Abstract
Flowering time is determined by florigens. These genes include, Heading date 3a (Hd3a) and Rice FT 1 (RFT1) in rice, which are specifically expressed in the vascular tissues of leaves at the floral transition stage. To study the cis-regulatory elements present in the promoter region of Hd3a, we generated transgenic plants carrying the 1.75-kb promoter fragment of Hd3a that was fused to the β-glucuronidase (GUS) reporter gene. Plants expressing this construct conferred a vascular cell-specific expression pattern for the reporter gene. However, GUS was expressed in leaves at all developmental stages, including the early seedling stage when Hd3a was not detected. Furthermore, the reporter was expressed in roots at all stages. This suggests that the 1.75-kb region lackings cis-elements that regulate leaf-specific expression at the appropriate developmental stages. Deletion analyses of the promoter region indicated that regulatory elements determining vascular cell-specific expression are present in the 200-bp region between -245 bp and -45 bp from the transcription initiation site. By transforming the Hd3a–GUS construct to rice cultivar ‘Taichung 65’ which is defective in Ehd1, we observed that Ehd1 is the major regulatory element that controls Hd3a promoter activity.
Keywords: β-glucuronidase (GUS), cis- elements, Ehd1, Hd3a, promoter, vascular bundle
INTRODUCTION
Flowering is an important biological process in plant reproduction. The responsible molecular mechanisms have been broadly studied in a long-day (LD) flowering plant, Arabidopsis (Arabidopsis thaliana) (Imaizumi and Kay, 2006; Tsuji et al., 2011), and in a short-day (SD) flowering plant, rice (Oryza sativa) (Imaizumi and Kay, 2006; Lee and An, 2015; Cho et al., 2017). In Arabidopsis, flowering time is determined by the florigen, FLOWERING LOCUS T (FT) (Takada and Goto, 2003; Notaguchi et al., 2008). Its ortholog, Hd3a, has been identified in rice through QTL mapping of a cross between ‘Nipponbare’ and ‘Kasalath’ cultivars (Yano and Sasaki, 1997). Whereas RNAi lines of Hd3a do not flower until 300 days after germination (DAG) (Komiya et al., 2008), overexpression of this gene causes flowering at the callus induction stage (Monna et al., 2002; Hori et al., 2013). This indicates that it has a major role in controlling flowering time. Hd3a is preferentially functional under SD conditions and its mRNA is diurnally expressed, showing a peak at approximately 2 h after sunrise (Izawa et al., 2002; Ryu et al., 2009; Lee et al., 2010; Yang et al., 2013).
Florigens are expressed in the leaf phloem and transported to the shoot apical meristem (SAM) to provide a flowering signal (Corbesier et al., 2007; Jaeger and Wigge, 2007; Lin et al., 2007; Tamaki et al., 2007). Hd3a protein interacts with 14-3-3 protein, generating a complex that moves to the nucleus and binds with OsFD1. This ‘florigen activation complex’ of Hd3a, OsFD1, and 14-3-3 activates expression of OsMADS15, a rice APETALA1 ortholog (Taoka et al., 2011).
Hd3a and florigens from other species have diverse roles in plant development (Pin and Nilsson, 2012). For example, FT stimulates the opening of stomata in Arabidopsis (Kinoshita et al., 2011). SINGLE FLOWER TRUSS (SFT), a florigen in Lycopersicon esculentum, regulates leaf complexity and inflorescence pattern (Lifschitz et al., 2006; Krieger et al., 2010). In rice, Hd3a promotes lateral branching (Tsuji et al., 2015), and its expression is induced by Ehd1 (Doi et al., 2004). Phosphorylation of the receiver domain of Ehd1 induces dimerization of the protein, which is required for Ehd1 to stimulate Hd3a expression (Cho et al., 2016). In addition, Hd1 appears to induce Hd3a expression directly (Yano et al., 2000).
Ehd1 is controlled by several upstream regulatory factors (Lee and An, 2015). During the early vegetative stages, its expression is inhibited by several repressors, including three CO-like proteins: Hd1, OsCOL4, and Ghd7 (Yano et al., 2000; Lee et al., 2010; Zhao et al., 2012). OsLFL1, SNB, and OsIDS1 are other repressors that also function upstream of Ehd1 (Lee et al., 2014). At later stages of development, expression of those repressors is reduced due to the function of regulatory genes farther upstream. For example, Ghd7 expression is inhibited by a chromatin remodeling factor, OsTrx1, and its interacting partner, Ehd3 (Matsubara et al., 2011; Choi et al., 2014). Another chromatin remodeling factor, OsVIL2, inhibits expression of OsLFL1 by forming a complex with polycomb repressive complex 2 (Yang et al., 2013). Expression of SNB and OsIDS1 is suppressed by increased expression of microRNA172 at the later stage of vegetative development (Lee et al., 2014).
Several genes involved in controlling flowering time are preferentially expressed in the vascular bundle cells. Another rice florigen gene, RFT1, is also expressed in vascular tissues (Komiya et al., 2009). An FT homolog from maize (Zea mays), CENTRORADIALIS8, is also specifically expressed in the leaf vascular cells (Meng et al., 2011). Similar to these florigen genes, Ehd1 is specifically expressed in the vascular bundle (An et al., 2004; Endo et al., 2005; Giakountis and Coupland, 2008). Ghd7 expression is high in the vascular cells of leaf blades but low in the cells of roots and leaf sheaths (Xue et al., 2008).
In addition to flowering genes, numerous others are preferentially expressed in vascular tissues (Hernandez-Garcia and Finer, 2014). They include various genes from viruses and bacteria that are preferentially expressed in the phloem. Genes from rice tungro bacilliform virus (RTBV) are specifically expressed in phloem cells (Yin and Beachy, 1995), while genes from commelina yellow mottle virus are expressed in vascular bundles (Medberry et al., 1992). The activity of a promoter from wheat dwarf geminivirus is specific to vascular cells (Dinant et al., 2004). Moreover, promoters from Agrobacterium genes rolB and rolC are expressed only in the vasculature (Schmülling et al., 1989).
Genes involved in nutrient transport show vascular specificity. For example, Arabidopsis thaliana Sucrose Transporter 2 (AtSUC2), which encodes a sucrose–H+ symporter needed for long-distance sucrose transport, is expressed in the phloem cells of photosynthetic leaves (Stadler and Sauer, 1996; Gottwald et al., 2000). In rice, OsSUT1 is expressed in the phloem cells of leaf blades and scutellar vascular bundles (Scofield et al., 2007). Other sugar transporters, i.e., sugars will eventually be exported transporters (SWEETs), are expressed in phloem cells, including AtSWEET11 and AtSWEET12 in the rosette leaves of Arabidopsis (Chen et al., 2012) and the OsSWEET11 promoter, which specifically induces the expression of β-glucuronidase (GUS) in the phloem cells of rice (Ma et al., 2017).
A promoter from glutamine synthetase 3A (GS3A) in Pisum sativum, encoding a protein that functions in ammonia assimilation, shows phloem specificity in transgenic Nicotiana tabacum and Medicago sativa (Brears et al., 1991; Pageau et al., 2006). A gene encoding Arabidopsis H+-ATPase isoform 3 (AHA3), a major target for 14-3-3 proteins, is expressed exclusively in the phloem cells of the leaf, root, stem, and flower at all developmental stages (DeWitt et al., 1991; Alsterfjord et al., 2004). Finally, two genes encoding rice phloem proteins RPP16 and RPP17, which are homologous to viral movement proteins, are specifically expressed in the phloem parenchyma cells of the leaf blade and root (Asano et al., 2002).
In this study, we identified a 200-bp region of the Hd3a promoter that directs the expression of the GUS marker gene within the parenchyma cells of vascular tissues.
MATERIALS AND METHODS
Plant materials and growth conditions
Plants of Oryza sativa ‘Longjing27’ and ‘Taichung65’ were used. Seeds of those cultivars were germinated either on a 1/2 Murashige and Skoog (MS) medium containing 3% sucrose or directly in soil as previously reported (Cho et al., 2016). The seedlings were then cultured in controlled growth rooms under SD conditions (12 h light at 28°C/12 h darkness at 22°C; 50% humidity).
Generation of transgenic plants carrying pHd3a–GUS fusions
The 1.75-kb fragment of the Hd3a promoter region was amplified with the pair of primers as previously reported (Ishikawa et al., 2005; Tamaki et al., 2007). Genomic DNA from ‘Dongjin’ rice was used as template. Three deletions from that fragment were generated using the primers listed in Supplementary Table S1 (Fig. 1A). The promoter fragments were inserted in the KpnI and BamHI restriction sites of the pGA3519 binary vector that carries the GUS coding region followed by the nopaline synthase terminator (Yoon et al., 2014). In the constructs, the Hd3a coding region located 153 bp from the ATG start codon was fused to the GUS coding region within the same reading frame. All constructs were confirmed by sequencing the insertion. The binary vectors were transformed into Agrobacterium tumefaciens LBA4404 (An et al., 1989). Transgenic rice plants were generated via the Agrobacterium-mediated co-cultivation method as previously reported (Lee et al., 1999), using embryonic calli derived from mature seeds of ‘Longjing27’ and ‘Taichung65’ rice. Plants carrying this Hd3a promoter–GUS chimeric gene were selected by PCR using genomic DNA isolated from hygromycin-resistant seedlings as templates and primers located in the Hd3a promoter and GUS coding region (Supplementary Table S1). These transgenics were cultured in the growth room until maturity. After their seeds were harvested, we selected two independent transgenic lines with high GUS expression.
Fig. 1. Schematic representation of Hd3a promoter-GUS reporter constructs and histochemical localization of GUS.
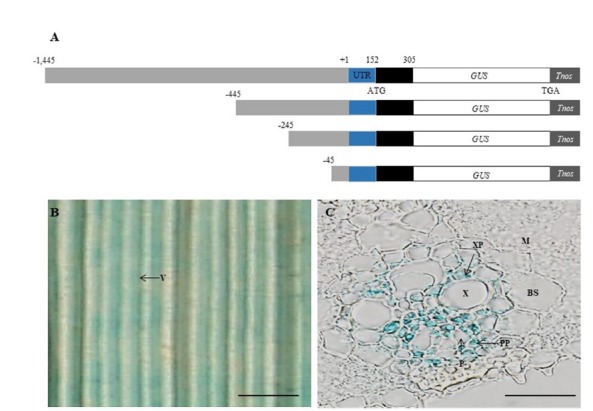
(A) Hd3a genomic fragments comprising −1,445, −445, −245, and −45 bp promoter regions (grey box), the 152-bp 5′ UTR (blue box) and 153-bp coding region of Hd3a (black box) were connected to GUS coding region and nopaline synthase terminator (Tnos). (B) Leaf blade of −1,445 Hd3a promoter–GUS transgenic at ZT 1 on 35 DAG. (C) Transverse section of leaf blade in Panel B. BS, bundle sheath cells; M, mesophyll cells; P, phloem; PP, phloem parenchyma; V, vascular bundle; X, xylem; XP, xylem parenchyma. Scale bars =1 mm (B) and 20 μm (C).
Histochemical GUS staining
Histochemical GUS staining was performed as previously described (Yoon et al., 2017). Seeds of the transgenic plants were germinated on an MS medium containing 50 μg L-1 of hygromycin and the seedlings were cultured in the controlled growth room under SD conditions. Leaf blades were collected and placed in a GUS-staining solution containing 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, 50 mM sodium phosphate (pH 7.0), 0.1% Triton X-100, 10 mM EDTA, 1% DMSO, and 1.0 mg mL−1 of 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid. After vacuum-infiltration for 30 min, the samples were incubated at 37°C for 10 h. Chlorophylls were removed by incubating in 70% ethanol at 65°C. The tissues were dehydrated in an ethanol series (50, 70, 90, and 100%) and then treated with tert-butyl alcohol. They were either observed immediately under a BX61 optical microscope (Olympus, http://www.olympus-global.com/en/) or fixed in paraffin and sectioned (10 μm thickness) with a microtome (Model 2165; Leica Microsystems, http://www.leica-microsystems.com/) before being observed under that microscope.
RNA isolation and quantitative RT-PCR analyses
Leaf blades and root tissues were harvested at 7, 14, 21, 28, and 35 DAG from SD-grown ‘Longjing27’ plants. Total RNA was isolated from the samples using RNAiso Plus (TaKaRa, Shiga, Japan; http://www.takarabio.com) and qualified by a Nano Nanodrop ND-2000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA; http://www.nanodrop.com) as described previously (Cho et al., 2016). Complementary DNA (cDNA) was made with 10 ng of the oligo (dT)18 primer, 2.5 mM deoxy ribonucleotide triphosphates, and Moloney murine leukemia virus reverse transcriptase. Quantitative real-time RT-PCR (qRT-PCR) was performed with the synthesized cDNAs as templates and primers listed in Supplementary Table S1, using SYBR Premix Ex Taq™ II (TaKaRa) and the Rotor-Gene 6000 instrument system (Corbett Research, Sydney, Australia; http://www.corbettlifescience.com). Rice Ubi1 served as an internal control. At least three biological replicates were analyzed. We used the data only when the melting curve showed a single sharp peak.
RESULTS
Expression patterns of the Hd3a promoter–GUS fusion
Transgenic plants containing the 1.75-kb Hd3a promoter showed reporter gene expression in the phloem and xylem parenchyma cells of the leaf blades (Hayama et al., 2003; Tamaki et al., 2007). However, the organ-preferential expression patterns of the construct were not examined. An identical promoter fragment was isolated that consisted of the 1,445-bp promoter region, the 152-bp 5′ untranslated region (UTR), and the 153-bp coding region of Hd3a (Fig. 1A). The promoter fragment was placed upstream of GUS in the same reading frame. Several transgenic rice plants expressing the fusion molecules were generated and two independent lines of the primary transgenics were grown under SD conditions. Because expression of Hd3a peaks at approximately 35 DAG under short days (Komiya et al., 2008; Ryu et al., 2009; Lee et al., 2010), we conducted a histochemical GUS assay of leaf blades from the transgenic plants at that time point and found that the GUS reporter gene was expressed (Fig. 1B). Cross-sectioning revealed GUS staining in the parenchyma cells within the vascular bundles (Fig. 1C). This pattern of tissue expression is similar to that previously reported for Hd3a in rice (Tamaki et al., 2007).
We also examined the developmental expression patterns of Hd3a and GUS in transgenic plants carrying the Hd3a promoter–GUS construct at 7, 14, 21, 28, and 35 DAG (Fig. 2). Transcript levels of Hd3a were low during the early developmental stages but began to increase at 21 DAG before reaching a high level at 35 DAG, as previously reported (Fig. 2A). However, the GUS reporter was constitutively expressed in the leaves throughout all five stages (Fig. 2B). Histochemical analyses also indicated that the transgenics expressed the reporter protein constitutively during their development (Figs. 3A). This demonstrated that the 1.75-kb fragment did not contain the regulatory element needed to determine a proper pattern of developmental expression for Hd3a. To examine whether Hd3a is preferentially expressed in a certain part of the leaf, we measured expression levels of the gene by qRT-PCR from three different positions. Transcript levels of this gene were similar among samples from the top, middle, and bottom portions of the leaf blade (Fig. 3I). Similar results were obtained for expression levels of the GUS reporter (Fig. 3J). Histochemical GUS analyses also revealed that the reporter was expressed at a similar level regardless of position (Figs. 3K–3M).
Fig. 2. Developmental expression patterns of Hd3a and GUS in leaf blade (LB) and root (R).
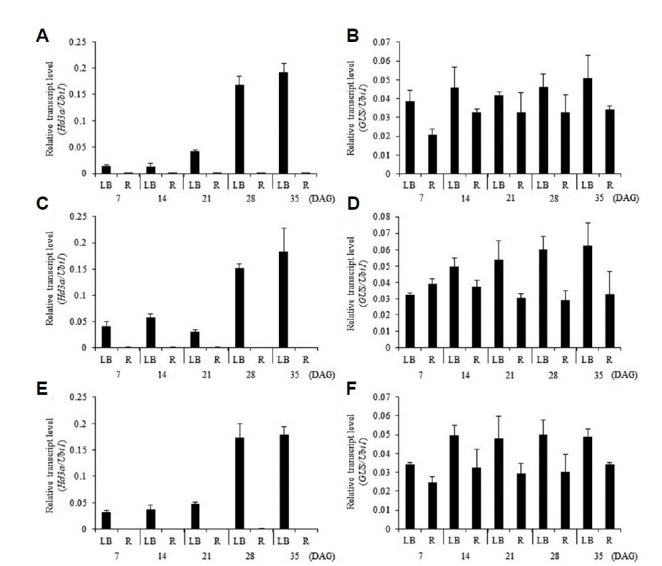
Quantitative real-time RT-PCR analyses of transcript abundance of Hd3a (A, C, and E) and GUS (B, D, and F) from −1,445 Hd3a promoter–GUS (A, B), −445 Hd3a promoter–GUS (C, D), and −245 Hd3a promoter–GUS (E, F). Error bars represent standard deviations. n = 6. DAG, days after germination.
Fig. 3. Histochemical localization of GUS activity in leaf blades at five developmental stages and expression pattern of Hd3a at 3 positions along leaf blade top, middle and bottom.
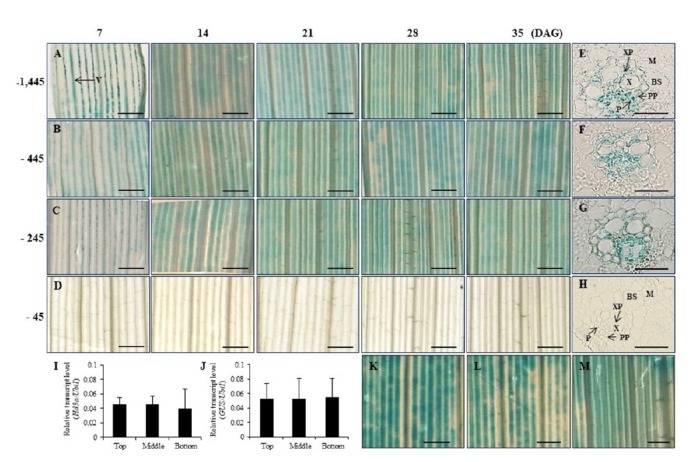
Leaf blades of transgenic rice plants carrying −1,445 Hd3a promoter–GUS (A), −445 Hd3a promoter–GUS (B), −245 Hd3a promoter–GUS (C), and −45 Hd3a promoter–GUS (D). Cross-sections of leaf blades at 35 DAG of −1,445 (E), −445 (F), −245 (G), and −45 (H) construct. Quantitative real-time RT-PCR analyses of Hd3a transcript (I) and −1,445 Hd3a promoter-driven GUS transcript (J) at top, middle, and bottom at ZT 1 on 35 DAG. Histochemical localization of GUS activity in leaf blades in Panel A at top (K), middle (L), and bottom (M). Mean values from two biological replicates are shown. Error bars represent standard deviation in (I and J). n = 6. Scale bars = 20 μm in Panels E, F, G and H; 1 mm in others. BS, bundle sheath cells; M, mesophyll cells; P, phloem; PP, phloem parenchyma; V, vascular bundle; X, xylem; XP, xylem parenchyma.
Transcript of Hd3a was not found in the roots at any time during the vegetative growth period (Fig. 2A). The same has been reported previously (Tamaki et al., 2007). However, GUS transcript was detectable in roots from transgenic plants expressing the reporter gene under the 1.75-kb Hd3a promoter (Fig. 2B), and a considerable amount of transcript accumulated in the roots during all five developmental stages. GUS staining also showed that the reporter was expressed in roots at all stages (Figs. 4A–4E). These results suggested that the promoter fragment did not contain the regulatory elements needed to suppress Hd3a expression in the roots.
Fig. 4. Histochemical localization of GUS activity in roots at five developmental stages.
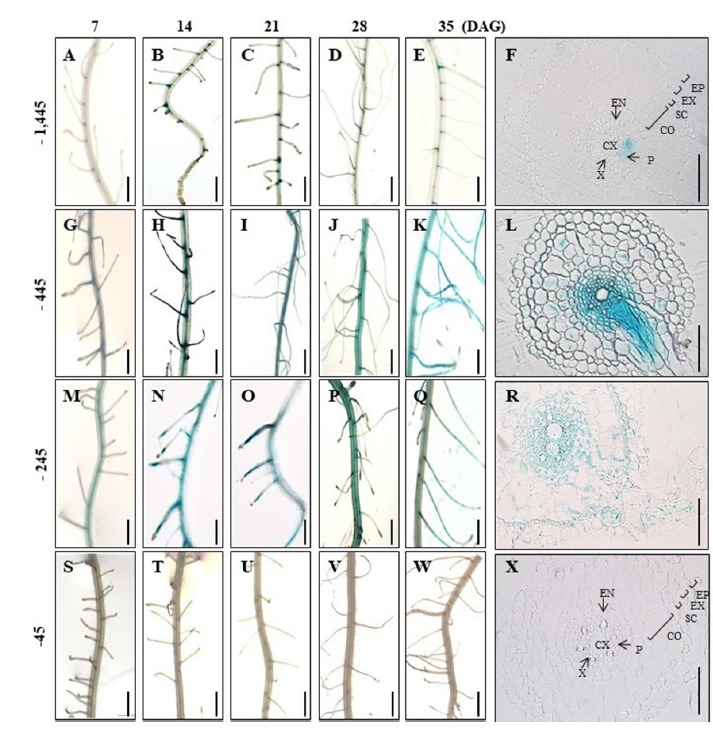
(A–E) Roots of transgenic plants carrying −1,445 Hd3a promoter–GUS at 7 (A), 14 (B), 21 (C), 28 (D), and 35 (E) DAG. (G–K) Roots of transgenic plants carrying −445 Hd3a promoter–GUS at 7 (G), 14 (H), 21 (I), 28 (J), and 35 (K) DAG. (M–Q) Roots of transgenic plants carrying −245 Hd3a promoter–GUS at 7 (M), 14 (N), 21 (O), 28 (P), and 35 (Q) DAG. (S–W) Roots of transgenic plants carrying −45 Hd3a promoter–GUS at 7 (S), 14 (T), 21 (U), 28 (V), and 35 (W) DAG. Cross-sections of roots at 35 DAG from −1,445 Hd3a promoter–GUS (F), −445 Hd3a promoter–GUS (L), −245 Hd3a promoter–GUS (R), and −45 Hd3a promoter–GUS (X). Co, cortex; CX; central xylem; En, endodermis, Ep, epidermis; Ex, exodermis; P, phloem; Sc, sclerenchyma; V, vascular bundle; X, xylem. Scale bars = 50 μm in Panels F, L, R, and X; 5 mm in others.
Identification of the region responsible for vascular cell-specific expression
Although the 1.75-kb fragment did not contain elements for organ and developmental specificities, it did carry those for vascular-tissue specificity. To find the responsible region, we made three deletion constructs (Fig. 1A). The first was a deletion of 1,000-bp upstream region from the 1.75-kb fragment, leaving the 445-bp promoter region. The second was a further 200-bp deletion that resulted in the 245-bp promoter region. The third construct was made by an additional 200-bp deletion from the second construct, leaving only the 45-bp promoter region. All three constructs retained the 152-bp UTR and 153-bp coding region of Hd3a.
From several transformed plants, we selected two independent plants from each deletion and stained for GUS activity. Their progeny were grown under SD conditions and their GUS expression patterns were studied at 7, 14, 21, 28, and 35 DAG. The first- and second-deletion constructs showed reporter gene expression that was specific to the vascular tissue in the leaves at all developmental stages (Figs. 3B and 3C). However, plants carrying the third-deletion construct did not have any detectable level of GUS activity (Figs. 3D). Cross-sections of the leaf blades revealed staining in both phloem parenchyma cells and xylem parenchyma cells within the vascular bundles of the three longer constructs (Figs. 3E, 3F, and 3G), but not from the smaller fragment (Fig. 3H). Mesophyll cells and bundle sheath cells were not stained. This indicated that elements responsible for vascular cell-specific expression in the leaves were located in the 200-bp region between -45 and -245 from the transcription initiation site.
Analyses of roots from the transgenic plants also showed that plants expressing the first- and second-deletion constructs presented vascular tissue-preferential expression at all stages (Figs. 4G–4K, 4M–4Q). However, GUS expression was not detected from plants carrying the third construct (Figs. 4S–4W). This demonstrated that cis-elements for vascular-preferential expression in the roots also resided within the 200-bp region identified from the leaf analyses.
Role of Ehd1 in expression of Hd3a–GUS
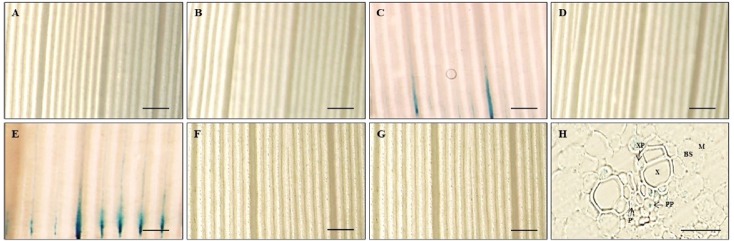
Hd3a functions immediately downstream of Ehd1 (Doi et al., 2004). To examine whether the expression of the 1.75-kb promoter fragment was dependent upon Ehd1, we transformed the 1.75-kb Hd3a promoter–GUS construct into ‘Taichung65’ rice, which carried a defective Ehd1. Analyses of seven independent transgenic plants showed that five displayed no GUS activity regardless of developmental stage (Figs. 5A, 5B, 5D, 5F and 5G). However, a low level of GUS staining was observed in vascular tissues from two plants (Figs. 5C and 5E). These findings indicated that Ehd1 is a major regulatory factor for phloem-specific expression of Hd3a.
Fig. 5. Histochemical analyses of GUS in ‘Taichung65’ rice carrying −1,445 Hd3a promoter-GUS.
(A–G) Leaf blades of 7 independent plants were analyzed at 45 DAG. (H) Cross-section of leaf blade in (E). Scale bars: 1 mm (A–G) or 20 μm (H). DAG, days after germination. BS, bundle sheath cells; M, mesophyll cells; P, phloem; PP, phloem parenchyma; V, vascular bundle; X, xylem; XP, xylem parenchyma.
DISCUSSION
We studied the 1.75-kb promoter fragment of Hd3a to investigate cis-regulatory elements present in the region. Our objective in using this fragment was to demonstrate that the promoter-driven GFP reporter protein is produced in leaf phloem and moves to the SAM. Our findings support earlier conclusions that Hd3a is a mobile signal that transmits the flowering signal from leaves to the shoot apex (Tamaki et al., 2007; Komiya et al., 2009). In this study, the -1.75-kb promoter-driven reporter gene was expressed in the vascular bundles of leaves and in the roots at all developmental stages, but no GUS activity was observed in the SAM (Supplementary Fig. S1). These results are quite comparable to the tissue specificity of FT expression in Arabidopsis (Takada and Goto, 2003). In Arabidopsis, the 5.7-kb FT promoter sequence upstream of the translation start site is required for mediating spatial and temporal expression of the gene (Adrian et al., 2010). Shortening the promoter to 4.0 kb obliterates that ability, indicating that a promoter region of that size is not large enough to determine promoter specificity. Therefore, all of these results suggest that the necessary regulatory elements occur in the region between 4 kb and 5.7 kb for FT promoter specificity.
Although the 1.75-kb Hd3a promoter fragment lacks elements controlling leaf specificity and the developmental expression pattern of the gene, it confers vascular cell-specific expression within the leaf blades. To identify the cis-regulatory elements involved in the latter, we generated truncated promoters and found that the removal from -1445 to -245 did not affect vasculature specificity. However further deletion of 200 bp abolished such specificity. This indicated that the 200-bp region between -245 and -45 carries the cis-elements needed for cell specificity.
Using the rice RTBV promoter, Yin et al. (1997) have identified the GCA repeat element GCATC (N)9 GCATC that is required for phloem-specific expression. Two similar elements, GCATA(N)10GCATA and GCACC(N)8GCAGC, have been identified from the phloem-specific promoters of Arabidopsis H+-ATPase isoform 3 (AHA3) and maize sucrose synthase-1 (Shl), respectively (Werr et al., 1985; DeWitt et al., 1991). The GCA repeat is located within the phloem-specific region at −193 GCAGA(N)9GCATG−175 of the Hd3a promoter region. Another phloem-specific element, the GATA motif, has been identified from the CaMV 35S promoter that is strongly expressed in vascular tissues (Lam and Chua, 1989). That element is also present in the AHA3 promoter as the GAGATGATA motif (Yin et al., 1997) and in the phloem-specific glutamine synthetase 3A (GS3A) promoter as the TAGTAGATA motif (Brears et al., 1991). The GATA motif exists within the phloem-specific region at −144 GAGATGATA −136 of the Hd3a promoter. Further study is needed to examine whether these elements indeed determine the vascular specificity of that promoter.
Supplementary data
ACKNOWLEDGMENTS
This work was supported in part by a grant from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center, No. PJ013210), Rural Development Administration, Republic of Korea. We thank Priscilla Licht for her critical proofreading of the manuscript.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F. Cis-regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22:1425–1440. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsterfjord M, Sehnke PC, Arkell A, Larsson H, Svennelid F, Rosenquist M, Ferl RJ, Sommarin M, Larsson C. Plasma membrane H+-ATPase and 14-3-3 isoforms of Arabidopsis leaves: evidence for isoform specificity in the 14-3-3/H+ -ATPase interaction. Plant Cell Physiol. 2004;45:1202–1210. doi: 10.1093/pcp/pch136. [DOI] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB. Plant Molecular Biology Manual. A3. Dordrecht: Kluwer Academic Publisher; 1989. Binary vectors; pp. 1–19. [Google Scholar]
- An HL, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- Asano T, Kusano H, Okuda T, Kubo N, Shimada H, Kadowaki K. Rpp16 and Rpp17, from a common origin, have different protein characteristics but both genes are predominantly expressed in rice phloem tissues. Plant Cell Physiol. 2002;43:668–674. doi: 10.1093/pcp/pcf083. [DOI] [PubMed] [Google Scholar]
- Brears T, Walker EL, Coruzzi GM. A promoter sequence involved in cell-specific expression of the pea glutamine synthetase GS3A gene in organs of transgenic tobacco and alfalfa. Plant J. 1991;1:235–244. doi: 10.1111/j.1365-313x.1991.00235.x. [DOI] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012;335:207–211. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- Cho LH, Yoon J, Pasriga R, An G. Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 2016;170:2159–2171. doi: 10.1104/pp.15.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho LH, Yoon J, An G. The control of flowering time by environmental factors. Plant J. 2017;90:708–719. doi: 10.1111/tpj.13461. [DOI] [PubMed] [Google Scholar]
- Choi SC, Lee S, Kim SR, Lee YS, Liu C, Cao X, An G. Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with Early heading date3. Plant Physiol. 2014;164:1326–1337. doi: 10.1104/pp.113.228049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- DeWitt ND, Harper JF, Sussman MR. Evidence for a plasma membrane proton pump in phloem cells of higher plants. Plant J. 1991;1:121–128. doi: 10.1111/j.1365-313x.1991.00121.x. [DOI] [PubMed] [Google Scholar]
- Dinant S, Ripoll C, Pieper M, David C. Phloem specific expression driven by wheat dwarf geminivirus V-sense promoter in transgenic dicotyledonous species. Physiol Plant. 2004;121:108–116. doi: 10.1111/j.0031-9317.2004.00296.x. [DOI] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-Iike gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A. Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell. 2005;17:1941–1952. doi: 10.1105/tpc.105.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giakountis A, Coupland G. Phloem transport of flowering signals. Curr Opin Plant Biol. 2008;11:687–694. doi: 10.1016/j.pbi.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA. 2000;97:13979–13984. doi: 10.1073/pnas.250473797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014;217:109–119. doi: 10.1016/j.plantsci.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Hori K, Ogiso-Tanaka E, Matsubara K, Yamanouchi U, Ebana K, Yano M. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 2013;76:36–46. doi: 10.1111/tpj.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Tamaki S, Yokoi S, Inagaki N, Shinomura T, Takano M, Shimamoto K. Suppression of the floral activator gene Hd3a is the principal cause of the night break effect in rice. Plant Cell. 2005;17:3326–3336. doi: 10.1105/tpc.105.037028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002;16:2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17:1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Ono N, Hayashi Y, Morimoto S, Nakamura S, Soda M, Kato Y, Ohnishi M, Nakano T, Inoue S, et al. Flowering locus T regulates stomatal opening. Curr Biol. 2011;21:1232–1238. doi: 10.1016/j.cub.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet. 2010;42:459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- Lam E, Chua NH. ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell. 1989;1:1147–1156. doi: 10.1105/tpc.1.12.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, An G. Regulation of flowering time in rice. J Plant Biol. 2015;58:353–360. [Google Scholar]
- Lee S, Jeon JS, Jung KH, An G. Binary vectors for efficient transformation of rice. J Plant Biol. 1999;42:310–316. [Google Scholar]
- Lee YS, Jeong DH, Lee DY, Yi J, Ryu CH, Kim SL, Jeong HJ, Choi SC, Jin P, Yang J, et al. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 2010;63:18–30. doi: 10.1111/j.1365-313X.2010.04226.x. [DOI] [PubMed] [Google Scholar]
- Lee YS, Lee DY, Cho LH, An G. Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice. 2014;7:31. doi: 10.1186/s12284-014-0031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MK, Belanger H, Lee YJ, Varkonyi-Gasic E, Taoka KI, Miura E, Xoconostle-Cázares B, Gendler K, Jorgensen RA, Phinney B, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhang D, Miao Q, Yang J, Xuan Y, Hu Y. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 2017;58:863–873. doi: 10.1093/pcp/pcx040. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 2011;66:603–612. doi: 10.1111/j.1365-313X.2011.04517.x. [DOI] [PubMed] [Google Scholar]
- Medberry SL, Lockhart BE, Olszewski NE. The Commelina yellow mottle virus promoter is a strong promoter in vascular and reproductive tissues. Plant Cell. 1992;4:185–192. doi: 10.1105/tpc.4.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskayaa ON. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell. 2011;23:942–960. doi: 10.1105/tpc.110.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monna L, Lin X, Kojima S, Sasaki T, Yano M. Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor Appl Genet. 2002;104:772–778. doi: 10.1007/s00122-001-0813-0. [DOI] [PubMed] [Google Scholar]
- Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 2008;49:1645–1658. doi: 10.1093/pcp/pcn154. [DOI] [PubMed] [Google Scholar]
- Pageau K, Reisdorf-Cren M, Morot-Gaudry JF, Masclaux-Daubresse C. The two senescence-related markers GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilisation are differentially regulated during pathogen attack, by stress hormones and reactive oxygen species in Nicotiana tabacum L. leaves J Exp Bot. 2006;57:547–557. doi: 10.1093/jxb/erj035. [DOI] [PubMed] [Google Scholar]
- Pin PA, Nilsson O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012;35:1742–1755. doi: 10.1111/j.1365-3040.2012.02558.x. [DOI] [PubMed] [Google Scholar]
- Ryu CH, Lee S, Cho LH, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009;32:1412–1427. doi: 10.1111/j.1365-3040.2009.02008.x. [DOI] [PubMed] [Google Scholar]
- Schmülling T, Schell J, Spena A. Promoters of the rolA, B, and C genes of Agrobacterium rhizogenes are differentially regulated in transgenic plants. Plant Cell. 1989;1:665–670. doi: 10.1105/tpc.1.7.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Sauer N. The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot Acta. 1996;109:299–306. [Google Scholar]
- Scofield GN, Aoki N, Hirose T, Takano M, Jenkins CLD, Furbank RT. The role of the sucrose transporter, OsSUT1, in germination and early seedling growth and development of rice plants. J Exp Bot. 2007;58:483–495. doi: 10.1093/jxb/erl217. [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol. 2011;14:45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Tsuji H, Tachibana C, Tamaki S, Taoka K, Kyozuka J, Shimamoto K. Hd3a promotes lateral branching in rice. Plant J. 2015;82:256–266. doi: 10.1111/tpj.12811. [DOI] [PubMed] [Google Scholar]
- Werr W, Frommer WB, Maas C, Starlinger P. Structure of the sucrose synthase gene on chromosome 9 of Zea mays L. EMBO J. 1985;4:1373–1380. doi: 10.1002/j.1460-2075.1985.tb03789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee S, Hang R, Kim SR, Lee YS, Cao X, Amasino R, An G. OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 2013;73:566–578. doi: 10.1111/tpj.12057. [DOI] [PubMed] [Google Scholar]
- Yano M, Sasaki T. Genetic and molecular dissection of quantitative traits in rice. Plant Mol Biol. 1997;35:145–153. [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Beachy RN. The regulatory regions of the rice tungro bacilliform virus promoter and interacting nuclear factors in rice (Oryza sativa L.) Plant J. 1995;7:969–980. doi: 10.1046/j.1365-313x.1995.07060969.x. [DOI] [PubMed] [Google Scholar]
- Yin Y, Chen L, Beachy RN. Promoter elements required for phloem-specific gene expression from the RTBV promoter in rice. Plant J. 1997;12:1179–1188. doi: 10.1046/j.1365-313x.1997.12051179.x. [DOI] [PubMed] [Google Scholar]
- Yoon J, Cho LH, Kim SL, Choi H, Koh HJ, An G. The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 2014;79:717–728. doi: 10.1111/tpj.12581. [DOI] [PubMed] [Google Scholar]
- Yoon J, Cho LH, Antt HW, Koh HJ, An G. KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol. 2017;174:312–325. doi: 10.1104/pp.17.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huang X, Ouyang X, Chen W, Chen W, Du A, Zhu L, Wang S, Deng XW, Li S. OsELF3-1, an ortholog of Arabidopsis EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering. PloS One. 2012;7:e43705. doi: 10.1371/journal.pone.0043705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.