Abstract
Cytoplasmic linker-associated proteins (CLASPs) are microtubule-associated proteins essential for microtubule regulation in many cellular processes. However, the molecular mechanisms underlying CLASP activity are not understood. Here, we use purified protein components and total internal reflection fluorescence microscopy to investigate the effects of human CLASP2 on microtubule dynamics in vitro. We demonstrate that CLASP2 suppresses microtubule catastrophe and promotes rescue without affecting the rates of microtubule growth or shrinkage. Strikingly, when CLASP2 is combined with EB1, a known binding partner, the effects on microtubule dynamics are strongly enhanced. We show that synergy between CLASP2 and EB1 is dependent on a direct interaction, since a truncated EB1 protein that lacks the CLASP2-binding domain does not enhance CLASP2 activity. Further, we find that EB1 targets CLASP2 to microtubules and increases the dwell time of CLASP2 at microtubule tips. Although the temporally averaged microtubule growth rates are unaffected by CLASP2, we find that microtubules grown with CLASP2 display greater variability in growth rates. Our results provide insight into the regulation of microtubule dynamics by CLASP proteins and highlight the importance of the functional interplay between regulatory proteins at dynamic microtubule ends.
INTRODUCTION
Cytoplasmic linker-associated proteins (CLASPs) are a conserved family of microtubule-associated proteins, essential for a broad range of cellular processes including cell division, motility, and neuronal development. In dividing cells, CLASPs organize spindle microtubules (Pasqualone and Huffaker, 1994; Inoue et al., 2000; Lemos et al., 2000; Maiato et al., 2002, 2003, 2005; Yin et al., 2002; Cheeseman et al., 2005; Hannak and Heald, 2006; Mimori-Kiyosue et al., 2006; Pereira et al., 2006; Ambrose et al., 2007; Bratman and Chang, 2007; Maffini et al., 2009; Maia et al., 2012; Funk et al., 2014); in migrating cells, CLASPs polarize the microtubule network (Akhmanova et al., 2001; Wittmann and Waterman-Storer, 2005; Drabek et al., 2006; Efimov et al., 2007; Miller et al., 2009; Stehbens et al., 2014); and, in neurons, CLASPs regulate microtubules in growth cones and synapses (Lee et al., 2004; Beffert et al., 2012; Schmidt et al., 2012; Marx et al., 2013; Dillon et al., 2017).
CLASPs localize at the tips and along the lattices of microtubules in cells (Akhmanova et al., 2001; Mimori-Kiyosue et al., 2005; Wittmann and Waterman-Storer, 2005; Kumar et al., 2009). Several structural features mediate the interaction between CLASPs and microtubules: multiple conserved tumor overexpressed gene (TOG) domains permit binding to tubulin (Al-Bassam et al., 2010; Leano et al., 2013; Funk et al., 2014), and a basic region mediates electrostatic interactions with the microtubule lattice (Al-Bassam et al., 2010; Patel et al., 2012). Depending on the species and isoform, CLASPs can possess canonical TOG domains, similar to those of the XMAP215/Dis1family of proteins, as well as cryptic TOG domains, which are structurally conserved but sequence divergent (Slep and Vale, 2007; Leano et al., 2013). In addition, Ser-x-Ile-Pro (SxIP) motifs mediate the binding of CLASPs to EB proteins (Patel et al., 2012; Maki et al., 2015), which target CLASPs to microtubule tips in cells (Mimori-Kiyosue et al., 2005).
CLASPs are known regulators of microtubule dynamics. Microtubules switch between phases of growth and shrinkage through transitions known as microtubule catastrophe and rescue (Mitchison and Kirschner, 1984). In cells, CLASPs have been reported to stabilize microtubules (Akhmanova et al., 2001; Mimori-Kiyosue et al., 2005; Drabek et al., 2006; Bratman and Chang, 2007; Sousa et al., 2007), and studies in vitro have found that CLASPs suppress catastrophe and promote rescue (Al-Bassam et al., 2010; Moriwaki and Goshima, 2016; Yu et al., 2016). Although the effects of CLASPs on catastrophe and rescue are known, their effects on growth and shrinkage are less consistent. Fission yeast CLASPCls1p was reported to increase growth rates and suppress shrinkage rates (Al-Bassam et al., 2010), while Drosophila CLASPMast/Orbit strongly suppressed both growth and shrinkage rates (Moriwaki and Goshima, 2016), revealing potential species-specific differences. Importantly, whether the modulation of microtubule growth and shrinkage underlies the anti-catastrophe and rescue activity of CLASPs is unclear. Furthermore, how CLASP activity is affected by its interaction with EB1 is not known. Here, we used total internal reflection fluorescence (TIRF) microscopy and purified human CLASP2γ protein to investigate the regulation of microtubule dynamics by CLASP2γ in the presence of EB1.
RESULTS AND DISCUSSION
Human CLASP2γ binds to microtubule ends, and polymeric and soluble tubulin
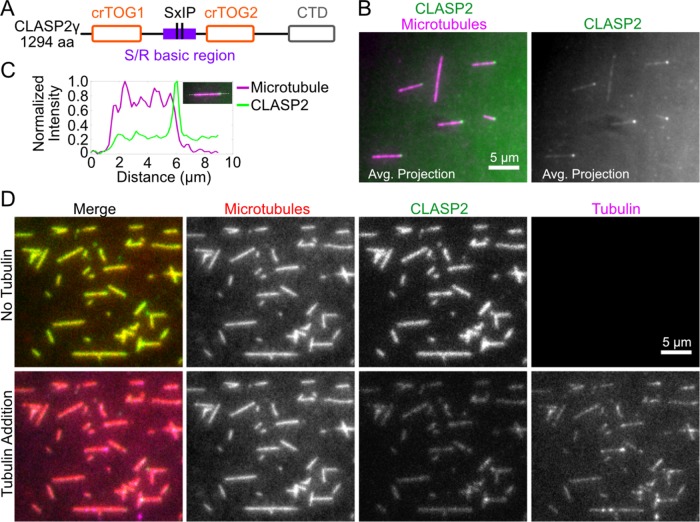
We expressed and purified full-length human CLASP2γ with and without a C-terminal eGFP tag using an insect cell expression system (Figure 1A and Supplemental Figure S1A). By performing single-molecule fluorescence analysis, we established that the purified CLASP2y is monomeric (Supplemental Figure S1, B and C). To investigate binding of CLASP2γ to nondynamic microtubules in the absence of soluble tubulin, we imaged human CLASP2γ-eGFP (100 pM) on guanosine-5′’-[(α,β)-methyleno]triphosphate (GMPCPP)-stabilized microtubules using TIRF microscopy (Gell et al., 2010). We observed enhanced binding of CLASP2γ to one of the two microtubule ends (Figure 1, B and C). We subsequently identified the microtubule polarity and determined that CLASP2γ specifically recognized the plus ends of the nondynamic microtubules (Supplemental Figure S1D).
FIGURE 1:
CLASP2γ binds to the ends of GMPCPP-stabilized microtubules and recruits soluble tubulin to the microtubule lattice. (A) Schematic of the domain structure of human CLASP2γ showing cryptic TOG domains (orange), SxIP motifs, S/R basic region (purple), and C-terminal domain (gray). (B) Representative average intensity projections of GMPCPP-stabilized microtubules incubated with 100 pM CLASP2γ-eGFP. (C) Representative intensity profile through a microtubule (inset) showing CLASP2γ-eGFP intensity enrichment at the microtubule tip. (D) Representative fields of view of GMPCPP-stabilized microtubules incubated with 50 nM CLASP2γ-eGFP before (top row) and after (bottom row) addition of 1 µM Cy5-labeled soluble tubulin.
A distinct structural feature of CLASPs are TOG domains, also found in the XMAP215/Dis1 family of microtubule polymerases. In XMAP215/Dis1 proteins, TOG domains encode the autonomous recognition of microtubule ends (Van Breugel et al., 2003; Slep and Vale, 2007; Brouhard et al., 2008; Podolski et al., 2014). In contrast, CLASPs are thought to interact with microtubules via electrostatic interactions mediated by a serine-arginine–rich basic region (Al-Bassam et al., 2010; Patel et al., 2012) (Figure 1A). Recently, CLASP1α, which contains a canonical TOG domain similar to those of XMAP215/Dis-1, has been shown to autonomously track growing microtubule plus ends in vitro (Yu et al., 2016). However, CLASP2γ lacks the canonical TOG domain, and tip recognition has not previously been observed. Our results indicate that the cryptic TOG domains of CLASP2γ may also confer the ability to recognize a specific structural feature of the microtubule plus end.
When microtubules were incubated with a higher concentration of CLASP2γ-eGFP (50 nM), we observed a uniform localization along the microtubule lattice (Figure 1D, top). Furthermore, a pull-down experiment revealed that human CLASP2γ can also directly bind to soluble tubulin (Supplemental Figure S1E). To test whether human CLASP2γ can simultaneously bind to polymerized and soluble tubulin, we introduced 1 µM Cy5-labeled soluble tubulin to microtubule lattices that were prebound with CLASP2γ (Figure 1D, bottom). Under these conditions, we observed that tubulin was recruited to the CLASP2γ-coated microtubule lattices. In contrast, soluble tubulin did not localize to microtubules in the absence of CLASP2γ (Supplemental Figure S1F). Thus, we conclude that, similarly to Schizosaccharomyces pombe CLASPCls1p (Al-Bassam et al., 2010), human CLASP2γ is able to simultaneously bind to polymerized and soluble tubulin.
EB1 increases the dwell time of CLASP2γ on microtubules
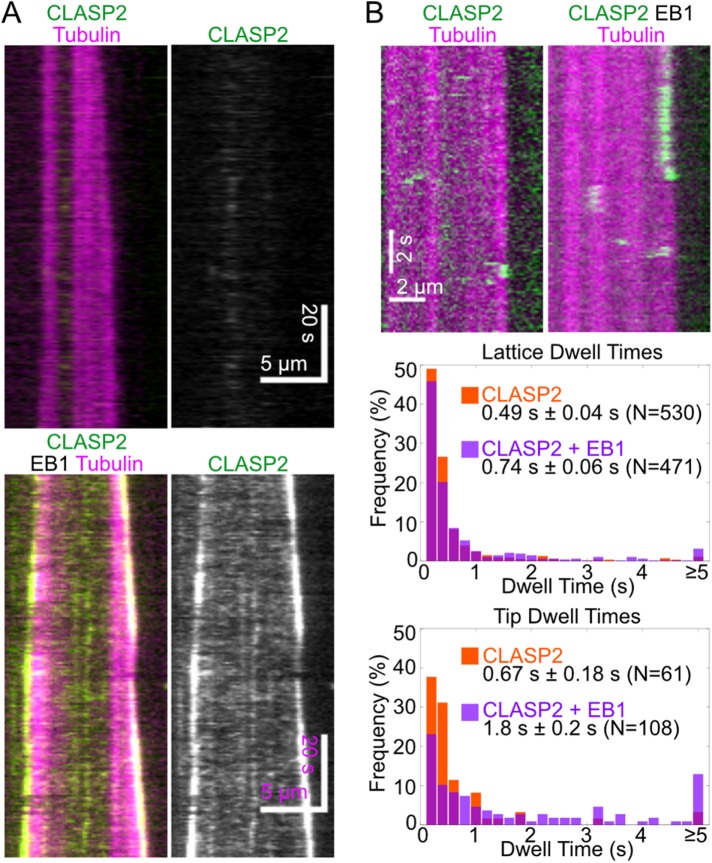
We next examined the localization of CLASP2γ on dynamic microtubules, grown using 10 µM tubulin in the presence of 15 nM CLASP2γ-eGFP. Under these conditions, CLASP2γ did not exhibit strong binding to microtubule lattices or ends (Figure 2A, top). Given that the concentration of soluble tubulin dimers in this assay is several orders of magnitude higher than that of microtubule tips (estimated to be in picomolar range), it is possible that the majority of CLASP2γ molecules are bound to tubulin in solution. In cells, CLASPs localize to growing microtubule ends by interaction with the tip-tracking EB proteins (Mimori-Kiyosue et al., 2005). Indeed, upon introduction of human EB1 into our assay, we observed recruitment of CLASP2γ-eGFP to the growing microtubule ends (Figure 2A, bottom), in agreement with previous reports (Patel et al., 2012; Moriwaki and Goshima, 2016; Yu et al., 2016). We found that CLASP2γ localized both to the plus and minus ends of growing microtubules, consistent with the indiscriminate localization of EB1 on microtubules grown in vitro (Bieling et al., 2007; Zanic et al., 2009).
FIGURE 2:
EB1 increases the dwell time of CLASP2γ at dynamic microtubule tips. (A) Representative kymographs showing the localization of 15 nM CLASP2γ-eGFP on microtubules grown with 10 µM Cy5-labeled tubulin in the presence and absence of 50 nM EB1. (B) Representative kymographs of CLASP2γ-eGFP (200 pM) binding events and distributions of dwell times on microtubules grown with 16 µM Cy5-labeled tubulin in the presence and absence of 10 nM EB1. Errors are SEM.
To investigate whether the duration of CLASP2γ microtubule binding events is affected by EB1, we measured the dwell times of CLASP2γ-eGFP molecules (200 pM) on growing microtubule ends and lattices in the presence and absence of 10 nM EB1 (dimeric concentration) (Figure 2B). Without EB1, CLASP2γ-eGFP binding events were transient, with a mean dwell time of 0.49 ± 0.04 s (SE, N = 530) on the microtubule lattice and 0.67 ± 0.18 s (SE, N = 61) on microtubule tips. The difference between the tips and lattice dwell times was not statistically significant (p = 0.1, Wilcoxon rank sum test). The addition of EB1 led to a 2.5-fold increase in the dwell time of CLASP2γ-eGFP molecules at growing microtubule tips (1.8 ± 0.2 s, SE, N = 108, p < 0.001 when compared with the CLASP2γ alone condition, Wilcoxon rank sum test) and a slight increase in the dwell times of lattice-binding events (0.74 ± 0.06 s, SE, N = 471; p = 0.03 when compared with the CLASP2γ alone condition, Wilcoxon rank sum test). Furthermore, EB1 increased the proportion of tip- over lattice-binding events (19% vs. 10% of all observed binding events) and also increased the tip association rate of CLASP2γ 1.6-fold, from 0.290 ± 0.007 s–1 nM–1 (SE, N = 61) in the CLASP2γ-alone condition to 0.53 ± 0.01 s–1 nM–1 (SE, N = 108, p < 0.001, Welch’s unpaired t test) in the presence of EB1. We conclude that EB1 specifically targets and extends the dwell time of CLASP2γ at growing microtubule ends; thus, EB1 defines CLASP2γ localization. Such spatial regulation of CLASP2 is likely to be of particular significance in cells, since CLASPs function at discrete cellular domains rather than throughout the cell volume (Maiato et al., 2003; Lansbergen et al., 2006; Stehbens et al., 2014).
The anti-catastrophe and rescue activities of CLASP2γ are enhanced by direct interaction with EB1
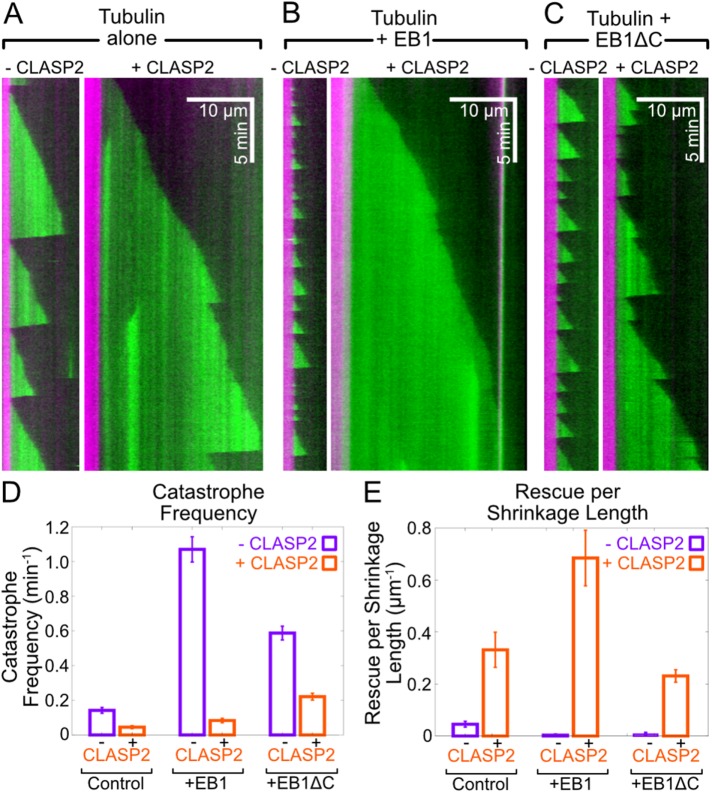
Next, we determined the effects of purified human CLASP2γ on plus-end microtubule dynamics alone and with equimolar amounts of EB1. The addition of 400 nM CLASP2γ to 8 µM tubulin (Figure 3A) resulted in more than a threefold suppression of microtubule catastrophe, from 0.14 ± 0.02 min–1 (SE, N = 73) to 0.04 ± 0.01 min–1 (SE, N = 26; Figure 3D), and a strong promotion of microtubule rescue, from 0.05 ± 0.01 µm–1 (SE, N = 17) to 0.33 ± 0.07 µm–1 (SE, N = 25) rescues per shrinkage length (Figure 3E). In agreement with previous reports, 200 nM dimeric EB1 highly increased the catastrophe frequency (Figure 3B) (Zanic et al., 2013). Strikingly, the combination of CLASP2γ with EB1 (Figure 3B) led to a 13-fold suppression of catastrophe from 1.07 ± 0.07 min–1 (SE, N = 216) to 0.08 ± 0.01 min–1 (SE, N = 42; Figure 3D), as well as a dramatic increase in rescue, from 0.003 ± 0.005 µm–1 (SE, N = 1) to 0.68 ± 0.11 µm–1 (SE, N = 42) rescues per shrinkage length (Figure 3E). Thus, CLASP2γ completely overrides the effects of EB1 on microtubule dynamics.
FIGURE 3:
CLASP2γ suppresses catastrophe and promotes rescue. Representative kymographs of microtubules grown with (A) 8 µM Alexa 488-labeled tubulin alone and in the presence of 400 nM CLASP2γ, (B) 8 µM Alexa 488-labeled tubulin and 200 nM EB1 ± 400 nM CLASP2γ, and (C) 8 µM Alexa 488-labeled tubulin and 200 nM EB1∆C ± 400 nM CLASP2γ. (D, E) Quantification of catastrophe frequency and rescue per shrinkage length of microtubules grown under the conditions described in A–C. Data are means ± SE from two independent experiments.
Synergistic effects on microtubule dynamics have previously been observed with the combination of EB1 and XMAP215; in this case, it was found that the synergy does not depend on a direct interaction between the two proteins (Zanic et al., 2013). To probe whether the observed synergy between CLASP2γ and EB1 requires a direct interaction, we used a truncated construct of EB1 lacking the last 20 amino acids from the C-terminus (EB1ΔC); this deletion disrupts the characteristic binding of EB1 to its partners (Honnappa et al., 2009). Similarly to full-length EB1, 200 nM EB1∆C strongly promoted microtubule catastrophe (Figure 3C). However, the addition of 400 nM CLASP2γ to EB1∆C had similar effects on microtubule dynamics as CLASP2γ alone. Specifically, microtubule catastrophe decreased threefold from 0.59 ± 0.04 min–1 (SE, N = 218) to 0.22 ± 0.02 min–1 (SE, N = 124; Figure 3D) and rescue increased from 0.007 ± 0.004 µm–1 (SE, N = 4) to 0.23 ± 0.02 µm–1 (SE, N = 94; Figure 3E). Therefore, we conclude that the enhancement of CLASP2γ activity observed in the presence of full-length EB1 is a consequence of a direct interaction between the two proteins.
CLASP2γ suppresses microtubule catastrophe without changing the microtubule growth rate
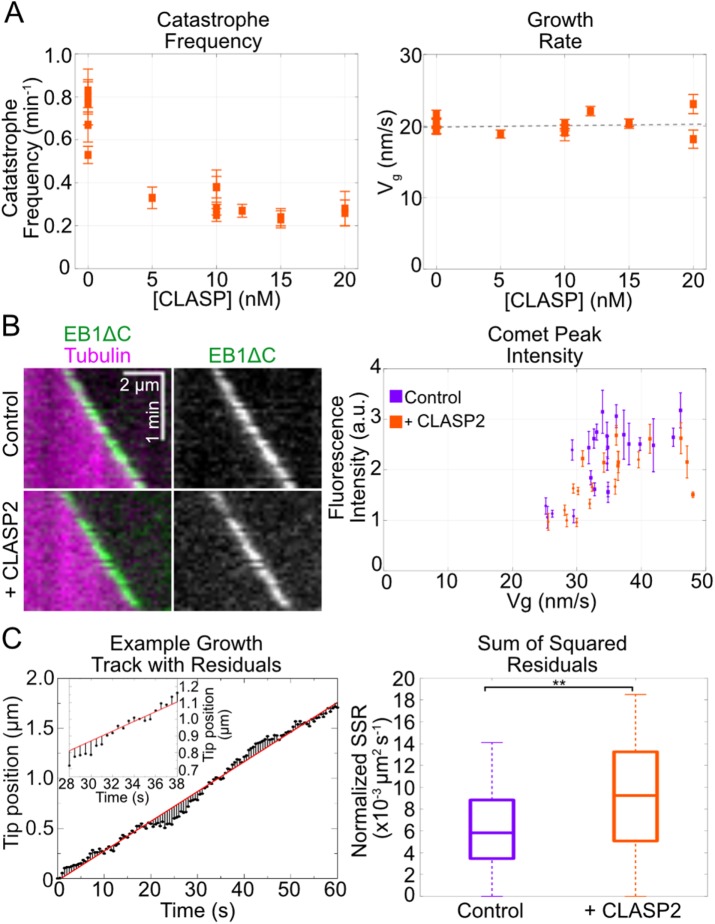
Previous reports on the effect of CLASPs on microtubule dynamics in vitro consistently found that CLASPs suppress microtubule catastrophe; however, the observed effects on microtubule growth rate have been varied (Al-Bassam et al., 2010; Moriwaki and Goshima, 2016; Yu et al., 2016). Whether the effects of CLASPs on catastrophe are related to their effects on growth rate is not clear. To determine the relationship between microtubule growth and catastrophe rates in the presence of human CLASP2γ, we performed titration experiments using 0 to 20 nM CLASP2γ in the background of 50 nM EB1 and 10 µM tubulin. We found that catastrophe was strongly suppressed in all investigated conditions (Figure 4A and Supplemental Figure S2A). Specifically, CLASP2γ reduced the catastrophe frequency from 0.65 ± 0.03 min–1 in the absence of CLASP2γ (weighted mean, SE, N = 5 independent experiments) to 0.29 ± 0.02 min–1 with as little as 10 nM CLASP2γ (weighted mean, SE, N = 4 independent experiments). Surprisingly, while the effect of 10 nM CLASP2γ on microtubule catastrophe was highly statistically significant compared with control (p < 0.001, Welch’s unpaired t test), we found that CLASP2γ had no concomitant effect on microtubule growth rate (see linear fit in Figure 4A; slope not significantly different from zero, 95% CI: [–0.07 nm s–1 nM–1, 0.13 nm s–1 nM–1]).
FIGURE 4:
CLASP2γ suppresses catastrophe without changing the growth rate or increasing EB1ΔC comets. (A) Quantification of catastrophe frequency and growth rate of microtubules grown with 10 µM Cy5-labeled tubulin; 50 nM EB1 and concentrations of CLASP2γ-eGFP ranging from 0 to 20 nM. Data are from six independent experiments, and error bars represent SE. The dotted line represents a linear fit to the data (slope not significantly different from zero, 95% CI: [–0.07 nm s–1 nM–1, 0.13 nm s–1 nM–1]). (B) Representative kymographs and quantification of peak EB1∆C-eGFP comet intensity on microtubules grown with 12 µM Alexa 647-labeled tubulin and 200 nM EB1∆C-eGFP with and without 1 µM CLASP2γ. Data are intensity means ± SE for individual comet measurements: circles and squares represent data from two independent experimental days. Mean comet intensity: 2.21 ± 0.14 a.u. for control (SE, N = 23), and 1.81 ± 0.13 a.u. for CLASP2γ (SE, N = 19); p = 0.04, Welch’s unpaired t test. (C) Left, An example microtubule growth episode showing the tip position as a function of time (points). Linear fit to the position over time trajectory (red line). The lines between the fitted curve and the data points represent the residuals. Inset is a zoom showing detail. Right, Distributions of normalized SSR of microtubules grown with 12 µM Alexa 488-labeled tubulin and 200 nM EB1 with or without 400 nM CLASP2γ. Whiskers are 1.5 IQR. Welch’s t test p = 0.004. N = 117 control tracks, N = 57 CLASP2γ tracks analyzed.
There are several ways in which microtubule growth and catastrophe rates could be related. Early in vitro reports found that an increase in microtubule growth rate correlates with suppression of microtubule catastrophe (Walker et al., 1988). This can be potentially explained by an increase in the size of the stabilizing GTP (guanosine triphosphate) cap with increasing growth rates (Duellberg et al., 2016). Conversely, suppression of growth rate has also been linked to microtubule stabilization through capping, as is the case for the γ-tubulin ring complex (Kollman et al., 2011), or, alternatively, through modulation of the structure of the growing end, as recently proposed for CAMSAPs at microtubule minus ends (Atherton et al., 2017). Our finding that CLASP2γ suppresses catastrophe without changing the growth rate shows that these two parameters are decoupled in the presence of CLASP2γ.
One possible mechanism by which suppression of catastrophe can be achieved while leaving the growth rate unchanged is through decreasing the rate of GTP hydrolysis, thus increasing the size of the protective GTP cap at microtubule ends. Such a mechanism has recently been suggested for the mitotic spindle assembly protein TPX2 (Zhang et al., 2017). To test whether CLASP2γ changes the GTP hydrolysis rate, we investigated the effects of CLASP2γ on EB1 localization at growing microtubule ends. Previous studies have established that EB proteins are sensitive to the nucleotide state of tubulin in the microtubule and that EB “comets” can thus be used as markers for the stabilizing nucleotide cap (Zanic et al., 2009; Maurer et al., 2012). To exclude the possibility that CLASP2γ is modifying EB1 comets through direct interaction, we investigated the localization of the truncated EB1∆C variant, which is expected to solely report on the state of the growing microtubule end. We measured the peak comet intensity and the comet decay length using 200 nM EB1∆C-eGFP with 12 µM tubulin, in the absence and presence of 1 µM CLASP2γ (Figure 4B and Supplemental Figure S2B). While this relatively high concentration of CLASP2γ significantly suppressed microtubule catastrophe (Supplemental Figure S2C), we found that neither the time-averaged intensity nor the length of the EB1∆C-eGFP comets increased in the presence of CLASP2γ (Figure 4B and Supplemental Figure S2D). Thus, we conclude that CLASP2γ does not suppress microtubule catastrophe by increasing the GTP cap at microtubule ends.
Although we did not observe significant differences between control and CLASP2γ conditions when comparing time-averaged growth rates, we observed temporal variability in growth rates of individual microtubules. To investigate the possibility that CLASP2γ affects fluctuations in microtubule growth, we tracked the growth phase of individual microtubules grown with and without CLASP2γ and imaged at higher spatiotemporal resolution. We then used linear regression analysis to determine the deviations from the mean track velocity for each individual growth track. Interestingly, we found that the sum of squared residuals (SSR) was statistically significantly higher for microtubules grown with CLASP2γ than for the control microtubules (p = 0.004; Welch’s unpaired t test) (Figure 4C). In other words, microtubules grown with CLASP2γ display a higher degree of variability in microtubule growth rates.
CLASP2γ promotes microtubule rescue without suppressing the microtubule shrinkage rate
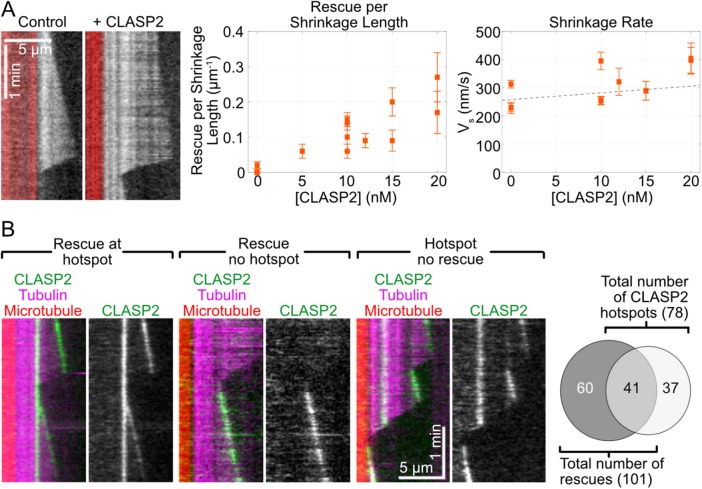
In addition to strongly suppressing microtubule catastrophe, we found that CLASP2γ strongly promoted microtubule rescue (Figure 5A). It has been proposed that suppression of microtubule shrinkage rate can increase the likelihood of rescue, as observed for other known rescue factors, including TPX2 (Roostalu et al., 2015; Wieczorek et al., 2015; Reid et al., 2016) and CLIP170 (Arnal et al., 2004). Previous in vitro studies have reported that CLASPs suppress microtubule shrinkage rate (Al-Bassam et al., 2010; Moriwaki and Goshima, 2016; Yu et al., 2016). Noticeably, in our experiments, increasing concentrations of CLASP2γ strongly promoted rescue without affecting microtubule shrinkage rate (Figure 5B; slope not significantly different from zero, 95% CI: [–3.8 nm s–1 nM–1, 8.6 nm s–1 nM–1]). Specifically, 10 nM CLASP2γ increased the number of rescues per microtubule shrinkage length from 0.001 ± 0.003 µm–1 in the absence of CLASP2γ (weighted mean, SE, N = 5 independent experiments, three observed rescues) to 0.10 ± 0.01 µm–1 in the presence of 10 nM CLASP2γ (weighted mean, SE, N = 4 independent experiments, 112 observed rescues; p < 0.001 between 0 and 10 nM CLASP2γ condition, Welch’s unpaired t test). The finding that human CLASP2γ strongly promotes microtubule rescue without suppressing the microtubule shrinkage rate indicates that CLASP2γ-mediated rescues do not occur through overall stabilization of microtubule polymer along the entirety of its lattice or by specific reduction of the GDP-tubulin off-rate.
FIGURE 5:
CLASP2γ promotes rescue without suppressing shrinkage rate and its localization does not predict sites of rescue. (A) Representative kymographs and quantification of rescue per shrinkage length and shrinkage rates on microtubules grown with 10 µM Cy5-labeled tubulin, 50 nM EB1 and concentrations of CLASP2γ-eGFP ranging from 0 to 20 nM. Data are from six independent experiments; errors bars represent SE. Dotted line represents a linear fit to the data (slope not significantly different from zero, 95% CI: [–3.8 nm s–1 nM–1, 8.6 nm s–1 nM–1]). (B) Representative kymographs and quantification of CLASP2γ-eGFP localization with respect to microtubule rescue events on microtubules grown with 10 µM Cy5-labeled tubulin, 50 nM EB1, and 10 or 15 nM CLASP2γ-eGFP. Data in the Venn diagram are total numbers of events observed from five independent experiments.
Previous reports found that fission yeast CLASPCls1p promotes microtubule rescue through local CLASPCls1p-dependent accumulation of tubulin in discrete sites on the microtubule lattice (Al-Bassam et al., 2010). To determine whether human CLASP2γ anticipates sites of rescue in this way, we assessed CLASP2γ-eGFP localization with respect to rescue events on microtubules grown in the presence of 10 µM tubulin and 50 nM EB1. While we did observe local sites of CLASP2γ enrichment (“hotspots”), which occasionally served as points of rescue, the majority of rescue events occurred at sites without local CLASP2γ accumulation (Figure 5B). Furthermore, we did not observe significant localization of CLASP2γ molecules on shrinking microtubule ends. Quantitative analysis of CLASP2γ-eGFP localization at rescue sites, in local hotspots and at growing microtubule tips confirmed that local enrichment of CLASP2γ-eGFP fluorescence intensity did not correlate with rescue and that most rescue sites contained less CLASP2γ than the growing tips (Supplemental Figure S3). Our results suggest that CLASP2γ can promote rescue without significant local accumulation on the shrinking microtubule ends or lattice.
Microtubule rescue is arguably the least well-understood aspect of microtubule dynamics (Gardner et al., 2013; Brouhard, 2015). Rescue in cells may occur at microtubule lattice sites with distinct structural features, such as stretches of GTP-tubulin (Dimitrov et al., 2008). It has recently been reported that GTP-tubulin incorporates at sites of microtubule lattice damage and that such sites of self-repair promote microtubule rescue (Schaedel et al., 2015; Aumeier et al., 2016). It is possible that microtubules grown in the presence of CLASP2γ might also incorporate distinct structural features that can subsequently serve as points of rescue. Indeed, previous observations in cells led to the hypothesis that CLASPs may modulate the microtubule lattice structure (Grimaldi et al., 2014, 2015). Although we do not always observe strong local accumulation of CLASP2γ at the onset of rescue, our results do not exclude the possibility that CLASP2γ encodes discrete microtubule rescue sites during the process of microtubule growth.
In conclusion, our results demonstrate that CLASP2γ possesses a unique ability to specifically modulate the switching transitions between microtubule growth and shrinkage without affecting the global growth and shrinkage rates. While it is widely accepted that microtubule catastrophe occurs through the loss of the stabilizing GTP-tubulin cap (Mitchison and Kirschner, 1984), the details of this process are still not understood and many models have been proposed (Bowne-Anderson et al., 2013). Naturally occurring fluctuations in microtubule growth affect the size of the GTP cap; nevertheless, the microtubule ends can withstand these fluctuations over relatively long periods of microtubule lifetime (Kerssemakers et al., 2006; Schek et al., 2007; Howard and Hyman, 2009; Gardner et al., 2011a; Rickman et al., 2017). Notably, the probability of catastrophe increases with microtubule age (Odde et al., 1995; Gardner et al., 2011b), due to conformational fluctuations or evolution of the microtubule end structure (Margolin et al., 2012; Bowne-Anderson et al., 2013; Coombes et al., 2013; Zakharov et al., 2015). It is possible that aging renders the microtubule end more susceptible to growth fluctuations. Indeed, slowdowns in growth have been observed prior to the onset of catastrophe (Maurer et al., 2014; Duellberg et al., 2016). In this context, we speculate that CLASP2γ rejuvenates microtubules by allowing them to withstand a greater degree of growth rate variability, which would otherwise lead to catastrophe. Finally, our finding that the interaction of CLASP2γ with EB1 potently boosts CLASP’s effect on microtubule dynamics is yet another example of synergistic regulation that arises through the combined effects of microtubule regulators. Understanding the individual capabilities of regulators, as well as their concerted actions, will ultimately allow us to unravel the complex microtubule network regulation at play within cellular milieus.
MATERIALS AND METHODS
DNA constructs and cloning
The cDNA encoding full-length human CLASP2γ (NCBI Accession: NM_001207044.1; a kind gift from I. Kaverina, Vanderbilt University) was PCR amplified using PfuX7 Polymerase (Nørholm, 2010) and subcloned into the following vectors: 1) a modified pHAT vector (a kind gift from S. Bechstedt and G. Brouhard, McGill University) containing an N-terminal 6xHis tag and a C-terminal eGFP and StrepII tag; and 2) a pFastBacHT vector (Invitrogen) containing an N-terminal 6xHis-tag. Cloning products were verified by DNA sequencing.
Tubulin preparation
Bovine brain tubulin was purified using the high-molarity method (Castoldi and Popov, 2003). Tubulin was labeled with TAMRA, Alexa Fluor 488, Alexa Fluor 647, and Cy5 dyes (Invitrogen) according to the standard protocols, as previously described (Gell et al., 2010). Fluorescent-labeled tubulin was typically used at ratio of between 5 and 10% of the total tubulin.
Protein preparation
EB1, EB1∆C, and EB1∆C-eGFP were purified as described previously (Zanic et al., 2013). His-CLASP2γ and His-CLASP2γ-eGFP-StrepII were expressed in baculovirus-infected Sf9 insect cells using the Bac-to-Bac system according to the manufacturer’s instructions (Invitrogen). After the first amplification, baculovirus-infected insect cells (BIIC) stocks were prepared as previously described (Wasilko and Lee, 2006; Wasilko et al., 2009). Briefly, Sf9 insect cells were infected at a density of 1 × 106 viable cells/ml with BIIC stocks at a ratio of 10–4 BIIC:total culture volume. Cells were harvested 5 d after infection. Cell pellets were lysed by one freeze–thaw cycle and Dounce homogenizing in lysis buffer containing protease inhibitors; genomic DNA was subsequently sheared by passing the lysate through an 18-gauge needle. The following lysis buffers were used: 50 mM PIPES (pH 6.8), 120 mM KCl, 2 mM MgCl2, 50 mM l-glutamine, 50 mM l-arginine, 10% glycerol, 0.1% Tween-20, and 1 mM dithiothreitol (DTT) for His-CLASP2γ and 50 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (pH 7.5), 120 mM KCl, 5% glycerol, 0.1% Tween-20, 2 mM MgCl2, 10 mM imidazole, and 1 mM DTT for His-CLASP2γ-eGFP-StrepII. Crude lysates were clarified by centrifugation for 20 min at 4°C and 35,000 rpm in a Beckman L90K Optima and 50.2 Ti rotor. Clarified lysates were applied to a HisTrapHP column (GE Lifesciences) according to the manufacturer’s protocol. His-tagged proteins were eluted with the following elution buffers: 50 mM PIPES (piperazine-N,N′-bis(2-ethanesulfonic acid)) (pH 6.8), 400 mM KCl, 5% glycerol, 0.1% Tween-20, 2 mM MgCl2, 10 mM imidazole, 1 mM DTT, 50 mM l-glutamine, 50 mM l-arginine, and 300 mM imidazole for His-CLASP2γ and 50 mM HEPES (pH 7.5), 120 mM KCl, 5% glycerol, 0.1% Tween-20, 2 mM MgCl2, 1 mM DTT, and 300 mM imidazole for His-CLASP2γ-eGFP-StrepII. Purified His-CLASP2γ was desalted into storage buffer (25 mM PIPES) [pH 6.8], 250 mM KCl, 5% glycerol, 0.1% Tween-20, 50 mM l-glutamine, 50 mM l-arginine, and 1 mM DTT) using an Amicon centrifugal filter and snap frozen in liquid nitrogen. Peak His-CLASP2γ-eGFP-StrepII elution fractions from the HisTrap were pooled and applied to a StrepTrapHP column (GE Lifesciences) according to the manufacturer’s protocol. Purified His-CLASP2γ-eGFP-StrepII was eluted with 2.5 mM desthiobiotin and snap frozen in liquid nitrogen.
TIRF microscopy
Imaging was performed using a Nikon Eclipse Ti microscope with a 100×/1.49 n.a. TIRF objective, Andor iXon Ultra EM-CCD (electron-multiplying charge-coupled device), and NEO sCMOS (complementary metal–oxide–semiconductor) cameras; 488-, 561-, and 640-nm solid-state lasers (Nikon Lu-NA); Finger Lakes Instruments HS-625 high speed emission filter wheel; and standard filter sets. An objective heater was used to maintain the sample at 35°C. Microscope chambers were constructed as previously described (Gell et al., 2010). In brief, 22 × 22 mm and 18 × 18 mm silanized coverslips were separated by strips of Parafilm to create a narrow channel for the exchange of solution (Gell et al., 2010). Images were acquired using NIS-Elements (Nikon).
Single-molecule fluorescence intensity analysis
To determine single-molecule fluorescence intensities, proteins were introduced to an unblocked, narrow flow cell (∼10 µl volume) at single-molecule concentrations to nonspecifically adhere single molecules to the glass surface similar to previous studies (Sturgill et al., 2016). Samples were diluted in imaging buffer consisting of BRB80 (80 mM PIPES adjusted to pH 6.8 with KOH, 1 mM MgCl2, and 1 mM EGTA), supplemented with 40 mM glucose, 40 µg/ml glucose oxidase, 16 µg/ml catalase, 0.5 mg/ml casein, 0–100 mM KCl and 10 mM DTT for 2 min. Flow cells were then washed twice with 40 µl imaging buffer to remove nonadsorbed fluorescent proteins, and the samples were then imaged by TIRF microscopy using 100-ms exposure time, maximum laser power, and an acquisition rate of 10 fps.
To obtain first-frame intensity values, a two-dimensional Gaussian fitting routine was implemented using the GDSC SMLM plug-in for ImageJ (Herbert, 2013). Local maxima were detected automatically, and the reported Gaussian intensity values (area under the curve) were reported after local background correction. For time-lapse intensity traces showing stepwise photobleaching, background-corrected intensity values were obtained by implementing the two-dimensional Gaussian fitting routine on the same x–y position in time. The intensity was reported as zero when the intensity fell below the required threshold for the fitting routine.
Pull-down assay
Purified proteins at equimolar concentrations (250 nM) were mixed in a total volume of 150 µl binding buffer (BRB80 supplemented with 50 mM KCl) on ice. Triple-washed PerfectPro Ni-NTA Agarose beads (100 µl) were added to the proteins and allowed to incubate with rotation overnight at 4°C. Beads were pelleted by centrifugation and washed three times in 500 µl binding buffer. Bound proteins were eluted with 300 mM imidazole, and samples were analyzed by SDS–PAGE.
Dynamic microtubule assay
Preparation of GMPCPP-stabilized microtubules was performed according to standard protocols (Hyman et al., 1992; Gell et al., 2010). Dynamic microtubule extensions were polymerized from surface-immobilized GMPCPP-stabilized templates as described previously (Gell et al., 2010). Imaging buffer containing concentrations of tubulin ranging from 8 to 16 µM tubulin, 1 mM GTP, and proteins at the concentrations indicated in the text were introduced into the imaging chamber. The imaging buffer consisted of BRB80 supplemented with 40 mM glucose, 40 µg/ml glucose oxidase, 16 µg/ml catalase, 0.5 mg/ml casein, 50–100 mM KCl, 10 mM DTT, and 0%–0.1% methylcellulose. To determine the polarity of the individual microtubule polymers, 12 µM soluble tubulin was introduced into the flow cell, which resulted in the growth of dynamic microtubule extensions from the stabilized microtubule templates. Polarity was established by measuring growth rates with faster-growing extensions indicating plus ends and slower-growing extensions indicating minus ends. Quantification of microtubule dynamics parameters was performed using kymographs generated in Fiji (Schindelin et al., 2012) as described previously (Zanic, 2016). Catastrophe frequency was calculated by dividing the number of catastrophes by the total time spent in the growth phase. Rescue was calculated by dividing the number of rescues observed by the total shrinkage length. The error for catastrophe frequency and rescue per shrinkage length are counting errors. Gamma distribution parameters were estimated from lifetime data for the control condition (10 µM tubulin and 50 nM EB1) and the CLASP2γ condition (10 µM tubulin, 50 nM EB1, and 12 nM CLASP2γ-eGFP) as described previously (Gardner et al., 2011b; Zanic, 2016).
Dwell-time analysis
Microtubules were polymerized using 16 µM tubulin in imaging buffer supplemented with 0.1% methylcellulose and imaged at 10 fps and high laser power. Microtubules were grown in the presence of either 200 pM CLASP2γ-eGFP alone or 200 pM CLASP2γ-eGFP in combination with 10 nM EB1. The duration of binding events were measured from kymographs generated as described above. Dwell times longer than 10 s in duration were identified as outliers and discarded from subsequent analysis.
EB1∆C comet analysis
Dynamic microtubules were grown with 12 µM Alexa 647-labeled tubulin and 200 nM EB1∆C-eGFP ± 400 nM CLASP2γ. Kymographs of EB1∆C-eGFP comets on growing microtubules were generated as described above. A custom Matlab (MathWorks) function was developed to determine the comet intensities and the comet lengths as follows: intensity profiles along the microtubule for each time frame were prealigned at the maximum peak intensity. Each intensity profile was then fitted to an exponential decay convolved with a Gaussian function given by
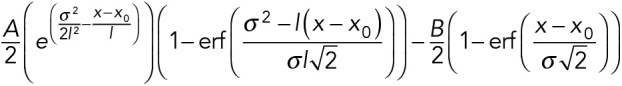 |
where A is the amplitude of the intensity profile, B is the background lattice intensity, σ is the point spread function estimated from intensity profiles of single molecule fluorophores, l is the characteristic decay length of the comets, and x0 is the predicted comet end position. The intensity profiles that had R-squared values below 0.8 were discarded. Remaining intensity profiles were then aligned with subpixel precision using x0 from the fit and averaged to obtain a single average intensity profile for a given kymograph. These averaged intensity profiles were further fitted to the above convolved function to determine the characteristic decay length of the EB comets. The average of peak intensity values for each growth event was determined by subtracting the lattice background intensity (measured 20–30 pixels away from the tip) from the maximum tip intensity for all intensity profiles that were not discarded by the initial fitting routine as described above.
Variability in microtubule growth rates
Growing microtubules were imaged at 2 fps using a 100× objective and an Andor Neo camera (pixel size of 70 nm). Dynamic microtubule tip positions as a function of time were tracked using KymographDirect and KymographClear (Mangeol et al., 2016). The analysis was restricted to growth episodes lasting at least 60 s using a custom Matlab (MathWorks) function in the following manner. A point xi is marked as the beginning of a catastrophe if the forward five consecutive instantaneous velocities, 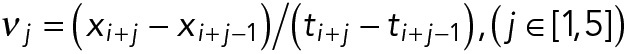 , are smaller than –100 nm/s. Similarly, the point is marked as a rescue point if
, are smaller than –100 nm/s. Similarly, the point is marked as a rescue point if 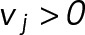 . Tracks that contained catastrophes were divided into segments to include only the growth episodes. A linear function was fitted to the first 60 s of the segments to find the mean growth velocity. In total, 117 tracks of control and 57 tracks of CLASP2 condition were analyzed. The deviation from the mean growth velocity within the growth episodes was characterized by the sum of squared residuals (SSR), normalized by the time interval of the window where the fit was performed (60 s), as follows:
. Tracks that contained catastrophes were divided into segments to include only the growth episodes. A linear function was fitted to the first 60 s of the segments to find the mean growth velocity. In total, 117 tracks of control and 57 tracks of CLASP2 condition were analyzed. The deviation from the mean growth velocity within the growth episodes was characterized by the sum of squared residuals (SSR), normalized by the time interval of the window where the fit was performed (60 s), as follows:
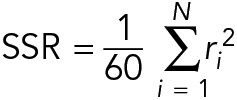 |
where ri is the residual for xi.
CLASP2γ fluorescence intensity at rescue sites, microtubule tips and hotspots
CLASP2γ-eGFP fluorescence intensity measurements were performed on kymographs of microtubules grown in the presence of 15 nM CLASP2γ-eGFP, generated as described above. Linescans (50 pixels long × 2 pixels wide) were centered on microtubule tips during 1) growth and 2) rescue and 3) when encountering CLASP2γ-eGFP hotspots. The solution and lattice intensities were determined by averaging the intensities of the first (solution) and last (lattice) 15 pixels of the linescans. All analyzed sites were measured from a single time-lapse movie. The tip intensities were determined by finding the local maxima in the four middle pixels of the linescan. The tip-to-lattice ratio, R, was calculated as
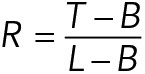 |
where T is tip intensity, B is solution intensity background, and L is lattice intensity. The error bars were determined from variability in lattice intensity values using error propagation. The R values were then sorted and plotted with color codes. A rescue site was classified as a CLASP2γ hotspot when the CLASP2γ intensity was higher than mean lattice intensity + two SDs.
Supplementary Material
Acknowledgments
We thank M. Braun (B CUBE, Dresden) for the kind gift of EB1∆C protein, I. Kaverina (Vanderbilt University) for CLASP2γ cDNA, and G. Brouhard and S. Bechstedt (McGill University) for the modified pHAT vector. We thank I. Kaverina, R. Ohi, S. Bechstedt, and J. Alper for comments on our manuscript and the members of the Zanic laboratory and the Vanderbilt Microtubules and Motors club for discussion and feedback. This work was supported by National Institutes of Health grant R35GM119552 to M. Zanic. M. Zanic acknowledges support from the Human Frontier Science Program and the Searle Scholars Program.
Abbreviations used:
- GMPCPP
guanosine-5′-[(α,β)-methyleno]triphosphate
- TIRF
total internal reflection fluorescence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-01-0016) on March 22, 2018.
REFERENCES
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, et al (2001). CLASPs are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell , 923–935. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F. (2010). CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell , 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose JC, Shoji T, Kotzer AM, Pighin JA, Wasteneys GO. (2007). The arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell Online , 2763–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnal I, Heichette C, Diamantopoulos GS, Chrétien D. (2004). CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. CurrBiol , 2086–2095. [DOI] [PubMed] [Google Scholar]
- Atherton J, Jiang K, Stangier MM, Luo Y, Hua S, Houben K, Van Hooff JJE, Joseph AP, Scarabelli G, Grant BJ, et al (2017). A structural model for microtubule minus-end recognition and protection by CAMSAP proteins. Nat Struct Mol Biol , 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumeier C, Schaedel L, Gaillard JJ, John K, Blanchoin L, Thery M, Théry M. (2016). Self-repair promotes microtubule rescue. Nat Cell Biol , 1054–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, Dillon GM, Sullivan JM, Stuart CE, Gilbert JP, Kambouris JA, Ho A. (2012). Microtubule plus-end tracking protein CLASP2 regulates neuronal polarity and synaptic function. J Neurosci , 13906–13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T. (2007). Reconstitution of a microtubule plus-end tracking system in vitro. Nature , 1100–1105. [DOI] [PubMed] [Google Scholar]
- Bowne-Anderson H, Zanic M, Kauer M, Howard J. (2013). Microtubule dynamic instability: a new model with coupled GTP hydrolysis and multistep catastrophe. BioEssays , 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratman SV, Chang F. (2007). Stabilization of overlapping microtubules by fission yeast CLASP. Dev. Cell , 812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ. (2015). Dynamic instability 30 years later: complexities in microtubule growth and catastrophe. Mol Biol Cell , 1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. (2008). XMAP215 is a processive icrotubule polymerase. Cell , 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Popov AV. (2003). Purification of brain tubulin through two cycles of polymerization- depolymerization in a high-molarity buffer. Protein Expr Purif , 83–88. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, MacLeod I, Yates JR, Oegema K, Desai A. (2005). The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr Biol , 771–777. [DOI] [PubMed] [Google Scholar]
- Coombes CE, Yamamoto A, Kenzie MR, Odde DJ, Gardner MK. (2013). Evolving tip structures can explain age-dependent microtubule catastrophe. Curr Biol , 1342–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon GM, Tyler WA, Omuro KC, Kambouris J, Tyminski C, Henry S, Haydar TF, Beffert U, Ho A. (2017). CLASP2 links reelin to the cytoskeleton during neocortical development. Neuron , 1344.e5–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov A, Quesnoit M, Moutel S, Cantaloube I, Poüs C, Perez F. (2008). Detection of GTP-tubulin conformation in vivo reveals a role for GTP remnants in microtubule rescues. Science , 1353–1356. [DOI] [PubMed] [Google Scholar]
- Drabek K, van Ham M, Stepanova T, Draegestein K, van Horssen R, Sayas CL, Akhmanova A, ten Hagen T, Smits R, Fodde R, et al (2006). Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr Biol , 2259–2264. [DOI] [PubMed] [Google Scholar]
- Duellberg C, Cade NI, Holmes D, Surrey T. (2016). The size of the EB cap determines instantaneous microtubule stability. Elife , 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia ARR, McLeod IX, et al (2007). Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell , 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C, Schmeiser V, Ortiz J, Lechner J. (2014). A TOGL domain specifically targets yeast CLASP to kinetochores to stabilize kinetochore microtubules. J Cell Biol , 555–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Charlebois BD, Janosi IM, Howard J, Hunt AJ, Odde DJ. (2011a). Rapid microtubule self-assembly kinetics. Cell , 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. (2011b). Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell , 1092–1103. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Howard J. (2013). Microtubule catastrophe and rescue. Curr Opin Cell Biol , 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J, et al (2010). Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol , 221–245. [DOI] [PubMed] [Google Scholar]
- Grimaldi AD, Maki T, Fitton BP, Roth D, Yampolsky D, Davidson MW, Svitkina T, Straube A, Hayashi I, Kaverina I. (2014). CLASPs are required for proper microtubule localization of end-binding proteins. Dev Cell , 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi AD, Zanic M, Kaverina I. (2015). Encoding the microtubule structure: allosteric interactions between the microtubule +TIP complex master regulators and TOG-domain proteins. Cell Cycle , 1375–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, Heald R. (2006). Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J Cell Biol , 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A. (2013). Single-molecule Plugins ImageJ. Available at www.sussex.ac.uk/gdsc/intranet/microscopy/imagej/smlm_plugins (accessed 4 January 2018)
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJA, Buey RM, Lawera A, et al (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell , 366–376. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. (2009). Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol , 569–574. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. (1992). Role of GTP hydrolysis in microtubule dynamics: information from a slowly hydrolyzable analogue, GMPCPP. Mol Biol Cell , 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue YH, Avides MDC, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, Matsukage A, Glover DM. (2000). Orbit, a novel microtubule-associated protein essential for mitosis in 3Drosophila melanogaster. J Cell Biol , 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerssemakers JWJ, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. (2006). Assembly dynamics of microtubules at molecular resolution. Nature , 709–712. [DOI] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. (2011). Microtubule nucleation by γ-tubulin complexes. Nat Rev Mol Cell Biol , 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Lyle KS, Gierke S, Matov A, Danuser G, Wittmann T. (2009). GSK33 phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J Cell Biol , 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G, Grigoriev I, Yuko M-K, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, et al (2006). CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell , 21–32. [DOI] [PubMed] [Google Scholar]
- Leano JB, Rogers SL, Slep KC. (2013). A cryptic TOG domain with a distinct architecture underlies CLASP-dependent bipolar spindle formation. Structure , 939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Engel U, Rusch J, Scherrer S, Sheard K, Van Vactor D. (2004). The microtubule plus end tracking protein orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron , 913–926. [DOI] [PubMed] [Google Scholar]
- Lemos CL, Sampaio P, Maiato H, Costa M, Omel’yanchuk LV, Liberal V, Sunkel CE. (2000). Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization. EMBO J , 3668–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffini S, Maia ARR, Manning AL, Maliga Z, Pereira AL, Junqueira M, Shevchenko A, Hyman A, Yates JR, Galjart N, et al (2009). Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr Biol , 1566–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia ARR, Garcia Z, Kabeche L, Barisic M, Maffini S, Macedo-Ribeiro S, Cheeseman IM, Compton DA, Kaverina I, Maiato H. (2012). Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J Cell Biol , 285–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Fairley EAL, Rieder CL, Swedlow JR, Sunkel CE, Earnshaw WC. (2003). Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell , 891–904. [DOI] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. (2005). Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol , 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Sampaio P, Lemos CL, Findlay J, Carmena M, Earnshaw WC, Sunkel CE. (2002). MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J Cell Biol , 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki T, Grimaldi AD, Fuchigami S, Kaverina I, Hayashi I. (2015). CLASP2 has two distinct TOG domains that contribute differently to microtubule dynamics. J Mol Biol , 2379–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeol P, Prevo B, Peterman EJG. (2016). KymographClear and KymographDirect: two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Mol Biol Cell , 1948–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin G, Gregoretti IV, Cickovski TM, Li C, Shi W, Alber MS, Goodson HV. (2012). The mechanisms of microtubule catastrophe and rescue: implications from analysis of a dimer-scale computational model. Mol Biol Cell , 642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A, Godinez WJ, Tsimashchuk V, Bankhead P, Rohr K, Engel U. (2013). Xenopus cytoplasmic linker-associated protein 1 (XCLASP1) promotes axon elongation and advance of pioneer microtubules. Mol Biol Cell , 1544–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Cade NI, Bohner G, Gustafsson N, Boutant E, Surrey T. (2014). EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr Biol , 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. (2012). EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell , 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia ARR, Efimova N, Efimov A, Kaverina I. (2009). Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol , 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, et al (2005). CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol , 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Sasaki H, Matsui C, Akhmanova A, Tsukita S, Vorobjev I. (2006). Mammalian CLASPs are required for mitotic spindle organization and kinetochore alignment. Genes Cells , 845–857. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. (1984). Dynamic instability of microtubule growth. Nature , 237–242. [DOI] [PubMed] [Google Scholar]
- Moriwaki T, Goshima G. (2016). Five factors can reconstitute all three phases of microtubule polymerization dynamics. J Cell Biol , 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørholm MHH. (2010). A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol , 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odde DJ, Cassimeris L, Buettner HM. (1995). Kinetics of microtubule catastrophe assessed by probabilistic analysis. Biophys J , 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualone D, Huffaker TC. (1994). STU1, a suppressor of a β-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol , 1973–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K, Nogales E, Heald R. (2012). Multiple domains of human CLASP contribute to microtubule dynamics and organization in vitro and in Xenopus egg extracts. Cytoskeleton , 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AL, Pereira AJ, Maia ARR, Drabek K, Sayas CL, Hergert PJ, Lince-Faria M, Matos I, Duque C, Stepanova T, et al (2006). Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol Biol Cell , 4526–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolski M, Mahamdeh M, Howard J. (2014). Stu2, the budding yeast XMAP215/Dis1 homolog, promotes assembly of yeast microtubules by increasing growth rate and decreasing catastrophe frequency. J Biol Chem , 28087–28093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid TA, Schuster BM, Mann BJ, Balchand SK, Plooster M, McClellan M, Coombes CE, Wadsworth P, Gardner MK. (2016). Suppression of microtubule assembly kinetics by the mitotic protein TPX2. J Cell Sci , 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman J, Duellberg C, Cade NI, Griffin LD, Surrey T. (2017). Steady-state EB cap size fluctuations are determined by stochastic microtubule growth and maturation. Proc Natl Acad Sci USA , 3427–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Cade NI, Surrey T. (2015). Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat Cell Biol , 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedel L, John K, Gaillard JJ, Nachury MV, Blanchoin L, Théry M, Thery M. (2015). Microtubules self-repair in response to mechanical stress. Nat Mater , 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schek HT, 3rd, Gardner MK, Cheng J, Odde DJ, Hunt AJ. (2007). Microtubule assembly dynamics at the nanoscale. Curr Biol , 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt N, Basu S, Sladecek S, Gatti S, van Haren J, Treves S, Pielage J, Galjart N, Brenner HR. (2012). Agrin regulates CLASP2-mediated capture of microtubules at the neuromuscular junction synaptic membrane. J Cell Biol , 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC, Vale RD. (2007). Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell , 976–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A, Reis R, Sampaio P, Sunkel CE. (2007). The Drosophila CLASP homologue, mast/orbit regulates the dynamic behaviour of interphase microtubules by promoting the pause state. Cell Motil Cytoskeleton , 605–620. [DOI] [PubMed] [Google Scholar]
- Stehbens SJ, Paszek M, Pemble H, Ettinger A, Gierke S, Wittmann T. (2014). CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat Cell Biol , 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Norris SR, Guo Y, Ohi R. (2016). Kinesin-5 inhibitor resistance is driven by kinesin-12. J Cell Biol , 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breugel M, Drechsel D, Hyman A. (2003). Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J Cell Biol , 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RA, Brien O, Pryer K, Soboeiro ME, Voter WA, Erickson HP, Salmon ED. (1988). Dynamic instability of individual microtubules. J Cell Biol , 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilko D, Lee SE. (2006). TIPS: titerless infected-cells preservation and scale-up. Bioprocess J , 29–32. [DOI] [PubMed] [Google Scholar]
- Wasilko DJ, Edward Lee S, Stutzman-Engwall KJ, Reitz BA, Emmons TL, Mathis KJ, Bienkowski MJ, Tomasselli AG, David Fischer H. (2009). The titerless infected-cells preservation and scale-up (TIPS) method for large-scale production of NO-sensitive human soluble guanylate cyclase (sGC) from insect cells infected with recombinant baculovirus. Protein Expr Purif , 122–132. [DOI] [PubMed] [Google Scholar]
- Wieczorek M, Bechstedt S, Chaaban S, Brouhard GJ. (2015). Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat Cell Biol , 907–916. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. (2005). Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3-beta in migrating epithelial cells. J Cell Biol , 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, You L, Pasqualone D, Kopski KM, Huffaker TC. (2002). Stu1p is physically associated with beta-tubulin and is required for structural integrity of the mitotic spindle. Mol Biol Cell , 1881–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Signorile L, Basu S, Ottema S, Lebbink JHGG, Leslie K, Smal I, Dekkers D, Demmers J, Galjart N. (2016). Isolation of functional tubulin dimers and of tubulin-associated proteins from mammalian cells. Curr Biol , 1728–1736. [DOI] [PubMed] [Google Scholar]
- Zakharov P, Gudimchuk N, Voevodin V, Tikhonravov A, Ataullakhanov FI, Grishchuk EL. (2015). Molecular and mechanical causes of microtubule catastrophe and aging. Biophys J , 2574–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M. (2016). Measuring the effects of microtubule-associated proteins on microtubule dynamics in vitro. In: Methods in Molecular Biology, ed. Chang P, Ohi R, New York: Springer New York, 47–61. [DOI] [PubMed] [Google Scholar]
- Zanic M, Stear JH, Hyman AA, Howard J. (2009). EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PLoS One , e7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M, Widlund PO, Hyman AA, Howard J. (2013). Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat Cell Biol , 688–693. [DOI] [PubMed] [Google Scholar]
- Zhang R, Roostalu J, Surrey T, Nogales E. (2017). Structural insight into TPX2-stimulated microtubule assembly. Elife , e30959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.