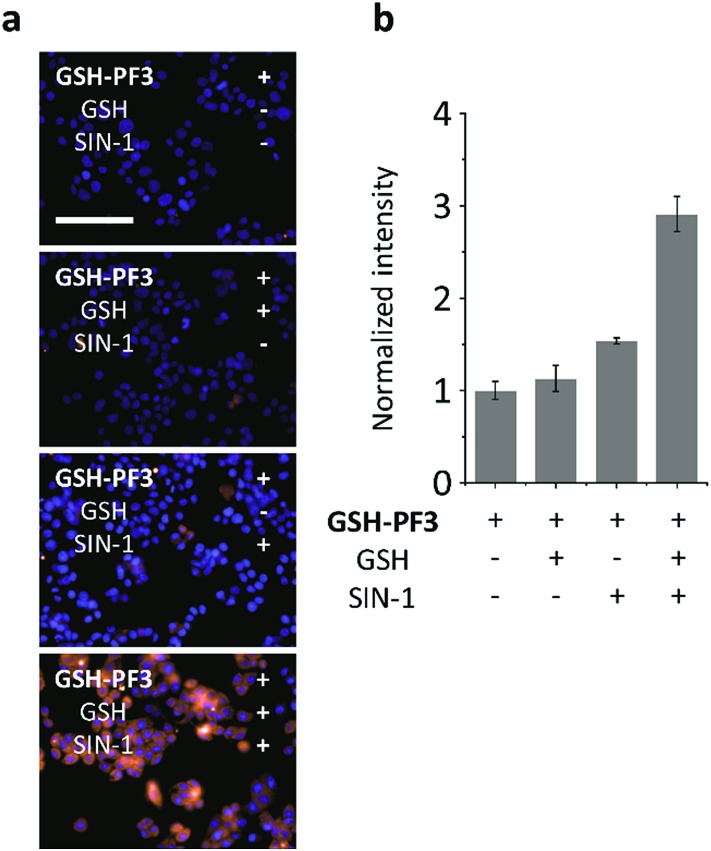
The macrophage cell line – RAW264.7 (ATCC® TIB-71™; obtained from ATCC [American Type Culture Collection]) – was used for cell imaging experiments. The cells were incubated with GSH-PF3, followed by either treatment of SIN-1 (an ONOO– donor)33 to produce intracellular ONOO– or by addition of exogenous GSH. As shown below in Fig. 3, addition of GSH separately resulted in no fluorescence response, whereas ONOO– led to a small fluorescence response in cells. This observation is believed to be due to the presence of a low levels of endogenous biological thiols in the cells, resulting in the activation of the probe's fluorescence. As predicted, treatment of cells with both GSH and SIN-1 led to a clear fluorescence increase in RAW264.7 cells with GSH-PF3, in clear agreement with the analytical data obtained on the fluorimeter.
Fig. 3. Fluorescence imaging (a) and quantification (b) of RAW264.7 cells with GSH-PF3 (10 μM) in the absence and presence of exogenously added GSH (50 μM) and/or SIN-1 (500 μM). Excitation channel = 460–490 nm, emission channel filtered = 530–590 nm. Cell nuclei were stained with Hoechst 33342. Scale bar = 100 μm. Error bars represent SD.
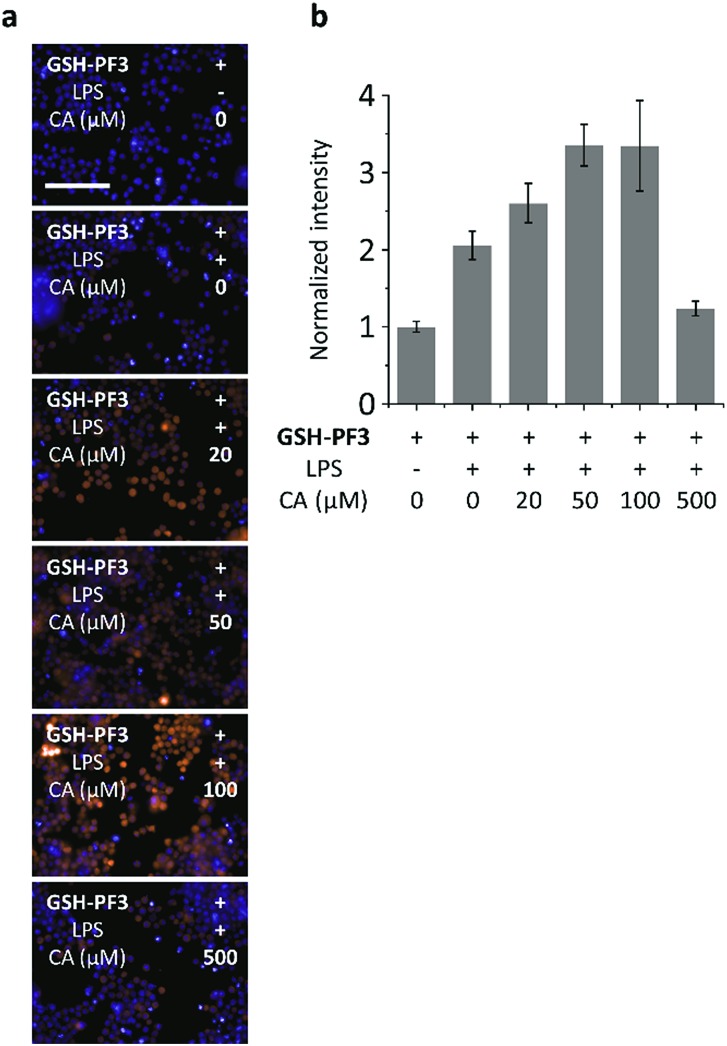
We then turned our attention to evaluate the ability of GSH-PF3 for monitoring the co-existence of metabolically produced ONOO– by lipopolysaccharide (LPS) simulation34 and GSH through a drug treatment.35 It has been reported that LPS can elicit ONOO– in macrophage, which is a signature of inflammation, whereas caffeic acid (CA) is commonly used to treat inflammation through the augmentation of intracellular GSH. Consequently, treatment with LPS, followed by the addition of increasing CA (0–100 μM) led to a gradual enhancement in the fluorescence intensity of GSH-PF3 (Fig. 4). This result is due to the co-existence of ONOO– produced by LPS stimulation as well as GSH elicited by CA. Interestingly, a further increase of the CA concentration (>100 μM) led to the suppression of probe's fluorescence, which is believed to be due to the production of an excess of GSH, resulting in quenching of the ONOO– (Fig. 4). To corroborate the presence of intracellular GSH in macrophage, a commercial GSH probe was used. The result suggested that both the exogenous addition of GSH and stimulation of metabolic GSH by treatment of CA/LPS activated the commercial probe's fluorescence (Fig. S5†). Furthermore, a cell proliferation assay indicated that GSH-PF3 is not toxic for RAW264.7 cells (Fig. S6†).
Fig. 4. Fluorescence imaging (a) and quantification (b) of RAW264.7 cells with GSH-PF3 (10 μM) in the absence and presence of LPS (1 μg mL–1), which elicits ONOO– and increasing caffeic acid (CA, a drug that elicits endogenous GSH to quench ONOO–). Excitation channel = 460–490 nm, emission channel filtered = 530–590 nm. Cell nuclei were stained with Hoechst 33342. Scale bar = 100 μm. Error bars represent SD.
These results clearly demonstrate the potential of AND logic based fluorescence imaging probes for optimizing drug dosage in the treatment of inflammation and various other diseases. In addition this system could be used to help monitor the treatment of Alzheimer's disease (AD), since it is known that high levels of ONOO– correlate with oxidative stress and the progression of AD, while, high levels of GSH have been implicated in AD therapy.36–39 Therefore, we believe that our GSH-PF3 probe, which can detect GSH “and” ONOO– could potentially be used to monitor cellular resistance towards ROS and map therapeutic improvements in AD victims.
In summary, GSH-PF3 is an easy-to-prepare fluorescence based probe providing a platform for the development of other novel AND logic based fluorescence imaging probes for use in medical diagnostics. We are currently exploring the use of 6-amino/carboxyfluorescein, which we believe could provide the opportunity to attach additional fluorophores in order to develop ratiometric fluorescence sensors40 or for the attachment of targeting or therapeutic units.41–43
Conflicts of interest
No conflicts of interest.
Supplementary Material
Acknowledgments
We would like to thank the EPSRC and the University of Bath for funding. ACS and JEG thank the EPSRC for studentships. X.-P. He thanks the National Natural Science Foundation of China (21722801 and 21572058), the Science and Technology Commission of Shanghai Municipality (15540723800), the Fundamental Research Funds for the Central Universities (222201717003) and the Shanghai Rising-Star Program (16QA1401400) for financial support. TDJ wishes to thank the Royal Society for a Wolfson Research Merit Award and ECUST for a guest professorship. The Catalysis and Sensing for our Environment (CASE) network is thanked for research exchange opportunities. NMR characterisation facilities were provided through the Chemical Characterisation and Analysis Facility (CCAF) at the University of Bath (http://www.bath.ac.uk/ccaf). The EPSRC UK National Mass Spectrometry Facility at Swansea University is thanked for analyses. All data supporting this study are provided as Electronic Supplementary Information (ESI) accompanying this paper.
Footnotes
†Electronic supplementary information (ESI) available: Additional figures, experimental section and original spectra of new compounds. See DOI: 10.1039/c8sc00733k
References
- Pacher P., Beckman J. S., Liaudet L. Physiol. Rev. 2007;87:315. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman J. S., Koppenol W. H. Am. J. Physiol. Cell Physiol. 1996;271:C1424. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- Weidinger A., Kozlov A. V. Biomolecules. 2015;5:472. doi: 10.3390/biom5020472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi P., di Masi A., Sciorati C., Clementi E. Biofactors. 2010;36:264. doi: 10.1002/biof.103. [DOI] [PubMed] [Google Scholar]
- Ischiropoulos H., Beckman J. S. J. Clin. Invest. 2003;111:163. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarchielli P., Galli F., Floridi A., Gallai V. Amino Acids. 2003;25:427. doi: 10.1007/s00726-003-0028-6. [DOI] [PubMed] [Google Scholar]
- Wink D. A., Vodovotz Y., Laval J., Laval F., Dewhirst M. W., Mitchell J. B. Carcinogenesis. 1998;19:711. doi: 10.1093/carcin/19.5.711. [DOI] [PubMed] [Google Scholar]
- Lu S. C. Mol. Aspect. Med. 2009;30:42. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall K. A., Reist R., Jenner P., Halliwell B. Free Radic. Biol. Med. 1999;27:515. doi: 10.1016/s0891-5849(99)00094-5. [DOI] [PubMed] [Google Scholar]
- Bolanos J. P., Heales S. J. R., Land J. M., Clark J. B. J. Neurochem. 1995;64:1965. doi: 10.1046/j.1471-4159.1995.64051965.x. [DOI] [PubMed] [Google Scholar]
- Li M., Wu X. M., Wang Y., Li Y. S., Zhu W. H., James T. D. Chem. Commun. 2014;50:1751. doi: 10.1039/c3cc48128j. [DOI] [PubMed] [Google Scholar]
- Palanisamy S., Wu P. Y., Wu S. C., Chen Y. J., Tzou S. C., Wang C. H., Chen C. Y., Wang Y. M. Biosens. Bioelectron. 2017;91:849. doi: 10.1016/j.bios.2017.01.027. [DOI] [PubMed] [Google Scholar]
- Cheng D., Pan Y., Wang L., Zeng Z. B., Yuan L., Zhang X. B., Chang Y. T. J. Am. Chem. Soc. 2017;139:285. doi: 10.1021/jacs.6b10508. [DOI] [PubMed] [Google Scholar]
- Sedgwick A. C., Sun X. L., Kim G., Yoon J., Bull S. D., James T. D. Chem. Commun. 2016;52:12350. doi: 10.1039/c6cc06829d. [DOI] [PubMed] [Google Scholar]
- Sun X., Xu Q., Kim G., Flower S. E., Lowe J. P., Yoon J., Fossey J. S., Qian X., Bull S. D., James T. D. Chem. Sci. 2014;5:3368. [Google Scholar]
- Yin J., Kwon Y., Kim D., Lee D., Kim G., Hu Y., Ryu J. H., Yoon J. J. Am. Chem. Soc. 2014;136:5351. doi: 10.1021/ja412628z. [DOI] [PubMed] [Google Scholar]
- Xu Q., Lee K.-A., Lee S., Lee K. M., Lee W.-J., Yoon J. J. Am. Chem. Soc. 2013;135:9944. doi: 10.1021/ja404649m. [DOI] [PubMed] [Google Scholar]
- Lei Z., Li X., Luo X., He H., Zheng J., Qian X., Yang Y. Angew. Chem., Int. Ed. 2017;56:2979. doi: 10.1002/anie.201612301. [DOI] [PubMed] [Google Scholar]
- Kuriki Y., Kamiya M., Kubo H., Komatsu T., Ueno T., Tachibana R., Hayashi K., Hanaoka K., Yamashita S., Ishizawa T., Kokudo N., Urano Y. J. Am. Chem. Soc. 2018;140:1767. doi: 10.1021/jacs.7b11014. [DOI] [PubMed] [Google Scholar]
- Romieu A. Org. Biomol. Chem. 2015;13:1294. doi: 10.1039/c4ob02076f. [DOI] [PubMed] [Google Scholar]
- Yu L., Wang S. L., Huang K. Z., Liu Z. G., Gao F., Zeng W. B. Tetrahedron. 2015;71:4679. [Google Scholar]
- Van de Bittner G. C., Bertozzi C. R., Chang C. J. J. Am. Chem. Soc. 2013;135:1783. doi: 10.1021/ja309078t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Lin W. Y., Xie Y. N., Chen B., Zhu S. S. J. Am. Chem. Soc. 2012;134:1305. doi: 10.1021/ja2100577. [DOI] [PubMed] [Google Scholar]
- Dai C. G., Liu X. L., Du X. J., Zhang Y., Song Q. H. ACS Sens. 2016;1:888. [Google Scholar]
- Debieu S., Romieu A. Org. Biomol. Chem. 2015;13:10348. doi: 10.1039/c5ob01624j. [DOI] [PubMed] [Google Scholar]
- Yi L., Wei L., Wang R. Y., Zhang C. Y., Zhang J., Tan T. W., Xi Z. Chem.–Eur. J. 2015;21:15167. doi: 10.1002/chem.201502832. [DOI] [PubMed] [Google Scholar]
- Ang C. Y., Tan S. Y., Wu S. J., Qu Q. Y., Wong M. F. E., Luo Z., Li P. Z., Selvan S. T., Zhao Y. L. J. Mater. Chem. C. 2016;4:2761. [Google Scholar]
- Erbas-Cakmak S., Kolemen S., Sedgwick A. C., Gunnlaugsson T., James T. D., Yoon J., Akkaya E. U. Chem. Soc. Rev. 2018 doi: 10.1039/c7cs00491e. [DOI] [PubMed] [Google Scholar]
- Yu F. B., Li P., Wang B. S., Han K. L. J. Am. Chem. Soc. 2013;135:7674. doi: 10.1021/ja401360a. [DOI] [PubMed] [Google Scholar]
- Yu F. B. A., Li P., Li G. Y., Zhao G. J., Chu T. S., Han K. L. J. Am. Chem. Soc. 2011;133:11030. doi: 10.1021/ja202582x. [DOI] [PubMed] [Google Scholar]
- Sikora A., Zielonka J., Lopez M., Joseph J., Kalyanaraman B. Free Radic. Biol. Med. 2009;47:1401. doi: 10.1016/j.freeradbiomed.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. Y., Fang Y. Z., Yang S., Lupton J. R., Turner N. D. J. Nutr. 2004;134:489. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- Li X., Tao R. R., Hong L. J., Cheng J., Jiang Q., Lu Y. M., Liao M. H., Ye W. F., Lu N. N., Han F., Hu Y. Z., Hu Y. H. J. Am. Chem. Soc. 2015;137:12296. doi: 10.1021/jacs.5b06865. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres A., Jones-Carson J., Balish E. Infect. Immun. 1996;64:3127. doi: 10.1128/iai.64.8.3127-3133.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moridani M. Y., Scobie H., Jamshidzadeh A., Salehi P., O'Brien P. J. Drug Metab. Dispos. 2001;29:1432. [PubMed] [Google Scholar]
- Smith M. A., Harris P. L. R., Sayre L. M., Beckman J. S., Perry G. J. Neurosci. 1997;17:2653. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Gutierrez E., Munoz-Arenas G., Trevino S., Espinosa B., Chavez R., Rojas K., Flores G., Diaz A., Guevara J. Synapse. 2017;71:e21990. doi: 10.1002/syn.21990. [DOI] [PubMed] [Google Scholar]
- Pocernich C. B., Butterfield D. A. Biochim. Biophys. Acta, Mol. Basis Dis. 2012;1822:625. doi: 10.1016/j.bbadis.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciatore I., Baldassarre L., Fornasari E., Mollica A., Pinnen F. Oxid. Med. Cell. Longevity. 2012:240146. doi: 10.1155/2012/240146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers A. E., Okreglak V. S., Chang C. J. J. Am. Chem. Soc. 2006;128:9640. doi: 10.1021/ja063308k. [DOI] [PubMed] [Google Scholar]
- Lee M. H., Kim J. Y., Han J. H., Bhuniya S., Sessler J. L., Kang C., Kim J. S. J. Am. Chem. Soc. 2012;134:12668. doi: 10.1021/ja303998y. [DOI] [PubMed] [Google Scholar]
- Bhuniya S., Maiti S., Kim E. J., Lee H., Sessler J. L., Hong K. S., Kim J. S. Angew. Chem., Int. Ed. 2014;53:4469. doi: 10.1002/anie.201311133. [DOI] [PubMed] [Google Scholar]
- He X.-P., Tian H. Chem. 2018;4:246. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.