Summary
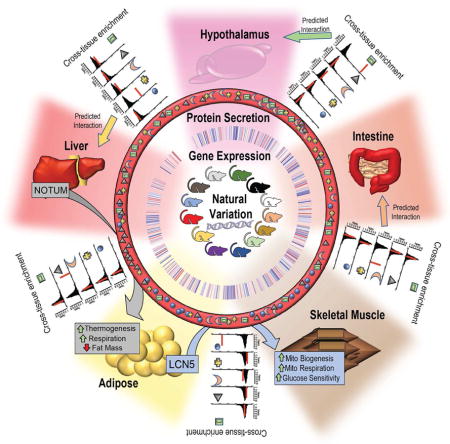
Inter-tissue communication via secreted proteins has been established as a vital mechanism for proper physiologic homeostasis. Here, we report a bioinformatics framework using a mouse reference population, the Hybrid Mouse Diversity Panel (HMDP), which integrates global multi-tissue expression data and publicly available resources to identify and functionally annotate novel circuits of tissue-tissue communication. We validate this method by showing that we can identify known as well as novel endocrine factors responsible for communication between tissues. We further show the utility of this approach by identification and mechanistic characterization of two new endocrine factors. Adipose-derived Lipocalin-5 is shown to enhance skeletal muscle mitochondrial function and liver-secreted Notum promotes browning of white adipose tissue, also known as “beiging”. We demonstrate the general applicability of the method by providing in vivo evidence for 3 additional novel molecules mediating tissue-tissue interactions.
Keywords: Endocrine, cross-tissue communication, secreted peptides, skeletal muscle respiration, mitochondria, adipocyte beiging, Lipocalin-5, Notum, SPARC Related Modular Calcium Binding 1, Inter-Alpha-Trypsin Inhibitor heavy chain H5, Pro-Platelet Basic Protein
eTOC Blurb
Seldin and colleagues use a bioinformatics-based approach to identify new inter-tissue endocrine circuits. Using this resource, they report on two novel endocrine factors -Adipose-derived Lipocalin-5, which promotes muscle mitochondrial respiration and liver-secreted Notum, which mediates adipose tissue beiging.
INTRODUCTION
Multicellular organisms have evolved dynamic means for communication between cell types to maintain proper physiologic homeostasis. In mammals, secreted peptides have been described to regulate nearly all aspects of physiology. Beginning with the discovery of insulin nearly a century ago (Williams, 1993), endocrine tissue cross-talk has been a primary focus for the detection and treatment of adverse metabolic perturbations. Since then, many secreted proteins have been identified which play crucial roles in metabolism (Friedman and Halaas, 1998; Kershaw and Flier, 2004; Salomon et al., 1989; Trujillo and Scherer, 2005), immune system function (Belardelli, 1995; Mosmann and Coffman, 1989), cardiovascular pathobiology (Nian et al., 2004; Tedgui and Mallat, 2006) and many other aspects of physiology. Previous studies have shown that substantial interactions between tissues persist across multiple transcripts in various physiologic states (Consortium, 2013; Price et al., 2011). Even for autonomous cells such as bacteria, mechanisms have been identified by which secreted components relay signals from one cell to another (Miller and Bassler, 2001). Regardless of the total cell number, size or complexity of species, nearly all organisms have maintained a capacity to communicate by means of secreted polypeptides throughout evolution. Current estimates in mice and humans predict that roughly ~3,000 unique proteins are secreted, accounting for ~16% of the entire protein coding genome (Lindskog, 2015; Uhlen et al., 2010), and recent studies have provided convincing evidence that miRNAs can also mediate inter-tissue communication (Thomou et al., 2017).
Advances in “omics” technologies such as shotgun proteomics and RNA-seq have enabled unbiased discovery of many additional systemic regulators of whole-body homeostasis (Civelek and Lusis, 2014; Gehlenborg et al., 2010; Joyce and Palsson, 2006; Kussmann et al., 2006). Substantial efforts have been dedicated to assessing single-cell or tissue-specific secretome through proteomics analysis of cultured media (Alvarez-Llamas et al., 2007; Hathout, 2007; Katz-Jaffe et al., 2009) and to filtering transcriptomic data for the presence of signal peptides for secretion (Hathout, 2007; Mosquera et al., 2009; Nielsen et al., 1997). Similarly, serum has been subjected to mass spectrometry proteomics in order to identify candidate molecules involved in inter-tissue communication, focusing on biomarker identification (Hathout, 2007; Lawlor et al., 2009; Makridakis and Vlahou, 2010; Petricoin and Liotta, 2004). More advanced approaches have shown differential networks of gene expression across tissues (Keller et al., 2008; Pierson et al., 2015), and even tissue-specific networks to suggest individual genes regulating these axes (Long et al., 2016). One important component lacking in these approaches, however, is the ability to gain insight about the function, mechanism of action and specific target tissue(s) of identified secreted proteins. In general, uncovering the physiologic functions of endocrine and paracrine polypeptides requires costly and time-consuming biochemical experimentation.
Here, we report a bioinformatics framework that uses natural variation in transcript levels across tissues in a population to identify and functionally annotate endocrine circuits. This method, termed Quantitative Endocrine Network Interaction Estimation (QENIE), can be applied to any collection of organs, tissues, or cell types sampled from a genetically diverse population and profiled using omics platforms. Many of the top-ranked candidates for each tissue-tissue axis here already have established mechanisms of endocrine communication, validating the approach. To prioritize putative endocrine factors, we consider relevant physiologic and clinical traits along with publicly available resources on tissue-specific gene expression. These efforts led to the identification of two new endocrine factors. The first is an adipose-derived protein which enhances skeletal muscle mitochondrial activity resulting in increased lean body mass and improved insulin sensitivity. The second involves a hepatic secreted protein which can promote adipose tissue “beiging”, resulting in improved resistance to cold and increased energy expenditure in vivo. To demonstrate the general applicability of the approach, we identify and validate in vivo 3 additional secreted molecules involving communication between tissues, adipose and heart (SMOC1 and IT1H5), and within aorta (PPBP). We also provide evidence for the applicability of the method to human omics data.
RESULTS
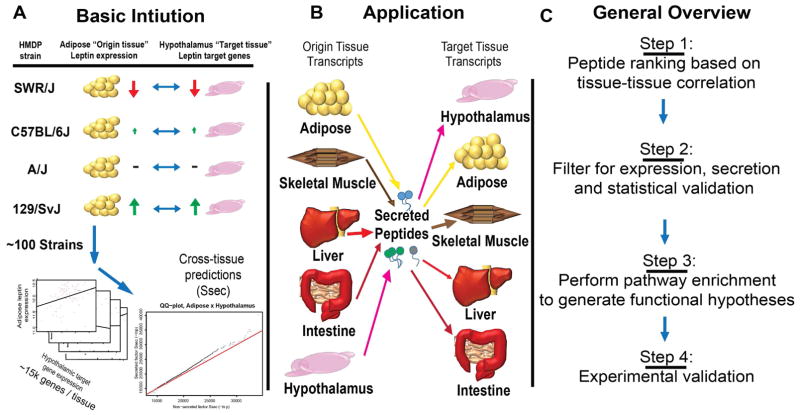
The basic intuition for QENIE is as follows: In a population exhibiting natural variation, transcript levels for most genes differ among individuals. Thus, the inter-organ communication between two tissues will vary in a coordinated manner, with differences in expression of an endocrine factor in one tissue showing correlation with the responder genes and pathways in a target tissue. To identify secreted endocrine factors, we screened for correlations by examining global transcript levels between pairs of tissues and filtering for origin (or sender) tissue secretion (Figure 1). Applying this approach to data from a large cohort of inbred strains of mice revealed many previously described endocrine interactions that were consistent with their published functions. For example, in several inbred mouse strains, genetic diversity and gene-by-environment interactions will drive adipose tissue variation in Leptin expression. Given the significant role of Leptin in regulating hypothalamus gene expression and function, we expect to observe variation in hypothalamic “target” genes in a manner consistent with the levels of Leptin in adipose (Figure 1A). By applying this intuition to a diverse population, we can assess strength of cross-tissue predictions for endocrine circuits, termed Ssec. Here, we also uncovered many novel potential circuits for inter-tissue communication (Figure 1B), several of which we experimentally validated.
Figure 1. Annotated schematic for Quantitative Estimation of Endocrine Network Interaction, (QENIE).
A, The basic intuition for our approach is illustrated for 4 strains where adipose tissue expression of Leptin varies in a manner consistent with hypothalamus target genes. When applied to ~100 HMDP strains, this correlation of gene expression is used for cross-tissue predictions. B, This framework was applied to 5 tissues, each examined for gene expression using ~20,000 probes corresponding to ~15,000 genes or RNA-seq in 106 inbred strains of mice. We assigned each axis for every combination of “origin” and “target” tissues. C, Generalized steps for method progression and the application, discussed on greater detail in the main text.
Transcriptomic screening for regulators of inter-tissue cross-talk in the HMDP
To uncover secreted proteins which regulate processes in an endocrine fashion, we utilized global transcript levels determined using expression arrays and RNA-seq from five tissues (liver, skeletal muscle, adipose, hypothalamus and intestine) in 106 common inbred strains of mice that constitute the Hybrid Mouse Diversity panel (HMDP). Our initial studies were performed with mice fed a high-fat/high-sucrose diet (Hasin-Brumshtein et al., 2016; Parks et al., 2013; Parks et al., 2015). A tutorial of our pipeline, pretreatment of data, datasets used, and R script implemented for each step of this pipeline is provided at: https://github.com/marcus-seldin/QENIE. For every tissue pair, cross-tissue bi-weight midcorrelation matrices were calculated across mouse strains. This coefficient, termed bicor is similar to a Pearson’s correlation coefficient (typically illustrated as r), but offers the advantage of more robustness due to removal of outliers in a dataset (Langfelder and Horvath, 2012). To assess the significance of the correlations in these matrices, Student correlation P-values were calculated using the bicor coefficient, which minimizes effects due to outliers (Langfelder and Horvath, 2008). For each matrix of tissue-by-tissue P-values of associations, individual organs were then designated “origin” or “target” tissues.
To identify proteins which act in an endocrine fashion for each matrix of correlations, we included only genes for each origin tissue that encode secreted proteins. This was done by overlaying gene symbols with those annotated to be secreted the Universal Protein Resource (UniProt) database (Leinonen et al., 2006; UniProt, 2015). In total, these annotations contain ~2,300 unique secreted proteins of which 1,047 to 1,213 (depending on the tissue) were retained in our expression data as origin tissue genes encoding a secreted protein. The P-value matrices of secreted genes were then ranked by the level of significance of association with all target tissues, listed as “Step 1- ranking” in Figure 1. This was accomplished by computing the sum of the −ln(p-value) for each origin secreted protein across all target tissue transcripts, listed further as Ssec. In order to make the Ssec values comparable between various endocrine circuits, this sum of the −ln(pvalue) was normalized by the total number of target tissue transcripts. We have illustrated an overview of the procedure (Figure S1).
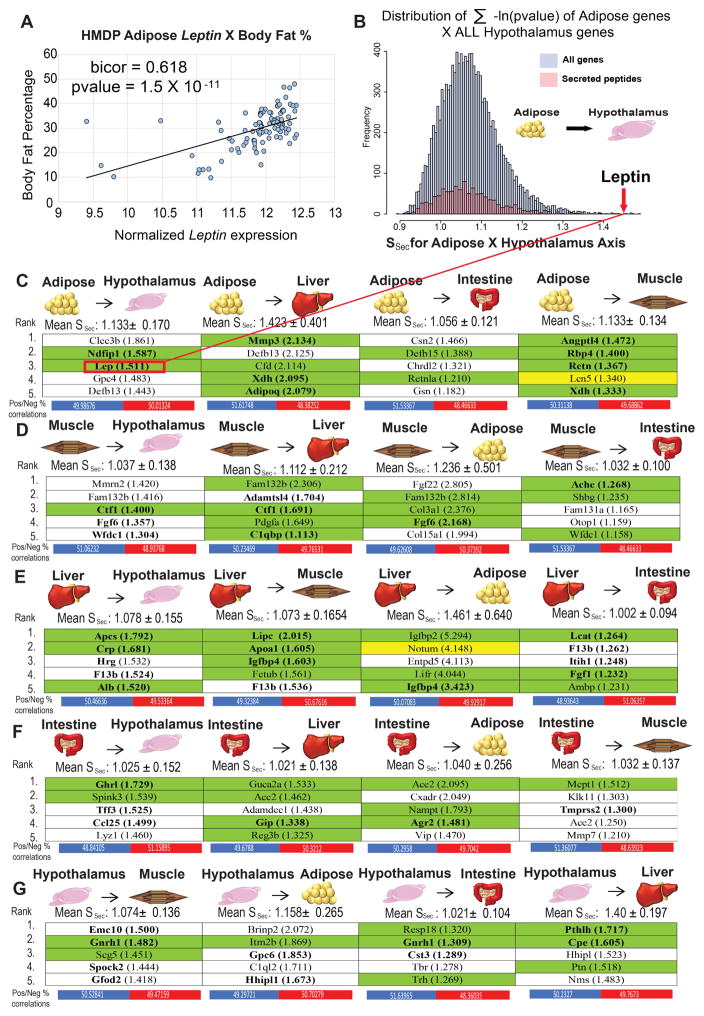
For example, in adipose-to-hypothalamus, the expression of Leptin in adipose tissue varies across the mouse population and shows significant correlation with body weight (Figure 2A), consistent with corresponding positive plasma correlations to body mass index across the human populations (Farooqi and O’Rahilly, 2009; Margetic et al., 2002). The Ssec scores for each individual adipose transcript across all hypothalamic transcripts had a skewed distribution towards higher values, indicating physiologically-significant interactions (Figure 2B). Leptin ranked at the extreme upper-end of all Ssec scores, consistent with many previous observations that Leptin exerts a robust impact on hypothalamic gene expression (Elmquist et al., 1998; Schwartz et al., 1996).
Figure 2. QENIE application to Hybrid Mouse Diversity Panel tissues uncovers known regulators of endocrine function.
A, Leptin adipose expression in the HMDP correlates with relevant clinical traits consistent with its known function(s). B, Distribution of significance score for all adipose genes (blue) and secreted peptides (pink) across all hypothalamic gene expression in 106 strains. Leptin (indicated by arrow) identified as a top-ranked peptide for this axis indicated by red line. C–G, Normalized average significance scores and corresponding standard deviations for each cross-organ combination. Arrows indicate directionality of secreted peptide genes assigned to “origin” tissues for adipose (C), skeletal muscle (D), liver (E) intestine (F) and hypothalamus (G) across all transcripts in each respective target tissue. Listed are the 5 highest ranked “origin” secreted proteins for each axis, with corresponding mean significance scores in parentheses. Below each axis are the percentiles of positively vs negatively significantly correlated target tissue genes (pvalue < 0.05) which contribute to all origin Ssec scores. Bolded names indicate a documented SNPs for the gene associating with a clinical trait and green backing shows peptides which have been shown to affect function of target tissues. The two genes highlighted with a yellow background are followed up in this study.
The Ssec rankings were then filtered ensuring that genes were primarily expressed by the origin tissue. This was accomplished by manual inspection of each gene in multi-tissue expression panels available via BioGPS (Su et al., 2002; Wu et al., 2009) in both mouse and humans (representative examples using BioGPS arrays shown in Figure S3). Genes not detected in BioGPS arrays were investigated for tissue specificity using the UniProt literature query for “tissue specificity”. We provide the top five secreted proteins remaining after the tissue-specific filtering for all pairs of adipose, skeletal muscle, hypothalamus, intestine and liver tissue (Figure 2C–G). A significant portion (44%) of the highest ranked secreted proteins have previously been implicated as functional contributors to physiology. In Figure 2C–G, bolded text indicates the secreted proteins with an association between single nucleotide polymorphisms (SNPs) and metabolism-related clinical traits in human. Also indicated are any published observations in which a secreted protein has been shown to affect its corresponding target tissue, highlighted with a green background. Several notable interactions were identified. For example, Ghrelin (gene symbol Ghrl) presents the strongest intestine-to-hypothalamus interaction (Figure 2F), consistent with this hormone functioning as one of the most robust satiety signals (Klok et al., 2007; Lazarczyk et al., 2003). In addition, many well-known adipokines were identified (Figure 2C), including Adiponectin (Adipoq) (Kadowaki et al., 2006; Scherer et al., 1995), Angiopoietin-Like 4 (Angptl4) (Kersten et al., 2000; Wozniak et al., 2009; Yoshida et al., 2002) Resistin (Res) (Kusminski et al., 2005; Steppan et al., 2001) and Retinol binding protein-4 (Rbp4) (Graham et al., 2006; Rocchi et al., 1989). To assess the average “level” of communication between tissues, we calculated the mean and standard deviation values for all secreted proteins in the origin tissue and their respective average cross-tissue Ssec scores (Figure 2C–G). Across all tissues, the highest significance of any secreted protein was found between liver and adipose, gauged by the mean and standard error of the Ssec scores (Figure 2C, E). Listed below each tissue-tissue axis in Figure 2, we also provide the number of positive and negative correlations contributing to each circuit. Given that our Ssec score is only one example of assessing cross-tissue communication, other strategies could include focusing of direction of correlation, as opposed to level of significance. It is also worth mentioning that among liver and adipose tissue arrays, we observed concordant cross-tissue Ssec scores between sexes amongst all secreted factors which was markedly reduced amongst the top 50 genes for each axis (Figure S3) where we expect the strongest predictive value for endocrine communication. These provide a generalized strategy to identify the strongest sets of tissue-tissue interactions within our dataset.
Top-ranked candidate peptide significance scores reside in the upper-limits of distributions of permuted Ssec calculations
Given that we did not know the distribution of the test statistic tissue-specific expression arrays, we ran permutation tests to estimate the levels of significance for each top-ranked gene for every tissue-tissue axis using an adaptive permutation procedure (O’Gorman, 2002), highlighted by green arrows in Figure S1. We permuted the target tissue strains and recalculated Ssec to construct the null distribution and maintain the gene-gene correlation structures in each tissue. These tests typically required 1000–1,000,000 permutations, listed as P-values in Table S1. The permutation tests were designed to specifically to allow generation of a statistic while maintaining correlation structure to limit the influence of inflation of cross-tissue gene correlations (discussed below). We note that in these tests the pvalues used reflect a placeholder for the correlation coefficient which does not require independence. Beyond permutation testing, we also compared various strategies to rank cross-tissue circuits. An alternative approach to Ssec scoring could include counting the number of significant correlations. For the two endocrine circuits explored in-depth (below), we compared the Ssec (sum −ln(pvalue)) to ranking systems generated by counting the total number of significant correlations at various pvalue cut-offs (Figure S4). These analyses also help to point out that Ssec is capturing more of an aggregate of many lower-strength but still significantly correlated, genes as opposed to being driven by fewer highly-corelated genes.
To further provide rationale for focusing on secreted proteins using the Ssec ranking system, we compared the relative enrichment of cross-tissue −ln(pvalue) for all secreted proteins vs genes coding for non-secreted factors. For nearly every inter-organ axis, we observed higher significance of pvalues for secreted factors vs non-secreted, most notably within the top percentiles of significance strength (Figure S5). One possible explanation for inter-tissue circuits which showed stronger significance amongst non-secreted factors (such as muscle-to-adipose) is that we are observing metabolic processes which generate small molecules which act between tissues. In these scenarios, one would expect strong correlations between genes involved in enzymatic production of a metabolite in the origin tissue and genes which utilize or sense the molecule in the target tissue. These observations indicate that while Ssec for secreted peptides vary across tissues, they are significant for the top-ranked candidates. Using this final list of ranked origin tissue expressed and secreted proteins and their subsequent target tissue level of statistically validated significance we next focused on functional assessment and specific hypothesis testing of several novel candidates.
Suggestive pathways targeted by inter-tissue communication and screening pipeline for validation
Ranked lists of genes targeted by potential endocrine proteins were next evaluated for pathway and process enrichment using The Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al., 2007; Sherman et al., 2007). The gene ranks consisted of the top-500 most significantly correlated genes in the target tissue (2–2.7% of probes on the arrays, discussed in Methods). In this way, we interrogated cross-tissue enrichment using the Gene Ontology (GO) Biological Process resource (Ashburner et al., 2000; Gene Ontology, 2015) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database (Du et al., 2014; Ogata et al., 1999). While generally providing broad biological descriptions, the cross-tissue enrichment results nonetheless yielded substantial insight into the likely functions of each secreted protein, thus allowing for effective hypothesis generation.
Implementing these steps generated lists of potential endocrine factors for each tissue-tissue circuit and testable hypotheses with which to follow up. Given that our criteria for validation was based on changes in gene expression in predicted target tissue pathways, we first generated lists of marker genes for each pathway. These genes were chosen based on 1) presence in the top correlated (absolute, positive or negative) genes that were used for pathway prediction and 2) the gene being annotated in the literature as a robust regulator of physiologic process and thus, appropriate surrogate for tested functional outcomes.
To functionally validate these factors, we implemented the following experimental approaches: Firstly, cDNA constructs (or GFP control) for each endocrine factor were cloned into an expression plasmid and over-expressed in HEK293 cells. Media harvested from these cells was used as treatment on target tissue cell types and validated using qPCR. Following up, an epitope-tagged protein from this system was purified from the media and subsequently used for cell culture treatment or injection into mice. This overall process is illustrated in Figure 3 and summarized from left-to-right panels. First, by screening predicted proteins in various mouse populations with a conditioned media approach we have observed a roughly 15–25% success rate depending on the population used. In total, we have screened 36 potential secreted factors via conditioned media on cell lines reflecting target tissue and observed 8 in which predicted target genes are significantly altered. While potential factors which could confound predictions are discussed in detail below, we note that a “negative result” with this artificial system does not mean that a bona fide interaction does not persist, just that gene expression effects were not observed in these specific contexts. A more thorough system to screen factors for physiologic impacts is presented in Figure 3. Here, we applied QENIE to two separate HMDP populations (as opposed to Figure 2), under basal chow conditions (Figure 3A–F) or a proatherosclerotic setting (Figure 3G–I) and uncovered 3 novel inter-tissue interactions. We note that the factors presented in Figure 3 do not necessarily show the strongest cross-tissue enrichment. These factors were chosen in part based on our own interests. The results suggest that the endocrine interactions persist beyond the upper-limits of the predicted distributions.
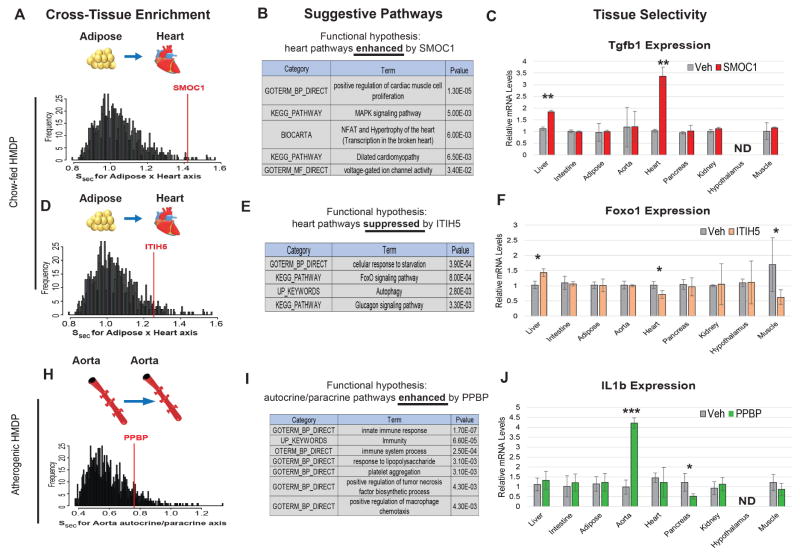
Figure 3. Application of QENIE is robust in identifying endocrine factors across various datasets.
Three additional endocrine factors were screened using the same approach, where the enrichment amoungst all secreted factors is shown for each tissue-tissue axis in various datasets (left), as well as suggestive pathway enrichment based on conditioned correlation coefficents (middle) and tissue specificity of action following injection of recombinant protein (right). By implementing data from a different HMDP cohort on a chow diet, QENIE uncovered SMOC1 (A–C) and ITIH5 (D–F) as adipose-derived regulators of heart Tgf1b and Foxo1, respectively. Data used from a atherosclerosis-prone mouse population was applied to uncover autocrine/paracrine regulators of aortic gene expression, where Ppbp was found to enhance IL1b gene expression (H–J). Tissue specificity screening utilized C57BL/6J mice on a chow diet, injected with veh (saline) or 0.1ug/gram body weight recombinant protein. ND = Not detected (assessed by a raw qPCR Ct value above 30). All data presented as mean ± SEM *p<0.05, **p<0.01, ***p<0.001
Initially, we uncovered the adipokine SPARC-related modular calcium-binding protein 1 (SMOC1) as a potential regulator of heart muscle proliferation and hypertrophy (Figure 3A–C). Therefore, we asked whether injection of recombinant SMOC1 could alter a key regulator of muscle cell development and proliferation, Transformation growth factor beta 1 (Tfgb1). Here we observed a significant induction of Tgfb1 in the hearts of SMOC1-injected mice (Figure 3C). We also investigated the potential regulation of other predicted pathways, specifically MAPK, hypertrophy and calcium-handling. While we observed changes in calcium-related gene expression (Cam2k) in the SMOC1-injected hearts, no changes were observed for Mapk1 (data not shown). SMOC1 has been demonstrated to regulate developmental processes, specifically BMP antagonism leading to limb malformations (Okada et al., 2011; Rainger et al., 2011), as well as endothelial TGFB signaling (Awwad et al., 2015) suggesting that it acts in tissues other than heart.
Another analysis of the same chow-fed dataset uncovered Inter-Alpha-Trypsin Inhibitor Heavy Chain Family Member 5 (ITIH5) as enriched across the cardiac gene expression profile (Figure 3D). GO showed enrichment for pathways involved in cellular starvation response with ITIH5 as a negative regulator (Figure 3E). In response to injection of recombinant ITIH5, tissue-wide expression analysis showed ITIH5 reduced Forkhead box O1 (Foxo1) gene expression in heart (Figure 3F). While we did not observe changes in AMPK (Prkaa1) in response to the protein injection, other downstream targets, such as Acetyl Co-A carboxylase 1 (Acc1) were significantly altered (data not shown). ITIH5 has been shown to impact myofibroblast differentiation (Martin et al., 2016) and implicated as a marker of obesity (Anveden et al., 2012) and several types of cancers (Dotsch et al., 2015; Himmelfarb et al., 2004; Kloten et al., 2014; Mai et al., 2014; Rose et al., 2014).
Finally, we sought to identify autocrine/paracrine regulators of vascular cell function in a pro-atherosclerotic setting (Figure 3H). Using aorta expression profiles from a HMDP study where mice were genetically sensitized for atherosclerosis (Bennett et al., 2015) we asked whether the same approach could be applied to a complex vascular milieu of cells to uncover autocrine/paracrine factors. We observed Platelet Basic Protein Precursor (PPBP) as an enriched secreted factor with QENIE (Figure 3H) and GO analysis of Ppbp expression suggested a role for the protein in enhancing inflammatory processes (Figure 3I). Consistent with the pathway enrichment, mice injected with the protein showed elevated levels of aortic Interleukin 1-beta (Figure 3J). We also observed similar trends in expression for Tumor necrosis factor alpha and Interferon gamma which impacted many additional tissues (data not shown) suggesting the protein robustly promotes inflammatory cascades in an autocrine/paracrine and endocrine fashion. Despite a small size (~120 amino acids), the PPBP protein is proteolytically cleaved to produce several functionally distinct peptides (Ehlert et al., 1998; Holt et al., 1986), one of which has been found to be increased in patients with acute coronary disease (Ma and Liew, 2003; Smith et al., 2006). While other studies have described distinct roles for these peptide products in various processes, we show PPBP acts in an autocrine/paracrine manner to enhance aortic inflammation. Collectively, these data provide evidence that our generalized method can be applied to additional populations to uncover new regulators of tissue-tissue communication.
Identification of Lipocalin-5 as an endocrine regulator of skeletal muscle metabolism
Since skeletal muscle is the most abundant organ in the body and is responsible for most fuel oxidation, we sought an endocrine factor which targets skeletal muscle to regulate metabolic processes and improve pathophysiologic metabolic status. Based on the pathway enrichment and high significance score of cross-organ analysis using the HF/HS panel shown in Figure 1 and Figure 2, we chose to focus on the adipose-expressed protein, Lipocalin-5 (LCN5). While little is known about the evolutionarily conserved function of Lipocalin-5, the protein belongs to a larger family of lipocalins, which have been implicated in many diverse physiologic roles (Du et al., 2015; Flower et al., 2000; Schiefner and Skerra, 2015). Expression of Lcn5 was approximately 3-fold higher in adipose tissue compared to others via qPCR (Figure 4A). Further, the BioGPS arrays used for tissue-specific expression filtering showed a much higher degree of expression in adipose tissue (Figure S2A). GO analysis showed highly significant enrichment for metabolic pathway regulation, specifically mitochondrial components in skeletal muscle (Figure 4B). As additional support for the metabolic relevance of the gene, we also observed a highly significant negative correlation amongst the 106 HMDP strains between body weight, various fat depot weights and total fat mass percentage with adipose Lcn5 expression (Figure 4C). Based on these observations, we hypothesized that LCN5 is an endocrine factor acting on skeletal muscle to regulate mitochondrial biogenesis and consequent oxidative capacity.
Figure 4. Functional validation of Lcn5 as a regulator of skeletal muscle metabolism in vitro.
A, qPCR analysis of Lcn5 expression across indicated tissues in C57BL6J mice. B, Suggestive pathway enrichment derived from skeletal muscle genes ranked by correlation with adipose Lcn5. C, Adipose Lcn5 expression correlation with clinical traits within the HMDP. D, C2C12 myotubes were treated overnight with media containing either PBS (veh) or recombinant LCN5 protein (1μg/mL) then subjected to qPCR analysis. E–H, The same treatment as in (D) was carried out for 30hrs, then cells were immunoblotted for respiratory complex abundance (E) or subjected to Seahorse bioanalyzer (F) to assess oxygen consumption in both basal (G) and maximal respiratory (H) conditions. For Seahorse experiments, oxygen consumption was normalized to total protein content. I–K, 3T3l1 preadipocytes infected with either AAV-GFP or AAV-LCN5 were differentiated and placed in cocultures with C2C12 myotubes (I), AML12 hepatocytes (J) or 3T3L1 adipocytes (J) for 24 hrs and qPCR-probed for Sdhc or Atp5f1. n=4–8 per group. All data presented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001
After identifying LCN5 as a potential adipose-derived regulator of skeletal muscle metabolism, we tested this connection in several models to assess mitochondrial function. To verify that signaling is mediated by LCN5 itself, we treated differentiated C2C12 myotubes, a commonly used model of skeletal muscle function, with LCN5 recombinant protein. LCN5 acted on C2C12 cells in a dose-dependent manner (Figure S6), and a dose of 1μg/mL purified LCN5 protein was sufficient to induce robust expression of genes associated with both mitochondrial biogenesis and oxidation (Figure 4D). Consistent with changes in gene expression, LCN5 treatment enhanced the protein content of mitochondrial electron transport chain complexes (Figure 4E). To show that changes in gene expression and protein levels of oxidative machinery leads to an enhanced capacity of LCN5 to elevate skeletal muscle oxidation directly, myotubes treated with the same dose of protein were assessed for cellular respiration using a Seahorse system (Brown et al., 2007). Consistent with changes in gene expression, recombinant LCN5 significantly enhanced both basal (Figure 4F, G) and maximal respiratory capacity (Figure 4F, H) in myotubes. While the differences under basal conditions appeared small (Figure 4F, G) relative to stimulated conditions, both remained significant across 3 independent experiments. To confirm that a direct physiologic mechanism persists whereby LCN5 derived from fat cells is capable of enhancing skeletal muscle oxidation, we performed experiments, whereby 3T3L1 adipocytes were infected with Adeno Associated Virus containing either GFP (AAV-GFP) or Lcn5 (AAV-LCN5) and cocultured with 3 cell lines (Figure 4I–K). AAV-LCN5 treatment lead to a ~10-fold increase in expression compared to GFP control (Figure S7). Consistent with observations above, two of the strongest genes induced by recombinant protein (Sdhc and Atp5f1) were also enhanced specifically when AAV-LCN5 infected adipocytes were cultured with myotubes (Figure 4I) and not liver (Figure 4J) or adipocytes (Figure 4K). These data show that LCN5 acts in a manner consistent with the inferred pathways to enhance mitochondrial respiratory activity in myotubes.
Overexpression of LCN5 in the mouse enhances muscle mitochondria complex function and improves glucose metabolism
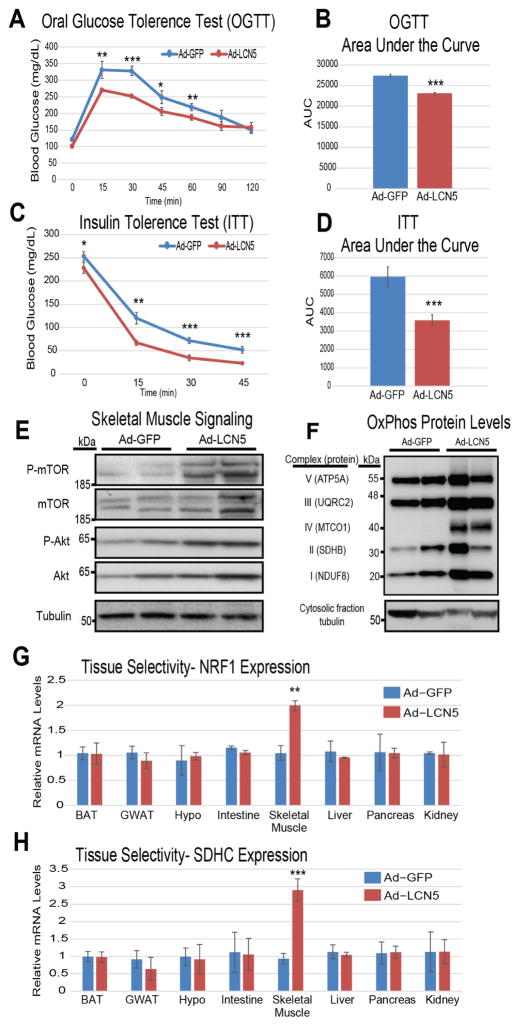
Our next aim was to evaluate the physiologic significance of elevated LCN5 levels in a mouse model of diet-induced insulin resistance. Mice administered a HF/HS diet for 6 weeks were injected with adenovirus encoding Lcn5 (Ad-LCN5) or control (Ad-GFP) where a majority of the overexpression was detected in liver and adipose tissue. One week following injection, mice were evaluated for various metabolic parameters. While no difference in body weight was observed in this short time period between groups, the cohort which received Ad-LCN5 showed significantly enhanced metabolic parameters. Specifically, an approximate 2.2-fold increase in plasma levels (Figure S8) was sufficient to enhance glucose and insulin sensitivity (Figure 5A–D). The effects of Ad-LCN5 in improving glucose regulation also persisted, albeit to a much lesser extent, under normal chow conditions (Figure S9). Consistent with the in vitro results, Ad-LCN5 significantly increased abundance of skeletal muscle insulin signaling components Protein Kinase B (Akt) and mammalian target of rapamycin (mTOR) (Figure 5E), as well as enhanced mitochondrial complex abundance (Figure 5F) in whole gastrocnemius muscle. To show that this physiologic circuit acts in a manner consistent with predictions, the same mice were interrogated for gene expression across relevant metabolic tissues. Here, we observed substantial skeletal muscle specificity for the action of LCN5 on both regulators of biogenesis (Figure 5G) and oxidation (Figure 5H). We also tested whether chronic expression of Lcn5 could impact diet-induced obese phenotypes by using an Adeno-Associated Virus (AAV) encoding LCN5 under the control of an adiponectin promoter. To increase the adipose tissue specificity of expression, this construct contains a 3′ micro-RNA122 binding motif, resulting in select degradation if expressed in the liver (O’Neill et al., 2014). While the induction of overexpression was modest in these mice (~15% increase in plasma concentration), LCN5 expression slightly, but significantly enhanced glucose- and insulin tolerance (Figure S10). Unlike the short-term adenoviral study, long-term expression of LCN5 using AAV enhanced lean mass (Figure S10B). Further, these observations were consistent whether mice were administered the AAV either at the onset (Figure S10A–E) or following (Figure S10F–J) metabolic perturbation via HF/HS diet. Consistent with this notion, activation of Akt has been demonstrated to promote skeletal muscle hypertrophy (Lai et al., 2004). Understanding whether the Akt pathway is directly involved in LCN5-mediated lean mass growth would be of interest. We next utilized the GFP-tagged LCN5 from these mice to infer site of tissue action, where skeletal muscle showed the highest amount of GFP. It is worth noting, there was greater relative amounts of GFP in slow-twitch oxidative fiber (Soleus) compared to fast glycolytic (Plantaris), suggesting LCN5 might preferentially bind oxidative fiber types (Figure S11). These observations illustrate the physiologic impacts of both acute and chronic overexpression of Lcn5.
Figure 5. Lcn5 over-expression reverses the effects of diet-induced metabolic syndrome by enhancing skeletal muscle mitochondrial abundance.
Plasma concentration LCN5 in the Ad-GFP or Ad-Lcn5 groups assessed by immunoblot standard curve of recombinant protein (A). Mice fed a HF/HS diet for 6 weeks were administered either Ad-GFP or Ad-Lcn5 then evaluated for Oral glucose tolerance (B–C), insulin tolerance (D–E), skeletal muscle insulin signaling cascades (F) or mitochondrial complex abundance (G). The same animals were subjected to qPCR analysis for predicted genes Nrf1 (H) and Sdhc (I) across various tissues. n=5 per group, each tolerance test was performed on a separate group of mice. All data presented as mean ± SEM *p<0.05, **p<0.01, ***p<0.001
Extension of LCN5/6 functional impact and cross-tissue scoring from mice to humans
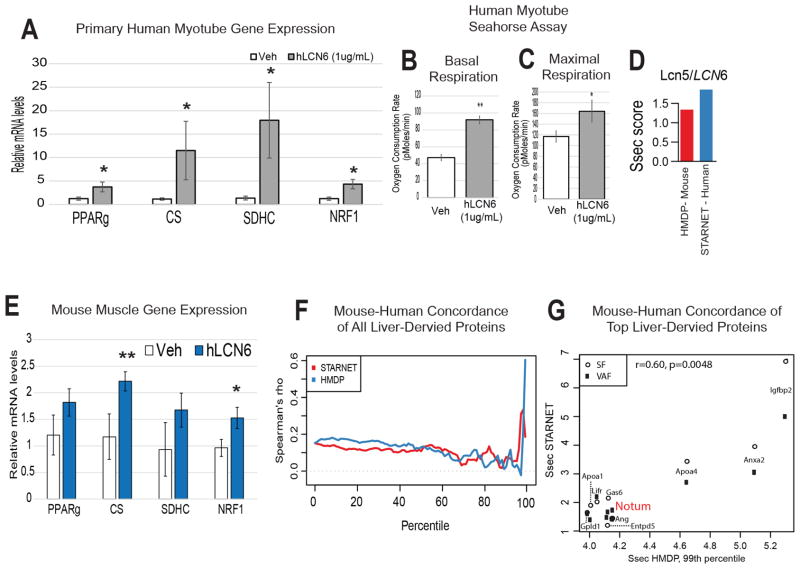
Initially, we sought to gauge the therapeutic relevance of these observations and test the application of LCN5 in human disease. The most conserved protein between species is another lipocalin family member, Lipocalin-6 (LCN6) (Hamil et al., 2003). To assess the functional impact of the LCN6 protein, primary human muscle cells harvested from healthy male donors were treated with recombinant human LCN6 protein (1μg/mL). Treatment resulted in similar induction of the expression of master mitochondrial biogenesis and oxidation regulators (Figure 6A), as well as enhanced basal (Figure 6B) and maximal (Figure 6C) respiratory capacity was observed. These observations indicate functional impacts of mouse LCN5 can be recapitulated in a human system using the orthologous protein, LCN6.
Figure 6. Concordance of mouse and human data using the QENIE pipeline.
As discussed in the text, Lcn6 is the predicted functional orthologue of mouse Lcn5. A, Primary human myotubes were treated overnight with PBS (Veh) or LCN6 (1μg/mL) and gene expression was evaluated using qPCR. n = 6 B–C, Primary human myotubes were treated for 30hrs with veh or LCN6 (1ug/mL) then subjected to Seahorse bioanalyzer system, where significantly enhanced basal (B) and maximal (C) respiratory capacity was observed. n = 4 per group. All data presented as mean ± SEM *p<0.05. D, Ssec scores for both mouse LCN5 (red) and human LCN6 in HMDP and STARNET, respectively. E, Gene expression from gastrocnemius in mice 8 hours after injection of PBS (veh) or 0.1ug/gram body weight recombinant human LCN6. N = 6. F, Spearman’s rho correlation between Ssec of all mouse and human orthologous peptides (y-axis) as a measure of Ssec score percentile within each dataset (x-axis) originating from liver. Note the sharp increase in rho at the tail end of the percentile distribution. G, The top 1% of the HMDP genes from were correlated against their Ssec in STARNET from liver across either visceral (VAF, black squares) or subcutaneous (SF, open circles) fat, with NOTUM highlighted in red.
Next, to gain an unbiased view of overlap between mouse and humans, we utilized a human dataset of RNA-seq from several tissues. These include visceral and subcutaneous adipose tissue, muscle and liver from the Stockholm-Tartu Atherosclerosis Reverse Networks Engineering Task study (STARNET) (Franzen et al., 2016). We first asked if application of QENIE to human tissues showed concordance between mouse and human orthologous genes. In comparing the two datasets using Spearman correlation, we observed a significant positive overall correlation (r = 0.121, pvalue = 1.85 × 10−23) between the Ssec from mice and humans from corresponding tissues (Figure S12). Also, in common with mouse LCN5, we found that LCN6 also scored highly and similarly in STARNET for visceral adipose to muscle tissue (Figure 6D), supporting experimental data (Figure 6A–C). To provide additional evidence for the orthologous functions of LCN5 and LCN6, mice were injected with recombinant human LCN6, where oberved similar induction of skeletal muscle gene expression (Figure 6E). These data show human LCN6 as an orthologue for mouse LCN5 at both levels of regulation, as well as function and illustrate that our generalized approach can be easily applied to human datasets.
Application of QENIE to uncover Notum as a liver-derived enhancer of white adipose “beiging” gene expression
In comparing the mouse and human populations, we next asked which inter-tissue circuits showed the strongest similarities between the two datasets. Since we assume that bona fide endocrine circuits will be found for a small fraction of genes in the tail end of cross-tissue significance distributions, we focused on the top subset of proteins for each axis. Therefore, individual genes were ranked from lowest-to-highest Ssec percentile within each dataset (x-axis) and plotted against their Spearman’s rho correlation between mouse and human orthologous gene scores (y-axis) from liver to adipose tissue (Figure 6F). Within the liver-derived endocrine factors, we observed striking concordance amongst orthologous gene Ssec scores at the top percentiles, where we expect to find proteins mediating inter-tissue communication (Figure 6F, G). Here, the top 1% of all HMDP Ssec scored secreted proteins from liver showed significant positive correlation (r = 0.6, pvalue = 0.0048) with adipose tissue Ssec in STARNET (Figure 6G).
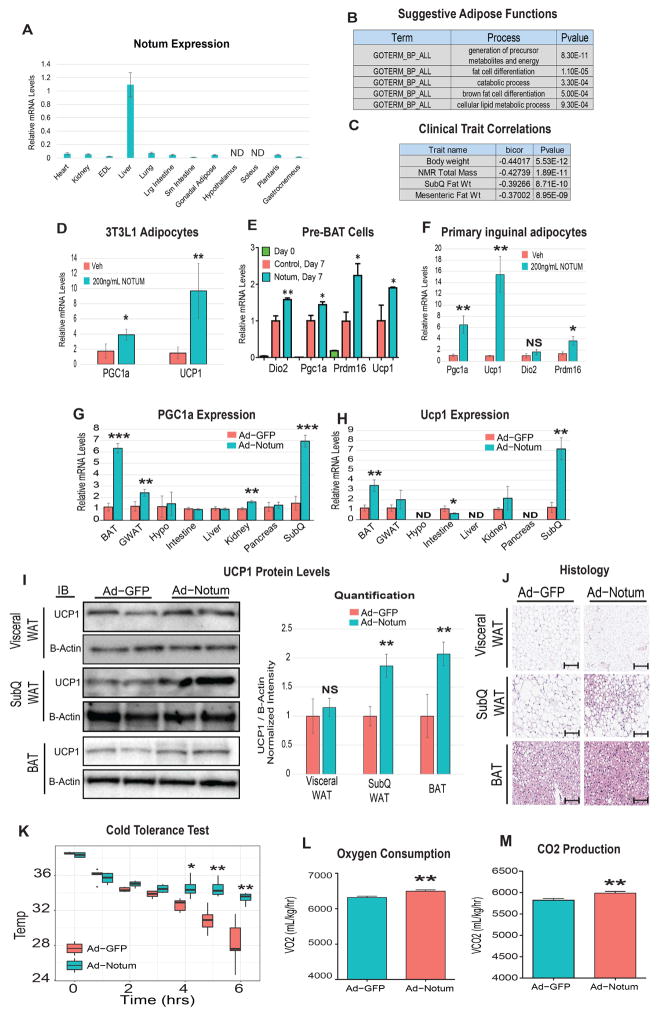
Within this top percentile of genes, we found Notum pectinacetylesterase homolog (NOTUM), suggested to regulate a liver to white adipose tissue axis (Table S1, Figure 2E, Figure 6F). While NOTUM has been extensively studied and acts in part through inhibition of Wnt signaling it has never been demonstrated to act on adipose tissue. The tissue distribution of Notum transcripts via qPCR and BioGPS shows that expression is highly restricted to liver (Figure 7A, Figure S2B) and pathway enrichment analysis suggested that NOTUM enhances catabolic and brown adipose tissue-like machinery (Figure 7B). Consistent with this, we observed negative correlations between adipose Notum expression and fat mass traits across our panel of inbred strains (Figure 7C). Based on these data, we hypothesized that liver-derived NOTUM acts on is a conserved enhancer of adipose tissue thermogenesis.
Figure 7. QENIE uncovers Notum as a liver-derived enhancer of adipose tissue thermogenesis.
A, qPCR analysis of Lcn5 expression across indicated tissues in C57BL6J mice. N = 6 B, Suggestive pathway enrichment using DAVID output for top white adipose genes correlating with liver Notum expression. C, Notum liver expression in the HMDP shows negative correlations with body weight and fat mass. D, 3T3L1 adipocytes reached confluency then subjected to differentiation using a standard cocktail containing PBS (veh) or 200ng/mL NOTUM for three days. Following this treatment, media was removed and protocol was carried out for 5 days whereby RNA was extracted, reverse transcribed and qPCR-investigated for gene expression (n=6). E, pre-BAT cells were incubated with recombinant NOTUM throughout differentiation and qPCR-evaluated for similar beiging markers. (n=6). F, Primary inguinal adipocytes were isolated and cultured, followed by treatment with veh or recombinant NOTUM protein for 4 days and analyzed for the same genes as in E. G–J, Mice were injected with adenovirus containing cDNA sequence for GFP or Notum, then tissues were qPCR-probed for Pgc1a (G) and Ucp1 (H) gene expression and adipose depots were immonblotted for UCP1 protein (I) and subjected to histological examination (J). To assess physiologic parameters, Ad-GFP or Ad-Notum mice underwent cold tolerance tests (K), as well as metabolic chamber analysis at 5 degrees (L–M) ND = Not detected (assessed by a raw qPCR Ct value above 30). All data presented as mean ± SEM *p<0.05, **p<0.01, ***p<0.001
To experimentally test the hypothesis, we treated 3T3L1 adipocytes for the first three days of brown differentiation with vehicle or 200ng/mL NOTUM, a protocol that has been previously used to test whether various pharmacologic agents could promote beiging of 3T3L1 adipocytes (Asano et al., 2014; Zhang et al., 2014). We observed increased expression of PGC1a and UCP1, markers of beiging, in cells treated with NOTUM (Figure 7D). Given that several other “beige” target genes, including iodothyronine deiodinase 2 (Dio2), and PR domain containing 16 (Prdm16) were below the detection limit of qPCR in 3T3L1 adipocytes, we adopted another in vitro model to test the hypothesis in which NOTUM promotes beige gene expression. Therefore, pre-BAT cells were differentiated into brown-like adipocytes as previously described (Rodriguez-Cuenca et al., 2007; Villanueva et al., 2013) in the presence of vehicle or 200mg/ml of NOTUM. Here, we also observed significantly enhanced gene expression of brown adipocyte markers (Figure 7E). Finally, we adopted a traditional culture model of browning, primary inguinal cultured cells. Following 4 days of treatment with recombinant NOTUM protein, we observed a marked induction of the same markers of browning (Figure 7F). Under these conditions, the average Ct values of browning genes lower than that of 3T3L1 adipocytes (~24 vs 30), suggesting a more relevant physiologic setting to explore the impacts of the protein.
To validate the functional impact of NOTUM in vivo, mice were injected with adenoviral vectors containing GFP (Ad-GFP) or Notum (Ad-Notum). Nine days following injection, where circulating NOTUM levels increased ~2-fold (Figure S8), we observed significant increases in the same two beige markers in subcutaneous adipose, brown adipose and to a lesser extent gonadal adipose tissue (Figure 7G, H). Here, a tissue-wide interrogation of Ucp1 and Pparg also showed that the capacity of NOTUM to enhance these two genes is specific for fat cells, consistent with the prediction based on the QENIE method. While the largest changes in Ucp1 and Pparg transcription were observed in adipose tissue depots, expression was also altered in other tissues such as kidney and intestine. These observations suggest Notum impacts other organs when overexpressed for 9 days, either through direct action of the circulating protein or as a secondary response from the impact of adipose tissue. We also observed increased UCP1 protein expression in subcutaneous and brown fat pads, but not visceral (Figure 7I).
Next, we asked whether the impact of beige gene expression and protein levels was sufficient to enhance whole-body functional outcomes. Histological examination showed smaller size and less abundance of adipocytes within the Ad-Notum group (Figure 7J) with the most notable difference in the subcutaneous fat. To assess the physiologic significance of Notum overexpression, mice were subjected to a cold tolerance test. During an acute cold challenge, mice expressing Ad-Notum showed a markedly enhanced capacity to sustain body heat (Figure 7K). Next, to gain additional insight as to the impact of NOTUM on whole-body physiology mice were studied in metabolic chambers. Ad-Notum mice kept at 5° showed increased oxygen consumption (Figure 7L) and produced more and carbon dioxide (Figure 7M), with no differences in food intake or Respiratory Exchange Ratio (RER) (Figure S13) compared to controls. While statistically significant respiratory differences were not observed at room temperature (despite a trend toward increases), all molecular phenotypes (Figure S13) persisted in the absence of a cold stress. Taken together, these data provide strong validation that NOTUM constitutes a novel physiologic mechanism in mice by which the liver can enhance thermogenesis in adipose tissue.
DISCUSSION
We present a framework for investigating endocrine circuits between tissues based on natural variation in gene expression populations of mice. The method requires global transcriptomic or proteomic data from the tissues of interest in a sufficient number of individuals to provide statistical power for the detection of correlation structure between the endocrine factor and its downstream effects in the target tissue. For this purpose, we have utilized a panel of approximately 100 inbred strains, the HMDP, maintained under defined environmental conditions. Our preliminary analysis of expression data from the STARNET population resource suggests that the method will be applicable to human populations as well. Future investigation of cross-tissue circuits in other human datasets, such as communication among inter-tissue signals in GTEX (Consortium, 2013) or identification of signaling between tumor and adjacent healthy tissue in TCGA offer the potential for greater translation of these factors to human disease and physiology. Undoubtedly, due to environmental factors, age differences, inability to cleanly access relevant tissues and lack of replications, the human data is expected to be much noisier than mouse. The validity of the approach is supported by the fact that many of the predicted endocrine circuits are already known. Another key aspect of the method is to utilize pathway enrichment to define the likely molecular targets of the endocrine factors. In the case of the HMDP resource, we were also able to utilize previous clinical and physiologic data that enabled us to formulate hypotheses regarding the targets of the putative endocrine factors. Finally, we have developed a straightforward pipeline to experimentally test the predictions of our bioinformatic analyses, involving expression of the endocrine factor for both in vitro and in vivo assessment of its impact. To demonstrate the utility of our approach for discovery, we report the physiologic impact of two novel endocrine factors, LCN5/6 and NOTUM, and in vivo experimental validation of three additional factors. Below, we discuss some consideration relating to the future application of our method and the implications of our findings relating to LCN5/6, NOTUM and the other validated endocrine circuits.
Mouse Lipocalin-5 was first described as a small-molecular weight protein secreted by the murine epididymis, where, unlike other closely conserved lipocalins, it did not show a strong affinity for binding spermatozoa (Hamil et al., 2003; Rankin et al., 1992b). It belongs to a larger family of proteins characterized by their ability to bind and carry small molecules via a conserved hydrophobic binding pocket in circulation (Flower et al., 2000; Schiefner and Skerra, 2015). LCN5 has been shown to bind cis- and trans-retinoic acid, but not retinol via application of labelled retinoic acid to a DEAE ion exchange chromatography (Rankin et al., 1992a). While a specific factor in which LCN5 binds has yet to be described, the protein could potentially promote uptake of a factor involved in oxidation into muscle. Some preliminary experiments from our group indicates that the protein alone can enhance gene expression in cultured myotubes. Additional mechanistic studies will be required to narrow a receptor-mediated pathway. Prior to this study there has been no description of the regulation and role of Lcn5 in adipose tissue. However, epididymal gene expression can be strongly induced by androgen receptor activation, a pathway which is suppressed by both castration (Lareyre et al., 1998) and forkhead box transcription factor 2 subclass A (Yu et al., 2006). Given the significant roles for forkhead box transcription factors and hormone signaling in impacting metabolic homeostasis, the mechanism of LCN5 could offer insight into how these components intertwine to mediate sex-specific differences in whole-body metabolism. While the half-life of LCN5 in vivo is not known, the turnover of family member Lipocalin 2 is fairly rapid (10–20min) (Axelsson et al., 1995), which has implications for the regulation and efficacy. Much less has been described as to the regulation and function of human LCN6, which was initially cloned based on cDNA sequence similarity to LCN5 and LCN8. The impact of LCN5 transcriptional components mediating mitochondrial abundance and function (Figure 4H, I) suggest a specific signaling axis, although we cannot exclude the possibility that other physiologic effects are so exclusive, requiring tissue-wide functional assays. The capacity of the protein to enhance whole-body glucose metabolism were markedly greater under HF/HS dietary conditions when compared to chow diet. While we do not have a clear explanation for these observations, it is worth noting that QENIE predictions for LCN5 function were made using a mouse population fed a HF/HS diet (Parks et al., 2013; Parks et al., 2015). Specifically, evaluation of mice expressing LCN5 and LCN6 using metabolic cages or similar measurements under different dietary conditions will be informative to the physiologic consequence to protein action, thus guiding mechanism-based investigation. Moreover, while acute changes in LCN5 expression (adenovirus) did not alter body mass composition, chronic overexpression (through AAV) was sufficient to promote lean mass (Figure S10). A possible mechanism would be that cycles of acute LCN5-mediated sensitization of Akt signaling cascades (Figure 4E) occurred over a prolonged period, whereby elevated Akt is driving muscle proliferation. Consistent with this notion, activation of Akt has been demonstrated to promote skeletal muscle hypertrophy (Lai et al., 2004), however proving the relationship to LCN5 function requires further exploration.
The NOTUM protein was first described in Drosophila as a secreted repressor of Wingless activity and a regulator of morphogenesis gradients (Giraldez et al., 2002). In human cells, NOTUM has been demonstrated to act as a hydrolase, releasing GPI-anchored proteins from the cell surface (Traister et al., 2008) and fine-tuning the Hedgehog pathway (Ayers et al., 2010; Ayers et al., 2012). A recent study also showed that NOTUM can act as a carboxylesterase, removing palmitoleate from key members of the Wnt pathway and blunting signaling capacity (Kakugawa et al., 2015). Recently, a liver-specific deletion of Notum was reported. The authors did not observe ablation of Wnt/β-catenin signaling but did document changes in hepatic glucose output (Canal et al., 2016). Our study is the first to document effects of NOTUM on adipose tissue. The relatively rapid response (~9 days) of subcutaneous beiging highlights the efficacy of the protein and offers insight into potential mechanisms. Several acute physiologic conditions have been shown to enhance thermogenic features in subcutaneous fat, including modulating angiogenesis (Park et al., 2017) or increasing catecholamine signaling (Collins, 2011). It is also noteworthy that we observe slight, but significant changes in kidney PGC1a following Notum overexpression. Both Wnt signaling (Logan and Nusse, 2004; Pulkkinen et al., 2008) and PGC1a (Lynch et al., 2017; Stadler et al., 2015) have been demonstrated to significantly impact kidney development and disease. Wnt signaling has been widely demonstrated to regulate adipose tissue function and differentiation and recently inhibition of Wnt has been linked to beiging of adipose tissue (Fulzele et al., 2017; Lo et al., 2016)
Our studies reveal novel potential endocrine functions for SMOC1, ITIH5 and PPBP based on changes in gene expression in pathways predicted using the QENIE method. Clearly, additional experiments are required to assess the physiologic impacts of these molecules. Our findings do, however, illustrate the broad potential applications of our approach to not only metabolism but also processes such as inflammation and growth regulation.
Limitations of Study
Our pipeline carries several caveats which should be considered. First, the process of identification eliminates secreted proteins that are ubiquitously expressed in many diverse cell types, but that could nonetheless still function as endocrine factors. Of note, several growth factor family members (fetal growth factors and insulin-like growth factors (Bottcher and Niehrs, 2005; Froesch et al., 1985; Jones and Clemmons, 1995; Nies et al., 2015)) and conserved family members based on sequence similarities to immune-acting peptides (Interleukins and C1Q superfamily (Cunningham and De Souza, 1993; Harbuz et al., 1992; Pedersen and Febbraio, 2008; Seldin et al., 2012; Seldin et al., 2014)) were consistently highly ranked, but were eliminated (Fig. 2C–G) after examination of tissue-specific expression. These proteins could of course be included in the analysis, but this would introduce doubt regarding the primary source of these proteins and therefore dilute subsequent interpretation based on enrichment of correlated gene expression in target tissues. In addition, the rate of bona fide interactions discovered using this method could be substantially influenced by the system used for screening (eg. in vitro vs in vivo) and the genetic architecture and size of the populations. For example, long-range disequilibrium due to population structure among the HMDP could result in correlation between a locus controlling a secreted factor and the genetic regulation of genes elsewhere in the genome that are expressed in a target tissue. Also, one might imagine if a secreted protein and a gene in a target tissue are both affected by the same genetic locus, it could lead to false correlations and higher Ssec values. While we cannot absolutely rule out all sources of confounders, these results from permutation testing (Figure 2 and Table S1) suggest that these factors have no more than a minor impact on the results. This conclusion is further supported by the finding that the method reveals many known endocrine factors. Another important consideration is that organism-wide expression patterns due to physiologic status (such as global metabolic state) could bias cross-tissue scores by acting through latent variables. In a scenario where both expression of origin tissue gene and target tissue pathways are regulated by insulin, significant cross-tissue enrichment could be observed not as a result of direct circuit of communication, but rather both the gene and target pathways are conditioned on the impact of insulin. For this reason, it is essential to experimentally confirm mechanisms of putative secreted proteins, as illustrated for the five proteins described above. Investigation of the upstream physiologic factors which regulate expression of these secreted factors (beyond natural variation) would be especially informative as to their conserved physiologic roles. It is also worth noting that while this pipeline identifies inter-tissue circuits based on expression in a single origin tissue, our screening process selects for proteins only sufficient to alter target tissue expression profiles where expression of the protein is not necessarily regulated by its physiologic source of production. Finally, the Ssec scores are based not only on the significance of correlations but also on the number of correlated genes. Hence, endocrine factors that affect pathways with feedback involving many cellular processes (eg. altering global cellular metabolism) or that affect large gene expression programs will tend to dominate the Ssec score ranking. In contrast, endocrine factors that regulate fewer genes will be ranked lower, but might still serve important endocrine roles. An alternative strategy which would compute significance scores of peptides on groups of genes divided into individual pathways (eg. immune system components) or co-correlated gene modules (Franzen et al., 2016) would also be a potential approach to target regulators of specific inter-tissue processes and perhaps enhance the resolution of detecting interactions
In conclusion, we have developed a population-based approach for identifying novel communication axes between tissues and cell types using correlation structure. We have utilized a well-characterized and diverse population of mice but our results suggest that other populations, including human cohorts, can be used. Endocrine functions have been identified for only a tiny fraction of secreted proteins, and our method provides a straightforward approach to expand our understanding. Clearly, such understanding may lead to novel therapeutic strategies. Our framework may be applicable to analyses of strong inter-tissue cross-talk beyond endocrine circuits, such as metabolite-mediated signals and microbiota-host interactions. We expect that as global proteomics data become available these will be particularly informative.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jake Lusis – jlusis@mednet.ucla.edu.
EXPERIMENTAL MODELS AND SUBJECTS
Animals
All animal experiments were approved by the University of California Los Angeles (UCLA) Animal Care and Use Committee, in accordance with Public Health Service guidelines. C57BL/6J mice were purchased from the Jackson Laboratory. Beginning at 6 weeks of age, mice were fed a standard High-fat/High Sucrose diet (35% kcal fat, Research diets cat # D12266B). Mice were administered AAV via intraperitoneal injection at a titer of approximately 1e12 per mouse. Oral glucose tolerance tests were performed following an overnight fast. Mice were gavaged with 10uL/gram body weight of a 10% (w/v) solution of glucose in PBS. Insulin tolerance tests were performed after a 4 hour fast, whereby animals administered 1U/kg body weight human recombinant insulin protein (Sigma). The cohort of mice testing LCN5 expression was administered a HF/HS diet prior to receiving adenovirus, while all other cohorts were fed a standard chow diet. For the recombinant protein injections, shown in Fig 3, mice were fasted overnight, then injected with 0.15ug/gram body weight recombinant protein in PBS. Tissus were harvested for gene expression 6 hours following injection.
Adeno-associated virus
Mouse lipocalin-5 cDNA or GFP was cloned into an AAV expression plasmid purchased from the University of Pennsylvania Gene Therapy program vector core (AAV8.hAdp.GFP.miR122.SV40) driven under an adiponectin promoter. The plasmid was used to synthesize AAV which was carried out at the University of Pennsylvania Gene Therapy program vector core.
Adenovirus
Recombinant adenovirus was generated using the AdEasy system as previously described (Bennett et al., 2013). Briefly, linearized shuttle vector containing full-length mouse cDNA for Notum and Lcn5 were transformed into E. coli BJ5183AD cells containing the adenoviral backbone plasmid pAdEasy-1 for homologous recombination. Positive recombinants were linearized and transfected into HEK293AD cells for virus packaging and propagation. Adenoviruses expressing the candidate gene were purified by CsCl banding and stored at −80°C until use. For adenoviral infection, 7-week-old male C57BL6J mice were injected with adenoviral construct (~2.5 × 109 PFUs diluted in 0.2 ml saline) Intraperitoneally. After overnight fasting, mice were sacrificed 9 days post injection, tissues were extracted, the expression gene expression was assessed by RT-PCR. The control group consisted of mice injected with adenoviral construct expressing the GFP gene.
Cell culture
C2C12 mouse myocytes were cultured as previously described 89,90. In brief, myocytes were maintained below 60% confluency in high glucose (4.5g/L) Debulcos Modified Eagles Medium (DMEM, Life Technologies) supplemented with 20% FBS (Altanta Biologicals), 100U/mL penicillin, 100μg/mL streptomycin, 1mmol/L sodium pyruvate and 4mmol/L L-Glutamine. For differentiation into myotubes, cells were grown to >95% confluency and serum starved (DMEM containing 2% Horse Serum (Life Technologies) and lacking sodium pyruvate) for a period of 6–8 days. Differentiation of myotubes was confirmed morphologically by formation of multinucleated fibers. Transfections for introduction of exogenous DNA into cells were performed using Lipofectamine 2000 (Life technologies) in accordance with the recommended protocol. For experiments involving concentrated media, cells transfected with indicated vectors were incubated in Opti-MEM Reduced Serum Media (Life Technologies) for 48 hours to allow sufficient amounts of protein to be secreted. Media was then harvested and concentrated using 10,000 molecular weight cut-off Centrifugal Filter Units (Millipore) as described in the product protocol.
Adipocyte Cell culture
3T3L1 cells were maintained in below 60% confluency in high glucose (4.5g/L) Debulcos Modified Eagles Medium (DMEM, Life Technologies) supplemented with 20% FBS (Altanta Biologicals), 100U/mL penicillin, 100μg/mL streptomycin, 1mmol/L sodium pyruvate and 4mmol/L L-Glutamine. Protocol for differentiation was carried out as previously described when cells reached >95% confluency (Asano et al., 2014; Zhang et al., 2014). The mouse brown preadipocyte cell line (pre-BAT) were isolated from the stromal vascular fraction of mouse interscapular brown adipose tissues and immortalized by infecting with retrovirus vector expressing the SV40 T antigen and was cultured as previously described (Asano et al., 2014; Zhang et al., 2014). The pre-BAT cells were grown in DMEM (high glucose) supplemented with 10% fetal calf serum and 100 U/ml of both penicillin and streptomycin (growth media). For differentiation experiments. The pre-BAT cells were plated in 6-well plate at 1× 10^6 cells per well. Three days after plating (day 0), when the cells reached nearly 100% confluency, the cells were treated with an induction media containing growth media supplemented with 5 μg/ml insulin, 1 nM 3,3′,5-Triiodo-L-thyronine (T3), 125 μM indomethacin, 2 μg/ml dexamethasone, 0.5 mM 3-Isobutyl-1-methylxanthine, and 0.5 μM Rosiglitazone for 48 hrs (day 0 to day 2). After 48 hrs, the cells were treated with a maintenance media containing the growth media supplemented with 5 μg/ml insulin, 1 nM 3,3′,5-Triiodo-L-thyronine (T3), and 0.5 μM Rosiglitazone, with a media change every 2–3 days until day 7 when RNA was isolated for gene expression analysis. For the NOTUM treatment group, 200 ng/ml of human recombinant NOTUM (Origene) were added to the induction media and the cells were treated with NOTUM from day 0 to day 2 for 48 hrs. RNA samples were also collected from cells at day 0 as a baseline control.
Primary Human Skeletal Muscle Cultures
Primary human muscle cells were isolated and cultured as previously described (Skrobuk et al., 2012). In general, experiments were performed from 5–9 passages. Myocyte cells were growth in DMEM/F-12 media containing 25mM glucose, GlutaMAX (Life Technologies), 20% FBS and antibiotic/antimycotic (Life Technologies) at low confluency. Differentiation treatments began when cells reached 80% confluency, where media was changed to DMEM/F-12 containing GlutaMAX (Life Technologies antibiotic/antimycotic (Life Technologies), 5mM glucose and 2% FBS. Differentiation was carried out 5–7 days and confirmed by the formations of multinucleated muscle fibers. All treatments were carried out in the media used for differentiation.
METHODS DETAILS
Coculture Experiments
Each cell line was cultured under conditions as described above. Experiments were carried out in a trasnwell system (corning # 07-200-170). 3T3L1 adipocytes were plated into the culture insert and allowed to achieve confluency. Cells were then treated with AAV (1E1010per mL) for 24 hours. Cells were then washed and differentiated for 8 days (listed above). Following differentiation, adipocytes inserts were added to each respective cell line for 24hours.
Immunoblot procedure and analysis
Cells and tissues were lysed in Whole Cell Extraction buffer (WCE) containing 62.5mM Tris-HCl (pH 6.8), 2% (wt/v) Sodium dodecyl sulfate, and 10% glycerol. Samples were then heated and diluted 1:5 in water and protein content measured using a BCA protein assay kit (Pierce). Total protein concentration was normalized to 3ug/uL and samples were then denatured in 4x LDS loading buffer (Life Technologies) with 10x reducing agent (Life Technologies) at 99°C for 20min. Samples were then loaded at 10uL/well into 4–12% Bis-Tris gels (Invitrogen) and separated out at 130 volts for 2hr. Protein was then transferred to PVDF membranes (Immobilon) for 1.5hr at 35 volts. Following transfer, membranes were washed with TBST, and then blocked in 5% skim milk (Gibco) in TBST for 1hr at room temperature. Membranes were then placed in primary antibodies (1:2000) on a shaker overnight at 4°C. Primary antibodies were used as follows rabbit polyclonal LCN5 (myBioSource # MBS2027564, 1:1000), rabbit monoclonal GFP (cell signaling # 2956S), rabbit polyclonal NOTUM (abcam # ab106448, 1:1000), rabbit polyclonal α-tubulin (Cell Signaling #2144, 1:2500), mouse anti-Noq7 (abcam #11083, 1:2000), Total OxPhos complex cocktail (abcam, # ab110413, 1:2000), rabbit UCP1 polyclonal (abcam # ab10983, 1:1000). The following day, membranes were washed 3X in TBST then placed in secondary antibodies (1:2000) for 1hr at room temperature. Blots were then washed 3X in TBST and placed Amersham ECL detection solution (GE health sciences). Blots were imaged using IMAGER and bands were quantified using Image J Software.
Plasma protein measurements
Quantification of plasma proteins shown in Suppl. Fig. 8 were performed using immunoblot analysis (above) using equal volumes of recombinant protein vs plasma. While size differences appear between the bands indicated when comparing the two sample types, we confirmed these observations using multiple antibodies targeting different regions of the proteins.
Mitochondrial preparation
Mitochondria lysate was prepared from either skeletal muscle or C2C12 myotubes using Fisher isolation kit (cat # 89874) according to manufacturer recommendations. For whole tissue, muscle was first trypsin digested then subjected to isolation procedure. Lysate was normalized to total cytosolic protein content then subjected to immunoblot procedure.
Metabolic cages
Indirect calorimetry was performed using a Columbus Instruments Comprehensive Lab Animal Monitoring System. Animals were placed individually in chambers for 8 hrs at cold temperature (53C) with 12-hour light/dark cycles. Animals had free access to food and water. Oxygen (VO2) and carbon dioxide (CO2) respiratory measurements were made in 20-minute intervals after an overnight acclimation period.
Cold tolerance test
To assess cold tolerance, C57BL6J mice were placed in 4 degrees and monitored for body temperature using a rodent rectal temperature probe (World Precision Instruments) every hour for a period of 6 hours.
Seahorse Biosystems
Real-time measurement of oxygen consumption rate (OCR) in cells was carried out using the XF24 Extracellular Flux Analyzer (Agilent). To directly assess mitochondrial metabolism, OCR measurements were made before and after the sequential injection of 0.4 μM oligomycin (ATP synthase inhibitor, which allows determination of ATP-linked respiration), 1.5 μM FCCP (an uncoupler, which allows determination of maximal mitochondrial respiratory capacity), and 2 μM rotenone/myxothiazol (inhibitors of complex I/III of the electron transport chain, which allows determination of non-mitochondrial respiration). OCR was normalized per protein content using a bradford assay.
Reagents and Chemicals
HIS-tagged mecombinant mouse Lipocalin-5 produced in HEK293 cells was purchased from creative biomart. mouse ITIH5 protein purified from HEK 293 was purchased from MyBioSource. Recombinant NOTUM (human), Lipocalin-6 (human) and SMOC1 (human) and purchased from Origene. Recombinant PPBP (mouse) was purchased from Lifespan Biosystems. The protein purity was verified as >90% by commassie stain following SDS-PAGE. Treatment of cells with recombinant protein was performed overnight for qPCR analysis or 30hours for Seahorse assay and protein quantification.
RNA extraction and reverse transcription
Cells or tissue were homogenized in Qiazol (Qiagen) and RNA extraction was carried out as recommended. Samples were suspended in 0.5mL Qiazol each then 100uL chloroform was added. After vortexing, phase separation was achieved with centrifugation at 13k rpm for 15min. The aqueous layer was then transferred to 1mL isopropanol, vortexed then centrifuged again. The remaining pellets were washed in 70% ethanol in water then air dried following centrifugation for 10 min. Purified RNA was then suspended in 30uL of water and assessed for purity and concentration using a Nanodrop ND-100 Spectophotometer. Two ug of total RNA per sample was reverse transcribed using a High-Capacity cDNA reverse transcription kit (Applied Biosystems) with random primers. Reverse-transcribed cDNA was then diluted in water for qPCR analysis.
Quantitative PCR
Quantitative PCR was carried out using a Kappa SYBR Fast qPCR kit as recommended by the manufacturer. Samples were ran on a LightCycler 480 II (Roche) and analyzed using the Roche LightCycler 1.5.0 Software. All qPCR targets were normalized to geometric mean of RPL13a and PPIA expression and quantified using the delta Ct method. All qPCR primer sequences were obtained from PrimerBank (http://pga.mgh.harvard.edu/primerbank). All Sequences to qPCR primers are listed below:
| Species | Gene | Sequence 5′ - > 3′ |
|---|---|---|
| mouse | SIRT1- F | GCTGACGACTTCGACGAC |
| mouse | SIRT1 - R | TCGGTCAACAGGAGGTTGTCT |
| mouse | NRF2 - F | CTTTAGTCAGCGACAGAAGGAC |
| mouse | NRF2 - R | AGGCATCTTGTTTGGGAATGTG |
| mouse | Foxo1 - F | AAGGATAAGGGCGACAGCAA |
| mouse | Foxo1 -R | TCTTGCCCAGACTGGAGAGAT |
| mouse | Rpl13 - F | CTGTGAAGGCATCAACATTTCTG |
| mouse | Rpl13 - R | GACCACCATCCGCTTTTTCTT |
| mouse | Sdhc - F | GCTGCGTTCTTGCTGAGACA |
| mouse | Sdhc - R | ATCTCCTCCTTAGCTGTGGTT |
| mouse | PPIA - F | GAGCTGTTTGCAGACAAAGTTC |
| mouse | PPIA - R | CCCTGGCACATGAATCCTGG |
| mouse | CS - F | GGACAATTTTCCAACCAATCTGC |
| mouse | CS - R | AGTCAATGGCTCCGATACTGC |
| mouse | Ppargc1a - F | TATGGAGTGACATAGAGTGTGCT |
| mouse | Ppargc1a - R | GTCGCTACACCACTTCAATCC |
| mouse | Atp5f1 - F | AGTTCCTTTACCCTAAGACTGGT |
| mouse | Atp5f1 - R | TTCATGCTCGACTGCTTTACTT |
| mouse | UCP1 - F | CGATGTCCATGTACACCAAGGA |
| mouse | UCP1 - R | TCGCAGAAAAGAAGCCACAA |
| mouse | LCN5 - F | CTCCAAGATGGGTGCATACGG |
| mouse | LCN5 - R | CCTCATTGTAATAGGTGGTGGTC |
| mouse | Cpt1a - F | CTCCGCCTGAGCCATGAAG |
| mouse | Cpt1a - R | CACCAGTGATGATGCCATTCT |
| mouse | Il1b - F | GCAACTGTTCCTGAACTCAACT |
| mouse | Il1b - R | ATCTTTTGGGGTCCGTCAACT |
| mouse | Tgfb1 - F | CTCCCGTGGCTTCTAGTGC |
| mouse | Tgfb1 - R | GCCTTAGTTTGGACAGGATCTG |
| human | PPARg - F | GGGATCAGCTCCGTGGATCT |
| human | PPARg - R | TGCACTTTGGTACTCTTGAAGTT |
| human | RPL13a - F | GCCATCGTGGCTAAACAGGTA |
| human | RPL13a - F | GTTGGTGTTCATCCGCTTGC |
| human | Cs - F | TGCTTCCTCCACGAATTTGAAA |
| human | CS - R | CCACCATACATCATGTCCACAG |
| human | PPIA - F | CCCACCGTGTTCTTCGACATT |
| human | PPIA - R | GGACCCGTATGCTTTAGGATGA |
| human | sdhc - F | CTGTTGCTGAGACACGTTGGT |
| human | sdhc - R | ACAGAGGACGGTTTGAACCTA |
| human | NRF1 -F | AGGAACACGGAGTGACCCAA |
| human | NRF1 - R | TATGCTCGGTGTAAGTAGCCA |
QUANTIFICATION AND STATISTCIAL ANALYSES
Statistical Analysis
All computational procedures were carried out using R statistical software. Correlations and associated p-values were calculated with the biweight midcorrelation, which is robust to outliers and associated pvalue (Langfelder and Horvath, 2008, 2012). Single comparisons between two groups were performed using two-tailed Student’s t tests with 95% confidence intervals. Comparisons involving multiple time points were assessed using a two-way ANOVA with Tukeys posthoc tests. Permutations of target tissue redistributions were carried out as described in Figure S2. Values were considered significant at p < 0.05. All data are presented as means ± S.E
Cross-tissue correlations and optimization of informatics resources
Cross tissue midweight bi-correlation coefficients and corresponding pvalues were calculated using the R package WGCNA (Langfelder and Horvath, 2008, 2012). To retrieve and overlay annotated secreted proteins, we used the list deposited in the Universal Protein resource (UniProt) as “secreted” localization annotations [SL-0243] for overlapping HUGO symbol in Mus musculus (Mouse) [10090].
DATA AND SOFTWARE AVAILABILITY
Github
As referenced in the text, the R script used to perform the pipeline, data pretreatment, sample datasets and example of qq-plots are available at: https://github.com/marcus-seldin/QENIE
Data Pretreatment
Mouse expression arrays for the original HF/HS HMDP studies were performed on a Affymetrix HT_MG-430A. The GEO Accession for arrays is: GSE64770 Our pipeline begins with gene expression arrays for liver and adipose tissue, where each gene is represented as an averaged value across probes and strains used in the study. These aggregate matrices are also provided in this repository. The arrays consisted of ~22,400 probes which were aggregated to averages for each gene (12,242). The expression values for each mouse were also averaged to reflect a single value per gene per strain (106). Therefore, each liver and adipose tissue expression matrix consists of 12,242 genes among 106 unique HMDP strains.
Datasets Used
The raw datasets used in this study (In addition to GSE64770) are available using the following GEO accessions: chow aorta - GSE38120; chow heart - GSE77263; chow liver - GSE16780; chow adipose - GSE42890 and HF/HS - hypothalamus: GSE79551. All other data is directly available on the Github.
Optimization of ranking lists for pathway enrichment
In an effort to provide optimal pathway enrichment using the target tissue correlation coefficents conditioned on each origin tissue secreted protein, we systematically screened these lists and assessed pvalues of GO biologic pathways. Here, we took all suggestive pathway lists for the top-ranked peptides within adipose, liver and muscle (targeting all others) and assessed DAVID output for the top 100, 200, 500, 1000 or 2000 target tissue genes ranked by positive or negative bicor or pvalue. The top 3 GO biologic pathways showed the lowest average pvalues (Benjamani) using the list of 500 target tissue genes, ranked from highest-to-lowest bicor. It is worth noting that this generally conditions our approach to pathways positively enhanced by the peptide. Alternatively, using the ranking system of lowest-to-highest bicor could potentially condition for pathways suppressed by origin tissue peptides or using the pvalue to infer “general processes engaged”. Examples of such are presented in Figure 3.
ADDITIONAL RESOURCES
None
Supplementary Material
Ssec and pvalue of permutation score for each top-ranked peptide, related to Figure 2
Highlights.
A population-based method to discover novel endocrine factors using expression data
Discovery that adipose-derived mouse LCN5/human LCN6 enhances muscle respiration
Discovery that liver-derived NOTUM promotes adipose tissue thermogenesis
Significant overlap of predictions between mouse and human endocrine interactions
Acknowledgments
We thank Rosa Chen for help in preparing the manuscript. We also thank members of the Vallim-Tarling lab for providing experimental expertise.
This work was supported by the following funding sources: NIH-T32HL007895 (MMS), NIH-T32HL69766 (MMS), NIH-HL28481 (AJL), NIH HL30568 (AJL), NIH-R00HL121172 (MC) and the Fondation Leducq (CADgenomics: Understanding CAD Genes, 12CVD02) (AJL and JLMB).
Footnotes
Declaration of Interest: The authors have no competing interests to declare
Author Contributions:
MMS, GMR, PR, LV, YM, MM, TP, EDK and RG performed experiments. MMS, PR, GMR, LV and DS analyzed raw data. SM and HAK provided human muscle cells and expertise with respect to interpretation of human experiments. NC, CP, PR, PT, MC, BWP, FN YHB, ML, CAD, KR and AJL reviewed the data and made substantial contributions to improving the studies. CP, RMC and AJL provided statistical consult on permutation analyses. MMS and AJL wrote the manuscript, which was reviewed by all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Llamas G, Szalowska E, de Vries MP, Weening D, Landman K, Hoek A, Wolffenbuttel BH, Roelofsen H, Vonk RJ. Characterization of the human visceral adipose tissue secretome. Mol Cell Proteomics. 2007;6:589–600. doi: 10.1074/mcp.M600265-MCP200. [DOI] [PubMed] [Google Scholar]
- Anveden A, Sjoholm K, Jacobson P, Palsdottir V, Walley AJ, Froguel P, Al-Daghri N, McTernan PG, Mejhert N, Arner P, et al. ITIH-5 expression in human adipose tissue is increased in obesity. Obesity (Silver Spring) 2012;20:708–714. doi: 10.1038/oby.2011.268. [DOI] [PubMed] [Google Scholar]
- Asano H, Kanamori Y, Higurashi S, Nara T, Kato K, Matsui T, Funaba M. Induction of beige-like adipocytes in 3T3-L1 cells. J Vet Med Sci. 2014;76:57–64. doi: 10.1292/jvms.13-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awwad K, Hu J, Shi L, Mangels N, Abdel Malik R, Zippel N, Fisslthaler B, Eble JA, Pfeilschifter J, Popp R, et al. Role of secreted modular calcium-binding protein 1 (SMOC1) in transforming growth factor beta signalling and angiogenesis. Cardiovasc Res. 2015;106:284–294. doi: 10.1093/cvr/cvv098. [DOI] [PubMed] [Google Scholar]
- Axelsson L, Bergenfeldt M, Ohlsson K. Studies of the release and turnover of a human neutrophil lipocalin. Scand J Clin Lab Invest. 1995;55:577–588. doi: 10.3109/00365519509110257. [DOI] [PubMed] [Google Scholar]
- Ayers KL, Gallet A, Staccini-Lavenant L, Therond PP. The long-range activity of Hedgehog is regulated in the apical extracellular space by the glypican Dally and the hydrolase Notum. Dev Cell. 2010;18:605–620. doi: 10.1016/j.devcel.2010.02.015. [DOI] [PubMed] [Google Scholar]
- Ayers KL, Mteirek R, Cervantes A, Lavenant-Staccini L, Therond PP, Gallet A. Dally and Notum regulate the switch between low and high level Hedgehog pathway signalling. Development. 2012;139:3168–3179. doi: 10.1242/dev.078402. [DOI] [PubMed] [Google Scholar]
- Belardelli F. Role of interferons and other cytokines in the regulation of the immune response. APMIS. 1995;103:161–179. doi: 10.1111/j.1699-0463.1995.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Davis RC, Civelek M, Orozco L, Wu J, Qi H, Pan C, Packard RR, Eskin E, Yan M, et al. Genetic Architecture of Atherosclerosis in Mice: A Systems Genetics Analysis of Common Inbred Strains. PLoS Genet. 2015;11:e1005711. doi: 10.1371/journal.pgen.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- Brown LD, Rodney GG, Hernandez-Ochoa E, Ward CW, Schneider MF. Ca2+ sparks and T tubule reorganization in dedifferentiating adult mouse skeletal muscle fibers. Am J Physiol Cell Physiol. 2007;292:C1156–1166. doi: 10.1152/ajpcell.00397.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canal F, Charawi S, Grimber G, Houbron C, Drouet V, Colnot S, Terris B, Cavard C, Perret C. Generation of Mice with Hepatocyte-Specific Conditional Deletion of Notum. PLoS One. 2016;11:e0150997. doi: 10.1371/journal.pone.0150997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. beta-Adrenoceptor Signaling Networks in Adipocytes for Recruiting Stored Fat and Energy Expenditure. Front Endocrinol (Lausanne) 2011;2:102. doi: 10.3389/fendo.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham ET, Jr, De Souza EB. Interleukin 1 receptors in the brain and endocrine tissues. Immunol Today. 1993;14:171–176. doi: 10.1016/0167-5699(93)90281-o. [DOI] [PubMed] [Google Scholar]
- Dotsch MM, Kloten V, Schlensog M, Heide T, Braunschweig T, Veeck J, Petersen I, Knuchel R, Dahl E. Low expression of ITIH5 in adenocarcinoma of the lung is associated with unfavorable patients’ outcome. Epigenetics. 2015;10:903–912. doi: 10.1080/15592294.2015.1078049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Yuan Z, Ma Z, Song J, Xie X, Chen Y. KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol Biosyst. 2014;10:2441–2447. doi: 10.1039/c4mb00287c. [DOI] [PubMed] [Google Scholar]
- Du ZP, Wu BL, Wu X, Lin XH, Qiu XY, Zhan XF, Wang SH, Shen JH, Zheng CP, Wu ZY, et al. A systematic analysis of human lipocalin family and its expression in esophageal carcinoma. Sci Rep. 2015;5:12010. doi: 10.1038/srep12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert JE, Gerdes J, Flad HD, Brandt E. Novel C-terminally truncated isoforms of the CXC chemokine beta-thromboglobulin and their impact on neutrophil functions. J Immunol. 1998;161:4975–4982. [PubMed] [Google Scholar]
- Elmquist JK, Maratos-Flier E, Saper CB, Flier JS. Unraveling the central nervous system pathways underlying responses to leptin. Nat Neurosci. 1998;1:445–450. doi: 10.1038/2164. [DOI] [PubMed] [Google Scholar]
- Farooqi IS, O’Rahilly S. Leptin: a pivotal regulator of human energy homeostasis. Am J Clin Nutr. 2009;89:980S–984S. doi: 10.3945/ajcn.2008.26788C. [DOI] [PubMed] [Google Scholar]
- Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- Franzen O, Ermel R, Cohain A, Akers NK, Di Narzo A, Talukdar HA, Foroughi-Asl H, Giambartolomei C, Fullard JF, Sukhavasi K, et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–830. doi: 10.1126/science.aad6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- Froesch ER, Schmid C, Schwander J, Zapf J. Actions of insulin-like growth factors. Annu Rev Physiol. 1985;47:443–467. doi: 10.1146/annurev.ph.47.030185.002303. [DOI] [PubMed] [Google Scholar]
- Fulzele K, Lai F, Dedic C, Saini V, Uda Y, Shi C, Tuck P, Aronson JL, Liu X, Spatz JM, et al. Osteocyte-Secreted Wnt Signaling Inhibitor Sclerostin Contributes to Beige Adipogenesis in Peripheral Fat Depots. J Bone Miner Res. 2017;32:373–384. doi: 10.1002/jbmr.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehlenborg N, O’Donoghue SI, Baliga NS, Goesmann A, Hibbs MA, Kitano H, Kohlbacher O, Neuweger H, Schneider R, Tenenbaum D, et al. Visualization of omics data for systems biology. Nat Methods. 2010;7:S56–68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Copley RR, Cohen SM. HSPG modification by the secreted enzyme Notum shapes the Wingless morphogen gradient. Dev Cell. 2002;2:667–676. doi: 10.1016/s1534-5807(02)00180-6. [DOI] [PubMed] [Google Scholar]
- Graham TE, Yang Q, Bluher M, Hammarstedt A, Ciaraldi TP, Henry RR, Wason CJ, Oberbach A, Jansson PA, Smith U, et al. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med. 2006;354:2552–2563. doi: 10.1056/NEJMoa054862. [DOI] [PubMed] [Google Scholar]
- Hamil KG, Liu Q, Sivashanmugam P, Anbalagan M, Yenugu S, Soundararajan R, Grossman G, Rao AJ, Birse CE, Ruben SM, et al. LCN6, a novel human epididymal lipocalin. Reprod Biol Endocrinol. 2003;1:112. doi: 10.1186/1477-7827-1-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbuz MS, Stephanou A, Sarlis N, Lightman SL. The effects of recombinant human interleukin (IL)-1 alpha, IL-1 beta or IL-6 on hypothalamo-pituitary-adrenal axis activation. J Endocrinol. 1992;133:349–355. doi: 10.1677/joe.0.1330349. [DOI] [PubMed] [Google Scholar]
- Hasin-Brumshtein Y, Khan AH, Hormozdiari F, Pan C, Parks BW, Petyuk VA, Piehowski PD, Brummer A, Pellegrini M, Xiao X, et al. Hypothalamic transcriptomes of 99 mouse strains reveal trans eQTL hotspots, splicing QTLs and novel non-coding genes. Elife. 2016;5 doi: 10.7554/eLife.15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathout Y. Approaches to the study of the cell secretome. Expert Rev Proteomics. 2007;4:239–248. doi: 10.1586/14789450.4.2.239. [DOI] [PubMed] [Google Scholar]
- Himmelfarb M, Klopocki E, Grube S, Staub E, Klaman I, Hinzmann B, Kristiansen G, Rosenthal A, Durst M, Dahl E. ITIH5, a novel member of the inter-alpha-trypsin inhibitor heavy chain family is downregulated in breast cancer. Cancer Lett. 2004;204:69–77. doi: 10.1016/j.canlet.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Holt JC, Harris ME, Holt AM, Lange E, Henschen A, Niewiarowski S. Characterization of human platelet basic protein, a precursor form of low-affinity platelet factor 4 and beta-thromboglobulin. Biochemistry. 1986;25:1988–1996. doi: 10.1021/bi00356a023. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev. 1995;16:3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- Joyce AR, Palsson BO. The model organism as a system: integrating ‘omics’ data sets. Nat Rev Mol Cell Biol. 2006;7:198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakugawa S, Langton PF, Zebisch M, Howell SA, Chang TH, Liu Y, Feizi T, Bineva G, O’Reilly N, Snijders AP, et al. Notum deacylates Wnt proteins to suppress signalling activity. Nature. 2015;519:187–192. doi: 10.1038/nature14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz-Jaffe MG, McReynolds S, Gardner DK, Schoolcraft WB. The role of proteomics in defining the human embryonic secretome. Mol Hum Reprod. 2009;15:271–277. doi: 10.1093/molehr/gap012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MP, Choi Y, Wang P, Davis DB, Rabaglia ME, Oler AT, Stapleton DS, Argmann C, Schueler KL, Edwards S, et al. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 2008;18:706–716. doi: 10.1101/gr.074914.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. J Clin Endocrinol Metab. 2004;89:2548–2556. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- Kersten S, Mandard S, Tan NS, Escher P, Metzger D, Chambon P, Gonzalez FJ, Desvergne B, Wahli W. Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. J Biol Chem. 2000;275:28488–28493. doi: 10.1074/jbc.M004029200. [DOI] [PubMed] [Google Scholar]
- Klok MD, Jakobsdottir S, Drent ML. The role of leptin and ghrelin in the regulation of food intake and body weight in humans: a review. Obes Rev. 2007;8:21–34. doi: 10.1111/j.1467-789X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- Kloten V, Rose M, Kaspar S, von Stillfried S, Knuchel R, Dahl E. Epigenetic inactivation of the novel candidate tumor suppressor gene ITIH5 in colon cancer predicts unfavorable overall survival in the CpG island methylator phenotype. Epigenetics. 2014;9:1290–1301. doi: 10.4161/epi.32089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski CM, McTernan PG, Kumar S. Role of resistin in obesity, insulin resistance and Type II diabetes. Clin Sci (Lond) 2005;109:243–256. doi: 10.1042/CS20050078. [DOI] [PubMed] [Google Scholar]
- Kussmann M, Raymond F, Affolter M. OMICS-driven biomarker discovery in nutrition and health. J Biotechnol. 2006;124:758–787. doi: 10.1016/j.jbiotec.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Lai KM, Gonzalez M, Poueymirou WT, Kline WO, Na E, Zlotchenko E, Stitt TN, Economides AN, Yancopoulos GD, Glass DJ. Conditional activation of akt in adult skeletal muscle induces rapid hypertrophy. Mol Cell Biol. 2004;24:9295–9304. doi: 10.1128/MCB.24.21.9295-9304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw. 2012;46 [PMC free article] [PubMed] [Google Scholar]
- Lareyre JJ, Zheng WL, Zhao GQ, Kasper S, Newcomer ME, Matusik RJ, Ong DE, Orgebin-Crist MC. Molecular cloning and hormonal regulation of a murine epididymal retinoic acid-binding protein messenger ribonucleic acid. Endocrinology. 1998;139:2971–2981. doi: 10.1210/endo.139.6.6074. [DOI] [PubMed] [Google Scholar]
- Lawlor K, Nazarian A, Lacomis L, Tempst P, Villanueva J. Pathway-based biomarker search by high-throughput proteomics profiling of secretomes. J Proteome Res. 2009;8:1489–1503. doi: 10.1021/pr8008572. [DOI] [PubMed] [Google Scholar]
- Lazarczyk MA, Lazarczyk M, Grzela T. Ghrelin: a recently discovered gut-brain peptide (review) Int J Mol Med. 2003;12:279–287. [PubMed] [Google Scholar]
- Leinonen R, Nardone F, Zhu W, Apweiler R. UniSave: the UniProtKB sequence/annotation version database. Bioinformatics. 2006;22:1284–1285. doi: 10.1093/bioinformatics/btl105. [DOI] [PubMed] [Google Scholar]
- Lindskog C. The potential clinical impact of the tissue-based map of the human proteome. Expert Rev Proteomics. 2015:1–3. doi: 10.1586/14789450.2015.1040771. [DOI] [PubMed] [Google Scholar]
- Lo KA, Ng PY, Kabiri Z, Virshup D, Sun L. Wnt inhibition enhances browning of mouse primary white adipocytes. Adipocyte. 2016;5:224–231. doi: 10.1080/21623945.2016.1148834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Long Q, Argmann C, Houten SM, Huang T, Peng S, Zhao Y, Tu Z, Zhu J Consortium GT. Inter-tissue coexpression network analysis reveals DPP4 as an important gene in heart to blood communication. Genome Med. 2016;8:15. doi: 10.1186/s13073-016-0268-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MR, Tran MT, Parikh SM. PGC1alpha in the kidney. Am J Physiol Renal Physiol. 2017 doi: 10.1152/ajprenal.00263.2017. ajprenal 00263 02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Liew CC. Gene profiling identifies secreted protein transcripts from peripheral blood cells in coronary artery disease. J Mol Cell Cardiol. 2003;35:993–998. doi: 10.1016/s0022-2828(03)00179-2. [DOI] [PubMed] [Google Scholar]
- Mai C, Zhao JJ, Tang XF, Wang W, Pan K, Pan QZ, Zhang XF, Jiang SS, Zhao BW, Li YF, et al. Decreased ITIH5 expression is associated with poor prognosis in primary gastric cancer. Med Oncol. 2014;31:53. doi: 10.1007/s12032-014-0053-1. [DOI] [PubMed] [Google Scholar]
- Makridakis M, Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J Proteomics. 2010;73:2291–2305. doi: 10.1016/j.jprot.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Margetic S, Gazzola C, Pegg GG, Hill RA. Leptin: a review of its peripheral actions and interactions. Int J Obes Relat Metab Disord. 2002;26:1407–1433. doi: 10.1038/sj.ijo.0802142. [DOI] [PubMed] [Google Scholar]
- Martin J, Midgley A, Meran S, Woods E, Bowen T, Phillips AO, Steadman R. Tumor Necrosis Factor-stimulated Gene 6 (TSG-6)-mediated Interactions with the Inter-alpha-inhibitor Heavy Chain 5 Facilitate Tumor Growth Factor beta1 (TGFbeta1)-dependent Fibroblast to Myofibroblast Differentiation. J Biol Chem. 2016;291:13789–13801. doi: 10.1074/jbc.M115.670521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- Mosquera G, Giraldo MC, Khang CH, Coughlan S, Valent B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as Biotrophy-associated secreted proteins in rice blast disease. Plant Cell. 2009;21:1273–1290. doi: 10.1105/tpc.107.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nian M, Lee P, Khaper N, Liu P. Inflammatory cytokines and postmyocardial infarction remodeling. Circ Res. 2004;94:1543–1553. doi: 10.1161/01.RES.0000130526.20854.fa. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nies VJ, Sancar G, Liu W, van Zutphen T, Struik D, Yu RT, Atkins AR, Evans RM, Jonker JW, Downes MR. Fibroblast Growth Factor Signaling in Metabolic Regulation. Front Endocrinol (Lausanne) 2015;6:193. doi: 10.3389/fendo.2015.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Gorman TW. An adaptive test of significance for a subset of regression coefficients. Stat Med. 2002;21:3527–3542. doi: 10.1002/sim.1278. [DOI] [PubMed] [Google Scholar]
- O’Neill SM, Hinkle C, Chen SJ, Sandhu A, Hovhannisyan R, Stephan S, Lagor WR, Ahima RS, Johnston JC, Reilly MP. Targeting adipose tissue via systemic gene therapy. Gene Ther. 2014;21:653–661. doi: 10.1038/gt.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada I, Hamanoue H, Terada K, Tohma T, Megarbane A, Chouery E, Abou-Ghoch J, Jalkh N, Cogulu O, Ozkinay F, et al. SMOC1 is essential for ocular and limb development in humans and mice. Am J Hum Genet. 2011;88:30–41. doi: 10.1016/j.ajhg.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim M, Sun K, An YA, Gu X, Scherer PE. VEGF-A-Expressing Adipose Tissue Shows Rapid Beiging and Enhanced Survival After Transplantation and Confers IL-4-Independent Metabolic Improvements. Diabetes. 2017;66:1479–1490. doi: 10.2337/db16-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17:141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Sallam T, Mehrabian M, Psychogios N, Hui ST, Norheim F, Castellani LW, Rau CD, Pan C, Phun J, et al. Genetic architecture of insulin resistance in the mouse. Cell Metab. 2015;21:334–346. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BK, Febbraio MA. Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiol Rev. 2008;88:1379–1406. doi: 10.1152/physrev.90100.2007. [DOI] [PubMed] [Google Scholar]
- Petricoin EF, Liotta LA. SELDI-TOF-based serum proteomic pattern diagnostics for early detection of cancer. Curr Opin Biotechnol. 2004;15:24–30. doi: 10.1016/j.copbio.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Pierson E, Koller D, Battle A, Mostafavi S, Ardlie KG, Getz G, Wright FA, Kellis M, Volpi S, et al. Consortium GT. Sharing and Specificity of Co-expression Networks across 35 Human Tissues. PLoS Comput Biol. 2015;11:e1004220. doi: 10.1371/journal.pcbi.1004220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Helgason A, Thorleifsson G, McCarroll SA, Kong A, Stefansson K. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 2011;7:e1001317. doi: 10.1371/journal.pgen.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen K, Murugan S, Vainio S. Wnt signaling in kidney development and disease. Organogenesis. 2008;4:55–59. doi: 10.4161/org.4.2.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainger J, van Beusekom E, Ramsay JK, McKie L, Al-Gazali L, Pallotta R, Saponari A, Branney P, Fisher M, Morrison H, et al. Loss of the BMP antagonist, SMOC-1, causes Ophthalmo-acromelic (Waardenburg Anophthalmia) syndrome in humans and mice. PLoS Genet. 2011;7:e1002114. doi: 10.1371/journal.pgen.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin TL, Ong DE, Orgebin-Crist MC. The 18-kDa mouse epididymal protein (MEP 10) binds retinoic acid. Biol Reprod. 1992a;46:767–771. doi: 10.1095/biolreprod46.5.767. [DOI] [PubMed] [Google Scholar]
- Rankin TL, Tsuruta KJ, Holland MK, Griswold MD, Orgebin-Crist MC. Isolation, immunolocalization, and sperm-association of three proteins of 18, 25, and 29 kilodaltons secreted by the mouse epididymis. Biol Reprod. 1992b;46:747–766. doi: 10.1095/biolreprod46.5.747. [DOI] [PubMed] [Google Scholar]
- Rocchi M, Covone A, Romeo G, Faraonio R, Colantuoni V. Regional mapping of RBP4 to 10q23----q24 and RBP1 to 3q21----q22 in man. Somat Cell Mol Genet. 1989;15:185–190. doi: 10.1007/BF01535081. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Cuenca S, Monjo M, Frontera M, Gianotti M, Proenza AM, Roca P. Sex steroid receptor expression profile in brown adipose tissue. Effects of hormonal status. Cell Physiol Biochem. 2007;20:877–886. doi: 10.1159/000110448. [DOI] [PubMed] [Google Scholar]
- Rose M, Gaisa NT, Antony P, Fiedler D, Heidenreich A, Otto W, Denzinger S, Bertz S, Hartmann A, Karl A, et al. Epigenetic inactivation of ITIH5 promotes bladder cancer progression and predicts early relapse of pT1 high-grade urothelial tumours. Carcinogenesis. 2014;35:727–736. doi: 10.1093/carcin/bgt375. [DOI] [PubMed] [Google Scholar]
- Salomon F, Cuneo RC, Hesp R, Sonksen PH. The effects of treatment with recombinant human growth hormone on body composition and metabolism in adults with growth hormone deficiency. N Engl J Med. 1989;321:1797–1803. doi: 10.1056/NEJM198912283212605. [DOI] [PubMed] [Google Scholar]
- Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem. 1995;270:26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- Schiefner A, Skerra A. The menagerie of human lipocalins: a natural protein scaffold for molecular recognition of physiological compounds. Acc Chem Res. 2015;48:976–985. doi: 10.1021/ar5003973. [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Seeley RJ, Campfield LA, Burn P, Baskin DG. Identification of targets of leptin action in rat hypothalamus. J Clin Invest. 1996;98:1101–1106. doi: 10.1172/JCI118891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MM, Peterson JM, Byerly MS, Wei Z, Wong GW. Myonectin (CTRP15), a novel myokine that links skeletal muscle to systemic lipid homeostasis. J Biol Chem. 2012;287:11968–11980. doi: 10.1074/jbc.M111.336834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MM, Tan SY, Wong GW. Metabolic function of the CTRP family of hormones. Rev Endocr Metab Disord. 2014;15:111–123. doi: 10.1007/s11154-013-9255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman BT, Huang da W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrobuk P, von Kraemer S, Semenova MM, Zitting A, Koistinen HA. Acute exposure to resveratrol inhibits AMPK activity in human skeletal muscle cells. Diabetologia. 2012;55:3051–3060. doi: 10.1007/s00125-012-2691-1. [DOI] [PubMed] [Google Scholar]
- Smith C, Damas JK, Otterdal K, Oie E, Sandberg WJ, Yndestad A, Waehre T, Scholz H, Endresen K, Olofsson PS, et al. Increased levels of neutrophil-activating peptide-2 in acute coronary syndromes: possible role of platelet-mediated vascular inflammation. J Am Coll Cardiol. 2006;48:1591–1599. doi: 10.1016/j.jacc.2006.06.060. [DOI] [PubMed] [Google Scholar]
- Stadler K, Goldberg IJ, Susztak K. The evolving understanding of the contribution of lipid metabolism to diabetic kidney disease. Curr Diab Rep. 2015;15:40. doi: 10.1007/s11892-015-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, Patel HR, Ahima RS, Lazar MA. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedgui A, Mallat Z. Cytokines in atherosclerosis: pathogenic and regulatory pathways. Physiol Rev. 2006;86:515–581. doi: 10.1152/physrev.00024.2005. [DOI] [PubMed] [Google Scholar]
- Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R, Grinspoon SK, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traister A, Shi W, Filmus J. Mammalian Notum induces the release of glypicans and other GPI-anchored proteins from the cell surface. Biochem J. 2008;410:503–511. doi: 10.1042/BJ20070511. [DOI] [PubMed] [Google Scholar]
- Trujillo ME, Scherer PE. Adiponectin--journey from an adipocyte secretory protein to biomarker of the metabolic syndrome. J Intern Med. 2005;257:167–175. doi: 10.1111/j.1365-2796.2004.01426.x. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva CJ, Vergnes L, Wang J, Drew BG, Hong C, Tu Y, Hu Y, Peng X, Xu F, Saez E, et al. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARgamma specifies lipid storage versus thermogenic gene programs. Cell Metab. 2013;17:423–435. doi: 10.1016/j.cmet.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MJ. J. J. R. Macleod: the co-discoverer of insulin. Proc R Coll Physicians Edinb. 1993;23:1–125. [PubMed] [Google Scholar]
- Wozniak SE, Gee LL, Wachtel MS, Frezza EE. Adipose tissue: the new endocrine organ? A review article. Dig Dis Sci. 2009;54:1847–1856. doi: 10.1007/s10620-008-0585-3. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Shimizugawa T, Ono M, Furukawa H. Angiopoietin-like protein 4 is a potent hyperlipidemia-inducing factor in mice and inhibitor of lipoprotein lipase. J Lipid Res. 2002;43:1770–1772. doi: 10.1194/jlr.c200010-jlr200. [DOI] [PubMed] [Google Scholar]
- Yu X, Suzuki K, Wang Y, Gupta A, Jin R, Orgebin-Crist MC, Matusik R. The role of forkhead box A2 to restrict androgen-regulated gene expression of lipocalin 5 in the mouse epididymis. Mol Endocrinol. 2006;20:2418–2431. doi: 10.1210/me.2006-0008. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li R, Meng Y, Li S, Donelan W, Zhao Y, Qi L, Zhang M, Wang X, Cui T, et al. Irisin stimulates browning of white adipocytes through mitogen-activated protein kinase p38 MAP kinase and ERK MAP kinase signaling. Diabetes. 2014;63:514–525. doi: 10.2337/db13-1106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ssec and pvalue of permutation score for each top-ranked peptide, related to Figure 2