Abstract
Background
Numerous studies have shown that miRNA levels are closely related to the survival time of patients with colon, rectal, or colorectal cancer (CRC). However, the outcomes of different investigations have been inconsistent. Accordingly, a meta-analysis was conducted to study associations among the three types of cancers.
Materials and methods
Studies published in English that estimated the expression levels of miRNAs with survival curves in CRC were identified until May 20, 2017 by online searches in PubMed, Embase, Web of Science, and the Cochrane Library by two independent authors. Pooled HRs with 95% CIs were used to estimate the correlation between miRNA expression and overall survival.
Results
A total of 63 relevant articles regarding 13 different miRNAs, with 10,254 patients were ultimately included. CRC patients with high expression of blood miR141 (HR 2.52, 95% CI 1.68–3.77), tissue miR21 (HR 1.31, 95% CI 1.12–1.53), miR181a (HR 1.52, 95% CI 1.26–1.83), or miR224 (HR 2.12, 95% CI 1.04–4.34), or low expression of tissue miR126 (HR 1.55, 95% CI 1.24–1.93) had significantly poor overall survival (P<0.05).
Conclusion
In general, blood miR141 and tissue miR21, miR181a, miR224, and miR126 had significant prognostic value. Among these, blood miR141 and tissue miR224 were strong biomarkers of prognosis for CRC.
Keywords: microRNA, colorectal cancer, prognosis, meta-analysis
Introduction
Numerous researchers have studied the associations between miRNA expression and the survival outcomes of colorectal cancer (CRC) patients.1–258 CRC has a 10% cancer incidence and mortality worldwide,259 and thus, it is one of the most serious diseases threatening human health. Despite great success in the treatment of CRC, the prognosis of CRC patients is still poor. Therefore, it is fundamental for the diagnosis, treatment, and prognosis of CRC patients to understand its emphasized molecular origin.260 Despite a comprehensive study about the mechanisms of CRC, there are still some challenges that require recognizing prognostic biomarkers with minimal invasion and sensitivity. Accordingly, it is of vital significance to improve the survival rate of CRC patients, utilizing rapid and reliable tumor-prognosis biomarkers.
miRNAs, small noncoding RNA gene products of approximately 22 nucleotides, are found in various types of organisms. They account for 2%–5% of the entire genome, number about 1,000, and regulate the expression of ≥20% of human genes.261 In addition, they play crucial roles in regulating the translation and degradation of mRNAs via base pairing to partially complementary sites, predominantly in the 3′-untranslated areas of mRNAs.262–264
In the study of CRC, a large number of articles have covered the fact that miRNAs are closely related to the survival time of patients.1–258 There were relatively small samples in these papers, and the present work aims to estimate the most accurate prognostic value between miRNA level and survival outcome of CRC patients, better to comprehend the miRNAs with prognostic pertinence that are potential candidates for clinical verification in the future.
Materials and methods
Search strategy
We used four online databases – PubMed, Embase, Web of Science, and the Cochrane Library – to find pertinent literature published until May 20, 2017. The combination term “miR and colorectal cancer” was employed for the literature search. Two authors (S Gao and ZY Zhao) independently performed this comprehensive online search.
Inclusion criteria
Articles qualified if they satisfied the following criteria: patients with colon/rectal cancer or CRC; miRNA levels in tissue, plasma, or serum and survival results were measured; at least one survival curve was measured of overall survival (OS), cause-specific survival (CSS), disease-free survival (DFS), recurrence-free survival (RFS), progression-free survival (PFS), and metastasis-free survival (MFS), with or without HRs/95% CIs; and full text published in English.
Exclusion criteria
Exclusion criteria were experimental studies, reviews, or letters without primary data and retracted papers; frequency of research evaluating prognostic value of miRNAs in tissue of four or less. Only the most comprehensive study was included for this meta-analysis if more than one paper had been published in the same research group.
Quality assessment
SG and ZY Zhao identified all qualifying studies analyzing the prognostic value of miRNAs in CRC, and YZ reevaluated uncertain data.
Study selection
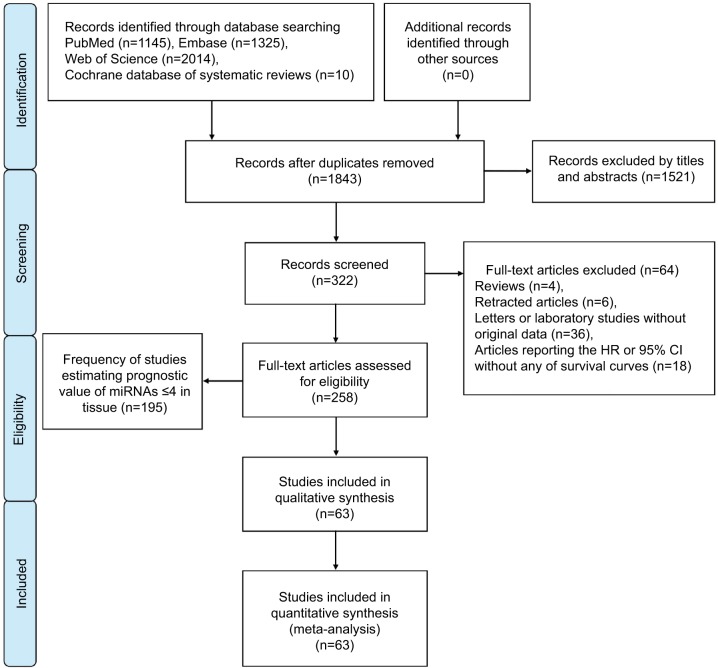
A flow diagram of the study selection process is presented in Figure 1. Our study found 1,843 articles for consideration within this meta-analysis, and 322 articles suitable for assessment of prognostic miRNA signatures in CRC and full-text papers were acquired by evaluating titles and abstracts. On elaborate review of research methodologies, 64 investigations were excluded, the details of which are shown in Figure 1. On the basis of the exclusion criteria, 63 studies were finally included in this meta-analysis.
Figure 1.
Flow diagram of literature search and selection.
Study frequency
The frequency of studies estimating the prognostic value of miRNAs in CRC are shown in Tables 1 (blood) and 2 (tissue), including miRNA name, number of studies estimating prognostic value, and references.
Table 1.
Frequency of studies estimating prognostic value of blood miRNA expression in colorectal cancer
| miR | n | Reference(s) | miR | n | Reference(s) | miR | n | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| 15b | 1 | 1 | 122 | 1 | 16 | 221 | 1 | 26 |
| 17-3p | 1 | 2 | 124-5p | 1 | 9 | 324-3p | 1 | 12 |
| 19a | 1 | 3 | 135 | 1 | 17 | 345 | 1 | 12 |
| 21 | 4 | 4–7 | 139-5p | 1 | 18 | 372 | 1 | 27 |
| 23b | 1 | 8 | 141 | 2 | 14, 19 | 628-5p | 1 | 12 |
| 26a | 1 | 9 | 143 | 1 | 12 | 885-5p | 1 | 28 |
| 29a | 1 | 10 | 155 | 1 | 20 | 886-3p | 1 | 12 |
| 29b | 1 | 11 | 183 | 1 | 21 | 1290 | 1 | 29 |
| 34a* | 1 | 12 | 194 | 1 | 11 | 4772-3p | 1 | 30 |
| 92a | 2 | 10, 13 | 196b | 1 | 22 | 6826 | 1 | 17 |
| 96 | 1 | 14 | 200b | 2 | 14, 16 | 6875 | 1 | 17 |
| 103 | 1 | 15 | 200c | 1 | 23 | |||
| 106a | 1 | 2 | 203 | 2 | 24, 25 |
Note: Highlighted studies were included in the present meta-analysis.
Study characteristics
Literature with Kaplan–Meier survival curves for CRC are detailed in Table 3. If data were not provided visually and merely as curves, they were extracted from the curves, and estimated HRs with 95% CIs were subsequently calculated using the method of Tierney et al265 with Engauge Digitizer version 4.1 software. In addition, if outcomes of both univariate and multiple covariates were covered, only the latter was chosen, because of adjustment for confounders.
Table 3.
Characteristics of studies included on colorectal cancer
| miRNA | Study | Country/source | Design | Sample | Number | Stage | Cutoff | Method | Follow-up (months) | Result | HR (L/H) | HR (H/L) | 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21 | Menéndez et al4 | Spain | P | Serum | 102 | I–IV | 1.00 | qRT-PCR | 36 | OSa | 0.50 | 0.25–1.02 | |
| DFSa | 0.51 | 0.25–1.06 | |||||||||||
| 21 | Toiyama et al5 | Japan | R | Serum | 188 | I–IV | <0.01 | RT-qPCR | 84 | OSa | 4.12 | 1.10–15.40 | |
| 21 | Monzo et al6 | Spain | R | Plasma | 52 | I–IV | Median | TaqMan | 48 | DFSb | 2.32 | 0.80–6.71 | |
| 21 | Tsukamoto et al7 | Japan | R | Plasma | 326 | I–IV | Median | qRT-PCR | 84 | OSa | 2.28 | 1.81–5.74 | |
| 259 | DFSa | 2.34 | 1.87–4.60 | ||||||||||
| 92a | Wang and Gu10 | China | R | Serum | 74 | II–IV | <0.06 | RT-qPCR | 35 | OSb | 1.17 | 0.70–1.97 | |
| 92a | Liu et al13 | China | R | Serum | 166 | I–IV | <0.01 | RT-qPCR | 53 | OSa | 4.36 | 1.64–11.57 | |
| 141 | Cheng et al19 | China, USA | R | Plasma | 258 | I–IV | Median | RT-qPCR | 96 | OSa | 2.40 | 1.18–4.86 | |
| 141 | Sun et al14 | USA | R | Plasma | 168 | I–IV | Mean | RT-qPCR | 96 | OSb | 2.58 | 1.58—4.21 | |
| 200b | Maierthaler et al16 | Germany I | R | Plasma | 308 | I–IV | Median | RT-qPCR | >72 | OSa | 0.77 | 0.57–1.05 | |
| Germany II | 219 | OSa | 1.21 | 0.98–1.50 | |||||||||
| 200b | Sun et al14 | USA | R | Plasma | 169 | I–IV | Mean | RT-qPCR | 96 | OSb | 2.46 | 1.57–3.85 | |
| 203 | Hur et al24 | Japan | R | Serum | 186 | I–IV | ROC | RT-qPCR | 70 | OSa | 2.14 | 1.09–4.21 | |
| 203 | Shi et al25 | China | R | Serum | 180 | II–IV | Median | RT-qPCR | 60 | OSb | 0.47 | 0.27–0.81 | |
| 21 | Kulda et al59 | Czech Republic | R | Frozen | 46 | I–IV | 8.10 | RT-qPCR | 56 | DFSb | 1.80 | 0.05–65.37 | |
| 21 | Shibuya et al60 | Japan | R | Frozen | 156 | I–IV | Mean | TaqMan | 84 | OSa | 1.95 | 1.05–4.48 | |
| 116 | DFSa | 2.53 | 1.15–5.59 | ||||||||||
| 21 | Nielsen et al61 | Denmark I | R | FFPE | 129 | II | 65% | ISH | >60 | OSb | 1.17 | 1.02–1.34 | |
| DFSa | 1.29 | 1.06–1.56 | |||||||||||
| Denmark II | 67 | OSb | 0.97 | 0.83–1.13 | |||||||||
| DFSa | 0.85 | 0.73–1.01 | |||||||||||
| 21 | Faltejskova et al62 | Czech Republic | R | Tissue | 44 | I–IV | Median | RT-qPCR | 86 | OSb | 2.72 | 0.63–11.83 | |
| 21 | Kjaer-Frifeldt et al63 | Denmark | P | FFPE | 520 | II | Mean | ISH | 84 | OSa | 1.08 | 0.97–1.22 | |
| RF-CSSa | 1.41 | 1.19–1.67 | |||||||||||
| 21 | Schee et al64 | Norway | P | Frozen | 193 | I–III | Median | qRT-PCR | >60 | MFSb | 1.17 | 0.59–2.32 | |
| 21 | Chen et al65 | China | R | Tissue | 195 | I–IV | Mean | RT-qPCR | >100 | OSa | 2.56 | 1.43–4.57 | |
| 21 | Toiyama et al5 | Japan | R | FFPE | 166 | I–IV | 3.70 | RT-qPCR | 84 | OSa | 0.59 | 0.21–1.63 | |
| 21 | Oue et al66 | Japan | R | FFPE | 87 | II–III | None | qRT-PCR | 60 | OSa | 3.13 | 1.20–8.17 | |
| Germany | 145 | II | OSa | 2.65 | 1.06–6.66 | ||||||||
| 21 | Bullock et al67 | UK | P | Frozen, FFPE | 50 | II | Mean | qRT-PCR | 96 | OSb | 2.47 | 1.19–5.55 | |
| DFSb | 2.68 | 1.21–5.93 | |||||||||||
| 21 | Fukushima et al68 | Japan | R | Frozen | 306 | I–IV | Mean | RT-qPCR | 90 | OSa | 2.88 | 1.70–5.08 | |
| 244 | DFSa | 2.94 | 1.68–5.36 | ||||||||||
| 21 | Kang et al69 | South Korea | R | FFPE | 277 | IIA–IIIC | Median | ISH | 80 | RFSa | 2.24 | 1.25–4.02 | |
| 21 | Caritg et al58 | Spain | R | Frozen | 69 | II | 2.04 | TaqMan | >140 | DFSb | 1.33 | 0.14–12.47 | |
| 21 | Feiersinger et al70 | Germany | R | FFPE | 29 | I–IV | Median | qRT-PCR | 205.15 | OSb | 1.45 | 0.39–5.43 | |
| DFSb | 1.76 | 0.75–4.11 | |||||||||||
| 21 | Iseki et al71 | Japan | R | FFPE | 32 | None | 8.10 | qRT-PCR | 63.2 | OSa | 2.52 | 0.65–8.34 | |
| PFSa | 4.93 | 1.08–20.81 | |||||||||||
| 21 | Lee et al72 | South Korea | R | FFPE | 170 | I–IV | Median | ISH | 105 | OSb | 0.93 | 0.54–1.60 | |
| 21 | Mima et al73 | USA I | P | FFPE | 190/192 | I–IV | 25% | RT-qPCR | 207.6 | OSa | 0.99 | 0.75–1.31 | |
| CSSa | 0.88 | 0.58–1.31 | |||||||||||
| USA II | 192/192 | 50% | OSa | 1.03 | 0.78–1.35 | ||||||||
| CSSa | 1.10 | 0.75–1.60 | |||||||||||
| USA III | 191/192 | 75% | OSa | 1.40 | 1.07–1.84 | ||||||||
| CSSa | 1.42 | 0.98–2.04 | |||||||||||
| 106a | Díaz et al49 | Spain | R | Frozen | 110 | I–IV | Median | RT-qPCR | 99 | OSa | 0.53 | 0.26–1.08 | |
| DFSa | 0.36 | 0.17–0.78 | |||||||||||
| 106a | Feng et al111 | China | R | Frozen | 28 | MB–NIB | Median | qRT-PCR | 60 | MFSb | 3.63 | 0.56–23.68 | |
| 106a | Schee et al64 | Norway | P | Frozen | 193 | I–III | Median | qRT-PCR | >60 | MFSb | 0.81 | 0.41–1.59 | |
| 106a | Ak et al112 | Turkey | R | FFPE | 40 | I–IV | None | qRT-PCR | >200 | OSb | 0.94 | 0.35–2.56 | |
| 106a | Bullock et al67 | UK | P | Frozen, FFPE | 50 | II | Mean | qRT-PCR | 96 | OSb | 2.25 | 1.00–5.04 | |
| DFSb | 2.91 | 1.32–6.42 | |||||||||||
| 106a | Hao et al113 | China | R | Tissue | 138 | I–IV | 66% | RT-qPCR | >60 | OSa | 1.87 | 1.13–3.09 | |
| DFSa | 1.22 | 0.70–2.12 | |||||||||||
| 106a | Hao et al114 | China | R | FFPE | 65 | I–IV | Median | qRT-PCR | >60 | OSb | 2.00 | 0.51–7.85 | |
| 125b | Nishida et al119 | Japan | R | Frozen | 89 | None | Median | RT-qPCR | >96 | OSb | 2.42 | 0.99–5.91 | |
| 125b | Ak et al112 | Turkey | R | FFPE | 40 | I–IV | None | qRT-PCR | >200 | OSb | 0.90 | 0.32–2.56 | |
| 125b | Cappuzzo et al35 | Italy | R | FFPE | 183 | None | None | None | 48 | OSa | 0.58 | 0.32–1.05 | |
| 125b | Rokavec et al106 | TCGA | R | Tissue | 438 | I–IV | None | Downloaded | >133 | OSb | 1.88 | 1.36–2.60 | |
| 125b | Sun et al33 | TCGA | R | Tissue | 107 | I–IV | Median | Downloaded | 141.1 | OSb | 2.29 | 1.33–3.92 | |
| 126 | Hansen et al120 | Denmark | R | FFPE | 89 | None | Median | ISH | 58 | OSb | 1.93 | 1.13–3.29 | |
| 83 | PFSb | 2.69 | 1.42–5.08 | ||||||||||
| 126 | Hansen et al121 | Sweden, Denmark | P | FFPE | 89 | None | Median | qRT-PCR | >30 | PFSa | 2.04 | 1.19–3.45 | |
| 126 | Hansen et al122 | DCCG | P | FFPE | 560 | II | Median | qRT-PCR | 84 | OSa | 1.32 | 1.00–1.72 | |
| RF-CSSa | 1.04 | 0.71–1.52 | |||||||||||
| 126 | Liu et al123 | China | R | Frozen | 92 | I–IV | None | qRT-PCR | 92 | OSb | 2.65 | 1.00–6.98 | |
| 126 | Ebrahimi et al124 | Australia | R | FFPE | 132 | I–IV | <l/>2 | qRT-PCR | >100 | OSb | 1.81 | 0.82–4.00 | |
| 126 | Yuan et al125 | China | R | Tissue | 75 | I–IV | 0/>0 | ISH | 68 | OSb | 2.35 | 0.91–6.06 | |
| 143 | Kulda et al59 | Czech Republic | R | Frozen | 46 | I–IV | 11.40 | RT-qPCR | 56 | DFSb | 0.45 | 0.07–2.78 | |
| 143 | Drebber et al150 | Germany | R | FFPE | 40 | I–IV | 1.00 | RT-qPCR | 76.8 | OSb | 1.52 | 0.32–7.22 | |
| 143 | Pichler et al151 | Austria | R | FFPE | 77 | II–IV | None | qRT-PCR | >125 | CSSa | 1.86 | 1.06–3.25 | |
| 52 | PFSb | 1.55 | 0.91–2.66 | ||||||||||
| 143 | Guo et al152 | China | R | Tissue | 79 | I–IV | Median | qRT-PCR | 122 | OSb | 1.45 | 0.69–3.07 | |
| 143 | Ak et al112 | Turkey | R | FFPE | 40 | I–IV | 1.76 | qRT-PCR | >200 | OSb | 2.69 | 0.80–9.08 | |
| 143 | Simmer et al153 | DCCG | P | FFPE | 55 | I–IV | Median | TaqMan | 42 | PFSa | 0.45 | 0.24–0.85 | |
| 145 | Drebber et al150 | Germany | R | FFPE | 40 | I–IV | 0.10 | RT-qPCR | 76.8 | OSb | 1.95 | 0.43–8.79 | |
| 145 | Schee et al64 | Norway | P | Frozen | 193 | I–III | Median | qRT-PCR | >60 | MFSb | 0.61 | 0.30–1.22 | |
| 145 | Pecqueux et al155 | Germany | R | Frozen | 47 | None | Median | RT-qPCR | >60 | OSb | 3.73 | 1.45–9.55 | |
| 145 | Zhou et al156 | China | R | Frozen | 60 | I–IV | Median | qRT-PCR | 80 | OSb | 2.57 | 1.12–5.90 | |
| DFSb | 2.58 | 1.12–5.94 | |||||||||||
| 145 | Sun et al33 | TCGA | R | Tissue | 107 | I–IV | Median | Downloaded | >144 | OSb | 0.52 | 0.30–0.77 | |
| 181a | Nishimura et al165 | Japan | R | Frozen | 162 | I–IV | Median | qRT-PCR | >144 | OSb | 2.00 | 1.05–3.80 | |
| DFSb | 2.26 | 1.10–4.61 | |||||||||||
| 181a | Ji et al166 | China I | R | Tissue | 137 | I–IV | Median | RT-qPCR | 100 | OSa | 1.87 | 1.08–3.25 | |
| China II | FFPE | 294 | 1.00 | ISH | OSa | 1.38 | 1.11–1.72 | ||||||
| 181a | Pichler et al167 | Austria | R | FFPE | 80 | II–IV | None | qRT-PCR | >125 | CSSa | 0.63 | 0.37–1.21 | |
| 54 | PFSb | 0.57 | 0.36–0.91 | ||||||||||
| 181a | Li et al168 | China | R | Frozen | 72 | I–IV | None | RT-qPCR | >60 | OSb | 2.06 | 1.00–4.23 | |
| 181a | Miyoshi et al18 | TCGA | R | Tissue | 93 | II–III | None | Downloaded | 135 | RFSb | 2.85 | 1.24–6.55 | |
| 224 | Liao et al209 | China | R | Frozen | 110 | I–IV | Median | qRT-PCR | 87 | OSb | 1.82 | 0.88–3.79 | |
| 224 | Yuan et al206 | China | R | Tissue | 108 | I–III | None | qRT-PCR | 60 | OSa | 0.27 | 0.14–0.51 | |
| 54 | DFSa | 0.07 | 0.02–0.25 | ||||||||||
| 224 | Zhang et al210 | China | R | Frozen | 108 | I–II | 25.72 | qRT-PCR | 62.5 | DFSb | 1.87 | 0.79–4.41 | |
| 224 | Adamopoulos et al211 | Greece | R | Frozen | 104 | I–IV | 56% | qRT-PCR | 120 | OSa | 4.41 | 1.72–11.34 | |
| 91 | DFSa | 4.61 | 1.41–15.09 | ||||||||||
| 224 | Ling et al212 | TCGA | R | Tissue | 143 | I–IV | None | Downloaded | 72 | OSa | 2.88 | 0.97–8.56 | |
| Italy I | 54 | qRT-PCR | 115 | OSa | 2.77 | 0.95–8.11 | |||||||
| Italy II | 68 | qRT-PCR | 115 | OSa | 4.14 | 0.96–17.76 | |||||||
| Romania | 38 | qRT-PCR | 70 | OSa | 1.76 | 0.36–8.64 | |||||||
| Austria | 74 | qRT-PCR | 130 | OSa | 2.36 | 1.32–4.21 | |||||||
| UK | 41 | qRT-PCR | 60 | OSb | 4.92 | 1.31–18.46 | |||||||
| MFSb | 6.51 | 1.97–21.51 | |||||||||||
| 429 | Li et al228 | China | R | Frozen | 107 | I–III | Median | qRT-PCR | 82 | OSb | 2.09 | 0.84–5.17 | |
| 429 | Diaz et al149 | Spain | R | Frozen | 127 | I–III | None | TaqMan | 113 | OSb | 0.35 | 0.16–0.77 | |
| 429 | Sun et al229 | China | R | Frozen | 84 | I–IV | None | qRT-PCR | 96 | OSb | 0.29 | 0.16–0.55 | |
| 429 | Dong et al230 | China | R | Frozen | 78 | I–IV | Median | qRT-PCR | 60 | OSb | 2.66 | 1.25–5.68 | |
| 429 | Han et al231 | China | R | Frozen | 71 | I–IV | Median | qRT-PCR | 60 | OSa | 1.85 | 1.02–3.33 |
Notes:
multiple-covariate analysis;
Univariate analysis;
Abbreviations: L/H, low versus high miRNA expression; H/L, high versus low miRNA expression; P, prospective; qRT-PCR, quantitative real-time polymerase chain reaction; OS, overall survival; DFS, disease-free survival; R, retrospective; RT-qPCR, reverse transcription qRT-PCR; RF-CSS, recurrence-free cause-specific survival; ROC, receiver-operating characteristic; FFPE, formalin-fixed, paraffin-embedded; ISH, in situ hybridization; MFS, metastasis-free survival; RFS, recurrence-free survival; PFS, progression-free survival; CSS, cause-specific survival; TCGA, the Cancer Genome Atlas; DCCG, Dutch Colorectal Cancer Group.
Statistical analyses
All analyses were performed utilizing Stata version 13.0 (StataCorp, College Station, TX, USA). Merged HRs were regarded as significant at the P<0.05 level if 95% CIs did not contain the value 1. Effect values for HRs were regarded as large if ≥2. HRs for OS were regarded as the prime reference standard if OS P-values were inconsistent with other survival outcomes with respect to the associated miRNA level. All analyses employed random-effect models instead of fixed-effect models, because there existed differences among the studies, including tissue detected (frozen or formalin-fixed, paraffin-embedded), blood (plasma or serum), tumor stage (I–IV), cutoff values, and miRNA-analysis methods. Publication bias was measured by Begg’s funnel plot, and a two-tailed P-value <0.05 was regarded as significant. The trim-and-fill method was performed if publication bias occurred. Sensitivity analysis was employed to weigh how powerful merged HRs were after a single study had been removed. An individual study was suspected of having excess of influence if the point estimation was outside the 95% CI after removal from the analysis.
Results
Meta-analysis
An overview of HRs appraised from comprehensive analysis of all the miRNAs is given in Table 4. Thirteen miRNAs were involved in this meta-analysis: miR21, miR92a, miR106a, miR125b, miR126, miR141, miR143, miR145, miR181a, miR200b, miR203, miR224, and miR429. Results of survival analyses of these miRNAs are given in Figures 2–8.
Table 4.
Meta-analysis results for miRNA expression in colorectal cancer
| miRNA | Survival analysis | Articles | Studies included | HR | 95% CI | Figure | P-value | Heterogeneity (Higgins’s I2) | Patients, n |
|---|---|---|---|---|---|---|---|---|---|
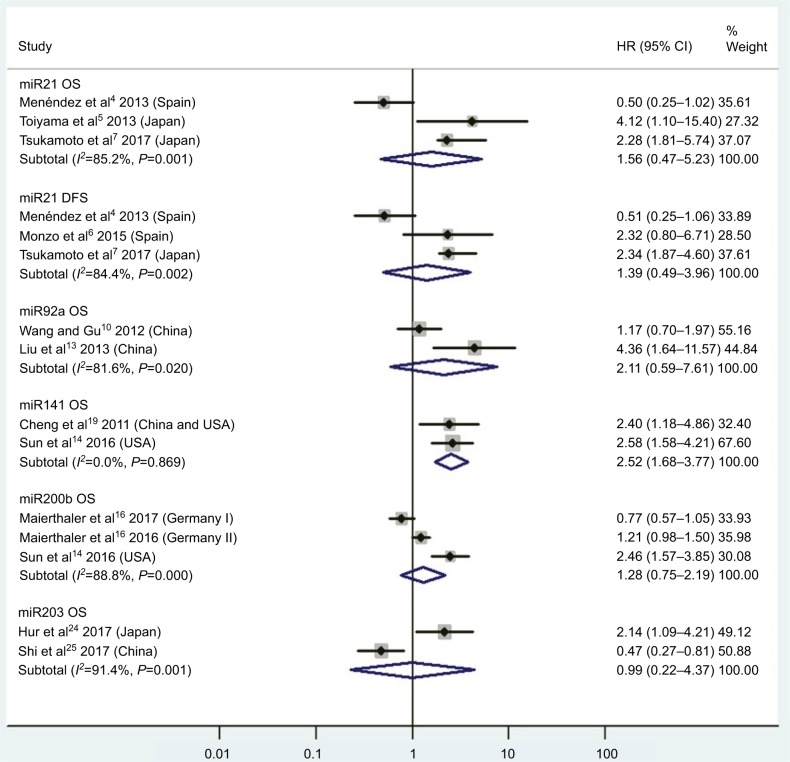
| High miR21 | OS | 3 | 4, 5, 7 | 1.56 | 0.47–5.23 | 2 | 0.47 | 85.2%, P<0.01 | 616 |
| High miR21 | DFS | 3 | 4, 6, 7 | 1.39 | 0.49–3.96 | 2 | 0.53 | 84.4%, P<0.01 | 480 |
| High miR92a | OS | 2 | 10, 13 | 2.11 | 0.59–7.61 | 2 | 0.25 | 81.6%, P=0.02 | 240 |
| High miR141 | OS | 2 | 14, 19 | 2.52 | 1.68–3.77 | 2 | <0.01 | 0.0%, P=0.87 | 426 |
| High miR200b | OS | 2 | 14, 16 | 1.28 | 0.75–2.19 | 2 | 0.36 | 88.8%, P<0.01 | 696 |
| High miR203 | OS | 2 | 24, 25 | 0.99 | 0.22–4.37 | 2 | 0.99 | 91.4%, P<0.01 | 366 |
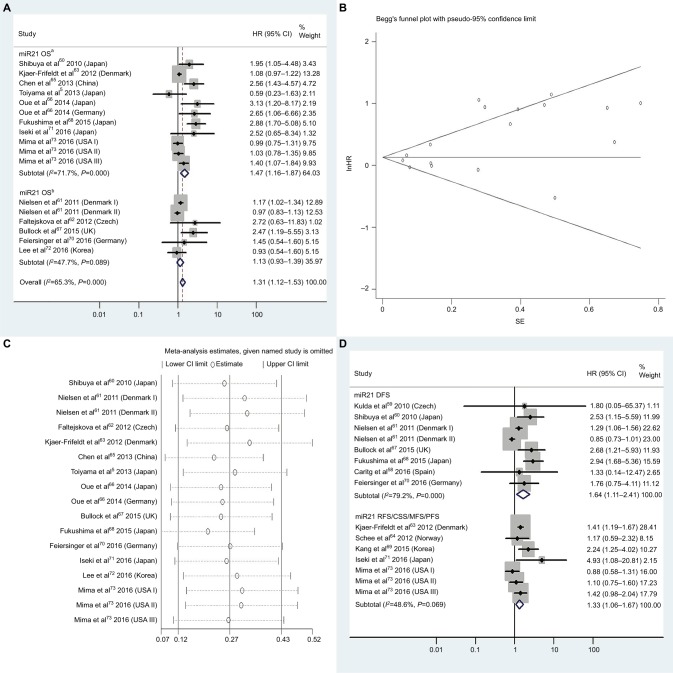
| High miR21 | OS | 13 | 5, 60–68, 70–73 | 1.31 | 1.12–1.53 | 3A | <0.01 | 65.3%, P<0.01 | 2,861 |
| High miR21 | OSa | 8 | 5, 60, 63, 65, 66, 68, 71, 73 | 1.47 | 1.16–1.87 | 3A | <0.01 | 71.7%, P<0.01 | 2,372 |
| High miR21 | DFS | 7 | 58–61, 67, 68, 70 | 1.64 | 1.11–2.41 | 3D | 0.01 | 79.2%, P<0.01 | 554 |
| High miR21 | RFS/CSS/MFS/PFS | 5 | 63, 64, 69, 71, 73 | 1.33 | 1.06–1.67 | 3D | 0.01 | 48.6%, P=0.07 | 1,787 |
| High miR21 | OS, adjustedb | 1.13 | 0.96–1.34 | 4B | 0.15 | 71.6%, P<0.01 | |||
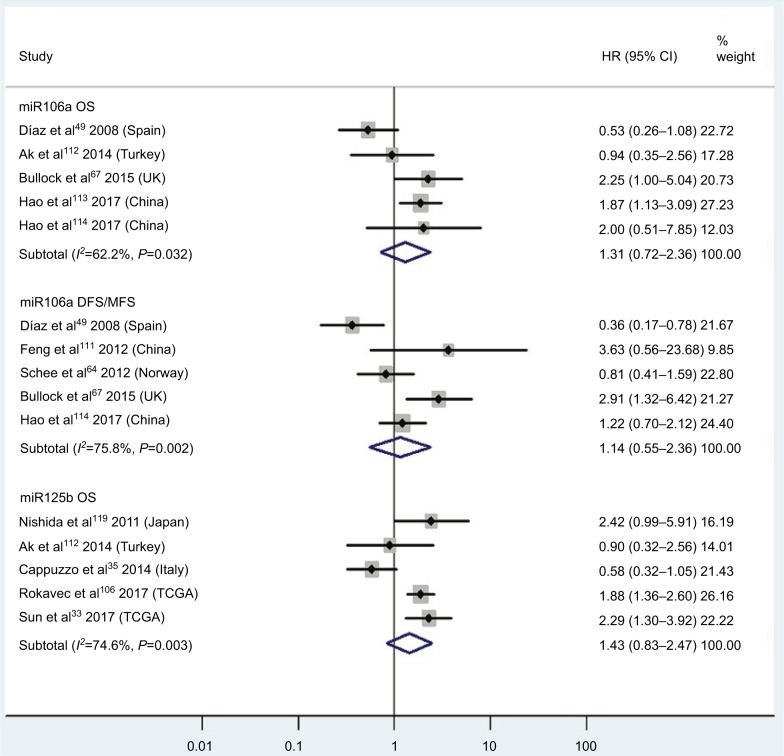
| High miR106a | OS | 5 | 49, 67, 112–114 | 1.31 | 0.72–2.36 | 5 | 0.38 | 62.2%, P=0.03 | 403 |
| High miR106a | DFS/MFS | 5 | 49, 64, 67, 11, 113 | 1.14 | 0.55–2.36 | 5 | 0.72 | 75.8%, P<0.01 | 519 |
| High miR125b | OS | 5 | 33, 35, 106, 112, 119 | 1.43 | 0.83–2.47 | 5 | 0.19 | 74.6%, P<0.01 | 857 |
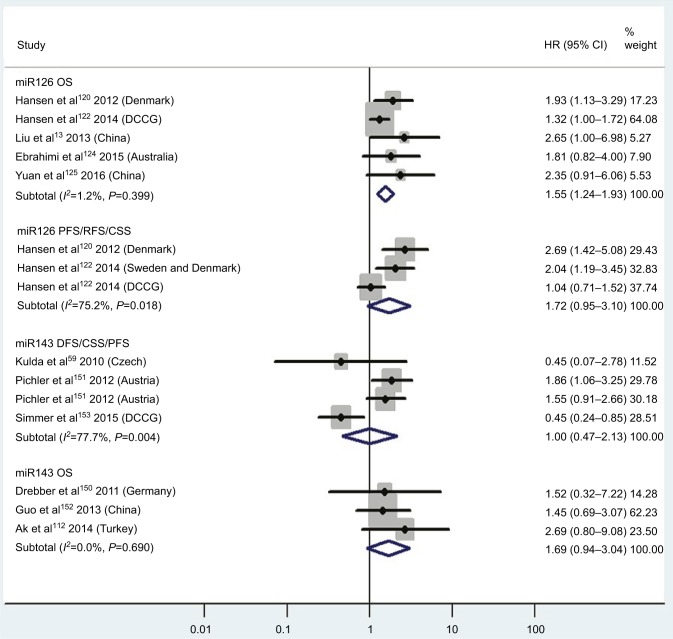
| Low miR126 | OS | 5 | 120, 122–125 | 1.55 | 1.24–1.93 | 6 | <0.01 | 1.2%, P=0.40 | 948 |
| Low miR126 | PFS/RFS/CSS | 3 | 120–122 | 1.72 | 0.95–3.10 | 6 | 0.07 | 75.2%, P=0.02 | 732 |
| Low miR143 | DFS/CSS/PFS | 3 | 59, 151, 153 | 1.00 | 0.47–2.13 | 6 | 1.00 | 77.7%, P<0.01 | 230 |
| Low miR143 | OS | 3 | 112, 150, 152 | 1.69 | 0.94–3.04 | 6 | 0.08 | 0.0%, P=0.69 | 159 |
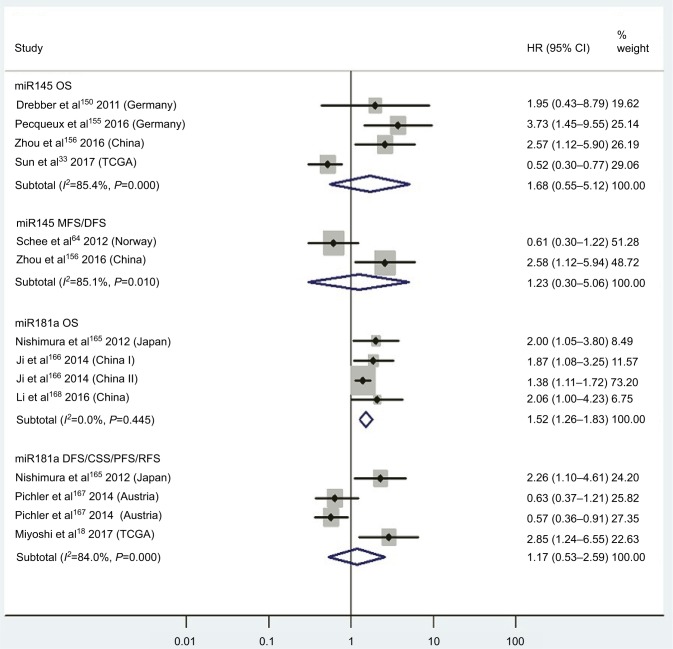
| Low miR145 | OS | 4 | 33, 150, 155, 156 | 1.68 | 0.55–5.12 | 7 | 0.36 | 85.4%, P<0.01 | 254 |
| Low miR145 | MFS/DFS | 2 | 64, 156 | 1.23 | 0.30–5.06 | 7 | 0.77 | 85.1%, P<0.01 | 253 |
| High miR181a | OS | 3 | 165, 166, 168 | 1.52 | 1.26–1.83 | 7 | <0.01 | 0.0%, P=0.45 | 665 |
| High miR181a | DFS/CSS/PFS/RFS | 3 | 18, 165, 67 | 1.17 | 0.53–2.59 | 7 | 0.69 | 84.0%, P<0.01 | 309 |
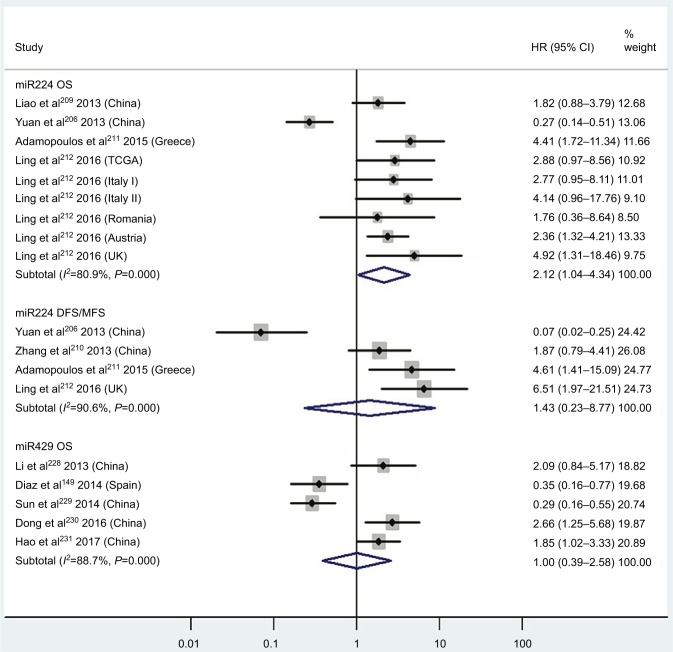
| High miR224 | OS | 4 | 206, 209, 211, 212 | 2.12 | 1.04–4.34 | 8 | 0.04 | 80.9%, P<0.01 | 740 |
| High miR224 | DFS/MFS | 4 | 206, 210–212 | 1.43 | 0.23–8.77 | 8 | 0.70 | 90.6%, P<0.01 | 294 |
| High miR429 | OS | 5 | 146, 228–231 | 1.00 | 0.39–2.58 | 8 | 1.00 | 88.7%, P<0.01 | 467 |
Notes:
Multiple-covariate analysis;
adjusted with trim-and-fill method.
Abbreviations: OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival; CSS, cause-specific survival; MFS, metastasis-free survival; PFS, progression-free survival.
Figure 2.
Pooled analyses of OS or DFS in association with high blood miR21-, miR92a-, miR141-, miR200b-, and miR203-expression levels.
Note: Weights are from random-effect analysis.
Abbreviations: OS, overall survival; DFS, disease-free survival.
Figure 3.
(A) Forest plots of pooled analyses of OS or OS (multiple-covariate analysis) in association with high tissue miR21-expression levels; (B) Begg’s funnel plot of publication bias for pooled analysis of OS in association with high tissue miR21-expression levels; (C) sensitivity analysis of pooled analysis of OS in association with high tissue miR21-expression levels; (D) forest plots of pooled analyses of DFS or RFS/CSS/MFS/PFS in association with high tissue miR21-expression levels. Weights are from random-effects analysis in A and D. aMultiple-covariate analysis; bunivariate analysis.
Abbreviations: OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival; CSS, cause-specific survival; MFS, metastasis-free survival; PFS, progression-free survival.
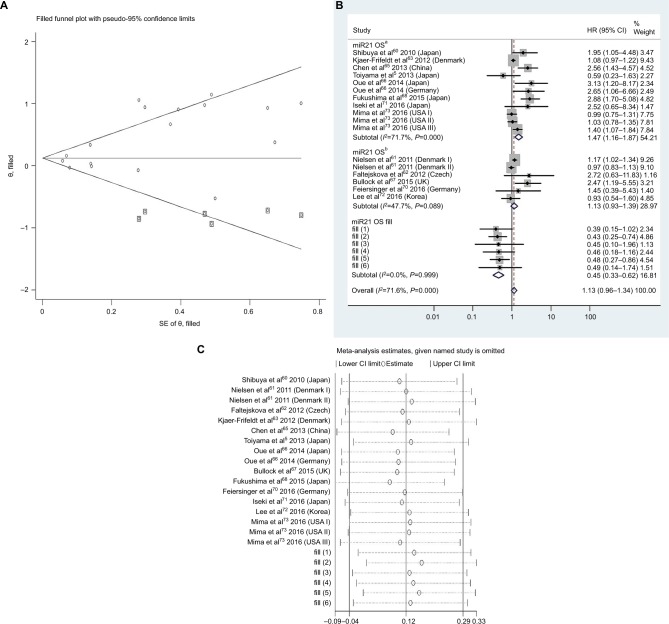
Figure 4.
(A) Funnel plot of pooled analysis adjusted with the trim-and-fill method of OS in association with high tissue miR21-expression levels. Circles, included studies; diamonds, presumed missing studies. (B) Forest plot of pooled analysis adjusted with the trim-and-fill method of OS in association with high tissue miR21-expression levels. (C) Sensitivity analysis of pooled analysis adjusted with the trim-and-fill method of OS in association with high tissue miR21-expression levels. Weights are from random-effects analysis. aMultiple-covariate analysis; bunivariate analysis.
Abbreviation: OS, overall survival.
Figure 5.
Pooled analyses of OS or DFS/MFS in association with high tissue miR106a- and miR125b-expression levels. Weights are from random-effects analysis.
Abbreviations: OS, overall survival; DFS, disease-free survival; MFS, metastasis-free survival; TCGA, the Cancer Genome Atlas.
Figure 6.
Pooled analyses of OS, PFS/RFS/CSS, or DFS/CSS/PFS in association with low tissue miR126- and miR143-expression levels. Weights are from random-effects analysis.
Abbreviations: OS, overall survival; DCCG, Dutch Colorectal Cancer Group; PFS, progression-free survival; RFS, recurrence-free survival; CSS, cause-specific survival; DFS, disease-free survival.
Figure 7.
Pooled analyses of OS, MFS/DFS or DFS/CSS/PFS/RFS in association with high tissue miR145-expression levels or low tissue miR181a-expression levels. Weights are from random-effects analysis.
Abbreviations: OS, overall survival; TCGA, the Cancer Genome Atlas; MFS, metastasis-free survival; DFS, disease-free survival; CSS, cause-specific survival; PFS, progression-free survival; RFS, recurrence-free survival.
Figure 8.
Pooled analyses of OS or DFS/MFS in association with high tissue miR224- and miR429-expression levels. Weights are from random-effects analysis.
Abbreviations: OS, overall survival; TCGA, the Cancer Genome Atlas; DFS, disease-free survival; MFS, metastasis-free survival.
CRC patients with high blood miR141, high tissue miR181a and miR224, or low tissue miR126 expression have significantly shorter OS
Two studies14,19 focused on associations between high blood miR141 levels and OS, indicating that CRC patients with high blood miR141 levels had significantly shorter OS than those with low miR141 expression (HR 2.52, 95% CI 1.68–3.77, P<0.01; Figure 2). Five papers120,122–125 stressed connections between low tissue miR126 levels and OS, suggesting that CRC patients with low expression of tissue miR126 levels had significantly poorer OS than those with high miR126 expression (HR 1.55, 95% CI 1.24–1.93, P<0.01; Figure 6).
Three articles concentrated on the relationship between high tissue miR181a levels and OS, demonstrating that CRC patients with high miR181a levels had significantly worse OS than those with low miR181a expression (HR 1.52, 95% CI 1.26–1.83, P<0.01; Figure 7). Four studies paid attention to correlations between high expression of tissue miR224 levels and OS, showing that CRC patients with high tissue miR224 levels had significantly shorter OS than those with low miR224 expression (HR 2.12, 95% CI 1.04–4.34, P=0.04; Figure 8).
There was no significant relationship between high expression levels of blood miR21, miR92a, miR200b, miR203, tissue miR106a, miR125b, or miR429 or low expression levels of tissue miR143 or miR145 and OS
High tissue miR21 expression forecasts poor OS
Thirteen investigations5,60–68,70–73 analyzed the connection between high tissue miR21 levels and OS, showing that CRC patients with high tissue miR21 levels had significantly worse OS than those with low miR21 expression (HR 1.31, 95% CI 1.12–1.53, P<0.01; Figure 3A).
Publication bias
To assess publications showing some degree of bias for OS of CRC patients with high tissue miR21 levels, our study used Begg’s funnel plot (Figure 3B). The P-value was less than 0.01, indicating the presence of publication bias. As such, the trim-and-fill method was performed and the pooled HR recalculated with presumed missing studies to estimate asymmetry in the funnel plot (Figure 4A), indicating no publication bias (P=0.73). The recalculated HR changed significance for OS (HR 1.13, 95% CI 0.96–1.34, P=0.15; Figure 4B).
Sensitivity analysis
For research on OS of CRC patients with high tissue miR21 levels, the sensitivity analysis did not manifest alterations during outcomes on the basis of the exclusion of any single investigation (Figure 3C), showing that no sole study significantly affected the merged HR or 95% CI. This also proved true for the outcome of OS adjusted with the trim-and-fill method (Figure 4C).
Key findings
We carried out a meta-analysis of 13 miRNAs and OS. Serving as the most investigated miRNA, miR21 (high tissue levels) in CRC showed significantly shorter OS than low tissue miR21 levels (P<0.05). However, there was no significant relationship between high blood miR21 levels and OS (P=0.47). The different detected sample types and relatively small sample capacity of the miR21 blood group (only three studies analyzing the relationship between blood miR21 levels and OS) may have been potential clinical reasons and caused the statistical significance between tissue and blood miR21 levels.
Encouragingly, the HR from analysis of the association between high tissue miR21 levels and OS (multiple-covariate analysis)5,60,63,65,66,68,71,73 was 1.47, which was greater than that reported in any of the 13 articles.5,60–78,70–73 Nevertheless, the significance did not remain in accordance with the forest plot, which was adjusted with the trim-and-fill method because publication bias existed (P<0.01; Figure 3B). This result indicated that the prognostic value of tissue miR21 was not stable in CRC patients. There were other miRNAs with significant prognostic value in CRC, including blood miR141 and tissue miR21, miR181a, miR224, and miR126 (P<0.05). Among these, blood miR141 and tissue miR224 were powerful prognostic candidates in CRC (HR ≥2).
Discussion
Present situation
Increasing numbers of studies have indicated that diverse miRNAs are connected with survival results in CRC patients.1–258 Nevertheless, no systematic review or meta-analysis has evaluated HRs between miRNA levels and survival outcomes of CRC patients. Therefore, it was of vital significance to launch a meta-analysis to comprehend the relationship between expression levels of miRNAs and prognoses of CRC patients.
Molecular mechanisms for miRNAs researched
An overview of miRNAs with dysregulated expression and their potential targets and pathways of entry is detailed in Figure 9. There was noticeable functional overlap and relationships among the miRNAs. Seven miRNAs (miR21, miR106a, miR126, miR143, miR181a, miR224, and miR429) touched upon cell functions, including cell apoptosis, cell cycle, and death. To sum up, these associations may refer to CRC progression.
Figure 9.
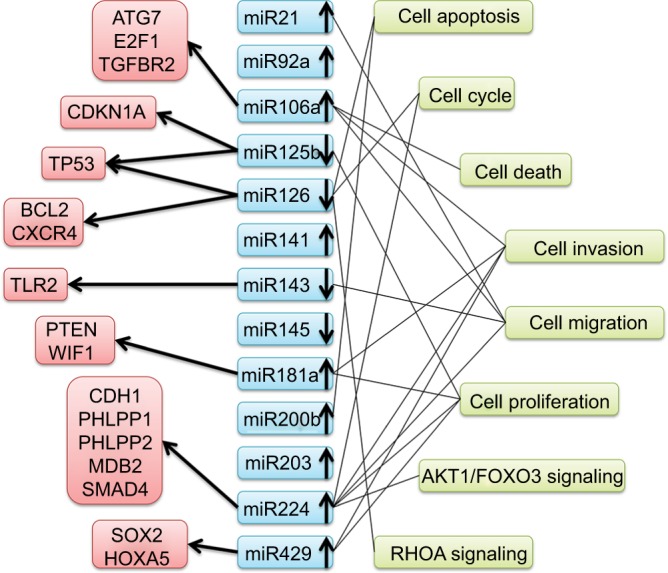
Summary of microRNAs with altered expression and potential targets and pathways entered in this study.
Abbreviations: ATG7, autophagy related 7; E2F1, E2F transcription factor 1; TGFBR2, transforming growth factor beta receptor 2; CDKN1A, cyclin dependent kinase inhibitor 1A; TP53, tumor protein p53; BCL2, apoptosis regulator; CXCR4, C-X-C motif chemokine receptor 4; TLR2, toll like receptor 2; PTEN, phosphatase and tensin homolog; WIF1, WNT inhibitory factor 1; CDH1, cadherin 1; PHLPP1, PH domain and leucine rich repeat protein phosphatase 1; PHLPP2, PH domain and leucine rich repeat protein phosphatase 2; MBD2, methyl-CpG binding domain protein 2; SMAD4, SMAD family member 4; SOX2, SRY-box 2; HOXA5, homeobox A5; AKT1, AKT serine/threonine kinase 1; FOXO3, forkhead box O3; RHOA, ras homolog family member A.
Other CRC molecular pathways
In addition to miRNAs, there are some other molecular data that can be confounders, related to mortalities, such as the chromosomal instability pathway, the DNA mismatch repair system, and microsatellite instability (MSI). Features of distinctive pathways are different models of genetic instability, succeeding clinical presentations, and features of pathological behavior. A majority of CRC follows the chromosomal instability pathway, features of which are extensive loss of heterozygosis and gross chromosomal abnormalities.266,267 Second, about 15% of CRC is due to the derangement of the DNA mismatch repair system and consequential MSI. The former is in charge of protein production, which identifies and directly repairs mononucleotide mismatches at MS sequences that escape the proofreading system of DNA polymerase. Furthermore, a previous meta-analysis indicated that MSI-high CRC patients had a 40% better OS rate compared with MS-stable CRC patients.268
Molecular pathological epidemiology (MPE)
MPE is a multidisciplinary research field of associations between endogenous and exogenous ingredients, molecular cancer biomarkers, and cancer progression and also a comprehensive interdisciplinary science on the strength of the characteristic principal and continuum theory of diseases.269,270 Other than miRNAs, DNA mutation and methylation and other diagnostics, such as blood tests, also play crucial roles in cancer prognosis and MPE, which deeply investigates environmental exposure, intermediate phenotypes, such as blood biomarkers, and molecular changes in cancer using molecular pathologic analyses. MPE helps precision medicine by providing robust evidence for exposure–outcome associations, such as with drugs.
Strengths
This study has some strengths. Almost all the articles with survival consequences in CRC patients with disparate miR-NAs were searched. Furthermore, the current expression profile of miRNAs is explicitly detailed in Tables 1 and 2 according to miRNAs and types of detected samples (blood or tissue). Papers assessing at least one of the survival curves of OS, CSS, DFS, RFS, PFS, and MFS were eventually included, and papers covering merely HRs or 95% CIs without any of the survival curves were excluded. Meta-analyses were performed on miRNAs investigated five or more times in CRC tissues. Virtually all the studies included had sample sizes ≥30 (except two),70,111 reinforcing the usability and enlarging the feasibility of consequences to CRC patients.
Table 2.
Frequency of studies estimating prognostic value of tissue-miRNA expression in colorectal cancer
| miR | n | Reference(s) | miR | n | Reference(s) | miR | n | Reference(s) | miR | n | Reference(s) | miR | n | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| let7a-5p | 1 | 31 | 34a-5p | 1 | 99 | 143 | 6 | 59, 112, 150–153 | 211 | 1 | 197 | 487b | 1 | 233 |
| let7a-2 | 1 | 32 | 34a | 1 | 100 | 144 | 1 | 154 | 212 | 1 | 198 | 490-3p | 1 | 234 |
| let7a | 1 | 33 | 92a | 3 | 64, 101, 102 | 145 | 5 | 33, 64, 150, 155, 156 | 214 | 1 | 199 | 491-5p | 1 | 205 |
| let7b | 1 | 34 | 93 | 2 | 34, 103 | 148a* | 1 | 157 | 215 | 4 | 58, 155, 179, 200 | 494 | 2 | 34, 235 |
| let7c | 1 | 35 | 96-5p | 1 | 104 | 148a | 3 | 81, 158, 159 | 217 | 2 | 201, 202 | 498 | 1 | 215 |
| let7e | 1 | 18 | 96 | 1 | 105 | 149 | 2 | 160, 161 | 218 | 2 | 203, 204 | 503 | 3 | 227, 236, 237 |
| let7g* | 1 | 36 | 99a-3p | 1 | 36 | 150 | 1 | 162 | 221-3p | 1 | 205 | 505* | 1 | 36 |
| let7g | 1 | 37 | 99a | 2 | 35, 106 | 153 | 1 | 163 | 221* | 1 | 206 | 505 | 1 | 32 |
| let7i | 1 | 28 | 99b-5p | 1 | 107 | 154 | 1 | 164 | 221 | 2 | 28, 207 | 506 | 2 | 238, 239 |
| 7 | 3 | 34, 39, 40 | 100 | 2 | 106, 108 | 155 | 1 | 60 | 223 | 1 | 208 | 515-5p | 1 | 134 |
| 9 | 1 | 41 | 101 | 1 | 64 | 181a-1 | 1 | 32 | 224 | 5 | 206, 209–212 | 517a | 1 | 240 |
| 10b | 4 | 28, 42–44 | 103a | 1 | 58 | 181a | 5 | 18, 165–168 | 229-5p | 1 | 36 | 542-3p | 2 | 241, 242 |
| 15a-5p | 1 | 45 | 103-1 | 1 | 81 | 181b | 2 | 18, 169 | 296 | 1 | 213 | 556 | 1 | 67 |
| 15a | 1 | 46 | 103 | 1 | 109 | 181 c | 1 | 170 | 320a | 1 | 28 | 570 | 1 | 157 |
| 16 | 3 | 46–48 | 106a-5p | 1 | 110 | 182 | 3 | 171–173 | 320e | 1 | 214 | 573 | 1 | 134 |
| 17-5p | 3 | 49–51 | 106a | 7 | 49, 64, 67, 111–114 | 183 | 1 | 174 | 320 | 1 | 215 | 579 | 1 | 134 |
| 17 | 1 | 52 | 106b | 3 | 58, 115, 116 | 185 | 1 | 133 | 326 | 1 | 216 | 590-5p | 2 | 243, 244 |
| 18a | 2 | 53, 54 | 107 | 1 | 36 | 187 | 2 | 175, 176 | 328 | 1 | 32 | 592 | 2 | 186, 245 |
| 19b | 1 | 38 | 124-5 p | 1 | 9 | 188-3p | 1 | 177 | 335 | 1 | 217 | 610 | 1 | 246 |
| 20a-5p | 2 | 55, 56 | 124 | 2 | 117, 118 | 191 | 1 | 178 | 337-5p | 1 | 36 | 625-3p | 1 | 247 |
| 20a | 2 | 57, 58 | 125b | 5 | 33, 35, 106, 112, 119 | 192 | 2 | 179, 180 | 338-3p | 1 | 218 | 625 | 1 | 248 |
| 21 | 17 | 5, 58–73 | 126 | 6 | 120–125 | 193a-5p | 1 | 181 | 340 | 1 | 219 | 630 | 1 | 249 |
| 22 | 2 | 74, 75 | 128 | 2 | 126, 127 | 193b | 1 | 182 | 342-3p | 1 | 205 | 638 | 1 | 250 |
| 23b | 2 | 76, 77 | 130a | 1 | 81 | 194 | 3 | 38, 183, 184 | 361-5p | 1 | 220 | 652 | 1 | 32 |
| 24-3p | 2 | 78, 79 | 130b | 1 | 128 | 195-5p | 1 | 33 | 362-3p | 1 | 157 | 664-3p | 1 | 186 |
| 25 | 1 | 80 | 132 | 2 | 129, 130 | 195 | 2 | 33, 34 | 365-1 | 1 | 76 | 720 | 1 | 251 |
| 26a-2 | 1 | 81 | 133a | 2 | 131, 132 | 196a | 1 | 185 | 365-2 | 1 | 76 | 802 | 1 | 134 |
| 26b | 1 | 82 | 133b | 2 | 133, 134 | 196b-5p | 1 | 186 | 365 | 1 | 221 | 875-5p | 1 | 252 |
| 29a | 2 | 83, 84 | 134 | 1 | 135 | 196b | 1 | 185 | 370 | 1 | 36 | 885-5p | 1 | 28 |
| 29b | 1 | 85 | 135b | 3 | 136–138 | 197 | 1 | 32 | 372 | 1 | 222 | 889 | 1 | 36 |
| 30a-5p | 1 | 86 | 137 | 2 | 139, 140 | 198 | 1 | 187 | 376a | 1 | 223 | 944 | 1 | 157 |
| 30a | 1 | 87 | 138-5p | 2 | 141, 142 | 199a-3p | 1 | 188 | 378a-3p | 1 | 224 | 1260b | 1 | 253 |
| 30b | 1 | 88 | 138 | 1 | 143 | 199b | 1 | 189 | 378a-5p | 1 | 224 | 1288 | 1 | 254 |
| 30d | 1 | 89 | 139-3p | 1 | 144 | 200a | 3 | 82, 149, 190 | 378 | 1 | 225 | 1290 | 1 | 29 |
| 31-3p | 2 | 90, 91 | 139-5p | 2 | 18, 145 | 200c | 3 | 23, 149, 191 | 422a | 2 | 141, 226 | 1292 | 1 | 227 |
| 31-5p | 3 | 91–93 | 139 | 1 | 146 | 203 | 2 | 24, 192 | 424-3p | 1 | 227 | 1297 | 1 | 255 |
| 31 | 4 | 64, 94–96 | 140-5p | 2 | 147, 148 | 204-5p | 2 | 193, 194 | 429 | 5 | 149, 228–231 | 1826 | 1 | 256 |
| 32 | 2 | 32, 97 | 141 | 2 | 34, 149 | 206 | 1 | 195 | 450b-5p | 1 | 232 | 4500 | 1 | 257 |
| 33b | 1 | 98 | 143-5p | 1 | 58 | 210 | 1 | 196 | 455-5p | 1 | 186 | 4775 | 1 | 258 |
Note: Highlighted studies were included in the present meta-analysis.
Limitations
Nevertheless, we cannot overemphasize the following limitations. There was much heterogeneity in designs of studies, and most of the outcomes from our meta-analyses contained high heterogeneity (I2≥50%). Statistical assessment of publication bias was suboptimal. There existed differences among the studies, including tissue-detected (frozen or formalin-fixed, paraffin-embedded), blood (plasma or serum), tumor stage (I–IV), cutoff values, and miRNA methods. The present meta-analysis simply included papers published in English, perhaps excluding potential studies published in other languages with respect to miRNA level and prognosis of CRC patients. Papers covering only HRs or 95% CIs without survival curves were excluded, lowering the sample sizes of the papers included. Because of the massive interrelation between papers and data about CRC, we subjectively and selectively included specific studies on the basis of the inclusion and exclusion criteria, bringing about the omission of several possible miRNAs with prognostic value and a relatively small number of included studies. The studies included contained three types of cancers (colon and rectal cancer and CRC), which blurred the division between tumor types. Some blood miRNAs were from cell-free RNA, while others were from exosome isolates. These were considered the same to some degree and may have caused some deviations in the final results.
Implications for prospective clinical and scientific study
It should be mentioned that the current meta-analysis is the first system assessment of the pertinence of miRNA level to the prognosis of CRC patients. This study presents foundations for prospective clinical and scientific study with respect to clinical staff and other health care providers, for whom simultaneous determination of miRNA expression is able greatly to reinforce assessment of life expectancy of CRC patients, thus enabling prompt therapy, and for scientific researchers. Current research progress and trends in connections between miRNAs and prognosis of CRC patients are shown in Tables 1 and 2. Selectively basic experiments can be conducted using these details (Figure 9). Conflicting results on the prognosis of miRNAs may be addressed based on the present meta-analysis.
Conclusion
In general, blood miR141 and tissue miR21, miR181a, miR224, and miR126 have significant prognostic value. Among these, blood miR141 and tissue miR224 are strong biomarkers of prognosis in CRC.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Li J, Chen Y, Guo X, et al. Inhibition of miR-15b decreases cell migration and metastasis in colorectal cancer. Tumour Biol. 2016;37(7):8765–8773. doi: 10.1007/s13277-015-4396-9. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Liu Y, Wang C, et al. Serum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma. Sci Rep. 2015;5:12921. doi: 10.1038/srep12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumura T, Sugimachi K, Iinuma H, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–281. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menéndez P, Padilla D, Villarejo P, et al. Prognostic implications of serum microRNA-21 in colorectal cancer. J Surg Oncol. 2013;108(6):369–373. doi: 10.1002/jso.23415. [DOI] [PubMed] [Google Scholar]
- 5.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monzo M, Martínez-Rodenas F, Moreno I, et al. Differential MIR-21 expression in plasma from mesenteric versus peripheral veins: an observational study of disease-free survival in surgically resected colon cancer patients. Medicine (Baltimore) 2015;94(1):e145. doi: 10.1097/MD.0000000000000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y. Circulating exosomal microRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology. 2017;92(6):360–370. doi: 10.1159/000463387. [DOI] [PubMed] [Google Scholar]
- 8.Kou CH, Zhou T, Han XL, Zhuang HJ, Qian HX. Downregulation of mir-23b in plasma is associated with poor prognosis in patients with colorectal cancer. Oncol Lett. 2016;12(6):4838–4844. doi: 10.3892/ol.2016.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinushi T, Shibayama Y, Kinoshita I, et al. Low expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4. Cancer Med. 2014;3(6):1544–1552. doi: 10.1002/cam4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012;36(1):e61–e67. doi: 10.1016/j.canep.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Basati G, Razavi AE, Pakzad I, Malayeri FA. Circulating levels of the miRNAs, miR-194, and miR-29b, as clinically useful biomarkers for colorectal cancer. Tumour Biol. 2016;37(2):1781–1788. doi: 10.1007/s13277-015-3967-0. [DOI] [PubMed] [Google Scholar]
- 12.Schou JV, Rossi S, Jensen BV, et al. miR-345 in metastatic colorectal cancer: a non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecan. PLoS One. 2014;9(6):e99886. doi: 10.1371/journal.pone.0099886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu GH, Zhou ZG, Chen R, et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34(4):2175–2181. doi: 10.1007/s13277-013-0753-8. [DOI] [PubMed] [Google Scholar]
- 14.Sun Y, Liu Y, Cogdell D, et al. Examining plasma microRNA markers for colorectal cancer at different stages. Oncotarget. 2016;7(10):11434–11449. doi: 10.18632/oncotarget.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao L, Feng W, Yu Y, Xu X. Serum expression of miRNA-103, a potential diagnostic and prognostic biomarker for colorectal cancer. Int J Clin Med. 2016;9(7):14212–14218. [Google Scholar]
- 16.Maierthaler M, Benner A, Hoffmeister M, et al. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int J Cancer. 2017;140(1):176–187. doi: 10.1002/ijc.30433. [DOI] [PubMed] [Google Scholar]
- 17.Kijima T, Hazama S, Tsunedomi R, et al. MicroRNA-6826 and-6875 in plasma are valuable non-invasive biomarkers that predict the efficacy of vaccine treatment against metastatic colorectal cancer. Oncol Rep. 2017;37(1):23–30. doi: 10.3892/or.2016.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyoshi J, Toden S, Yoshida K, et al. MiR-139-5p as a novel serum biomarker for recurrence and metastasis in colorectal cancer. Sci Rep. 2017;7:43393. doi: 10.1038/srep43393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng H, Zhang L, Cogdell DE, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6(3):e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv ZC, Fan YS, Chen HB, Zhao DW. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumour Biol. 2015;36(3):1619–1625. doi: 10.1007/s13277-014-2760-9. [DOI] [PubMed] [Google Scholar]
- 21.Yuan D, Li K, Zhu K, Yan R, Dang C. Plasma miR-183 predicts recurrence and prognosis in patients with colorectal cancer. Cancer Biol Ther. 2015;16(2):268–275. doi: 10.1080/15384047.2014.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C, Gu L. The diagnostic effect of serum miR-196b as biomarker in colorectal cancer. Biomed Rep. 2017;6(1):39–45. doi: 10.3892/br.2016.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toiyama Y, Hur K, Tanaka K, et al. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259(4):735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hur K, Toiyama Y, Okugawa Y, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66(4):654–665. doi: 10.1136/gutjnl-2014-308737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi SQ, Ke JJ, Wu WQ, Xu QS. Serum miRNA-203 expression is associated with chemo-response to standard FOLFOX treatment of patients with colorectal cancer. Int J Clin Exp Pathol. 2017;10(1):105–116. [Google Scholar]
- 26.Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25(10):1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, Jin L, Jiang L, et al. Serum miR-372 is a diagnostic and prognostic biomarker in patients with early colorectal cancer. Anticancer Agents Med Chem. 2016;16(4):424–431. doi: 10.2174/1871520615666150716110406. [DOI] [PubMed] [Google Scholar]
- 28.Hur K, Toiyama Y, Schetter AJ, et al. Identification of a metastasis-specific microRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015;107(3):dju492. doi: 10.1093/jnci/dju492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imaoka H, Toiyama Y, Fujikawa H, et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol. 2016;27(10):1879–1886. doi: 10.1093/annonc/mdw279. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Eng C, Shen J, et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget. 2016;7(46):76250–76260. doi: 10.18632/oncotarget.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu TP, Huang CC, Yeh KT, et al. Down-regulation of let-7a-5p predicts lymph node metastasis and prognosis in colorectal cancer: implications for chemotherapy. Surg Oncol. 2016;25(4):429–434. doi: 10.1016/j.suronc.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Xu M, Kuang Y, Wang M, Han X, Yang Q. A microRNA expression signature as a predictor of survival for colon adenocarcinoma. Neoplasma. 2017;64(1):56–64. doi: 10.4149/neo_2017_107. [DOI] [PubMed] [Google Scholar]
- 33.Sun M, Song H, Wang S, et al. Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J Hematol Oncol. 2017;10(1):79. doi: 10.1186/s13045-017-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang IP, Tsai HL, Miao ZF, et al. Development of a deregulating microRNA panel for the detection of early relapse in postoperative colorectal cancer patients. J Transl Med. 2016;14(1):108. doi: 10.1186/s12967-016-0856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cappuzzo F, Sacconi A, Landi L, et al. MicroRNA signature in metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodies. Clin Colorectal Cancer. 2014;13(1):37.e4–45.e4. doi: 10.1016/j.clcc.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Molina-Pinelo S, Carnero A, Rivera F, et al. MiR-107 and miR-99a-3p predict chemotherapy response in patients with advanced colorectal cancer. BMC Cancer. 2014;14:656. doi: 10.1186/1471-2407-14-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salendo J, Spitzner M, Kramer F, et al. Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g. Radiother Oncol. 2013;108(3):451–457. doi: 10.1016/j.radonc.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 38.Kahlert C, Klupp F, Brand K, et al. Invasion front-specific expression and prognostic significance of microRNA in colorectal liver metastases. Cancer Sci. 2011;102(10):1799–1807. doi: 10.1111/j.1349-7006.2011.02023.x. [DOI] [PubMed] [Google Scholar]
- 39.Suto T, Yokobori T, Yajima R, et al. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. 2015;36(3):338–345. doi: 10.1093/carcin/bgu242. [DOI] [PubMed] [Google Scholar]
- 40.Nagano Y, Toiyama Y, Okugawa Y, et al. MicroRNA-7 is associated with malignant potential and poor prognosis in human colorectal cancer. Anticancer Res. 2016;36(12):6521–6526. doi: 10.21873/anticanres.11253. [DOI] [PubMed] [Google Scholar]
- 41.Zhu M, Xu Y, Ge M, Gui Z, Yan F. Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell proliferation and apoptosis. Cancer Sci. 2015;106(7):833–839. doi: 10.1111/cas.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishida N, Yamashita S, Mimori K, et al. MicroRNA-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann Surg Oncol. 2012;19(9):3065–3071. doi: 10.1245/s10434-012-2246-1. [DOI] [PubMed] [Google Scholar]
- 43.Pizzini S, Bisognin A, Mandruzzato S, et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics. 2013;14:589. doi: 10.1186/1471-2164-14-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H, Liu J, Chen Y, Ma C, Li B, Hao T. Up-regulation of mir-10b predicate advanced clinicopathological features and liver metastasis in colorectal cancer. Cancer Med. 2016;5(10):2932–2941. doi: 10.1002/cam4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kontos CK, Tsiakanikas P, Avgeris M, Papadopoulos IN, Scorilas A. miR-15a-5p, a novel prognostic biomarker, predicting recurrent colorectal adenocarcinoma. Mol Diagn Ther. 2017;21(4):453–464. doi: 10.1007/s40291-017-0270-3. [DOI] [PubMed] [Google Scholar]
- 46.Xiao G, Tang H, Wei W, Li J, Ji L, Ge J. Aberrant expression of microRNA-15a and microRNA-16 synergistically associates with tumor progression and prognosis in patients with colorectal cancer. Gastroenterol Res Pract. 2014;2014:364549. doi: 10.1155/2014/364549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian J, Jiang B, Li M, Chen J, Fang M. Prognostic significance of microRNA-16 expression in human colorectal cancer. World J Surg. 2013;37(12):2944–2949. doi: 10.1007/s00268-013-2205-4. [DOI] [PubMed] [Google Scholar]
- 48.Diamantopoulos MA, Kontos CK, Kerimis D, Papadopoulos IN, Scorilas A. Upregulated miR-16 expression is an independent indicator of relapse and poor overall survival of colorectal adenocarcinoma patients. Clin Chem Lab Med. 2017;55(5):737–747. doi: 10.1515/cclm-2016-0756. [DOI] [PubMed] [Google Scholar]
- 49.Díaz R, Silva J, García JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47(9):794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 50.Ma Y, Zhang P, Wang F, et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3:1291. doi: 10.1038/ncomms2276. [DOI] [PubMed] [Google Scholar]
- 51.Fang L, Li H, Wang L, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5(10):2974–2987. doi: 10.18632/oncotarget.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu G, Tang JQ, Tian ML, et al. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol. 2012;106(3):232–237. doi: 10.1002/jso.22138. [DOI] [PubMed] [Google Scholar]
- 53.Motoyama K, Inoue H, Takatsuno Y, et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34(4):1069–1075. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 54.Wu CW, Dong YJ, Liang QY, et al. MicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangi-ectasia mutated in colorectal cancer. PLoS One. 2013;8(2):e57036. doi: 10.1371/journal.pone.0057036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Shi K, Wang Y, et al. Clinical value of integrated-signature miRNAs in colorectal cancer: miRNA expression profiling analysis and experimental validation. Oncotarget. 2015;6(35):37544–37556. doi: 10.18632/oncotarget.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng D, Zhao S, Tang H, et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget. 2016;7(29):45199–45213. doi: 10.18632/oncotarget.9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang GJ, Li Y, Zhou H, Xiao HX, Zhou T. miR-20a is an independent prognostic factor in colorectal cancer and is involved in cell metastasis. Mol Med Rep. 2014;10(1):283–291. doi: 10.3892/mmr.2014.2144. [DOI] [PubMed] [Google Scholar]
- 58.Caritg O, Navarro A, Moreno I, et al. Identifying high-risk stage II colon cancer patients: a three-microRNA-based score as a prognostic biomarker. Clin Colorectal Cancer. 2016;15(4):e175–e182. doi: 10.1016/j.clcc.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 59.Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200(2):154–160. doi: 10.1016/j.cancergencyto.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 60.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79(3–4):313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 61.Nielsen BS, Jørgensen S, Fog JU, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28(1):27–38. doi: 10.1007/s10585-010-9355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Faltejskova P, Besse A, Sevcikova S, et al. Clinical correlations of miR-21 expression in colorectal cancer patients and effects of its inhibition on DLD1 colon cancer cells. Int J Colorectal Dis. 2012;27(11):1401–1408. doi: 10.1007/s00384-012-1461-3. [DOI] [PubMed] [Google Scholar]
- 63.Kjaer-Frifeldt S, Hansen TF, Nielsen BS, et al. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br J Cancer. 2012;107(7):1169–1174. doi: 10.1038/bjc.2012.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schee K, Boye K, Abrahamsen TW, Fodstad Ø, Flatmark K. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer. 2012;12:505. doi: 10.1186/1471-2407-12-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen TH, Chang SW, Huang CC, et al. The prognostic significance of APC gene mutation and miR-21 expression in advanced-stage colorectal cancer. Colorectal Dis. 2013;15(11):1367–1374. doi: 10.1111/codi.12318. [DOI] [PubMed] [Google Scholar]
- 66.Oue N, Anami K, Schetter AJ, et al. High miR-21 expression from FFPE tissues is associated with poor survival and response to adjuvant chemotherapy in colon cancer. Int J Cancer. 2014;134(8):1926–1934. doi: 10.1002/ijc.28522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bullock MD, Pickard K, Mitter R, et al. Stratifying risk of recurrence in stage II colorectal cancer using deregulated stromal and epithelial microRNAs. Oncotarget. 2015;6(9):7262–7279. doi: 10.18632/oncotarget.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukushima Y, Iinuma H, Tsukamoto M, Matsuda K, Hashiguchi Y. Clinical significance of microRNA-21 as a biomarker in each Dukes’ stage of colorectal cancer. Oncol Rep. 2015;33(2):573–582. doi: 10.3892/or.2014.3614. [DOI] [PubMed] [Google Scholar]
- 69.Kang WK, Lee JK, Oh ST, Lee SH, Jung CK. Stromal expression of miR-21 in T3-4a colorectal cancer is an independent predictor of early tumor relapse. BMC Gastroenterol. 2015;15:2. doi: 10.1186/s12876-015-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feiersinger F, Nolte E, Wach S, et al. MiRNA-21 expression decreases from primary tumors to liver metastases in colorectal carcinoma. PLoS One. 2016;11(2):e0148580. doi: 10.1371/journal.pone.0148580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iseki Y, Shibutani M, Maeda K, et al. Prognostic significance of microRNA-21 expression in patients with unresectable metastatic colon cancer. Anticancer Res. 2016;36(10):5145–5151. doi: 10.21873/anticanres.11084. [DOI] [PubMed] [Google Scholar]
- 72.Lee KS, Nam SK, Koh J, et al. Stromal expression of microRNA-21 in advanced colorectal cancer patients with distant metastases. J Pathol Transl Med. 2016;50(4):270–277. doi: 10.4132/jptm.2016.03.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mima K, Nishihara R, Yang J, et al. MicroRNA MIR21 (miR-21) and PTGS2 expression in colorectal cancer and patient survival. Clin Cancer Res. 2016;22(15):3841–3848. doi: 10.1158/1078-0432.CCR-15-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang G, Xia S, Tian H, Liu Z, Zhou T. Clinical significance of miR-22 expression in patients with colorectal cancer. Med Oncol. 2012;29(5):3108–3112. doi: 10.1007/s12032-012-0233-9. [DOI] [PubMed] [Google Scholar]
- 75.Li B, Li B, Sun H, Zhang H. The predicted target gene validation, function, and prognosis studies of miRNA-22 in colorectal cancer tissue. Tumour Biol. 2017;39(3) doi: 10.1177/1010428317692257. 1010428317692257. [DOI] [PubMed] [Google Scholar]
- 76.Zhou X, Xu X, Wang J, Lin J, Chen W. Identifying miRNA/mRNA negative regulation pairs in colorectal cancer. Sci Rep. 2015;5:12995. doi: 10.1038/srep12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu S, Bai L, Li ZF, Li QG, Xie J, Jian B. Clinical significance and prognostic value of microRNA-23b expression level in colon cancer. Int J Clin Exp Pathol. 2016;9(10):10587–10592. [Google Scholar]
- 78.Gao Y, Liu Y, Du L, et al. Down-regulation of miR-24–3p in colorectal cancer is associated with malignant behavior. Med Oncol. 2015;32(1):362. doi: 10.1007/s12032-014-0362-4. [DOI] [PubMed] [Google Scholar]
- 79.Kerimis D, Kontos CK, Christodoulou S, Papadopoulos IN, Scorilas A. Elevated expression of miR-24-3p is a potentially adverse prognostic factor in colorectal adenocarcinoma. Clin Biochem. 2017;50(6):285–292. doi: 10.1016/j.clinbiochem.2016.11.034. [DOI] [PubMed] [Google Scholar]
- 80.Li X, Yang C, Wang X, Zhang J, Zhang R, Liu R. The expression of miR-25 is increased in colorectal cancer and is associated with patient prognosis. Med Oncol. 2014;31(1):781. doi: 10.1007/s12032-013-0781-7. [DOI] [PubMed] [Google Scholar]
- 81.Xu J, Zhao J, Zhang R. Four microRNAs signature for survival prognosis in colon cancer using TCGA data. Sci Rep. 2016;6:38306. doi: 10.1038/srep38306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Robertis M, Loiacono L, Fusilli C, et al. Dysregulation of EGFR pathway in EphA2 cell subpopulation significantly associates with poor prognosis in colorectal cancer. Clin Cancer Res. 2017;23(1):159–170. doi: 10.1158/1078-0432.CCR-16-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weissmann-Brenner A, Kushnir M, Yanai GL, et al. Tumor microRNA-29a expression and the risk of recurrence in stage II colon cancer. Int J Oncol. 2012;40(6):2097–2103. doi: 10.3892/ijo.2012.1403. [DOI] [PubMed] [Google Scholar]
- 84.Tang W, Zhu Y, Gao J, et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br J Cancer. 2014;110(2):450–458. doi: 10.1038/bjc.2013.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inoue A, Yamamoto H, Uemura M, et al. MicroRNA-29b is a novel prognostic marker in colorectal cancer. Ann Surg Oncol. 2015;22(Suppl 3):S1410–S1418. doi: 10.1245/s10434-014-4255-8. [DOI] [PubMed] [Google Scholar]
- 86.Yang LH, Yin SY, He RQ, et al. Prospective target genes and pathways of miR-30a-5p in colorectal cancer: an investigation using TCGA and bioinformatics analysis. Int J Clin Exp Med. 2017;10(3):4373–4385. [Google Scholar]
- 87.Zhang Q, Tang Q, Qin D, et al. Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis. Mol Cell Biol. 2015;35(6):988–1000. doi: 10.1128/MCB.01242-14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Liao WT, Ye YP, Zhang NJ, et al. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol. 2014;232(4):415–427. doi: 10.1002/path.4309. [DOI] [PubMed] [Google Scholar]
- 89.Yan L, Qiu J, Yao J. Downregulation of microRNA-30d promotes cell proliferation and invasion by targeting LRH-1 in colorectal carcinoma. Int J Mol Med. 2017;39(6):1371–1380. doi: 10.3892/ijmm.2017.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Manceau G, Imbeaud S, Thiébaut R, et al. Hsa-miR-31-3p expression is linked to progression-free survival in patients with KRAS wild-type metastatic colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res. 2014;20(12):3338–3347. doi: 10.1158/1078-0432.CCR-13-2750. [DOI] [PubMed] [Google Scholar]
- 91.Mlcochova J, Faltejskova-Vychytilova P, Ferracin M, et al. MicroRNA expression profiling identifies miR-31-5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximab. Oncotarget. 2015;6(36):38695–38704. doi: 10.18632/oncotarget.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Igarashi H, Kurihara H, Mitsuhashi K, et al. Association of microRNA-31-5p with clinical efficacy of anti-EGFR therapy in patients with metastatic colorectal cancer. Ann Surg Oncol. 2015;22(8):2640–2648. doi: 10.1245/s10434-014-4264-7. [DOI] [PubMed] [Google Scholar]
- 93.Kiss I, Mlcochova J, Bortlicek Z, et al. Efficacy and toxicity of panitumumab after progression on cetuximab and predictive value of MiR-31-5p in metastatic wild-type KRAS colorectal cancer patients. Anticancer Res. 2016;36(9):4955–4959. doi: 10.21873/anticanres.11063. [DOI] [PubMed] [Google Scholar]
- 94.Yang MH, Yu J, Chen N, et al. Elevated microRNA-31 expression regulates colorectal cancer progression by repressing its target gene SATB2. PLoS One. 2013;8(12):e85353. doi: 10.1371/journal.pone.0085353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen T, Yao LQ, Shi Q, et al. MicroRNA-31 contributes to colorectal cancer development by targeting factor inhibiting HIF-1α (FIH-1) Cancer Biol Ther. 2014;15(5):516–523. doi: 10.4161/cbt.28017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nosho K, Igarashi H, Nojima M, et al. Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis. 2014;35(4):776–783. doi: 10.1093/carcin/bgt374. [DOI] [PubMed] [Google Scholar]
- 97.Wu W, Yang P, Feng X, et al. The relationship between and clinical significance of microRNA-32 and phosphatase and tensin homologue. Genes Chromosomes Cancer. 2013;52(12):1133–1140. doi: 10.1002/gcc.22108. [DOI] [PubMed] [Google Scholar]
- 98.Liao W, Gu C, Huang A, Yao J, Sun R. MicroRNA-33b inhibits tumor cell growth and is associated with prognosis in colorectal cancer patients. Clin Transl Oncol. 2016;18(5):449–456. doi: 10.1007/s12094-015-1388-6. [DOI] [PubMed] [Google Scholar]
- 99.Gao J, Li N, Dong Y, et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorec-tal cancer. Oncogene. 2015;34(31):4142–4152. doi: 10.1038/onc.2014.348. [DOI] [PubMed] [Google Scholar]
- 100.Hiyoshi Y, Schetter AJ, Okayama H, et al. Increased microRNA-34b and-34c predominantly expressed in stromal tissues is associated with poor prognosis in human colon cancer. PLoS One. 2015;10(4):e0124899. doi: 10.1371/journal.pone.0124899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou T, Zhang G, Liu Z, Xia S, Tian H. Overexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Int J Colorectal Dis. 2013;28(1):19–24. doi: 10.1007/s00384-012-1528-1. [DOI] [PubMed] [Google Scholar]
- 102.Ke TW, Wei PL, Yeh KT, Chen WT, Cheng YW. MiR-92a promotes cell metastasis of colorectal cancer through PTEN-mediated PI3K/AKT pathway. Ann Surg Oncol. 2015;22(8):2649–2655. doi: 10.1245/s10434-014-4305-2. [DOI] [PubMed] [Google Scholar]
- 103.Xiao ZG, Deng ZS, Zhang YD, Zhang Y, Huang ZC. Clinical significance of microRNA-93 downregulation in human colon cancer. Eur J Gastroenterol Hepatol. 2013;25(3):296–301. doi: 10.1097/MEG.0b013e32835c077a. [DOI] [PubMed] [Google Scholar]
- 104.Ress AL, Stiegelbauer V, Winter E, et al. MiR-96-5p influences cellular growth and is associated with poor survival in colorectal cancer patients. Mol Carcinog. 2015;54(11):1442–1450. doi: 10.1002/mc.22218. [DOI] [PubMed] [Google Scholar]
- 105.Rapti SM, Kontos CK, Papadopoulos IN, Scorilas A. High miR-96 levels in colorectal adenocarcinoma predict poor prognosis, particularly in patients without distant metastasis at the time of initial diagnosis. Tumour Biol. 2016;37(9):11815–11824. doi: 10.1007/s13277-016-5023-0. [DOI] [PubMed] [Google Scholar]
- 106.Rokavec M, Horst D, Hermeking H. Cellular model of colon cancer progression reveals signatures of mRNAs, miRNA, lncRNAs, and epigenetic modifications associated with metastasis. Cancer Res. 2017;77(8):1854–1867. doi: 10.1158/0008-5472.CAN-16-3236. [DOI] [PubMed] [Google Scholar]
- 107.Li W, Chang J, Wang S, et al. miRNA-99b-5p suppresses liver metastasis of colorectal cancer by down-regulating mTOR. Oncotarget. 2015;6(27):24448–24462. doi: 10.18632/oncotarget.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen P, Xi Q, Wang Q, Wei P. Downregulation of microRNA-100 correlates with tumor progression and poor prognosis in colorectal cancer. Med Oncol. 2014;31(10):235. doi: 10.1007/s12032-014-0235-x. [DOI] [PubMed] [Google Scholar]
- 109.Zheng YB, Xiao K, Xiao GC, et al. MicroRNA-103 promotes tumor growth and metastasis in colorectal cancer by directly targeting LATS2. Oncol Lett. 2016;12(3):2194–2200. doi: 10.3892/ol.2016.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yue B, Sun B, Liu C, et al. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106(10):1323–1332. doi: 10.1111/cas.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feng B, Dong TT, Wang LL, et al. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One. 2012;7(8):e43452. doi: 10.1371/journal.pone.0043452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ak S, Tunca B, Tezcan G, et al. MicroRNA expression patterns of tumors in early-onset colorectal cancer patients. J Surg Res. 2014;191(1):113–122. doi: 10.1016/j.jss.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 113.Hao H, Liu L, Zhang D, et al. Diagnostic and prognostic value of miR-106a in colorectal cancer. Oncotarget. 2017;8(3):5038–5047. doi: 10.18632/oncotarget.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hao H, Xia G, Wang C, Zhong F, Liu L, Zhang D. miR-106a suppresses tumor cells death in colorectal cancer through targeting ATG7. Med Mol Morphol. 2017;50(2):76–85. doi: 10.1007/s00795-016-0150-7. [DOI] [PubMed] [Google Scholar]
- 115.Wang YX, Lang F, Liu YX, Yang CQ, Gao HJ. In situ hybridization analysis of the expression of miR-106b in colonic cancer. Int J Clin Exp Pathol. 2015;8(1):786–792. [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang GJ, Li JS, Zhou H, Xiao HX, Li Y, Zhou T. MicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. J Exp Clin Cancer Res. 2015;34:73. doi: 10.1186/s13046-015-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang MJ, Li Y, Wang R, et al. Downregulation of microRNA-124 is an independent prognostic factor in patients with colorectal cancer. Int J Colorectal Dis. 2013;28(2):183–189. doi: 10.1007/s00384-012-1550-3. [DOI] [PubMed] [Google Scholar]
- 118.Qiu Z, Guo W, Wang Q, et al. MicroRNA-124 reduces the pen-tose phosphate pathway and proliferation by targeting PRPS1 and RPIA mRNAs in human colorectal cancer cells. Gastroenterology. 2015;149(6):1587.e11–1598.e11. doi: 10.1053/j.gastro.2015.07.050. [DOI] [PubMed] [Google Scholar]
- 119.Nishida N, Yokobori T, Mimori K, et al. MicroRNA miR-1205b is a prognostic marker in human colorectal cancer. Int J Oncol. 2011;38(5):1437–1443. doi: 10.3892/ijo.2011.969. [DOI] [PubMed] [Google Scholar]
- 120.Hansen TF, Sørensen FB, Lindebjerg J, Jakobsen A. The predictive value of microRNA-126 in relation to first line treatment with capecitabine and oxaliplatin in patients with metastatic colorectal cancer. BMC Cancer. 2012;12:83. doi: 10.1186/1471-2407-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hansen TF, Christensen RD, Andersen RF, Sørensen FB, Johnsson A, Jakobsen A. MicroRNA-126 and epidermal growth factor-like domain 7-an angiogenic couple of importance in metastatic colorectal cancer: results from the Nordic ACT trial. Br J Cancer. 2013;109(5):1243–1251. doi: 10.1038/bjc.2013.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hansen TF, Kjær-Frifeldt S, Morgenthaler S, et al. The prognostic value of microRNA-126 and microvessel density in patients with stage II colon cancer: results from a population cohort. J Transl Med. 2014;12:254. doi: 10.1186/s12967-014-0254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu Y, Zhou Y, Feng X, et al. Low expression of microRNA-126 is associated with poor prognosis in colorectal cancer. Genes Chromosomes Cancer. 2014;53(4):358–365. doi: 10.1002/gcc.22146. [DOI] [PubMed] [Google Scholar]
- 124.Ebrahimi F, Gopalan V, Wahab R, Lu CT, Smith RA, Lam AK. Deregulation of miR-126 expression in colorectal cancer pathogenesis and its clinical significance. Exp Cell Res. 2015;339(2):333–341. doi: 10.1016/j.yexcr.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 125.Yuan W, Guo YQ, Li XY, et al. MicroRNA-126 inhibits colon cancer cell proliferation and invasion by targeting the chemokine (C-X-C motif) receptor 4. Oncotarget. 2016;7(37):60230–60244. doi: 10.18632/oncotarget.11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takahashi Y, Iwaya T, Sawada G, et al. Up-regulation of NEK2 by microRNA-128 methylation is associated with poor prognosis in colorectal cancer. Ann Surg Oncol. 2014;21(1):205–212. doi: 10.1245/s10434-013-3264-3. [DOI] [PubMed] [Google Scholar]
- 127.Lu W, Wang J, Yang G, et al. Posttranscriptional regulation of galectin-3 by miR-128 contributes to colorectal cancer progression. Oncotarget. 2017;8(9):15242–15251. doi: 10.18632/oncotarget.14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Colangelo T, Fucci A, Votino C, et al. MicroRNA-130b promotes tumor development and is associated with poor prognosis in colorectal cancer. Neoplasia. 2013;15(9):1086–1099. doi: 10.1593/neo.13998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zheng YB, Luo HP, Shi Q, et al. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J Gastroenterol. 2014;20(21):6515–6522. doi: 10.3748/wjg.v20.i21.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mokutani Y, Uemura M, Munakata K, et al. Down-regulation of microRNA-132 is associated with poor prognosis of colorectal cancer. Ann Surg Oncol. 2016;23(Suppl 5):599–608. doi: 10.1245/s10434-016-5133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wan TM, Lam CS, Ng L, et al. The clinicopathological significance of miR-133a in colorectal cancer. Dis Markers. 2014;2014:919283. doi: 10.1155/2014/919283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang LL, Du LT, Li J, et al. Decreased expression of miR-133a correlates with poor prognosis in colorectal cancer patients. World J Gastroenterol. 2014;20(32):11340–11346. doi: 10.3748/wjg.v20.i32.11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Akçakaya P, Ekelund S, Kolosenko I, et al. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol. 2011;39(2):311–318. doi: 10.3892/ijo.2011.1043. [DOI] [PubMed] [Google Scholar]
- 134.Azizian A, Epping I, Kramer F, et al. Prognostic value of microRNAs in preoperative treated rectal cancer. Int J Mol Sci. 2016;17(4):568. doi: 10.3390/ijms17040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xie Y, Song J, Zong Q, et al. Decreased expression of miR-134 and its clinical significance in human colorectal cancer. Hepatogastroenterology. 2015;62(139):615–619. [PubMed] [Google Scholar]
- 136.Gaedcke J, Grade M, Camps J, et al. The rectal cancer microRNAome: microRNA expression in rectal cancer and matched normal mucosa. Clin Cancer Res. 2012;18(18):4919–4130. doi: 10.1158/1078-0432.CCR-12-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Valeri N, Braconi C, Gasparini P, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kan SF, Yang JS, Sun GX, Sun JJ. MicroRNA-135b is associated with tumor progression in colorectal cancer. Int J Clin Exp Med. 2016;9(3):6533–6538. [Google Scholar]
- 139.Chen DL, Wang DS, Wu WJ, et al. Overexpression of paxillin induced by miR-137 suppression promotes tumor progression and metastasis in colorectal cancer. Carcinogenesis. 2013;34(4):803–811. doi: 10.1093/carcin/bgs400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Smith AR, Marquez RT, Tsao WC, et al. Tumor suppressive microRNA-137 negatively regulates Musashi-1 and colorectal cancer progression. Oncotarget. 2015;6(14):12558–12573. doi: 10.18632/oncotarget.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Qin YZ, Xie XC, Liu HZ, Lai H, Qiu H, Ge LY. Screening and preliminary validation of miRNAs with the regulation of hTERT in colorectal cancer. Oncol Rep. 2015;33(6):2728–2736. doi: 10.3892/or.2015.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhao L, Yu H, Yi S, et al. The tumor suppressor miR-138–5p targets PD-L1 in colorectal cancer. Oncotarget. 2016;7(29):45370–45384. doi: 10.18632/oncotarget.9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Long L, Huang G, Zhu H, Guo Y, Liu Y, Huo J. Down-regulation of miR-138 promotes colorectal cancer metastasis via directly targeting TWIST2. J Transl Med. 2013;11:275. doi: 10.1186/1479-5876-11-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu X, Duan B, Dong Y, et al. MicroRNA-139-3p indicates a poor prognosis of colon cancer. Int J Clin Exp Pathol. 2014;7(11):8046–8052. [PMC free article] [PubMed] [Google Scholar]
- 145.Song M, Yin Y, Zhang J, et al. MiR-139-5p inhibits migration and invasion of colorectal cancer by downregulating AMFR and NOTCH1. Protein Cell. 2014;5(11):851–861. doi: 10.1007/s13238-014-0093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guo H, Hu X, Ge S, Qian G, Zhang J. Regulation of RAP1B by miR-139 suppresses human colorectal carcinoma cell proliferation. Int J Biochem Cell Biol. 2012;44(9):1465–1472. doi: 10.1016/j.biocel.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 147.Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget. 2015;6(23):19735–19746. doi: 10.18632/oncotarget.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang W, Zou C, Pan L, et al. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37(3):1123–1133. doi: 10.1159/000430237. [DOI] [PubMed] [Google Scholar]
- 149.Diaz T, Tejero R, Moreno I, et al. Role of miR-200 family members in survival of colorectal cancer patients treated with fluoropyrimidines. J Surg Oncol. 2014;109(7):676–683. doi: 10.1002/jso.23572. [DOI] [PubMed] [Google Scholar]
- 150.Drebber U, Lay M, Wedemeyer I, et al. Altered levels of the onco-microRNA 21 and the tumor-suppressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int J Oncol. 2011;39(2):409–415. doi: 10.3892/ijo.2011.1036. [DOI] [PubMed] [Google Scholar]
- 151.Pichler M, Winter E, Stotz M, et al. Down-regulation of KRAS-interacting miRNA-143 predicts poor prognosis but not response to EGFR-targeted agents in colorectal cancer. Br J Cancer. 2012;106(11):1826–1832. doi: 10.1038/bjc.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guo H, Chen Y, Hu X, Qian G, Ge S, Zhang J. The regulation of Toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol Cancer. 2013;12:77. doi: 10.1186/1476-4598-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Simmer F, Venderbosch S, Dijkstra JR, et al. MicroRNA-143 is a putative predictive factor for the response to fluoropyrimidine-based chemotherapy in patients with metastatic colorectal cancer. Oncotarget. 2015;6(26):22996–23007. doi: 10.18632/oncotarget.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Iwaya T, Yokobori T, Nishida N, et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33(12):2391–2397. doi: 10.1093/carcin/bgs288. [DOI] [PubMed] [Google Scholar]
- 155.Pecqueux M, Liebetrau I, Werft W, et al. A comprehensive microRNA expression profile of liver and lung metastases of colorectal cancer with their corresponding host tissue and its prognostic impact on survival. Int J Mol Sci. 2016;17(10):E1755. doi: 10.3390/ijms17101755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhou P, Sun L, Liu D, Liu C, Sun L. Long non-coding RNA lincRNA-ROR promotes the progression of colon cancer and holds prognostic value by associating with miR-145. Pathol Oncol Res. 2016;22(4):733–740. doi: 10.1007/s12253-016-0061-x. [DOI] [PubMed] [Google Scholar]
- 157.Christensen LL, Tobiasen H, Holm A, et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int J Cancer. 2013;133(1):67–78. doi: 10.1002/ijc.28010. [DOI] [PubMed] [Google Scholar]
- 158.Takahashi M, Cuatrecasas M, Balaguer F, et al. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PLoS One. 2012;7(10):e46684. doi: 10.1371/journal.pone.0046684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hibino Y, Sakamoto N, Naito Y, et al. Significance of miR-148a in colorectal neoplasia: downregulation of miR-148a contributes to the carcinogenesis and cell invasion of colorectal cancer. Pathobiology. 2015;82(5):233–241. doi: 10.1159/000438826. [DOI] [PubMed] [Google Scholar]
- 160.Wang F, Ma YL, Zhang P, et al. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol. 2013;229(1):12–24. doi: 10.1002/path.4078. [DOI] [PubMed] [Google Scholar]
- 161.Xu K, Liu X, Mao X, et al. MicroRNA-149 suppresses colorectal cancer cell migration and invasion by directly targeting forkhead box transcription factor FOXM1. Cell Physiol Biochem. 2015;35(2):499–515. doi: 10.1159/000369715. [DOI] [PubMed] [Google Scholar]
- 162.Ma Y, Zhang P, Wang F, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61(10):1447–1453. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 163.Zhang L, Pickard K, Jenei V, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73(21):6435–6447. doi: 10.1158/0008-5472.CAN-12-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kai Y, Qiang C, Xinxin P, Miaomiao Z, Kuailu L. Decreased miR-154 expression and its clinical significance in human colorectal cancer. World J Surg Oncol. 2015;13:195. doi: 10.1186/s12957-015-0607-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 165.Nishimura J, Handa R, Yamamoto H, et al. microRNA-181a is associated with poor prognosis of colorectal cancer. Oncol Rep. 2012;28(6):2221–2226. doi: 10.3892/or.2012.2059. [DOI] [PubMed] [Google Scholar]
- 166.Ji D, Chen Z, Li M, et al. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer. 2014;13:86. doi: 10.1186/1476-4598-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pichler M, Winter E, Ress AL, et al. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. J Clin Pathol. 2014;67(3):198–203. doi: 10.1136/jclinpath-2013-201904. [DOI] [PubMed] [Google Scholar]
- 168.Li Z, Wang H, Xu Z, Sun Y, Han J. Expression and mechanism of microRNA-181A on incidence and survival in late liver metastases of colorectal cancer. Oncol Rep. 2016;35(3):1403–1408. doi: 10.3892/or.2016.4546. [DOI] [PubMed] [Google Scholar]
- 169.Bovell LC, Shanmugam C, Putcha BD, et al. The prognostic value of microRNAs varies with patient race/ethnicity and stage of colorectal cancer. Clin Cancer Res. 2013;19(14):3955–3965. doi: 10.1158/1078-0432.CCR-12-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yamazaki N, Koga Y, Taniguchi H, et al. High expression of miR-181c as a predictive marker of recurrence in stage II colorectal cancer. Oncotarget. 2017;8(4):6970–6983. doi: 10.18632/oncotarget.14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu H, Du L, Wen Z, et al. Up-regulation of miR-182 expression in colorectal cancer tissues and its prognostic value. Int J Colorectal Dis. 2013;28(5):697–703. doi: 10.1007/s00384-013-1674-0. [DOI] [PubMed] [Google Scholar]
- 172.Rapti SM, Kontos CK, Papadopoulos IN, Scorilas A. Enhanced miR-182 transcription is a predictor of poor overall survival in colorectal adenocarcinoma patients. Clin Chem Lab Med. 2014;52(8):1217–1227. doi: 10.1515/cclm-2013-0950. [DOI] [PubMed] [Google Scholar]
- 173.Wang S, Yang MH, Wang XY, Lin J, Ding YQ. Increased expression of miRNA-182 in colorectal carcinoma: an independent and tissue-specific prognostic factor. Int J Clin Exp Pathol. 2014;7(6):3498–3503. [PMC free article] [PubMed] [Google Scholar]
- 174.Zhou T, Zhang GJ, Zhou H, Xiao HX, Li Y. Overexpression of microRNA-183 in human colorectal cancer and its clinical significance. Eur J Gastroenterol Hepatol. 2014;26(2):229–233. doi: 10.1097/MEG.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 175.Wang ZS, Zhong M, Bian YH, et al. MicroRNA-187 inhibits tumor growth and invasion by directly targeting CD276 in colorectal cancer. Oncotarget. 2016;7(28):44266–44276. doi: 10.18632/oncotarget.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang F, Luo Y, Shao Z, et al. MicroRNA-187, a downstream effector of TGFβ pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 2016;373(2):203–213. doi: 10.1016/j.canlet.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 177.Pichler M, Stiegelbauer V, Vychytilova-Faltejskova P, et al. Genome-wide microRNA analysis identifies miR-188-3p as a novel prognostic marker and molecular factor involved in colorectal carcinogenesis. Clin Cancer Res. 2017;23(5):1323–1333. doi: 10.1158/1078-0432.CCR-16-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Qin S, Zhu Y, Ai F, et al. MicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3. Neoplasma. 2014;61(1):27–34. [PubMed] [Google Scholar]
- 179.Karaayvaz M, Pal T, Song B, et al. Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer. 2011;10(4):340–347. doi: 10.1016/j.clcc.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shan B, Chen P, Li S, Xu L, Yu H. Decreased expression of microRNA-192 correlates with tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2017;10(1):595–602. [Google Scholar]
- 181.Zhang P, Ji DB, Han HB, Shi YF, Du CZ, Gu J. Downregulation of miR-193a-5p correlates with lymph node metastasis and poor prognosis in colorectal cancer. World J Gastroenterol. 2014;20(34):12241–12248. doi: 10.3748/wjg.v20.i34.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Guo F, Luo Y, Mu YF, et al. miR-193b directly targets STMN1 and inhibits the malignant phenotype in colorectal cancer. Am J Cancer Res. 2016;6(11):2463–2475. [PMC free article] [PubMed] [Google Scholar]
- 183.Zhao HJ, Ren LL, Wang ZH, et al. MiR-194 deregulation contributes to colorectal carcinogenesis via targeting AKT2 pathway. Theranostics. 2014;4(12):1193–1208. doi: 10.7150/thno.8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang B, Shen ZL, Gao ZD, et al. MiR-194, commonly repressed in colorectal cancer, suppresses tumor growth by regulating the MAP4K4/c-Jun/MDM2 signaling pathway. Cell Cycle. 2015;14(7):1046–1058. doi: 10.1080/15384101.2015.1007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ge J, Chen Z, Li R, Lu T, Xiao G. Upregulation of microRNA-196a and microRNA-196b cooperatively correlate with aggressive progression and unfavorable prognosis in patients with colorectal cancer. Cancer Cell Int. 2014;14(1):128. doi: 10.1186/s12935-014-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Boisen MK, Dehlendorff C, Linnemann D, et al. Tissue microRNAs as predictors of outcome in patients with metastatic colorectal cancer treated with first line capecitabine and oxaliplatin with or without bevacizumab. PLoS One. 2014;9(10):e109430. doi: 10.1371/journal.pone.0109430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang M, Wang J, Kong X, et al. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyltransferase 8. Sci Rep. 2014;4:6145. doi: 10.1038/srep06145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wan D, He S, Xie B, et al. Aberrant expression of miR-199a-3p and its clinical significance in colorectal cancers. Med Oncol. 2013;30(1):378. doi: 10.1007/s12032-012-0378-6. [DOI] [PubMed] [Google Scholar]
- 189.Shen ZL, Wang B, Jiang KW, et al. Downregulation of miR-199b is associated with distant metastasis in colorectal cancer via activation of SIRT1 and inhibition of CREB/KISS1 signaling. Oncotarget. 2016;7(23):35092–35105. doi: 10.18632/oncotarget.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pichler M, Ress AL, Winter E, et al. MiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patients. Br J Cancer. 2014;110(6):1614–1621. doi: 10.1038/bjc.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xi Y, Formentini A, Chien M, et al. Prognostic values of microRNAs in colorectal cancer. Biomark Insights. 2006;2:113–121. [PMC free article] [PubMed] [Google Scholar]
- 192.Deng B, Wang B, Fang J, et al. MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Sci Rep. 2016;6:28301. doi: 10.1038/srep28301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sümbül AT, Göğebakan B, Ergün S, et al. miR-204-5p expression in colorectal cancer: an autophagy-associated gene. Tumour Biol. 2014;35(12):12713–12719. doi: 10.1007/s13277-014-2596-3. [DOI] [PubMed] [Google Scholar]
- 194.Yin Y, Zhang B, Wang W, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorec-tal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20(23):6187–6199. doi: 10.1158/1078-0432.CCR-14-1030. [DOI] [PubMed] [Google Scholar]
- 195.Sun P, Sun D, Wang X, Liu T, Ma Z, Duan L. miR-206 is an independent prognostic factor and inhibits tumor invasion and migration in colorectal cancer. Cancer Biomark. 2015;15(4):391–396. doi: 10.3233/CBM-150489. [DOI] [PubMed] [Google Scholar]
- 196.Qu A, Du L, Yang Y, et al. Hypoxia-inducible MiR-210 is an independent prognostic factor and contributes to metastasis in colorectal cancer. PLoS One. 2014;9(3):e90952. doi: 10.1371/journal.pone.0090952. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 197.Sümbül AT, Göğebakan B, Bayram S, Batmacı CY, Öztuzcu S. MicroRNA 211 expression is upregulated and associated with poor prognosis in colorectal cancer: a case-control study. Tumour Biol. 2015;36(12):9703–9709. doi: 10.1007/s13277-015-3708-4. [DOI] [PubMed] [Google Scholar]
- 198.Meng X, Wu J, Pan C, et al. Genetic and epigenetic down-regulation of microRNA-212 promotes colorectal tumor metastasis via dysregulation of MnSOD. Gastroenterology. 2013;145(2):426.e1-e6–436.e1-e6. doi: 10.1053/j.gastro.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 199.Chen DL, Wang ZQ, Zeng ZL, et al. Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology. 2014;60(2):598–609. doi: 10.1002/hep.27118. [DOI] [PubMed] [Google Scholar]
- 200.Li S, Gao J, Gu J, Yuan J, Hua D, Shen L. MicroRNA-215 inhibits relapse of colorectal cancer patients following radical surgery. Med Oncol. 2013;30(2):549. doi: 10.1007/s12032-013-0549-0. [DOI] [PubMed] [Google Scholar]
- 201.Wang B, Shen ZL, Jiang KW, et al. MicroRNA-217 functions as a prognosis predictor and inhibits colorectal cancer cell proliferation and invasion via an AEG-1 dependent mechanism. BMC Cancer. 2015;15:437. doi: 10.1186/s12885-015-1438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhang N, Lu C, Chen L. miR-217 regulates tumor growth and apoptosis by targeting the MAPK signaling pathway in colorectal cancer. Oncol Lett. 2016;12(6):4589–4597. doi: 10.3892/ol.2016.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yu H, Gao G, Jiang L, et al. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2013;6(12):2904–2911. [PMC free article] [PubMed] [Google Scholar]
- 204.Li PL, Zhang X, Wang LL, et al. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36(12):1484–1493. doi: 10.1093/carcin/bgv145. [DOI] [PubMed] [Google Scholar]
- 205.Tao K, Yang J, Guo Z, et al. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6(4):391–401. [PMC free article] [PubMed] [Google Scholar]
- 206.Yuan K, Xie K, Fox J, et al. Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology. 2013;145(4):853.e9–864.e9. doi: 10.1053/j.gastro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Cai K, Shen F, Cui JH, Yu Y, Pan HQ. Expression of miR-221 in colon cancer correlates with prognosis. Int J Clin Exp Med. 2015;8(2):2794–2798. [PMC free article] [PubMed] [Google Scholar]
- 208.Li ZW, Yang YM, Du LT, et al. Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):256. doi: 10.1007/s12032-014-0256-5. [DOI] [PubMed] [Google Scholar]
- 209.Liao WT, Li TT, Wang ZG, et al. microRNA-224 promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2. Clin Cancer Res. 2013;19(17):4662–4672. doi: 10.1158/1078-0432.CCR-13-0244. [DOI] [PubMed] [Google Scholar]
- 210.Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. Up-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancer. Cancer Cell Int. 2013;13(1):104. doi: 10.1186/1475-2867-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Adamopoulos PG, Kontos CK, Rapti SM, Papadopoulos IN, Scorilas A. miR-224 overexpression is a strong and independent prognosticator of short-term relapse and poor overall survival in colorectal adenocarcinoma. Int J Oncol. 2015;46(2):849–859. doi: 10.3892/ijo.2014.2775. [DOI] [PubMed] [Google Scholar]
- 212.Ling H, Pickard K, Ivan C, et al. The clinical and biological significance of miR-224 expression in colorectal cancer metastasis. Gut. 2016;65(6):977–989. doi: 10.1136/gutjnl-2015-309372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.He Z, Yu L, Luo S, et al. miR-296 inhibits the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100A4. BMC Cancer. 2017;17(1):140. doi: 10.1186/s12885-017-3121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Perez-Carbonell L, Sinicrope FA, Alberts SR, et al. MiR-320e is a novel prognostic biomarker in colorectal cancer. Br J Cancer. 2015;113(1):83–90. doi: 10.1038/bjc.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68(15):6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 216.Wu L, Hui H, Wang LJ, Wang H, Liu QF, Han SX. MicroRNA-326 functions as a tumor suppressor in colorectal cancer by targeting the Nin one binding protein. Oncol Rep. 2015;33(5):2309–2318. doi: 10.3892/or.2015.3840. [DOI] [PubMed] [Google Scholar]
- 217.Sun Z, Zhang Z, Liu Z, Qiu B, Liu K, Dong G. MicroRNA-335 inhibits invasion and metastasis of colorectal cancer by targeting ZEB2. Med Oncol. 2014;31(6):982. doi: 10.1007/s12032-014-0982-8. [DOI] [PubMed] [Google Scholar]
- 218.Sun K, Su G, Deng H, Dong J, Lei S, Li G. Relationship between miRNA-338-3p expression and progression and prognosis of human colorectal carcinoma. Chin Med J (Engl) 2014;127(10):1884–1890. [PubMed] [Google Scholar]
- 219.Takeyama H, Yamamoto H, Yamashita S, et al. Decreased miR-340 expression in bone marrow is associated with liver metastasis of colorectal cancer. Mol Cancer Ther. 2014;13(4):976–985. doi: 10.1158/1535-7163.MCT-13-0571. [DOI] [PubMed] [Google Scholar]
- 220.Ma F, Song H, Guo B, et al. MiR-361-5p inhibits colorectal and gastric cancer growth and metastasis by targeting staphylococcal nuclease domain containing-1. Oncotarget. 2015;6(19):17404–17416. doi: 10.18632/oncotarget.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nie J, Liu L, Zheng W, et al. MicroRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting cyclin D1 and Bcl-2. Carcinogenesis. 2012;33(1):220–225. doi: 10.1093/carcin/bgr245. [DOI] [PubMed] [Google Scholar]
- 222.Yamashita S, Yamamoto H, Mimori K, et al. MicroRNA-372 is associated with poor prognosis in colorectal cancer. Oncology. 2012;82(4):205–212. doi: 10.1159/000336809. [DOI] [PubMed] [Google Scholar]
- 223.Mo ZH, Wu XD, Li S, Fei BY, Zhang B. Expression and clinical significance of microRNA-376a in colorectal cancer. Asian Pac J Cancer Prev. 2014;15(21):9523–9527. doi: 10.7314/apjcp.2014.15.21.9523. [DOI] [PubMed] [Google Scholar]
- 224.Li H, Dai S, Zhen T, et al. Clinical and biological significance of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur J Cancer. 2014;50(6):1207–1221. doi: 10.1016/j.ejca.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 225.Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer. 2014;14:109. doi: 10.1186/1471-2407-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zheng GX, Qu AL, Yang YM, Zhang X, Zhang SC, Wang CX. miR-422a is an independent prognostic factor and functions as a potential tumor suppressor in colorectal cancer. World J Gastroenterol. 2016;22(24):5589–5597. doi: 10.3748/wjg.v22.i24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Torres S, Garcia-Palmero I, Bartolomé RA, et al. Combined miRNA profiling and proteomics demonstrates that different miRNAs target a common set of proteins to promote colorectal cancer metastasis. J Pathol. 2017;242(1):39–51. doi: 10.1002/path.4874. [DOI] [PubMed] [Google Scholar]
- 228.Li J, Du L, Yang Y, et al. MiR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2. Cancer Lett. 2013;329(1):84–90. doi: 10.1016/j.canlet.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 229.Sun Y, Shen S, Tang H, et al. miR-429 identified by dynamic transcriptome analysis is a new candidate biomarker for colorectal cancer prognosis. OMICS. 2014;18(1):54–64. doi: 10.1089/omi.2012.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Dong SJ, Cai XJ, Li SJ. The clinical significance of miR-429 as a predictive biomarker in colorectal cancer patients receiving 5-fluorouracil treatment. Med Sci Monit. 2016;22:3352–3361. doi: 10.12659/MSM.900674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Han Y, Zhao Q, Zhou J, Shi R. miR-429 mediates tumor growth and metastasis in colorectal cancer. Am J Cancer Res. 2017;7(2):218–233. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 232.Ye YP, Wu P, Gu CC, et al. miR-450b-5p induced by oncogenic KRAS is required for colorectal cancer progression. Oncotarget. 2016;7(38):61312–61324. doi: 10.18632/oncotarget.11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hata T, Mokutani Y, Takahashi H, et al. Identification of microRNA-487b as a negative regulator of liver metastasis by regulation of KRAS in colorectal cancer. Int J Oncol. 2017;50(2):487–496. doi: 10.3892/ijo.2016.3813. [DOI] [PubMed] [Google Scholar]
- 234.Xu X, Chen R, Li Z, et al. MicroRNA-490-3p inhibits colorectal cancer metastasis by targeting TGFβR1. BMC Cancer. 2015;15:1023. doi: 10.1186/s12885-015-2032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sun HB, Chen X, Ji H, et al. miR-494 is an independent prognostic factor and promotes cell migration and invasion in colorectal cancer by directly targeting PTEN. Int J Oncol. 2014;45(6):2486–2494. doi: 10.3892/ijo.2014.2665. [DOI] [PubMed] [Google Scholar]
- 236.Li L, Sarver AL, Khatri R, et al. Sequential expression of miR-182 and miR-503 cooperatively targets FBXW7, contributing to the malignant transformation of colon adenoma to adenocarcinoma. J Pathol. 2014;234(4):488–501. doi: 10.1002/path.4407. [DOI] [PubMed] [Google Scholar]
- 237.Noguchi T, Toiyama Y, Kitajima T, et al. miRNA-503 promotes tumor progression and is associated with early recurrence and poor prognosis in human colorectal cancer. Oncology. 2016;90(4):221–231. doi: 10.1159/000444493. [DOI] [PubMed] [Google Scholar]
- 238.Zhang Y, Lin C, Liao G, et al. MicroRNA-506 suppresses tumor proliferation and metastasis in colon cancer by directly targeting the oncogene EZH2. Oncotarget. 2015;6(32):32586–32601. doi: 10.18632/oncotarget.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhou H, Lin C, Zhang Y, et al. miR-506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing MDR1/P-gp expression. Cell Prolif. 2017;50(3):e12341. doi: 10.1111/cpr.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ma W, Yu Q, Jiang J, et al. miR-517a is an independent prognostic marker and contributes to cell migration and invasion in human colorectal cancer. Oncol Lett. 2016;11(4):2583–2589. doi: 10.3892/ol.2016.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ye C, Yue G, Shen Z, et al. miR-542-3p suppresses colorectal cancer progression through targeting survivin. Transl Cancer Res. 2016;5(6):817–826. [Google Scholar]
- 242.Yuan L, Yuan P, Yuan H, et al. miR-542-3p inhibits colorectal cancer cell proliferation, migration and invasion by targeting OTUB1. Am J Cancer Res. 2017;7(1):159–172. [PMC free article] [PubMed] [Google Scholar]
- 243.Zhou Q, Zhu Y, Wei X, et al. MiR-590-5p inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor A axis. Cell Death Dis. 2016;7(10):e2413. doi: 10.1038/cddis.2016.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ou C, Sun Z, Li X, et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53–63. doi: 10.1016/j.canlet.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 245.Liu M, Zhi Q, Wang W, Zhang Q, Fang T, Ma Q. Up-regulation of miR-592 correlates with tumor progression and poor prognosis in patients with colorectal cancer. Biomed Pharmacother. 2015;69:214–220. doi: 10.1016/j.biopha.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 246.Shi F, Li R, Guo P. Expression and prognostic value of miR-610 in patients with colorectal cancer. Biomed Res. 2017;28(3):1321–1324. [Google Scholar]
- 247.Rasmussen MH, Jensen NF, Tarpgaard LS, et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol Oncol. 2013;7(3):637–646. doi: 10.1016/j.molonc.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lou X, Qi X, Zhang Y, Long H, Yang J. Decreased expression of microRNA-625 is associated with tumor metastasis and poor prognosis in patients. J Surg Oncol. 2013;108(4):230–235. doi: 10.1002/jso.23380. [DOI] [PubMed] [Google Scholar]
- 249.Chu D, Zheng J, Li J, et al. MicroRNA-630 is a prognostic marker for patients with colorectal cancer. Tumour Biol. 2014;35(10):9787–9792. doi: 10.1007/s13277-014-2223-3. [DOI] [PubMed] [Google Scholar]
- 250.Zhang J, Fei B, Wang Q, et al. MicroRNA-638 inhibits cell proliferation, invasion and regulates cell cycle by targeting tetraspanin 1 in human colorectal carcinoma. Oncotarget. 2014;5(23):12083–12096. doi: 10.18632/oncotarget.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang X, Kuang Y, Shen X, et al. Evaluation of miR-720 prognostic significance in patients with colorectal cancer. Tumour Biol. 2015;36(2):719–727. doi: 10.1007/s13277-014-2697-z. [DOI] [PubMed] [Google Scholar]
- 252.Zhang T, Cai X, Li Q, et al. Hsa-miR-875-5p exerts tumor suppressor function through down-regulation of EGFR in colorectal carcinoma (CRC) Oncotarget. 2016;7(27):42225–42240. doi: 10.18632/oncotarget.9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Liu DR, Guan QL, Gao MT, Jiang L, Kang HX. miR-1260b is a potential prognostic biomarker in colorectal cancer. Med Sci Monit. 2016;22:2417–2423. doi: 10.12659/MSM.898733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gopalan V, Pillai S, Ebrahimi F, et al. Regulation of microRNA-1288 in colorectal cancer: altered expression and its clinicopathological. Mol Carcinog. 2014;53(Suppl 1):E36–E44. doi: 10.1002/mc.21993. [DOI] [PubMed] [Google Scholar]
- 255.Ju HQ, Lu YX, Chen DL, et al. Redox regulation of stem-like cells through the CD44v-xCT Axis in colorectal cancer: mechanisms and therapeutic implications. Theranostics. 2016;6(8):1160–1175. doi: 10.7150/thno.14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hu Y, Yi B, He S, et al. Clinical significance of miR-1826 as a novel prognostic biomarker in colorectal cancer. Anticancer Agents Med Chem. 2016;16(9):1109–1116. doi: 10.2174/1871520615666150507122434. [DOI] [PubMed] [Google Scholar]
- 257.Yu FY, Tu Y, Deng Y, et al. MiR-4500 is epigenetically downregulated in colorectal cancer and functions as a novel tumor suppressor by regulating HMGA2. Cancer Biol Ther. 2016;17(11):1149–1157. doi: 10.1080/15384047.2016.1235661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zhao S, Sun H, Jiang W, et al. miR-4775 promotes colorectal cancer invasion and metastasis via the Smad7/TGFβ-mediated epithelial to mesenchymal transition. Mol Cancer. 2017;16(1):12. doi: 10.1186/s12943-017-0585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 260.Valeri N, Croce CM, Fabbri M. Pathogenetic and clinical relevance of microRNAs in colorectal cancer. Cancer Genomics Proteomics. 2009;6(4):195–204. [PubMed] [Google Scholar]
- 261.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105(39):14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 263.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 264.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 265.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lin JK, Chang SC, Yang YC, Li AF. Loss of heterozygosity and DNA aneuploidy in colorectal adenocarcinoma. Ann Surg Oncol. 2003;10(9):1086–1094. doi: 10.1245/aso.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 267.Leary RJ, Lin JC, Cummins J, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105(42):16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46(15):2788–2798. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 269.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ogino S, Nishihara R, van der Weele TJ, et al. The role of molecular pathological epidemiology in the study of neoplastic and non-neoplastic diseases in the era of precision medicine. Epidemiology. 2016;27(4):602–611. doi: 10.1097/EDE.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]