Summary
We describe a novel virus, TPV1 (Thermococcus prieurii virus 1), which was discovered in a hyperthermophilic euryarchaeote isolated from a deep-sea hydrothermal chimney sample collected at a depth of 2700 m at the East Pacific Rise. TPV1 is the first virus isolated and characterized from the hyperthermophilic euryarchaeal genus Thermococcus. TPV1 particles have a lemon-shaped morphology (140 nm × 80 nm) similar to the structures previously reported for Fuselloviruses and for the unclassified virus-like particle PAV1 (Pyrococcus abyssi virus 1). The infection with TPV1 does not cause host lysis and viral replication can be induced by UV irradiation. TPV1 contains a double-stranded circular DNA of 21.5 kb, which is also present in high copy number in a free form in the host cell. The TPV1 genome encompasses 28 predicted genes; the protein sequences encoded in 16 of these genes show no significant similarity to proteins in public databases. Proteins predicted to be involved in genome replication were identified as well as transcriptional regulators. TPV1 encodes also a predicted integrase of the tyrosine recombinase family. The only two genes that are homologous between TPV1 and PAV1 are TPV1-22 and TPV1-23, which encode proteins containing a concanavalin A-like lectin/glucanase domain that might be involved in virus–host recognition.
Introduction
Since the discovery of Archaea, the third domain of life, by Woese and Fox (1977), extensive studies have been carried out on their extra-chromosomal genetic elements (viruses and plasmids). To date about 50 archaeal viruses have been described in detail. Archaeal viruses have been detected in such extreme habitats as hypersaline environments (Porter et al., 2007), hot and acidic springs (Rachel et al., 2002), and hydrothermal deep-sea vents (Geslin et al., 2003a,b). The majority of archaeal viruses was isolated from hot terrestrial and coastal environments and infects members of the phylum Crenarchaeota (one of the two major archaeal phyla), in particular the genera Sulfolobus, Thermoproteus, Acidianus, Pyrobaculum, Stygiolobus and Aeropyrum. These viruses show unexpected diversity at the morphological and genomic levels and have been classified into eight new viral families (reviewed by Prangishvili et al., 2006; Mochizuki et al., 2010).
The second major archaeal phylum, Euryarchaeota, includes methanogens, halophiles, thermophilic and extremely acidophilic microorganisms of the order Thermoplasmatales and hyperthermophilic microorganisms of the orders Thermococcales and Archaeoglobales and show much greater physiological and metabolical diversity than Crenarchaeota. Conversely, viruses described to date in Euryarchaeota are much less diverse than viruses of Crenarchaeota. Most of the isolated euryarchaeal viruses have been isolated from mesophilic organisms and display morphologies similar to those of head-and-tail bacteriophages. The few viruses isolated from methanogens such as ‘ΨM1-like viruses’ belong to the Siphoviridae family (Pfister et al., 1998). Haloarchaeal viruses such as phiCH1, phiH, HF2, BJ1, belong to the Myoviridae or Siphoviridae families (Schnabel et al., 1982; Nuttall and Dyall-Smith, 1993; Witte et al., 1997; Pagaling et al., 2007), except for His 1 and His 2, which infect Haloarcula hispanica and are classified in a novel virus group the Salterprovirus (Bath et al., 2006). His 1 and His 2 are lytic lemon-shaped haloviruses with linear dsDNA genomes of 14.4 kb and 16 kb respectively. Another exception is HRPV-1, a non-lytic pleomorphic halovirus, which is the first characterized archaeal virus with a single-stranded DNA genome (Pietilä et al., 2009).
No virus has been so far isolated from thermophilic or hyperthermophilic euryarchaea of the orders Thermoplasmatales and Archaeoglobales, and only one virus-like particle (VLP) has been described from the order Thermococcales (Geslin et al., 2003a). Thermococcales is one of the predominant groups of hyperthermophilic microorganisms isolated from deep-sea hydrothermal vents (Prieur et al., 2006). These ecosystems represent one of the most extreme environments on Earth. They are characterized by strong physicochemical gradients such as temperature from 2°C to more than 350°C and high hydrostatic pressure, lack of solar energy and prevalence of chemosynthesis. The order Thermococcales is represented by three genera: Thermococcus, Pyrococcus and Paleococcus (Stetter, 1986; Achenbach-Richter, 1988; Takai et al., 2000), and includes obligate anaerobic, sulfur-metabolizing hyperthermophiles.
The widespread Thermococcales have become popular model organisms for studies on microbial adaptation to extreme temperature and ionizing radiation, DNA replication mechanisms, metabolism, phylogeny and genome evolution (Prieur et al., 2004; Leigh et al., 2011; Soler et al., 2011). Accordingly, intensive effort has been made to sequence diverse genomes of members of this order, resulting in the availability of seven genomes of Thermococcus (T. kodakaraensis, T. gammatolerans, T. onnurineus, T. sibiricus, T. barophilus, Thermococcus sp. AM4, Thermococcus sp. 4557) and five genomes of Pyrococcus (P. horikoshii, P. furiosus, P. abyssi, P. yayanosii, Pyrococcus sp. NA2).
The availability of all these data makes Thermococcales a good model to explore the hyperthermophilic euryarchaeal virome that currently remains largely uncharacterized. Several proviruses have been detected in genomes of Thermococcales (TKV1 to TKV4 in T. kodakaraensis (Fukui et al., 2005) (TGV1 and TGV2 in T. gammatolerans) (Zivanovic et al., 2009) (PHV1 in P. horikoshii genome) (Soler et al., 2010). To investigate the diversity of VLPs from deep-sea vents, a systematic search was carried out on samples collected in various geographically distant hydrothermal sites located on the East Pacific Rise (EPR 9° N and 13° N) and Mid-Atlantic Ridge (MAR 36° N and 37° N). This study revealed striking similarity between VLP morphologies in deep-sea and terrestrial hot environments (Geslin et al., 2003b).
All this effort notwithstanding PAV1, isolated from P. abyssi (Geslin et al., 2003a), is the only VLP described in Thermococcales. The PAV1 virion is lemon-shaped (120 nm × 80 nm) and is morphologically similar to the members (SSV1-7, SSVk1, SSVrh, ASV1) of the Fuselloviridae family (Palm et al., 1991; Schleper et al., 1992; Stedman et al., 2003; Wiedenheft et al., 2004; Peng, 2008; Redder et al., 2009) and Salterprovirus genus (His1 and His2) (Bath et al., 2006). The viral particle persists in the host strain in a stable carrier state. The genome of PAV1 consists of a double-stranded circular DNA of 18 kb and is also present at a high copy number in a free form in the host cell. The genome of PAV1 displays unique genetic features and does not show affinity to any known virus family.
Isolation of new viruses from Thermococcales is the first step required to characterize viral diversity in deep-sea hydrothermal environments and eventually the role of viruses in these extreme ecosystems.
Here we report the characterization of a virus, Thermococcus prieurii virus 1 (TPV1), from a new species of Thermococcus isolated from a hydrothermal deep-sea vent. To date TPV1 and PAV1 are the only viruses isolated from described marine hyperthermophilic archaea (Euryarchaeota). TPV1 is the first virus isolated and described from the hyperthermophilic Thermococcus genus. We show that TPV1 is UV inducible and causes a growth retardation of species of Thermococcus, in contrast to PAV1 for which infectivity has not been shown. These properties make TPV1 a promising element to study host–virus interactions in Thermococcales. We further present the results of analysis of the complete sequence of TPV1 genome (21.5 kb), which shows very limited similarity to PAV1 indicating that TPV1 might become a founding member of a new virus family.
Results
Viral morphology
Particles of TPV1 were lemon-shaped, about 140 nm long and 80 nm wide, with a 15 nm – tail terminated by caudal fibres (Fig. 1A), which are likely to participate in virus adsorption on the host cells. Clusters of virus particles forming rosette-like aggregates were also observed (Fig. 1B).
Fig. 1.
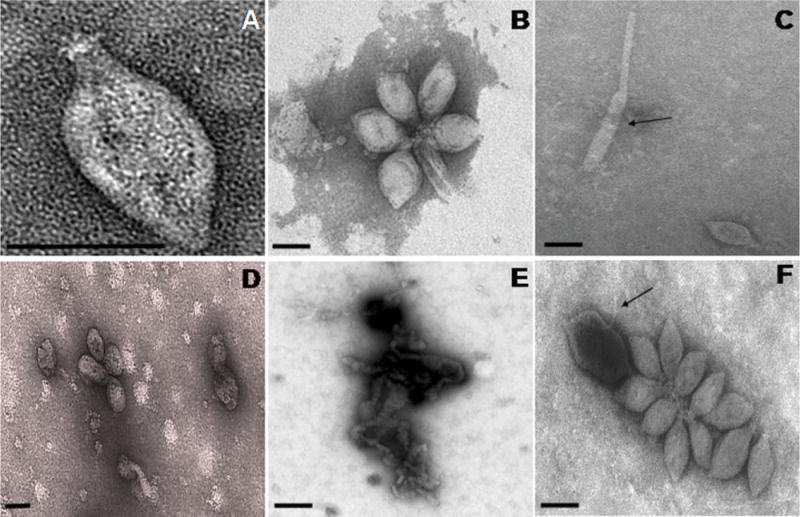
A. Electron micrographs of TPV1 negatively stained with 2% uranyl acetate. Lemon shaped particle with a tail terminated by fibres.
B. Clusters of virus particles forming rosette-like aggregates.
C. Elongated virus particles (arrow).
D. Effect of exposure to chloroform on virus particle structure.
E. Disrupted virions caused by exposure to proteinase K.
F. Effect of acidic pH on virions structure (arrow). Scale bars represent 120 nm.
In CsCl2 solution, TPV1 had a density of 1.35 g ml−1 and in iodixanol solution, TPV1 had a density of 1.20 g ml−1. TPV1 particles seemed not to be sensitive to purification procedures, e.g. high-speed centrifugation or suspension in iodixanol or CsCl2 solutions, indicating that they were osmotically resistant. Infrequently virus particles appeared to be flexible and a few elongated forms were observed indicating plasticity of the particle shape (Fig. 1C).
The structural stability of TPV1 was assessed by exposing purified virions to organic solvents, detergents and proteolytic degradation. The viral particles appear to be sensitive to chloroform (25% wt/vol) because only spherical particles were observed in treated samples (Fig. 1D). The particles were also disrupted by treatment with 0.3% Triton X-100 (data not shown) as well as by incubation with 0.1% sodium dodecyl sulfate (SDS) (data not shown). Treatment with proteinase K (1 mg ml−1) resulted in complete destruction of virions (Fig. 1E). These preliminary results suggest that TPV1 is covered by an envelope composed of proteins and lipids. The purified virions were tolerant to all temperatures (4°C–85°C) and pH (4–11) tested (Fig. 1F). Thus, TPV1 seems to be psychro/thermotolerant and stable under acidic and basic conditions.
Virus–host relationship
Viruses appeared to be released continuously and spontaneously in the extracellular medium throughout the growth of T. prieurii (Fig. 2). Due to the lack of a plaque assay for Thermococcales, quantification of TPV1 was performed by epifluorescence microscopy. The viral count reached its maximum after 18 h of incubation (2 × 108 virus particles ml−1). During growth of the infected host, there was neither a decrease of cellular density nor formation of cell debris. This observation indicated that viral production was not accompanied by lysis of the host cells. Attempts were made to increase the viral production by induction using different chemical or physical treatments and physiological stresses. Induction was never observed under any stress condition except for UV. UV irradiation of growing host cells induced the production and release of TPV1. The viral production began almost immediately after UV irradiation, followed by an increase of the viral titre, which reached about 1011 virus particles ml−1 15 h after induction. UV exposure for 1 h was required to obtain the higher production of TPV1. A significant decrease in host cell counts and concomitant increase in cell debris were observed after 9 h incubation post irradiation (Fig. 2). Despite the presence of cell debris, no extensive lysis of the host was detected suggesting that TPV1 is a temperate virus.
Fig. 2.
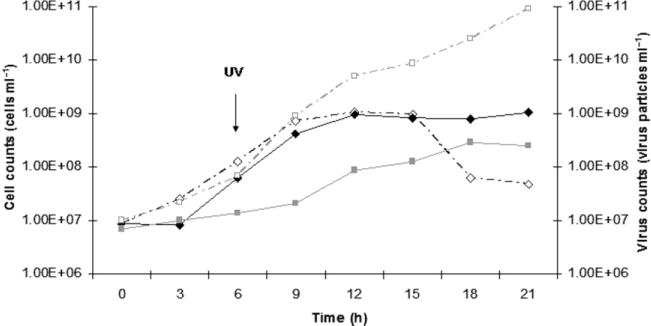
Typical growth curve (black diamonds) of the host and production of TPV1 (grey squares). UV induction. Cultures were grown and irradiated for 1 h (white diamonds) and the virus counts were estimated by epifluorescence microscopy (white squares).
Figure S1 shows many viruses protruding through the envelope, obviously during extrusion. Protrusions of particles through the cell envelope were an indication of viral production and were never observed in unirradiated cultures (Fig. S1).
Host range
The host range of TPV1 was tested in liquid medium by adding iodixanol purified viruses to growing cultures of potential hosts: Thermococcus siculi, T. barophilus, T. gorgonarius, T. kodakaraensis, T. celer, T. pacificus and T. fumicolans. TPV1 production in infected cultures was monitored by epifluorescence microscopy. Of the potential host strains tested, TPV1 was able to cause, in liquid medium, a substantial delay of T. barophilus, T. celer, T. kodakaraensis and T. gorgonarius growth during approximately 20 h after infection (Figs 3 and S2).
Fig. 3.
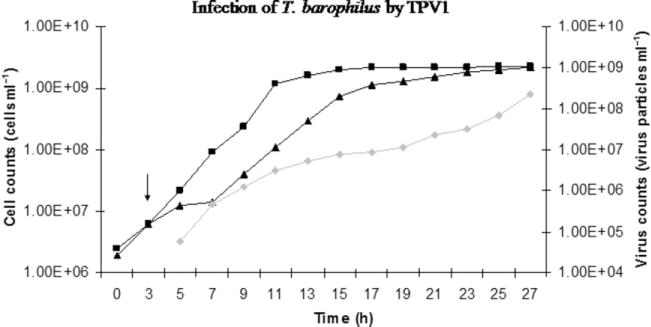
Infection of T. barophilus by TPV1. Squares: cells of T. barophilus non-infected. Triangles: cells of T. barophilus infected with TPV1. Diamonds: free TPV1 virions. The arrow indicates the addition of TPV1 virions in the culture.
To confirm that the viral production occurred inside the infected host, aliquots of T. barophilus infected with TPV1 were collected after 1 h and 12 h of incubation. Total DNA was extracted, and using specific primers, a viral DNA fragment of 600 bp was amplified by PCR from the DNA extracted after 12 h of incubation but not after 1 h of incubation (Fig. S3).
Viral genome
Three extra-chromosomal elements were isolated from Thermococcus prieurii by the alkaline lysis method (Fig. S4A) (A. Gorlas and C. Geslin, unpublished). We suspected that one of these elements could be the free form of the TPV1 genome. To test this hypothesis, the viral genome was extracted from purified particles and was compared with cccDNAs (covalently closed circular DNA) isolated from the host strain (Fig. S4B). The same viral band was found in both cases. Samples of viral DNA extracted from the virions were heated for 10 min at 70°C and chilled on ice before loading on gels to determine if the viral DNA was in fact not circular in the virions, but rather linear with cohesive ends that cause circularization. No change in the band pattern was observed (Fig. S4C). Nucleic acid extracted from purified virus particles was digested by a number of type II restriction enzymes and was not sensitive to RNase. We conclude that the viral genome is a double-stranded, circular DNA that is not affected by Dcm or Dam-like methylation. The genome size was estimated at 21.5 kb (Fig. S4D).
The copy number of the episomal form of the viral genome in the host was evaluated by real-time PCR. A viral fragment of 160 bp and 160 bp of the 16S rDNA of the host were amplified by qPCR. There was a linear correlation between the threshold cycle (ct) and the initial quantity of DNA. The standard curves (for viral and chromosomal fragments) showed slopes of −3.27 and −3.13, respectively, indicating high effectiveness of PCR. The assays for viral and host DNA showed a strong correlation (R2 ≥ 0.98). The result indicated an intracellular copy number of TPV1 genome of approximately 20 copies per host chromosome (Figs S5 and S11).
The circular, dsDNA genome of TPV1 was totally sequenced using a combination of overlapping clone library and primer walking.
Genome analysis of TPV1
The complete nucleotide sequence of the TPV1 genome was determined on both strands as outlined in Experimental procedures with a minimum threefold coverage. The genome size of TPV1 determined by sequencing is 21 592 bp. Sequence analysis in the six possible frames resulted in the prediction of 28 protein-coding genes each of which encoded a putative protein product of at least 62 amino acids; collectively, these predicted genes covered 90.6% of the genome sequence (Fig. 4). The positions and main features of the predicted genes are listed in Table 1. The majority of the putative genes (24) have the same orientation and four are present on the complementary strand. The overall G + C content of the TPV1 genome (49.9%) is comparable with the overall G + C content of the host genome (53.6%) (e.g. A. Gorlas, unpublished). All predicted coding regions were preceded by putative ribosome-binding sites (see Table S1 for details). The predominant start codon is ATG (85.5 %) and three ORFs start from GTG.
Fig. 4.
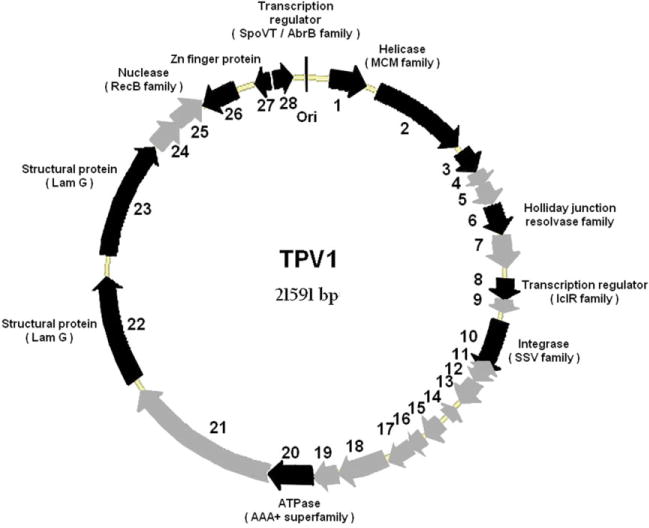
TPV1 genome map. Predicted genes are presented by thick arrows; grey arrows indicate ORFs with no similarity or unassigned function and black arrows indicate either conserved hypothetical ORFs or ORFs with an attributed function. The approximate location of the origin (Ori) of replication is also indicated.
Table 1.
General features of the predicted genes (ORFs) of TPV1 virus.
| Predicted gene | Predicted protein size (aa) | Closest homologue: e-value, % identity, taxonomy | Homologues: taxonomic range | Conserved motifs/domains and predicted function(s) | Comments |
|---|---|---|---|---|---|
| TPV1-1 | 200 | TK_1361. 2e-45; 44% TKV4 (T. kodakaraensis) |
TKV4 (T. kodakaraensis) only | None | N-terminal part of the MCM2-like helicase in TKV4 but a separate protein in TPV1 |
| TPV1-2 | 561 | TK_1361. 7e-88; 36% TKV4 (T. kodakaraensis) |
All archaea and eukaryotes | All conserved motifs of the MCM family within the AAA+ ATPase superfamily. MCM2-like helicase involved in DNA replication initiation (licensing factor). | C-terminal part of the MCM2-like helicase in TKV4 but a separate protein in TPV1; contains all elements required for licensing factor activity |
| TPV1-3 | 136 | None | None | MutL-transducer domain (e-value 1.7e-03, RPS-BLAST); possible involvement in replication, interaction with TPV1-2. | In DNA gyrase, involved in transduction of structural change from ATP-binding site to breakage-rejoining Toprim domain |
| TPV1-4 | 83 | None | None | None | |
| TPV1-5 | 133 | TGAM_0671. 1e-47; 61% TGV1 (T. gammatolerans) |
T. gammatolerans provirus and T. barophilus plasmid | None | |
| TPV1-6 | 174 | TGAM_0672. 8e-119; 94% TGV1 (T. gammatolerans) |
Most archaea, some bacteria, some archaeal and bacterial viruses | Archaeal-type Holliday junction resolvase, all catalytic motifs conserved | Member of a distinct family of Holliday junction conserved in Thermococcales and Archaeoglobales |
| TPV1-7 | 187 | None | None | None | |
| TPV1-8 | 117 | TK0576/TK0420. 1e-54; 81% TKV3/TKV2 (T. kodakaraensis) |
Many archaea and bacteria | Winged helix–turn–helix domain; transcription regulator of the IclR family | |
| TPV1-9 | 71 | None | None | Predicted integral membrane protein | |
| TPV1-10 | 318 | TK0381. 9e-66; 45% TKV2 (T. kodakaraensis) |
Many archaeal viruses and proviruses integrated in archaeal genomes | SSV1-type integrase | |
| TPV1-11 (−) | 87 | None | None | None | |
| TPV1-12 | 119 | None | None | Predicted integral membrane protein | |
| TPV1-13 | 136 | None | None | Predicted integral membrane protein | |
| TPV1-14 (−) | 62 | None | None | None | |
| TPV1-15 | 112 | None | None | Predicted integral membrane protein | |
| TPV1-16 | 89 | None | None | Predicted integral membrane protein | |
| TPV1-17 | 152 | None | None | None | |
| TPV1-18 | 285 | None | None | Predicted integral membrane protein | |
| TPV1-19 | 147 | None | None | None | |
| TPV1-20 | 258 | MA2591. 0.026; 26% Methanosarcina acetivorans |
Distant homologues in all Archaea, bacteria and eukaryotes, some archaeal viruses | A distinct AAA+ superfamily ATPase; all motifs characteristic of active ATPases conserved | AAA+ ATPases with low sequence similarity but similar size and arrangement of conserved motifs found in several crenarchaeal viruses (Fig. S10) |
| TPV1-21 | 884 | None | None | Predicted non-globular protein | Large non-globular proteins are common among bacteriophage tail subunits |
| TPV1-22 | 623 | PAV1_ORF676. 7e-15; 23% PAV1 |
Distant homologues in many Archaea, bacteria and eukaryotes | Concanavalin A-lectin/glucanase domain (jelly roll fold); two predicted transmembrane helices | Potential role in adsorption |
| TPV1-23 | 688 | PAV1_ORF678. 3e-91; 36% | Distant homologues in many Archaea, bacteria and eukaryotes | Concanavalin A-lectin/glucanase domain (jelly roll fold); three predicted transmembrane helices | Potential role in adsorption |
| TPV1-24 | 183 | None | None | None | |
| TPV1-25 | 182 | PNA2_1322. 0.002; 34% Pyrococcus sp (predicted provirus) |
TKV3 (TK_0597) and other putative proviruses in Thermococcales | None | |
| TPV1-26 (−) | 234 | PNA2_0680. 7e-30; 37% Pyrococcus sp. |
Distant homologues in most archaea and bacteria | A distinct subfamily of RecB family nucleases; restriction-like endonuclease superfamily motifs | |
| TPV1-27 (−) | 83 | Arcve_1816. 6e-18; 71% Archaeoglobus veneficus |
Archaeoglobus veneficus, A. fulgidus, Pyrococcus sp.; more distant homologues in all eukaryotes | C2H2 Zn finger | |
| TPV1-28 | 106 | PNA2_0681. 6e-12; 33% Pyrococcus sp. |
Most Archaea (including TKV2/3), many bacteria | Helix–turn–helix domain; transcription regulator of the SpoVT/AbrB family |
In an attempt to identify the origin of replication, a cumulative GC skew analysis was performed (Fig. S6). However, the results of this analysis failed to clearly pinpoint the origin. One inflection point, between positions 19 160 and 21 080 bp, was evident, indicating a change in the base composition bias. A region containing AT-rich repeats was detected at positions 19 770 to 19 850 bp, and the DNA sequence between positions 20 675 and 21 592 bp contained the largest intergenic region, 917 bp. Furthermore, helical stability analysis (Huang and Kowalski, 2003) indicates that positions 21 241 bp to 21 341 bp correspond to the lowest helical stability region, suggesting that this region contains a DNA unwinding element (DUE).
To identify homologous relationships and predict functions of the putative proteins of TPV1, their amino acid sequences were compared with those in the public sequence databases using the PSI-BLAST and HHPred methods (Table 1). For more than half of the predicted proteins (16 out of 28), no significant similarity with any sequences in the public databases was detected. However, six of these ‘ORFans’ encode predicted membrane proteins (Table 1). Notably, genes encoding the predicted integral membrane proteins form a cluster in the middle of the TPV1 genome suggesting that these proteins are coexpressed and might form distinct, membrane-spanning complexes in TPV1 virions.
The TPV1-2 gene encodes a predicted helicase of the minichromosome maintenance (MCM) family within the AAA+ superfamily of P-loop ATPases (Koonin, 1993; Iyer et al., 2004). The closest homologue of this TPV1 protein was found in the provirus TKV4 (TK1351), with 35% identity (Krupovic and Bamford, 2008). The MCM protein of TPV1 possesses all the conserved motifs that are known to be required for the ATPase and helicase activities (Walters and Chong, 2010) (Fig. S7). Phylogenetic tree analysis placed the TPV1 MCM protein into a strongly supported group of proviral and archaeal proteins from Thermococcales (Fig. S8). The MCM proteins are so-called licensing factors that are involved in the initiation of DNA replication in all archaea and eukaryotes (Sakakibara et al., 2009), and a similar role in viral genome replication can be inferred for TPV1-2.
TPV1-20 encodes another predicted AAA+ superfamily ATPase (Iyer et al., 2004) that has no closely related homologues but, in terms of size and spacing of conserved motifs implicated in ATPase activity resembles, ATPases encoded by several viruses (e.g. SSV1) and plasmids of hyperthermophilic Crenarchaeota (Table 1 and Fig. S10). These proteins are in some cases described as DnaA-like but their similarity to DnaA, the protein involved in DNA replication initiation in all bacteria, is remote.
TPV1-6 encodes a predicted archaeal-type Holliday junction resolvase (Aravind et al., 2000), with the closest homologue is encoded in the TGV1 provirus integrated in the genome of T. gammatolerans (Zivanovic et al., 2009). TPV1-28 encodes another predicted nuclease that belongs to the RecB family (Aravind et al., 1999), with the closest homologue present in the genome of Pyrococcus sp. NA2 (Lee et al., 2011).
TPV1-10 encodes an integrase (SSV-type) that belongs to the tyrosine recombinase family. The closest homologue of this protein is present in the TKV2 provirus (Fukui et al., 2005), and more distant homologues are found in numerous archaeal viruses and integrated virus-like elements. The multiple alignment of SSV-type integrase sequences and TPV1-10 shows the conservation of the tyrosine recombinase superfamily catalytic site motif R … KXXR … Y (She et al., 2004) (Fig. S9).
Two genes of TPV1 encode predicted helix–turn–helix transcription regulators, one of the IclR family (TPV1-8) and the other of the SpoVT/AbrB family (TPV1-28). Highly conserved homologues of both these proteins are present in the TKV3 genome (Table 1). SpoVT/AbrB-like transcriptional regulators have also been detected in the genomes of two crenarchaeal viruses, STSV1 and SIFV (Prangishvili et al., 2006; Krupovic and Bamford, 2008), and in the pT26-2 plasmid from Thermococcus sp. (Soler et al., 2010). These transcriptional regulators seem to represent a distinct family of SpoVT/AbrB-like protein that is specific to archeoviruses.
On the complementary strand, TPV1-27 encodes a C2H2-type Zn-finger protein that is highly similar to small proteins from Pyrococcus sp. and two Archaeoglobus species. The phylogenetic spread of this protein family is unusual in that, except for TPV1 and three archaeal species, highly conserved homologues are present only in eukaryotes. This protein potentially could be an additional regulator of TPV1 gene expression.
The domain architectures of the predicted proteins encoded by tandem genes TPV1-22 and TPV1-23 are similar and include, respectively, two and three predicted transmembrane segments, in the C-terminal portion and a concanavalin A like lectin/glucanase (LamGL) domain in the N-terminal portion. These predicted proteins are homologous to two proteins (ORF676 and ORF678) of PAV1 (Geslin et al., 2007). Similarities with LamGL domain-containing protein encoded in the genomes of crenarchaeal lipothrixviruses (AFV3, AFV6 and AFV8) were also detected. Many concanavalin A-like domains are contained in proteins involved in cell recognition and adhesion (Crennell et al., 1994; Tisi et al., 2000).
Discussion
Approximately 40 archaeal species of the order Thermococcales have been described (Prieur et al., 2006) but despite the prominence of this euryarchaeal order in marine thermophilic ecosystems, little is known about their viruses. It is unclear whether this lack of described viruses is due to the low abundance of viruses in Thermococcales or insufficient screening, or both. An unexpected morphological diversity of VLPs was observed in high temperature enrichment cultures from deep-sea hydrothermal samples where Thermococcales are well represented. Among the viral morphotypes observed, the lemon-shaped type prevailed (Geslin et al., 2003b). Thus, it seems likely that viruses are as common in Thermococcales as they are in hyperthermophilic Crenarchaeota, and the main reason for the current lack of isolated viral forms could be insufficient experimental effort. A major goal of future research will be to combine omics and cultural approaches mimicking as close as possible environmental conditions (i.e. a multiplicity of physical and chemical gradients) to explore the virome in deep-sea hydrothermal environments.
Only one viral particle, PAV1 (Pyrococcus abyssi virus 1), has been described from hyperthermophilic Euryarchaeota (Geslin et al., 2003a; 2007). Here, we discuss the characterization of the second lemon-shaped virus (TPV1: Thermococcus prieurii virus 1) discovered in a hyperthermophilic Euryarchaeota isolated from a deep-sea hydrothermal vent.
TPV1, which is not yet classified, shows morphological similarities with Fuselloviruses (Peng, 2008), His1/His2 of the Salterprovirus group (Bath et al., 2006) and to the unclassified PAV1 (Geslin et al., 2003a). This lemon morphology is commonly observed among archaeal viruses isolated from the environments inhabited by hyperthermophilic, acidophilic and halophilic archaea (Zillig et al., 1994; Oren et al., 1997; Rice et al., 2001; Rachel et al., 2002). With regard to the taxonomic status of the viruses of Thermococcales, TPV1 was found to infect a Thermococcus strain and PAV1 a Pyrococcus strain; these strains were isolated from two distinct hydrothermal deep-sea vents, respectively, in the North East-Pacific Rise and in the North Fiji Basin. The two euryarchaeal viruses, PAV1 and TPV1, have similar lemon-shaped morphologies but share only two homologous genes detectable at the sequence level. The lectin domain-containing proteins encoded by these paralogous genes are exposed on the surface of the enveloped virus and are likely to be involved in host cell recognition. These two viruses might become prototypes for two novel viral families or possibly two genera within the same family.
TPV1 causes growth retardation of several species of Thermococcus like T. barophilus, T. celer, T. gorgonarius and T. kodakaraensis isolated respectively from a hydrothermal vent site on the Mid-Atlantic Ridge (Marteinsson et al., 1999), from solfataric marine water in Italy (Zillig et al., 1983), from a New Zealand submarine hot vent (Miroshnichenko et al., 1998) and from a solfatara in Japan. The fact that TPV1 presents a wide host range with a broad geographic distribution and delays the growth of T. kodakaraensis, which is a hyperthermophilic model organism for genetic studies, makes TPV1 a promising tool – to study virus–host interactions in Thermococcales.
TPV1 is released during all phases of host growth. Virions extrude from cells without causing host lysis. In contrast to PAV1, TPV1 viral production can be induced with UV irradiation, and this production is accompanied by a decrease in cell density. In UV-induced cultures, viral titre peaked at > 1011 particles ml−1. It is notable that TPV1 is UV inducible whereas PAV1 is not although the implications of this difference for the biology of virus–host interactions remain unclear. It will be important to address this question in follow-up studies, and one approach would involve characterization of the transcription cycle of the UV inducible TPV1 by Northern analysis. Such analysis has been performed for SSV1, and it has been shown that SSV1 exhibits temporal regulation of transcription upon UV irradiation and that the cycle starts with a small UV-specific transcript (Fröls et al., 2007).
Little is known about the modes of entry or release of archaeal viruses (Bize et al., 2009). Attempts to analyse the processes of TPV1 adsorption and extrusion by visualization of thin sections by electron microscopy have, thus far, been unsuccessful. Like many archaea, T. prieurii has only a thin, glycoprotein surface layer (S-layer) protecting the cell membrane, which means that release of virions does not necessarily require breakdown of the cell wall and subsequent cell lysis. TPV1 seems to extrude without cells disruption, so it should have a release mechanism similar to that of Fuselloviruses. Indeed, similarly to PAV1 or Fuselloviruses, TPV1 appears to be a non-lytic virus. The host cells produce virions constitutively, consistent with an equilibrium existing between viral replication and host cell multiplication. Moreover, these viruses persist in cells in a stable state and are not lost during continuous growth of infected cell cultures (Prangishvili and Garrett, 2005).
In contrast to PAV1, TPV1 is likely to integrate into the host chromosome. Viral integration is a process widely shared in viruses present in the three domains of life. In the extreme environments, proviruses could contribute to the fitness of the host strains whereas for the virus, the integrated state represents a means to avoid the harsh conditions of these ecosystems (Williamson et al., 2008). The genome of TPV1 contains a gene encoding a tyrosine integrase homologous to the integrases of Fuselloviruses and putative proviruses (TKV, TGV, PHV) as well as the plasmid pT26-2 isolated from Thermococcales (Fukui et al., 2005; Zivanovic et al., 2009; Soler et al., 2010). A characteristic feature of the SSV-type integrase is that the gene is partitioned into longer Int (C) and shorter Int (N) fragments after integration in the host chromosome (She et al., 2004). Integration of Fuselloviruses occurs within specific tRNA genes (for example: tRNAArg for SSV1, tRNAgly for SSV2 and SSV7, tRNAglu for SSV4) (Redder et al., 2009). In the case of TPV1, additional analyses remain to be done in order to identify potential viral integration into the host chromosome.
All archaeal viruses described so far possess dsDNA genomes, with the sole exception of HRPV-1, a recently isolated haloarchaeal virus with a ssDNA genome (Pietilä et al., 2009). The 21.5 kb dsDNA genome of TPV1 is packaged into viral particles and an episomal form is present in the host cells. The genome of TPV1 is circular like the genomes of PAV1 and HRPV-1. All other known euryarchaeal viruses have a linear genome (Dyall-Smith et al., 2003; Porter et al., 2005).
Functions and/or biochemical activities were assigned to approximately half of the predicted TPV1 proteins on the basis of homology to functionally characterized proteins, conservation of functional motifs and prediction of structural features. A substantial fraction of TPV1 genes (eight out of 28) encodes predicted membrane proteins that most likely are components of the viral envelope. In contrast, the identity of the major capsid protein of TPV1 remains unknown; detailed searches indicate that TPV1 does not encode a homologue of the icosahedral capsid protein found in TKV4 (Krupovic and Bamford, 2008). The TPV1 genome encodes several proteins implicated in genome replication, such as a minichromosome maintenance protein (TPV1-2). MCM proteins are DNA-dependent ATPases involved in the initiation of archaeal and eukaryal DNA replication (Sakakibara et al., 2009; Krupovic et al., 2010). MCM proteins have been identified in one Bacillus phage genome (Krupovic and Bamford, 2008), in one halovirus BJ1 (Pagaling et al., 2007) and in the provirus TKV4 (Krupovic and Bamford, 2008). Phylogenetic analysis clearly shows the common origin of the MCM proteins of TPV1 and TKV4. The TPV1-20 encodes a putative AAA+ superfamily ATPase that has no highly conserved homologues, but in size and location of conserved motifs, resembles predicted ATPases encoded by many crenarchaeal viruses. Thus, TPV1 encodes an unusual combination of two ATPases that are both likely to be involved in genome replication initiation. Involvement in genome replication and/or recombination and repair is predicted also for the Holliday junction resolvase (TPV1-6) and the RecB-family nuclease (TPV1-26) and encoded in the TPV1 genome.
Genomic alignment shows that TPV1 shares more homologous genes with autonomous genetic elements (proviruses and plasmids) from Thermococcus species than with PAV1, the only other known virus for hyperthermophilic euryarchaea (Fig. 5). On the whole, the genomes of all these elements show a patchy pattern of gene sharing, resembling that revealed by genomic comparison of Crenarchaeal viruses (Prangishvili et al., 2006). The SSV-type integrase that is missing only in PAV1 seems to be a signature of viruses and virus-like elements of hyperthermophilic euryarchaea and could be used as a marker to explore the viral diversity in the Thermococcales order. Moreover, the frequent occurrence of homologous resolvases and transcription regulators is notable as well (Fig. 5).
Fig. 5.
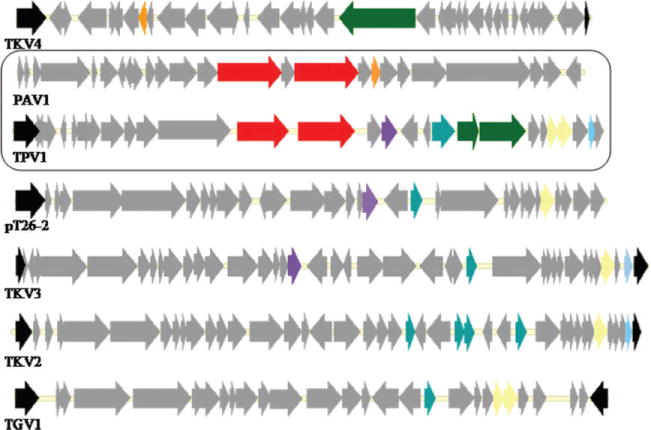
Alignment of the genomes of TKV2-4, pT26-2, TGV1, PAV1 and TPV1, with a focus on genes present in the TPV1 and/or PAV1 genomes, and shared with other euryarchaeal genes are a colour coded as follows: integrase in black, the LamG domain protein in red, the MCM protein in green, transcription regulator SpoVT/AbrB in dark blue and the resolvase in yellow. Orange, light blue and mauve ORFs represent genes encoding uncharacterized conserved proteins.
Although horizontal gene transfer occurs in Thermococcales (Soler et al., 2011), the mechanisms of gene disper sion and its frequency remain unknown. Nevertheless, it is evident that the identified genetic elements including viruses, proviruses and plasmids share a common pool of genes and probably play an important role in the evolution of Thermococcales.
Experimental procedures
Thermococcales strains
Thermococcus celer JCM (Japan Collection of Microorganisms) 8558T, T. siculi DSM (Deutsche Sammlung von Mikroorganismen) 12349T, T. barophilus DSM 11836T, T. kodakaraensis JCM 12380T, Thermococcus gorgonarius DSM 10395T, Thermococcus pacificus DSM 10394T, Thermococcus fumicolans DSM 12820T were used as reference strains and provided by the Brittany culture collection.
The new species T. prieurii strain Bio-pl-0405IT2 (JCM16307T = LMM3069) is the host of the virus TPV1, and carries also two other extra-chromosomal elements in a free form in the cytoplasm. Thermococcus prieurii was isolated from a hydrothermal chimney sample collected from the East Pacific Rise at 2700 m depth on the ‘Sarah Spring’ area (7°25′24 S, 107°47′66 W).
Culture conditions
Thermococcales strains were cultured in Ravot medium as previously described (Geslin et al., 2003a) with minor modifications as described in Supporting information.
Isolation of TPV1
A total of 150 novel hyperthermophilic and anaerobic microorganisms isolated from deep-sea hydrothermal vents in the East Pacific Rise were screened for genetic elements.
From these 150 isolates, cccDNA were extracted by the alkaline lysis method allowing us to detect presence of extra- chromosomal DNA, in free form in the host cytoplasm. From positive samples, epifluorescence microscopy was performed and isolates scored positive were checked for the presence of virus particles by transmission electron microscopy. One of them, the new species T. prieurii was discovered to be the host of TPV1.
Purification of TPV1
The viral suspension was purified by ultracentrifugation in a linear Iodixanol gradient [OptiPrep, 30–45% diluted in a buffer (10 mM Tris-HCL, 100 mM NaCl, 5 mM CaCl2)] at 180 000 g for 6 h (Beckman Optima LE-80 K 70.1Ti rotor). Following ultracentrifugation, the opaque virus band was recovered and stored at 4°C until use.
TPV1 particles were also purified by centrifugation in a CsCl buoyant density gradient (Beckman Optima LE-80 K 70.1Ti rotor) at 180 000 g for 6 h. Fractions containing the nucleic acids were detected at 254 nm and collected using a density gradient fractionator (model 185, ISCO). Fractions were then dialysed by using nitrocellulose filters (MF Membrane Filter, 0.025 μm, Millipore) placed above a large volume of buffer given above. Viral solutions dialysed were recovered on the surface of the filter.
Virus counts by epifluoresence microscopy
The virus counts were determined by epifluorescence microscopy according to Noble and Fuhrman (Noble and Fuhrman, 1998) and as described in supporting information. At 1000 × magnification 20–100 positive particles were counted in each of 20 random fields. The average number of viruses per millitre was calculated according to Suttle (Suttle and Fuhrman, 2010).
Transmission electronic microscopy
A droplet of a viral solution dialysed and resuspended in a buffer (10 mM Tris-HCL, 100 mM NaCl, 5 mM CaCl2), was adsorbed onto a carbon-coated copper grid for 2 min. After removed the excess of liquid, the sample was negatively stained with 2% uranyl acetate for 45 s, as described by Geslin and colleagues (2003a). Specimens were examined using a JEOL electron microscope, JEM 100 CX II.
Virus sensitivity and stability
All assays were performed with viral suspensions purified by ultracentrifugation in CsCl, dialysed and resuspended in the buffer containing 10 mM Tris-HCL, 100 mM NaCl, 5 mM CaCl2.
A suspension of purified TPV1 was exposed to chloroform (25 % wt/vol) for 1, 2 and 5 min at room temperature with constant agitation. TPV1 was also exposed to 0.3% (wt/vol) Triton X-100 for 1 and 3 min at room temperature, and to 0.1 % (wt/vol) SDS for 3 min at 50°C. TPV1 was also incubated with proteinase K (1 mg ml−1) for 1 h at 56°C in a reaction buffer containing 10 mM Tris (pH 7.8), 5 mM EDTA and 0.5% SDS.
Finally purified viruses were incubated at different temperatures (4°C, 20°C, 37°C, 50°C, 75°C and 85°C for 1 and 15 h in the buffer used to resuspend TPV1) and different pH (4 and 11.5 for 1 h and 15 h) in acid or basic buffer. To determine viral stability, all samples obtained from the different conditions tested were examined under a transmission electron microscope (JEM 100 CX II).
Induction assays
Attempts were made to increase the viral production by induction using different chemical or physical treatments that damage DNA (antibiotics, UV) and exposure of the T. prieurii host cells to physiological stresses (aerobiosis, hydrostatic pressure, sulfur depletion and freezing). In this part only the UV induction is described, informations about the other treatments are given in the Supporting information.
All UV experiments were carried out in an anaerobic chamber under 93% N2 – 7% H2 atmosphere and in semidarkness to avoid possible photoreactivation. A culture in a mid-exponential-phase (108 cells ml−1), was transferred in glass Petri dishes by paying attention to avoid the presence of sulfur. UV irradiation (254 nm) was performed with an ultraviolet lamp (typical small short wave UV light, model UVGL-25, UVP, Upland, CA, emitting essentially monochromatic 254-nm UV at a maximal intensity less than 750 μW cm−2) at a distance of 3 cm from the cell suspension at different exposure times: 15 min, 30 min, 1 h and 2 h. After irradiation, the cell suspension was placed in sterile vials containing elemental sulfur (1%), and incubated at 85°C. Aliquots (8 ml) were collected 4, 8 and 15 h after UV treatment to determine the optimal UV exposure time. When the optimal condition was established, aliquots (2 ml) were collected every 3 h. A nonirradiated culture, treated identically, was used as control.
Aliquots were collected to determine the microbial growth by direct cell counting by using a phase-contrast microscope and a modified Thoma chamber (depth 10 μm). The viral counts were estimated by epifluorescence microscopy as previously described. All the induction assays were performed in duplicate.
Host range
Despite several attempts, we never obtained a cured T. prieurii strain. TPV1 is very stable in its host, as it has never been lost after many transfers in subcultures and was found in all colony clones checked. Reference marine strains belonging to the Thermococcales order were screened for extra-chromosomal genetic elements and strains scored negative were used as potential hosts.
Infection tests were done in liquid culture. Potential host cells were cultured in 50 ml of Ravot medium at 85°C. 10 μl of a viral suspension purified by ultracentrifugation in a linear Iodixanol gradient (106 viruses) were added 3 h after the inoculation (at 1%) of the host cells and incubations were continued at 85°C. Aliquots were collected every 2 or 3 h until 27 h. The microbial growth and the viral abundance were determinated as previously described. Controls were performed to verify that iodixanol and the other buffers used did not present an inhibitory effect on the host growth.
To confirm that the viral production occurred inside the infected host, aliquots of T. barophilus infected with TPV1 were collected after 1 and 12 h of incubation and total DNA were extracted. A PCR using forward (5′-GGC GAT ATT TAC CTC GTC ATC-3′) and reverse (5′-ATG GGC GCA ACA TTC AAC-3′) primers specific to TPV1 was performed. The reaction was performed in a volume of 25 μl containing 50 ng template, 10 mM of each primer, 10 mM dNTPs, 25 mM MgCl2, 1 × buffer and 1 U polymerase (Taq Core, Qiagen). The PCR products were purified by PCR QIAquick (Qiagen) kit and deposited on an agarose gel electrophoresis.
Total DNA, cccDNA and viral DNA extraction
Total DNA from T. prieurii was prepared as previously described (Charbonnier et al., 1992; Geslin et al., 2003a).
cccDNA (Covalently Closed Circular DNA) was extracted from cells in exponential growth phase, by the alkaline lysis method (Birnboim and Doly, 1979; Geslin et al., 2003a).
Viral DNA was extracted from purified viral fractions. After a treatment of the viral fraction with DNase I (final concentration 50 μg ml−1) and RNase I (final concentration 100 μg ml−1), the viral DNA was extracted by the same procedure than that used to obtain total DNA as previously described.
Determination of the nucleotide sequence
Purified TPV1 DNA was completely digested with HindIII and SmaI respectively and all the fragments obtained were cloned in the corresponding sites of pUC18 to obtain an overlapping clone library of TPV1 genome. Sequencing reactions were carried out with the kit ‘BigDye Terminator’ (Applied Biosystems) and analysed at the ‘Platforme Biogenouest’ (Roscoff, France, http://www.sb-roscoff.fr/plateformes-techniques/genomique-sbr.html) on an ABI prismTM 3100 GA. Each insert was sequenced from both ends using the M13 forward and M13 reverse primers. Gaps in the sequence were filled by using specific primers directly for sequencing on library clones (GATC Biotech AG, Konstanz – Germany). The sequences were trimmed and assembled using the SeqMan, Lasergene 8.0 program (DNASTAR, Madison, USA) with both strands completely sequenced and with a threefold coverage.
Sequence analysis and annotation
Glimmer (Delcher et al., 1999) and RBS finder (Suzek et al., 2001) were used to predict protein-coding genes. The amino acid sequence of each predicted protein was searched against the NCBI non-redundant protein database (Altschul et al., 1997), and compared with the database of known protein structures using the HHPred program (Söding et al., 2005). Membrane-spanning region in ORFs were predicted using the TMHMM program (Krogh et al., 2001). Multiple amino acid sequence alignments were constructed using the MUSCLE program (Edgar, 2004). Maximum likelihood phylogenetic trees were constructed using the FastTree program (Price et al., 2010). Cumulative GC skew analysis was performed using GC Skew Tool (http://bioinformatics.upmc.edu/SKEW/ index.html). It was calculated according to the following formula: Σ(G−C)/(G + C), using a sliding window of twenty nucleotides.
Analysis of the Helical stability is computed using the nearest-neighbour-thermodynamics algorithm (Huang and Kowalski, 2003) using the web-based program WEB-THERMODYN (http://wings.buffalo.edu/gsa/dna/dk/WEBTHERMODYN/).
Nucleotide sequence accession number
The sequence data described here have been deposited in GenBank under Accession No. JQ010983.
Supplementary Material
Fig. S1. Electron micrographs of UV-irradiated Thermococcus prieurii cells negatively stained with 2% uranyl acetate. Extrusion of TPV1 outside the host cell (A, B). Scale bars represent 300 nm.
Fig. S10. Multiple amino acid sequence alignment of the putative AAA+ ATPase of TPV1 and the homologous proteins from crenarchaeal viruses was constructed. All these proteins are about the same size and show the location of conserved motifs in the ATPase domain.
Fig. S11. Agarose gel electrophoresis of the total DNA extracted from T. prieurii and digested with HindIII. The arrows indicate overstoichiometric bands of viral DNA. Marker: Smart ladder.
Fig. S2. Infection of Thermococcales by TPV1.
A. Infection of T. celer by TPV1. Squares: cells of T. celer non-infected. Triangles: cells of T. celer infected with TPV1. Diamonds: free TPV1 virions. The arrow indicates the addition of TPV1 virions in the culture.
B. Infection of T. kodakaraensis by TPV1. Squares: cells of T. kodakaraensis non-infected. Triangles: cells of T. kodakaraensis infected with TPV1. Diamonds: free TPV1 virions. The arrow indicates the addition of TPV1 virions in the culture.
C. Infection of T. gorgonarius by TPV1. Squares: cells of T. gorgonarius non-infected. Triangles: cells of T. gorgonarius infected with TPV1. Diamonds: free TPV1 virions. The arrow indicates the addition of TPV1 virions in the culture.
Fig. S3. Amplification of a specific viral fragment from T. barophilus infected with TPV1. Viral DNA of TPV1 (lane 1); Total DNA of T. barophilus infected by TPV1 and extracted 1 hour after infection (lane 2); Total DNA of T. barophilus infected by TPV1 and extracted 12 h after infection (lane 3); Total DNA of T. barophilus non-infected by TPV1 and extracted 1 h after incubation (lane 4);. Total DNA of T. barophilus non-infected by TPV1 and extracted 12 h after incubation (lane 5); M: Smart Ladder.
Fig. S4. Agarose gel electrophoresis of the cccDNA extracted from T. prieurii.
A. cccDNAs (extra-chromosomal elements) extracted from a culture of T. prieurii. The arrows indicate the three elements (lane 2).
B. cccDNAs (extra-chromosomal elements) extracted from a culture of T. prieurii (lane 1); viral DNA extracted from purified TPV1 particles (lane 2).
C. Uncut viral DNA (lane 1); viral DNA digested with HindIII (lane 2); viral DNA heated (70°C) and digested with HindIII (lane 3).
D. Uncut viral DNA (lane 1); viral DNA digested with HindIII (lane 2); viral DNA digested with SmaI (lane 3); viral DNA digested with EcoRI (lane 4); viral DNA digested with MboI (lane 5); viral DNA digested with DpnI (lane 6); viral DNA digested with DpnII (lane 7). M: Smart Ladder.
Fig. S5. Results of qPCR.
A. Dissociation curve. The products obtained had identical melting points of 86°C. Agarose gel electrophoresis (0.8%) of the PCR products show expected product length of 160 bp, M: Smart Ladder.
B. A representative standard curve for the quantification of viral samples.
Fig. S6. Cumulative GC skew (window size of 20 bp).
Fig. S7. Alignment of MCM proteins and homologues related to the MCM encoded by TPV1. DNA replication licensing factor MCM6 (Saccharomyces cerevisia) (1); MCM encoded by TPV1 (2); MCM4 (Encephalitozoon cuniculi) (3); MCM2 (Methanosarcina acetivorans) (4); ATPase (Methanopyrus kandleri) (5); Hypothetical protein PAB0384 (Pyrococcus abyssi GE5) (6); Cell division control protein (Pyrococcus abyssi GE5) (7); DNA replication licensing factor (Pyrobaculum aerophilum) (8); ATPase involved in replication control (Thermoplasma volcanium) (9); DNA replication licensing factor MCM related protein (Thermoplasma acidophilum) (10). The four characteristic motifs were observed.
Fig. S8. A ML phylogenetic tree of archaeal MCM proteins. 44 archaeal homologues were retrieved by blastp from NCBI nr database and clustered by 70% identical amino acids. Sequences were aligned by MUSCLE; the tree was constructed by Fasttree using default parameters. Branches supported by bootstrap values less that 50 were collapsed. Sequence abbreviations: Acibo, Aciduliprofundum boonei T469; Acisa, Acidilobus saccharovorans 345–15; Aerpe, Aeropyrum pernix K1; Arcfu, Archaeoglobus fulgidus; Arcpr, Archaeoglobus profundus DSM 5631; Calma, Caldivirga maquilingensis IC-167; CanKo, Candidatus Korarchaeum cryptofilum OPF8; CanMi, Candidatus Micrarchaeum acidiphilum ARMAN-2; CanNa, Candidatus Nanosalinarum sp. J07AB56; CanPa, Candidatus Parvarchaeum acidophilus ARMAN-5; Deska, Desulfurococcus kamchatkensis 1221n; Ferac, Ferroplasma acidarmanus fer1; Halvo, Haloferax volcanii DS2; Hypbu, Hyperthermus butylicus DSM 5456; Ignag, Ignisphaera aggregans DSM 17230; Ignho, Ignicoccus hospitalis KIN4/I; Metac, Methanosarcina acetivorans C2A; Metco, Methanosaeta concilii GP6; Metfe, Methanocaldococcus fervens AG86; Methu, Methanospirillum hungatei JF-1; Metig, Methanotorris igneus Kol 5; Metin, methanocaldococcus infernus ME; Metla, Methanocorpusculum labreanum Z; Metma, Methanococcus maripaludis C6; Metpa, Methanocella paludicola SANAE; Metpe, Methanoplanus petrolearius DSM 11571; Metst, Methanosphaera stadtmanae DSM 3091; Metvo, Methanococcus voltae A3; Naneq, Nanoarchaeum equitans Kin4-M; Pyrfu, Pyrolobus fumarii 1A; Pyrsp, Pyrococcus sp. NA2; Sulis, Sulfolobus islandicus M.14.25; Theba, Thermococcus barophilus MP; Thega, Thermococcus gammatolerans EJ3; Theko, Thermococcus kodakarensis KOD1; Thene, Thermoproteus neutrophilus V24Sta; Theon, Thermococcus onnurineus NA1; Thepe, Thermofilum pendens Hrk 5; Thesp, Thermococcus sp. 4557; Thevo, Thermoplasma volcanium GSS1; Vuldi, Vulcanisaeta distributa DSM 14429; unceu, uncultured euryarchaeote Alv-FOS1.
Fig. S9. A. Alignment of the SSV-type integrases from crenarchaeal viruses and TPV1-10.
B. Alignment of the SSV-type integrases from TKV1 (TK0073), TKV2 (TK0361), TKV3 (TK0614), TKV4 (TK1342), pT26-2 (ORF1) and TPV1-10. The consensus sequence was observed inside the case.
Table S1. Specific features of the predicted genes of TPV1 (proteins size; RBS and start codons).
Acknowledgments
We thank the captain and crew of the NO L’Atalante, the pilots and support crew of the submersible Nautile, the chief scientist D. Jollivet, and M. Gaillard for helping us to collect deep-sea hydrothermal vent samples during the BioSpeedo oceanographic cruise. We thank G. Sinquin for assistance in electron microscopy and Natalya Yutin for help with phylogenetic analysis. Reviewers are also greatly acknowledged. This work was financially supported by a MENRT grant for A. Gorlas; CNRS ‘PID Origines des Planètes et de la Vie’ programs (2007–2009); The Foundation for Research on Biodiversity (FRB) (2009) and the ANR Deep Oases.
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Achenbach-Richter CR. Rooting the archaebacterial tree: the pivotal role of Thermococcus celer in archaebacterial evolution. Syst Appl Microbiol. 1988;10:231–240. doi: 10.1016/s0723-2020(88)80007-9. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;17:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;5:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;18:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath C, Cukalac T, Porter K, Dyall-Smith ML. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology. 2006;350:228–239. doi: 10.1016/j.virol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Birnboim H, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bize A, Karlsson EA, Ekefjärd K, Quax TEF, Pina M, Prevost MC, et al. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci USA. 2009;106:11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier F, Erauso G, Barbeyron T, Prieur D, Forterre P. Evidence that a plasmid from a hyper- thermophilic arachaebacterium is relaxed at physiologyical temperatures. J Bacteriol. 1992;174:6103–6108. doi: 10.1128/jb.174.19.6103-6108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crennell S, Garman E, Laver G, Vimr E, Taylor G. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure. 1994;2:535–544. doi: 10.1016/s0969-2126(00)00053-8. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith M, Tang SL, Bath C. Haloarchaeal viruses: how diverse are they? Res Microbiol. 2003;154:309–313. doi: 10.1016/S0923-2508(03)00076-7. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröls S, Gordon PMK, Panlilio MA, Schleper C, Sensen CW. Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology. 2007;365:48–59. doi: 10.1016/j.virol.2007.03.033. [DOI] [PubMed] [Google Scholar]
- Fukui T, Atomi H, Kanai H, Matsumi R, Fujiwara S, Imanaka T. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 2005;15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geslin C, Le Romancer M, Erauso G, Gaillard M, Perrot G, Prieur D. PAV1, the first virus-like particle isolated from a hyperthermophilic Euryarchaeota: Pyrococcus abyssi. J Bacteriol. 2003a;185:3888–3894. doi: 10.1128/JB.185.13.3888-3894.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geslin C, Le Romancer M, Gaillard M, Erauso G, Prieur D. Observation of virus-like particles in high temperature enrichment cultures from deep-sea hydrothermal vents. Res Microbiol. 2003b;154:303–307. doi: 10.1016/S0923-2508(03)00075-5. [DOI] [PubMed] [Google Scholar]
- Geslin C, Gaillard M, Flament D, Rouault K, LeRo-mancer M, Prieur D, Erauso G. Analysis of the first genome of a hyperthermophilic marine virus-like particle, PAV1, isolated from Pyrococcus abyssi. J Bacteriol. 2007;189:4510–4519. doi: 10.1128/JB.01896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Kowalski D. WEB-THERMODYN: sequence analysis software for profiling DNA helical stability. Nucleic Acids Res. 2003;31:3819–3821. doi: 10.1093/nar/gkg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Bamford DH. Archaeal proviruses TKV4 and MVV extend the PRD1 adenovirus lineage to the phylum Euryarchaeota. Virology. 2008;375:292–300. doi: 10.1016/j.virol.2008.01.043. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Gribaldo S, Bamford DH, Forterre P. The evolutionary history of archaeal MCM helicases: a case study of vertical evolution combined with hitchhiking of mobile genetic elements. Mol Biol Evol. 2010;27:2716–2732. doi: 10.1093/molbev/msq161. [DOI] [PubMed] [Google Scholar]
- Lee HS, Bae SS, Kim MS, Kwon KK, Kang SG, Lee JH. Complete genome sequence of hyper- thermophilic Pyrococcus sp. strain NA2, isolated from a deep-sea hydrothermal vent area. J Bacteriol. 2011;193:3666–3667. doi: 10.1128/JB.05150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Albers SV, Atomi H, Allers T. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol Rev. 2011;35:577–608. doi: 10.1111/j.1574-6976.2011.00265.x. [DOI] [PubMed] [Google Scholar]
- Marteinsson VT, Birrien JL, Reysenbach AL, Vernet M, Marie D, Gambarcota A, et al. Thermococcus barophilus sp. nov., a new barophilic and hyperthermophilic archaeon isolated under high hydrostatic pressure from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 1999;49:351–359. doi: 10.1099/00207713-49-2-351. [DOI] [PubMed] [Google Scholar]
- Miroshnichenko ML, Gongadze GM, Rainey FA, Kostyukova AS, Lysenko AM, Chernyh NA, Bonch-Osmolovskaya EA. Thermococcus gorgonarius sp. nov. and Thermococcus pacificus sp. nov.: heterotrophic extremely thermophilic archaea from New Zealand submarine hot vents. Int J Syst Bacteriol. 1998;48:23–29. doi: 10.1099/00207713-48-1-23. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Yoshida T, Tanaka R, Forterre P, Sako Y, Prangishvili D. Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology. 2010;402:347–354. doi: 10.1016/j.virol.2010.03.046. [DOI] [PubMed] [Google Scholar]
- Noble R, Fuhrman J. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- Nuttall SD, Dyall-Smith ML. HF1 and HF2: Novel bacteriophages of halophilic archaea. Virology. 1993;197:678–684. doi: 10.1006/viro.1993.1643. [DOI] [PubMed] [Google Scholar]
- Oren A, Bratbak G, Heldal M. Occurrence of virus-like particles in the Dead Sea. Extremophiles. 1997;1:143–149. doi: 10.1007/s007920050027. [DOI] [PubMed] [Google Scholar]
- Pagaling E, Haigh RD, Grant WD, Cowan DA, Jones BE, Ma Y, et al. Sequence analysis of an Archaeal virus isolated from a hypersaline lake in Inner Mongolia, China. BMC Genomics. 2007;8:410–423. doi: 10.1186/1471-2164-8-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter W, Zillig W. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology. 1991;185:242–250. doi: 10.1016/0042-6822(91)90771-3. [DOI] [PubMed] [Google Scholar]
- Peng X. Evidence for the horizontal transfer of an integrase gene from a fusellovirus to a pRN-like plasmid within a single strain of Sulfolobus and the implications for plasmid survival. Microbiology. 2008;154:383–391. doi: 10.1099/mic.0.2007/012963-0. [DOI] [PubMed] [Google Scholar]
- Pfister P, Wasserfallen A, Stettler R, Leisinger T. Molecular analysis of Methanobacterium phage psiM2. Mol Microbiol. 1998;30:233–244. doi: 10.1046/j.1365-2958.1998.01073.x. [DOI] [PubMed] [Google Scholar]
- Pietilä MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol Microbiol. 2009;72:307–319. doi: 10.1111/j.1365-2958.2009.06642.x. [DOI] [PubMed] [Google Scholar]
- Porter K, Kukkaro P, Bamford JKH, Bath C, Kivelä HM, Dyall-Smith ML, Bamford DH. SH1: A novel, spherical halovirus isolated from an Australian hypersaline lake. Virology. 2005;335:22–33. doi: 10.1016/j.virol.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Porter K, Russ BE, Dyall-Smith ML. Virus– host interactions in salt lakes. Curr Opin Microbiol. 2007;4:418–424. doi: 10.1016/j.mib.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Garrett RA. Viruses of hyperthermophilic Crenarchaea. Trends Microbiol. 2005;13:535–542. doi: 10.1016/j.tim.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat Rev Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieur D, Erauso G, Geslin C, Lucas S, Gaillard M, Bidault A, et al. Genetic elements of Thermococcales. Biochem Soc Trans. 2004;32:184–187. doi: 10.1042/bst0320184. [DOI] [PubMed] [Google Scholar]
- Prieur D, Erauso G, Flament D, Gaillard M, Geslin C, Gonnet M, et al. Deep-sea Thermococcales and their genetic elements: plamids and viruses. Methods Microbiol. 2006;35:253–278. [Google Scholar]
- Rachel R, Bettstetter M, Hedlund BP, Haring M, Kessler A, Stetter KO, Prangishvili D. Remarkable morphological diversity of viruses and virus-like particles in hot terrestrial environments. Arch Virol. 2002;147:2419–2429. doi: 10.1007/s00705-002-0895-2. [DOI] [PubMed] [Google Scholar]
- Redder P, Peng X, Brügger K, Shah SA, Roesch F, Greve B, et al. Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ Microbiol. 2009;11:2849–2862. doi: 10.1111/j.1462-2920.2009.02009.x. [DOI] [PubMed] [Google Scholar]
- Rice G, Stedman KM, Snyder J, Wiedenheft B, Brumfield S, McDermott T, Young M. Novel viruses from extreme thermal environments. Proc Natl Acad Sci USA. 2001;98:13341–13345. doi: 10.1073/pnas.231170198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara N, Kelman LM, Kelman Z. How is the archaeal MCM helicase assembled at the origin ? Possible mechanisms. Biochem Soc Trans. 2009;37:7–11. doi: 10.1042/BST0370007. [DOI] [PubMed] [Google Scholar]
- Schleper C, Kubo K, Zillig W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci USA. 1992;89:7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel H, Zillig W, Pfaffle M, Schnabel R, Michel H, Delius H. Halobacterium halobium phage ΦH. EMBO J. 1982;1:87–92. doi: 10.1002/j.1460-2075.1982.tb01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q, Shen B, Chen L. Archaeal integrases and mechanisms of gene capture. Biochem Soc Trans. 2004;32:222–226. doi: 10.1042/bst0320222. [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;1:33. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler N, Marguet E, Cortez D, Desnoues N, Keller J, Van Tilbeurgh H, et al. Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins. Nucleic Acids Res. 2010;38:5088–5104. doi: 10.1093/nar/gkq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler N, Gaudin M, Marguet E, Forterre P. Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem Soc Trans. 2011;39:36–44. doi: 10.1042/BST0390036. [DOI] [PubMed] [Google Scholar]
- Stedman KM, She Q, Phan H, Arnold HP, Holz I, Garrett RA, Zillig W. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res Microbiol. 2003;154:295–302. doi: 10.1016/S0923-2508(03)00074-3. [DOI] [PubMed] [Google Scholar]
- Stetter KO. Diversity of extremely thermophilic archaebacteria. In: Brock TD, editor. Thermophiles: General, Molecular and Applied Microbiology. New York, NY, USA: John Wiley and Sons; 1986. pp. 39–74. [Google Scholar]
- Suttle CA, Fuhrman JA. Enumeration of virus particles in aquatic or sediment samples by epifluorescence microscopy. In: Wilhelm SW, Weinbauer MG, Suttle CA, editors. Manual of Aquatic Viral Ecology. Waco, TX, USA: American Society of Limnology and Oceanography; 2010. pp. 145–153. Chapter 15. [Google Scholar]
- Suzek BE, Ermolaeva MD, Schreiber M, Salzberg S. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics. 2001;17:1123–1130. doi: 10.1093/bioinformatics/17.12.1123. [DOI] [PubMed] [Google Scholar]
- Takai K, Sugai A, Itoh T, Horikoshi K. Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int J Syst Evol Microbiol. 2000;2:489–500. doi: 10.1099/00207713-50-2-489. [DOI] [PubMed] [Google Scholar]
- Tisi D, Talts JF, Timpl R, Hohenester E. Structure of the C-terminal laminin G-like domain pair of the laminin alpha2 chain harbouring binding sites for alphadystroglycan and heparin. EMBO J. 2000;19:1432–1440. doi: 10.1093/emboj/19.7.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters AD, Chong JPJ. An archaeal order with multiple minichromosome maintenance genes. Microbiology. 2010;156:1405–1414. doi: 10.1099/mic.0.036707-0. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Stedman K, Roberto F, Willits D, Gleske AK, Zoeller L, et al. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J Virol. 2004;78:1954–1961. doi: 10.1128/JVI.78.4.1954-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SJ, Cary SC, Williamson KE, Helton RR, Bench SR, Winget D, Wommack KE. Lysogenic virus–host interactions predominate at deep-sea diffuse-flow hydrothermal vents. Int J Syst Evol Microbiol. 2008;10:1–10. doi: 10.1038/ismej.2008.73. [DOI] [PubMed] [Google Scholar]
- Witte A, Baranyi U, Klein R, Sulzner M, Luo C, Wanner G, et al. Characterization of Natronobacterium magadii phage phi Ch1, a unique archaeal phage containing DNA and RNA. Mol Microbiol. 1997;23:603–616. doi: 10.1046/j.1365-2958.1997.d01-1879.x. [DOI] [PubMed] [Google Scholar]
- Woese CR, Fox GE. Phylogenetic structure of the procaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W, Holz I, Janekovic D, Schäfer W, Reiter WD. The archaebacterium Thermococcus celer represents a novel genus within the thermophilic branch of the archaebacteria. Syst Appl Microbiol. 1983;4:88–94. doi: 10.1016/S0723-2020(83)80036-8. [DOI] [PubMed] [Google Scholar]
- Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, et al. Screening for Sulfolobales, their plasmids and their viruses in icelandic solfatars. Syst Appl Microbiol. 1994;16:609–628. [Google Scholar]
- Zivanovic Y, Armengaud J, Lagorce A, Leplat C, Guérin P, Dutertre M, et al. Genome analysis and genome-wide proteomics of Thermococcus gammatolerans, the most radioresistant organism known amongst the Archaea. Genome Biol. 2009;10:1–23. doi: 10.1186/gb-2009-10-6-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Electron micrographs of UV-irradiated Thermococcus prieurii cells negatively stained with 2% uranyl acetate. Extrusion of TPV1 outside the host cell (A, B). Scale bars represent 300 nm.
Fig. S10. Multiple amino acid sequence alignment of the putative AAA+ ATPase of TPV1 and the homologous proteins from crenarchaeal viruses was constructed. All these proteins are about the same size and show the location of conserved motifs in the ATPase domain.
Fig. S11. Agarose gel electrophoresis of the total DNA extracted from T. prieurii and digested with HindIII. The arrows indicate overstoichiometric bands of viral DNA. Marker: Smart ladder.
Fig. S2. Infection of Thermococcales by TPV1.
A. Infection of T. celer by TPV1. Squares: cells of T. celer non-infected. Triangles: cells of T. celer infected with TPV1. Diamonds: free TPV1 virions. The arrow indicates the addition of TPV1 virions in the culture.
B. Infection of T. kodakaraensis by TPV1. Squares: cells of T. kodakaraensis non-infected. Triangles: cells of T. kodakaraensis infected with TPV1. Diamonds: free TPV1 virions. The arrow indicates the addition of TPV1 virions in the culture.
C. Infection of T. gorgonarius by TPV1. Squares: cells of T. gorgonarius non-infected. Triangles: cells of T. gorgonarius infected with TPV1. Diamonds: free TPV1 virions. The arrow indicates the addition of TPV1 virions in the culture.
Fig. S3. Amplification of a specific viral fragment from T. barophilus infected with TPV1. Viral DNA of TPV1 (lane 1); Total DNA of T. barophilus infected by TPV1 and extracted 1 hour after infection (lane 2); Total DNA of T. barophilus infected by TPV1 and extracted 12 h after infection (lane 3); Total DNA of T. barophilus non-infected by TPV1 and extracted 1 h after incubation (lane 4);. Total DNA of T. barophilus non-infected by TPV1 and extracted 12 h after incubation (lane 5); M: Smart Ladder.
Fig. S4. Agarose gel electrophoresis of the cccDNA extracted from T. prieurii.
A. cccDNAs (extra-chromosomal elements) extracted from a culture of T. prieurii. The arrows indicate the three elements (lane 2).
B. cccDNAs (extra-chromosomal elements) extracted from a culture of T. prieurii (lane 1); viral DNA extracted from purified TPV1 particles (lane 2).
C. Uncut viral DNA (lane 1); viral DNA digested with HindIII (lane 2); viral DNA heated (70°C) and digested with HindIII (lane 3).
D. Uncut viral DNA (lane 1); viral DNA digested with HindIII (lane 2); viral DNA digested with SmaI (lane 3); viral DNA digested with EcoRI (lane 4); viral DNA digested with MboI (lane 5); viral DNA digested with DpnI (lane 6); viral DNA digested with DpnII (lane 7). M: Smart Ladder.
Fig. S5. Results of qPCR.
A. Dissociation curve. The products obtained had identical melting points of 86°C. Agarose gel electrophoresis (0.8%) of the PCR products show expected product length of 160 bp, M: Smart Ladder.
B. A representative standard curve for the quantification of viral samples.
Fig. S6. Cumulative GC skew (window size of 20 bp).
Fig. S7. Alignment of MCM proteins and homologues related to the MCM encoded by TPV1. DNA replication licensing factor MCM6 (Saccharomyces cerevisia) (1); MCM encoded by TPV1 (2); MCM4 (Encephalitozoon cuniculi) (3); MCM2 (Methanosarcina acetivorans) (4); ATPase (Methanopyrus kandleri) (5); Hypothetical protein PAB0384 (Pyrococcus abyssi GE5) (6); Cell division control protein (Pyrococcus abyssi GE5) (7); DNA replication licensing factor (Pyrobaculum aerophilum) (8); ATPase involved in replication control (Thermoplasma volcanium) (9); DNA replication licensing factor MCM related protein (Thermoplasma acidophilum) (10). The four characteristic motifs were observed.
Fig. S8. A ML phylogenetic tree of archaeal MCM proteins. 44 archaeal homologues were retrieved by blastp from NCBI nr database and clustered by 70% identical amino acids. Sequences were aligned by MUSCLE; the tree was constructed by Fasttree using default parameters. Branches supported by bootstrap values less that 50 were collapsed. Sequence abbreviations: Acibo, Aciduliprofundum boonei T469; Acisa, Acidilobus saccharovorans 345–15; Aerpe, Aeropyrum pernix K1; Arcfu, Archaeoglobus fulgidus; Arcpr, Archaeoglobus profundus DSM 5631; Calma, Caldivirga maquilingensis IC-167; CanKo, Candidatus Korarchaeum cryptofilum OPF8; CanMi, Candidatus Micrarchaeum acidiphilum ARMAN-2; CanNa, Candidatus Nanosalinarum sp. J07AB56; CanPa, Candidatus Parvarchaeum acidophilus ARMAN-5; Deska, Desulfurococcus kamchatkensis 1221n; Ferac, Ferroplasma acidarmanus fer1; Halvo, Haloferax volcanii DS2; Hypbu, Hyperthermus butylicus DSM 5456; Ignag, Ignisphaera aggregans DSM 17230; Ignho, Ignicoccus hospitalis KIN4/I; Metac, Methanosarcina acetivorans C2A; Metco, Methanosaeta concilii GP6; Metfe, Methanocaldococcus fervens AG86; Methu, Methanospirillum hungatei JF-1; Metig, Methanotorris igneus Kol 5; Metin, methanocaldococcus infernus ME; Metla, Methanocorpusculum labreanum Z; Metma, Methanococcus maripaludis C6; Metpa, Methanocella paludicola SANAE; Metpe, Methanoplanus petrolearius DSM 11571; Metst, Methanosphaera stadtmanae DSM 3091; Metvo, Methanococcus voltae A3; Naneq, Nanoarchaeum equitans Kin4-M; Pyrfu, Pyrolobus fumarii 1A; Pyrsp, Pyrococcus sp. NA2; Sulis, Sulfolobus islandicus M.14.25; Theba, Thermococcus barophilus MP; Thega, Thermococcus gammatolerans EJ3; Theko, Thermococcus kodakarensis KOD1; Thene, Thermoproteus neutrophilus V24Sta; Theon, Thermococcus onnurineus NA1; Thepe, Thermofilum pendens Hrk 5; Thesp, Thermococcus sp. 4557; Thevo, Thermoplasma volcanium GSS1; Vuldi, Vulcanisaeta distributa DSM 14429; unceu, uncultured euryarchaeote Alv-FOS1.
Fig. S9. A. Alignment of the SSV-type integrases from crenarchaeal viruses and TPV1-10.
B. Alignment of the SSV-type integrases from TKV1 (TK0073), TKV2 (TK0361), TKV3 (TK0614), TKV4 (TK1342), pT26-2 (ORF1) and TPV1-10. The consensus sequence was observed inside the case.
Table S1. Specific features of the predicted genes of TPV1 (proteins size; RBS and start codons).


