Abstract
Coronary artery disease is the leading global cause of mortality. Long recognized to be heritable, recent advances have started to unravel the genetic architecture of the disease. Common variant association studies have linked about 60 genetic loci to coronary risk. Large-scale gene sequencing efforts and functional studies have facilitated a better understanding of causal risk factors, elucidated underlying biology and informed the development of new therapeutics. Moving forward, genetic testing could enable precision medicine approaches, by identifying subgroups of patients at increased risk of CAD or those with a specific driving pathophysiology in whom a therapeutic or preventive approach is most useful.
Introduction
Observational epidemiology and translational research efforts have led to significant progress in improving the understanding of the pathophysiology underlying coronary artery disease (CAD) (Box 1). Prevention and treatment strategies developed on the basis of this knowledge led to a >50% decrease in age-adjusted CAD mortality rate in the United States of America between 1980 and 2000.1 However, despite these advances, CAD remains the leading global cause of mortality.2 More than 900,000 individuals in the United States will suffer a myocardial infarction (‘heart attack’) or die of CAD this year.3
Box 1 | The pathophysiology and treatment of coronary artery disease.
Coronary artery disease (CAD) refers to the build-up of atherosclerotic plaque in the blood vessels that supply oxygen and nutrients to the heart (reviewed in REF 127). The complex process of atherosclerosis begins early in life and is thought to initiate with dysfunction of endothelial cells that line the coronary arteries; these cells are no longer able to appropriately regulate vascular tone (narrowing or constriction of the vessels) with nitric oxide signalling. Progressive infiltration of the vessel wall by lipoprotein particles carrying cholesterol propagates an inflammatory response by cholesterol-loaded macrophage ‘foam cells.’ Smooth muscle cells underlying the vessel wall proliferate and lead to remodelling of the vessel that ultimately can lead to a narrowing of the vessel that obstructs blood flow. A myocardial infarction (‘heart attack’) is typically caused when a blood clot is incited by a rupture in the surface of the plaque – this deprives the heart muscle downstream of the blood clot of adequate blood flow and leads to cell death.
Epidemiologic studies of CAD demonstrated that age, male gender, smoking, elevated blood pressure, diabetes, obesity, and a sedentary lifestyle each lead to an increased risk of suffering from a myocardial infarction. Similarly, increased concentrations of circulating low-density lipoprotein (LDL) cholesterol, increased triglyceride-rich lipoproteins (a form of fat storage), or decreased high-density lipoprotein (HDL) cholesterol are associated with risk of CAD. In clinical practice, these risk factors can be combined to identify subsets of the population at increased risk of CAD who would most benefit from preventive therapies.
Efforts to prevent CAD (reviewed in REF 128) begin with encouraging adherence to a healthy lifestyle — for example, not smoking, avoiding obesity, a healthy diet and regular exercise — in the population. Higher-risk individuals benefit from additional medications to reduce LDL cholesterol (for example, statins), lower blood pressure, or help prevent formation of a blood clot (for example, aspirin). Should an individual ultimately suffer a myocardial infarction, blood flow can be restored via a procedure to place a stent in the narrowed vessel or bypass it via open-heart surgery. Because of a substantial risk of recurrence, medical therapy is intensified in these individuals.
As with most complex diseases, an individual’s risk of developing CAD is modulated by an interplay between genetic and lifestyle factors.4 Clinical observations dating back to the 1950’s have supported the notion that risk of CAD is heritable.5 A study of over 20,000 Swedish twins subsequently confirmed this finding of increased risk for CAD among close relatives and estimated a heritability of ≈ 50% for fatal CAD.6,7 An analysis that quantified heritability using updated genome-wide approaches similarly estimated the heritability of CAD at 40 to 50%.8 In the Framingham Heart Study, a family history of cardiovascular disease in a parent or sibling was a strong predictor of incident disease.9,10 These seminal studies laid the foundation for the application of emerging human genetics tools to understand the underlying genetic architecture, uncover novel biology, and translate these findings into clinical practice.
Substantial progress in understanding the genetic underpinnings of CAD has been made since the topic was last reviewed in this forum in 2006.11 First reported in 2007, genetic association studies have identified roughly 60 genetic variants with robust links to risk of CAD. Sequencing studies of rare variants have highlighted pathways underlying CAD and, in several cases, directly catalysed the development of a novel therapeutic strategies. Large-scale biobanks containing both genetic and clinical information have enabled assessment of relationships between a given variant and a broad range of human disease states.
In this Review, we outline: research efforts to understand the genetic drivers of CAD, the role of human genetics in catalyzing CAD drug discovery efforts, and the promises and challenges of integrating genetic information into routine clinical practice.
Gene discovery for CAD
Moving from a recognition of familial patterns to discovery of the discrete genetic drivers has been the primary focus of gene discovery efforts in CAD. Sequencing of the human genome (which was completed in 2003), rapid declines in the costs of genotyping, and data-sharing via multi-national collaborations have each played a key role in these efforts.
Family-based studies
Detailed studies of families with a predisposition to early-onset CAD, as classically performed via linkage analysis, provided the first opportunity to gain insight into monogenic causes of CAD. A hereditary pattern of increased cholesterol and CAD, now known as familial hypercholesterolemia, was first described among six patients with xanthomata (skin nodules reflecting deposition of excess cholesterol) in 1938.12 In 1985, a 5 kb deletion in LDLR, the gene encoding the low-density lipoprotein (LDL) receptor, was identified in a patient with familial hypercholesterolemia and his mother. This was the first demonstration of a molecular defect in a single gene can drive CAD risk.13 Impaired receptor-mediated hepatic uptake of LDL leads to substantially increased levels of circulating cholesterol and premature CAD. Family-based studies similarly identified mutations in APOB (which encodes apolipoprotein B) and gain-of-function mutations in PCSK9 (which encodes proprotein convertase subtilisin/kexin type 9) as additional causes of familial hypercholesterolemia by preventing the binding of the LDL particle to LDL receptors for uptake or promoting LDL receptor catabolism respectively.14,15 The genetic underpinnings of autosomal recessive hypercholesterolemia and were linked to null mutations in the genes encoding low-density lipoprotein receptor adapter protein 1 (LDLRAP1) and ATP-binding cassete sub-family G members 5 or 8 (ABCG5, ABCG8) respectively.16,17
Beyond familial hypercholesterolemia and related conditions, the use of family studies to identify monogenic aetiologies of CAD has proven challenging. A 2003 analysis of a large family implicated a locus at chromosome 15q26 as an autosomal dominant aetiology. Sequencing of MEF2A, a gene encoding the transcription factor myocyte enhancer factor 2A, which is expressed in the vasculature, identified a 21 bp deletion in each of the affected family members.18 However, despite biological plausibility, subsequent efforts have failed to confirm an association between loss of function variants in MEF2A and CAD,19 suggesting this initial report may have been related to a chance finding. Additional linkage analyses of families with early-onset CAD and metabolic risk factors have noted potentially causal missense mutations in the genes encoding LDL receptor-related protein 6 (LRP6) and Dual Specificity Tyrosine Phosphorylation Regulated Kinase 1B (DYRK1B).20,21 Due to the rarity of these genotypes, definitive confirmation of these findings in the general population has not been possible to date.
The careful study of families with extreme phenotypes (for example, onset of CAD at a young age in the absence of traditional risk factors) may still prove useful in gene discovery; however, prior to assuming causality for any given finding, rigorous replication that places the genetic, bioinformatics and experimental results into a broader context is required.22
Common variant association studies
Despite the tendency to cluster in families, CAD is a complex and common disorder. Genotyping chips designed to capture the majority of common inter-individual genetic variation provided the foundation for common variant association studies (CVAS), also termed genome-wide association studies (GWAS). Common variants occur often enough that it is practical to test each variant individually by comparing its frequency in disease cases and disease-free controls.23 One operational definition of ‘common’ is variation with frequency ≥0.5% (one carrier per 100 individuals).24
The first CVAS for CAD were published in 2007, when three independent groups reported common variants at the 9p21 locus associated with a ~30% increased risk of CAD per copy of the risk allele.25–27 Subsequent studies have both replicated this finding and extended the assocation to other vascular phenotypes, including carotid atherosclerosis,28 peripheral arterial disease,29 and stroke.30 Preliminary evidence suggests that the 9p21 risk variants alter expression of the noncoding RNA ANRIL, thereby altering activity of two nearby cyclin dependent kinase inhibitors (CDKN2A and CDKN2B) involved in regulation of the cell cycle and cellular proliferation.31,32 Furthermore, inflammatory signalling mediated by interferon-γ may alter long-range DNA interactions, linking the 9p21 risk alleles to CKDN2A and CDNK2B expression.33 However, despite scrutiny over the last 10 years, the precise mechanism underlying the 9p21 association remains elusive.
Since 2007, progressively larger sample sizes have been used to interrogate the genetic architecture of CAD, resulting in nearly 60 distinct genetic loci for CAD.34–39 The results of this cumulative experience permit several conclusions. First, the vast majority of these variants have minor allele frequency >5% in the population, are associated with modest increases in CAD risk (for example, <20% change in risk per allele), and cumulatively explain 30–40% of CAD heritability.39,40 By contrast, 15 low-frequency variants (minor allele frequency <5%) identified using a false-discovery rate threshold explained only 2% of CAD heritability.39 This pattern of results for CAD is similar to other complex diseases, including type 2 diabetes mellitus and schizophrenia.41,42
Second, most of the variants identified to date are located outside of protein-coding regions. Using a genotyping chip designed to capture the vast majority of coding variants in individuals of European ancestry, a recent CVAS identified robust associations for coding variants at only four loci.43 Rather, a significant enrichment of variants in regulatory regions has highlighted a predominant impact of CAD risk variants in altering gene expression.39,44
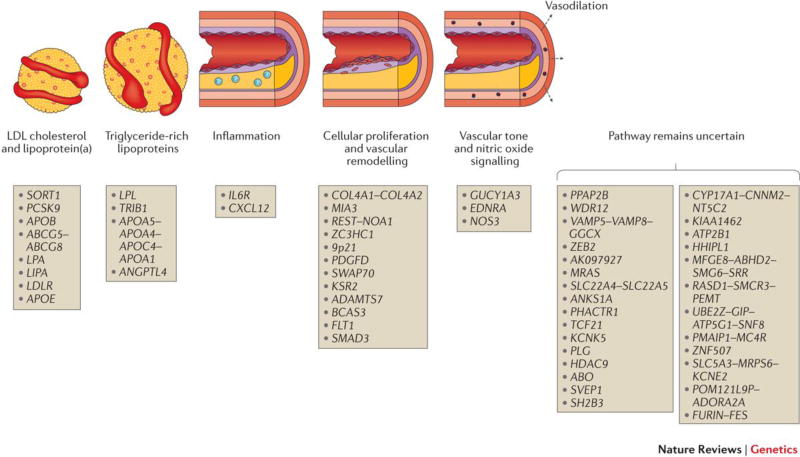
Third, the genetic loci identified to date have highlighted the biology underlying CAD risk (Figure 1). About 20% of the loci are located near genes with known roles in metabolism of low-density lipoproteins (LDL), triglyceride-rich lipoproteins (TRL), or lipoprotein(a) (a modified LDL particle), reinforcing the key role for these pathways in the development of CAD and providing internal validation of CVAS findings. An additional 5–10% of the loci relate to blood pressure, a known and modifiable causal risk factor for CAD. For example, Guanylate Cyclase 1, Soluble, Alpha 3 (GUCY1A3) and Nitric Oxide Synthase 3 (NOS3) are key regulators of vascular tone and platelet aggregation. Common DNA sequence variants at the GUCY1A3 and NOS3 loci have been associated with both blood pressure and CAD.39,45 Furthermore, loss-of-function mutations in GUCY1A3 were associated with risk of myocardial infarction in a large family enriched for premature CAD.46 The equivalent mutations in mice were shown to accelerate thrombus formation in the microcirculation following local trauma.46 These data highlight a role of nitric oxide signalling in protection from CAD.
Figure 1. Physiologic pathways related to genetic loci associated with coronary artery disease.
Genetic loci identified to date are displayed along with presumed relation to causal pathway. Loci are labelled based on nearest genes because the causal genes and variants have not been definitively identified for most loci; this commonly used form of annotation may prove incorrect in some cases. Adapted from REF. 129.
Rare variant association studies
A rapid decline in price has facilitated large-scale genetic sequencing efforts that enable rare variant association studies (RVAS). Such variants are not typically included on genotyping chips (which are designed to capture variation that is common in the population). Furthermore, rare variants do not occur with sufficient frequency to allow for association tests of individual variants. Rather, rare variants in a given gene are aggregated into sets allowing for a comparison of the aggregate frequency across disease strata.47 Ongoing and future studies that perform whole-exome or whole-genome sequencing in a large number of individuals will provide additional statistical power for RVAS.
Studies to date have identified at least 9 genes for which an aggregation of rare mutations alters risk of CAD (Table 1). A whole-exome sequencing study comparing ~5,000 cases with early-onset CAD to CAD-free controls was used to scan each gene for association with CAD in an unbiased fashion.48 Unsurprisingly, the strongest signal was noted for damaging mutations in LDLR, which was associated with a four-fold increase in risk of CAD; about 2% of individuals with early-onset CAD harboured a mutation. A second finding related to inactivating mutations in PCSK9. Unlike the gain-of-function mutations causative for familial hypercholesterolemia discussed above, gene sequencing revealed two damaging PCSK9 mutations with an aggregate frequency of 2% of individuals with African ancestry.49 Carriers of either of these two mutations had substantially lower LDL cholesterol and risk of CAD.50 Most recently, a whole-genome sequencing study of Icelandic individuals identified a 12-bp deletion that leads to inactivation of ASGR1 (which encodes asialoglycoprotein receptor).51 Heterozygous carriers of this mutation had decreased levels of LDL cholesterol and triglycerides, translating into a decreased risk of CAD.
Table 1.
Summary of results from gene sequencing studies for coronary artery disease.
| Gene | Carrier frequency |
Intermediate phenotype |
CAD risk | Therapy to mimic protective variants |
Refs |
|---|---|---|---|---|---|
| Inactivating mutations confer increased risk | |||||
| LDLR | 1 in 221 (0.5%) | ↑LDL cholesterol | ↑ 320% | na | 48 |
| LPL | 1 in 249 (0.4%) | ↑Triglyceride-rich lipoproteins | ↑ 84% | na | 52 |
| APOA5 | 1 in 216 (0.5%) | ↑Triglyceride-rich lipoproteins | ↑ 120% | na | 48 |
| Inactivating mutations confer decreased risk | |||||
| PCSK9 | 1 in 50 (2%)* | ↓LDL cholesterol | ↓ 88% | Alirocumab, Evolucumab (FDA & EMA approved) | 50 |
| NPC1L1 | 1 in 650 (0.2%) | ↓LDL cholesterol | ↓ 53% | Ezetemibe (FDA & EMA approved) | 72 |
| ASGR1 | 1 in 120 (0.8%) | ↓LDL cholesterol ↓Triglyceride-rich lipoproteins | ↓ 34% | None | 51 |
| APOC3 | 1 in 150 (0.7%) | ↓Triglyceride-rich lipoproteins | ↓ 40% | Volanesorsen (Phase III trials) | 53,54 |
| ANGPTL4 | 1 in 360 (0.3%) | ↓Triglyceride-rich lipoproteins | ↓ 53% | REGN1001 (Preclinical development) | 55,56 |
| LPA | 1 in 285 (0.4%) | ↓Lipoprotein(a) | ↓ 24 | IONIS-ANGPTL3-LRx (Phase II trials) | 91 |
Damaging mutations in at least nine genes have been robustly associated with risk of coronary artery disease; in each case, identified genes disrupt pathways related to LDL cholesterol, triglyceride-rich lipoproteins, or lipoprotein(a) metabolism. Pharmacologic therapies are in current use or development to mimic the protective variants for five of the six genes in which inhibition of the related protein would be predicted to reduce risk. FDA: Food and Drug Administration. EMA: European Medicines Agency.
Prevalence estimate based on individuals of African ancestry. Carrier frequency substantially lower in other racial/ethnic groups. CAD, coronary artery disease; na, not applicable.
RVAS have also provided evidence linking genes related to the metabolism of triglyceride-rich lipoproteins, in particular those in the lipoprotein lipase (LPL) pathway, with risk of CAD. The enzymatic activity of LPL serves as the rate-determining step in the clearance of dietary fat from the circulation. Individuals heterozygous for a damaging mutation in LPL have increased circulating triglycerides as well as risk of CAD.52 The activity of LPL is regulated by the protein products of multiple additional genes, several of which have been similarly associated with CAD. Damaging mutations in APOA5, which encodes a protein that enhances LPL activity, is associated with increased CAD risk.48 By contrast, rare mutations in APOC3 and ANGPTL4, the protein products of which inhibit lipoprotein lipase, are associated with decreased CAD risk.53–56
Biologic underpinnings of CAD
From association to mechanism
The numerous genetic loci robustly associated with CAD using the CVAS approach have exposed new mechanisms leading to atherosclerosis. For example, a systematic series of experiments for a novel genetic locus at 1p13 associated with increased LDL cholesterol and risk of CAD identified the causal variant that affected expression of the SORT1 gene.34,57,58 Functional studies confirmed a role for sortilin, the protein product of SORT1, in both the secretion of apolipoprotein B (the major protein component of LDL particles) and LDL catabolism.59,60 Similar lines of work hold substantial promise for the identification of novel therapeutic targets, particularly for loci that are not linked to pathways (for example, LDL metabolism) that are currently targeted by available drugs.
ADAMTS7 – a nonlipid cause of CAD identified by human genetics studies
A 2010 CVAS identified a variant located in an intronic region of disintegrin and metalloproteinase with thrombospondin motifs-7 (ADAMTS7), which was associated with a 19% increased risk of obstructive CAD (>50% narrowing of a major coronary vessel).61 This finding led to substantial interest in elucidating the underlying mechanism, particularly because the variant was not associated with blood lipid levels or other known CAD risk factors. ADAMTS7 belongs to a family of proteins involved in proteolysis (the cleavage of substate proteins) and remodelling of the walls of blood vessels. The risk variant at ADAMTS7 is associated with increased gene expression, proteolytic activity, and the ability to promote vascular smooth muscle migration in vitro.62 ADAMTS7 knockout mice were noted to have significantly reduced atherosclerosis burden.63 Furthermore, mice deficient in ADAMTS7 demonstrated decreased cellular proliferation and enhanced endothelial cell repair in response to vascular injury.63,64 These results suggest that a pharmacologic strategy of ADAMTS7 inhibition could prove useful in attenuating the cellular proliferation that plays a key role in progression of CAD.
Catalysing drug development for CAD
The development of novel, efficacious and safe therapeutic strategies for CAD remains a major public health need. However, the cost of developing a new drug has continued to increase – a recent analysis estimated direct costs of $1.4 billion per new compound approved.65 Much of this cost relates to the vast majority (>95%) of compounds that fail in clinical development, largely due to inadequate therapeutic efficacy or unanticipated toxicity.66 Human genetics may provide a key opportunity to improve drug development efforts by confirming the physiologic and causal relevance of a given target in humans and anticipating the full range of efficacy and safety consequences of pharmacologic modulation (Figure 2).67
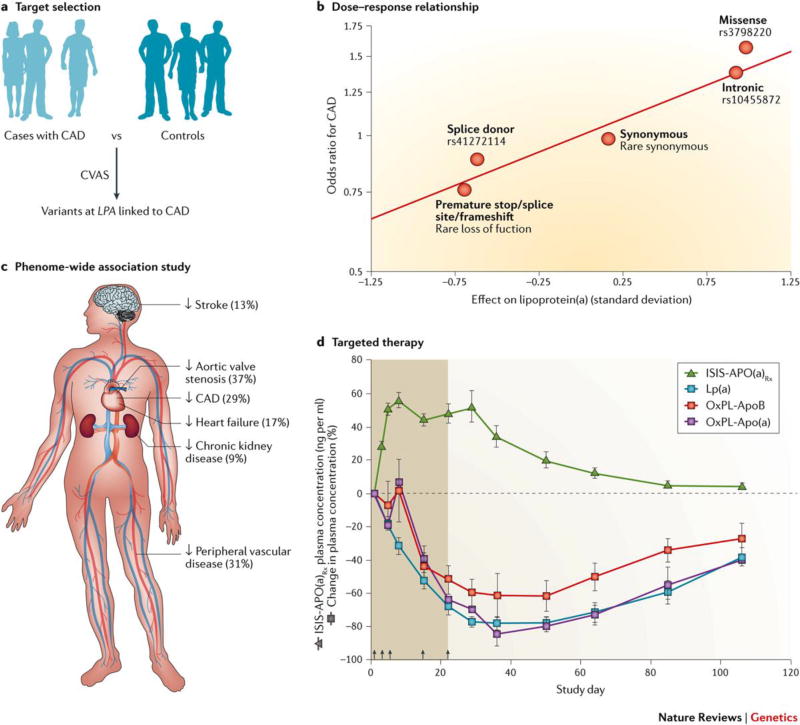
Figure 2. Human genetics to facilitate drug development – Lipoprotein.
(a). Human genetics data serves as the foundation for ongoing efforts to develop therapies to reduce lipoprotein(a) levels, a causal risk factor for coronary artery disease. a | The selection of lipoprotein(a) as a therapeutic target was supported by a 2009 genome-wide association study comparing 1,145 CAD cases to 3,352 controls, noting a robust association between variants near the LPA gene, levels of circulating lipoprotein(a), and risk of coronary artery disease.70 b | A dose-response relationship was noted, such that a given variant’s impact on circulating lipoprotein(a) levels was predictive of the association with CAD (Reproduced with permission from REF 91) c | In order to anticipate the full spectrum of phenotypic consequences of lipoprotein(a) reduction, a phenome-wide association study was performed among participants of the UK Biobank. A genetically-mediated one standard deviation decrease in levels of lipoprotein(a) was associated with a reduced risk of six distinct diseases (Reproduced with permission from REF 91). d | An antisense oligonucleotide targeting hepatic production of lipoprotein(a) was associated with a >80% decrease in circulating levels, providing proof-of-principle that targeting this causal pathway in a highly specific fashion (Reproduced with permission from REF 83).
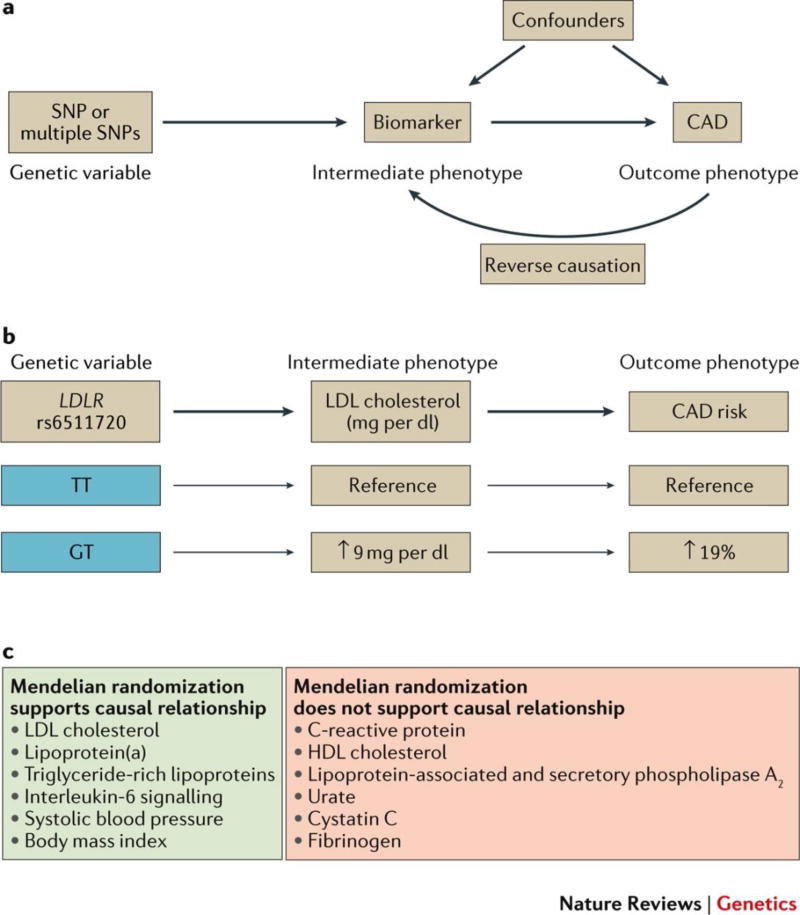
Selection of therapeutic targets by Mendelian randomization
Human genetics can complement existing preclinical prioritization strategies by providing confidence that a given target is a root cause of CAD in humans. Mendelian randomization studies have emerged as a technique grounded in human genetics to help infer causality between a given biomarker and risk of CAD (Figure 3). For example, the large number of common genetic variants linked to lipid levels (>150) provided an opportunity to test the causality of lipid biomarkers using Mendelian randomization.68 As expected, variants affecting LDL cholesterol levels had robust associations with risk of CAD in a concordant fashion.69 A similar finding was noted for LPA variants affecting the levels of lipoprotein(a).70 In combination with the sequencing-based analyses of genes related to the lipoprotein lipase pathway discussed above, Mendelian randomization studies have supported a causal relationship between triglyceride-rich lipoproteins and CAD and have reinvigorated interest in targeting triglyceride metabolism for therapeutic gain.71 By contrast, no consistent relationship of variants affecting HDL cholesterol levels and CAD was observed, raising the possibility that the well-established inverse association between HDL cholesterol levels and risk of CAD is not reflective of a causal relationship.69
Figure 3. Mendelian randomization to identify causal biomarkers for risk of coronary artery disease.
a | Mendelian randomization analyses require that: 1) a genetic variant (or group of variants) is robustly associated with the biomarker of interest; and 2) the genetic variant is independent of confounders that influence the biomarker or risk of CAD; and 3) any impact of the genetic variant on risk of CAD is mediated by the biomarker (as opposed to other pleiotropic genetic effects). Because genetic variants are assorted within the population at time of conception largely at random, these analyses are less susceptible to the issues of confounding or reverse causality that commonly limit causal inference from observational epidemiology. An important limitation of this study is that it requires a robust relationship between the genetic variant and a biomarker – a small effect size mandates a large number of individuals afflicted with CAD to achieve adequate power. b | For example, rs6511720 is an intronic variant in the LDL receptor (LDLR) gene. Each copy of the ‘T’ allele is associated with a lower LDL cholesterol and an decreased risk of CAD.69 Among biomarkers linked with coronary artery disease in the population, studies performed to date have supported a causal relationship for some (e.g. LDL cholesterol, triglyceride-rich lipoproteins, lipoprotein(a)) and a non-causal relationship for others (e.g., HDL cholesterol, C-Reactive Protein).
Early examples suggest that Mendelian randomization studies may predict the outcome of large clinical trials. For example, inactivating mutations in the NPC1L1 gene were associated with decreased LDL cholesterol and risk of CAD.72 A subsequent report from a randomized controlled trial of the drug ezetemibe, which inhibits the protein product of NPC1L1 to decrease cholesterol absorption, confirmed efficacy in cardiovascular event reduction.73 By contrast, despite promising observational epidemiology and animal model evidence,74,75 lipoprotein-associated phospholipase A2 (Lp-PLA2) inhibitors proved ineffective in multiple trials involving over 28,000 patients.76,77 Genetic studies of both common and rare variants in the gene encoding Lp-PLA2 (PLA2G7) were shown to have no impact on the risk of CAD, raising the possibility that the null clinical trial results might have been foreseen.78,79
Developing therapeutics to mimic protective variants
The examples of NPC1L1 and PLA2G7 were based on targets with drug development programs already in place. However, genetics evidence that individuals with inactivating mutations in PCSK9 have decreased levels of circulating LDL cholesterol and reduced risk of coronary disease fostered intense interest in the development of PCSK9 inhibitors.50 In 2015, a mere 12 years after the initial discovery of PCSK9,15 two monoclonal antibodies that inhibit PCSK9 were approved by the US Food and Drug Administration. Each of the drugs led to a ~50% decrease in circulating LDL cholesterol in initial clinical trials and seemed to decrease risk of cardivascular events – ongoing large clinical trials seek to confirm this preliminary signal for efficacy.80,81 Similarly, antisense oligonucleotides designed to mimic the protective mutations noted in APOC3 or LPA demonstrated a ~70% reduction in triglyceride levels and 80% reduction in circulating lipoprotein(a) levels, respectively, in early-phase studies.82,83 Future studies will seek to demonstrate favourable impact on clinical outcomes.
Phenome-wide association studies
Population-based analyses of variants related to a putative drug target can anticipate the full range of phenotypic consequences that might be expected by pharmacologic modulation. For example, a common variant in the gene encoding HMG-coenzyme A reductase, HMGCR, the target of statin therapy was associated with decreased LDL cholesterol levels but an increased risk of type 2 diabetes.84 This finding is well-aligned with an adverse effect of statins in increasing risk of diabetes, which was discovered only after decades of clinical trial experience.85 Furthermore, an ongoing trial is seeking to determine whether anacetrapib, a cholesteryl ester transfer protein (CETP) inhibitor known to increase HDL cholesterol, can decrease risk of cardiovascular disease (ClinicalTrials.gov number, NCT01252953).86 Genetic variants within the CETP gene associated with increased HDL cholesterol were recently linked to an increased risk of age-related macular degeneration, a leading cause of blindness.87–89 Had this genetic finding been discovered earlier, trial sponsors or regulatory agencies may have requested inclusion of a surveillance program to determine whether pharmacologic inhibition would similarly lead to the unanticipated toxicity predicted by human genetics.
These examples focus on relationships between genetic variants and candidate phenotypes selected a priori for investigation. However, this concept can be generalized using phenome-wide association studies (PheWAS) which enable the unbiased interrogation of relationships between a given variant and a broad range of human disease states.90 The utility of this approach is likely to increase in coming years, as large biobanks link genotype information with electronic health records. For example, our group recently leveraged data from >100,000 participants of the UK Biobank to assess the phenotypic consequences of genetically-lowered lipoprotein(a) levels on a range of 36 different disease states.91 Beyond confirming an expected association with decreased risk of CAD and calcific aortic valve stenosis, this PheWAS uncovered relationships with reduced risk of stroke, peripheral vascular disease, congestive heart failure, and chronic kidney disease. By contrast, genetic analysis did not replicate toxicity concerns based on observational epidemiology that linked reduced lipoprotein(a) levels to an increased risk of type 2 diabetes or cancer.92,93
Recall by genotype studies
Deep phenotyping of individuals with a genetic variant related to a drug target provides an opportunity for testing of specific hypotheses in a rigorous fashion. Human genetics has traditionally employed a ‘phenotype-first’ paradigm, in which participants or families were ascertained on the basis of a phenotype of interest (for example, early-onset CAD) and the genetic underpinnings determined. The availability of genetic information in large populations allows for a complementary ‘genotype-first’ approach, in which individuals with a genotype of interest (e.g. a large-effect inactivating mutation in a CAD-related gene) are called back for additional hypothesis-driven phenotyping.
For ANGPTL4, a CAD target validated through human genetics, the identification of a potential toxicity in animal models has raised concern about whether it is an appropriate pharmacologic target. A monoclonal antibody-based approach designed to inhibit ANGPTL4, thus mimicking the variants protective against CAD, led to a substantial decrease in triglyceride-rich lipoproteins in both mice and non-human primates.56 However, these compounds also led to abnormal lipid accumulation in abdominal lymph nodes in these model systems, a finding previously observed in both a murine model of ANGPTL4 inhibition and knockout mice.94,95 Phenotyping humans with a lifelong genetic deficiency in ANGPTL4 would provide one way to assess whether this toxicity has relevance in humans as well. For example, magnetic resonance imaging can be used to characterize the size and tissue characteristics of abdominal lymph nodes. A comparison of such features between wild-type controls and those homozygous for an inactivating mutation in ANGPTL4 may prove informative.
Genomic medicine
The substantial progress in understanding the genetic underpinnings of CAD in the population has laid the groundwork for the integration of genetic data into routine clinical practice - ‘genomic medicine.’ If treating providers had widespread access to their patients’ genetic data, as may well be the case in coming years, what are the potential implications for the prevention and treatment of CAD?
A longstanding debate in the field of complex traits genetics pits the ‘common disease-common variant’ and ‘common disease-rare variant’ hypotheses against one another.96 In the latter, the manifestation of CAD may be reflective of a different monogenic disease in each individual.97 Rare but large-effect variation could be used to stratify CAD into multiple disease ‘subtypes,’ and treatment directed accordingly. However, while likely to be the case for rare monogenic conditions such as familial hypercholesterolemia, studies to date have instead suggested that the majority of heritable risk for CAD risk is predicated on common variants. These variants, in combination with environmental factors, lead to a quantitative blend of multiple driving processes in each individual.
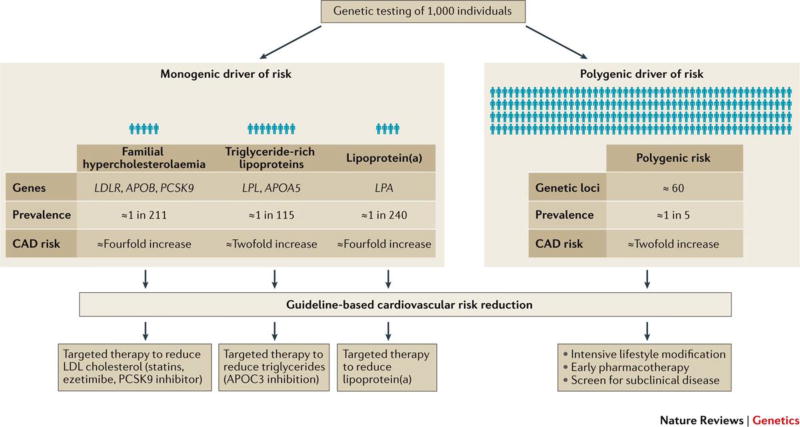
Here, we describe the potential utility in identifying individuals with either monogenic or polygenic increased risk for CAD (Figure 4). Recent reviews have highlighted an additional potential use of predicting drug efficacy or toxicity based on human genetics, often termed ‘pharmacogenetics’.98,99
Figure 4. Precision medicine guided by human genetics.
Genetic testing of 1,000 people would identify 1–2% of individuals with a monogenic driver of risk. For example, individuals with familial hypercholesterolemia102 may derive particular benefit from lowering of LDL cholesterol levels with PCSK9 inhibitors. Second, damaging mutations in LPL52 or APOA548 increase CAD risk by preventing the clearance of dietary fat and triglyceride-rich lipoproteins from the bloodstream – this risk might be directly attenuated through use of inhibition of APOC3. Thirdly, those with a genetic predisposition to increased lipoprotein(a), i.e. at least two risk variants (at SNP sites rs10455872 or rs3798220) as previously characterized,70 might be offered an antisense oligonucleotide to decrease circulating levels. Importantly, any such therapies would likely be incorporated in addition to guideline-based cardiovascular risk reduction therapeutics. Furthermore, confirmation that targeted PCSK9 inhibition, APOC3 inhibition, or Lp(a) reduction leads to reduction in CAD events in clinical trials is needed prior to widespread implementation. By contrast, 20% of individuals tested would be in the top quintile of a polygenic risk score – this population might be targeted for aggressive lifestyle or pharmacologic therapies beyond current treatment guidelines.
Lessons learned from familial hypercholesterolemia
In principle, familial hypercholesterolemia, the primary monogenic driver of CAD identified to date, represents a major opportunity to use human genetics to improve cardiovascular health. A national program in the Netherlands to perform genetic testing in individuals with features suggestive of familial hypercholesterolemia – including family history, physical exam findings, and severely elevated LDL cholesterol – has identified and provided treatment to thousands of individuals since its initiation in 1994.100 In practice, implementation of programmes to systematically identify and treat individuals with familial hypercholesterolemia at scale in the population has proven difficult. Although the condition remains both underdiagnosed and undertreated, efforts at both the national and institutional level to improve recognition are ongoing.101
Largely due to cost and logistical concerns, few previous efforts have sought to combine gene-sequencing data and observed LDL cholesterol levels in the prediction of CAD. We recently found that, for any given observed LDL cholesterol, the risk of CAD is substantially higher among those who harbour a familial hypercholesterolemia mutation compared to those who do not.102 Direct comparison of serial LDL cholesterol levels between individuals with versus without a familial hypercholesterolemia mutation confirmed higher cumulative exposure among those with a mutation. This observation supports the hypothesis that the increased lifelong exposure to higher LDL cholesterol levels is a key driver of the CAD risk and provides evidence for the potential clinical utility of knowing an individual’s mutation status.
The study of familial hypercholesterolemia additionally has highlighted concepts of broad relevance to CAD population genetics – penetrance, phenocopy, and variant annotation. Incomplete penetrance refers to the finding that not all individuals with a familial hypercholesterolemia mutation manifest severely elevated cholesterol levels. For example, despite a significant increase in average LDL cholesterol among carriers of a familial hypercholesterolemia mutation, 27% of these individuals had a normal concentration, reflective of significant heterogeneity both within and across specific mutations.102 Secondly, the majority of cases of severely elevated LDL cholesterol cannot be explained by familial hypercholesterolemia – a ‘phenocopy’ of severe hypercholesterolemia can occur due to lifestyle or polygenic causes as well.103–104 Among individuals with severe hypercholesterolemia (defined as LDL cholesterol >190 mg/dl), only 2% harboured a familial hypercholesterolemia mutation.102 Thirdly, assessing the functional impact of rare missense variants (‘variant annotation’) is not always straightforward, particularly if a variant has not been seen before. Efforts to centralize annotation based on previous observations, refine computer prediction algorithms, and high-throughput functional screening of identified variants are likely to improve efficiency and consistency of this process in coming years.
Human genetics to guide precision medicine therapeutics
For a subset of the genes with validated associations with CAD, pharmacologic therapies are available or in development to specifically target a pathway and provide an additional opportunity for risk reduction beyond standard lifestyle and pharamacotherapy recommendations (Figure 4). Importantly, although these ‘precision medicine therapeutics’ are likely to be initially used in populations with abnormalities in the pathway, they may well prove useful for the population at large as well. For example, the efficacy and safety of statin therapy was initially demonstrated in 1994 by a clinical trial of participants with markedly elevated cholesterol levels and manifest CAD.105 Numerous trials in ensuing years have demonstrated clinical benefit across a broad range of lower baseline cholesterol and CAD risk, albeit with lower absolute risk reductions.106
However, only a small proportion of individuals harbour a rare, large-effect mutation in one of the genes discussed above. A more common aetiology of an increased genetic risk related to a polygenic cause – specifically the inheritance of a large number of smaller impact, more common risk alleles. For any given individual, a weighted score of risk variants can be calculated to provide a continuous and quantitative measure of genetic risk.107 This approach was first implemented for CAD in 2008 using DNA sequence variants associated with circulating cholesterol levels and extended in 2009 to nine validated CAD risk variants.34,108 As compared to those in the lowest quintile of this polygenic risk score, those in the top quintile were at a more than two-fold increased risk of early-onset CAD.34 This finding has subsequently been confirmed by multiple groups using progressively more variants.109–112
Because DNA is stable over the course of the lifetime, genetic risk can be ascertained from the time of birth. Polygenic risk scores may thus be particularly useful in risk prediction among younger patients (for example, children or young adults) in whom the cumulative impact of lifestyle factors is less pronounced. This early knowledge may facilitate an intervention to attenuate this risk. For example, adherence to a healthy lifestyle may be particularly important in these high-risk individuals. A recent analysis of individuals at high genetic risk noted a 46% attenuation of risk of incident CAD in those with a favourable versus unfavourable lifestyle.4 Similarly, among those with a high genetic risk, statin therapy was associated with a nearly 50% reduction in CAD risk.112
These examples provide key evidence that, rather than operating in a deterministic fashion, high genetic risk is indeed modifiable. However, both lifestyle and statin therapy are safe and effective across a broad range of individual-level risk. The approach of targeted therapy by genetic risk might be of more utility in the allocation of therapies with significant adverse effects (e.g. antiplatelet therapies associated with bleeding) or cost (e.g. antibody-based therapies).
A key goal of genomic medicine is to disclose genetic risk to individuals and their providers to enable behavioural changes or pharmacologic therapy to attenuate risk prior to onset of disease. For example, the MI-GENES study demonstrated that the incorporation of a genetic risk score into shared decision making sessions with patients and providers led to a modest increase in statin utilization and lower LDL cholesterol levels in those with high genetic risk.113 Ongoing and future studies are needed to determine the optimal approach for genetic risk disclosure and assess whether this approach can improve clinical outcomes.
Future applications of human genetics findings
The study of the genetic architecture of human CAD has led to substantial progress in gene discovery, informing drug development via an improved understanding of human pathophysiology, and laying the foundation for genomic medicine. Despite substantial progress, a number of key questions regarding the genetics of CAD remain. Among them: 1) Can human genetics highlight a novel pathway, independent of LDL, TRL, and Lp(a) metabolism, that underlies CAD risk? 2) Can genetics facilitate an understanding of the phenotypic consequences of each gene as they relate to CAD?; 3) Can the information gained about protective mutations be used to develop a therapy that would effectively cure atherosclerosis in this century?
Functional genomics to highlight novel causal pathways
The CVAS approach has been highly successful in validating a large number of genetic loci linked to CAD. However, for the majority of loci, key questions regarding the mechanisms underlying these associations remain unclear – specifically the causal DNA variant, the causal gene under regulation, the mechanism by which the variant affects the gene, and the mechanism by which the gene influences the risk of CAD. This line of work has proven challenging; because the majority of the variants lie in non-coding intergenic regions,39 even the initial step of identifying the gene of interest (i.e., the effector transcript) is often not straightforward.
Successful approaches in functional genomics are likely to require close partnerships between computational and experimental biologists. Deep sequencing of the areas near CVAS loci and Bayesian approaches to prioritize variants can help in identifying the causal variant.59,114 Insights into the mechanism of gene regulation can be gained from publically available databases, including the Encyclopedia of DNA Elements (ENCODE) project, which provide details on histone marks and transcription factor binding sites in the region.115 The relationship of a given variant with tissue-specific expression of candidate genes (‘expression quantitative trait loci’) can be evaluated using data from the Genotype-Tissue Expression (GTEx) project, thus facilitating integrative network analyses.116 Epigenetic, metabolomic or proteomic data have become increasingly available and may similarly prove useful. From a functional standpoint, >100 distinct genes have been shown to influence atherosclerosis in validated mouse models.117 Collaborative efforts to demonstrate the relevance of such signals using human genetics analyses and, conversely, to use human genetic signals to prioritize candidate genes for animal model validation represent a key opportunity for moving the field forward. Importantly, viral vectors that facilitate gene overexpression or knockdown in animal models are readily available. Substantial progress in gene editing approaches has dramatically decreased the time needed to engineer mouse models with a variant of interest. Similarly, induced pluripotent stem cells can be isolated from humans with a genotype of interest or engineered into existing cell lines for additional study. This rapidly expanding functional genomics toolkit is likely to accelerate the process of moving from genomic localization to mechanistic discovery in coming years with the ultimate aim of identifying a novel pathway that can be therapeutically targeted.118
Genetics at population-scale to understand genome–phenome relationships
A robust understanding of the phenotypic consequences of inactivating mutations in a given gene will require very large matrices that link an individual’s genetic information with phenotypes (e.g., phenotypes from electronic health records), several of which are currently in use or in development (Table 2). Recent gene sequencing efforts have identified individuals homozygous for rare inactivating mutations (‘human knockouts’) in >1,000 genes; this phenomenon occurs with substantially greater frequency in populations with higher rates of consanguinity.119,120 Just as the targeted deletion of each gene in the mouse (Knockout Mouse Project) has been an invaluable resource to understand gene function, one might imagine a Human Knockout Project involving a systematic effort to phenotype humans who naturally lack a given gene.
Table 2.
Current and emerging biobanks to facilitate genome–phenome interrogation.
| Biobank | Enrollment locations |
Initial enrollment |
Enrollment to date |
Target enrollment |
|---|---|---|---|---|
| Commercial funding | ||||
| deCODE Genetics (Amgen) (http://www.decode.com/) | Iceland | 1996 | >200,000 | Unknown |
| Geisinger MyCode® Community Health (Regeneron Pharmaceuticals and Others) | Geisinger Health System (Danville, PA) | 2007 | >50,000 | Unknown |
| Government funding | ||||
| China Kadoorie Biobank (http://www.ckbiobank.org/site/) | China | 2004 | >500,000 | Enrollment Completed |
| UK Biobank (https://www.ukbiobank.ac.uk/) | United Kingdom | 2006 | >500,000 | Enrollment Completed |
| Electronic Medical Records and Genomics (eMERGE) Network (https://emerge.mc.vanderbilt.edu/about-emerge/) | United States Hospital Sites | 2007 | >50,000 | Unknown |
| Million Veterans Program (http://www.research.va.gov/mvp/) | Veterans Affairs Hospital | 2011 | >500,000 | ~1,000,000 |
| Precision Medicine Initiative (https://www.nih.gov/precision-medicine-initiative-cohort-program) | United States | Early 2017 | -- | ~1,000,000 |
| Institutional funding | ||||
| BioVu Biorepository (https://victr.vanderbilt.edu/pub/biovu/) | Vanderbilt University Medical Center (Nashville, TN) | 2007 | >215,000 | Unknown |
| Kaiser Permanente Research Bank (http://researchbank.kaiserpermanente.org/) | United States | 2016 | >250,000 | ~500,000 |
| Partners Healthcare Biobank (https://biobank.partners.org/) | Partners Health Care (Boston, MA) | 2010 | >50,000 | ~100,000 |
Biobanks that link genetic and phenotypic information in a large number of individuals will provide a key resource for additional research in coming years. These biobanks will be most useful when linked with infrastructure to support hypothesis-based deep phenotyping among those with a genotype of interest. A representative example of such biobanks is included.
As proof-of-concept, a recent exome sequencing analysis identified an individual homozygous for a damaging mutation in APOC3, confirmed to have near-absent levels of plasma APOC3.120 A subsequent call-back study of 27 additional family members confirmed the overall health of human APOC3 knockouts and a dramatic reduction in triglyceride-rich lipoproteins, particularly pronounced after a meal rich in fat. The identification of healthy individuals with an inactivating mutation in both copies of a given gene, effectively ‘human knockouts,’ can provide some reassurance that pharmacologic inhibition of the gene’s product would be tolerated in humans. Additional mechanistic and lipoprotein kinetics studies in these individuals are likely to have direct relevance to APOC3 inhibition as a therapeutic strategy for CAD.
Genome editing as curative therapy
Despite demonstrated efficacy of currently available therapies, adherence and adverse effects limit their impact in clinical practice. For example, only 39% of individuals reported adherence to statin therapy in the year following a heart attack, even when the medicine was provided free of charge.121 As noted above, rare genetic mutations in several genes have been noted to confer lifelong resistance to the development of CAD without detectable toxicity. A gene editing based therapeutic that introduced such mutations via a one-time injection could extend these protective effects into the population at large.
This approach would leverage one of the most significant scientific advances in recent decades, namely use of a CRISPR RNA-guided endonuclease system to cleave sequences of the human genome in a highly specific fashion.122,123 A single injection of a viral vector designed to target hepatic PCSK9 using CRISPR/CAS9 led to mutations in ~50% of hepatocytes and a 40% reduction in cholesterol levels in a mouse model.124 Similar such therapeutics could be designed to decrease circulating triglyceride-rich lipoproteins, lipoprotein(a), or other causal risk factors for CAD identified in coming years.
Because this gene editing approach would lead to irreversible change, an abundance of caution is needed prior to clinical implementation. Ongoing research seeks to attenuate the immune reaction by the viral vectors used to introduce the therapeutic, increase efficiency and specificity in targeting the desired gene, and ensure that no other genes are affected (so-called off-target effects). With regard to target selection, careful phenotyping of ‘human knockouts’ may help ensure that there are no unanticipated side effects of gene inactivation. Gene editing to decrease a gene product’s expression or function is more tractable with current technology. However, for genes in which damaging mutations confer increased risk of CAD (e.g. LDLR), future advances may enable upregulation or potentiation of gene activity. Furthermore, alteration of an individual’s DNA raises a host of ethical and social questions that will need to be fully explored.125 However, at present, the majority of individuals in the United States approve of gene editing with the express purpose of improving health or preventing disease.126
Conclusions
Here, we reviewed the substantial progress in understanding the genetic underpinnings of CAD. Moving forward, the price of genetic sequencing will continue to decrease – an increased emphasis on variant interpretation, functional validation, and integration with large-scale phenotyping efforts will become paramount. Over the next ten years, we are hopeful that human genetics will prove useful in identifying novel root causes of CAD, guiding drug development efforts in anticipating the safety and efficacy profile of a given therapeutic, and providing patients and their providers with genetic data that will aid in CAD prevention and treatment.
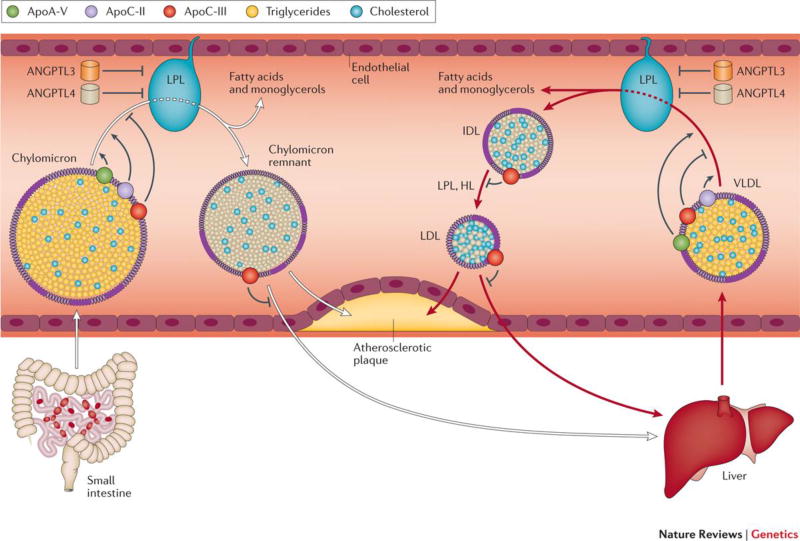
Figure 5.
Lipoprotein lipase (LPL) is an enzyme anchored to the endothelial cells lining blood capillaries. Dietary fat is absorbed by the small intestine and enters the blood stream as triglyceride-rich lipoprotein particles known as chylomicrons. These chylomicrons are hydrolysed by LPL to provide free fatty acids (used for energy by muscle tissue or deposited into fat stores) and chylomicron remnant particles. LPL plays an additional role in hydrolysing very-low density lipoprotein (VLDL) particles secreted by the liver to produce intermediate density lipoprotein (IDL), subsequently degraded into low-density lipoprotein (LDL) particles by hepatic lipase (HL). Both chylomicron remnants and LDL can penetrate the vessel wall and propagate atherosclerotic plaque. LPL activity is the rate-determining step in clearance of dietary fat from the circulation and is highly regulated in the body – apolipoprotein A-V and apolipoprotein C-II activate LPL and apolipoprotein C-III (APOC-III), Angiopoietin-like 4, and angiopoietin-like 3 (ANGPTL3) each inhibit LPL activity. Rare variant association studies have supported a link between several of the proteins involved in the LPL pathway and CAD, including LPL itself, apoC-III, apoA-V, and ANGPTL4 (Table 1).
Key points.
Coronary artery disease is a heritable disorder that, despite advances in treatment and prevention strategies, remains the leading cause of global mortality. Human genetics studies have started to unravel the genetic underpinnings of the disorder.
Gene discovery efforts have rapidly transitioned from family-based studies (for example, those that led to the discovery of familial hypercholesterolemia) to large cohorts that facilitate both common and rare variant association studies.
Common variant association studies have confirmed about 60 genetic loci with a robust association with coronary disease, the majority of which are of modest effect size and in noncoding regions. Rare variant association studies have linked inactivating mutations in at least 9 genes with risk of coronary artery disease.
Human genetics and large-scale biobanks can facilitate drug development for coronary artery disease by highlighting causal biology and helping to understand the phenotypic consequences of lifelong deficiency of a given protein.
Genomic medicine may enable providing patients and their providers with genetic data that will aid in coronary artery disease prevention and treatment.
Genome editing to introduce mutations protective against coronary artery disease into the population could prove curative with a one-time injection, although substantial additional work is needed to confirm efficacy, safety, and address the underlying ethics.
Acknowledgments
Dr. Khera’s work on this manuscript supported by a John S. LaDue Memorial Fellowship in Cardiology and KL2/Catalyst Medical Research Investigator Training award (an appointed KL2 award) from Harvard Catalyst. Dr. Kathiresan is supported by an Ofer and Shelley Nemirovsky MGH Research Scholar Award and by the following awards from the National Institutes of Health: HL127564 and UM1HG008895.
Glossary
- heritable
Capable of being transmitted from parent to offspring via genetic variation
- genetic architecture
The full spectrum of common and rare genetic variation that contributes to a trait of interest
- sitosterolemia
A recessive condition characterized by marked elevations in dietary sterols and increased LDL cholesterol
- linkage analysis
Systematic localization of a genetic region that is co-inherited with a trait of interest in members of a family
- monogenic cause
Variation in a single gene dictates the observed variation in a trait of interest; also referred to as ‘Mendelian’ disorders
- allele frequency
The relative frequency of an allele (specific genetic variant) in the population; typically reported as the proportion of all chromosomes in the population that carry an allele
- inactivating mutations
Variants that disrupt the ability of a given gene to produce its protein product, i.e. due to premature truncation, scrambling of the amino acid code, or disrupting gene splicing
- mendelian randomization
A human genetics tool that leverages the random assortment of genetic variants at time of conception to assess causality of observed associations
- consanguinity
Production of offspring by related individuals (e.g. second cousins or closer)
Biographies
Dr. Amit V. Khera is a cardiologist at Massachusetts General Hospital and an Instructor in Medicine at Harvard Medical School. He started a postdoctoral research fellowship with Dr. Sekar Kathiresan in 2015. His research focuses on the use of human genetics to understand cardiovascular disease, facilitate drug development, and inform clinical care.
Dr. Sekar Kathiresan is a physician scientist and a human geneticist, is the Director of the Center for Human Genetic Research at Massachusetts General Hospital, Director of the Cardiovascular Disease Initiative at the Broad institute, and an Associate Professor of Medicine at Harvard Medical School. Dr. Kathiresan leverages human genetics to understand the root causes of heart attack and to improve preventive cardiac care. Among his scientific contributions, he has helped highlight new biological mechanisms underlying heart attack, discovered mutations that protect against heart attack risk, and developed a genetic test for personalized heart attack prevention.
Footnotes
Competing interests
A. V. K. reports consulting fees from Amarin Pharmaceuticals and Merck & Co. S. K. reports grant support from Bayer Healthcare and Amarin, equity in San Therapeutics and Catabasis, and receiving personal fees for scientific advisory board participation for Bayer Healthcare, Catabasis, Regeneron Genetics Center, Merck, Celera, Genomics PLC, Novartis, Sanofi, AstraZeneca, Alnylam, Eli Lilly, Leerink Partners, Noble Insights, and Ionis.
Subject categories
Health sciences / Cardiology / Cardiovascular biology / Cardiovascular genetics [URI /692/4019/592/2727]
Biological sciences / Genetics / Clinical genetics / Disease genetics [URI /631/208/2489/144]
Biological sciences / Genetics / Genetic association study / Genome-wide association studies [URI /631/208/205/2138]
Biological sciences / Genetics / Genome / Genetic variation [URI /631/208/726/649]
References
- 1.Ford ES, et al. Explaining the decrease in U.S. deaths from coronary disease, 1980–2000. N Engl J Med. 2007;356:2388–2398. doi: 10.1056/NEJMsa053935. [DOI] [PubMed] [Google Scholar]
- 2.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mozaffarian D, et al. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133:e38–60. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 4.Khera AV, et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Risk of Coronary Disease. NEJM. doi: 10.1056/NEJMoa1605086. (online ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gertler MM, Garn SM, White PD. Young candidates for coronary heart disease. J Am Med Assoc. 1951;147:621–625. doi: 10.1001/jama.1951.03670240005002. [DOI] [PubMed] [Google Scholar]
- 6.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. This was the first large-scale prospective study of twins to confirm an increased risk of early-onset CAD among highly related individuals. [DOI] [PubMed] [Google Scholar]
- 7.Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, De Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med. 2002;252:247–254. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 8.Won HH, et al. Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease. PLoS Genet. 2015;11:e1005622. doi: 10.1371/journal.pgen.1005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd-Jones DM, et al. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204–2211. doi: 10.1001/jama.291.18.2204. [DOI] [PubMed] [Google Scholar]
- 10.Murabito JM, et al. Sibling cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults. JAMA. 2005;294:3117–3123. doi: 10.1001/jama.294.24.3117. [DOI] [PubMed] [Google Scholar]
- 11.Watkins H, Farrall M. Genetic susceptibility to coronary artery disease: from promise to progress. Nat Rev Genet. 2006;7:163–173. doi: 10.1038/nrg1805. [DOI] [PubMed] [Google Scholar]
- 12.Muller C. Xanthomata, hypercholesterolemia, angina pectoris. Acta Med Scand. 1938;89:75–84. [Google Scholar]
- 13.Lehrman MA, Schneider WJ, Südhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. The first family-based study to identify a discrete mutation in a single gene predisposing to CAD (familial hypercholesterolemia). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci. U S A. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 16.Garcia CK, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–1398. doi: 10.1126/science.1060458. [DOI] [PubMed] [Google Scholar]
- 17.Berge KE, et al. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–1775. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Fan C, Topol SE, Topol EJ, Wang Q. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science. 2003;302:1578–1581. doi: 10.1126/science.1088477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieb W, et al. Lack of association between the MEF2A gene and myocardial infarction. Circulation. 2008;117:185–191. doi: 10.1161/CIRCULATIONAHA.107.728485. [DOI] [PubMed] [Google Scholar]
- 20.Mani A, et al. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keramati AR, et al. A form of the metabolic syndrome associated with mutations in DYRK1B. N Engl J Med. 2014;370:1909–1919. doi: 10.1056/NEJMoa1301824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacArthur DG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. A framework for systematic assessment of a potentially causal relationship between a given genetic variant and human disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuk O, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111:E455–64. doi: 10.1073/pnas.1322563111. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 27.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye S, Willeit J, Kronenberg F, Xu Q, Kiechl S. Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. J Am Coll Cardiol. 2008;52:378–384. doi: 10.1016/j.jacc.2007.11.087. [DOI] [PubMed] [Google Scholar]
- 29.Helgadottir A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 30.Smith JG, et al. Common genetic variants on chromosome 9p21 confers risk of ischemic stroke: a large-scale genetic association study. Circ Cardiovasc Genet. 2009;2:159–164. doi: 10.1161/CIRCGENETICS.108.835173. [DOI] [PubMed] [Google Scholar]
- 31.Jarinova, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 32.Holdt LM, et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 33.Harismendy O, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myocardial Infarction Genetics Consortium et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2010;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erdmann J, Grosshennig A, Braund PS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coronary Artery Disease Genetics (C4D) Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 37.IBC 50K CAD Consortium. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 2011;7:e1002260. doi: 10.1371/journal.pgen.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.CARDIoGRAMplusC4D Consortium. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikpay M, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- 41.Flannick J, Florez JC. Type 2 diabetes: genetic data sharing to advance complex disease research. Nat Rev Genet. 2016;17:535–59. doi: 10.1038/nrg.2016.56. [DOI] [PubMed] [Google Scholar]
- 42.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374:1134–1144. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehret GB, Ferreira T, Chasman DI, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48:1171–1184. doi: 10.1038/ng.3667. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erdmann J, et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature. 2013;504:432–436. doi: 10.1038/nature12722. [DOI] [PubMed] [Google Scholar]
- 47.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Do R, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. The first large study to use whole-exome sequencing to examine the relationship of rare variants in each gene with CAD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 50.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. This paper linked inactivating mutations in PCSK9 with significantly reduced LDL cholesterol and risk of incident CAD. [DOI] [PubMed] [Google Scholar]
- 51.Nioi P, et al. Variant ASGR1 Associated with a Reduced Risk of Coronary Artery Disease. N Engl J Med. 2016;374:2131–2141. doi: 10.1056/NEJMoa1508419. [DOI] [PubMed] [Google Scholar]
- 52.Khera AV, et al. Damaging Mutations in Lipoprotein Lipase and Risk for Coronary Artery Disease. JAMA. (under revision) [Google Scholar]
- 53.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 54.Crosby J, Peloso GM, Auer PL, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374:1134–1144. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dewey FE, et al. Inactivating Variants in ANGPTL4 and Risk of Coronary Artery Disease. N Engl J Med. 2016;374:1123–1133. doi: 10.1056/NEJMoa1510926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. This paper was the first to characterize the mechanism linking a noncoding variant with changes in gene regulation related to LDL cholesterol metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strong A, et al. Hepatic sortilin regulates both apolipoprotein B secretion and LDL catabolism. J Clin Invest. 2012;122:2807–16. doi: 10.1172/JCI63563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reilly MP, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pu X, et al. ADAMTS7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. Am J Hum Genet. 2013;92:366–374. doi: 10.1016/j.ajhg.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bauer RC, et al. Knockout of Adamts7, a novel coronary artery disease locus in humans, reduces atherosclerosis in mice. Circulation. 2015;131:1202–1213. doi: 10.1161/CIRCULATIONAHA.114.012669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kessler T, et al. ADAMTS-7 inhibits re-endothelialization of injured arteries and promotes vascular remodeling through cleavage of thrombospondin-1. Circulation. 2015;131:1191–1201. doi: 10.1161/CIRCULATIONAHA.114.014072. [DOI] [PubMed] [Google Scholar]
- 65.DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: New estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 66.Paul SM, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nature Rev. Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 67.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. Review article describing the potential utility of human genetics to expedite drug development. [DOI] [PubMed] [Google Scholar]
- 68.Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voight BF. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clarke R, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 71.Do R, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Myocardial Infarction Genetics Consortium Investigators Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371:2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cannon CP, et al. Ezetimibe Added to Statin Therapy after Acute Coronary Syndromes. N Engl J Med. 2015;372:2387–97. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 74.Lp-PLA(2) Studies Collaboration et al. Lipoprotein-associated phospholipase A(2) and risk of coronary disease, stroke, and mortality: collaborative analysis of 32 prospective studies. Lancet. 2010;375:1536–1544. doi: 10.1016/S0140-6736(10)60319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilensky RL, et al. Inhibition of lipoprotein-associated phospholipase A2 reduces complex coronary atherosclerotic plaque development. Nat Med. 2008;14:1059–1066. doi: 10.1038/nm.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.STABILITY Investigators et al. Darapladib for preventing ischemic events in stable coronary heart disease. N Engl J Med. 2014;370:1702–1711. doi: 10.1056/NEJMoa1315878. [DOI] [PubMed] [Google Scholar]
- 77.O’Donoghue ML, et al. Effect of darapladib on major coronary events after an acute coronary syndrome: the SOLID-TIMI 52 randomized clinical trial. JAMA. 2014;312:1006–1015. doi: 10.1001/jama.2014.11061. [DOI] [PubMed] [Google Scholar]
- 78.Polfus LM, et al. Coronary heart disease and genetic variants with low phospholipase A2 activity. N Engl J Med. 2015;372:295–296. doi: 10.1056/NEJMc1409673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Casas JP, et al. PLA2G7 genotype, lipoprotein-associated phospholipase A2 activity, and coronary heart disease risk in 10 494 cases and 15 624 controls of European Ancestry. Circulation. 2010;121:2284–2293. doi: 10.1161/CIRCULATIONAHA.109.923383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson JG, et al. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- 81.Sabatine MS. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1500–1509. doi: 10.1056/NEJMoa1500858. [DOI] [PubMed] [Google Scholar]
- 82.Gaudet D, et al. Antisense Inhibition of Apolipoprotein C-III in Patients with Hypertriglyceridemia. N Engl J Med. 2015;373:438–447. doi: 10.1056/NEJMoa1400283. [DOI] [PubMed] [Google Scholar]
- 83.Tsimikas S, et al. Antisense therapy targeting apolipoprotein(a): a randomised, double-blind, placebo-controlled phase 1 study. Lancet. 2015;386:1472–1483. doi: 10.1016/S0140-6736(15)61252-1. [DOI] [PubMed] [Google Scholar]
- 84.Swerdlow DI, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. 2015;385:351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sattar N, et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet. 2010;375:735–742. doi: 10.1016/S0140-6736(09)61965-6. [DOI] [PubMed] [Google Scholar]
- 86.US National Library of Medicine. ClinicalTrials.gov. 2016 https://clinicaltrials.gov/ct2/show/ NCT01252953.
- 87.Neale BM, et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC) Proc Natl Acad Sci U S A. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen W, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107:7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng CY, et al. New loci and coding variants confer risk for age-related macular degeneration in East Asians. Nat Commun. 2015;6:6063. doi: 10.1038/ncomms7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bush WS, Oetjens MT, Crawford DC. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet. 2016;17:129–145. doi: 10.1038/nrg.2015.36. [DOI] [PubMed] [Google Scholar]
- 91.Emdin CA, et al. Phenotypic Characterization of Genetically Lowered Human Lipoprotein(a) Levels. JACC. (in press) [Google Scholar]
- 92.Sawabe M, et al. Low lipoprotein(a) concentration is associated with cancer and all-cause deaths: a population-based cohort study (the JMS cohort study) PLoS ONE. 2012;7:e31954. doi: 10.1371/journal.pone.0031954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mora S, Kamstrup PR, Rifai N, Nordestgaard BG, Buring JE, Ridker PM. Lipoprotein(a) and risk of type 2 diabetes. Clin Chem. 2010;56:1252–1260. doi: 10.1373/clinchem.2010.146779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lichtenstein L, et al. Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell Metab. 2010;12:580–592. doi: 10.1016/j.cmet.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Desai U, et al. Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid phenotype found in angiopoietin-like 4 knockout mice. Proc Natl Acad Sci U S A. 2007;104:11766–11771. doi: 10.1073/pnas.0705041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 13:135–45. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–7. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 98.Nelson MR, et al. The genetics of drug efficacy: opportunities and challenges. Nat Rev Genet. 2016;17:197–206. doi: 10.1038/nrg.2016.12. [DOI] [PubMed] [Google Scholar]
- 99.Paynter NP, Ridker PM, Chasman DI. Are Genetic Tests for Atherosclerosis Ready for Routine Clinical Use? Circ Res. 2016;118:607–619. doi: 10.1161/CIRCRESAHA.115.306360. [DOI] [PubMed] [Google Scholar]
- 100.Umans-Eckenhausen MA, Defesche JC, Sijbrands EJ, Scheerder RL, Kastelein JJ. Review of first 5 years of screening for familial hypercholesterolaemia in the Netherlands. Lancet. 2001;357:165–168. doi: 10.1016/S0140-6736(00)03587-X. [DOI] [PubMed] [Google Scholar]
- 101.Nordestgaard BG. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 2013;34:3478–3490a. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khera AV, et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J Am Coll Cardiol. 2016;67:2578–2589. doi: 10.1016/j.jacc.2016.03.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Talmud PJ, et al. Use of low-density lipoprotein cholesterol gene score to distinguish patients with polygenic and monogenic familial hypercholesterolaemia: a case-control study. Lancet. 2013;381:1293–1301. doi: 10.1016/S0140-6736(12)62127-8. [DOI] [PubMed] [Google Scholar]
- 105.Scandinavian Simvastatin Survival Study Group. Randomized trial of cholesterol lowering in 4444 patients with coronary heart disease: the Scandinavian Simvastatin Survival Study (4S) Lancet. 1994;344:1383–1138. [PubMed] [Google Scholar]
- 106.Cholesterol Treatment Trialists’ (CTT) Collaborators. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet. 2012;380:581–590. doi: 10.1016/S0140-6736(12)60367-5. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kathiresan S, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 109.Ripatti S, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tada H, et al. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur Heart J. 2016;37:561–567. doi: 10.1093/eurheartj/ehv462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abraham G, et al. Genomic prediction of coronary heart disease. Eur Heart J. 2016 Sep 21; doi: 10.1093/eurheartj/ehw450. [Epub ahead of print] PubMed PMID: 27655226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mega JL, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kullo IJ, et al. Incorporating a Genetic Risk Score Into Coronary Heart Disease Risk Estimates: Effect on Low-Density Lipoprotein Cholesterol Levels (the MI-GENES Clinical Trial) Circulation. 2016;133:1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wellcome Trust Case Control Consortium et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 116.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stylianou IM, Bauer RC, Reilly MP, Rader DJ. Genetic basis of atherosclerosis: insights from mice and humans. Circ Res. 2012;110:337–355. doi: 10.1161/CIRCRESAHA.110.230854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nurnberg ST, et al. From Loci to Biology: Functional Genomics of Genome-Wide Association for Coronary Disease. Circ Res. 2016;118:586–606. doi: 10.1161/CIRCRESAHA.115.306464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Narasimhan VM, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352:474–477. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Saleheen D, et al. Human knockouts in a cohort with a high rate of consanguinity. [(Accessed June 2016)];bioRxiv. http://dx.doi.org/10.1101/031518 Identification of humans with inactivating mutations (‘knockouts’) in ~1,000 genes and genotype-based callback to understand relevant physiology.
- 121.Choudhry NK, et al. Full coverage for preventive medications after myocardial infarction. N Engl J Med. 2011;365:2088–2097. doi: 10.1056/NEJMsa1107913. [DOI] [PubMed] [Google Scholar]
- 122.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ding Q, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. Proof-of-concept in a mouse model that permanent disruption of PCSK9 using gene editing can decrease LDL cholesterol and atherosclerosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lander ES. Brave New Genome. N Engl J Med. 2015;373:5–8. doi: 10.1056/NEJMp1506446. [DOI] [PubMed] [Google Scholar]
- 126.Blendon RJ, Gorski MT, Benson JM. The Public and the Gene-Editing Revolution. N Engl J Med. 2016;374:1406–1411. doi: 10.1056/NEJMp1602010. [DOI] [PubMed] [Google Scholar]
- 127.Libby P. The Vascular Biology of Atherosclerosis. In: Bonor RO, Mann DL, Zipes DP, Libby P, editors. Braunwald’s Heart Disease: A Textbook of Cardiovascular Medicine. 10. Atlanta, GA: Elsevier Health Sciences; 2014. pp. 873–890. [Google Scholar]
- 128.Ridker PM, Libby P, Buring JE. Risk Markers and the Primary Prevention of Cardiovascular Disease. In: Bonor RO, Mann DL, Zipes DP, Libby P, editors. Braunwald’s Heart Disease: A Textbook of Cardiovascular Medicine. 10. Atlanta, GA: Elsevier Health Sciences; 2014. pp. 891–933. [Google Scholar]
- 129.Kessler T, Vilne B, Schunkert H. The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol Med. 2016;8:688–701. doi: 10.15252/emmm.201506174. [DOI] [PMC free article] [PubMed] [Google Scholar]