Abstract
The gaseous hormone ethylene influences many aspects of plant growth, development, and responses to a variety of stresses. The biosynthesis of ethylene is tightly regulated by various internal and external stimuli, and the primary target of the regulation is the enzyme 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS), which catalyzes the rate-limiting step of ethylene biosynthesis. We have previously demonstrated that the regulation of ethylene biosynthesis is a common feature of most of the phytohormones in etiolated Arabidopsis seedlings via the modulation of the protein stability of ACS. Here, we show that various phytohormones also regulate ethylene biosynthesis from etiolated rice seedlings in a similar manner to those in Arabidopsis. Cytokinin, brassinosteroids, and gibberellic acid increase ethylene biosynthesis without changing the transcript levels of neither OsACS nor ACC oxidases (OsACO), a family of enzymes catalyzing the final step of the ethylene biosynthetic pathway. Likewise, salicylic acid and abscisic acid do not alter the gene expression of OsACS, but both hormones downregulate the transcript levels of a subset of ACO genes, resulting in a decrease in ethylene biosynthesis. In addition, we show that the treatment of the phytohormones results in distinct etiolated seedling phenotypes, some of which resemble ethylene-responsive phenotypes, while others display ethylene-independent morphologies, indicating a complicated hormone crosstalk in rice. Together, our study brings a new insight into crosstalk between ethylene biosynthesis and other phytohormones, and provides evidence that rice ethylene biosynthesis could be regulated by the post-transcriptional regulation of ACS proteins.
Keywords: ethylene biosynthesis, hormone, OsACO, OsACS, post-transcriptional regulation, rice
INTRODUCTION
Rice is one of the most important food crops, and it feeds more than half of the world’s population. While rice has evolved elaborate mechanisms to adapt various stress conditions including hypoxia (Ma et al., 2010), its productivity and sustainability are significantly threatened by many abiotic and biotic stresses such as drought, chilling, submergence, and salinity (Sahi et al., 2006; Shimamoto, 1999). During these stress conditions, ethylene plays a primary role in acclimating plants to unfavorable surroundings (Abeles, 1973; Morgan and Drew, 1997). Many studies have shown that the biosynthesis of ethylene is highly regulated transcriptionally and post-transcriptionally (Argueso et al., 2007; Yoon, 2015). The biosynthesis of ethylene consists of three simple steps, starting with the amino acid methionine in both monocot and dicot plants (Yang and Hoffman, 1984). In the first step, methionine is converted to S-adenosyl methionine (SAM), which is subsequently converted to 1-aminocyclopropane-1-carboxylic acid (ACC) by a family of enzymes known as ACC synthases (ACS). Conversion of SAM to ACC by ACS is the first committed and generally rate-limiting step in the pathway. In the last step, ACC is finally converted to ethylene by ACC oxidases (ACO), a member of the oxygenase superfamily member (Bidonde et al., 1998). The Arabidopsis genome contains 12 putative ACS-like genes (e.g., ACS1 to 12). Among the ACS genes, ACS3 is a pseudogene with a short sequence, and ACS10 and ACS12 encode an aminotransferase without the catalytic activity of ACS (Yamagami et al., 2003). The other nine ACS genes encode a group of ACS proteins that can be classified into 3 types, named type-1, type-2, and type-3, based on the presence or absence of putative phosphorylation sites at the C-terminal domain of the proteins (Chae and Kieber, 2005). Type-1 ACS proteins contain putative phosphorylation target sites for both mitogen-activated protein kinase (MAPK) and calcium-dependent protein kinase (CDPK). Type-2 ACS proteins have only a putative target site for CDPK, but they also contain a unique regulatory motif called a Target of ETO1 (TOE) at their C-termini. TOE is a binding site for ETHYLENE OVERPRODUCER 1 (ETO1) (Wang et al., 2004), which is a Broad complex/Tramtrack/Bric-a-brac (BTB) domain substrate adaptor for CULLIN-3 E3 (CUL3) ubiquitin ligase. ETO1 and its two paralogs, ETO1-like 1 (EOL1) and EOL2 specifically interact with type-2 ACS proteins via the TOE motif, and this interaction further leads to the degradation of type-2 ACS proteins via the 26S proteasome (Yoshida et al., 2005; 2006).
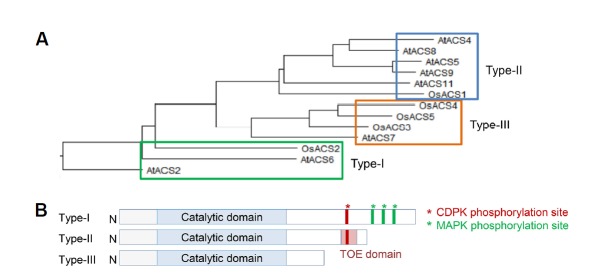
Compared to Arabidopsis, rice possesses a relatively small number of ACS genes. It contains only 5 putative ACS genes (OsACS1 to 5) which share 50 to 80% of similarity of amino acid sequence among the isoforms (Supplementary Table S1) (Zarembinski and Theologis, 1997). All OsACS genes possess seven conserved domains that were found in Arabidopsis and other plant species, including all 11 invariants amino acid residues found in ACS and aminotransferases (Yamagami et al., 2003). A phylogenetic tree of ACS proteins from Arabidopsis and rice and the analysis of ACS protein structure indicate that OsACS proteins also fall into three different types (Figs. 1A and 1B). OsACS2 protein exhibits a similar structure to Arabidopsis type-1 ACS2 and 6 proteins. It has the longest C-terminal domain among OsACS proteins with putative phosphorylation sites for MAPK and CDPK. OsACS1 was closely related to Arabidopsis type-2 ACS, and its C-terminus contains a potential CDPK target site and a putative TOE domain (Yoshida et al., 2006). While type-2 ACS proteins were the dominant form of ACS proteins in Arabidopsis, type-3 ACS is the most abundant ACS proteins in rice, with three isoforms of type-3 ACS. Similar to the Arabidopsis type-3 ACS, type-3 OsACS proteins have a short C-terminal domain and lack of any known regulatory sites. The structural similarity and the conserved regulatory sites found in both ACS proteins from Arabidopsis and rice imply the existence of an evolutionally conserved mechanism that underlies the regulation of ethylene biosynthesis in these two plant species.
Fig. 1. Phylogenetic tree of ACC synthases from Arabidopsis and rice and cartoon representing the structure of different types of ACS proteins.
The amino acid sequences of Arabidopsis and rice ACS proteins were aligned using ClustalW and the phylogenetic tree was generated using DNAstaw software (A). The structure of ACS proteins (B). The conserved catalytic domain is shown with blue rectangle boxes. The target serine residues for calcium-dependent protein kinase (CDPK) and mitogen-activated protein kinase (MAPK) are shown as a red or green asterisk, respectively. Target of ETO1 (TOE) motif in type-2 ACS is highlighted in red box. Type-3 ACS does not contain any regulatory motifs.
Given the diverse roles of ethylene in plant growth and physiological responses to various external and internal signals, ethylene biosynthesis is highly regulated at both transcriptional and post-transcriptional levels (Argueso et al., 2007). Regulation of the transcript levels of ACS genes appears to be a key control mechanism of ethylene production during stress conditions (Argueso et al., 2007; Zarembinski and Theologis, 1994). In Arabidopsis, the transcript levels of a subset of ACS have been shown to be under the control of biotic and abiotic stresses such as salinity, chilling, submergence, drought, and pathogen invasion (Argueso et al., 2007). Similarly, the transcript levels of OsACS genes were also affected by various stimuli, including submergence, herbivore attack, and fungus infection, resulting in altered ethylene production (Hazman et al., 2016; Iwai et al., 2006; Lu et al., 2014). In addition to the transcriptional control of ACS, several studies have provided compelling evidence that post-translational modifications, such as phosphorylation and ubiquitination, serve as an important mechanism to regulate the stability of Arabidopsis ACS proteins, thus controlling the levels of ethylene in plants (Chae and Kieber, 2005; Liu and Zhang, 2004; Wang et al., 2004; Yoon and Kieber, 2013).
ACC oxidase (ACO) is another enzyme involved in the final step of ethylene biosynthesis, converting ACC to ethylene (Argueso et al., 2007; Yang and Hoffman, 1984). ACO genes have been identified from various plant species, including but not limited to Arabidopsis (Lin et al., 2009), rice (Iwai et al., 2006), petunia (Tang et al., 1993), tomato (Barry et al., 1996), potato (Nie et al., 2002), and apple (Binnie and McManus, 2009). In most plant species investigated, ACO isoforms are encoded by a multigene family of ACO genes. For example, Arabidopsis and rice possess six and seven ACO genes, respectively (Iwai et al., 2006; Lin et al., 2009). Similar to the ACS, the gene expression of ACO from different species is also correlated to the rate of ethylene biosynthesis, and the transcript levels of multiple ACO genes are under the control of developmental and stress conditions (Gómez-Jiménez et al., 1998; Linkies and Leubner-Metzger, 2012; Matillaa and Matilla-Vázquezb, 2008). It has been shown that OsACO1 plays a role in the internode elongation of deepwater rice; submergence increases the levels of OsACO1 mRNA and ACO enzyme activity (Iwamoto et al., 2010; Mekhedov and Kende, 1996). The gene expression of OsACO2 and OsACO3 in etiolated rice seedlings is also shown to be differentially regulated by ethylene and auxin (Chae et al., 2000). Additionally, the transcripts of a subset of OsACO (OsACO2 and OsACO7) were highly induced upon infection with blast fungus, leading to an increase in ethylene production, which ultimately contributes to the disease resistance of rice to blast fungus (Iwai et al., 2006). Compared to the transcriptional regulation of OsACO genes, less is known about the post-transcriptional control of OsACO proteins.
Hormonal crosstalk plays a large role in enabling plants to respond to a given developmental or environmental input with plasticity (Depuydt and Hardtke, 2011). Ethylene interacts with many phytohormones and the resulting crosstalk regulates many important developmental processes in both rice and Arabidopsis. We have recently revealed that several phytohormones regulate ethylene biosynthesis in etiolated Arabidopsis seedlings, indicating that ethylene biosynthesis is a central crosstalk point among phytohormones (Lee et al., 2017). Phytohormones, including cytokinin, GA, BR, IAA, SA, methyl jasmonate (MJ), and ABA control ethylene biosynthesis either through the modulation of the turnover of ACS proteins or the transcript levels of ACS genes, or both (Lee et al., 2017). Cytokinin, BR, and GA mainly increase the stability of type-1 and type-2 ACS proteins without changing the transcript levels of ACS genes. SA, MJ, and ABA influence ethylene production from etiolated seedlings by affecting both the transcript levels of ACS genes and protein turnover rate of type-1 and 2 ACS proteins (Lee et al., 2017). Contrary to the previous known role of auxin in ethylene biosynthesis, IAA promotes ethylene production by regulating both ACS transcript levels and protein turnover of ACS proteins (Abeles et al., 1992; Tsuchisaka and Theologis, 2004; Wang et al., 2005; Yang and Hoffman, 1984). Interestingly, unlike type-1 and type-2 ACS, there is no effect on type-3 ACS7 transcript levels or ACS7 protein turnover by these phytohormones (Lee et al., 2017).
Here we report that various phytohormones control ethylene biosynthesis in rice. We examine the effects of phytohormones in controlling ethylene production in etiolated rice seedlings. We demonstrate that nearly all phytohormone-induced changes in ethylene biosynthesis are not regulated by ACS transcript changes in etiolated rice seedlings. However, contrary to the ACS, we found that some fractions of the phytohormones affect the transcript levels of ACO genes resulting in a decrease in ethylene biosynthesis. In addition, we show that various phytohormones induce distinctive morphological changes in rice seedlings, some of which resemble ethylene-responsive phenotypes, suggesting a possible role of these plant hormones in ethylene biosynthesis regulation. Together, our results provide new insights into the role of phytohormones in ethylene biosynthesis and the hormonal crosstalk between ethylene and other phytohormones in rice etiolated seedlings.
MATERIALS AND METHODS
Plant materials and growth conditions
Rice (Oryza sativa L. cv. Nipponbare) seeds were dehusked and sterilized with 30% sodium hypochlorite for 10 min, followed by rinsing with deionized water. The sterilized seeds were subsequently placed onto a sterile sieve, placed in a liquid MS media in a magenta box. The seedlings were grown in a growth chamber at 28°C in the dark.
Measurements of ethylene production
Ethylene measurements were performed as previously described (Hansen et al., 2009) with a brief modification. Surface-sterilized seeds were germinated in 22-ml gas chromatography vials containing 3 ml MS with 1% sucrose, 1% bactoagar, and with and without indicated concentration of hormones. Following 2 days of dark treatment, the vials were capped and incubated at 28°C for 3 days in the dark. The accumulated ethylene was measured by a gas chromatography using a Shimadzu GC2010 Plus capillary gas chromatography system with a HS-20 headspace autosampler. All treatments were measured from at least three replicates.
RNA extraction and quantitative RT-PCR
Total RNA was extracted by using a RNeasy Plus kit (Qiagen). cDNA was prepared from the total RNA using SuperScript II reverse transcriptase (Invitrogen) as described by the manufacturer. Quantitative RT-PCR was performed using PowerUP™ SYBR® green Master Mix (Applied Biosystems) and CFX Connect™ Real-Time System (Bio-Rad). Relative expression of OsACS or OsACO genes were normalized to OsActin1 as a reference gene and was calculated using 2ΔΔCT method. Primers used are listed in Supplementary Table S2.
Analysis of etiolated rice seedling morphology in response to phytohormones
The surface-sterilized wild-type seedlings were grown in liquid MS media in a growth chamber at 28°C in the dark. 2-day-old germinated rice seedlings were gently transferred onto a sieve that was placed on 1/2 MS solution with or without various phytohormones (10 μM ACC, 10 μM synthetic cytokinin BA, 10 μM BR, 10 μM GA3, 10 μM IAA, 5 mM SA, 5 μM MJ or 1 μM ABA), and further incubate them at 28°C in the dark for additional 3 days.
RESULTS
Phytohormones differentially regulate the biosynthesis of ethylene from etiolated rice seedlings
The biosynthesis of ethylene is regulated by many factors, including developmental cues and environmental stimuli, both of which can be integrated into the cells via the action of phytohormones in plants. We have previously demonstrated that most of the phytohormones influence the rate of ethylene biosynthesis in Arabidopsis thaliana via the alteration of mRNA levels or protein stability of ethylene biosynthesis components such as 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) and ACC oxidase (ACO) (Lee et al., 2017). To examine whether phytohormones affect ethylene biosynthesis of rice seedlings in a similar manner to that of Arabidopsis seedlings, we measured ethylene production from wild-type etiolated rice seedlings (Oryza sativa L. cv. Nipponbare) grown in the presence of various phytohormones, including cytokinin, BR, GA, IAA, SA, MJ, and ABA (Fig. 2). Cytokinin, BR, IAA, and GA, increased ethylene production in a concentration-dependent manner (Fig. 2). This is consistent with a previous report that these hormones promote ethylene production of Arabidopsis etiolated seedlings over-expressing type-1 or type-2 ACS protein (Lee et al., 2017). By contrast, the growth in the presence of either SA or ABA caused a reduction in the levels of ethylene production from etiolated rice seedlings. These results also agree with that both SA and ABA decrease ethylene production of Arabidopsis seedlings over-expressing ACS proteins (Lee et al., 2017). This is consistent with the observation that SA reduces ethylene production from detached rice leaves resulted from the inhibition of ACC formation and the conversion of ACC to ethylene (Huang et al., 1993). Unlike other phytohormones, MJ appeared not to alter the level of ethylene in etiolated rice seedlings. Together, these results indicate that most phytohormones tested in this study influence the level of ethylene biosynthesis from etiolated rice seedlings, and the alteration of ethylene biosynthesis is a common feature of many phytohormones in both rice and Arabidopsis.
Fig. 2. Ethylene production by 3-day-old etiolated seedlings in response to various phytohormones.
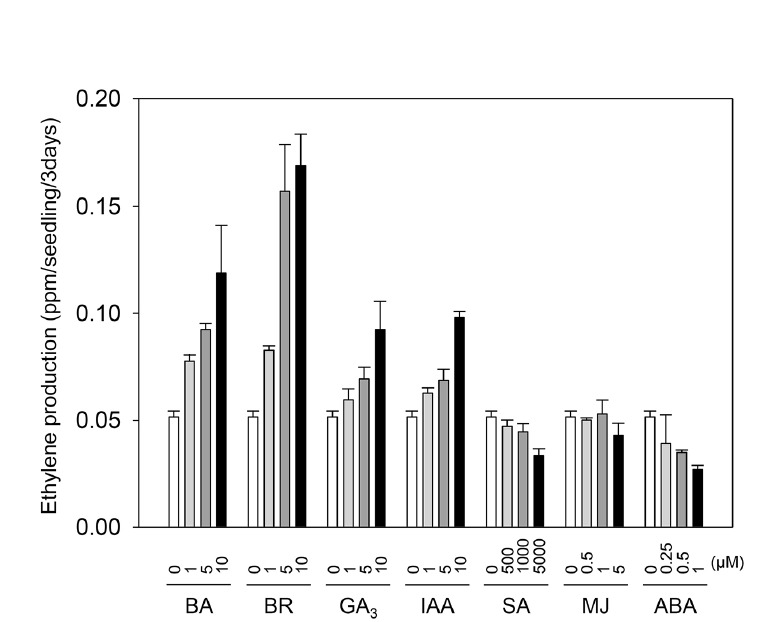
Wild-type rice seedlings were grown in MS media with the indicated concentration of phytohormones, capped for 3 days in the dark, and the accumulated levels of ethylene were measured at day 3 using gas chromatography. Error bars indicate SD; n = 3.
Most of the phytohormones, except IAA, do not alter the transcript levels of OsACS in etiolated rice seedlings
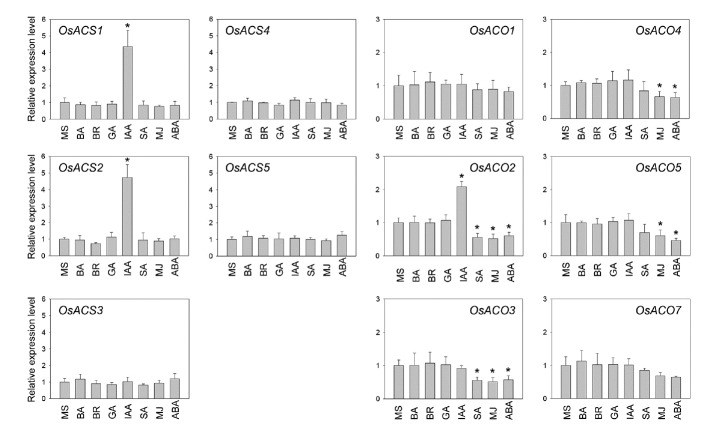
The changes in the levels of ethylene production from rice etiolated seedlings in response to the phytohormones indicate that there must be corresponding changes either at the levels of gene expression or protein stability or/and activity of ethylene biosynthesis components. To get further insight into the molecular basis underlying the phytohormone-regulated ethylene biosynthesis in etiolated rice seedlings, we first examined the effects of phytohormones on the transcript levels of different types of rice ACS genes, OsACS1 (type-2), OsACS2 (type-1), and OsACS3, 4, 5 (type-3) using qRT-PCR (Fig. 3). Most phytohormones did not alter the transcript levels of the majority of OsACS genes (Fig. 3). One exception was IAA, which caused a significant increase in the transcript levels of both OsACS1 and OsACS2 compared to its mock treatment. This is in agreement with previous studies showing that auxin induces the transcript levels of Arabidopsis type-1 and type-2 ACS (Lee et al., 2017). Interestingly, IAA did not change the transcript levels of OsACS3, OsACS4, and OsACS5, all of which belong to the type-3 ACS family. These results are also consistent with the prior studies showing that Arabidopsis type-3 ACS7 transcript levels are unchanged by IAA treatment in both etiolated and light-grown Arabidopsis seedlings.
Fig. 3. Phytohormone-induced transcriptional regulation of OsACS and OsACO genes.
Quantitative RT-PCR expression analysis of OsACS and OsACO genes in rice etiolated seedlings grown in the presence of indicated hormones (10 μM BA, 10 μM BR, 10 μM GA3, 10 μM IAA, 5 mM SA, 5 μM MeJA or 1 μM ABA) for 3 days. Expression of OsActin1 was used as an internal control. Error bars represent SD from three biological replicates. *P < 0.05, Student’s t-test.
Together, these results showed that most phytohormones, except IAA, do not alter the transcript levels of ACS genes in etiolated rice seedlings, suggesting a possible role for the post-transcriptional regulation of ACS proteins in the phytohormone-mediated ethylene biosynthesis in rice.
A subset of the OsACO transcript levels are regulated by phytohormones
Unlike the effects of cytokinin, BR, IAA, and GA, treatment of rice etiolated seedlings with SA and ABA led to a reduced ethylene production in a manner dependent on the concentration of the phytohormones used (Fig. 2). Given that the transcript levels of most of the OsACS genes were not affected by both SA and ABA, we postulated that the reduction of ethylene biosynthesis in response to SA and ABA could be due to changes in the transcript levels of ACC oxidases (OsACO). To this end, we determined the transcript levels of six OsACO genes from wild-type etiolated rice seedlings treated with various phytohormones (Fig. 3). OsACO6 was not included in the assay as it is a pseudogene. In general, the treatment of etiolated rice seedlings with ABA reduced the expression of most of the OsACO. Likewise, SA treatment also decreased the expression of a subset of the OsACO genes, OsACO2 and OsACO3. MJ treatment reduced the transcript levels of OsACO2, OsACO3, OsACO4, and OsACO5. Contrary to ABA, SA, and MJ, IAA increased the transcript levels of OsACO2, as previously described (Chae et al., 2000). Unlike SA, MJ, and ABA, cytokinin, GA, and BR did not affect the transcript levels of most of the OsACO. These results suggest that the reduced transcript levels of OsACO in response to ABA, SA, and MJ likely contribute to the decreased ethylene biosynthesis observed in etiolated rice seedlings treated with these hormones. It is still possible that these three hormones may also regulate ACO protein stability and/or activity and ACC conjugation.
Various phytohormones differently influence the growth of etiolated rice seedlings
Dark-grown Arabidopsis seedlings exposed to ethylene produce distinctive ethylene-responsive phenotypes, which are referred as the triple response (Guzman and Ecker, 1990). The triple response of etiolated seedlings includes the formation of an exaggerated apical hook, thickened and shortened hypocotyl, and inhibited root growth. Treatment of dark-grown Arabidopsis seedlings with cytokinin also causes the triple response due to a role of cytokinin in promoting ethylene biosynthesis (Chae et al., 2003). Unlike Arabidopsis, etiolated rice seedlings do not have the typical triple response. Instead, rice seedlings display unique ethylene-responsive phenotypes, namely, an increased growth of the coleoptile and inhibited root growth (Fig. 4A) (Kim et al., 2012; Ma et al., 2010). To determine the effect of the phytohormones in rice seedling growth in the dark, we observed the phenotypes of etiolated rice seedlings after treatment with the phytohormones. Consistent with previous studies, treatment of etiolated rice seedlings with ACC, a direct precursor of ethylene, led to the ethylene responsive phenotypes of elongated coleoptile and shortened root (Fig. 4A). Similar morphological changes were observed in etiolated rice seedlings treated with cytokinin and BR, indicating that these hormones play a role in promoting either ethylene biosynthesis or its signaling. However, IAA, SA, MJ, and ABA only caused the changes in the root growth of etiolated rice seedlings with varying degrees of root growth inhibition (Fig. 4B). The root growth inhibition caused by SA, MJ, and ABA was somewhat moderate compared to those from IAA or ACC-treated seedlings. In contrast, etiolated rice seedlings treated with GA displayed both an elongated coleoptile and root, consistent with the growth-promoting role of this phytohormone in plants. Together, these results revealed that the role of phytohormones in controlling ethylene biosynthesis in etiolated rice seedlings, either positively or negatively, is not always comparably reflected in morphological changes, highlighting the complicated nature of phytohormone crosstalk networks in plants.
Fig. 4. Morphological analysis of 3-day-old etiolated rice seedlings in the presence of phytohormones.
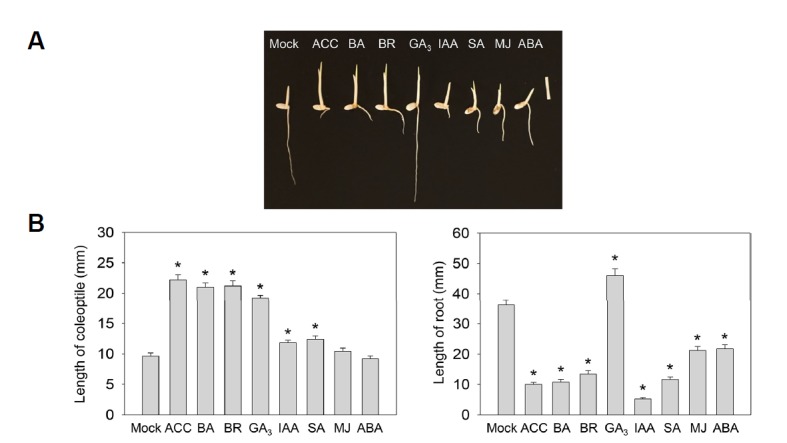
(A) A representative image of wild-type etiolated seedling that were grown on MS media supplemented with or without various phytohormones with the indicated concentration (10 μM ACC, 10 μM BA, 10 μM BR, 10 μM GA3, 10 μM IAA, 5 mM SA, 5 μM MJ or 1 μM ABA). Scale bar indicates 10 mm. (B) Measurement of the coleoptile (left) or root (right) length of etiolated rice seedlings grown with various phytohormones. Error bars indicate SD; n = 5, *P < 0.005, Student’s t-test.
DISCUSSION
Rice suffers from various biotic and abiotic stresses, resulting in a dramatic loss of yield and productivity. Ethylene plays a pivotal role in adapting the growth of rice in those harsh growth conditions. While the transcriptional regulation of ethylene biosynthesis is relatively well established, not much is known about the post-transcriptional regulation of ethylene biosynthesis in rice to date. However, multiple pieces of evidence support the idea that rice ethylene biosynthesis is in part regulated by the post-transcriptional regulation of ACS proteins, in a manner similar to what is observed in Arabidopsis. Such evidence includes the existence of ETO1-like E3 ubiquitin ligases orthologs in the rice genome and the negative role of OsETOL1 in ethylene biosynthesis regulation (Du et al., 2014). Du et al., showed that OsETOL1 modulates drought and submergence tolerance by down-regulating ethylene biosynthesis. OsETOL1 directly interacts with OsACS2, a homolog of Arabidopsis ACS protein. Moreover, overexpression of OsETOL1 resulted in a reduction in ethylene biosynthesis, which validates the negative role of OsETOL1 in ethylene biosynthesis. Further, rice contains 8 isoforms of 14-3-3 proteins, highly conserved regulatory proteins that regulate diverse physiological processes by phosphorylation-dependent protein–protein interactions (Jaspert et al., 2011). A recent study showed that Arabidopsis 14-3-3 protein acts as a positive regulator of ethylene biosynthesis by enhancing the protein stability of ACS proteins through the interaction with ACS proteins (Yoon and Kieber, 2013). Interestingly, Yao et al, demonstrated that rice 14-3-3 binds to OsACS1 using a yeast two hybrid, but the detailed biochemical analysis of this interaction has not been demonstrated (Yao et al., 2007). Together, these evidences strongly suggest that rice ethylene biosynthesis is likely under the control of the post-transcriptional regulation of ACS proteins.
Compared to the ACS proteins, the post-transcriptional control of ACO has not been clearly demonstrated. In this study, we found that BA, BR, and GA did not affect the transcript levels of all ACO genes tested, yet increased ethylene production in etiolated rice seedlings (Fig. 3). These results indicate the possible role of the post-transcriptional control of ACO proteins. Interestingly, a recent study suggested that the turnover of Arabidopsis ACO protein is likely responsible for altering ethylene production in etiolated seedlings. In Arabidopsis, the mutation of Related to Ubiquitin 1 (RUB)-conjugating enzyme (RCE1), an enzyme that catalyzes the covalent modification of RUB1 to SCF-E3 ligase complex, results in an increased activity of ACO, hence elevated ethylene production (Larsen and Cancel, 2004). Despite the widespread knowledge that ACS is the rate-limiting enzyme in ethylene biosynthesis, some evidence supports that ACO could also contribute to sustaining the high levels of ethylene production during some developmental processes such as pollination-mediated senescence and seedling germination (Nadeau et al., 1993; Petruzzelli et al., 2000). During these processes, plants elevate ethylene production by increasing ACO activity which is regulated by either ACO expression or the rate of ACO protein turnover, or both. In rice, the role of a subset of ACO has been implicated in submergence response and hormone crosstalk. Especially, the expression of OsACO1 and its enzymatic activity are greatly increased upon submergence stress to maintain the high level of ethylene production (Iwamoto et al., 2010; Mekhedov and Kende, 1996). This suggests the post-transcriptional control of ACO in addition to the transcriptional regulation of ACO gene expression. The interaction between ethylene and other phytohormones including GA during submergence stress may also contribute to an increase in ethylene biosynthesis via the modulation of the ACO activity (Fukao and Bailey-Serres, 2008; Jackson, 2008).
Phytohormone-mediated changes in the morphologies of etiolated rice seedlings suggest a complicated hormonal crosstalk in rice. ACC-treated etiolated rice seedlings showed unique ethylene-responsive phenotypes of an elongated coleoptile and inhibited root growth. Cytokinin and BR induce ethylene-responsive phenotypes in rice seedlings, which is consistent with their role in increasing ethylene biosynthesis. Interestingly, unlike what is observed in rice, BR does not trigger the triple response in dark-grown Arabidopsis seedlings, implying different actions of BR in rice and Arabidopsis, despite its common role in enhancing ethylene biosynthesis. The action of GA in rice seedlings is somewhat different from cytokinin and BR, although all of three hormones increase ethylene production. Unlike cytokinin and BR, GA promotes the growth of both coleoptiles and roots indicating that the ethylene-GA crosstalk may preferentially occur in the roots, or that ethylene differently works to promote root growth in the presence of GA. In fact, several studies showed that ethylene increases the growth of submerged roots of deep water rice by working together with GA in a synergistic manner (Jackson, 2008; Vriezen et al., 2003). Similarly, the synergistic interaction between GA and ethylene in coleoptiles has been shown to be an important adaptive feature of deep water rice to grow out of the water and survive flooding (Miro and Ismail, 2013; Watanabe et al., 2007). Interestingly, SA and ABA, which decreased ethylene biosynthesis in rice seedlings, promote enhanced ethylene-responses in roots, and have no effects on ethylene-responsive phenotypes in the coleoptiles of etiolated seedlings. In Arabidopsis, ABA inhibits the root growth of light-grown Arabidopsis seedlings by promoting ethylene biosynthesis via a calcium-dependent protein kinase-mediated phosphorylation of type-2 ACS6 (Luo et al., 2014). This result is somewhat different from the results from our previous study that ABA stabilizes type-1 and type-2 ACS, but the overall ethylene biosynthesis from ABA-treated seedlings is reduced due to a decrease in ACO mRNA levels. This discrepancy could be related to the growth condition of seedlings (light vs. dark), which might differently affect ACO gene expression. Similar to Arabidopsis etiolated seedlings, ABA decreased ethylene biosynthesis from rice etiolated seedlings likely through a reduction in a subset of ACO transcripts in rice (Fig. 3). However, surprisingly, ABA inhibits root growth without a concurrent increase in ethylene levels, indicating a different hormonal crosstalk between ethylene and ABA in rice plants. Likewise, SA appears to control the root elongation of rice through an ethylene-independent pathway, as the SA-induced reduction of ethylene did not influence root growth.
In summary, we show that the major phytohormones, with exception of IAA, influence ethylene biosynthesis in etiolated rice seedlings, through mechanisms that do not include the transcriptional changes of ACS genes, thus indicating the possibility of a post-transcriptional regulation of ACS proteins. In a similar manner to the ACS, the transcript levels of ACO were not altered by a subset of phytohormones including cytokinin, BR, and GA, implicating the potential involvement of the post-transcriptional control of ACO proteins. Moreover, we demonstrate that different phytohormones induced distinctive phenotypes in etiolated rice seedlings, which may or may not be directly related to the action of ethylene, despite their role in ethylene biosynthesis. Further understanding of the roles of phytohormones on the activity or stability of both ACS and ACO proteins will provide more insight into the regulatory mechanisms underlying ethylene biosynthesis in rice.
Supplementary data
ACKNOWLEDGMENTS
This work was supported by Purdue University start-up funds to GM.Y. We thank Cris Argueso for critical reading of the article.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Abeles FB. Ethylene in plant biology. Vol. 302 Academic; New York: 1973. [Google Scholar]
- Abeles FB, Morgan PW, Saltveit MEJ. Ethylene in plant biology. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Argueso CT, Hansen M, Kieber JJ. Regulation of ethylene biosynthesis. J Plant Growth Regul. 2007;262:92–105. [Google Scholar]
- Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 1996;9:525–535. doi: 10.1046/j.1365-313x.1996.09040525.x. [DOI] [PubMed] [Google Scholar]
- Bidonde S, Ferrer MA, Zegzouti H, Ramassamy S, Latche A, Pech JC, Hamilton AJ, Grierson D, Bouzayen M. Expression and characterization of three tomato 1-aminocyclopropane-1-carboxylate oxidase cDNAs in yeast. Eur J Biochem. 1998;253:20–26. doi: 10.1046/j.1432-1327.1998.2530020.x. [DOI] [PubMed] [Google Scholar]
- Binnie JE, McManus MT. Characterization of the 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase multigene family of Malus domestica Borkh. Phytochemistry. 2009;70:348–360. doi: 10.1016/j.phytochem.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Chae HS, Cho YG, Park MY, Lee MC, Eun MY, Kang BG, Kim WT. Hormonal cross-talk between auxin and ethylene differentially regulates the expression of two members of the 1-aminocyclopropane-1-carboxylate oxidase gene family in rice (Oryza sativa L.) Plant Cell Physiol. 2000;41:354–362. doi: 10.1093/pcp/41.3.354. [DOI] [PubMed] [Google Scholar]
- Chae HS, Kieber JJ. Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trend Plant Sci. 2005;10:291–296. doi: 10.1016/j.tplants.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell. 2003;15:545–559. doi: 10.1105/tpc.006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 2011;21:R365–373. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Du H, Wu N, Cui F, You L, Li X, Xiong L. A homolog of ETHYLENE OVERPRODUCER, OsETOL1, differentially modulates drought and submergence tolerance in rice. Plant J. 2014;78:834–849. doi: 10.1111/tpj.12508. [DOI] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci USA. 2008;105:16814–16819. doi: 10.1073/pnas.0807821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Jiménez MC, Matilla AJ, Garrido D. Isolation and characterization of a cDNA encoding an ACC oxidase from Cicer arietinum and its expression during embryogenesis and seed germination. Australian J Plant Physiol. 1998;25:765–773. [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chae HS, Kieber JJ. Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 2009;57:606–614. doi: 10.1111/j.1365-313X.2008.03711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazman M, Hause B, Eiche E, Riemann M, Nick P. Different forms of osmotic stress evoke qualitatively different responses in rice. J Plant Physiol. 2016;202:45–56. doi: 10.1016/j.jplph.2016.05.027. [DOI] [PubMed] [Google Scholar]
- Huang YF, Chen CT, Kao CH. Salicylic acid inhibits the biosynthesis of ethylene in detached rice leaves. Plant Growth Regul. 1993;12:79–82. [Google Scholar]
- Iwai T, Miyasaka A, Seo S, Ohashi Y. Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 2006;142:1202–1215. doi: 10.1104/pp.106.085258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Baba-Kasai A, Kiyota S, Hara N, Takano M. ACO1, a gene for aminocyclopropane-1-carboxylate oxidase: effects on internode elongation at the heading stage in rice. Plant Cell Environ. 2010;33:805–815. doi: 10.1111/j.1365-3040.2009.02106.x. [DOI] [PubMed] [Google Scholar]
- Jackson MB. Ethylene-promoted elongation: an adaptation to submergence stress. Ann Bot. 2008;101:229–248. doi: 10.1093/aob/mcm237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspert N, Throm C, Oecking C. Arabidopsis 14–3-3 proteins: fascinating and less fascinating aspects. Front Plant Sci. 2011;2:96. doi: 10.3389/fpls.2011.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wilson RL, Case JB, Binder BM. A comparative study of ethylene growth response kinetics in eudicots and monocots reveals a role for gibberellin in growth inhibition and recovery. Plant Physiol. 2012;160:1567–1580. doi: 10.1104/pp.112.205799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Cancel JD. A recessive mutation in the RUB1-conjugating enzyme, RCE1, reveals a requirement for RUB modification for control of ethylene biosynthesis and proper induction of basic chitinase and PDF1.2 in Arabidopsis. Plant J. 2004;38:626–638. doi: 10.1111/j.1365-313X.2004.02068.x. [DOI] [PubMed] [Google Scholar]
- Lee HY, Chen YC, Kieber JJ, Yoon GM. Regulation of the turnover of ACC synthases by phytohormones and heterodimerization in Arabidopsis. Plant J. 2017;91:491–504. doi: 10.1111/tpj.13585. [DOI] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J Exp Bot. 2009;60:3311–3336. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- Linkies A, Leubner-Metzger G. Beyond gibberellins and abscisic acid: how ethylene and jasmonates control seed germination. Plant Cell Rep. 2012;31:253–270. doi: 10.1007/s00299-011-1180-1. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li J, Ju H, Liu X, Erb M, Wang X, Lou Y. Contrasting effects of ethylene biosynthesis on induced plant resistance against a chewing and a piercing-sucking herbivore in rice. Mol Plant. 2014;7:1670–1682. doi: 10.1093/mp/ssu085. [DOI] [PubMed] [Google Scholar]
- Ma B, Chen SY, Zhang JS. Ethylene signaling in rice. Chinese Sci Bull. 2010;55:2204–2210. [Google Scholar]
- Matillaa AJ, Matilla-Vázquezb MA. Involvement of ethylene in seed physiology. Plant Sci. 2008;176:87–97. [Google Scholar]
- Mekhedov SI, Kende H. Submergence enhances expression of a gene encoding 1-aminocyclopropane-1-carboxylate oxidase in deepwater rice. Plant Cell Physiol. 1996;37:531–537. doi: 10.1093/oxfordjournals.pcp.a028976. [DOI] [PubMed] [Google Scholar]
- Miro B, Ismail AM. Tolerance of anaerobic conditions caused by flooding during germination and early growth in rice (Oryza sativa L.) Front Plant Sci. 2013;4:269. doi: 10.3389/fpls.2013.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PW, Drew CD. Ethylene and plant responses to stress. Physiologia Plantarum. 1997;100:620–630. [Google Scholar]
- Nadeau JA, Zhang XS, Nair H, O’Neill SD. Temporal and spatial regulation of 1-aminocyclopropane-1-carboxylate oxidase in the pollination-induced senescence of orchid flowers. Plant Physiol. 1993;103:31–39. doi: 10.1104/pp.103.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X, Singh RP, Tai GC. Molecular characterization and expression analysis of 1-aminocyclopropane-1-carboxylate oxidase homologs from potato under abiotic and biotic stresses. Genome. 2002;45:905–913. doi: 10.1139/g02-062. [DOI] [PubMed] [Google Scholar]
- Petruzzelli L, Coraggio I, Leubner-Metzger G. Ethylene promotes ethylene biosynthesis during pea seed germination by positive feedback regulation of 1-aminocyclopropane-1-carboxylic acid oxidase. Planta. 2000;211:144–149. doi: 10.1007/s004250000274. [DOI] [PubMed] [Google Scholar]
- Sahi C, Singh A, Kumar K, Blumwald E, Grover A. Salt stress response in rice: genetics, molecular biology, and comparative genomics. Funct Integr Genomics. 2006;6:263–284. doi: 10.1007/s10142-006-0032-5. [DOI] [PubMed] [Google Scholar]
- Shimamoto K. Molecular biology of rice. Springer-Verlag; Tokyo: 1999. [Google Scholar]
- Tang X, Wang H, Brandt AS, Woodson WR. Organization and structure of the 1-aminocyclopropane-1-carboxylate oxidase gene family from Petunia hybrida. Plant Mol Biol. 1993;23:1151–1164. doi: 10.1007/BF00042349. [DOI] [PubMed] [Google Scholar]
- Tsuchisaka A, Theologis A. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 2004;136:2982–3000. doi: 10.1104/pp.104.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen WH, Zhou Z, Van Der Straeten D. Regulation of submergence-induced enhanced shoot elongation in Oryza sativa L. Ann Bot. 2003;91 Spec No:263–270. doi: 10.1093/aob/mcf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature. 2004;428:945–950. doi: 10.1038/nature02516. [DOI] [PubMed] [Google Scholar]
- Wang NN, Shih MC, Li N. The GUS reporter-aided analysis of the promoter activities of Arabidopsis ACC synthase genes AtACS4, AtACS5, and AtACS7 induced by hormones and stresses. J Exp Bot. 2005;56:909–920. doi: 10.1093/jxb/eri083. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Hase S, Saigusa M. The effect of ethylene and other regulators on coleoptile growth of rice under anoxia Plant Prod. Sci. 2007;10:468–472. [Google Scholar]
- Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J Biol Chem. 2003;278:49102–49112. doi: 10.1074/jbc.M308297200. [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Ann Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- Yao Y, Du Y, Jiang L, Liu JY. Interaction between ACC synthase 1 and 14-3-3 proteins in rice: a new insight. Biochemistry. 2007;72:1003–1007. doi: 10.1134/s000629790709012x. [DOI] [PubMed] [Google Scholar]
- Yoon GM. New insights into the protein turnover regulation in ethylene biosynthesis. Mol Cells. 2015;38:597–603. doi: 10.14348/molcells.2015.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon GM, Kieber JJ. 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell. 2013;25:1016–1028. doi: 10.1105/tpc.113.110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 2005;5:14. doi: 10.1186/1471-2229-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Wang KL, Chang CM, Mori K, Uchida E, Ecker JR. The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Mol Biol. 2006;62:427–437. doi: 10.1007/s11103-006-9029-7. [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A. Ethylene biosynthesis and action: a case of conservation. Plant Mol Biol. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A. Expression characteristics of OS-ACS1 and OSACS2, two members of the 1-aminocyclopropane-1-carboxylate synthase gene family in rice (Oryza sativa L. cv. Habiganj Aman II) during partial submergence. Plant Mol Biol. 1997;33:71–77. doi: 10.1023/b:plan.0000009693.26740.c3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.