Abstract
L-pipecolic acid is a non-protein amino acid commonly found in plants, animals, and microorganisms. It is a well-known precursor to numerous microbial secondary metabolites and pharmaceuticals, including anticancer agents, immunosuppressants, and several antibiotics. Lysine cyclodeaminase (LCD) catalyzes β-deamination of L-lysine into L-pipecolic acid using β-nicotinamide adenine dinucleotide as a cofactor. Expression of a human homolog of LCD, μ-crystallin, is elevated in prostate cancer patients. To understand the structural features and catalytic mechanisms of LCD, we determined the crystal structures of Streptomyces pristinaespiralis LCD (SpLCD) in (i) a binary complex with NAD+, (ii) a ternary complex with NAD+ and L-pipecolic acid, (iii) a ternary complex with NAD+ and L-proline, and (iv) a ternary complex with NAD+ and L-2,4-diamino butyric acid. The overall structure of SpLCD was similar to that of ornithine cyclodeaminase from Pseudomonas putida. In addition, SpLCD recognized L-lysine, L-ornithine, and L-2,4-diamino butyric acid despite differences in the active site, including differences in hydrogen bonding by Asp236, which corresponds with Asp228 from Pseudomonas putida ornithine cyclodeaminase. The substrate binding pocket of SpLCD allowed substrates smaller than lysine to bind, thus enabling binding to ornithine and L-2,4-diamino butyric acid. Our structural and biochemical data facilitate a detailed understanding of substrate and product recognition, thus providing evidence for a reaction mechanism for SpLCD. The proposed mechanism is unusual in that NAD+ is initially converted into NADH and then reverted back into NAD+ at a late stage of the reaction.
Keywords: crystal structure, L-lysine cyclodeaminase, Streptomyces pristinaespiralis
INTRODUCTION
L-Pipecolic acid (L-Pip), also known as L-homoproline and piperidine-2-carboxylic acid, is a well-known precursor of various secondary metabolites in plants, animals, and micro-organisms (Durzan, 1983; Zacharius et al., 1954). Lysine cyclodeaminase (LCD) catalyzes β-deamination of L-lysine into L-Pip using β-nicotinamide adenine dinucleotide as a cofactor. In humans, L-Pip is synthesized in the brain and accumulates in the physiological fluids of patients with peroxisomal disorders, such as hyperpipecolic academia, Zellweger syndrome, and infantile Refsum disease (Mihalik et al., 1989; Rao and Chang, 1992). In these diseases, peroxisomes, which are the sites of L-Pip oxidation, are reduced in size and number and many peroxisomal enzymes present with low activity (Mihalik et al., 1989; Rao and Chang, 1992). Expression of a human LCD homolog, μ-crystallin, is elevated in prostate cancer patients (Malinowska et al., 2009). In plants, L-Pip acts as an osmoprotectant, inducer of flowering, indicator of abnormal amino acid metabolism, and has a critical role in plant immunity (Fujioka and Sakurai, 1992; Moulin et al., 2006; Návarová et al., 2012; Pálfi and Dézsi, 1968). In bacteria, L-Pip is synthesized during L-lysine metabolism and acts as an osmoprotectant, thus enabling bacteria to grow in hyperosmotic environments (Gouesbet et al., 1994; Neshich et al., 2013).
L-Pip is a useful precursor of several pharmaceuticals, including the immunosuppressant FK506, rapamycin, the anti-cancer agent VX710, and the antibiotics demethoxyrapamycin, sandramycin, and meridamycin (Bis et al., 2015; Boger et al., 1996; Gatto et al., 2006; Germann et al., 1997; Kadouri-Puchot and Comesse, 2005; Watanabe et al., 2005). As the importance and utility of L-Pip derivatives have become increasingly well-known, chemists have been developing different methods by which to synthesize L-Pip (Couty, 1999; Eichhorn et al., 1997; Lemire and Charette, 2010; Wilkinson et al., 2000). In particular, enzymatic production of L-Pip using the L-lysine metabolic pathway has been extensively investigated (Broquist, 1991; Byun et al., 2015; Fujii et al., 2002; Garcia et al., 2016; Gupta and Spenser, 1969; Miller and Rodwell, 1971; Tani et al., 2015; Ying et al., 2015; 2017a), because the precursor L-lysine is abundant in nature and enzymatic synthesis of L-Pip yields a product with better chiral purity than chemical synthesis. Two metabolic pathways have previously been investigated for use in the enzymatic synthesis of L-Pip (Broquist, 1991; Gupta and Spenser, 1969; Miller and Rodwell, 1971). However, both of these metabolic routes require two or more enzymes and involve several steps to produce L-Pip from L-lysine, which results in problematic byproducts and high purification costs (He, 2006).
By contrast, it has been shown that LCD can directly convert L-lysine into L-Pip in one step using NAD+ as a cofactor (Gatto et al., 2006; Tsotsou and Barbirato, 2007), which is more efficient than other methods used for L-Pip biosynthesis. Recently, conditions for L-Pip biosynthesis using LCD from Streptomyces pristinaespiralis (SpLCD) were optimized to increase catalytic efficiency (Byun et al., 2015; Tsotsou and Barbirato, 2007; Ying et al., 2015; 2017a). In addition, the crystal structures of SpLCD in complex with L-lysine and L-Pip have been determined and their modes of binding described (Ying et al., 2017b). Interestingly, SpLCD has a range of substrates with carbon lengths ranging from 4–6 carbons. However, it is currently unknown how SpLCD recognizes and acts on this diverse range of substrates, including how L-lysine and L-ornithine are converted into L-Pip and L-Proline (L-Pro), respectively. L-lysine has been demonstrated to be the best substrate for SpLCD, of all the known substrates such as L-2,4-diamino butyric acid (L-DABA), L-ornithine, and L-lysine, which have four, five, and six carbon chains, respectively (Byun et al., 2015).
The crystal structure and binding of ornithine cyclodeaminase (OCD) in complex with ornithine have been described (Goodman et al., 2004). Based on OCD X-ray crystallographic and mass spectroscopic data, a reaction mechanism has been proposed for OCD (Gatto et al., 2006; Goodman et al., 2004). The proposed mechanism is unusual in that NAD+ is initially converted into NADH and then reverted back into NAD+ at a late stage, where the pyridine nucleotide acts as a cosubstrate during most enzymatic reactions that utilize NAD(P)+ and NAD(P)H (Gatto et al., 2006). Similar use of NAD(P)+ as a catalytic cofactor has been proposed for other enzymes, including UDP-galactose 4-epimerase (Frey, 1996), S-adenosylhomocysteine hydrolase (Hu et al., 2001), and myo-inositol-1-phosphate synthase (Gatto et al., 2006; Jin and Geiger, 2003). Mechanisms similar to those utilized by OCD have been proposed for LCD. However, the LCD substrate L-lysine contains one carbon more than ornithine. Therefore, it would be of interest to determine how LCD and OCD active sites recognize their substrates and how their reaction mechanisms differ.
Because of the unusual reaction mechanism suggested for LCD and the importance of the reaction product L-Pip, we undertook a crystallographic study of LCD derived from S. pristinaespiralis. This provided a structural basis for determining how SpLCD recognizes several substrates, including L-lysine, L-ornithine, and L-DABA. Here, we report the crystal structures of SpLCD in four ligand-bound states: SpLCD/ NAD+, SpLCD/NAD+/L-DABA, SpLCD/NAD+/L-Pro, and SpLCD/ NAD+/L-Pip. By comparing the binding pockets of LCD and OCD, we found the hydrogen bonding residues, including Asp236, allow recognition of lysine as a substrate. Our structural and biochemical data facilitate a detailed understanding of substrate and product recognition by SpLCD, thus informing a proposal for the structure-based reaction mechanism for SpLCD.
MATERIALS AND METHODS
Cloning and protein preparation
L-lysine cyclodeaminase from S. pristinaespiralis (amino acids 1–355) was cloned into the pET21b (+) expression vector. This construct encoded an N-terminal His6 tag and the SpLCD gene under the control of the T7 promoter. The SpLCD protein was produced in Escherichia coli strain BL21 (DE3) by inducing expression with 0.5 mM isopropyl-β-D-1-thiogalactopyranoside and incubating at 25°C for 16 h. To lyse the cells, the cells were passed through a microfluidizer in lysis buffer A (20 mM Tris-HCl, pH 8.0, and 200 mM NaCl) containing 1 mM phe-nylmethylsulfonyl fluoride and 5% (w/v) glycerol. The lysed cells were centrifuged at 4,611 g (Vision V506CA rotor) for 30 min at 277 K to pellet cellular debris. The resulting supernatant was run through Ni-sepharose affinity resin (GE Healthcare). Proteins were eluted with buffer A containing 100–300 mM imidazole, further purified by size exclusion chromatography (HiLoad 16/600 Superdex 200 prep grade, GE Healthcare), and then concentrated to 30 mg/ml by ultra-filtration. The extinction coefficient of SpLCD was calculated using ProtParam (http://web.expasy.org/protparam).
Crystallization, determination of structure, and refinement
Crystals of SpLCD in complex with NAD+ were grown at 298 K using the sitting drop method Briefly, 1 μl of a 10 mg/ml solution of SpLCD in buffer A was mixed with 1 μl of reservoir solution (100 mM CAPSO buffer at pH 9.6, 0.2 M Li2SO4, 0.9 M Na-K tartrate, and 2% polyethylene glycol 3350) in the presence of 10 mM NAD+. For the SpLCD/NAD+/L-DABA, SpLCD/NAD+/L-Pro, and SpLCD/NAD+/L-Pip crystals, SpLCD was crystallized in the presence of 10 mM L-DABA, 10 mM L-ornithine, or 10 mM L-lysine, respectively. For cryoprotection, the crystals were transferred to reservoir solution containing 25% glycerol. Data was collected at 100 K in 1° oscillations at the 7A beamline of the Pohang Light Source and BL26B1 beamline of the SPring-8.
Crystals of SpLCD/NAD+, SpLCD/NAD+/L-Pip, SpLCD/ NAD+/L-Pro, and SpLCD/NAD+/L-DABA complexes were diffracted to a resolution of 1.92 Å, 1.92 Å, 1.79 Å, and 2.14 Å, respectively. The diffraction data were processed and scaled using the HKL2000 software package (Otwinowski and Minor, 1997). The SpLCD/NAD+ crystal belonged to space group C2 and had unit cell parameters of a = 270.5 Å, b = 64.4 Å, and c = 106.4 Å. SpLCD/NAD+/L-Pip crystal also belonged to the C2 space group and had unit cell parameters of a = 271.4 Å, b = 64.5 Å, and c = 106.3 Å. The SpLCD/NAD+/L-Pro belonged to space group C2 and had unit cell parameters of a = 271.5 Å, b = 64.8 Å, and c = 106.9 Å. SpLCD/NAD+/L-DABA crystal belonged to the C2 space group and had unit cell parameters of a = 271.1 Å, b = 64.8 Å, and c = 106.9. The structures were solved using the molecular replacement method using the Archaeoglobus fulgidus alanine dehydrogenase model, Protein Data Bank (PDB) entry 1OMO, as a probe (Gallagher et al., 2004). Cross-rotational and then translational searches were performed using the PHASER program (McCoy et al., 2007). Subsequent manual model building was performed using the COOT program (Emsley and Cowtan, 2004) and restrained refinement was carried out using the PHENIX program (Adams et al., 2002). Several rounds of model building, simulated annealing, positional refinement, and individual B-factor refinement were performed using the COOT and PHENIX programs. Table 1 lists the refinement statistics. The atomic coordinates and structural factors for SpLCD/NAD+, SpLCD/NAD+/L-Pip, SpLCD/NAD+/L-Pro, and SpLCD/NAD+/L-DABA complexes were deposited in PDB (accession codes 5YU0, 5YU1, 5YU3, and 5YU4).
Table 1.
Statistics for data collection and refinement
| Data set | SpLCD with NAD+ | SpLCD with NAD+ and L-Pip | SpLCD with NAD+ and L-Pro | SpLCD with NAD+ and DABA |
|---|---|---|---|---|
| A. Data collection | ||||
| X-ray source | PLS7A | PLS7A | SPring-8 BL26B1 | SPring-8 BL26B1 |
| X-ray wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Space group | C2 | C2 | C2 | C2 |
| Unit cell length (a, b, c, Å) | 270.5, 64.4, 106.4 | 271.4, 64.5, 106.3 | 271.5, 64.8, 106.9 | 271.1, 64.8, 106.9 |
| Unit cell angle (Å, β, γ, °) | 90, 104.1, 90 | 90, 104.2, 90 | 90, 104.2, 90 | 90, 104.1, 90 |
| Resolution range (Å) | 50.0–1.92 (1.95–1.92)a | 50–1.92 (1.95–1.92)a | 50–1.79 (1.82–1.79)a | 50–2.14 (2.18–2.14)a |
| Total / unique reflections | 955,775 / 136,288 | 905,894 / 134,717 | 1,267,763/ 169,178 | 689,619 / 97,978 |
| Completeness (%) | 99.6 (98.8)a | 99.3 (98.6)a | 99.9 (98.7)a | 100.0 (100.0)a |
| Average I/σ (I) | 28.6 (4.1)a | 39.6 (12.0)a | 42.2 (4.7)a | 33.3 (4.5)a |
| Rmergeb (%) | 8.2 (50.1)a | 8.9 (42.6)a | 8.0 (50.9)a | 11.3 (75.1)a |
| B. Model refinement | ||||
| Resolution range (Å) | 50.0–1.92 | 50.0–1.92 | 50.0–1.79 | 50.0–2.14 |
| Rwork / Rfreec (%) | 16.8 / 20.0 | 16.7 / 20.3 | 16.4 / 19.3 | 15.9 / 19.7 |
| Monomers per asymmetric unit | 4 | 4 | 4 | 4 |
| Number of non-hydrogen atoms / average B-factor (Å2) | ||||
| Protein | 10,292 / 18.1 | 10,292 / 20.2 | 10,292 / 21.6 | 10,292 / 34.3 |
| Water oxygen | 1,229 / 28.4 | 1,234 / 31.7 | 1,385 / 33.0 | 884 / 41.0 |
| Na+ | 4 / 18.6 | 4 / 22.5 | 4 / 15.6 | 4 / 31.6 |
| NAD+ | 4 / 13.4 | 4 / 14.4 | 4 / 16.3 | 4 / 28.6 |
| L-Pip | - | 4 / 17.9 | - | - |
| L-Pro | - | - | 4 / 25.5 | - |
| DABA | - | - | - | 4 / 42.0 |
| R.m.s. deviations from ideal geometry | ||||
| Bond lengths (Å) / bond angles (°) | 0.008 / 1.39 | 0.008 / 1.41 | 0.008 / 1.48 | 0.008 / 1.39 |
| Protein-geometry analysis | ||||
| Ramachandran favored (%) | 96.8 | 97.5 | 96.8 | 96.8 |
| Ramachandran allowed (%) | 2.9 | 2.2 | 2.9 | 3.1 |
| Ramachandran outliers (%) | 0.3 | 0.3 | 0.3 | 0.1 |
Values in parentheses refer to the highest resolution shell.
Rmerge = ∑h ∑i | I(h)i – < I(h) > | / ∑h ∑i I(h)i, where I(h) is the intensity of reflection h, ∑h is the sum over all reflections, and ∑i is the sum over i measurements of reflection h.
R = ∑hkl | ··· |Fobs| – |Fcalc| | / ∑hkl |Fobs|, where Rfree was calculated for a randomly chosen 5% of reflections, which were not used for structure refinement and Rwork was calculated for the remaining.
Analytical gel filtration
Purified SpLCD was subjected to analytical gel filtration chromatography using a Superdex 200 (10/300 GL) column with buffer A flowing at a constant rate of 0.5 ml/min (Choi et al., 2016). The standard curve was created using molecular weight markers (Sigma). The Stokes radii of β-amylase, alcohol dehydrogenase, carbonic anhydrase, and cytochrome C were calculated from the crystal structures of each protein (PDB entries 1FA2, 2HCY, 1V9E, and 1HRC, respectively) using the HYDROPRO program (Garcia De La Torre et al., 2000).
Alignment of sequences
The sequence of S. pristinaespiralis LCD was compared to the sequences of other LCDs and OCDs. The UniProtKB/ Swiss-Prot accession numbers of the sequences used were D9UBW0 (SpLCD), Q8RR22 (SvLCD) A0A0A0NQJ4 (SrLCD), E9KTB3 (SkLCD), U5VYY1 (AfLCD), Q9R468 (AtOCD), A0A179QU84 (PpOCD), A9WVR4 (BsOCD), C4IUX0 (BaOCD), and A0A0T7GIJ3 (NgOCD). Sequences were aligned using ClustalX (Thompson et al., 1997), and secondary structure elements were assigned using PyMOL (PyMOL Molecular Graphics System, http://www.pymol.org).
RESULTS AND DISCUSSION
Determination of the crystal structure of S. pristinaespiralis LCD
Crystal structures of SpLCD were determined using the molecular replacement method based on the A. fulgidus alanine dehydrogenase model (PDB entry 1OMO) (Gallagher et al., 2004) (Figs. 1A and 1B). The refined models for SpLCD/NAD+, SpLCD/NAD+/L-Pip, SpLCD/NAD+/ L-Pro, and SpLCD/ NAD+/L-DABA had Rwork/Rfree values of 16.8/20.0% for 50-1.92 Å, 16.7/20.3% for 50-1.92 Å, 16.4/19.3% for 50-1.79 Å, and 15.9/19.7% for 50-2.14 Å, respectively (Table 1). The last eleven disordered residues (345PLPLPLAAPAH355) of the C-termini are not shown for these structures. These residues are not conserved among LCDs, suggesting they are not critical for enzymatic functions. The refined model of SpLCD/NAD+ includes residues 1–344 in an asymmetric unit with 1,229 water molecules. Ramachandran plot analysis of the refined SpLCD/NAD+ model showed that 96.8, 2.9, and 0.3% of the non-glycine residues were in the most favored, allowed, and disallowed regions, respectively (Table 1). The crystal structure of SpLCD/NAD+ primarily consisted of fourteen strands and fifteen helices (Fig. 1A). The structure of SpLCD was comprised of a dimer of identical subunits, which corroborates with a previous study on PpOCD (Goodman et al., 2004) (Fig. 1B). Each subunit consisted of two domains that function in substrate binding, oligomerization, and cofactor binding (Fig. 1B). Single dinucleotide bonds in each subunit were formed via a Rossmann fold (Fig. 1B). The overall molecular dimension of the LCD dimer was 50 × 80 × 45 Å with the two longest dimensions oriented in the plane presented in Fig. 1B.
Fig. 1. Structure of Streptomyces pristinaespiralis LCD in complex with NAD+.
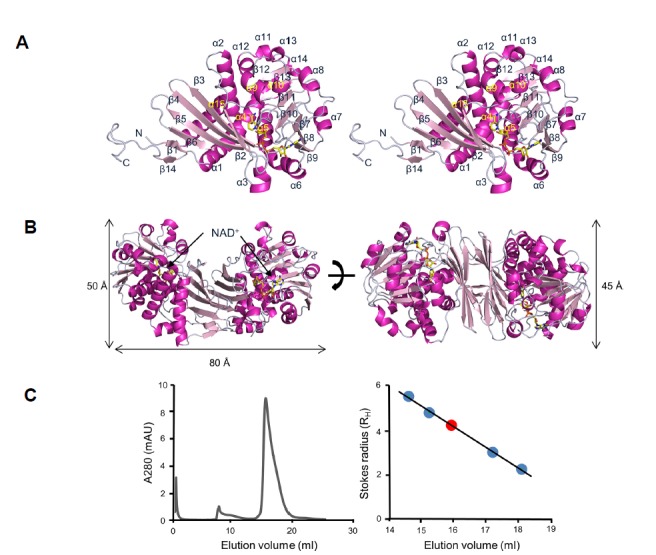
(A) Stereo ribbon diagram of S. pristinaespiralis LCD (SpLCD)/NAD+ monomer. Helices are light pink, strands are magenta, and loops are grey. The yellow stick represents NAD+. All figures, including protein structures, were drawn using PyMOL software (The PyMOL Molecular Graphics System, http://www.pymol.org). (B) Ribbon diagram of SpLCD/NAD+ dimer. The height of the structure was measured after rotating the structure 90 degrees. (C) Analyti-calgel filtration profile of SpLCD. Standard curve generated using molecular weight markers. The positions of the molecular weight markers for β-amylase, alcohol dehydrogenase, carbonic anhydrase, and cytochrome C are indicated with blue dots. The position of SpLCD is marked with a red dot.
Overall structure and structural comparisons
The overall structure of SpLCD resembled PpOCD, alanine dehydrogenase (AlaDH), and human μ-crystallin (CRYM). A search for structural similarity with SpLCD using the DALI server (Holm and Rosenstrom, 2010) identified crystal structures of OCD (PDB entry 1U7H) (Goodman et al., 2004), AlaDH (PDB entry 1OMO) (Gallagher et al., 2004), and CRYM (PDB entry 2I99) (Cheng et al., 2007). When the structure of SpLCD/NAD+ was overlaid with structures of OCD from Pseudomonas putida, AlaDH from A. fulgidus, and CRYM from humans, the r.m.s. deviations were 1.9 Å, 2.0 Å, and 2.2 Å for the 335, 313, and 306 Cα atoms, respectively. The PpOCD structure had the highest Z-score of 43.8 when superimposed with SpLCD despite differences in the NAD+ binding site. To analyze the oligomeric state of SpLCD in solution, analytical gel filtration was performed using a Superdex 200 (10/300 GL) column. The Stokes radius of SpLCD was estimated to be 3.91 nm, which is very similar to the Stokes radius calculated for the SpLCD dimer (3.43 nm) (Fig. 1C), suggesting that the SpLCD/NAD+ complex exists as a homodimer in solution. The solvent-accessible surface area buried at the interface between the two monomers measured 3132.8 Å2 (21.1% of the SpLCD monomer surface). Approximately 54% of the nonhydrogen atoms in the interface between the two monomers were polar, as indicated by the calculated proximal isovelocity surface areas (Krissinel and Henrick, 2007), which implied the interaction was mainly hydrophilic.
NAD+ binding of SpLCD
In general, the residues involved in NAD+ coordination at the active-site (Thr93, Arg121, Ala148, Gln149, Val233, and Ser301) were conserved between LCD and OCD (Figs. 2 and 3). However, there were some differences in the residues that interact with cofactors and substrates (Figs. 2 and 3). In the SpLCD/NAD+ complex, the main chain of Ala235 formed a hydrogen bond with the O3 atom of NAD+ (3.07 Å), while the side chain of Lys232 in PpOCD was involved in hydrogen bonding with the O3 atom of NAD+ (2.95 Å) (Fig. 3). The Lys232 side chain of PpOCD interacted with the O2 atom of NAD+ (2.75 Å) (Fig. 3). For hydrogen bonding with the O8 and O9 atoms of NAD+, PpOCD used the side chain of Lys331 (2.93 Å and 3.12 Å, respectively), while SpLCD used the side chain of His175 and two water molecules. The side chain of His175 formed a hydrogen bond with the O8 atom of NAD+ (2.85 Å), while two water molecules interacted with the O9 atom of NAD+. These two water molecules were sustained through hydrogen bonding with the Thr171 and Asp172 side chains (not shown) (Fig. 3). To create a hydrogen bond with O10 of NAD+, SpLCD used the main chain of Ser210 (2.32 Å), while PpOCD used Ala203 (3.34 Å) (Fig. 3). In the SpLCD/NAD+ complex, residues in SpLCD hydrogen bonds with water molecules indirectly coordinated with NAD+. The side chain of Glu307 indirectly interacted with the O1 atom of NAD+ through a water molecule (Fig. 2). In addition, the O4 atom of NAD+ interacted with Ile94 via a water molecule. The O5 atom of NAD+ interacted with a water molecule that then coordinated with three residues of SpLCD, Gly147, Ala150, and Ala208 (2.95 Å, 3.21 Å, and 2.95 Å, respectively) (Fig. 2). The main chain of Pro83 formed a hydrogen bond with a water molecule (3.08 Å) that interacted with the O7 atom of NAD+ (2.68 Å) and the side chain of Ser210 (2.77 Å). Finally, the main chain of Val218 interacted with the N4 atom of NAD+ (2.84 Å) through a water molecule (2.70 Å) (Fig. 2). These direct and indirect hydrogen bonds between NAD+ and SpLCD residues create a stable SpLCD/NAD+ complex for catalysis.
Fig. 2. Sequence alignment of SpLCD with LCDs and OCDs from other species.
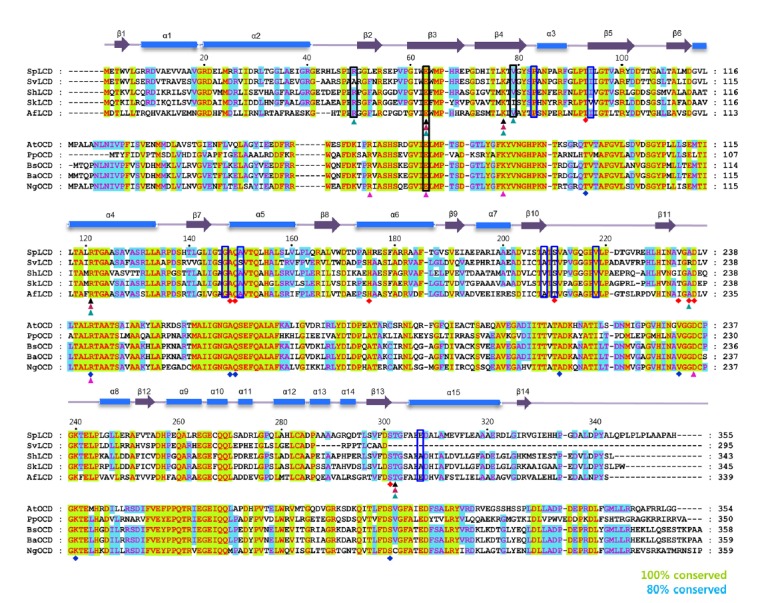
Multi-alignment of SpLCD (UniProtKB/Swiss-Prot accession numberD9UBW0) against LCD from Streptomyces virginiae(UniProtKB/Swiss-Prot accession numberQ8RR22), Streptomyces hygroscopicus(UniProtKB/Swiss-Prot accession numberA0A0A0NQJ4), Streptomyceskanamyceticus (UniProtKB/Swiss-Prot accession numberE9KTB3), Actinoplanes friuliensis(UniProtKB/Swiss-Prot accession numberU5VYY1), Agrobacterium tumefaciens (UniProtKB/Swiss-Prot accession numberQ9R468), Pseudomonas putida(UniProtKB/Swiss-Prot accession numberA0A179QU84), Brucella suis biovar 1(UniProtKB/Swiss-Prot accession numberA9WVR4), Brucella abortus biovar 1 (UniProtKB/Swiss-Prot accession numberC4IUX0), and Neorhizobium galegae by officinallis (UniProtKB/Swiss-Prot accession number A0A0T7GIJ3). Secondary structural elements were assigned using PyMOL and every twentieth residue is marked with a black dot. Strictly (100%) and semi-conserved residues (80%) are highlighted in green and light blue, respectively. Cylinders and arrows above the sequences denote α-helices and β-strands, respectively. Black, magenta, and cyan triangles below the LCD sequences denote L-pipecolic acid, L-proline, and L-DABA, respectively. Black boxes indicate residues interacting with water molecules when LCD or OCD bound its ligands and blue boxes denote residues interact with water molecules when LCD bound its cofactor; NAD+. Red and blue diamonds represent residues in the structures of SpLCD and Pseudomonas putida ornithine cyclodeaminase (PpOCD) that coordinate with NAD+ and NADH, respectively. Residues interacting with L-ornithine in OCD are shown in a pink triangle.
Fig. 3. A comparison of residues involved in coordination of NAD+ and NADH with SpLCD-NAD+ and PpOCD-NADH.
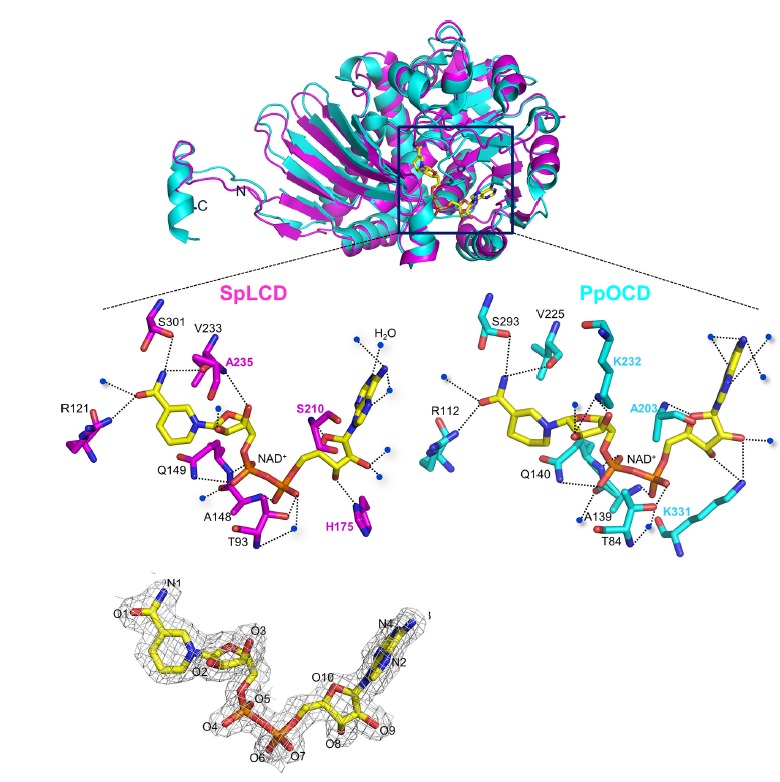
The residues involved in coordination of NAD+ and NADH with SpLCD-NAD+ (PDB entry 5YU0, magenta) and PpOCD-NADH (PDB entry 1U7H, cyan) were compared. Dashed lines represent hydrogen bonds. The residues interacting with NAD+ and NADH are shown in magenta for SpLCD and cyan for PpOCD. Blue dots represent water molecules. A 2Fo-Fc electron density map (1.0 σ) of the NAD+ molecule is presented at the bottom of the figure.
Structural insights into ligand recognition by SpLCD
To obtain information on the binding between SpLCD and L-lysine, we attempted to co-crystallize SpLCD with L-lysine. However, we instead obtained the electron-density map of L-Pip, the catalytic product of SpLCD (Fig. 4A), suggesting the reaction occurred prior to crystallization. Similarly, the electron density map of L-Pro was obtained when SpLCD was co-crystallized with L-ornithine (Fig. 4B). When we attempted to co-crystallize SpLCD with L-DABA, L-DABA was unable to be transformed in ring form. Instead, the electron density associated with L-DABA was observed (Fig. 4C). These results suggest that SpLCD has catalytic activity for L-lysine and L-ornithine and L-DABA is too short for ring formation. This is consistent with previous studies demonstrating SpLCD is able to perform catalysis on a range of substrates of varying carbon lengths (Byun et al., 2015; Tsotsou and Barbirato, 2007). Structural comparisons of SpLCD in complex with L-Pip, L-Pro, and L-DABA revealed that binding of the carboxyl group attached to the C2 carbon was similar for each substrate (Fig. 4), suggesting this binding significantly contributes to substrate recognition.
Fig. 4. Stereo diagrams of SpLCD active sites around L-pipecolic acid, L-proline, and L-DABA.
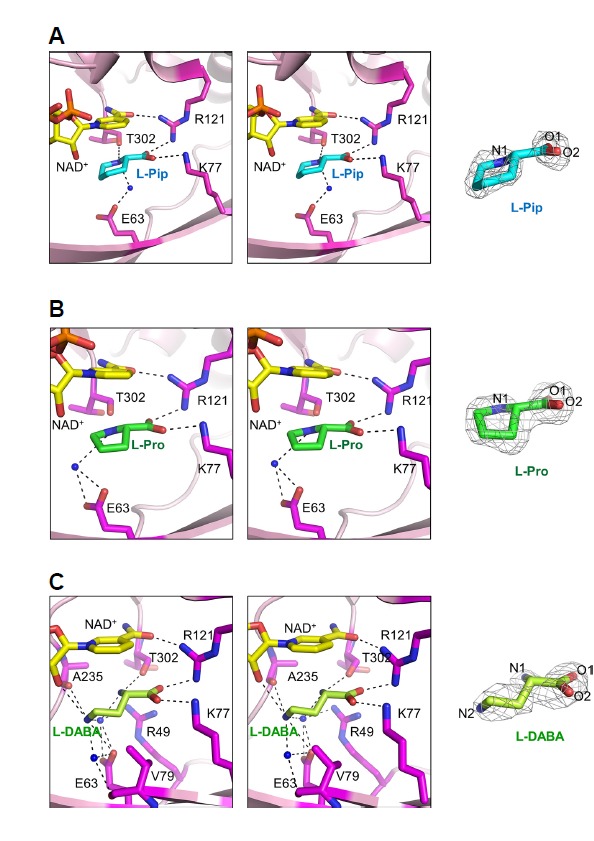
Stereo diagrams of the SpLCD active sites surrounding ligands (A) L-pipecolic acid, (B) L-proline, and (C) L-DABA. Dashed lines represent hydrogen bonds and blue dots indicate water molecules. A 2Fo-Fc electron density map (1.0 σ) for each ligand is shown to the right of each stereo diagram.
Next, we analyzed the binding of L-Pip and L-Pro to the active site of SpLCD. The side chains of Glu63, Lys77, and Arg121, and the main chain of Thr302, interacted with enzymatic products L-Pip and L-Pro (Figs. 4A and 4B). Interestingly, the binding positions and orientations of L-Pip and L-Pro were similar (Figs. 4A and 4B). In the SpLCD/L-Pip structure, the side chains of Lys77 and Arg121 interacted with the O1 (2.70 Å) and O2 atoms (2.91 Å) of L-Pip, respectively (Fig. 4A). In addition, the N1 atom of L-Pip interacted with the side chain of Thr302 (2.87 Å) (Fig. 4A). The side chain of Glu63 indirectly interacted with the N1 atom of L-Pip via one water molecule (Fig. 4A). For the SpLCD/L-Pro structure, the three residues Lys77, Arg121, and Thr302 were directly involved in hydrogen bonding with L-Pro (2.67 Å, 2.74 Å, and 3.20 Å, respectively), while the side chain of Glu63 indirectly interacted with the N1 atom of L-Pro via one water molecule in a manner similar to the SpLCD/L-Pip structure (Fig. 4B).
To interact with L-DABA, additional side chains were required to stabilize L-DABA in the active site. The side chains of Glu63, Lys77, and Arg121, and the main chain of Thr302, formed hydrogen bonds with the N2 (3.25 Å), O2 (3.01 Å), O1 (2.81 Å), and N1 (2.93 Å) atoms of L-DABA, respectively (Fig. 4C). The carbonyl oxygen of Ala235 (3.20 Å) and O2 atom of NAD+ (3.25 Å) formed hydrogen bonds with the N2 atom of L-DABA (Fig. 4C). In addition, two water molecules formed hydrogen bonds, where one interacted with the Val79 (3.14 Å), Glu63 (2.79 Å), and N2 atoms of L-DABA (2.86 Å) and the other interacted with Arg49 (2.83 Å) and Glu63 (3.05 Å and 2.85 Å) and the N1 (2.71 Å) and N2 (3.02 Å) atoms of NAD+ (Fig. 4C).
To examine what makes L-Lys the optimal substrate for SpLCD, a composite model was created by incorporating the L-ornithine molecule from the PpOCD/L-ornithine structure into our SpLCD structure (Fig. 5A). The major difference between SpLCD and PpOCD recognition of L-lysine and L-ornithine, respectively, is the positioning of Asp236 in the substrate binding sites. In PpOCD, the Asp228 residue (residue corresponding to SpLCD Asp236) interacted with Lys232 (residue corresponding to SpLCD Lys240) with an additional hydrogen bond with O2 atom of NAD+, creating a space optimal for binding of L-ornithine. By contrast, the Asp236 residue of SpLCD was in a slightly different position in SpLCD and formed a hydrogen bond with the O3 atom of NAD+. In addition, the side chain of SpLCD Lys240 was involved in different hydrogen bonding than PpOCD Lys232 (Fig. 5A). The different hydrogen bonds of Asp236, Lys240, and NAD+ in SpLCD created a space that accommodated L-lysine, which has one carbon more than L-ornithine.
Fig. 5. Hypothesized mechanism of catalysis by SpLCD.
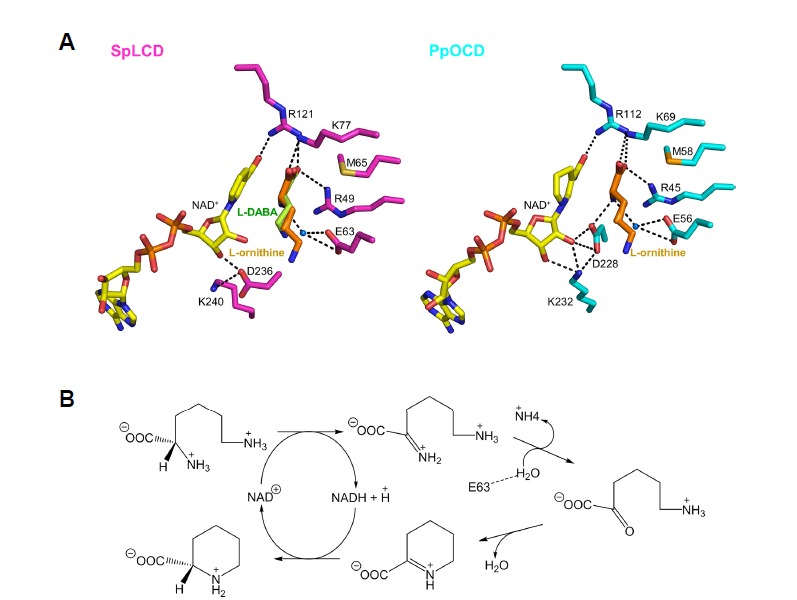
(A) The active sites of SpLCD/NAD+/L-DABA and PpOCD/NADH/L-ornithine are shown. The L-ornithine molecule with PpOCD was visualized in the active sites of SpLCD/NAD+/L-DABA by superimposing SpLCD and PpOCD. (B) Proposed catalytic mechanism of SpLCD. Dashed line represents the hydrogen bond between side chain of E63 and water molecule.
Proposed reaction mechanism of SpLCD
The reaction mechanism of LCD has previously been extensively studied using OCD and two distinct catalytic mechanisms have been proposed (Graupner and White, 2001). In one proposal, the reaction proceeds through direct attack, where a water molecule becomes an imino intermediate (Graupner and White, 2001). The other proposal suggested that the δ-amino group directly attacks the C2 position of L-ornithine without prior hydrolysis with the α-amino group subsequently leaving the intermediate molecule (Goodman et al., 2004).
Despite the presence of two amino groups, α and δ, in L-ornithine, an elegant study using 15N-radiolabled ornithine determined that the α-amino group at the C2 position is the amino group involved in cyclization (Graupner and White, 2001). Of the two proposals, the latter is plausible based on structural studies of PpOCD and SpLCD (Goodman et al., 2004; Ying et al., 2017b). In PpOCD, one water molecule resides near the C2 position of L-ornithine and binds the side chain of Glu56 and the α-amino group. However, the position of the water is not suitable for directly attacking the C2 carbon (Goodman et al., 2004). Despite extensive studies on the mechanism of catalysis of OCD, no structures of OCD in complex with its product, L-Pro, have been reported. In this study, we solved the crystal structure of SpLCD in complex with several reaction products, including L-Pro and L-Pip, enabling us to confirm the previously proposed mechanism of catalysis. The LCD substrate L-lysine contains one carbon more than the OCD substrate L-ornithine. Therefore, the reaction mechanism may be similar. By elucidating the structure of SpLCD and comparing it to the PpOCD structure, we developed the following mechanism. When L-lysine binds to the active site, the hydride ion is transferred to the NAD+ cofactor, resulting in an imino substitution (Fig. 5B). This process is facilitated by a neighboring general base. In OCD, Asp228 has been suggested to be this general base because it is directly coordinated with an α-amino group (Goodman et al., 2004). However, Asp236 of SpLCD, the residue corresponding with Asp236, is located far from the α-amino group (Fig. 5A). Rather, a water molecule forms a hydrogen bond with the α-amino group and the side chain of Glu63 (Fig. 5A), suggesting that the water molecule may serve as general base by donating a proton to Glu63. The next step is direct attack of the δ-amino group to the C2 position of the intermediate molecule in order to form the six-membered ring of L-Pip (Fig. 5B). The leaving group, ammonia, goes away and the electrophilic C2 center is susceptible to hydride ion transfer from NADH, yielding L-Pip and NAD+ as products.
This proposed mechanism is similar to that proposed for PpOCD and is based on an unusual NAD+/NADH cycle. In canonical NAD+ (or NADH) bound enzymes, NAD+ (or NADH) is unidirectionally converted into NADH (or NAD+) to mediate oxidation/reduction reactions. However, in SpLCD, NAD+ is converted into NADH at the intermediate step and then reverted back to NAD+. This mechanism could be beneficial for producing L-Pip, which is a useful precursor in pharmaceutical synthesis, because it does not require NADH molecules to be continually supplied during synthesis.
ACKNOWLEDGMENTS
The authors thank the staff of beamline 7A at the Pohang Light Source and BL26B1 beamline of the SPring-8 for their assistance during X-ray experiments. This study was supported by a grant from the National Research Foundation (NRF) of Korea funded by the Korean government (2015R1A5A1008958 and 2015R1C1A1A01054842) and a grant from Institute for Basic Science (IBS-R021-D1) to H.H.L.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bis DM, Ban YH, James ED, Alqahtani N, Viswanathan R, Lane AL. Characterization of the nocardiopsin biosynthetic gene cluster reveals similarities to and differences from the rapamycin and FK-506 pathways. Chembiochem. 2015;16:990–997. doi: 10.1002/cbic.201500007. [DOI] [PubMed] [Google Scholar]
- Boger DL, Chen J-H, Saionz KW. (−)-Sandramycin: total synthesis and characterization of DNA binding Properties. J Am Chem Soc. 1996;118:1629–1644. [Google Scholar]
- Broquist HP. Lysine-pipecolic acid metabolic relationships in microbes and mammals. Annu Rev Nutr. 1991;11:435–448. doi: 10.1146/annurev.nu.11.070191.002251. [DOI] [PubMed] [Google Scholar]
- Byun SM, Jeong SW, Cho DH, Kim YH. Optimized conversion of L-lysine to L-pipecolic acid using recombinant lysine cyclodeaminase from Streptomyces pristinaespiralis. Biotechnol Biopoc E. 2015;20:73–78. [Google Scholar]
- Cheng Z, Sun L, He J, Gong W. Crystal structure of human micro-crystallin complexed with NADPH. Protein Sci. 2007;16:329–335. doi: 10.1110/ps.062556907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Min K, Mikami B, Yoon HJ, Lee HH. Structural and biochemical studies reveal a putative FtsZ recognition site on the Z-ring stabilizer ZapD. Mol Cells. 2016;39:814–820. doi: 10.14348/molcells.2016.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couty F. Asymmetric syntheses of pipecolic acid and derivatives. Amino Acids. 1999;16:297–320. doi: 10.1007/BF01388174. [DOI] [PubMed] [Google Scholar]
- Durzan DJ. Plant nonprotein amino and imino acids: biological, biochemical, and toxicological properties. Gerald A Rosenthal The Quarterly Rev Biol. 1983;58:260. [Google Scholar]
- Eichhorn E, Roduit J-P, Shaw N, Heinzmann K, Kiener A. Preparation of (S)-piperazine-2-carboxylic acid, (R)-piperazine-2-carboxylic acid, and (S)-piperidine-2-carboxylic acid by kinetic resolution of the corresponding racemic carboxamides with stereoselective amidases in whole bacterial cells. Tetrahedron: Asymmetry. 1997;8:2533–2536. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- Fujii T, Mukaihara M, Tsunekawa H. Biotransformation of l-lysine to l-pipecolic acid catalyzed by l-lysine 6-aminotransferase and pyrroline-5-carboxylate reductase. Biosci Biotechnol Biochem. 2002;66:622–627. doi: 10.1271/bbb.66.622. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Sakurai A. Effect of L-pipecolic acid on flowering in Lemna paucicostata and Lemna gibba. Plant Cell Physiol. 1992;33:419–426. [Google Scholar]
- Gallagher DT, Monbouquette HG, Schroder I, Robinson H, Holden MJ, Smith NN. Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin. J Mol Biol. 2004;342:119–130. doi: 10.1016/j.jmb.2004.06.090. [DOI] [PubMed] [Google Scholar]
- Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PF, Wendisch PP, Wendisch VF. Engineering Corynebacterium glutamicum for fast production of l-lysine and l-pipecolic acid. Appl Microbiol Biotechnol. 2016;100:8075–8090. doi: 10.1007/s00253-016-7682-6. [DOI] [PubMed] [Google Scholar]
- Gatto GJ, Jr, Boyne MT.2nd Kelleher NL, Walsh CT. Biosynthesis of pipecolic acid by RapL, a lysine cyclodeaminase encoded in the rapamycin gene cluster. J Am Chem Soc. 2006;128:3838–3847. doi: 10.1021/ja0587603. [DOI] [PubMed] [Google Scholar]
- Germann UA, Shlyakhter D, Mason VS, Zelle RE, Duffy JP, Galullo V, Armistead DM, Saunders JO, Boger J, Harding MW. Cellular and biochemical characterization of VX-710 as a chemosensitizer: reversal of P-glycoprotein-mediated multidrug resistance in vitro. Anticancer Drugs. 1997;8:125–140. doi: 10.1097/00001813-199702000-00004. [DOI] [PubMed] [Google Scholar]
- Goodman JL, Wang S, Alam S, Ruzicka FJ, Frey PA, Wedekind JE. Ornithine cyclodeaminase: structure, mechanism of action, and implications for the mu-crystallin family. Biochemistry. 2004;43:13883–13891. doi: 10.1021/bi048207i. [DOI] [PubMed] [Google Scholar]
- Gouesbet G, Jebbar M, Talibart R, Bernard T, Blanco C. Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiology. 1994;140:2415–2422. doi: 10.1099/13500872-140-9-2415. [DOI] [PubMed] [Google Scholar]
- Graupner M, White RH. Methanococcus jannaschii generates L-proline by cyclization of L-ornithine. J Bacteriol. 2001;183:5203–5205. doi: 10.1128/JB.183.17.5203-5205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RN, Spenser ID. Biosynthesis of the piperidine nucleus. The mode of incorporation of lysine into pipecolic acid and into piperidine alkaloids. J Biol Chem. 1969;244:88–94. [PubMed] [Google Scholar]
- He M. Pipecolic acid in microbes: biosynthetic routes and enzymes. J Ind Microbiol Biotechnol. 2006;33:401–407. doi: 10.1007/s10295-006-0078-3. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yang X, Yin DH, Mahadevan J, Kuczera K, Schowen RL, Borchardt RT. Computational characterization of substrate binding and catalysis in S-adenosylhomocysteine hydrolase. Biochemistry. 2001;40:15143–15152. doi: 10.1021/bi015690d. [DOI] [PubMed] [Google Scholar]
- Jin X, Geiger JH. Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase. Acta Crystallogr D Biol Crystallogr. 2003;59:1154–1164. doi: 10.1107/s0907444903008205. [DOI] [PubMed] [Google Scholar]
- Kadouri-Puchot C, Comesse S. Recent advances in asymmetric synthesis of pipecolic acid and derivatives. Amino Acids. 2005;29:101–130. doi: 10.1007/s00726-005-0193-x. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. JMolBiol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lemire A, Charette AB. Stereoselective syntheses of l-pipecolic acid and (2S,3S)-3-hydroxypipecolic acid from a chiral N-imino-2-phenyl-1,2-dihydropyridine intermediate. J Org Chem. 2010;75:2077–2080. doi: 10.1021/jo902527s. [DOI] [PubMed] [Google Scholar]
- Malinowska K, Cavarretta IT, Susani M, Wrulich OA, Uberall F, Kenner L, Culig Z. Identification of mu-crystallin as an androgen-regulated gene in human prostate cancer. Prostate. 2009;69:1109–1118. doi: 10.1002/pros.20956. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalik SJ, Moser HW, Watkins PA, Danks DM, Poulos A, Rhead WJ. Peroxisomal L-pipecolic acid oxidation is deficient in liver from zellweger syndrome patients. Pediatr Res. 1989;25:548–552. doi: 10.1203/00006450-198905000-00024. [DOI] [PubMed] [Google Scholar]
- Miller DL, Rodwell VW. Metabolism of basic amino acids in Pseudomonas putida. Catabolism of lysine by cyclic and acyclic intermediates. J Biol Chem. 1971;246:2758–2764. [PubMed] [Google Scholar]
- Moulin M, Deleu C, Larher F, Bouchereau A. The lysine-ketoglutarate reductase-saccharopine dehydrogenase is involved in the osmo-induced synthesis of pipecolic acid in rapeseed leaf tissues. Plant Physiol Biochem. 2006;44:474–482. doi: 10.1016/j.plaphy.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring A-C, Zeier J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell. 2012;24:5123–5141. doi: 10.1105/tpc.112.103564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neshich IAP, Kiyota E, Arruda P. Genome-wide analysis of lysine catabolism in bacteria reveals new connections with osmotic stress resistance. ISME J. 2013;7:2400–2410. doi: 10.1038/ismej.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pálfi G, Dézsi L. Pipecolic acid as an indicator of abnormal protein metabolism in diseased plants. Plant and Soil. 1968;29:285–291. [Google Scholar]
- Rao VV, Chang YF. Assay for L-pipecolate oxidase activity in human liver: detection of enzyme deficiency in hyperpipecolic acidaemia. Biochim Biophys Acta. 1992;1139:189–195. doi: 10.1016/0925-4439(92)90133-8. [DOI] [PubMed] [Google Scholar]
- Tani Y, Miyake R, Mihara H. Functional expression of l-lysine α-oxidase from Scomber japonicus in Escherichia coli for one-pot synthesis of l-pipecolic acid from dl-lysine. Appl Microbiol Biotechnol. 2015;99:5045–5054. doi: 10.1007/s00253-014-6308-0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsotsou GE, Barbirato F. Biochemical characterisation of recombinant Streptomyces pristinaespiralis L-lysine cyclodeaminase. Biochimie. 2007;89:591–604. doi: 10.1016/j.biochi.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Watanabe LA, Haranaka S, Jose B, Yoshida M, Kato T, Moriguchi M, Soda K, Nishino N. An efficient access to both enantiomers of pipecolic acid. Tetrahedron: Asymmetry. 2005;16:903–908. [Google Scholar]
- Wilkinson TJ, Stehle NW, Beak P. Enantioselective syntheses of 2-Alkyl- and 2,6-dialkylpiperidine alkaloids: preparations of the hydrochlorides of (−)-coniine, (−)-solenopsin A, and (−)-dihydropinidine. Organic Lett. 2000;2:155–158. doi: 10.1021/ol9912534. [DOI] [PubMed] [Google Scholar]
- Ying HX, Wang J, Chen KQ. Enhanced conversion of l-lysine to l-pipecolic acid using a recombinant Escherichia coli containing lysine cyclodeaminase as whole-cell biocatalyst. J Mol Catal B-Enzym. 2015;117:75–80. [Google Scholar]
- Ying H, Tao S, Wang J, Ma W, Chen K, Wang X, Ouyang P. Expanding metabolic pathway for de novo biosynthesis of the chiral pharmaceutical intermediate l-pipecolic acid in Escherichia coli. Microb Cell Fact. 2017a;16:52. doi: 10.1186/s12934-017-0666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Wang J, Shi T, Zhao Y, Wang X, Ouyang P, Chen K. Studies of lysine cyclodeaminase from Streptomyces pristinaespiralis: Insights into the complex transition NAD+ state. Biochem Biophys Res Commun. 2017b;S0006-291X:32211–32218. doi: 10.1016/j.bbrc.2017.11.034. [DOI] [PubMed] [Google Scholar]
- Zacharius RM, Thompson JF, Steward FC. The detection, isolation and identification of L(−)pipecolic acid in the non-protein fraction of beans (Phascolus vulgaris)1,2. J Am Chem Soc. 1954;76:2908–2912. [Google Scholar]


