Abstract
PTEN-induced putative kinase 1 (PINK1) is a mitochondria-targeted kinase whose mutations are a cause of Parkinson’s disease. We set out to better understand PINK1’s effects on mitochondrial proteins in vivo. Using an unbiased phosphoproteomic screen in Drosophila, we found that PINK1 mediates the phosphorylation of MCAD, a mitochondrial matrix protein critical to fatty acid metabolism. By mimicking phosphorylation of this protein in a PINK1 null background, we restored PINK1 null’s climbing, flight, thorax, and wing deficiencies. Owing to MCAD’s role in fatty acid metabolism, we examined the metabolic profile of PINK1 null flies, where we uncovered significant disruptions in both acylcarnitines and amino acids. Some of these disruptions were rescued by phosphorylation of MCAD, consistent with MCAD’s rescue of PINK1 null’s organismal phenotypes. Our work validates and extends the current knowledge of PINK1, identifies a novel function of MCAD, and illuminates the need for and effectiveness of metabolic profiling in models of neurodegenerative disease.
INTRODUCTION
PTEN-induced putative kinase 1 (PINK1) is a mitochondria-targeted serine/threonine kinase that mediates several pathways to maintain mitochondrial health and function. When mitochondria are damaged or under stress, PINK1 is stabilized on the outer mitochondrial membrane where it phosphorylates its substrates to initiate mitochondrial clearance via mitophagy (Narendra et al., 2008, 2010; Zhou et al., 2008; Whitworth and Pallanck, 2009; Geisler et al., 2010; Vives-Bauza and Przedborski, 2011; Wang et al., 2011; Kondapalli et al., 2012; Chen and Dorn, 2013; Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014; Whitworth and Pallanck, 2017). PINK1 also affects mitochondrial morphology by interacting with fission-and-fusion machinery and promotes the activity of complex I in the electron transport chain (Poole et al., 2008, 2010; Yang et al., 2008; Ziviani et al., 2010; Liu et al., 2011; Vilain et al., 2012; Vos et al., 2012, 2017; Morais et al., 2014; Pogson et al., 2014; Gehrke et al., 2015). The loss of PINK1 in flies leads to a host of phenotypes, including compromised climbing and flight ability, deformed thoraxes, drooped/held-up wings, and reduced ATP production (Clark et al., 2006; Park et al., 2006; Yang et al., 2006). In humans, mutations in PINK1 underlie some cases of recessive early-onset Parkinson’s disease (Valente et al., 2004a, b). Thus, PINK1 plays a critical role in mitochondrial homeostasis, and its disruption has severe effects at the cellular and organismal level.
Of a mitochondrion’s myriad responsibilities, its most vital is the production of energy through metabolism, including fatty acid and amino acid metabolism. Fatty acids are broken down by β-oxidation and ultimately generate the most energy per gram of any metabolite. While amino acids do not deliver as much energy per gram as fatty acids, their metabolism boasts the widest variety of pathways. Inborn errors of either fatty acid metabolism or amino acid metabolism can have devastating consequences, including early childhood death (Rinaldo et al., 2002; Wu, 2009; Houten et al., 2016), which illustrates how critical these processes are to cell survival.
We show here that in addition to its other mitochondrial functions, PINK1 plays a role in fatty acid and amino acid metabolism. We find that an enzyme critical to fatty acid β-oxidation, medium-chain acyl-coenzyme A dehydrogenase (MCAD), is phosphorylated in a PINK1-dependent manner. MCAD’s canonical function is to break down medium-chain fatty acids (C6-12), and its deficiency is the most common inborn error of β-oxidation in humans (Rinaldo et al., 2002). Mimicking phosphorylation of MCAD in PINK1 null flies rescues the flies’ deformed thoraxes, drooped/held-up wings, and flight and climbing deficiencies. We also show that the loss of PINK1 leads to substantial disruptions in both acylcarnitines and amino acids and that mimicking phosphorylation of MCAD rescues some of these disruptions.
RESULTS
Phosphorylation of MCAD S347 is dependent on PINK1 in Drosophila
To identify potential novel targets of PINK1, we conducted an unbiased proteomic screen for phosphorylated residues in the mitochondrial fractions of PINK1 null flies and their genetic controls (Supplemental Figure S1) (Clark et al., 2006). Since PINK1 is a serine/threonine kinase, we searched for phosphorylated peptides whose abundances were decreased with the loss of PINK1. One phosphopeptide of Marf (the Drosophila homologue of mitofusin) was dramatically decreased in PINK1 null flies (Supplemental Figure S2 and Supplemental Table S1). This finding indicates that Marf phosphorylation is dependent on PINK1 in vivo and supports previous findings in multiple systems that Marf/mitofusin is a target of PINK1 (Poole et al., 2008, 2010; Yang et al., 2008; Tanaka et al., 2010; Chen and Dorn, 2013).
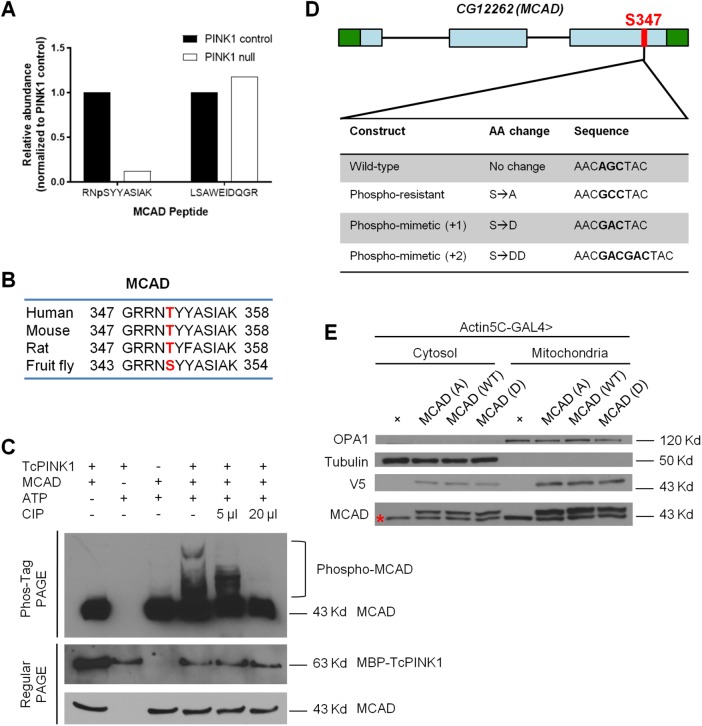
In addition to the Marf phosphopeptide, we identified another phosphopeptide with considerably lower abundance in PINK1 null flies. This phosphopeptide belongs to MCAD, an enzyme critical to the oxidation of medium-chain fatty acids (Rinaldo et al., 2002; Houten et al., 2016). The abundance of this phosphopeptide was reduced in PINK1 null flies to 12.4% of the level found in controls, while the abundance of another detected unphosphorylated MCAD peptide remained the same in both genotypes (Figure 1A and Supplemental Figure S2). Within the identified phosphopeptide, there was one serine site with a high probability of phosphorylation, verified by manual analysis of the MS/MS spectrum: serine 347 (Supplemental Figure S3 and Supplemental Table S1). This serine site is functionally conserved across species and correlates to threonine 351 in mammals (Figure 1B). Phosphorylation of this site was previously confirmed in humans and mice (Possemato, 2010; Grimsrud et al., 2012; Mulhern, 2012; Humphrey et al., 2013; Bian et al., 2014).
FIGURE 1:
Phosphorylation of MCAD S347 is dependent on PINK1 in Drosophila. (A) Phosphopeptide analysis showing relative levels of phosphorylated (left) and unphosphorylated (right) MCAD peptide in mitochondrial fractions from 5-d-old control and PINK1 null male flies. (B) Sequence alignment of human, mouse, rat, and fly MCAD. S347/T351 is marked in red. (C) Phos-tag and regular PAGE gels showing in vitro kinase assay for MCAD and TcPINK1. Phos-tag gel is probed with anti-MCAD to detect both unphosphorylated and the slower-migrating phosphorylated MCAD. Regular PAGE gels are probed with anti-MCAD and anti-MBP to detect MCAD and TcPINK1, respectively, in the in vitro kinase assays. CIP is calf intestinal alkaline phosphatase. The same results were repeated three times (D) Schematic depiction of Drosophila gene CG12262 (MCAD) detailing sequences used to generate wild-type, phosphoresistant and phosphomimetic S347. Green boxes are 5′ and 3′ UTRs; blue boxes are exons. (E) Representative Western blot of mitochondrial and cytosolic fractions from Actin5C-GAL4 alone (control) and Actin5C-GAL4 driving MCAD (A), MCAD (WT), and MCAD (D) transgenic constructs. Samples were probed with anti-tubulin as a cytosolic loading control, anti-OPA1 as a mitochondrial loading control, anti-MCAD to identify endogenous and transgenic localization, and anti-V5 to verify transgenic localization. Note that endogenous MCAD (red asterisk) migrates faster than V5-tagged MCAD in the anti-MCAD blot. The slightly shifted V5-tagged transgenes are verified in the separate anti-V5 blot. n = 25 flies lysed per sample for each experiment; three biological replicates per genotype.
Since MCAD S347 phosphopeptide abundance followed the same pattern as a known PINK1 substrate, Marf, we hypothesized that PINK1 mediates the phosphorylation of MCAD at serine 347. We confirmed that PINK1 can directly phosphorylate MCAD by coupling an in vitro PINK1 kinase assay with Phos-tag acrylamide analysis—Phos-tag ligands bind phosphorylated proteins and slow their migration. We used bacterially expressed Tribolium castaneum PINK1 (TcPINK1), which remains active in vitro (Woodroof et al., 2011), and human MCAD (Figure 1C). Collectively, we have provided evidence that PINK1 mediates the phosphorylation of MCAD both in vivo and in vitro.
Generating phosphoresistant and phosphomimetic MCAD S347 transgenic flies
Because phosphorylation of MCAD S347 is reduced in PINK1 null flies, we reasoned that mimicking phosphorylation at this site could rescue some of PINK1 null’s phenotypes. To test this hypothesis, we generated flies carrying wild-type (Ser347), phosphoresistant (Ser347Ala), and two phosphomimetic (Ser347Asp, Ser347AspAsp) MCAD transgenes downstream of an upstream activation sequence (UAS). We refer to these transgenes as MCAD (WT), MCAD (A), MCAD (D), and MCAD (DD), respectively (Figure 1D). We used two different phosphomimetic transgenes since it has been observed that sometimes a pair of acidic amino acids serves as a better mimic of phosphorylation than a lone acidic amino acid (Strickfaden et al., 2007; Pearlman et al., 2011). We also included a V5 tag at the C-terminal of these transgenes, so that we could evaluate their expression levels. All transgenes were inserted into the same genomic site using the PhiC31 integrase-mediated transgenesis system to ensure that they had the same genomic regulation (Bischof et al., 2007; Markstein et al., 2008).
To determine that neither the V5 tag nor the phosphorylation state affected localization of MCAD, we probed mitochondrial and cytosolic fractions of fly lysates for both endogenous and transgenic MCAD expression. We verified that an MCAD antibody generated against mammalian MCAD recognized endogenous fly MCAD (Supplemental Figure S4A) using MCAD deficient flies (described later). All V5-tagged MCAD transgenes were expressed at similar levels when driven by the ubiquitous Actin5C-GAL4 driver (Brand and Perrimon, 1993) (Figure 1E, V5 blot), and their expression levels were comparable to those of endogenous MCAD (Figure 1E, MCAD blot). Neither the V5 tag nor the phosphorylation states of the transgenes affected their localization, and both endogenous and transgenic MCAD protein localized to the mitochondria as well as the cytosol, consistent with a previous report in mammals (Du et al., 2013) (Figure 1E).
We then expressed these transgenic forms of MCAD in PINK1 null flies carrying Actin5C-GAL4 (Park et al., 2006). Importantly, we crossed our PINK1null; Actin5C-GAL4 flies with flies carrying the same attP40 landing site into which the MCAD transgenes were integrated to eliminate bias caused by differing genetic backgrounds. The protein expression levels of all transgenes were comparable, including in a PINK1 null background (Supplemental Figure S4, B and C). Thus, these transgenes allowed us to observe the effects of mimicking phosphorylation of MCAD S347 in a PINK1 null background, while controlling for any other genetic factors.
Phosphomimetic MCAD S347 rescues behavioral and morphological deficiencies in PINK1 null flies
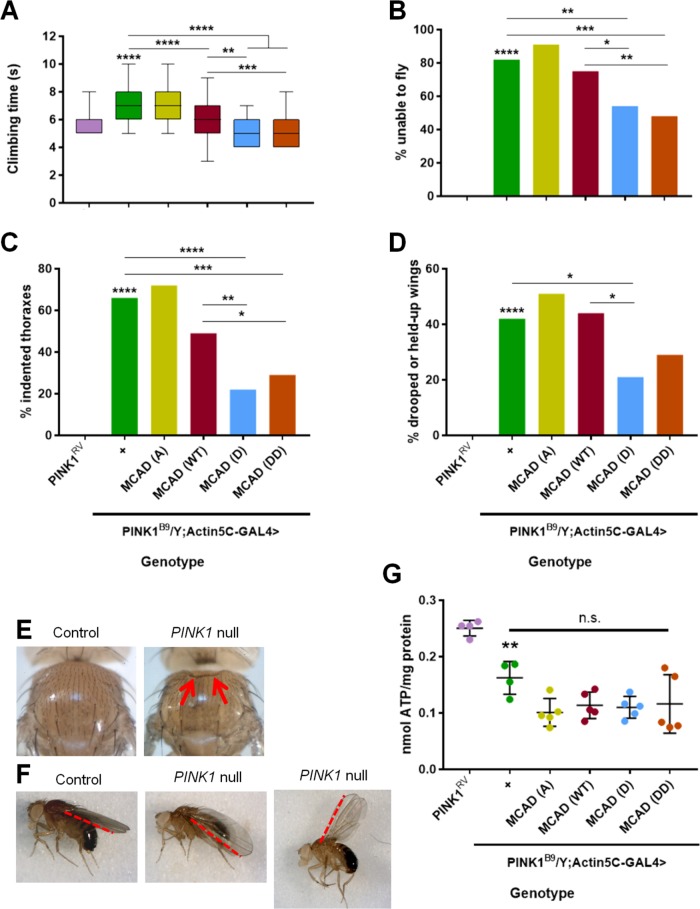
We first used these wild-type, phosphoresistant, and phosphomimetic MCAD transgenic flies to determine whether mimicking phosphorylation at MCAD S347 could rescue either of PINK1 null’s established climbing and flight phenotypes (Clark et al., 2006; Park et al., 2006). Our PINK1 null flies took significantly longer to climb 8 cm than control flies, and this climbing latency was fully rescued by MCAD (D) and MCAD (DD), partially rescued by MCAD (WT), and not rescued by MCAD (A). Notably, MCAD (D) and MCAD (DD) rescued the phenotype significantly better than MCAD (WT) (Figure 2A). Our PINK1 null flies also exhibited a striking flight deficit— ∼80% were unable to fly—which was significantly rescued by MCAD (D) and MCAD (DD) and not rescued by MCAD (A) or MCAD (WT) (Figure 2B). Thus, mimicking phosphorylation of MCAD at serine 347 rescued PINK1 null’s climbing and flight deficits.
FIGURE 2:
Phosphomimetic MCAD S347 rescues PINK1 null phenotypes. (A–F) All assays were conducted in 3- to 4-d-old male flies. n = 50–55 flies. For all panels, comparison is to PINK1RV unless otherwise indicated. All lines showing significance represent two-group comparisons. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 for all figures. (A) Time flies take to climb upward 8 cm. One-way ANOVA, followed by Tukey’s multiple comparisons. Middle line is the median, outer edges of the box are 25th and 75th percentiles, and whiskers are the minimum and maximum values. (B) Percentage of flies that cannot fly. (C) Percentage of flies with indented thoraxes. (D) Percentage of flies with drooped or held-up wings. (B–D) Chi-square test, followed by Fisher’s exact tests. (E) Representative images of a healthy PINK1RV (control) thorax (left), and the indented thorax of a PINK1B9(null) fly (right). Red arrows indicate indentation. (F) Representative images of healthy PINK1RV wings (left), as well as drooped (middle) and held-up (right) wings in PINK1B9flies. Red dotted lines emphasize the varying angles of the wings. (G) ATP levels divided by total protein levels, n = 4–5 samples, four flies per sample. One-way ANOVA, followed by Tukey’s multiple comparisons; n.s., not significant.
We next explored whether phosphomimetic MCAD could rescue either of PINK1 null’s established indented thorax or drooped/held-up wing phenotypes (Clark et al., 2006; Park et al., 2006). We observed that ∼65% of our PINK1 null flies displayed indented thoraxes, which are indicative of muscle degeneration (Clark et al., 2006; Park et al., 2006). This number was significantly decreased by expression of MCAD (D) and MCAD (DD) and not by expression of MCAD (A) or MCAD (WT) (Figure 2, C and E). Similarly, we found that ∼40% of our PINK1 null flies exhibited drooped or held-up wings. This number was significantly decreased by expression of MCAD (D), and not by expression of MCAD (A) or MCAD (WT). Rescue by MCAD (DD) did not reach significance, but MCAD (D) and MCAD (DD) were not significantly different (Figure 2, D and F). Overall, mimicking phosphorylation at MCAD S347 significantly improved PINK1 null’s thorax and wing posture phenotypes.
MCAD’s rescue of PINK1 null flies is independent of ATP production
Mitochondrial ATP production is compromised in PINK1 null flies, and is also influenced by MCAD enzymatic activity (Clark et al., 2006; Park et al., 2006; Vilain et al., 2012; Vos et al., 2012, 2017; Houten et al., 2016), so we tested whether MCAD (D)/(DD) restores PINK1 null’s organismal phenotypes by improving mitochondrial ATP generation. We found that neither MCAD (D) nor (DD) rescued the ATP deficit in PINK1 null flies, and that their ATP levels were indistinguishable from those of MCAD (A) and (WT) (Figure 2G). Therefore, it is likely that MCAD (D)/(DD) promotes mitochondrial functions other than ATP production to rescue PINK1 null’s organismal phenotypes.
MCAD’s rescue of PINK1 null flies is independent of its conventional role in β-oxidation
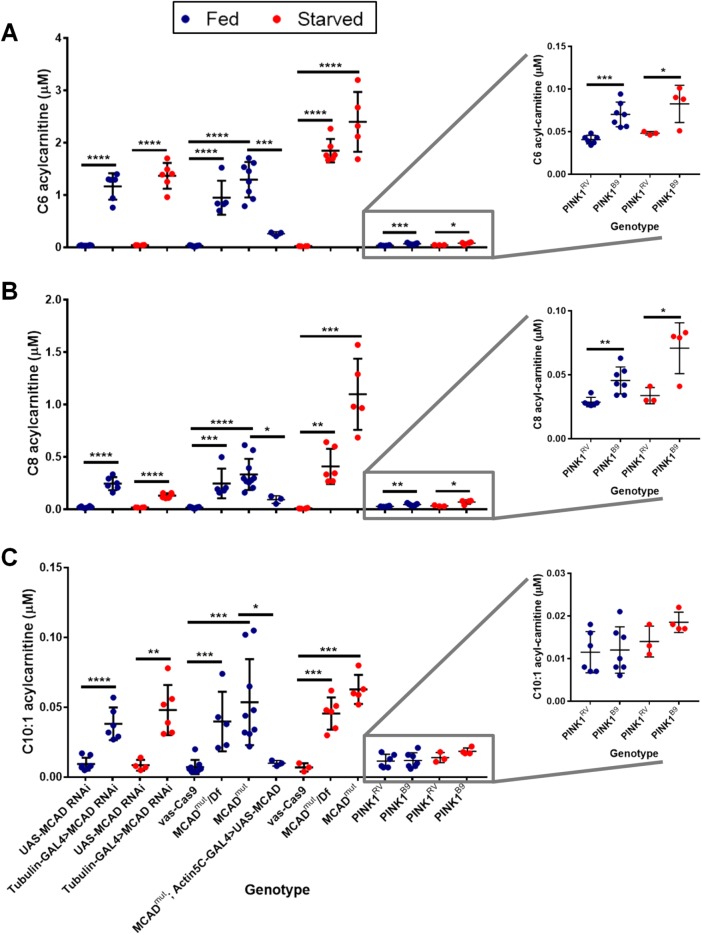
We next asked whether the observed organismal rescue could be due to MCAD’s role in β-oxidation. To answer this question, we analyzed the flies’ acylcarnitine profiles. This profile analysis is a well-established method by which newborns and patients are tested for defects in fatty acid oxidation, including MCAD enzymatic deficiency (Rinaldo et al., 2008). In addition to PINK1 null flies and their controls, we assayed MCAD deficient flies to validate that this analysis works in flies as it does in humans. Because there was no MCAD knockout fly available, we generated a fly model of MCAD deficiency by deleting a ∼1.5 kb fragment of the MCAD gene using CRISPR/Cas9 (Gratz et al., 2013), which we will call MCADmut. We also drove a UAS-MCAD RNAi line with Tubulin-GAL4 as an MCAD knockdown (Supplemental Figure S4A). Our homozygous MCADmut, transheterozygous MCADmut over a deficiency (Df) locus covering MCAD, and ubiquitous MCAD RNAi flies all exhibited significant elevations in medium-chain acylcarnitines characteristic of human MCAD deficiency: C6, C8, and C10:1 acylcarnitines. As expected, this pattern was exacerbated by starvation, because lowered glucose levels promote fatty acid oxidation (Houten et al., 2016). The elevations observed in MCADmut flies were fully rescued by expression of MCAD (WT), confirming that these elevations were specifically due to lack of MCAD (Figure 3). This profile is, to our knowledge, the first time that fly MCAD deficiency has been tested for similarity to human MCAD deficiency and validates our protocol.
FIGURE 3:
Acylcarnitine analysis in PINK1 null and MCAD mutant flies. Acylcarnitine analysis in lysates from 6- to 7-d-old male flies. “Starved” condition means flies were switched to vials containing only water for the final 24 h. Student’s t tests, n = 3–10 samples, five flies per sample, two technical replicates averaged per sample. Abundance of (A) C6, (B) C8, and (C) C10:1 acylcarnitines in MCAD deficient flies and PINK1 null flies.
We did not find that PINK1 null flies exhibited the same, stereotypical MCAD deficiency. Although C6 and C8 acylcarnitines increased slightly under both fed and starved conditions, their increase was nominal compared with the remarkable elevations observed in MCAD deficient flies (Figure 3, inset). Additionally, PINK1 null flies did not exhibit a significant C10:1 acylcarnitine increase (Figure 3 and Supplemental Table S2), suggesting that MCAD is still enzymatically active as an acyl-CoA dehydrogenase in PINK1 null flies. Expression of MCAD (D)/(DD) in PINK1 null flies did not alter their acylcarnitine profiles (Supplemental Figure S5). Therefore, phosphomimetic MCAD S347 likely rescues PINK1 null’s organismal phenotypes independently of MCAD’s acyl-CoA dehydrogenase activity.
PINK1 null flies have acylcarnitine and amino acid disruptions, which are selectively rescued by phosphomimetic MCAD S347
In addition to MCAD enzymatic deficiency, we used our acylcarnitine profile to determine whether other parts of the fatty acid oxidation pathway were affected by loss of PINK1. Our PINK1 null flies showed significant decreases in free carnitine and C2 and C3 acylcarnitines. In addition to C6 and C8 acylcarnitine (Figure 3, A and B), PINK1 null flies showed significant increases in four medium-chain acylcarnitines (whose elevations are not prominent characteristics of traditional MCAD deficiency), as well as nine long-chain acylcarnitines (Supplemental Table S2). Owing to increasing evidence that PINK1 plays a role in lipid and amino acid metabolic homeostasis (Senyilmaz et al., 2015; Stauch et al., 2016; Vos et al., 2017), we also measured free amino acids in PINK1 null flies. Of 40 amino acids tested, the flies were significantly deficient in 10 and had elevations in five (Supplemental Table S3).
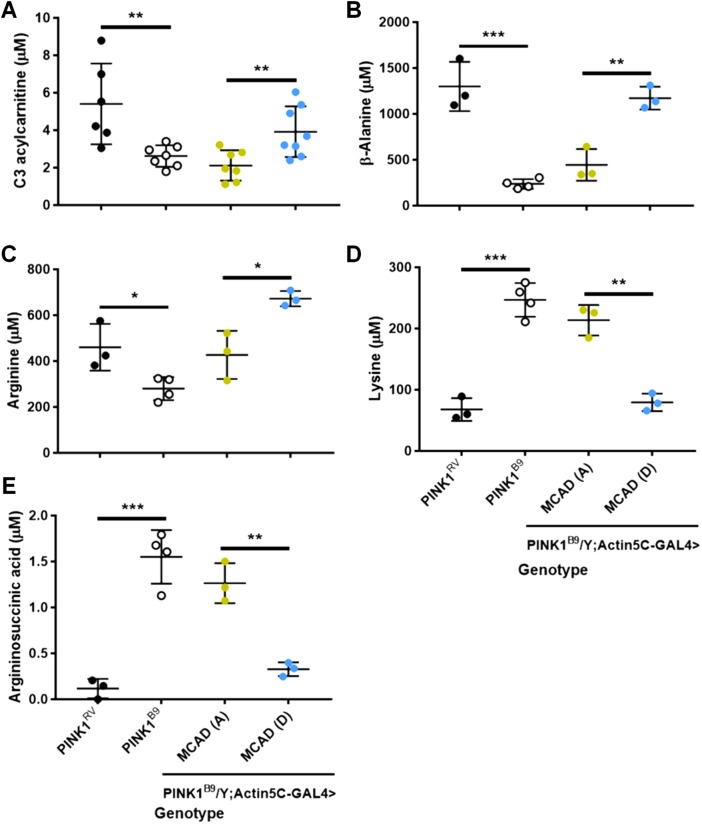
After identifying metabolites that were altered in PINK1 null flies, we wanted to know which might be involved in MCAD’s rescue of PINK1 null’s phenotypes. Among the metabolite deficiencies we observed in PINK1 null flies were C3 acylcarnitine, β-alanine, and arginine, which were rescued by expression of our phosphomimetic MCAD, as compared with our phosphoresistant MCAD. Similarly, two metabolites that were significantly elevated in PINK1 null flies were lysine and argininosuccininc acid, which were rescued by expression of our phosphomimetic MCAD, as compared with our phosphoresistant MCAD (Figure 4). Because the rescue of these metabolites by phosphomimetic MCAD are consistent with the rescue of PINK1 null’s flight, climbing, thorax, and wing posture phenotypes, phosphorylation of MCAD S347 may restore PINK1 null’s organismal function in part by restoring these acylcarnitine and amino acid abnormalities.
FIGURE 4:
MCAD S347D selectively rescues PINK1 null’s acylcarnitine and amino acid deficiencies. Comparing selected acylcarnitines and amino acids between PINK1RV and PINK1B9, and PINK1B9; Actin5C-GAL4>MCAD (A) and PINK1B9; Actin5C-GAL4>MCAD (D). Student’s t tests, n = 3–8 samples, five flies per sample, two technical replicates averaged per sample.
DISCUSSION
In this study, we found that the phosphorylation of MCAD at site S347 is dependent on PINK1 and that mimicking phosphorylation at this site in PINK1 null flies can rescue their flight, climbing, thorax, and wing posture phenotypes. To better understand why this phosphorylation is beneficial, we examined canonical functions of PINK1 and MCAD and found that the mechanism of rescue is divorced from both PINK1’s role in ATP generation, as well as MCAD’s role in β-oxidation. Instead, mimicking phosphorylation of MCAD S347 in a PINK1 null background corresponds to rescuing the altered C3 acylcarnitine, β alanine, arginine, lysine, and argininosuccininc acid levels found in PINK1 null flies. In addition, we established the first MCAD deficiency fly model and identified significant metabolite alterations in PINK1 null flies: 15 amino acids belonging to numerous metabolic pathways and 17 acylcarnitines of widely varying chain lengths. Thus, PINK1 plays a role in fatty acid and amino acid metabolism and mediates a novel function of the β-oxidation enzyme MCAD.
Our findings validate and expand on PINK1’s role in metabolic homeostasis, revealing a sweeping array of amino acids and acylcarnitines affected. While PINK1 null flies likely do not have an MCAD enzymatic deficiency, their acylcarnitine profiles reveal other β-oxidation defects. The pattern of disrupted short-, medium-, and long-chain acylcarnitines is suggestive of multiple acyl-CoA dehydrogenase deficiency (MADD) and does share some similarities to a MADD fly model (Alves et al., 2012). PINK1 knockout rats also show significantly decreased levels of both α and β subunits of electron transfer flavoprotein (ETF), which is consistent with our finding, since disruption of either of these subunits is a cause of MADD (Stauch et al., 2016). The elevations in long-chain acylcarnitines are particularly notable, and prominent peaks at C16 and C18:1 acylcarnitines are reminiscent of those found in human deficiencies in carnitine palmitoyltransferase 2 (CPT2) and/or carnitine acyl-CoA translocase (CACT). This observation is consistent with the recent finding that PINK1 knockout rats show significantly decreased levels of CPT2, as well as the finding that decreased PINK1 protein levels correlate to decreased very long-chain acyl-CoA dehydrogenase (VLCAD) levels in human brains (Choi et al., 2014; Stauch et al., 2016). Both of these deficiencies could lead to the elevations in long-chain acylcarnitines that we observed. The free amino acids disrupted in PINK1 null flies are wide-ranging and include essential, nonessential, and conditional amino acids. Because recent research in the PINK1 field indicates the need for more thorough metabolic analysis (Senyilmaz et al., 2015; Vos et al., 2017), our methods and results provide a template for future validation.
In the process of developing our metabolic profiles, we generated the first MCAD deficiency fly model, confirmed the canonical role of Drosophila MCAD in medium-chain fatty acid β-oxidation, and established that the fly MCAD deficiency acylcarnitine profile looks strikingly similar to the human profile, suggesting that the fly could be a good model for studying this and other inborn errors of metabolism. We also identified a new role for MCAD through PINK1-mediated phosphorylation of S347, since it rescues PINK1 null’s disruptions in C3 acylcarnitine, β alanine, arginine, lysine, and argininosuccininc acid without rescuing its dehydrogenase activity. These final three amino acids are metabolically linked in that argininosuccinic acid is a precursor of arginine, and arginine and lysine compete for the same transport proteins. Because we found that some fly MCAD localizes to the cytosol (Figure 1E), and PINK1 has been recently reported to have kinase activity there (Lee et al., 2017), there are several possibilities as to where PINK1 and MCAD interact and influence downstream targets. Phosphorylation of MCAD at S347 could cause a conformational change or alter protein binding to influence amino acids and acylcarnitines nearby or trigger a long-range signal to affect other metabolic pathways.
Because we assayed the PINK1 null flies under physiological conditions, we have further expanded PINK1’s role beyond stress-induced mitophagy. As fly and human PINK1 are functionally conserved (Clark et al., 2006), our observations of PINK1’s role in fly metabolic homeostasis could be relevant to human PINK1 as well. Recessive mutations in human PINK1 cause Parkinson’s disease, which is the second most common neurodegenerative disorder, as well as the second most common movement disorder. This study suggests that metabolic imbalance may constitute a part of pathogenesis in human neurodegenerative diseases.
Metabolism is a keystone of cellular function and must be tightly controlled, especially in cells with high energy demands. Our work links PINK1 to the β-oxidation enzyme MCAD and to lipid and amino acid metabolism and is one example of how metabolic profiling can be a rapid and effective way to study molecular and cellular functions of disease-causing genes. It also serves as a reminder of the underappreciated roles of known enzymes in metabolic activity and illustrates the power of coupling metabolic profiling with Drosophila genetics to identify those roles.
MATERIALS AND METHODS
Fly stocks
Fly stocks used were PE704, PINK15 (Clark et al., 2006), PINK1RV, PINK1B9 (Park et al., 2006), Actin5C-GAL4, Tubulin-GAL4, UAS-MCAD RNAiKK (VDRC 107820), y1 v1; P{CaryP}attP40 (BDSC 36304), M{vas-Cas9}ZH-2A (BDSC 51323), and Df(3L)BSC631 (BDSC 25722). All flies were maintained at 25°C and flipped into new food every 3–4 d. For details on the creation of transgenic flies and use of CRISPR/Cas9, please see the Supplemental Experimental Procedures.
Western blots and kinase assay
Please see the Supplemental Experimental Procedures.
Behavioral assays
To assess climbing ability, 3- to 4-d-old male flies were placed in a vial one at a time, allowed to acclimate for 1 min, gently tapped to the bottom, and then the time the fly took to climb 8 cm was recorded. To assess flight ability, flies were tapped one at a time out of a vial held upside down 1 ft over a bench top. If the fly flew away, then it was recorded as “1,” and if it was unable to fly, then it was recorded as “0.”
Wing and thorax assays
Wing posture was assessed in 3- to 4-d-old male flies by viewing a vial of five flies each under a dissecting scope, and recording whether wings were held normally (“0″) or drooped or held-up (“1″). Thorax indentations were observed by placing flies on a CO2 pad and viewing their thoraxes under a dissecting microscope. A healthy thorax was recorded as “0,” while an indented thorax was recorded as “1.”
ATP determination
ATP levels from lysates of four 30- to 31-d-old male flies per sample were measured using an ATP Determination Kit (Molecular Probes A22066). Flies were homogenized in 100 μl lysis buffer (6M guanidine-HCL, 100 mM Tris, 4 mM EDTA, pH 7.8 [ Park et al., 2006]), boiled for 5 min, placed on ice for 5 min, and centrifuged at 17,000 × g for 15 min. The supernatant was diluted 1:500 with kit reaction buffer, luciferase was added for 3 min, and then luminescence was measured on a GloMax Jr. luminometer (Promega). Values were normalized to protein levels, which were determined by diluting samples 1:1 and using a BCA Protein Assay Kit (Pierce 23225).
Phosphopeptide, acylcarnitine, and amino acid analysis
Please see the Supplemental Experimental Procedures.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7.03. To first confirm that the data were normally distributed, the D’Agostino-Pearson omnibus K2 test was used for N ≥ 8, and the Shapiro–Wilk test was used if N < 8. Student’s t tests were used to compare groups of two with Gaussian distributions. One-way analyses of variance (ANOVAs) were used to compare groups greater than two with Gaussian distributions, followed by Tukey’s multiple comparisons tests if the ANOVA gave p < 0.05. A chi-square test was used on groups of binary data, followed by Fisher’s exact tests if the chi-square gave p < 0.05.
Supplementary Material
Acknowledgments
We thank Bingwei Lu, Ming Guo, and Jongkyeong Chung for providing fly lines; Paul Valdmanis, Min Joo Kim, and Pei-I Tsai for technical support; and Vafa Bayat and Casey Guenthner for comments on the manuscript. This work was supported by the National Institutes of Health (NIH) (R00 NS067066) (X.W.), the Alfred P. Sloan Foundation (X.W.), the Shurl and Kay Curci Foundation (X.W.), the Klingenstein Foundation (X.W.), the National Science Foundation Graduate Research Fellowship Program (M.M.C. and A.M.P.), the Glenn/AFAR Scholarship for Research in the Biology of Aging (M.M.C.), and the BONFOR program of the University Clinic Bonn (C.S. and D.W.). The Drosophila Genomics Resource Center is supported by NIH grant 2P40OD010949.
Abbreviations used:
- ATP
adenosine triphosphate
- MCAD
medium-chain acyl-coenzyme A dehydrogenase
- PINK1
PTEN-induced putative kinase 1
- PTEN
phosphatase and tensin homologue
- UAS
upstream activation sequence
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-03-0155) on March 22, 2018.
REFERENCES
- Alves E, Henriques BJ, Rodrigues JV, Prudêncio P, Rocha H, Vilarinho L, Martinho RG, Gomes CM. (2012). Mutations at the flavin binding site of ETF: QO yield a MADD-like severe phenotype in Drosophila. Biochim Biophys Acta , 1284–1292. [DOI] [PubMed] [Google Scholar]
- Bian Y, Song C, Cheng K, Dong M, Wang F, Huang J, Sun D, Wang L, Ye M, Zou H. (2014). An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J Proteomics , 253–262. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific C31 integrases. Proc Natl Acad Sci USA , 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development , 401–415. [DOI] [PubMed] [Google Scholar]
- Chen Y, Dorn GW. (2013). PINK1-Phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science , 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Ravipati A, Nimmagadda V, Schubert M, Castellani RJ, Russell JW. (2014). Potential roles of PINK1 for increased PGC-1α-mediated mitochondrial fatty acid oxidation and their associations with Alzheimer disease and diabetes. Mitochondrion , 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. (2006). Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature , 1162–1166. [DOI] [PubMed] [Google Scholar]
- Du J, Li Z, Li QZ, Guan T, Yang Q, Xu H, Pritchard KA, Camara AK, Shi Y. (2013). Enoyl coenzyme A hydratase domain-containing 2, a potential novel regulator of myocardial ischemia injury. J Am Heart Assoc , e000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke S, Wu Z, Klinkenberg M, Sun Y, Auburger G, Guo S, Lu B. (2015). PINK1 and Parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab , 1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. (2010). PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol , 119–131. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics , 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsrud PA, Carson JJ, Hebert AS, Hubler SL, Niemi NM, Bailey DJ, Jochem A, Stapleton DS, Keller MP, Westphall MS, et al (2012). A quantitative map of the liver mitochondrial phosphoproteome reveals posttranslational control of ketogenesis. Cell Metab , 672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Violante S, Ventura FV, Wanders RJA. (2016). The biochemistry and physiology of mitochondrial fatty acid β-oxidation and its genetic disorders. Annu Rev Physiol , 23–44. [DOI] [PubMed] [Google Scholar]
- Humphrey SJ, Yang G, Yang P, Fazakerley DJ, Stöckli J, Yang JY, James DE. (2013). Dynamic adipocyte phosphoproteome reveals that akt directly regulates mTORC2. Cell Metab , 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. (2014). PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J Cell Biol , 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MMK. (2014). Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J , 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, et al (2012). PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating serine 65. Open Biol , 120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al (2014). Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature , 162–166. [DOI] [PubMed] [Google Scholar]
- Lee Y, Stevens DA, Kang S, Dawson VL, Shin J, Dawson TM. (2017). PINK1 primes Parkin-mediated ubiquitination of PARIS in dopaminergic neuronal survival. Cell Rep , 918–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Acín-Peréz R, Geghman KD, Manfredi G, Lu B, Li C. (2011). Pink1 regulates the oxidative phosphorylation machinery via mitochondrial fission. Proc Natl Acad Sci USA , 12920–12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. (2008). Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet , 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais VA, Haddad D, Craessaerts K, Bock P, De, Swerts J, Vilain S, Aerts L, Overbergh L, Seibler P, Klein C, et al (2014). PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science , 203–208. [DOI] [PubMed] [Google Scholar]
- Mulhern D. (2012). CST Curation Set: 13828; Year: 2012; biosample/treatment: cell line, Jurkat/calyculin_A & pervanadate; disease: T cell leukemia; SILAC: -; specificities of antibodies used to purify peptides prior to LCMS: (K/R)XpSX(K/R).
- Narendra D, Tanaka A, Suen D-F, Youle RJ. (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol , 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen D-F, Gautier CA, Shen J, Cookson MR, Youle RJ. (2010). PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol , e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim J-M, et al (2006). Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature , 1157–1161. [DOI] [PubMed] [Google Scholar]
- Pearlman SM, Serber Z, Ferrell JE. (2011). A mechanism for the evolution of phosphorylation sites. Cell , 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson JH, Ivatt RM, Sanchez-martinez A, Tufi R, Wilson E, Mortiboys H, Whitworth AJ. (2014). The complex I subunit NDUFA10 selectively rescues Drosophila pink1 mutants through a mechanism independent of mitophagy. PLoS Genet , e1004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. (2008). The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA , 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. (2010). The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/Parkin pathway. PLoS One , e10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato A. (2010). CST Curation Set: 9289; Year: 2010; biosample/treatment: cell line, Jurkat/pervanadate; disease: T cell leukemia; SILAC: -; specificities of antibodies used to purify peptides prior to LCMS: pTXR.
- Rinaldo P, Cowan TM, Matern D. (2008). Acylcarnitine profile analysis. Genet. Med , 151–156. [DOI] [PubMed] [Google Scholar]
- Rinaldo P, Matern D, Bennett MJ. (2002). Fatty acid oxidation disorders. Annu Rev Physiol , 477–502. [DOI] [PubMed] [Google Scholar]
- Senyilmaz D, Virtue S, Xu X, Tan CY, Griffin JL, Miller AK, Vidal-Puig A, Teleman AA. (2015). Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature , 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauch KL, Villeneuve LM, Purnell PR, Ottemann BM, Emanuel K, Fox HS. (2016). Loss of Pink1 modulates synaptic mitochondrial bioenergetics in the rat striatum prior to motor symptoms: concomitant complex I respiratory defects and increased complex II-mediated respiration. Proteomics Clin Appl , 1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. (2007). A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell , 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. (2010). Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol , 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MMK, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al (2004a). Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science , 1158–1160. [DOI] [PubMed] [Google Scholar]
- Valente EM, Salvi S, Ialongo T, Marongiu R, Elia AE, Caputo V, Romito L, Albanese A, Dallapiccola B, Bentivoglio AR. (2004b). PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann Neurol , 336–341. [DOI] [PubMed] [Google Scholar]
- Vilain S, Esposito G, Haddad D, Schaap O, Dobreva MP, Vos M, Van Meensel S, Morais VA, De Strooper B, Verstreken P. (2012). The yeast complex I equivalent NADH dehydrogenase rescues pink1 mutants. PLoS Genet , e1002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Przedborski S. (2011). Mitophagy: the latest problem for Parkinson’s disease. Trends Mol Med , 158–165. [DOI] [PubMed] [Google Scholar]
- Vos M, Esposito G, Edirisinghe JN, Vilain S, Haddad DM, Slabbaert JR, Van Meensel S, Schaap O, De Strooper B, Meganathan R, et al (2012). Vitamin K2 is a mitochondrial electron carrier that rescues pink1 deficiency. Science , 1306–1310. [DOI] [PubMed] [Google Scholar]
- Vos M, Geens A, Böhm C, Deaulmerie L, Swerts J, Rossi M, Craessaerts K, Leites EP, Seibler P, Rakovic A, et al (2017). Cardiolipin promotes electron transport between ubiquinone and complex I to rescue PINK1 deficiency. J Cell Biol , 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. (2011). PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell , 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, Pallanck LJ. (2009). The PINK1/Parkin pathway: a mitochondrial quality control system. J Bioenerg Biomembr , 499–503. [DOI] [PubMed] [Google Scholar]
- Whitworth AJ, Pallanck LJ. (2017). PINK1/Parkin mitophagy and neurodegeneration—what do we really know in vivo. Curr Opin Genet Dev , 47–53. [DOI] [PubMed] [Google Scholar]
- Woodroof HI, Pogson JH, Begley M, Cantley C, Deak M, Campbell DG, Van DMF, Whitworth AJ, Alessi DR, Muqit MK. (2011). Discovery of catalytically active orthologues of the Parkinson’s disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol , 110012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. (2009). Amino acids: metabolism, functions, and nutrition. Amino Acids , 1–17. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang J-W, Yang L, Beal MF, Vogel H, Lu B. (2006). Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci USA , 10793–10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. (2008). Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci USA , 7070–7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. (2008). The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci USA , 12022–12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. (2010). Drosophila Parkin requires PINK1 for mitochondrial translocation and ubiquitinates Mitofusin. Proc Natl Acad Sci USA , 5018–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.