Abstract
Background
MicroRNAs (miRNAs) are involved in many biological processes, including tumor suppression. Multiple studies have shown an association between the miRNA-196a2 rs11614913 and miRNA-146a rs2910164 polymorphisms and cancer risk. However, the implications of the reported data are debatable and inconclusive.
Materials and methods
Relevant articles were retrieved from the PubMed, EMBASE, China National Knowledge Infrastructure, and WanFang databases from January 1, 2007, to April 30, 2017. Studies were assessed based on designated inclusion and exclusion criteria, and data were manually extracted from relevant studies by two investigators. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to explore the association between two single-nucleotide polymorphisms (SNPs) in miRNAs and lung cancer susceptibility.
Results
Nine eligible articles were included, consisting of 3,101 cancer cases and 3,234 controls for miRNA-196a2 rs11614913, and 3,483 cases and 3,578 controls for miRNA-146a rs2910164. For studies evaluating miRNA-196a2 rs11614913, significant associations with lung cancer risk were discovered. Overall, the pooled analysis showed that miRNA-196a2 rs11614913 was associated with a decreased cancer risk (CC vs TT: OR = 1.25, 95% CI: 1.09–1.44; CT vs TT: OR = 1.26, 95% CI: 1.03–1.53). For miRNA-146a rs2910164, only the CC genotype was found to be associated with high lung cancer risk (OR = 1.30, 95% CI: 1.13–1.49). Subgroup analyses based on ethnicity, source of control group, and country indicated that there were strong associations between miRNA-146a rs2910164 and cancer risk.
Conclusion
The results indicated that lung cancer risk was significantly associated with miRNA-196a2 rs11614913 and miRNA-146a rs2910164. These two common SNPs in miRNAs may be potential biomarkers of lung cancer.
Keywords: microRNA, lung cancer, single nucleotide polymorphisms, susceptibility, meta-analysis
Introduction
Lung cancer is the most commonly diagnosed type of cancer and has the highest mortality rate, leading to approximately 1.38 million deaths in the world per year.1 Studies show that smoking is the primary risk factor for lung cancer. However, although more than 80% of lung cancer patients have smoked, only 15% of smokers will eventually be diagnosed with lung cancer, indicating that other factors such as genetic predisposition, contribute to the development of lung cancer.2
MicroRNAs (miRNAs) are an abundant category of endogenous small noncoding RNAs of about 18–25 nucleotides in length, which negatively regulate their target mRNAs via posttranscriptional gene silencing.3 Studies have shown that about 70% of mammalian miRNAs are located in transcription units, mostly in intron regions.4 miRNAs are present in various forms, and mature miRNAs exert a broad range of biological effects by binding to the mRNA of the target gene. Research suggests that miRNA regulates up to 30% of human genes,5 and accumulating evidence supports a significant association between miRNAs and the risk and prognosis of lung cancer.6,7 Takamizawa et al reported that lower expression of the let-7 miRNAs aggravated lung cancer,8 while Hayashita et al found that upregulation of the miRNA-17-92 may decrease lung cancer risk.9
Single-nucleotide polymorphisms (SNPs) can change the properties of miRNAs, thereby influencing an individual’s susceptibility to cancers.10–12 The miRNA-146a rs291016413 and miRNA-196a2 rs11614913 polymorphisms,14 in particular, have been found to be relevant to lung cancer susceptibility. However, the conclusions of the published studies are often in conflict.15,16 For example, Volpe et al found that miRNA-196a2 regulated the HOX gene family, which is associated with lung cancer metastasis. miRNA-196a2 rs11614913 polymorphisms regulated the expression of miRNA-196a2 itself and influenced the risk of lung cancer.15 However, Vinci et al did not find any connection between miRNA-196a2 rs11614913 polymorphisms and the risk of lung cancer, although they did find that miRNA-146a rs2910164 polymorphisms had a role in enhancing the risk of lung cancer.16
Therefore, in this study, a meta-analysis of previously and newly published data was performed to further explore the correlation between SNPs of miRNA-146a rs2910164 and miRNA-196a2 rs11614913 and lung cancer risk.
Materials and methods
Literature retrieval method
We searched the relevant published literature from January 1, 2007, to April 30, 2017, in English or Chinese, using the PubMed, EMBASE, China National Knowledge Infrastructure, and WanFang databases, with the following keywords: “lung cancer”, “miRNA-146a”, “miRNA-196a2”, “polymorphism”, “rs2910164”, and “rs11614913”. References of the retrieved articles were published in the primary literature and had no distinct overlap of population with other studies. The inclusion criteria were as follows: 1) case-control study; 2) case group for the diagnosis of lung cancer patients and control group for non-lung cancer population; and 3) data on the miRNA-196a2 rs11614913 and miRNA-146a rs2910164 polymorphisms. We excluded reports with the same data or overlapping data by the same authors.
Data extraction
Two reviewers extracted data from eligible studies in duplicate with a standard data collection form and reached a consensus on each item. The following information was extracted for each study: first author, publication date, country, ethnicity, source of control, genotype of miRNA-196a2 rs11614913, and miRNA-146a rs2910164 polymorphism.
Statistical analysis
Odds ratios (ORs) and 95% confidence intervals (CIs) were used to measure the degree of correlation between the polymorphism of rs2910164 or rs11614913 and the genetic susceptibility to lung cancer. All analyses used the wild genotype as a control. The combined OR values for CC vs TT, CC vs GG, CT vs TT, and CG vs GG, respectively, were calculated.
The combined OR value was calculated using a z-test with a significance level of P = 0.05. The Q value and I2 value were used to evaluate the heterogeneity between the results of the study. The calibrated Q-test based on the chi-square test was used to evaluate whether the distribution of effects of each study had significant heterogeneity. With α = 0.10 as the demarcation standard, the heterogeneity of the effect of the study was not statistically significant at P > 0.1, and the study results were merged by the Mantel-Haenszel method. When P ≤ 0.1, a random-effects model (inverse-variance method) was used to combine the results. I2 was used to describe the proportion of the variation between the studies accounting for total variation.
Publication bias was assessed by visual inspection of the Begg’s funnel plot. If the funnel plot was symmetrical, it was considered that there was no publication bias in the literature being considered; otherwise, it was considered biased. Our Begg’s funnel plot presented a symmetrical and funnel-like distribution of points, indicating that the publication bias risk was low in this analysis. Subgroup analyses were conducted to investigate the association of the subgroups and to identify any heterogeneity.
Results
Study characteristics
We searched the online databases using search criteria related to miRNA-196a2 rs11614913 and miRNA-146a rs2910164 SNPs and the risk of lung cancer. Thirteen published articles were initially collected. Four studies were excluded after duplicate removal, primary screening by reading titles and abstracts, and secondary screening by intensive reading. One of these was written by Tian and its data overlapped.17 One only reported survival times,18 one performed a meta-analysis,19 and another did not report sufficient related data.20 In total, nine studies met our inclusion criteria. Of these, seven were about miRNA-196a2 rs11614913, with a cumulative case group of 3,101 participants and a control group of 3,234. Six studies were about miRNA-146a rs2910164, with 3,483 and 3,578 participants in the case and control groups, respectively (Table 1).
Table 1.
Characteristics of researches included in the meta-analysis
| Study | Country (ethnicity) | Source of control | Methods | miRNA SNP | Number of subjects | Case
|
Control
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TT/GG | CT/CG | CC/CC | TT/GG | CT/CG | CC/CC | ||||||
| Yin et al21 | Chinese (Asian) | H | TaqMan PCR | 196a2 rs11614913 | 1,183 | 149 | 298 | 128 | 178 | 297 | 133 |
| Shang et al22 | Chinese (Asian) | P | PCR-RFLP | 196a2 rs11614913 | 116 | 7 | 17 | 8 | 48 | 26 | 10 |
| Yin et al23 | Chinese (Asian) | H | TaqMan PCR | 196a2 rs11614913 | 568 | 67 | 141 | 50 | 97 | 150 | 63 |
| Vinci et al16 | Italian (Caucasian) | H | PCR-HRMA | 196a2 rs11614913 | 230 | 12 | 54 | 35 | 10 | 61 | 58 |
| Hong et al24 | Korean (Asian) | P | TaqMan PCR | 196a2 rs11614913 | 834 | 96 | 224 | 86 | 134 | 198 | 96 |
| Kim et al25 | Korean (Asian) | H | Fluorescence | 196a2 rs11614913 | 1,294 | 162 | 305 | 187 | 185 | 300 | 155 |
| Tian et al26 | Chinese (Asian) | P | PCR-RFLP | 196a2 rs11614913 | 2,093 | 293 | 512 | 253 | 307 | 519 | 209 |
| Yin et al21 | Chinese (Asian) | H | TaqMan PCR | 146a rs2910164 | 1,183 | 97 | 280 | 198 | 127 | 313 | 168 |
| Yin et al23 | Chinese (Asian) | H | TaqMan PCR | 146a rs2910164 | 568 | 45 | 134 | 79 | 63 | 160 | 87 |
| Jia et al27 | Chinese (Asian) | H | PCR-RFLP | 146a rs2910164 | 800 | 64 | 182 | 154 | 76 | 200 | 124 |
| Jeon et al28 | Korean (Asian) | P | PCR-RFLP | 146a rs2910164 | 2,187 | 223 | 500 | 368 | 244 | 540 | 312 |
| Vinci et al16 | Italian (Caucasian) | H | PCR-HRMA | 146a rs2910164 | 230 | 44 | 48 | 9 | 73 | 45 | 11 |
| Tian et al26 | Chinese (Asian) | P | PCR-RFLP | 146a rs2910164 | 2,093 | 360 | 510 | 188 | 364 | 502 | 169 |
Abbreviations: miRNA, microRNA; SNP, single-nucleotide polymorphism; P, population; H, hospital; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; HRMA, high resolution melting analysis.
Meta-analysis statistic results of miRNA-196a2 rs11614913
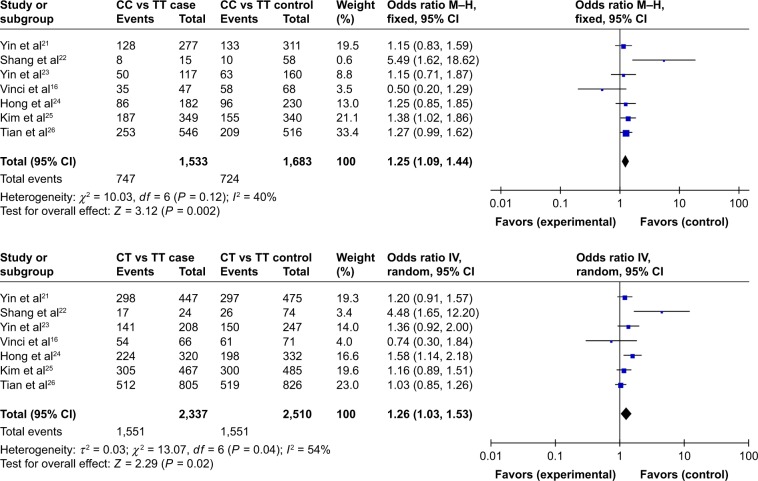
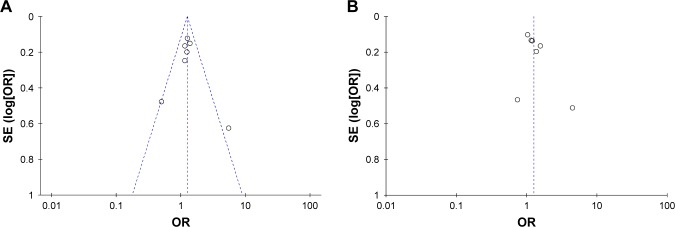
In the CC vs TT group, the heterogeneity test demonstrated the following: χ2 = 10.03, P = 0.12. The heterogeneity was not significant. The OR was 1.25, with a 95% CI of 1.09–1.44, and P = 0.002 using the fixed-effects model. The results showed that people with the CC genotype had an increased risk of lung cancer, and that the difference was statistically significant. For the CT vs TT group, the heterogeneity test demonstrated the following: χ2 = 13.07, P = 0.04, I2 = 54%, with moderate to high heterogeneity. The OR was 1.26, with a 95% CI of 1.03–1.53, and P = 0.02 according to the random-effects model. The results showed that individuals with the CT genotype had an increased risk of lung cancer, and that the difference was statistically significant (Figure 1). A Begg’s funnel plot was constructed to evaluate the publication bias of the literature; the results indicated that no obvious publication bias affected this meta-analytic review (Figure 2).
Figure 1.
Forest plot for association between the polymorphisms of miRNA-196a2 rs11614913 and the genetic susceptibility to lung cancer.
Abbreviations: M–H, Mantel–Haenszel; IV, inverse-variance.
Figure 2.
Begg’s funnel plot of miRNA-196a2 rs11614913 and genetic susceptibility to lung cancer.
Notes: (A) For CC vs TT. (B) For CT vs TT.
Meta-analysis statistic results of miRNA-146a rs2910164
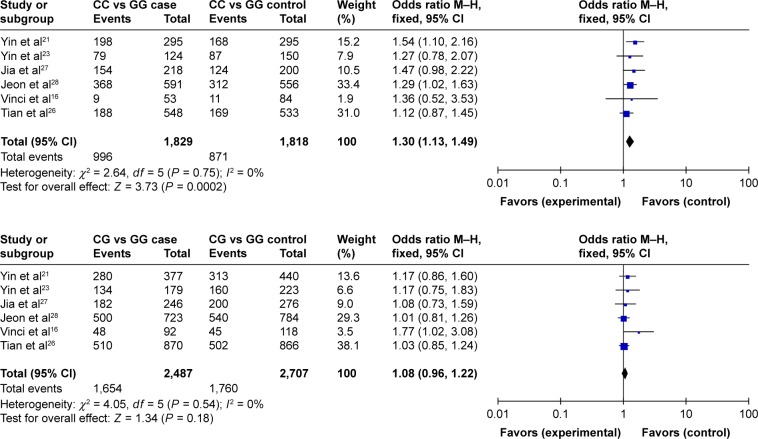
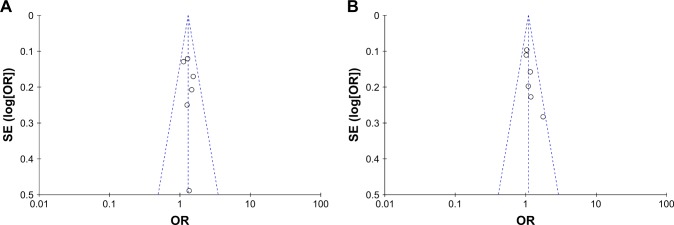
For the CC vs GG group, the heterogeneity test showed that χ2 = 2.64, P = 0.75, and that the heterogeneity was not significant. The OR was 1.30, with a 95% CI of 1.13–1.49, and P = 0.0002 according to the fixed-effects model, and the difference was statistically significant. The results showed that individuals with the CC genotype had an increased risk of lung cancer, and the difference was statistically significant. For the CG vs GG group, the heterogeneity test showed that χ2 = 4.05, P = 0.54, and the heterogeneity was not significant. The OR was 1.08, with a 95% CI of 0.96–1.22, and P = 0.18 according to the fixed-effects model (Figure 3). A Begg’s funnel plot was constructed; again, there was no observed publication bias in this study (Figure 4).
Figure 3.
Forest plot for association between the polymorphisms of miRNA-146a rs2910164 and the genetic susceptibility to lung cancer.
Abbreviation: M–H, Mantel–Haenszel.
Figure 4.
Begg’s funnel plot of miRNA-146a rs2910164 and genetic susceptibility to lung cancer.
Notes: (A) For CC vs GG. (B) For CG vs GG.
Results of subgroup analysis by ethnicity
Stratified analyses significantly reduced the heterogeneity of the subgroups. The results of subgroup analysis by ethnicity are shown in Table 2. There was only one study of ethnicity for Caucasians, and so no heterogeneity test was performed for Caucasian subgroups.
Table 2.
Subgroup analysis of correlation between polymorphisms and ethnicity
| miRNA SNP | Ethnicity | Comparison | χ2 | Phetero | I2 (%) | P-value | OR | 95% CI |
|---|---|---|---|---|---|---|---|---|
| 196a2 rs11614913 | Asian | CC vs TT | 6.31 | 0.28 | 21 | 0.003 | 1.29 | 1.09–1.52 |
| CT vs TT | 11.97 | 0.04 | 58 | 0.01 | 1.29 | 1.05–1.58 | ||
| Caucasian | CC vs TT | – | – | – | 0.15 | 0.50 | 0.20–1.29 | |
| CT vs TT | – | – | – | 0.51 | 0.74 | 0.30–1.84 | ||
| 146a rs2910164 | Asian | CC vs GG | 2.63 | 0.62 | 0 | 0.0002 | 1.30 | 1.13–1.49 |
| CG vs GG | 0.87 | 0.93 | 0 | 0.35 | 1.06 | 0.94–1.19 | ||
| Caucasian | CC vs GG | – | – | – | 0.53 | 1.36 | 0.52–3.53 | |
| CG vs GG | – | – | – | 0.04 | 1.77 | 1.02–3.08 |
Note: Bold values are statistically significant.
Abbreviations: miRNA, microRNA; SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
A significant increased risk of lung cancer based on ethnicity was found. For miRNA-196a2 rs11614913, the CC and CT genotypes were associated with increased susceptibility to lung cancer in the Asian population compared with the TT genotype, with a statistically significant difference; however, in the Caucasian population, no polymorphism of this locus was observed to be connected with susceptibility to lung cancer.
For miRNA-146a rs2910164, compared with the GG genotype, the CC genotype was associated with an increased susceptibility to lung cancer in the Asian population, with a statistically significant difference. Based on the current sample size, there was no correlation in Caucasians. The CC genotype was not associated with susceptibility to lung cancer in the Asian population, but CG genotype of Caucasians was associated with an increased risk of lung cancer, with a statistically significant difference.
Results of subgroup analysis by country
The results of subgroup analysis by country are shown in Table 3.
Table 3.
Subgroup analysis of correlation between polymorphisms and countries
| miRNA SNP | Country | Comparison | χ2 | Phetero | I2 (%) | P-value | OR | 95% CI |
|---|---|---|---|---|---|---|---|---|
| 196a2 rs11614913 | Chinese | CC vs TT | 6.01 | 0.11 | 50 | 0.08 | 1.30 | 0.97–1.75 |
| CT vs TT | 8.98 | 0.03 | 67 | 0.09 | 1.30 | 0.96–1.75 | ||
| Korean | CC vs TT | 0.15 | 0.70 | 0 | 0.02 | 1.33 | 1.05–1.69 | |
| CT vs TT | 2.08 | 0.15 | 52 | 0.06 | 1.33 | 0.99–1.80 | ||
| Italian | CC vs TT | – | – | – | 0.15 | 0.50 | 0.20–1.29 | |
| CT vs TT | – | – | – | 0.51 | 0.74 | 0.30–1.84 | ||
| 146a rs2910164 | Chinese | CC vs GG | 2.63 | 0.45 | 0 | 0.003 | 1.30 | 1.10–1.54 |
| CG vs GG | 0.66 | 0.88 | 0 | 0.30 | 1.08 | 0.93–1.24 | ||
| Korean | CC vs GG | – | – | – | 0.03 | 1.29 | 1.02–1.63 | |
| CG vs GG | – | – | – | 0.91 | 1.01 | 0.81–1.26 | ||
| Italian | CC vs GG | – | – | – | 0.53 | 1.36 | 0.52–3.53 | |
| CG vs GG | – | – | – | 0.04 | 1.77 | 1.02–3.08 |
Note: Bold values are statistically significant.
Abbreviations: miRNA, microRNA; SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
For miRNA-196a2 rs11614913, the CC genotype was not associated with an increased susceptibility to lung cancer compared with the TT genotype in either the Chinese or the Italian subgroup, whereas in the Korean subgroup, an association was found. The CT genotype was not connected with lung cancer risk in any country subgroup.
For miRNA-146a rs2910164, the CC genotype was associated with an increased susceptibility to lung cancer compared with the GG genotype in both the Chinese and Korean subgroups, and the difference was statistically significant. In the Italian subgroup, the CC genotype was not associated with lung cancer susceptibility. However, the CG genotype was associated with an increased susceptibility to lung cancer only in the Italian subgroup.
Results of subgroup analysis by source of control group
The association between SNPs and genetic susceptibility to lung cancer may also depend on the source of the control group. The results of the subgroup analysis according to the source of control group are shown in Table 4.
Table 4.
Subgroup analysis of correlation between polymorphisms and source of control
| miRNA SNP | Source of control | Comparison | χ2 | Phetero | I2 (%) | P-value | OR | 95% CI |
|---|---|---|---|---|---|---|---|---|
| 196a2 rs11614913 | Population | CC vs TT | 5.40 | 0.07 | 63 | 0.08 | 1.47 | 0.95–2.28 |
| CT vs TT | 11.51 | 0.003 | 83 | 0.09 | 1.58 | 0.93–2.70 | ||
| Hospital | CC vs TT | 4.21 | 0.24 | 29 | 0.22 | 1.17 | 0.91–1.50 | |
| CT vs TT | 1.55 | 0.67 | 0 | 0.04 | 1.19 | 1.01–1.41 | ||
| 146a rs2910164 | Population | CC vs GG | 0.60 | 0.44 | 0 | 0.03 | 1.21 | 1.02–1.44 |
| CG vs GG | 0.01 | 0.93 | 0 | 0.78 | 1.02 | 0.88–1.18 | ||
| Hospital | CC vs GG | 0.44 | 0.93 | 0 | 0.001 | 1.45 | 1.16–1.81 | |
| CG vs GG | 2.21 | 0.53 | 0 | 0.06 | 1.21 | 0.99–1.48 |
Note: Bold values are statistically significant.
Abbreviations: miRNA, microRNA; SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
For miRNA-196a2 rs11614913, the CC genotype was not associated with an elevated susceptibility to lung cancer compared with TT genotype in either the community or hospital subgroups. The CT genotype was associated with an increased susceptibility to lung cancer in the hospital control subgroup, but not in the community subgroup, and the difference was statistically significant.
For miRNA-146a rs2910164, the CC genotype was associated with an increased susceptibility to lung cancer compared with the GG genotype in both the community and hospital control groups, and the difference was statistically significant. However, the CG genotype was not connected with an increased susceptibility to lung cancer in either subgroup.
Discussion
Lung cancer is the most commonly diagnosed cancer, as well as the chief cause of cancer deaths globally. In addition to environmental exposure, SNPs can make an individual more susceptible to lung cancer. Therefore, there is a need to seek out valid biomarkers to predict the risk of the disease.
miRNAs are endogenous small RNAs of about 18–25 nucleotides in length. Previous studies have shown that miRNAs have important roles in key biological processes and are considered a key factor in tumorigenesis.29
Many researchers have aimed to understand the role of SNPs that occur in precursor and mature miRNAs, and their effects on gene function or expression, cancer susceptibility, and progression of various diseases.30 Identifying SNPs is important in predicting the risks of individuals and understanding the pathogenesis of cancer. Many studies have explored the common SNPs of miRNAs and their associations with risk of various cancers, including lung cancer.31–36 However, the mechanisms of carcinogenesis are complex, and the investigations on single genetic polymorphisms and studies with small sample sizes that have been performed so far have not provided a means to reliably predict the risk of cancer. In addition, the conclusions of previous studies have sometimes been inconsistent and conflicted with each other.16,22,37,38 To address this, we carried out a meta-analysis by gathering all of the studies that reported a connection between the miRNA-196a2 rs11614913 and miRNA-146a rs2910164 SNPs and the risk of lung cancer.
For the miRNA-196a2 rs11614913 locus, we found that the risk of lung cancer in the CC and CT genotype individuals was higher than in the TT genotype individuals. For the miRNA-146a rs2910164 locus, individuals with the CC genotype exhibited a higher risk of lung cancer than individuals with the GG genotype. Thus, the miRNA-196a2 rs11614913 and miRNA-146a rs2910164 loci can serve as potential candidate sites for lung cancer susceptibility screening.
To identify any differences between individuals of Caucasian and Asian ethnicities, we carried out a subgroup analysis. This showed that different ethnicity, to some extent, was linked to differences in the association between genetic polymorphisms and lung cancer risk. On this basis, a further subgroup analysis by country was performed – this was, to our knowledge, the first attempt to explore whether the susceptibility to lung cancer varies across different countries. We considered that the dietary and health-related habits of different countries might influence the risk of lung cancer. As there was only one study of Caucasian individuals and the participants in this study were Italian, the Italian subgroup was identical to the Caucasian subgroup. Our results indicated that the association between the miRNA-196a2 rs11614913 polymorphism and the risk of lung cancer only could be found in Asian populations. Our initial results indicated that the predictive value of miRNA-196a2 rs11614913 polymorphism was valid in Asian people. However, in the subgroup analysis of correlations between polymorphisms and countries, this relationship was found only in Korean individuals. Differences owing to ethnicity have also been found in other types of cancer.39 Wang et al39 found that the miRNA-196a2 rs11614913 CC genotype was significantly associated with breast cancer risk in Chinese and Indian individuals, but not in Caucasians. For the miRNA-146a rs2910164 locus, only the CC genotype was related to an increased susceptibility of lung cancer compared with the GG genotype in the Asian population, whereas the CG genotype of the Caucasian population was associated with an increased susceptibility of lung cancer compared with the GG genotype. After subgroup analysis based on the country, the CC genotype was associated with an increased susceptibility to lung cancer compared with GG genotype in both the Chinese and Korean subgroups. As there was only one report involving Caucasian population,16 and the sample size of this study was relatively small, further research is needed before any firm conclusions can be drawn regarding this population.
According to the subgroup analysis of the control source, the miRNA-196a2 rs11614913 CT genotype was associated with an increased susceptibility compared with the TT genotype in the hospital subgroup. By contrast, the miRNA-146a rs2910164 CC genotype was associated with a higher risk of lung cancer compared with the GG genotype in both community and hospital subgroups.
There were some advantages of the comprehensive analysis that we have performed here, such as the use of the funnel plot to evaluate publication bias, which showed good funnel plot symmetry. In addition, all of the nine articles in our study were of high quality. However, some limitations still need to be addressed. First, and most importantly, the results of the meta-analysis were limited by the available literature, such as the number of documents, quality, and sample size, which may affect its validity. For example, there were too few Caucasian studies available, and the heterogeneity among different studies cannot be ignored. Furthermore, we did not explore the association of SNP-SNP interactions. Such gene–gene interactions in cancers have been reported in the literature.40–43 In addition, lung cancer is a result of gene–environment interactions. In this study, only the influence of genetic factors was taken into account, and environmental factors, such as smoking and air pollution, were not explored. Future work examining these gene–gene and gene–environment interactions and their association with lung cancer is needed.
In summary, our meta-analysis suggested that the miRNA-196a2 rs11614913 CC and CT genotypes, and the miRNA-146a rs2910164 CC genotype, may increase the risk of lung cancer, especially in Asian populations. Owing to the low number of studies included and the relatively small sample size of the single Caucasian study, the negative results in Caucasian individuals need to be confirmed by further study. Overall, it will be necessary to confirm all these findings using well-designed and larger sample size studies. In addition, the interaction of genetic and environmental factors should be considered.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No 81660550), the Province Natural Science Foundation of Hainan (No 814292), and the Foundation for Fostering Talents of Hainan Medical College (No HY2014-025).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Yin Z, Cui Z, Li H, et al. Polymorphisms in miR-135a-2, miR-219-2 and miR-211 as well as their interaction with cooking oil fume exposure on the risk of lung cancer in Chinese nonsmoking females: a case-control study. BMC Cancer. 2016;16(1):751. doi: 10.1186/s12885-016-2784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhan X, Wu W, Han B, et al. Hsa-miR-196a2 functional SNP is associated with severe toxicity after platinum-based chemotherapy of advanced nonsmall cell lung cancer patients in a Chinese population. J Clin Lab Anal. 2012;26(6):441–446. doi: 10.1002/jcla.21544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22(3):165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortinovis D, Monica V, Pietrantonio F, Ceresoli GL, La Spina CM, Wannesson L. MicroRNAs in non-small cell lung cancer: current status and future therapeutic promises. Curr Pharm Des. 2014;20(24):3982–3990. doi: 10.2174/13816128113196660755. [DOI] [PubMed] [Google Scholar]
- 7.Jakopovic M, Thomas A, Balasubramaniam S, Schrump D, Giaccone G, Bates SE. Targeting the epigenome in lung cancer: expanding approaches to epigenetic therapy. Front Oncol. 2013;3:261. doi: 10.3389/fonc.2013.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 9.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 11.Loktionov A. Common gene polymorphisms, cancer progression and prognosis. Cancer Lett. 2004;208(1):1–33. doi: 10.1016/j.canlet.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69(4):1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon KA, Yoon H, Park S, et al. The prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non-small cell lung cancer. J Thorac Cardiovasc Surg. 2012;144(4):794–807. doi: 10.1016/j.jtcvs.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 15.Volpe MV, Pham L, Lessin M, et al. Expression of Hoxb-5 during human lung development and in congenital lung malformations. Birth Defects Res A Clin Mol Teratol. 2003;67(8):550–556. doi: 10.1002/bdra.10086. [DOI] [PubMed] [Google Scholar]
- 16.Vinci S, Gelmini S, Pratesi N, et al. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin Chem Lab Med. 2011;49(12):2073–2080. doi: 10.1515/CCLM.2011.708. [DOI] [PubMed] [Google Scholar]
- 17.Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 18.Hong MJ, Choi YY, Jang JA, et al. Association between genetic variants in pre-microRNAs and survival of early-stage NSCLC. J Thorac Oncol. 2013;8(6):703–710. doi: 10.1097/JTO.0b013e318288dc0a. [DOI] [PubMed] [Google Scholar]
- 19.Yuan Z, Zeng X, Yang D, Wang W, Liu Z. Effects of common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer risk. PLoS One. 2013;8(4):e61047. doi: 10.1371/journal.pone.0061047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parlayan C, Ikeda S, Sato N, Sawabe M, Muramatsu M, Arai T. Association analysis of single nucleotide polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer in elderly Japanese: a case-control study. Asian Pac J Cancer Prev. 2014;15(5):2101–2107. doi: 10.7314/apjcp.2014.15.5.2101. [DOI] [PubMed] [Google Scholar]
- 21.Yin Z, Cui Z, Ren Y, et al. Association between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female population. Lung Cancer. 2016;94:15–21. doi: 10.1016/j.lungcan.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Shang W, Pang M, Liu Y. High resolution melting analysis of the correlation of the miRNA-196a2 gene polymorphism and lung cancer. BMU J. 2016;(2):85–88. Chinese. [Google Scholar]
- 23.Yin Z, Cui Z, Guan P, et al. Interaction between polymorphisms in Pre-MiRNA genes and cooking oil fume exposure on the risk of lung cancer in Chinese non-smoking female population. PLoS One. 2015;10(6):e0128572. doi: 10.1371/journal.pone.0128572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong YS, Kang HJ, Kwak JY, et al. Association between microRNA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean population. J Prev Med Public Health. 2011;44(3):125–130. doi: 10.3961/jpmph.2011.44.3.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MJ, Yoo SS, Choi YY, Park JY. A functional polymorphism in the pre-microRNA-196a2 and the risk of lung cancer in a Korean population. Lung Cancer. 2010;69(1):127–129. doi: 10.1016/j.lungcan.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 27.Jia Y, Zang A, Shang Y, et al. MicroRNA-146a rs2910164 polymorphism is associated with susceptibility to non-small cell lung cancer in the Chinese population. Med Oncol. 2014;31(10):194. doi: 10.1007/s12032-014-0194-2. [DOI] [PubMed] [Google Scholar]
- 28.Jeon HS, Lee YH, Lee SY, et al. A common polymorphism in pre-microRNA-146a is associated with lung cancer risk in a Korean population. Gene. 2014;534(1):66–71. doi: 10.1016/j.gene.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Cheng C, Fu X, Alves P, Gerstein M. mRNA expression profiles show differential regulatory effects of microRNAs between estrogen receptor-positive and estrogen receptor-negative breast cancer. Genome Biol. 2009;10(9):R90. doi: 10.1186/gb-2009-10-9-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He F, Lin J, Yu T, et al. Interaction research on smoking and microRNA genes SNP related to lung cancer in Fujian Han population. Zhonghua Yu Fang Yi Xue Za Zhi. 2016;50(2):168–174. doi: 10.3760/cma.j.issn.0253-9624.2016.02.013. Chinese [with English abstract] [DOI] [PubMed] [Google Scholar]
- 31.Lelli D, Pedone C, Majeed M, Sahebkar A. Curcumin and lung cancer: the role of microRNAs. Curr Pharm Des. 2017;23(23):3440–3444. doi: 10.2174/1381612823666170109144818. [DOI] [PubMed] [Google Scholar]
- 32.Li T, Ding ZL, Zheng YL, Wang W. MiR-484 promotes non-small-cell lung cancer (NSCLC) progression through inhibiting Apaf-1 associated with the suppression of apoptosis. Biomed Pharmacother. 2017;96:153–164. doi: 10.1016/j.biopha.2017.09.102. [DOI] [PubMed] [Google Scholar]
- 33.Lu C, Xie Z, Peng Q. MiRNA-107 enhances chemosensitivity to paclitaxel by targeting antiapoptotic factor Bcl-w in non small cell lung cancer. Am J Cancer Res. 2017;7(9):1863–1873. eCollection 2017. [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez S, Risolino M, Mandia N, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34(25):3240–3250. doi: 10.1038/onc.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong C, Ding S, Xu Y, Huang H. MicroRNA-222 promotes human non-small cell lung cancer H460 growth by targeting p27. Int J Clin Exp Med. 2015;8(4):5534–5540. eCollection 2015. [PMC free article] [PubMed] [Google Scholar]
- 36.Bai X, Meng L, Sun H, Li Z, Zhang X, Hua S. MicroRNA-196b inhibits cell growth and metastasis of lung cancer cells by targeting runx2. Cell Physiol Biochem. 2017;43(2):757–767. doi: 10.1159/000481559. [DOI] [PubMed] [Google Scholar]
- 37.He B, Pan Y, Cho WC, et al. The association between four genetic variants in microRNAs (rs11614913, rs2910164, rs3746444, rs2292832) and cancer risk: evidence from published studies. PLoS One. 2012;7(11):e49032. doi: 10.1371/journal.pone.0049032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia L, Ren Y, Fang X, et al. Prognostic role of common microRNA polymorphisms in cancers: evidence from a meta-analysis. PLoS One. 2014;9(10):e106799. doi: 10.1371/journal.pone.0106799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F, Ma YL, Zhang P, et al. A genetic variant in microRNA-196a2 is associated with increased cancer risk: a meta-analysis. Mol Biol Rep. 2012;39(1):269–275. doi: 10.1007/s11033-011-0735-0. [DOI] [PubMed] [Google Scholar]
- 40.Cui JJ, Wang LY, Zhu T, et al. Gene-gene and gene-environment interactions influence platinum-based chemotherapy response and toxicity in non-small cell lung cancer patients. Sci Rep. 2017;7(1):5082. doi: 10.1038/s41598-017-05246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcus MW, Raji OY, Duffy SW, Young RP, Hopkins RJ, Field JK. Incorporating epistasis interaction of genetic susceptibility single nucleotide polymorphisms in a lung cancer risk prediction model. Int J Oncol. 2016;49(1):361–370. doi: 10.3892/ijo.2016.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin GT, Tseng HF, Yang CH, et al. Combinational polymorphisms of seven CXCL12-related genes are protective against breast cancer in Taiwan. OMICS. 2009;13(2):165–172. doi: 10.1089/omi.2008.0050. [DOI] [PubMed] [Google Scholar]
- 43.Yen CY, Liu SY, Chen CH, et al. Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med. 2008;37(5):271–277. doi: 10.1111/j.1600-0714.2007.00608.x. [DOI] [PubMed] [Google Scholar]