Abstract
IFIT1 (also known as ISG56) is a member of the interferon-inducible protein with tetratricopeptide repeats (IFITs) family. IFITs are strongly induced by type I interferon (IFN), double-stranded RNA and virus infection. Here, we investigated IFIT1 expression in human umbilical vein endothelial cells (HUVECs) and in human bronchus epithelial cells (BEAS-2Bs) induced by the H9N2 virus and inactivated viral particle at different time points. We also investigated the effect of H9N2 virus and viral particle infection on IFN-α/β production, and assessed whether hemagglutinin or neuraminidase protein induced IFIT1 expression. Results showed that both H9N2 virus infection and viral particle inoculation induced the expression of IFIT1 at mRNA and protein levels in the two cell lines. Hemagglutinin or neuraminidase protein binding alone is not sufficient to induce IFIT1 expression. Surprisingly, the expression patterns of IFIT1 in response to H9N2 virus and viral particles in the two cell lines were opposite, and production kinetics of IFN-α/β also differed. An additional finding was that induction of IFIT1 in response to H9N2 virus infection or viral particle inoculation was more sensitive in HUVECs than in BEAS-2Bs. Our data offers new insight into the innate immune response of endothelial cells to H9N2 virus infection.
Keywords: endothelial cells, H9N2 virus, IFIT1 expression pattern, IFN-α/β
INTRODUCTION
Pathogen infection can be recognized by pattern recognition receptors, which trigger activation of downstream transcript factors that control production of cytokines such as type I interferon (type I IFN). IFN activates Janus tyrosine kinases signal pathway (JAK-STAT) resulting in initiation of the expression of more than 300 interferon-stimulated genes (ISGs) (Metz et al., 2012). The products of these ISGs are critical proteins that regulate immune function and dominate antiviral response (Randall and Goodbourn, 2008; von Recum-Knepper et al., 2015). Interferon-inducible protein with tetratricopeptide repeats (IFITs) is one product of ISGs that plays an important role in resistance to virus invasion through a variety of mechanisms (Diamond and Farzan, 2013). To date, IFITs have been identified in various species including mammals, birds, reptiles, amphibians and bony fish. The number and composition of the genes varies greatly from species to species. In humans, there are four IFIT genes comprising IFIT1 (ISG56), IFIT2 (ISG54), IFIT3 (ISG60 or IFIT4) and IFIT5 (ISG58) (Fensterl and Sen, 2011). Most types of mammalian cells, except some myeloid cell subtypes, do not express IFIT proteins under basal conditions (Daffis et al., 2007). Viral infection can rapidly induce IFIT gene transcription to high levels in many cell types (Sarkar and Sen, 2004).
The avian H9N2 influenza virus occurred predominantly in Asia, while the Middle East, Africa and other non-Asian countries experienced an outbreak of lesser magnitude subsequently (Davidson et al., 2013; Parker et al., 2012; Tombari et al., 2013). Recent reports showed that the H9N2 virus was spread from birds to mammals (Yu et al., 2011; Zhang et al., 2013), with China experiencing reports of human infection (Cheng et al., 2011). Importantly, the H9N2 virus shares internal genes with other avian influenza virus (AIV) types to form new H7N9, H10N8 and H5N6 viruses that could infect and kill humans (Bi et al., 2016; Chen et al., 2014; Liu et al., 2013). Therefore, H9N2 virus is a potential threat to human health that should be investigated to identify the innate immune response of human cells to the infection.
Generally, influenza virus prefers to bind to epithelial cells because of the expression of α-2, 6- or α-2, 3-linked sialic acid receptors on these cells in the respiratory tract (Ibricevic et al., 2006; Nicholls et al., 2007). However, increasing evidence indicates that endothelial cells might play an important role in response to influenza virus infection. To date, two types of receptors have been detected in most types of endothelial cells that may account for the multiple organ involvement in influenza infection (Teijaro et al., 2011; Yao et al., 2008). Studies have shown that endothelial cells support influenza virus replication (Viemann et al., 2011; Zeng et al., 2012). H5N1 and H7N9 influenza viruses have been proven to directly infect human lung microvascular endothelial cells and replicate in endothelial cell lines (Zeng et al., 2012; 2015). Recent studies demonstrated that endothelial cells not only participate in regulating cytokine amplification (Teijaro et al., 2011), but also help initiate innate immune response by activating type I IFN response during influenza virus infection (Zeng et al., 2012). Earlier studies showed that IFIT genes could be induced by infection with viruses (Broz and Monack, 2013; Jakobsen et al., 2013; Lam et al., 2014; Siednienko et al., 2011), including influenza. H1N1 virus infection increases expression of IFIT1 in epithelial cells at both the mRNA and protein level (Kim et al., 2015). Our preliminary microarray analysis results in human umbilical vein endothelial cells showed that H9N2 virus infection induced a 34-fold increase at the transcriptional level compared with the control group. Our findings also showed that viral particle inoculation (inactivated by β-propionolactone) induced a higher level of IFIT1 than was produced by H9N2 virus infection (Wang et al., 2015). However, the levels of IFIT1 still need to be quantified by Real-Time PCR (RT-PCR) and western blot. Further, little is known about the expression patterns of IFIT1 induced by H9N2 virus or viral particles in endothelial and epithelial cells. The present study aimed to investigate IFIT1 expression induced by H9N2 virus and viral particles in human endothelial and epithelial cells, and to ascertain whether IFN-α/β production was induced during IFIT1 expression.
MATERIALS AND METHODS
Cell cultures
Human umbilical vein endothelial cells (HUVECs, CRL-1730, ATCC), human bronchus epithelial cells (BEAS-2Bs, CRL-9609, ATCC) and Madin-Darby canine kidney (MDCK, CCL-34, ATCC) are commonly used cell lines for evaluation of interactions between viruses and mammalian cells. In this study, HUVECs and BEAS-2Bs were used for detecting IFIT1 expression and IFN-α/β secretion. MDCK cells were used for the plaque assay. HUVECs, BEAS-2Bs and MDCK were cultured in DMEM medium (Gibco, USA) supplemented with FBS 10% (Gibco, USA), penicillin G 100 U/ml (Gibco, USA) and streptomycin 100 g/mL (Gibco, USA) in a CO2 5% incubator at 37°C. The cells 5–8 passages were seeded into six-well plates and cultured for 24 h before each experiment.
Virus and inactivated viral particle
The H9N2 virus used in this study was A/Chicken/Hebei/4/2008 (Ck/HB/4/08). The complete genome sequence of the virus is available from GenBank under the accession numbers FJ499463–FJ499470. H9N2 virus was propagated in 9-day-old embryonated eggs from specific-pathogen-free (SPF) hens at 37°C for 60 to 72 h. Virus titers were determined by plaque assay. To evaluate H9N2 virus infection, cells were washed with phosphate buffered saline (PBS), inoculated with virus and incubated for 1 h. Cells were then washed with PBS and added to DMEM supplemented with TPCK-treated trypsin 0.25 μg/ml. To investigate the expression of IFIT1 induced by viral particles, we created inactivated H9N2 viral particles with β-propionolactone 0.094% (BPL; SERVA Electrophoresis, Germany) according to a previously described protocol (Jonges et al., 2010).
Plaque assay
Plaque assay performed on MDCK cells was used to detect virus titers as described previously (Tobita et al., 1975). Briefly, MDCK cells were seeded into 6-well plates. Confluent monolayers were washed with PBS and infected with H9N2 virus or cell culture supernatants. The inoculum was discarded after incubation for 1 h and the remaining cells were washed with PBS. An overlay consisting of a mixture of aga-rose 1.6% (Lonza, USA) and double-strength DMEM with TPCK-trypsin 0.25 μg/ml (Worthington, USA) was added to the above cells and incubated at 37°C for 72 h. Plaques were stained with crystal violet 0.1% and counted.
RT-PCR analysis
RT-PCR analysis was used to investigate the expression of IFIT1 at the mRNA level. Cells were divided into three groups: control, H9N2 virus and viral particle. According to earlier work (Wang et al., 2015), we chose a multiplicity of infection (MOI) of 5 to infect or inoculate cells in this experiment. Cells in the H9N2 virus group were infected with H9N2 viruses at a MOI of 5, cells in viral particle group were incubated with viral particles at a MOI of 5 and cells in control group were inoculated with free media. To detect the effect of hemagglutinin (HA) or neuraminidase (NA) (Sino Biological, China) on IFIT1 mRNA expression, HUVECs or BEAS-2Bs were divided into control, low concentration and high concentration groups. Cells in the control group were inoculated with free media, while cells in the low and high concentration groups were treated with 0.1 μg/ml and 1 μg/ml HA or NA, respectively (Dirix et al., 2012; Shi et al., 2014). Total RNA in each group was isolated using TRIzol reagent (Invitrogen, USA) at 6, 12 and 24 h after treatment. cDNA was synthesized using random hexamer and a superscript III reverse transcriptase kit (Invitrogen, USA). RT-PCR was performed on the cDNA for RNA quantification using Ex TaqMix (Takara Bio, China) and Eva green (Biotium, USA) for IFIT1. GAPDH was amplified in parallel with the target genes as an endogenous control. All samples were analyzed in triplicate. The fold changes of specific mRNA from the different groups was determined via comparison to the control group. The primer sequences used were as follows: GAPDH, F (5′ to 3′): ACAACTTTGGTATCGT GGAAGGAC and R (5′ to 3′): AGGGATGATGTTCTGGAGA GCC, IFIT1, F (5′ to 3′): TGA CTCTTTTGCCTCTTTCTTCTAA and R (5′ to 3′): TTCTTGGGGT GCTCTGTGG.
Western blot
Expression of IFIT1 at the protein level was detected by western blotting. As described above, cells were divided into three groups and treated following the same procedure used in the RT-PCR analysis. Cells in each group were lysed with RIPA lysis buffer (Cell Signaling Technology, USA) at 12, 24 and 36 h, and the quantity of total proteins measured by BCA kit. The cell lysates were centrifuged and supernatants separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) before transfer onto polyvinylidene difluoride (PVDF) membranes (Roche, China). The membranes were blocked with 5% skim milk and probed with a monoclonal antibody to β-actin (Santa Cruz Biotechnology, USA), IFIT1 (ORIGENE, USA), after a further incubation with peroxidase conjugated secondary antibody (ORIGENE, USA). Proteins were visualized using enhanced chemiluminescence reagent (Cell Signaling Technology, USA). The relative protein levels of IFIT1 to β-actin were performed by Image J software.
ELISA assay
To evaluate the levels of IFN-α/β induced by H9N2 virus or viral particles, cells were divided into four groups: control, H9N2 virus, viral particle and positive control. Cells in the H9N2 virus group were infected with H9N2 viruses at a MOI of 5, cells in the viral particle group were incubated with viral particles at a MOI of 5, cells in the control group were inoculated with free media and the positive control group was treated with poly-I:C 10 μg/mL (Sigma Aldrich, China) (Herbert et al., 2014; Siednienko et al., 2011). Supernatant in each group was collected at 6, 12 and 24 h post-infection and detected using human IFN-α/β ELISA kits (R&D system Inc., USA).
Immunofluorescence
To locate H9N2 viral particles in HUVECs and BEAS-2Bs, we detected HA using immunofluorescence. Cells were inoculated with viral particles at a MOI of 5 and incubated for 1 h. They were then washed with PBS and added to DMEM. At 8 h after inoculation, viral particles were labeled with HA-specific antibody. According to previous work (Zeng et al., 2015), cells in each group were fixed directly in paraformaldehyde 4% and permeabilized with Triton X-100 0.5%. Cells were then blocked with PBS containing bovine serum albumin (BSA) 5% for 1 h at 37°C. Subsequently, cells were incubated with HA-specific antibody (Sino Biological Inc., China) overnight at 4°C followed by FITC conjugated secondary antibody (ORIGENE, USA) incubated for 1 h at 37°C. To count the number of stained cells, cell nuclei were labeled with 4, 6-diamidino-2-phenylindole hydrochloride (DAPI, Cell Signaling Technology, USA). Pictures were captured by OLYMPUS fluorescence microscope.
Antiviral response stimulated by viral particle
To investigate the effect of viral particle inoculation on the antiviral response in HUVECs and BEAS-2Bs, cells were divided into control and viral particle groups. Cells in the viral particle group were inoculated with viral particle at a MOI of 5 and incubated for 1 h. Cells in the control group were treated with free media and incubated for 1 h. At 24 h after inoculation with viral particles, cells in each group were infected with H9N2 virus at a MOI of 5. The supernatant from each group was collected at 24 h post-infection and virus titers detected using plaque assay.
IFIT1 CRISPR activation plasmid transfection
For overexpression assay, cells were transfected with control plasmid or IFIT1 CRISPR activation plasmid (Santa Cruz Bio-technology, USA) using Lipofectamine 3000 transfection reagent kit (Invitrogen, USA). The IFIT1 CRISPER activating plasmid is a synergy activation medium (SAM) transcriptional activation system designed to specifically upregulate gene expression. According to instructions of products, we firstly prepared mixtures (2 μg plasmid and 7.5 μl Lipofectamine 3000) of plasmid and Lipofectamine 3000 transfection reagents using Opti-MEM Media. Then cells were covered with the mixtures for 36 h at 37°C in a 5% CO2 incubator. The overexpression level of IFIT1 was detected by Western blot at 36 h after transfection.
Statistical analysis
Results were expressed as means ± standard deviations (SD). The statistical significance of differences was determined using the Student’s t-test or one-way analysis of variance (ANOVA) as appropriate. Comparison between two groups was performed using the unpaired Student’s t-test. All the statistical tests were performed using GraphPad Prism software 6.0 (GraphPad software, USA). A p-value < 0.05 was considered statistically significant.
RESULTS
Infection and replication of H9N2 virus in BEAS-2Bs
Our previous study indicated that the HUVECs used in this study support H9N2 virus infection and replication (Wang et al., 2015). Here, we investigated whether BEAS-2Bs support the replication of H9N2 virus. The virus titers in BEAS-2Bs culture supernatant were measured by plaque assay. First, replication of H9N2 virus with different MOI (0.2, 2, 5 and 10) was observed in BEAS-2Bs grown in 6-well plates. When BEAS-2Bs were infected with H9N2 virus at different MOIs for 24 h, a dose-dependent increment of virus titers was observed (Fig. 1A). Subsequently, the kinetics of H9N2 virus replication was detected at 6, 12, 24, 36, 48 and 60 h post-infection at MOI of 5 and 0.2. Productive replication of H9N2 virus was detected at 6 h post-infection and reached a peak titer of 9.07 × 105 PFU/ml at 24 h at a MOI of 5 (Fig. 1B). At a MOI of 0.2, virus titers were observed at 12 h post-infection and reached a peak of 5.69 × 105 PFU/mL at 36 h (Fig. 1B). Our data showed that the BEAS-2Bs used in the present study supported replication of H9N2 virus.
Fig. 1. Replication of H9N2 virus in BEAS-2Bs.
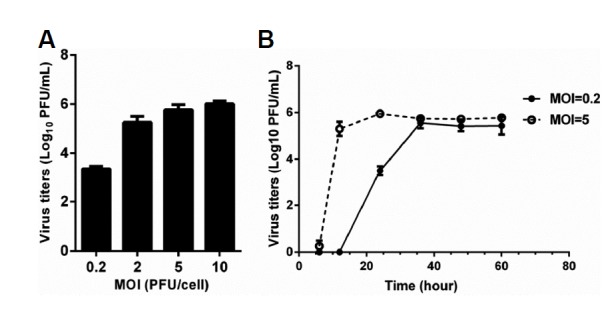
BEAS-2Bs grown on six-well plates were infected with H9N2 virus at MOIs. Culture supernatants were collected at a given time, and viral titers were determined by plaque-forming units. Values represent the means from three independent experiments plus standard deviations. (A) Replication of H9N2 virus at 24 h postinfection with different MOIs. (B) Replication of H9N2 virus at different time points after infection with MOI of 5 and 0.2, respectively.
H9N2 virus infection induced higher IFIT1 expression in BEAS-2Bs
To investigate the effect of H9N2 virus infection and viral particle inoculation on expression of IFIT1 in BEAS-2Bs, cells were analyzed by RT-PCR and western blot at different time points. As shown in Fig. 2, both H9N2 virus infection and viral particle inoculation rapidly induced the expression of IFIT1. Compared to the particle group, H9N2 virus infection induced higher expression of IFIT1 at the mRNA level at 6, 12 and 24 h post-infection (Fig. 2A). Protein levels induced by H9N2 virus infection were also higher than that induced by viral particle inoculation (Figs. 2B and 2C). The results suggested that expression of IFITl in BEAS-2Bs is more effectively induced by H9N2 virus infection than by viral particle inoculation.
Fig. 2. IFIT1 expression induced by H9N2 virus infection and viral particle inoculation in BEAS-2Bs.
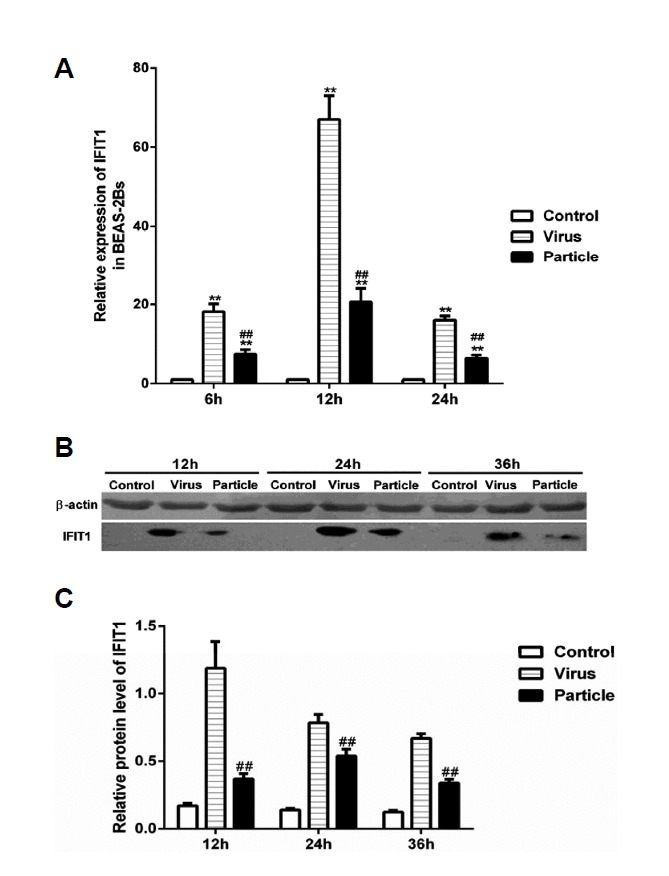
BEAS-2Bs were infected with H9N2 virus (i.e., Virus) and inoculated with inactivated viral particle (i.e., Particle) at a MOI of 5, cells used for RT-PCR assay were collected at 6 h, 12 h and 24 h postinfection, cells used for western blot assay were collected at 12 h, 24 h and 36 h postinfection. Values represent the means from three independent experiments plus standard deviations. (A) Expression of IFIT1 at mRNA level. (B) Expression of IFIT1 at protein level. (C) The relative protein level of IFIT1 compared to β-actin. *means particle group and virus group compared with control group (*P < 0.05, **P < 0.01, ANOVE). # means particle group compared with virus group (#P < 0.05, ##P < 0.01, t-test).
Viral particle inoculation induced higher IFIT1 expression in HUVECs
To investigate the distinct expression pattern of IFIT1 between epithelial and endothelial cells, we determined the expression of IFIT1 at mRNA and protein levels in HUVECs. Consistent with BEAS-2Bs, H9N2 virus infection and viral particle inoculation induced IFIT1 expression in HUVECs at both mRNA and protein levels (Fig. 3). However, the expression of IFIT1 induced by H9N2 virus infection was lower than with viral particle inoculation. At 12 h post-infection, H9N2 virus infection induced a 450-fold increase in IFIT1 at the mRNA level, and viral particle inoculation induced up to a 700-fold increase compared to the control group (Fig. 3A). The IFIT1 protein level induced by viral particles was also higher than that induced by H9N2 virus at 24 h (Figs. 3B and 3C). The results suggest that viral particles are more effective than H9N2 virus at inducing IFITl expression in HUVECs. Interestingly, H9N2 virus and viral particle induction of IFITl expression at the mRNA level was much higher in endothelial cells (450- and 700-fold, respectively) than epithelial cells (70- and 20-fold, respectively).
Fig. 3. IFIT1 expression induced by H9N2 virus infection and viral particle inoculation in HUVECs.
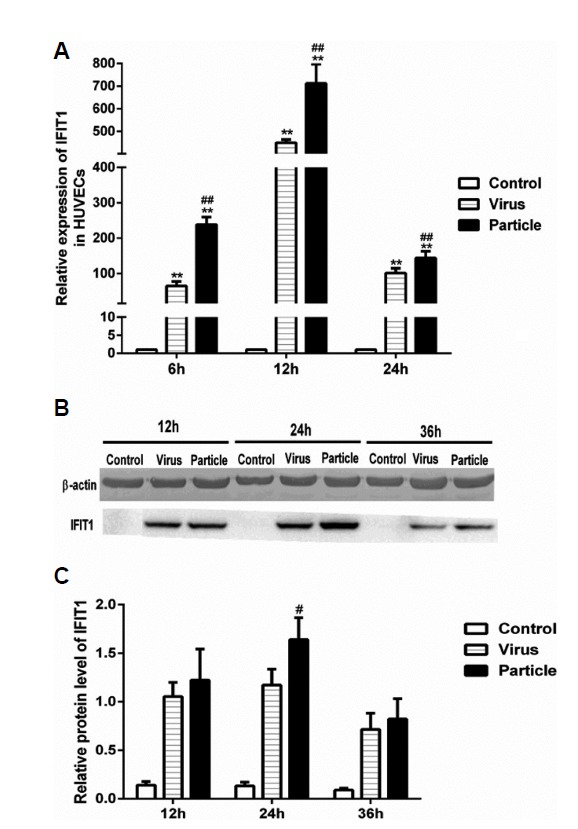
HUVECs were infected with H9N2 virus (i.e., Virus) and inoculated with inactivated viral particle (i.e., Particle) at a MOI of 5, cells used for RT-PCR assay were collected at 6 h, 12 h and 24 h postinfection, cells used for western blot assay were collected at 12, 24 and 36 h postinfection. Values represent the means from three independent experiments plus standard deviations. (A) Expression of IFIT1 at mRNA level. (B) Expression of IFIT1 at protein level. (C) The relative protein level of IFIT1 compared to β-actin. *means particle group and virus group compared with control group (*P < 0.05, **P < 0.01, ANOVE). # means particle group compared with virus group (#P < 0.05, ##P < 0.01, t-test).
IFN-α/β levels in HUVECs and BEAS-2Bs
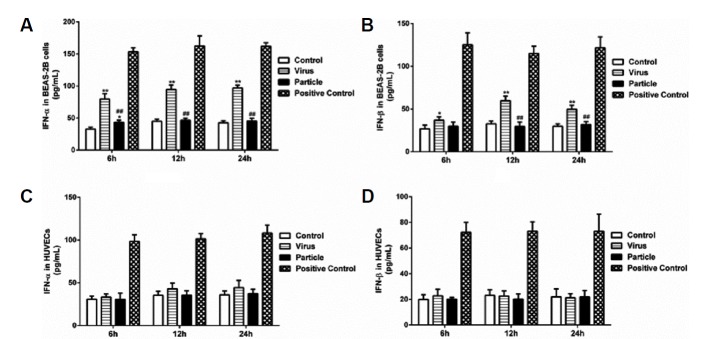
As described earlier, type I IFN can induce IFIT1 expression, and influenza virus infection can stimulate production of type I IFN. Therefore, we investigated whether IFN-α/β was induced during H9N2 virus and viral particle induced IFIT1 expression. We treated BEAS-2Bs and HUVECs with H9N2 virus or viral particles at a MOI of 5, and the supernatants were collected at 6, 12 and 24 h. The results showed that H9N2 virus infection significantly increased levels of IFN-α/β in BEAS-2Bs at 6, 12 and 24 h, whereas viral particle inoculation only increased IFN-α/β at 6 h (Figs. 4A and 4B). In contrast, secretion of IFN-α/β in HUVECs was not significantly changed after infection with H9N2 virus or viral particles (Figs. 4C and 4D). The results suggested that IFN-α/β might be involved in H9N2 virus and viral particle induced expression of IFIT1 in BEAS-2Bs. However, this did not appear to be the case in HUVECs.
Fig. 4. Evaluation of IFN-α/β production in HUVECs and BEAS-2Bs inoculated with H9N2 virus or viral particle.
HUVECs and BEAS-2Bs were infected with H9N2 virus (i.e., Virus) and inoculated with inactivated viral particle (i.e., Particle) at a MOI of 5, and then supernatants were collected at 6 h, 12 h and 24 h postinfection. Levels of IFN-α/β were detected using human ELISA kits. Values represent the means from three independent experiments plus standard deviations. (A) IFN-α levels in BEAS-2Bs. (B) IFN-β levels in BEAS-2B cells. (C) IFN-α levels in HUVECs. (D) IFN-β levels in HUVECs. *means particle group and virus group compared with control group (*P < 0.05, **P < 0.01, ANOVE). # means particle group compared with virus group (#P < 0.05, ##P < 0.01, t-test).
HA and NA proteins had no effect on the expression of IFIT1
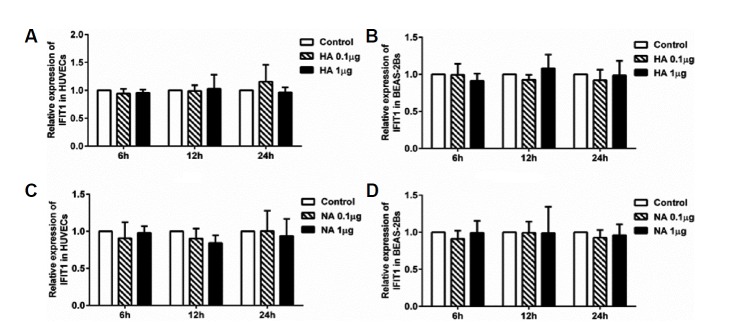
Inactivated viral particle inoculation significantly increased the expression of IFIT1 independently of IFN-α/β in HUVECs and in BEAS-2Bs. Thus, we investigated whether virus envelope proteins were involved in IFIT1 expression. We incubated HUVECs and BEAS-2Bs with HA or NA protein at different concentrations. Cells in each group were extracted for RT-PCR and Western blot analysis. The results showed that IFIT1 mRNA levels were not significantly increased at 6, 12 and 24 h (p > 0.05, ANOVA) in either cell type (Fig. 5). This suggested that HA or NA protein binding alone is not sufficient to induce IFIT1 expression in HUVECs or BEAS-2Bs.
Fig. 5. IFIT1 mRNA levels induced by HA or NA protein in HUVECs and BEAS-2Bs.
HUVECs and BEAS-2Bs were incubated with HA or NA at concentrations of 0.1 or 1 μg/ml. Cells used for RT-PCR analysis were collected at 6 h, 12 h and 24 h postinfection. (A) IFIT1 mRNA levels induced by HA in HUVECs. (B) IFIT1 mRNA levels induced by HA in BEAS-2Bs. (C) IFIT1 mRNA levels induced by NA in HUVECs. (d) IFIT1 mRNA levels induced by NA in BEAS-2Bs.
Location of viral particles in HUVECs and BEAS-2Bs
Viral particle inoculation induced IFIT1 expression in HUVECs via a mechanism that was independent of IFN. Envelope protein binding could not induce IFIT1 expression. We speculated that the effect of viral particles on IFIT1 expression was generated after cellular entry. Therefore, we located viral particles within cells using immunofluorescence. The results showed that HA-positive cells were observed at 8 h after H9N2 infection in both HUVECs and BEAS-2Bs (Fig. 6). The results indicated that cellular interaction between HU-VECs and viral particles might be involved in the induction of IFIT1 expression.
Fig. 6. Localization of viral particles in HUVECs and BEAS-2Bs.
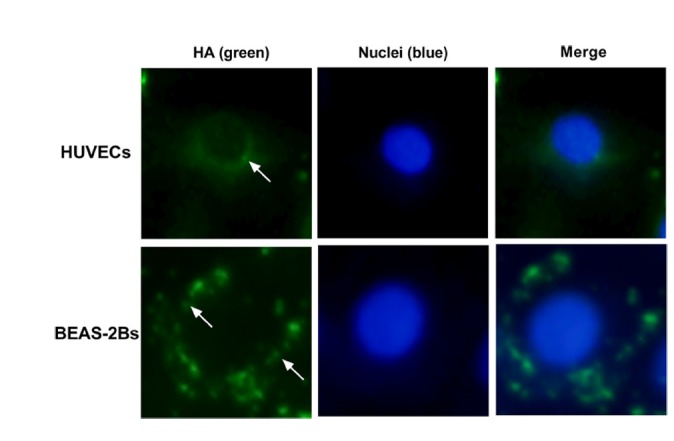
BEAS-2Bs and HUVECs were inoculated with viral particles at a MOI of 5 for 24 h, then cells were double-stained with an anti-HA antibody (green) and 4′, 6-diamidino-2-phenylindole (DAPI, blue).
Viral particle inoculation decreased the virus titers in HUVECs and BEAS-2Bs
H9N2 viral particle inoculation induces IFIT1 expression at mRNA and protein levels in HUVECs and in BEAS-2Bs. We investigated the effect of viral particle inoculation on antiviral response in these two cell lines. As shown in Fig. 7, virus titers in the particle group were significantly reduced in both HUVECs and BEAS-2Bs. Compared to the control group, virus titers in the particle group decreased by 40.49 ± 4.90% in HUVECs and 55.02 ± 3.88% in BEAS-2Bs.
Fig. 7. Antiviral response induced by viral particle inoculation in BEAS-2B2 and HUVECs.
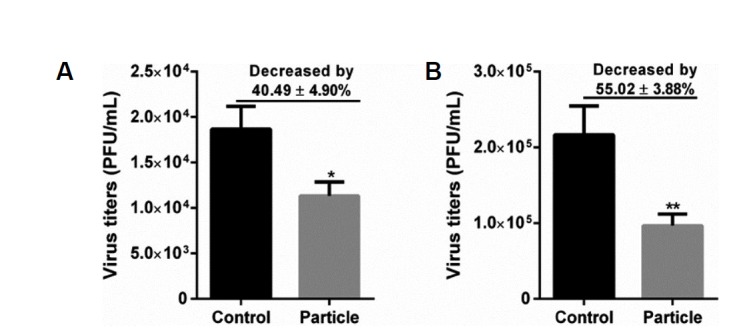
HUVECs and BEAS-2Bs cells were pretreated inactivated viral particle (i.e., Particle) at a MOI of 5 for 24 h, and then HUVECs and BEAS-2Bs were infected with H9N2 virus at a MOI of 5. At 24 h postinfection, supernatant in each group was collected, and viral titers were determined by plaque-forming units. Values represent the means from three independent experiments plus standard deviations. (A) Virus titers in HUVECs treated with viral particles. (B) Virus titers in BEAS-2Bs treated with viral particles. *means viral particle group compared with control group (*P < 0.05, t-test).
Our study showed that both H9N2 virus and inactivated viral particles could induce the expression of IFIT1 in BEAS-2Bs and HUVECs, and that IFIT1 expression patterns are different between these two cell types. In addition, IFN-α/β is not involved in H9N2 virus and viral particle induced expression of IFIT1 in HUVECs. These data provide further insight into the innate immune response of endothelial cells to H9N2 influenza virus.
Overexpression of IFIT1 could not significantly enhance the antiviral state
HUVECs or BEAS-2Bs were transfected with plasmid for 36 h before infected with H9N2 virus. The results showed that transfection with IFIT1 CRISPR activation plasmid upregulated the expression of IFIT1 in HUVECs and BEAS-2Bs (Fig. 8A). Compared to control group, the virus titers were significantly decreased by 9.72 ± 2.53% (P > 0.05, t-test) in BEAS-2Bs transfected with IFIT1 CRISPR activation plasmid, the virus titers were significantly decreased by 7.76 ± 1.14% (P > 0.05, t-test) in HUVECs transfected with IFIT1 CRISPR activation plasmid (Fig. 8B).
Fig. 8. Overexpression of IFIT1 could not significantly reduced virus titers in HUVECs and BEAS-2Bs.
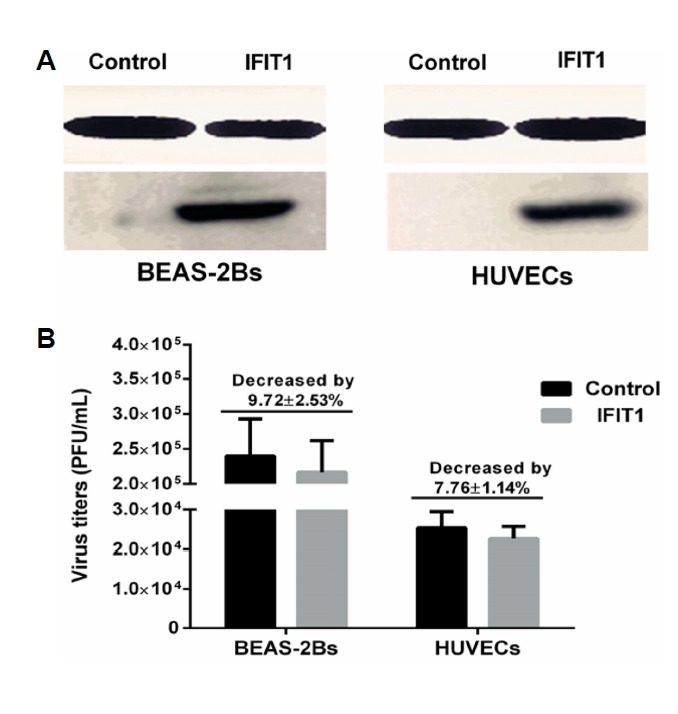
HUVECs and BEAS-2Bs were transfected with control plasmid (Control) or IFIT1 CRISPR activation plasmid (IFIT1) for 36 h, then cells were infected with H9N2 virus at MOI of 5. Virus titer of each group was detected by plaque assay at 36 h postinfection. The overexpression of IFIT1 was detected by Western blot. (A) IFIT1 protein levels after transfected with plasmid in BEAS-2Bs and HUVECs. (B) Virus titers in BEAS-2Bs and HUVECs transfected with control plasmid or IFIT1 CRISPR activation plasmid.
DISCUSSION
Proteins encoded by ISGs are major components of host antiviral effectors, and include the IFIT family. In humans, this family comprises four members with IFIT1 the first of these to be discovered (Chebath et al., 1983). IFIT1 has broad spectrum antiviral activity and has been shown to inhibit replication of a variety of viruses in mammalian cells (Raychoudhuri et al., 2011; Reynaud et al., 2015). We investigated the effect of inactivated viral particle inoculation on antiviral response in HUVECs. Results showed that virus titers were significantly decreased after treatment by viral particles at a MOI of 5 (Fig. 7). Although results showed that overexpressed IFIT1 could not decrease the H9N2 virus titers in HUVECs and BEAS-2Bs (Fig. 8). The results suggest that inactivated viral particle inoculation may confer protection against H9N2 virus infection in endothelial cells. In addition, influenza virus prefers to target epithelial cells located in the respiratory tract in vivo (van Riel et al., 2006; 2007; 2010). The basal lamina, with an average thickness of 1 μm, is the only structure that separates epithelial and endothelial cells in the alveolar wall (Weibel and Knight, 1964). A previous study demonstrated that cytokines produced in alveolar epithelial cells could further activate neighboring endothelial cells during influenza virus infection (Chan et al., 2009). So it is conceivable that interferon released by infected epithelial cells could readily spread to endothelial cells. Thus, regardless of whether the influenza virus could directly infect endothelial cells in vivo, endothelial cells may be involved in the innate immune response of the host during influenza virus infection.
A recent study showed that influenza virus infection could induce IFIT1 expression in respiratory tract epithelial cells (Kim et al., 2015). Our results also showed that H9N2 virus infection increased IFIT1 expression at mRNA and protein levels in BEAS-2Bs (Fig. 2). However, the expression pattern of IFIT1 induced by H9N2 virus in endothelial cells has not been described. The results showed that H9N2 virus infection induced the expression of IFIT1 in HUVECs (Fig. 3). Furthermore, we found that inactivated viral particle inoculation also significantly increased IFIT1expression in both HUVECs and BEAS-2B, and H9N2 viral particles were more potent than H9N2 viruses at inducing IFIT1 expression in HUVECs. The expression patterns of IFIT1 in the two cell lines were completely opposite. The levels of IFIT1 induced by H9N2 virus and viral particles were higher in endothelial cells than in epithelial cells (Figs. 2 and 3), indicating that endothelial cells may be more sensitive than epithelial cells in response to influenza virus infection. These results may further reveal the role of endothelial cells in the innate immune response during influenza virus infection.
Generally, most types of cells do not basally express IFIT proteins (Daffis et al., 2007). However, IFIT expression could be induced by virus and bacterial infection, and strongly induced by type I IFN (Kohli et al., 2012; Sarkar and Sen, 2004; Zhou and Amar, 2007). This is particularly obvious with IFIT1, which could be induced by exogenous IFN-α within 2 h (Bluyssen et al., 1994). Thus, we investigated the levels of IFN-α/β induced by H9N2 virus infection and viral particle inoculation. It is well known that influenza virus infection can induce production of type I IFN in respiratory epithelial cells (Xing et al., 2011). Under our experimental conditions, we found that both H9N2 virus infection and viral particle inoculation increased the levels of IFN-α/β in BEAS-2Bs (Fig. 4). This suggests that H9N2 virus or viral particles may induce the expression of IFIT1 via increasing type I IFN in BEAS-2Bs. Previous studies indicated that H5N1 virus infection could upregulate the expression of type I IFN genes in human pulmonary microvascular endothelial cells at 24 h post-infection. H5N1 virus infection in HUVECs could also induce high levels of IFN-β at earlier time points (Schmolke et al., 2009; Zeng et al., 2012). However, neither H9N2 virus infection nor viral particle inoculation significantly increased the levels of IFN-α/β in HUVECs in the present study (Fig. 4). However, both H9N2 virus infection and viral particle inoculation induced the expression of IFIT1 in HUVECs. We speculate that induction of ISGs expression depends mainly on the interaction between viral particles and HUVECs, but less so on viral replication. In our investigation of whether HA or NA protein binding induced IFIT1 expression, envelope protein binding alone was insufficient to induce IFIT1 expression (Fig. 5).
Recent studies indicated that IFIT gene expression could be induced by signals generated after the ligation of pattern-recognition receptors by pathogenic pattern molecules (Diamond and Farzan, 2013). Several studies demonstrated that IFN-regulatory factors could also directly stimulate IFIT gene expression through pathways that remain less well defined (Barnes et al., 2004; Grandvaux et al., 2002; Lou et al., 2009; Ogawa et al., 2005). Moreover, subsets of IFIT genes are selectively induced by virus infection in some cells (Fensterl et al., 2008; Terenzi et al., 2006; 2007; Wacher et al., 2007). These previous studies provide possible mechanisms for IFIT1 expression induced by H9N2 virus infection in HUVECs. However, these studies do not reveal the precise mechanism for viral particle induced IFIT1 expression. Therefore, we speculate that the effect of viral particles on IFIT1 expression is generated after cell entry. In addition, a previous study indicated that replication defective human cytomegalovirus could induce the expression of ISGs and that this is likely mediated by cellular interactions with the envelope glycoprotein B (Boehme et al., 2004). We used immunofluorescence to examine whether inactivated H9N2 viral particles could enter cells, with results showing that viral particles could enter HUVECs (Fig. 6). Taken together, we speculated that intracellular interaction might be involved in the induction of IFIT1 in HUVECs.
It is generally believed that virus infection can induce cellular antiviral responses, our results suggest that H9N2 virus infection and inactivated viral particle inoculation could induce the expression of IFIT1. Although we could not clearly clarify the innate immune response of cells to H9N2 virus or inactivated viral particle with our existing results, we can draw the following inferences. Endothelial cells and epithelial cells are two major cell types in the lung, and are believed to play important roles in response to virus infection. On the one hand, regardless of whether H9N2 virus infects endothelial cells in vivo, according to the results of this study, inactivated H9N2 virus may induce antiviral response in endothelial cells. According to the present results, we could inoculate inactivated viral particles through the respiratory tract to induce the antiviral state of host. Virus RNA activates the cellular antiviral response has been widely reported. In the present study, we did not investigate the effect of RNA of H9N2 virus on the antiviral response in HUVECs and BEAS-2Bs. However, we inoculated endothelial cells and epithelial cells with inactivated viral particles at a MOI of 5, and the RNA destroyed by β-propionolactone could not be replicated. Therefore, we hypothesize that such a low level of RNA could not be a major factor in inducing IFIT1 expression. And we will continue to investigate the effects of various components of the H9N2 virus on antiviral responses of endothelial cells in subsequent studies.
It is of course that further studies are necessary to go beyond our present results. For example, further investigation needs to be done for revealing the precise mechanism of distinct expression patterns of IFIT1 in epithelial cells and endothelial cells. In addition, the distinct expression patterns of IFIT1 between epithelial cells and endothelial cells needs to be validated in vivo. In particular, whether inactivated viral particle inoculation enhances the antiviral state of host.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant No. 31672597, 31572558), Natural Science Foundation of Beijing (KZ201510020020) and Special Fund for Agro-scientific Research in the Public Interest (Grant No. 201403051-10).
REFERENCES
- Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem. 2004;279:45194–45207. doi: 10.1074/jbc.M400726200. [DOI] [PubMed] [Google Scholar]
- Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, et al. Genesis, Evolution and Prevalence of H5N6 Avian Influenza Viruses in China. Cell Host Microbe. 2016;20:810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- Bluyssen HA, Vlietstra RJ, Faber PW, Smit EM, Hagemeijer A, Trapman J. Structure, chromosome localization, and regulation of expression of the interferon-regulated mouse Ifi54/Ifi56 gene family. Genomics. 1994;24:137–148. doi: 10.1006/geno.1994.1591. [DOI] [PubMed] [Google Scholar]
- Boehme KW, Singh J, Perry ST, Compton T. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J Virol. 2004;78:1202–1211. doi: 10.1128/JVI.78.3.1202-1211.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Monack DM. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol. 2013;13:551–565. doi: 10.1038/nri3479. [DOI] [PubMed] [Google Scholar]
- Chan MC, Chan RW, Yu WC, Ho CC, Chui WH, Lo CK, Yuen KM, Guan YI, Nicholls JM, Peiris JS. Influenza H5N1 virus infection of polarized human alveolar epithelial cells and lung microvascular endothelial cells. Respir Res. 2009;10:102. doi: 10.1186/1465-9921-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebath J, Merlin G, Metz R, Benech P, Revel M. Interferon-induced 56,000 Mr protein and its mRNA in human cells: molecular cloning and partial sequence of the cDNA. Nucleic Acids Res. 1983;11:1213–1226. doi: 10.1093/nar/11.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- Cheng VC, Chan JF, Wen X, Wu WL, Que TL, Chen H, Chan KH, Yuen KY. Infection of immunocompromised patients by avian H9N2 influenza A virus. J Infect. 2011;62:394–399. doi: 10.1016/j.jinf.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Daffis S, Samuel MA, Keller BC, Gale MJ, Diamond MS. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. Plos Pathog. 2007;3:e106. doi: 10.1371/journal.ppat.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson I, Shkoda I, Golender N, Perk S, Lapin K, Khinich Y, Panshin A. Genetic characterization of HA gene of low pathogenic H9N2 influenza viruses isolated in Israel during 2006–2012 periods. Vir Genes. 2013;46:255–263. doi: 10.1007/s11262-012-0852-4. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirix V, Verscheure V, Vermeulen F, De Schutter I, Goetghebuer T, Locht C, Mascart F. Both CD4(+) and CD8(+) lymphocytes participate in the IFN-gamma response to filamentous hemagglutinin from Bordetella pertussis in infants, children, and adults. Clin Dev Immunol. 2012;2012:795958. doi: 10.1155/2012/795958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V, Sen GC. The ISG56/IFIT1 gene family. J Interferon Cytokine Res. 2011;31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V, White CL, Yamashita M, Sen GC. Novel characteristics of the function and induction of murine p56 family proteins. J Virol. 2008;82:11045–11053. doi: 10.1128/JVI.01593-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandvaux N, Servant MJ, TenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert C, Zeng QX, Shanmugasundaram R, Garthwaite L, Oliver BG, Kumar RK. Response of airway epithelial cells to double-stranded RNA in an allergic environment. Transl Respir Med. 2014;2:11. doi: 10.1186/s40247-014-0011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol. 2006;80:7469–7480. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Tengchuan J, Laustsen A, Hansen K, Ostergaard L, Fitzgerald KA, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci USA. 2013;110:E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonges M, Liu WM, van der Vries E, Jacobi R, Pronk I, Boog C, Koopmans M, Meijer A, Soethout E. Influenza virus inactivation for studies of antigenicity and phenotypic neuraminidase inhibitor resistance profiling. J Clin Microbiol. 2010;48:928–940. doi: 10.1128/JCM.02045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim MJ, Park DY, Chung HJ, Kim CH, Yoon JH, Kim HJ. Mitochondrial reactive oxygen species modulate innate immune response to influenza A virus in human nasal epithelium. Antiviral Res. 2015;119:78–83. doi: 10.1016/j.antiviral.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Kohli A, Zhang X, Yang J, Russell RS, Donnelly RP, Sheikh F, Sherman A, Young H, Imamichi T, Lempicki RA, et al. Distinct and overlapping genomic profiles and antiviral effects of Interferon-lambda and -alpha on HCV-infected and noninfected hepatoma cells. J Viral Hepat. 2012;19:843–853. doi: 10.1111/j.1365-2893.2012.01610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Stein S, Falck-Pedersen E. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J Virol. 2014;88:974–981. doi: 10.1128/JVI.02702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- Lou YJ, Pan XR, Jia PM, Li D, Xiao S, Zhang ZL, Chen SJ, Chen Z, Tong JH. IRF-9/STAT2 [corrected] functional interaction drives retinoic acid-induced gene G expression independently of STAT1. Cancer Res. 2009;69:3673–3680. doi: 10.1158/0008-5472.CAN-08-4922. [DOI] [PubMed] [Google Scholar]
- Metz P, Dazert E, Ruggieri A, Mazur J, Kaderali L, Kaul A, Zeuge U, Windisch MP, Trippler M, Lohmann V, et al. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology. 2012;56:2082–2093. doi: 10.1002/hep.25908. [DOI] [PubMed] [Google Scholar]
- Nicholls JM, Bourne AJ, Chen H, Guan Y, Peiris JS. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. 2007;8:73. doi: 10.1186/1465-9921-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CD, Reid SM, Ball A, Cox WJ, Essen SC, Hanna A, Mahmood S, Slomka MJ, Irvine RM, Brown IH. First reported detection of a low pathogenicity avian influenza virus subtype H9 infection in domestic fowl in England. Vet Rec. 2012;171:372. doi: 10.1136/vr.100558. [DOI] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Raychoudhuri A, Shrivastava S, Steele R, Kim H, Ray R, Ray RB. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud JM, Kim DY, Atasheva S, Rasalouskaya A, White JP, Diamond MS, Weaver SC, Frolova EI, Frolov I. IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon. Plos Pathog. 2015;11:e1004863. doi: 10.1371/journal.ppat.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar SN, Sen GC. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther. 2004;103:245–259. doi: 10.1016/j.pharmthera.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Schmolke M, Viemann D, Roth J, Ludwig S. Essential impact of NF-kappaB signaling on the H5N1 influenza A virus-induced transcriptome. J Immunol. 2009;183:5180–5189. doi: 10.4049/jimmunol.0804198. [DOI] [PubMed] [Google Scholar]
- Shi SH, Yang WT, Yang GL, Cong YL, Huang HB, Wang Q, Cai RP, Ye LP, Hu JT, Zhou JY, et al. Immunoprotection against influenza virus H9N2 by the oral administration of recombinant Lactobacillus plantarumNC8 expressing hemagglutinin in BALB/c mice. Virology. 2014;464–465:166–176. doi: 10.1016/j.virol.2014.07.011. [DOI] [PubMed] [Google Scholar]
- Siednienko J, Maratha A, Yang S, Mitkiewicz M, Miggin SM, Moynagh PN. Nuclear factor kappaB subunits RelB and cRel negatively regulate Toll-like receptor 3-mediated beta-interferon production via induction of transcriptional repressor protein YY1. J Biol Chem. 2011;286:44750–44763. doi: 10.1074/jbc.M111.250894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR, Walsh KB, Cahalan S, Fremgen DM, Roberts E, Scott F, Martinborough E, Peach R, Oldstone MB, Rosen H. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146:980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi F, Hui DJ, Merrick WC, Sen GC. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J Biol Chem. 2006;281:34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- Terenzi F, White C, Pal S, Williams BR, Sen GC. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J Virol. 2007;81:8656–8665. doi: 10.1128/JVI.00322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobita K, Sugiura A, Enomote C, Furuyama M. Plaque assay and primary isolation of influenza A viruses in an established line of canine kidney cells (MDCK) in the presence of trypsin. Med Microbiol Immunol. 1975;162:9–14. doi: 10.1007/BF02123572. [DOI] [PubMed] [Google Scholar]
- Tombari W, Paul M, Bettaieb J, Larbi I, Nsiri J, Elbehi I, Gribaa L, Ghram A. Risk factors and characteristics of low pathogenic avian influenza virus isolated from commercial poultry in Tunisia. Plos One. 2013;8:e53524. doi: 10.1371/journal.pone.0053524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. H5N1 virus attachment to lower respiratory tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D, den Bakker MA, Leijten LM, Chutinimitkul S, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am J Pathol. 2010;176:1614–1618. doi: 10.2353/ajpath.2010.090949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viemann D, Schmolke M, Lueken A, Boergeling Y, Friesenhagen J, Wittkowski H, Ludwig S, Roth J. H5N1 virus activates signaling pathways in human endothelial cells resulting in a specific imbalanced inflammatory response. J Immunol. 2011;186:164–173. doi: 10.4049/jimmunol.0904170. [DOI] [PubMed] [Google Scholar]
- von Recum-Knepper J, Sadewasser A, Weinheimer VK, Wolff T. Fluorescence-activated cell sorting-based analysis reveals an asymmetric induction of interferon-stimulated genes in response to seasonal influenza A virus. J Virol. 2015;89:6982–6993. doi: 10.1128/JVI.00857-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacher C, Muller M, Hofer MJ, Getts DR, Zabaras R, Ousman SS, Terenzi F, Sen GC, King NJ, Campbell IL. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J Virol. 2007;81:860–871. doi: 10.1128/JVI.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Mu X, Zhao L, Wang J, Chu Y, Feng X, Feng B, Wang X, Zhang J, Qiao J. Transcriptional response of human umbilical vein endothelial cell to H9N2 influenza virus infection. Virology. 2015;482:117–127. doi: 10.1016/j.virol.2015.03.037. [DOI] [PubMed] [Google Scholar]
- Weibel ER, Knight BW. A morphometric study on the thickness of the pulmonary air-blood barrier. J Cell Biol. 1964;21:367–396. doi: 10.1083/jcb.21.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z, Harper R, Anunciacion J, Yang Z, Gao W, Qu B, Guan Y, Cardona CJ. Host immune and apoptotic responses to avian influenza virus H9N2 in human tracheobronchial epithelial cells. Am J Respir Cell Mol Biol. 2011;44:24–33. doi: 10.1165/rcmb.2009-0120OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Korteweg C, Hsueh W, Gu J. Avian influenza receptor expression in H5N1-infected and noninfected human tissues. FASEB J. 2008;22:733–740. doi: 10.1096/fj.06-7880com. [DOI] [PubMed] [Google Scholar]
- Yu H, Zhou YJ, Li GX, Ma JH, Yan LP, Wang B, Yang FR, Huang M, Tong GZ. Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Vet Microbiol. 2011;149:254–261. doi: 10.1016/j.vetmic.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Zeng H, Pappas C, Belser JA, Houser KV, Zhong W, Wadford DA, Stevens T, Balczon R, Katz JM, Tumpey TM. Human pulmonary microvascular endothelial cells support productive replication of highly pathogenic avian influenza viruses: possible involvement in the pathogenesis of human H5N1 virus infection. J Virol. 2012;86:667–678. doi: 10.1128/JVI.06348-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Belser JA, Goldsmith CS, Gustin KM, Veguilla V, Katz JM, Tumpey TM. A(H7N9) virus results in early induction of proinflammatory cytokine responses in both human lung epithelial and endothelial cells and shows increased human adaptation compared with avian H5N1 virus. J Virol. 2015;89:4655–4667. doi: 10.1128/JVI.03095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Xu W, Zhang Z, Wang T, Sang X, Cheng K, Yu Z, Zheng X, Wang H, Zhao Y, et al. Experimental infection of non-human primates with avian influenza virus (H9N2) Arch Virol. 2013;158:2127–2134. doi: 10.1007/s00705-013-1721-8. [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhang Z, Yu Z, Li L, Cheng K, Wang T, Huang G, Yang S, Zhao Y, Feng N, et al. Domestic cats and dogs are susceptible to H9N2 avian influenza virus. Virus Res. 2013;175:52–57. doi: 10.1016/j.virusres.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Amar S. Identification of signaling pathways in macrophage exposed to Porphyromonas gingivalis or to its purified cell wall components. J Immunol. 2007;179:7777–7790. doi: 10.4049/jimmunol.179.11.7777. [DOI] [PubMed] [Google Scholar]