Abstract
CRISPR screening is a powerful toolset for investigating diverse biological processes. Most CRISPR screens to date have been performed with in vitro cultures or cellular transplant models. To interrogate cancer in animal models that more closely recapitulate the human disease, autochthonous direct in vivo CRISPR screens have recently been developed that can identify causative drivers in the native tissue microenvironment. By empowering multiplexed mutagenesis in fully immunocompetent animals, direct in vivo CRISPR screens enable the rapid generation of patient-specific avatars that can guide precision medicine. This review discusses the current status of in vivo CRISPR screens in cancer, and offers perspectives on future applications.
Keywords: CRISPR screen, in vivo, functional genomics, cancer
CRISPR-mediated genome editing meets the complexity of the cancer genome
Over the past decade, tremendous efforts have been devoted to profiling patient cancers. Multi-institutional consortia such as The Cancer Genome Atlas have now profiled over 10,000 tumors, generating petabytes of high-dimensional data that illuminate the complexities of several cancer types [1]. Although the molecular portraits of cancer are now in higher resolution than ever before, the path to clinical translation for many cancer types still remains largely unexplored.
The central issue is that cancer genomics can inform what mutations are present, but has limited power to indicate which ones are functionally important [2,3]. Individual tumors often have hundreds, if not thousands, of molecular aberrations. While some of these alterations are found in well-established oncogenes and tumor suppressors, many novel aberrations occur in previously uncharacterized or even unannotated regions, making it difficult to reliably discern whether they are actually driving the progression of a given cancer. Furthermore, different patients can present with unique combinations of these various mutations that can drastically influence a cancer’s growth pattern, tendency to metastasize, and susceptibility to therapy.
CRISPR-mediated genome editing has become a powerful tool in cancer biology due to its programmability and flexibility [4]. In addition to its canonical use for targeted gene knockouts [5,6], CRISPR has also been reengineered for a variety of purposes including transcriptional activation [7,8], transcriptional repression [9], histone modification [10], base editing [11,12], DNA methylation [13–15], and genome architecture manipulation [16]. Following in the footsteps of RNA interference (RNAi) screens, the modularity of CRISPR has naturally lent itself to high-throughput screening approaches [17]. By designing custom libraries of single guide RNAs (sgRNAs), one can simultaneously screen large collections of genomic elements (coding genes, regulatory elements such as enhancers, non-coding RNAs, or other non-coding features) in a given biological context [18]. Here, we review the development and application of in vivo CRISPR screens.
In vivo CRISPR screens to interrogate the complexities of cancer
CRISPR screens were first developed by using pooled sgRNA libraries to target all annotated genes in the human genome [19,20]. High-throughput, and particularly genome-wide, CRISPR screens combine the power of forward genetics and reverse genetics. Such approaches offer an unbiased, yet precisely targeted method of identifying genes that contribute to a phenotype, such as cancer progression. Compared to random mutagenesis, CRISPR screens use customized sgRNA libraries, while simultaneously preserving the “randomness” during selection. Unlike RNAi screens, CRISPR generates precise mutations or complete knockouts instead of partial gene knockdown or silencing, which has been shown to facilitate higher between-construct concordance and lower off-target rates [21,22]. CRISPR screens have thus become the state-of-the-art approach for discovery of genetic drivers and phenotypic modulators in many biological contexts.
In cancer, CRISPR screens have been performed to identify genes involved in a wide variety of processes [23], including regulators of drug resistance [24–28], synergistic and synthetic lethal interactions [29,30], regulators of PD-L1 expression [31], and essential genes [32–34] (Figure 1). While in vitro studies are valuable for identifying cell-intrinsic properties of cancer cells and potential therapeutic windows (Figure 2A, Key Figure), they cannot address problems involving complex interactions between multiple cell types that reflect the bona fide nature of cancer as an organ, instead of a collection of isolated tumor cells [35]. As highlighted by recent advances in cancer immunotherapy, the microenvironmental milieu is constantly engaged in conversation with tumor cells, with significant clinical consequences [36–40]. To more faithfully recapitulate the human disease for enhanced translational accuracy, it is essential to investigate these phenomena in vivo where cancers can develop within the context of a tissue microenvironment.
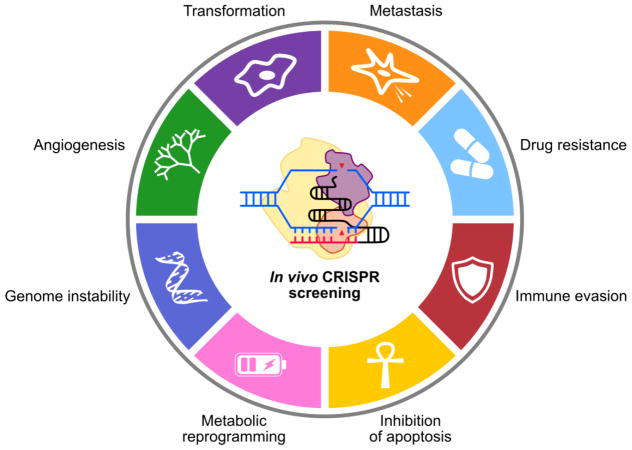
Figure 1. Functional cancer genomics with in vivo CRISPR screening.
In vivo CRISPR screening is a powerful, flexible tool to dissect important processes in cancer. Genome-wide CRISPR screens have illuminated novel regulators of metastasis, malignant transformation, immune evasion, and drug resistance. Moving forward, in vivo CRISPR screens may improve our understanding of other processes, such as angiogenesis, genome instability, and metabolic programming.
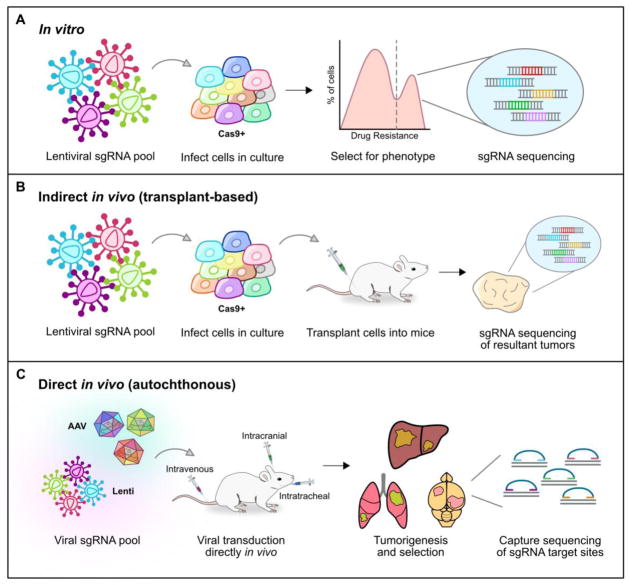
Figure 2 Key Figure. Three modes of CRISPR screening.
A. To perform an in vitro CRISPR screen, the desired sgRNA library must first be cloned into expression vectors. Lentiviral vectors are commonly used, as they can stably integrate into the host genome. After a selection phase to enrich for a desired phenotype, the sgRNA cassettes are amplified from genomic DNA and sequenced to identify the top candidate genes.
B. Indirect in vivo screens follow the same steps asin vitro studies, but the selection phase occurs within a recipient animal. Following transplantation of the mutagenized cell pool into mice, different mutants will become enriched. In the case of a tumorigenesis screen, highly abundant sgRNAs within the resultant tumors would be brought forward as candidate tumor suppressors.
C. For direct in vivo screens, CRISPR mutagenesis occurs at the autochthonous target organ site instead of in culture. Lentiviral and adeno-associated viral (AAV) approaches have both been successfully used for multiplexed direct in vivo mutagenesis. Intravenous, intracranial, and intratracheal viral injections can drive tumorigenesis from the liver, brain, and lung, respectively. Since AAVs do not integrate into the genome, capture sequencing must be performed to readout the results of the screen (i.e. highly abundant indel variants).
To date, virtually all in vivo CRISPR screens have aimed to investigate phenotypes of cancer, directly demonstrating the power and simplicity of this technology in oncology. The first in vivo CRISPR screen investigated annotated genes in the mammalian genome and their potential to promote tumor growth and metastasis upon mutagenesis [41]. In this study, the authors introduced a genome-wide sgRNA library into a non-metastatic cancer cell line. The pool-mutagenized cell library was then subcutaneously transplanted into the back skins of nude mice, and monitored for metastasis to the lung. By sequencing the sgRNAs present in the metastases, the authors identified and subsequently validated a panel of hits from the initial screen that functionally drove lung metastasis. Multiple in vivo CRISPR screens have since been performed to identify tumor suppressors [42–44], oncogenes [45], synthetically lethal genes [46], and regulators of cancer immunotherapy, both in two-cell type (2-CT) co-culture systems [47] and transplant tumor models [48], among others. A common thread in these studies is their two-step workflow: the sgRNA library is first introduced to cells in culture, followed by transplantation into mice to assess phenotypes in vivo. After a selection phase (for instance, expression of a reporter, outgrowth of a tumor, resistance to therapy), the cells are sequenced to identify enriched and/or depleted sgRNAs(Figure 2B).
Limitations of transplant-based in vivo CRISPR screens
By virtue of having a microenvironment, in comparison with in vitro CRISPR screens, it is anticipated that in vivo studies more faithfully model carcinogenesis as it occurs in humans. However, transplant-based in vivo screens have significant limitations. First, introducing large numbers of cancer cells into mice clearly does not resemble the normal process of tumorigenesis. Second, transplants are commonly performed subcutaneously, rather than orthotopically in the relevant organs. In addition, immunodeficient mice are often used to promote the grafting efficiency of the mutagenized cell library, limiting the applicability of these models for investigating cancer-immune interactions. Moreover, the engraftment efficiency of transferred cells can vary from cell line to cell line, and from host to host. Furthermore, there exist multiple cellular bottlenecks of tumor evolution in vivo, such as the circulation limit during the metastasis cascade. Finally, the native organ microenvironment casts numerous constraints on transplanted cells even in orthotopic settings. These limitations can affect the preclinical utility of cancer-cell transplant-based in vivo CRISPR screens.
Direct in vivo CRISPR screens to more faithfully recapitulate human cancers
To better model the natural context of cancer as it occurs in humans, the following features can provide guiding principles for more accurate in vivo CRISPR screen in cancer: 1) the tumors being modeled derive from the endogenous target tissue; 2) the immune system remains intact; and 3) the corresponding tissue microenvironment is preserved. These challenges can be overcome by directly mutagenizing target tissues in vivo, rather than using a transplant approach.
A number of studies have demonstrated efficient CRISPR-mediated in vivo mutagenesis directly at the target organ site. In the liver, hydrodynamic injection of sgRNA-containing plasmids into the tail veins of Cas9 mice was sufficient to induce multiplexed mutagenesis in hepatocytes [49], while in the lung, intratracheal delivery of sgRNA-carrying lentiviruses was able to induce mutagenesis directly in Cas9-expressing lung epithelial cells [50]. Through the use of adeno-associated viruses (AAVs), direct mutagenesis in the mouse brain was also shown to be feasible [51]. Because Cas9 and other genome-editing RNA-guided nucleases (RGNs) are large proteins, generation of Cas9 transgenic animals [52–54] simplifies the delivery of CRISPR components and facilitates direct in vivo mutagenesis.
Extending on these direct in vivo CRISPR techniques, autochthonous CRISPR screens of varying scales have now been performed in a few organ systems (Figure 2C). Hydrodynamic injection of plasmids with sgRNA and Cas9 expression cassettes flanked by Sleeping Beauty (SB) inverted repeats, along with a SB-transposase vector, has been utilized to screen 10 sgRNAs for their ability to induce CRISPR-mediated tumorigenesis in a KrasG12D-sensitized mouse liver [55]. As another approach, lentiviral pools were utilized to screen 11 sgRNAs for their ability to drive lung tumorigenesis in Cre-driven Cas9 mice [56]. Finally, stereotaxic delivery of an AAV sgRNA library directly into the mouse brain efficiently induced glioblastomas that recapitulate the pathologic features of the human disease [57], which enabled direct and quantitative assessment of the tumorigenicity of 280 sgRNAs targeting 56 genes. Of note, the relative mutant frequencies of these genes in the AAV-based model significantly correlated with the mutant frequencies observed in human glioblastoma cohorts. Delivery of AAV-CRISPR sgRNA pools intravenously followed by captured sequencing using customized probesets [57] or molecular inversion probe sequencing [58] allows autochthonous mapping of causative functional variants directly in the targeted genomic loci, as demonstrated in the brain [57] and liver [58]. Facilitated by the high efficiency of AAV-mediated transduction, various significantly co-occurring pairs were identified, thereby pinpointing synergistic driver pairs in tumorigenesis.
Nevertheless, direct in vivo approaches have their share of limitations. Optimal technical parameters that are easily achievable within in vitro or cell line transplant settings (i.e. “indirect” in vivo) such as library size, coverage and multiplicity of infection (MOI), are much more challenging with a direct in vivo approach. Therefore, the size of an sgRNA library must be controlled to ensure adequate coverage in vivo. With an oversized library, random sampling errors from low viral transduction rates will invariably lead to many spurious positive and negative “hits”. Other key constraints include the number of tumor-originating cells in the native organ, accessibility of such cells due to complexity of cellular organization in endogenous tissues, uncharacterized or unknown cell-cell interactions, viral transduction efficiency, and immune rejection. To this end, it is worth noting that AAV-mediated approaches have the major advantage of higher titer, higher direct in vivo transduction efficiencies, and provoke minimal immune reactions [57], thus enabling larger CRISPR libraries to be screened more efficiently in an autochthonous setting. Since AAVs usually do not integrate into the genome, the relative abundance of sgRNAs within tumors cannot be ascertained by sgRNA cassette sequencing as is commonly done for lentiviral-based screens. Instead, the AAV-mediated approach requires capture sequencing of the sgRNA target regions in order to functionally extract the mutational signatures of the tumors.
Direct in vivo CRISPR pooled mutagenesis technologies for precision medicine
With further improvements and modifications, we envision that direct in vivo CRISPR screens could be widely applied in multiple facets of precision medicine. Using the genomic information from a patient’s tumor, a personalized CRISPR library could be readily designed to directly generate disease mimics, or cancers encompassing the same set of mutations, for investigating the behaviors of those mutations in mice (Figure 3A). As a proof-of-principle, high-titer AAV libraries have been successfully used to infect single cells at high MOI in vivo, leading to genetically complex tumors with several mutations [57,58]. This “mouse avatar” approach would enable robust enumeration of the tumorigenic potential of each mutation present within a patient’s tumor. More importantly, rapid generation of such disease mimics allow robust therapeutic testing of approved or novel treatments in genetically matched tumors. This type of approach could then be utilized to predict the outcomes of treatments in patients, to potentially anticipate which specific tumors would exhibit sensitivity or resistance to chemo-, targeted- or immune- therapies, and thus be leveraged for prioritizing therapies.
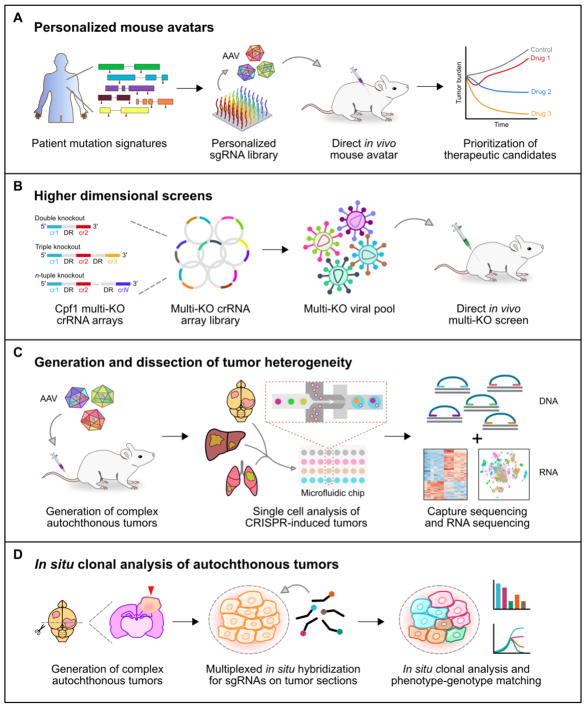
Figure 3. Applications and extensions of direct in vivo CRISPR screens.
A. Direct in vivo CRISPR screens can be readily used to generate personalized mouse avatars. CRISPR libraries can be customized to the mutations present within a given patient’s tumor. Following autochthonous mutagenesis, therapeutic candidates can be evaluated using the mouse avatars, informing clinical decision-making.
B. Higher dimensional screens (i.e. double, triple, n-tuple knockouts) using CRISPR-Cpf1 offer an elegant high-throughput approach to investigate genetic interactions. Such studies could uncover synergistic driver mutations and synthetically lethal combinations, which may help to inform patient prognostication and to identify novel therapeutic vulnerabilities.
C. Direct in vivo CRISPR mutagenesis drives the formation of genetically complex multi-clonal tumors. Coupled with single-cell capture sequencing and RNA sequencing, direct in vivo CRISPR screens offer a powerful approach for the generation and subsequent dissection of tumor heterogeneity.
D. In vivo CRISPR screens thus far have lacked spatial resolution, as sgRNA sequencing and/or capture sequencing is performed on genomic DNA extracted from dissociated cell suspensions. By combining direct in vivo CRISPR mutagenesis with multiplexed in situ hybridization, it may be feasible to perform clonal analysis and phenotype-genotype matching on tissue sections.
It was recently demonstrated that patient-derived tumor xenografts (PDXs) adopt divergent evolutionary courses from the human primary tumor [59]. While this effect is likely due in large part to intrinsic differences between mice and humans, a key contributing factor may lie in the method, as xenografts intrinsically require immunodeficient hosts for successful engraftment. The development and application of humanized mice [60] has helped to bridge this gap in part, particularly with regards to innate immunity, though a complete humanization of the full immune system remains to be seen. In contrast, direct in vivo models are designed to originate from the autochthonous tissue site in fully immunocompetent mice. For these reasons, we anticipate that direct in vivo CRISPR screens will more faithfully recapitulate the behavior and evolutionary course of human cancers.
Intratumoral heterogeneity is increasingly being recognized as an important feature of human cancers, potentially contributing to drug resistance and relapse [61,62]. In addition to cell-type heterogeneity and microenvironmental variations, intratumoral heterogeneity is often characterized by distinct subclonal mutation signatures that differentially influence cellular behavior [63]. To this end, AAV-mediated direct in vivo CRISPR screens are particularly well equipped to model the heterogeneity present within human cancers. As demonstrated in the brain and liver, AAV-CRISPR approaches can rapidly create genetically complex multi-clonal tumors within individual mice [57,58]. This immense heterogeneity inherent to AAV-mediated CRISPR screens could be exploited to investigate clonal dynamics in tumors, serving as a discovery platform for how tumor subclones interact in controlled experimental settings in otherwise wildtype organisms.
Concluding Remarks
Recent development and applications of in vivo CRISPR screens have showcased their power for unbiased identification of functional genetic elements. Particularly, direct in vivo CRISPR screens offer a high-throughput strategy to interrogate candidate genes for their ability to drive tumorigenesis from the autochthonous tissue. Unlike in vitro or transplant-based approaches, direct in vivo CRISPR studies retain the endogenous tissue microenvironment and can be performed in fully immunocompetent animals at high efficiency. Future studies will need to explicitly evaluate whether direct in vivo CRISPR models better recapitulate human disease (see outstanding questions). A key experiment would be to compare the in vivo evolutionary trajectory of a primary patient tumor and the corresponding mouse avatar, with or without therapeutic selection pressure.
Outstanding Questions.
Does a corresponding CRISPR-engineered mouse avatar mimic the drug response of a genetically matched tumor from a patient?
Can in vivo CRISPR screens be effectively leveraged for the study of complex genetic interactions in cancer?
How can direct in vivo CRISPR screens be efficiently coupled with other high-throughput approaches such as single-cell sequencing and high-content imaging to enable the dissection of highly heterogeneous cancers?
Can direct in vivo CRISPR screens be utilized to deduce mutations that are associated with sensitivity to immunotherapy, as well as those conferring primary or acquired resistance?
How can in vivo CRISPR screens be effectively used in immune cells to identify novel regulators of the tumor microenvironment?
The design philosophy behind most CRISPR screens is to study the effects of single gene mutations on a desired phenotype. However, many biological processes are driven by interactions between multiple genes. In cancer, the precise combinations of mutations within individual tumors can lead to strikingly diverse behaviors, influencing tumor aggressiveness and clinical response. In this regard, in vivo CRISPR screens can be readily applied to identify phenotypic modulators on specific sensitized backgrounds. For instance, direct in vivo CRISPR screens could be used to identify factors that influence the metastatic properties of mutant Kras-driven tumors. An exciting avenue for further work is the adaptation of Cpf1 (CRISPR-associated endonuclease in Prevotella and Francisella 1) [64,65] to in vivo cancer modeling. Like Cas9, the Cpf1 RGN can be used for precisely targeted genome editing, yet it has unique multiplexing capabilities due to its independence from a tracrRNA [64,65]. Cpf1 has just begun to emerge for higher-dimensional genetic screening of tumor growth and metastasis in vivo [66], and yet to be more broadly applied for other aspects of cancer. The development of this technology would enable high-dimensional screening of different mutation combinations, potentially leading to the identification of novel synergistic or synthetically lethal genetic interactions in cancer (Figure 3B).
Additionally, the application of dCas9-activator mice could enable the functional identification of oncogenes directly in vivo, providing an orthogonal perspective to preexisting tumor suppressors screens [67]. Similarly, mice engineered to express CRISPR-targeted base editors might offer an approach to screen specific point mutations that drive tumorigenesis from the autochthonous tissue. It would also be interesting to apply single cell RNA-seq (scRNA-seq) to tumors generated by direct in vivo CRISPR screens [68–70]. Together, these technologies would enable the creation and subsequent dissection of cancer heterogeneity at ultra-high resolution (Figure 3C). To further provide spatial information, multiplexed fluorescent in situ hybridization (i.e. MERFISH) [71] could potentially be adapted for the purpose of sequencing sgRNA pools directly in tissue sections. These data would allow for matched comparisons between CRISPR-induced mutational signatures and histopathological phenotypes (Figure 3D).
Finally, direct in vivo CRISPR screens can be readily applied for studies in cancer immunity, as these models are fully functional in immunocompetent animals. A key question on the forefront of immunooncology is to understand why only a fraction of patients respond to immunotherapy, such as checkpoint inhibitors. Multiplexed AAV-CRISPR screens could be applied in the context of checkpoint blockade in order to deduce which mutations are associated with sensitivity to immunotherapy, as well as those conferring primary or acquired resistance. With further technological development, direct in vivo screens on primary immune cell populations may also become feasible, allowing for high-throughput interrogation of factors that regulate immune responses against tumors.
As these technologies continue to develop and mature, they can be adapted for personalized cancer modeling, tumor driver profiling, as well as the identification of novel and relevant therapeutic targets. Emerging new technologies such as direct in vivo CRISPR screens continue to transform oncology discovery.
Trends Box.
In vivo CRISPR screens enable high-throughput interrogation of complex processes in cancer.
Direct autochthonous models recapitulate human cancer by maintaining the native microenvironment.
Direct in vivo CRISPR technologies can empower patient-specific cancer modeling for precision medicine.
Acknowledgments
We thank all members in the Chen laboratory for their support. S.C. is supported by Yale SBI/Genetics Startup Fund, Damon Runyon (DRG-2117-12; DFS-13-15), Melanoma Research Alliance (412806, 16-003524), St-Baldrick’s Foundation (426685), Breast Cancer Alliance, Cancer Research Institute (CLIP), AACR (499395), The Mary Kay Foundation (017-81), The V Foundation (V2017-022), DoD (W81XWH-17-1-0235) and NIH/NCI (1U54CA209992, 5P50CA196530-A10805, 4P50CA121974-A08306). R.D.C. is supported by the NIH MSTP training grant (T32GM007205).
Glossary Box
- Genome editing
Genome editing, or genome editing with engineered nucleases (GEEN), is a type of genetic engineering in which DNA is inserted, deleted or replaced in the genome of an organism using engineered nucleases, or “molecular scissors.”
- CRISPR
Clustered regularly interspaced short palindromic repeats (CRISPR) are segments of prokaryotic DNA containing short repetitions of base sequences. Each repetition is followed by short segments of “spacer DNA” from previous exposures to a bacteriophage virus or plasmid. The CRISPR/Cas system is a prokaryotic immune system that confers resistance to foreign genetic elements such as those present within plasmids and phages. The Cas protein(s) use the CRISPR spacers to recognize and cut these exogenous genetic elements in a manner analogous to RNA interference in eukaryotic organisms. By delivering the Cas9 nuclease and appropriate guide RNAs into a cell, the cell’s genome can be cut at a desired location, allowing existing genes to be removed and/or new ones added.
- Cas9
An endonuclease that is directed to specific sites in the genome by CRISPR spacers, where it induces double stranded breaks in the target DNA.
- dCas9
Catalytically dead Cas9 nuclease that cannot generate double stranded breaks in DNA. dCas9 is commonly tethered to other proteins to enable programmable targeting of the tethered enzymes.
- gRNA
Guide RNA, a short synthetic RNA composed of a “scaffold” sequence necessary for Cas9-binding and a user-defined ~20 nucleotide “spacer” or “targeting” sequence which defines the genomic target to be modified.
- sgRNA
Single-guide RNA, a term generally interchangeable with gRNA in genome editing with CRISPR/Cas9 system.
- crRNA
CRISPR RNA, the short RNA sequence that directly guides CRISPR nucleases to target sites in the genome that is independent of the scaffold sequence present in full-length sgRNAs.
- Cpf1: CRISPR-associated endonuclease in Prevotella and Francisella 1
Cpf1 (also known as Cas12a) is a CRISPR-guided endonuclease that can be utilized for targeted genome editing in diverse species. Unlike Cas9, Cpf1 does not require a tracrRNA for DNA cleavage, and has the capability to autonomously process crRNA arrays.
- In vivo CRISPR screen
By generating libraries of sgRNAs targeting different genes, a CRISPR screen can be performed to assess the importance of these genes towards a given phenotype. With in vivo CRISPR screens, the selection phase occurs inside of a living organism, for instance a mouse.
- Driver mutation
A driver mutation is causally implicated in oncogenesis. It confers a growth advantage on the cancer cell and is favored in the microenvironment of the tissue in which the cancer arises.
- Oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, oncogenes are often mutated or expressed at high levels.
- Tumor suppressor gene
A tumor suppressor gene, or anti-oncogene, is a gene that protects a cell from one step on the path to cancer. Mutations that inactivate tumor suppressors will contribute to cancer progression.
- Double knockout CRISPR screen
A CRISPR screen in which two genes are simultaneously interrogated, rather than a single gene.
- High-dimensional genetic screen
A CRISPR screen in which three or more genes are simultaneously interrogated, enabling the investigation of complex multigenic phenotypes.
- Reporter gene
A gene that researchers attach to a regulatory sequence to facilitate readout of gene regulatory activity. Common examples include green fluorescent protein (GFP) and firefly luciferase.
- Synthetic lethality
Synthetic lethality arises when a combination of mutations in two or more genes leads to cell death, whereas a mutation in only one of these genes does not. In a synthetic lethal genetic screen, it is necessary to begin with a mutation that does not kill the cell, although may confer a phenotype (for example, slow growth), and then systematically test other mutations at additional loci to determine which confer lethality. Synthetic lethality indicates functional relationships between genes.
- Tumor microenvironment
The stroma and supporting milieu surrounding the tumor, usually consisting of fibroblasts, immune cells and endothelial cells.
- Tumor immunity
Tumor immunity refers to the interaction between the host immune system and a tumor. In different contexts, the immune system can have opposing roles towards cancer progression. Cancer immunotherapies aim to tip the scales towards immune-mediated elimination of the tumor.
Footnotes
Glossary Notes
Certain definitions are general concepts, modified from original definitions.
Conflicts of Interest
The authors have no conflicts of interest related to this review.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vogelstein B, et al. Cancer Genome Landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin L, et al. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Hsu PD, et al. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert LA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi LS, et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komor AC, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudelli NM, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu XS, et al. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167:233–247. e17. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita S, et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol. 2016;34:1060–1065. doi: 10.1038/nbt.3658. [DOI] [PubMed] [Google Scholar]
- 15.Vojta A, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan SL, et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun. 2017:8. doi: 10.1038/ncomms15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalem O, et al. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montalbano A, et al. High-Throughput Approaches to Pinpoint Function within the Noncoding Genome. Mol Cell. 2017;68:44–59. doi: 10.1016/j.molcel.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, et al. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evers B, et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34:631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Rivera FJ, Jacks T. Applications of the CRISPR–Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:nrc3950. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou P, et al. A Genome-Wide CRISPR Screen Identifies Genes Critical for Resistance to FLT3 Inhibitor AC220. Cancer Res. 2017;77:4402–4413. doi: 10.1158/0008-5472.CAN-16-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong X, et al. Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature. 2017;550:270–274. doi: 10.1038/nature24037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz S, et al. A Genome-wide CRISPR Screen Identifies CDC25A as a Determinant of Sensitivity to ATR Inhibitors. Mol Cell. 2016;62:307–313. doi: 10.1016/j.molcel.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjana NE, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han K, et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol. 2017;35:463–474. doi: 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen JP, et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat Methods. 2017;14:573–576. doi: 10.1038/nmeth.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burr ML, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T, et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 2017;168:890–903. e15. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quail D, Joyce J. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma P, et al. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 39.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 40.Gordon SR, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katigbak A, et al. A CRISPR/Cas9 Functional Screen Identifies Rare Tumor Suppressors. Sci Rep. 2016;6:38968. doi: 10.1038/srep38968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song CQ, et al. Genome-Wide CRISPR Screen Identifies Regulators of Mitogen-Activated Protein Kinase as Suppressors of Liver Tumors in Mice. Gastroenterology. 2017;152:1161–1173. e1. doi: 10.1053/j.gastro.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kodama M, et al. In vivo loss-of-function screens identify KPNB1 as a new druggable oncogene in epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2017;114:E7301–E7310. doi: 10.1073/pnas.1705441114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braun CJ, et al. Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proc Natl Acad Sci U S A. 2016;113:E3892–3900. doi: 10.1073/pnas.1600582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rana TM, et al. Genome-wide CRISPR screen for essential cell growth mediators in mutant KRAS colorectal cancers. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-17-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel SJ, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manguso RT, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue W, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez-Rivera FJ, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swiech L, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dow LE, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33:390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu VT, et al. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 2016;16:4. doi: 10.1186/s12896-016-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber J, et al. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc Natl Acad Sci. 2015;112:13982–13987. doi: 10.1073/pnas.1512392112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogers ZN, et al. A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression. in vivo Nat Methods. 2017;14:nmeth.4297. doi: 10.1038/nmeth.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chow RD, et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci. 2017;20:1329–1341. doi: 10.1038/nn.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang G, et al. Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using AAV-CRISPR–mediated direct in vivo screening. Sci Adv. 2018;4:eaao5508. doi: 10.1126/sciadv.aao5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ben-David U, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49:ng.3967. doi: 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rongvaux A, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32:364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 62.Schmitt MW, et al. The influence of subclonal resistance mutations on targeted cancer therapy. Nat Rev Clin Oncol. 2016;13:335–347. doi: 10.1038/nrclinonc.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGranahan N, Swanton C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 64.Zetsche B, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zetsche B, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chow RD, et al. Mapping in vivo genetic interactomics through Cpf1 crRNA array screening. bioRxiv. 2017 doi: 10.1101/153486. [DOI] [Google Scholar]
- 67.Zhou H, et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR–dCas9-activator transgenic mice. Nat Neurosci. 2018 doi: 10.1038/s41593-017-0060-6. [DOI] [PubMed] [Google Scholar]
- 68.Dixit A, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866. e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaitin DA, et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896. e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 70.Datlinger P, et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14:nmeth.4177. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen KH, et al. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]