Abstract
Clinical isolates of influenza virus produce pleomorphic virus particles, including extremely long filamentous virions. In contrast, strains of influenza that have adapted to laboratory growth typically produce only spherical virions. As a result, the filamentous phenotype has been overlooked in most influenza virus research. Recent advances in imaging and improved animal models have highlighted the distinct structure and functional relevance of filamentous virions. In this review we summarise what is currently known about these strikingly elongated virus particles and discuss their possible roles in clinical infections.
Introduction
Influenza viruses are serious human and animal pathogens which have been intensively studied for decades. Despite such close attention, one of the most striking features of influenza infections is typically overlooked. Although influenza viruses are often described as producing spherical virions, natural infections are characterised by the additional presence of filaments: extremely elongated virions which can reach microns in length.
This oversight can be explained by the ease with which influenza viruses adapt to laboratory culture, a trait which has allowed so many other advances in their study. Passage in embryonated chicken eggs, which has been used to produce many commonly studied strains of influenza virus, rapidly selects against the production of filaments (Fig 1) (Choppin, 1963; Hayase et al., 1995; Seladi-Schulman et al., 2013). Filaments are also less physically robust during laboratory purification methods than spherical virions, further complicating their characterisation (Ada & Perry, 1958; Burnet & Lind, 1957; Roberts et al., 1998; Valentine & Isaacs, 1957; Vijayakrishnan et al., 2013). As a result, although influenza filaments have been recognised since 1946 (Mosley & Wyckoff, 1946), their study has until recently been sporadic.
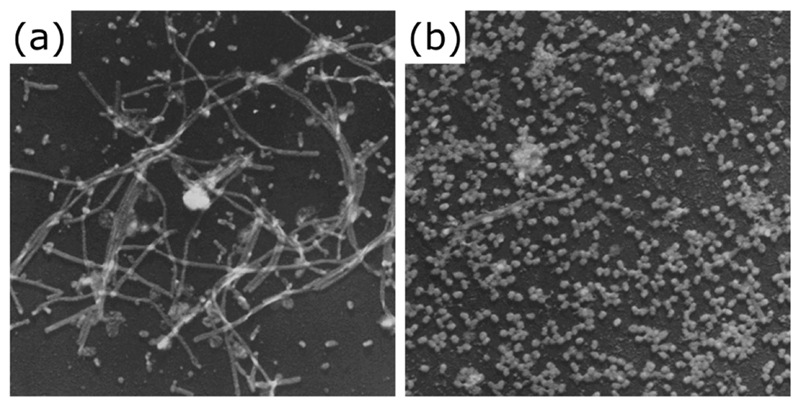
Figure 1. Filamentous influenza virions are lost on laboratory passage.
Filamentous influenza virions are clearly visible after two passages of the clinical isolate influenza A/Rockefeller Institute/1/1957 (H2N2) virus in embryonated chicken eggs (a) but are lost following twelve passages (b). Electron micrographs © Choppin et al., 1960.
Originally published in THE JOURNAL OF EXPERIMENTAL MEDICINE. 112:945-52.
It is now clear that mixtures of spherical and filamentous virions can be produced by influenza A, B and C viruses (Chu et al., 1949; Mosley & Wyckoff, 1946; Nishimura et al., 1990). Filament production has been repeatedly observed with low-passage clinical and veterinary influenza A virus isolates (Basu et al., 2012; Choppin et al., 1960; Chu et al., 1949; Elton et al., 2013; Hayase et al., 1995; Itoh et al., 2009; Kilbourne & Murphy, 1960; Lang et al., 1968; Seladi-Schulman et al., 2013; Shortridge et al., 1998) as well as in lung sections from a fatal human case (Nakajima et al., 2010). A similar mixture of filaments and spherical virions has been observed for other orthomyxoviruses: in a low-passage isolate from a fatal human thogotovirus infection (Kosoy et al., 2015) and for infectious salmon anaemia viruses in tissue cultures and the tissues of infected fish (Crane & Hyatt, 2011; Kibenge et al., 2001; Koren & Nylund, 1997). This suggests that the ability to produce filaments may be a general feature of the orthomyxovirus family.
Many observations of filament structure have been limited by the need to use electron microscopy (EM) methods such as negative staining, metal shadowing and ultrathin sectioning of resin embedded material. Although informative these depend on heavy metal contrasting agents, and often chemical fixation, and are therefore prone to artefacts including sample deformation and shrinkage. Following the development of cryo EM, it has been possible to determine the structure of filaments to higher resolution in a close-to-native environment without sample preparation artefacts (Calder et al., 2010; Vijayakrishnan et al., 2013; Wasilewski et al., 2012). This has shown that filaments have a distinctive and highly ordered ultrastructure.
The importance of filaments in natural infections is highlighted by recent experimental studies with influenza A viruses in animal models. These showed that despite being selected against in egg passage, filament production is selected for during serial intranasal passage of a highly laboratory-adapted spherical strain in guinea pigs (Seladi-Schulman et al., 2013). Furthermore, filament formation correlates with transmissibility between co-housed guinea pigs and by respiratory droplets in ferrets (Campbell et al., 2014a; Lakdawala et al., 2011). Taken together with the many observations of recently-isolated strains producing filaments, these data indicate that the filamentous phenotype is an important but neglected feature of natural influenza infections.
In this review we summarise seven decades of work on influenza virus filaments, with a particular emphasis on recent structural and molecular biology studies. We also discuss possible functions of this often neglected trait in the virus lifecycle.
Filament Structure
Influenza infections do not produce virions of a single, well-defined size. However, the virions produced by laboratory-adapted ‘spherical’ influenza viruses have broadly consistent dimensions. The majority (typically 65-75%) are spherical (axial ratio < 1.2), with a mean outer diameter of 120 nm (Harris et al., 2006; Yamaguchi et al., 2008). Irregularly-shaped virions are often observed (Almeida & Waterson, 1967a; b; Harris et al., 2006; Ruigrok et al., 1986; Stevenson & Biddle, 1966; Wrigley, 1979), but it appears that many of these result from damage during ultracentrifugation, storage and sample preparation for electron microscopy (Noda, 2011; Sugita et al., 2011). ‘Spherical’ strains also produce a minority of well-preserved but elongated virions, which for the most part are still less than 250 nm in length – too short to be described as filaments (Calder et al., 2010; Harris et al., 2006; Wasilewski et al., 2012; Yamaguchi et al., 2008). These intermediate-length virions, which often appear to be ellipsoidal, capsular or kidney-bean shaped, have been described as bacilliform (Vijayakrishnan et al., 2013).
Influenza viruses that retain their natural morphology produce not only spherical and bacilliform virions, but also a class of highly elongated virions, or filaments (Figs 1, 2) (Ada et al., 1958; Calder et al., 2010; Roberts et al., 1998; Vijayakrishnan et al., 2013). These striking structures are typically more than 250 nm in length and can reach many microns (Fig 3). Filaments reaching or exceeding 30 μm in length have been reported (Cox et al., 1980; Roberts et al., 1998), though their exact range of size is hard to determine as they are fragile (Burnet & Lind, 1957; Valentine & Isaacs, 1957), comparatively hard to purify (Ada et al., 1958; Sugita et al., 2011), prone to aggregation (Cox et al., 1980) and are hard to capture complete when cutting thin sections for transmission EM. The proportion of filaments in a given sample varies widely and depends on the virus strain used, tissues infected and the handling of virions (Bourmakina & Garcia-Sastre, 2003; Rossman et al., 2012; Seladi-Schulman et al., 2013; Vijayakrishnan et al., 2013).
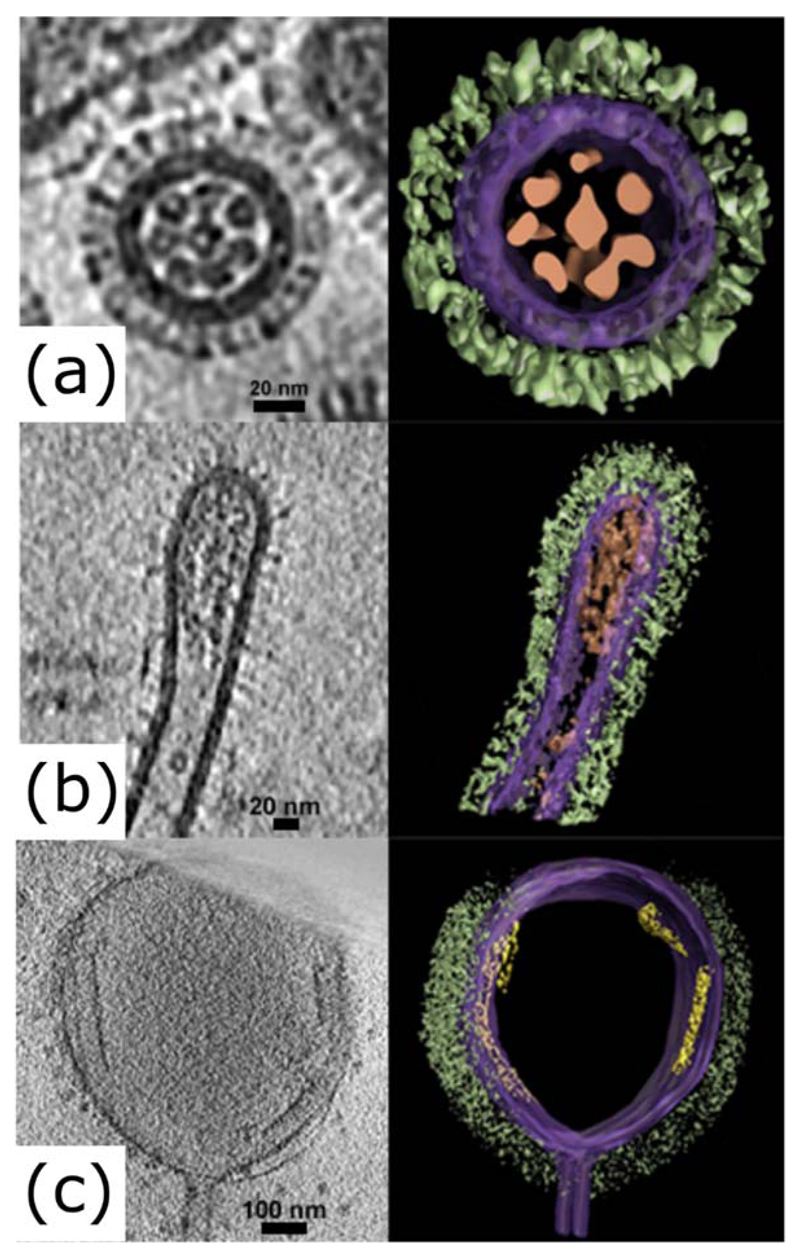
Figure 2. Bacilliform and filamentous influenza virions at high resolution.
Electron tomograms of influenza virions, showing slices (left panels) and segmented images (right panels) of (a) a transverse section of a bacilliform virion, (b) a longitudinal section of the tip of a filamentous virion and (c) a longitudinal section of an Archetti body at the end of a filamentous virion. Images were manually segmented and coloured to show viral glycoproteins (green), membrane and associated matrix (purple), genome (brown) and putative free M1 sheets (yellow). Tomograms were obtained as part of a previous study (Vijayakrishnan et al., 2013) and manually segmented using Amira (TGS).
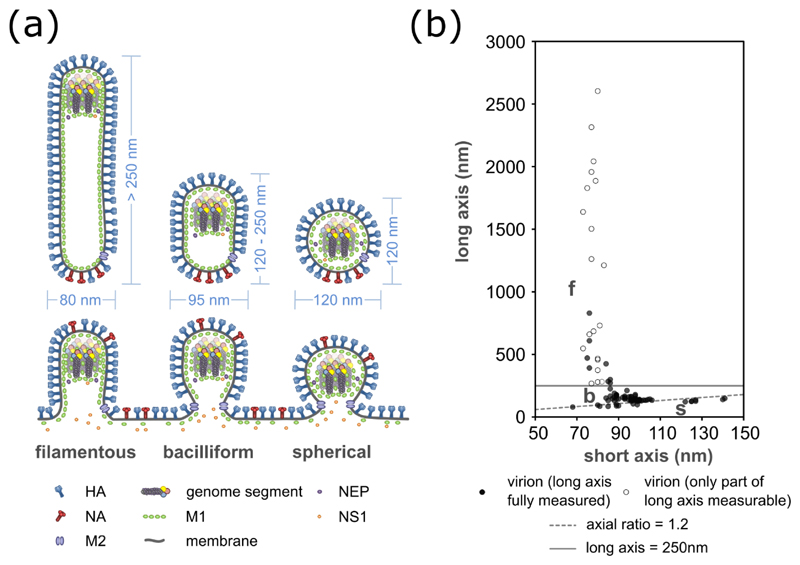
Figure 3. Dimensions of influenza virions.
The dimensions of influenza virions, shown (a) as a schematic of budding and released virions, with typical sizes indicated, and (b) as measurements of purified influenza A/Udorn/72 virions. For (a) it should be noted that the incorporation of NS1 and NEP has so far only been examined in spherical virions, and their general incorporation is inferred from this. For (b) measurements of 96 virions were taken by cryoelectron microscopy (data replotted from Vijayakrishnan et al., 2013). Open circles indicate filaments which extended beyond the field of view and so are longer than measured. Spherical virions (s) are distinguished from bacilliform virions (b) by having an axial ratio less than 1.2 (dashed line); filaments (f) are distinguished from bacilliform virions by having a length greater than 250 nm (solid line).
During the budding of filamentous influenza A and C viruses cord-like associations of multiple filaments have often been observed (Beale et al., 2014; Bialas et al., 2014; Bruce et al., 2010; Elton et al., 2013; Morgan et al., 1956; Muraki et al., 2007; Muraki et al., 2004; Nishimura et al., 1990; Simpson-Holley et al., 2002). End-to-end association of filaments has also been reported (Calder et al., 2010; Vijayakrishnan et al., 2013), though it is unclear if this is due to separate filaments associating, concatemers of filaments arising from incomplete budding, or a single filament fragmenting.
Filaments can be distinguished from spherical virions not just by their great length, but because their width, around 80 nm, is less than that of spherical virions (Fig 3a; Morgan et al., 1956; Vijayakrishnan et al., 2013). Infectious salmon anaemia virus, another orthomyxovirus, also produces narrower filamentous and wider spherical virions (Koren & Nylund, 1997). Bacilliform virions have an intermediate width of around 95 nm (Calder et al., 2010; Harris et al., 2006; Vijayakrishnan et al., 2013; Wasilewski et al., 2012; Yamaguchi et al., 2008), and show an inverse correlation between length and diameter (Fig 3b) (Vijayakrishnan et al., 2013). Particles can therefore be categorised based on their axial ratio (< 1.2 for spherical virions, > 1.2 for bacilliform virions and filaments) and length (> 250 nm for filaments; Fig 3). Particle dimensions do not provide sharp distinctions between these categories but they are useful, as closer examination shows that each category of virion has a characteristic composition and structure.
Filament Composition
Viral Components
Haemagglutinin (HA) and neuraminidase (NA) are the two major viral glycoproteins present in the envelope of influenza virus. HA binds to sialic acid, the viral receptor, and is required for entry of the virus into the host cell. NA mediates the release of viral progeny from the cell by cleaving sialic acid from cell surface proteins. The glycoproteins have a characteristic fringe-like appearance in electron micrographs of virions (Fig 2) and tomography shows that both spherical virions and filaments incorporate an abundance of HA along with smaller quantities of NA (Calder et al., 2010; Harris et al., 2006). NA forms clusters, which in bacilliform and filamentous virions tend to be at the pole proximal to the budding site on the host cell surface, at the opposite end of the virion to the viral genome (Fig 3a; Calder et al., 2010; Chlanda et al., 2015; Harris et al., 2006; Murti & Webster, 1986; Wasilewski et al., 2012). It is possible that this clustering of NA may play a role in the formation of cord-like bundles of budding filaments – with NA sequestered at the poles, sufficient sialic acid may remain attached to surface proteins along the length of the filaments to allow HA on adjacent filaments to bind. The viral glycoproteins appear to be more regularly distributed on filaments than on spherical virions, suggesting interactions with a more ordered matrix layer beneath (Wasilewski et al., 2012).
The matrix layer, which is bound to the internal surface of the viral membrane, is made of the M1 protein. M1 multimerises to form a helical matrix, the organisation of which appears to influence virion morphology (Calder et al., 2010). Multimerised M1 can form lattices with a range of curvatures: along the length of filaments it forms a rigid cylindrical helix, whereas in spherical particles and at the poles of filaments it appears to form a less-ordered spherical spiral (Calder et al., 2010). The poles of filaments sometimes form enlarged oval structures. Where these enlarged structures have a diameter greater than 200 nm they are termed Archetti bodies (Fig 2c) (Archetti, 1955; Vijayakrishnan et al., 2013). Archetti bodies retain a contiguous matrix layer and can sometimes contain coils of M1-like material that are not membrane-associated (Vijayakrishnan et al., 2013). Similar coils are observed within filaments as their structure transforms and becomes disorganised at low pH (Calder et al., 2010), a change which mimics the fragmentation of filaments in acidifying endosomes during viral entry (Rossman et al., 2012), suggesting that Archetti bodies may arise from a partial breakdown of filament structure.
The genome of influenza viruses consists of segments of viral RNA bound to the viral polymerase proteins (PB2, PB1 and PA/P3) and nucleoprotein (NP). It can be clearly visualised in spherical virions as a complex of rod-shaped segments, the longest spanning the internal diameter of the virion (Fig 3a; Calder et al., 2010; Noda et al., 2006). Studies of mutant viruses suggest that genome packaging is not strictly necessary for virion assembly, although it can make assembly more efficient (Gavazzi et al., 2013; Hutchinson et al., 2008), and in practice most virions do not have a full complement of functionally active genome segments (Brooke et al., 2014; Brooke et al., 2013; Heldt et al., 2015). Images of the genome have been obtained in bacilliform virions (Fig 2a) and occasionally in filaments, particularly shorter ones (Fig 2b) (Calder et al., 2010; Noda et al., 2006; Vijayakrishnan et al., 2013; Wasilewski et al., 2012). In such cases, the viral genome appears to remain associated with one pole of the virion (Calder et al., 2010; Vijayakrishnan et al., 2013; Wasilewski et al., 2012) – the distal tip when virions bud from the cell membrane (Fig 3a; Noda et al., 2006). The rod-shaped genome segments associate with the virion pole through their tips, and in filaments they appear to remain closely associated in a parallel array (Calder et al., 2010; Vijayakrishnan et al., 2013). In spherical virions this ordered clustering of genome segments can also be observed, though disordered arrangements of the genome appear to be more common (Harris et al., 2006).
At present the efficiency with which filaments package the viral genome is unclear. Early observations suggested that filaments might incorporate more viral genome, and be more infectious, than spherical virions (Ada & Perry, 1958; Ada et al., 1958; Ada et al., 1957; Burleigh et al., 2005; Roberts et al., 1998), a hypothesis consistent with the greater resistance of filament-containing stocks to ultraviolet inactivation (Smirnov et al., 1991), and recalling the polyploid filamentous virions of Ebola virus (Beniac et al., 2012). However, these observations could also be explained by the tendency of multiple filaments to form cord-like bundles. Other negative data do not support the hypothesis that filaments can package multiple copies of the genome: clear images of the genome have only been obtained in a minority of longer filaments, multiple copies of the genome have not been clearly visualised within a single virion (Morgan et al., 1956; Vijayakrishnan et al., 2013), and fragmentation of filaments using a number of methods does not increase the infectious titre (Ada & Perry, 1958; Burnet & Lind, 1957; Donald & Isaacs, 1954; Valentine & Isaacs, 1957).
The ion channel M2 has not been detected by immunofluorescence in filaments, suggesting that it is not an abundant component (Rossman et al., 2010a). However, its presence can be inferred as an M2-binding antibody causes filaments to fragment, while an M2 inhibitor allows filaments to resist fragmentation at low pH (Rossman et al., 2010a; Rossman et al., 2012). NS1 and NEP are known to be present at low levels in spherical virions (Hutchinson et al., 2014), but their presence in filaments has not been assessed.
Host Components
All influenza virions incorporate membrane from the host cell. As with spherical virions, the envelopes of filaments are resistant to low-temperature nonionic detergent extraction and contain material with a low buoyant density, implying the incorporation of lipid rafts (Simpson-Holley et al., 2002). Cholesterol also appears to be important for filament stability (Rossman et al., 2010a). Spherical virions incorporate a substantial quantity of host-encoded proteins, resembling those incorporated into exosomes (Hutchinson et al., 2014; Shaw et al., 2008). It is reasonable to assume that filaments also incorporate such host proteins – quite possibly more than spherical virions, due to their larger membrane area and internal volume – but this has not been assessed in detail. Fibrillar material, which does not have the appearance of the viral genome, has been observed inside filamentous particles but its identity is still unclear (Vijayakrishnan et al., 2013).
Filament Formation
General Requirements for Virion Formation
All influenza virions are formed at the cell surface in a concerted process which requires both viral and host components (Fig 3a; Hutchinson & Fodor, 2013; Noda et al., 2006; Rossman & Lamb, 2011). Viral glycoproteins accumulate at the apical plasma membrane and are individually sufficient to cause budding when over-expressed (Chen et al., 2007; Chlanda et al., 2015). M1 is neither necessary nor sufficient for the production of virus-like particles, but it does appear to be required for the production of infectious virions (Chen et al., 2007). This may be due to its interactions with genome segments: genome packaging increases the efficiency of budding, though the importance of this appears to depend on cell type (Hutchinson et al., 2008). Finally, abscission of the budded virion is mediated by M2 in an ESCRT-independent process (Rossman et al., 2010b). While defects in normal virion assembly can produce irregular virions which superficially resemble filaments – for example the elongated and distended virions resulting from mutations in the cytoplasmic tails of NA or HA (Jin et al., 1997; Mitnaul et al., 1996) or the beads-on-string structures due to M2 scission mutants (Rossman et al., 2010b) – ‘well-formed’ filaments appear to result from a process which involves a number of host and viral determinants.
Host Determinants of Filament Formation
Virions require host processes to assemble and interfering with these processes can impair filament formation. Drugs targeting the actin cytoskeleton, depletion of Rab11-family interacting protein 3 (FIP3, which regulates actin dynamics and membrane trafficking) and cholesterol depletion have all been shown to specifically reduce filament formation, while depletion of or mutation of Rab11 (a GTPase involved in endocytic recycling) affects both spherical and filamentous particle production (Bruce et al., 2010; Roberts et al., 1998; Rossman et al., 2010a; Simpson-Holley et al., 2002). It has been posited that all of these processes affect the supply of lipid-raft enriched membrane required to form the extensive surfaces of filaments. In addition, mutation of an LC3-interacting region (LIR) in the viral M2 protein, a motif which allows interaction with autophagosomal membranes via LC3, or depletion of ATG16L1, which is required for LC3 activation, reduces filament formation and decreases the stability of filamentous virions. This suggests that autophagosomal membranes are recruited to support filament formation (Beale et al., 2014).
Even without active intervention, some cell lines are less permissive to filament formation than others. The same strain of virus can have different morphologies in different cell lines (Al-Mubarak et al., 2015; Bialas et al., 2012; Itoh et al., 2009; Lakdawala et al., 2011), though whether a particular cell line is permissive for filaments can vary between different strains of the virus (Al-Mubarak et al., 2015). Generally, polarised cell types are more permissive to filament formation compared to nonpolarised cells, consistent with a role for the cytoskeleton in determining viral morphology (Roberts et al., 1998).
Viral determinants of filament formation
Filament formation is a heritable trait. Spherical virions purified from filament-forming stocks can form filamentous progeny (Chu et al., 1949) and the selection against filament formation during passage in embryonated chicken eggs can be slowed by passaging only the minimal amount of virus necessary for an infection, thereby excluding low-frequency mutants from the stock (Burnet & Lind, 1957). Although viral genes clearly contribute to filament formation the trait is complex. There appear to be multiple pathways to filament formation, with some loci relevant only in particular genetic backgrounds while others suppress filament formation in genotypes which would normally support it. A summary of loci in influenza A virus genes known to affect filament formation is given in Table 1.
Table 1.
Influenza A virus mutations known to influence filament production.
| Segment (gene) | Residues | Phenotype | Context | Reference |
|---|---|---|---|---|
| 7 (M1) | R95K E204D | Reduces filament formation | WSN with Ud M1 gene; residues changed to match WSN. | (Bourmakina & Garcia-Sastre, 2003) |
| A41V | Reduces filament formation | Ud, selected for resistance to an anti-M2 antibody. | (Roberts et al., 1998) | |
| P41A | Reduces filament length | SNP04 and PR8:SPN04 reassortants | (Campbell et al., 2014b) | |
| V41A+K95R+T2 18A | Confers filamentous morphology | Vic with WSN M1 gene; residues changed to match Ud. | (Elleman & Barclay, 2004) | |
| S85N N231D | Confers filamentous morphology | PR8 with Miami M1 gene; residues changed to match Nkt. | (Elton et al., 2013) | |
| K102A | Confers filamentous morphology | WSN | (Burleigh et al., 2005) | |
| 7 (M2) | S71A+M72A+R7 3A | Reduces filament formation | Ud | (Rossman et al., 2012) |
| 5 (NP) | R214K and I217S F253I | Reduces filament formation | WSN with Aichi M1 gene; NP residues changed to match Aichi. | (Bialas et al., 2014) |
Morphology (filamentous or spherical) is as originally defined in the cited studies.
Abbreviations: Aichi: A/Aichi/2/68; Miami: A/equine/Miami/63; Nkt: A/equine/Newmarket/11/03; PR8: A/PR8/34; SPN04: A/swine/Spain/53207/2004; Udorn: A/Udorn/72; Vic: A/Victoria/3/75; WSN: A/WSN/33.
Unsurprisingly, considering its role in maintaining virion structure (Calder et al., 2010), the majority of mutations affecting filament formation have been mapped to M1. Reverse-genetic studies using reassortant viruses showed that the M1 gene of influenza A/Udorn/301/72 virus (Udorn), one of the few strains to retain filament-forming ability after laboratory passage (Roberts et al., 1998), can confer filament-forming ability on spherical strains such as the influenza A/WSN/33 (WSN) or A/Puerto Rico/8/1934 (PR8) viruses (Bourmakina & Garcia-Sastre, 2003; Noton et al., 2007). Reciprocally, the M1 gene from the spherical WSN strain abrogated filament production by the normally filament-forming influenza A/Victoria/3/75 virus (Elleman & Barclay, 2004). While most studies of filament formation have considered influenza A viruses, a key role for M1 has also been identified in an influenza C virus (Muraki et al., 2007).
To date, mapping filament determinants in M1 has not produced a clear mechanistic model of filament formation. Difficulties in interpretation arise from the multiple structural roles of M1, which include membrane binding, homo-oligomerisation and interactions with other viral proteins (Burleigh et al., 2005), as well as from the overlap of the M1 gene with other viral genes, notably for the ion channel M2. For example M1 residue 41 was one of the first residues to be experimentally associated with filament formation (Campbell et al., 2014b; Roberts et al., 1998; Zebedee & Lamb, 1989); it is also known to have mutated during adaptation of the WSN strain, which is now spherical, to mouse brain passage (Ward, 1995). However, mutations affecting this position have also been shown to create a splice-variant form of the M2 protein, whose role in morphology remains to be determined (Wise et al., 2012).
One mechanism underlying filament formation can be inferred from studies of helix six, a basic alpha helix in M1. Scanning alanine mutagenesis shows that a number of residues in this region are required for the production of regularly-shaped virions and one mutation, M1 K102A, confers filament-forming abilities on spherical viruses (Burleigh et al., 2005). This mutation, which is adjacent to a proposed M1-M1 interaction site (Harris et al., 2001), causes M1 in virions to form a helix with similar symmetry to that observed in the filamentous Udorn strain, emphasising the importance of an ordered M1 helix in maintaining filament structure (Calder et al., 2010).
Other loci influencing filament formation have been mapped to the HA, NP, NA and M2 proteins (Table 1). Most strikingly, an appropriate NA can enhance filament formation in a laboratory-adapted virus and even induce limited filament formation when overexpressed alone (Campbell et al., 2014a; Chlanda et al., 2015). The influence of these proteins on filament formation has generally been attributed to altered interactions with M1 (Bialas et al., 2014; Chen et al., 2008; Liu et al., 2002) or by their influence on processes upstream of virion assembly. For example, it has been suggested that mutations in M2 and NA influence filament production by altering the recruitment of lipid rafts required for the virion membrane (Enami & Enami, 1996; Jin et al., 1997; Mitnaul et al., 1996; Rossman et al., 2010a; Zhang et al., 2000).
Filament Function
Decades of observational work, and more recent experimental studies in animal transmission models (Campbell et al., 2014a; Lakdawala et al., 2011; Seladi-Schulman et al., 2013), clearly show that filament-forming viruses have a selective advantage in natural influenza infections. While it is possible that filament production itself is a ‘spandrel’ – a conspicuous by-product of some underlying trait (Gould & Lewontin, 1979) – the more obvious explanation is that filaments act directly to increase viral fitness in their natural hosts. However, the particular functions of filaments have been difficult to determine due to the difficulty in completely separating spherical and bacilliform virions from filaments during analysis, and as filaments do not typically provide an advantage in embryonated eggs or in the tissue culture systems most suited to detailed functional studies. Despite these difficulties, a number of properties of filaments have been identified which suggest functional roles.
The Costs of Making Filaments
Filamentous strains have a clear selective disadvantage in embryonated chicken eggs (Seladi-Schulman et al., 2013). This may be due to the specific constraints of egg passage. For example, virions grown in eggs incorporate a different profile of host proteins to those grown in mammalian cells, notably by utilising different members of the tetraspanin family of membrane proteins (Hutchinson et al., 2014). It is possible that these egg-specific proteins, which are relatively abundant in virions, may increase the costs of filament production.
Alternatively, it may be that filaments have intrinsic costs which are not compensated for during laboratory passage. This also applies to passage in tissue cultures, in which the rate at which filamentous strains replicate varies, but does not typically exceed that of spherical strains. Filaments require a greater amount of membrane and viral proteins to form each virion and to the route of filament entry is different to that of spherical virions. Although filaments have receptor binding activity (Ada et al., 1958; Burnet & Lind, 1957; Chu et al., 1949; Donald & Isaacs, 1954; Seladi-Schulman et al., 2014), they are too large to enter cells through the canonical clathrin-mediated endocytic pathway that can take up spherical and bacilliform virions. Instead, filaments undergo delayed uptake through macropinocytosis, and break apart into smaller fragments as endosomal acidification triggers conformational changes in the virion (Rossman et al., 2012; Sieczkarski & Whittaker, 2005). The comparative efficiency of this process is unclear. While these issues may account for some of the fitness cost of filaments in laboratory culture, the structure of filaments suggests two advantages which may overcome these costs during a natural infection.
Filaments May Be More Robustly Infectious
Firstly, some studies suggest that filaments may have higher specific infectivities than spherical virions (Ada & Perry, 1958; Ada et al., 1958; Ada et al., 1957; Burleigh et al., 2005; Roberts et al., 1998). As discussed above, the implication that this is due to packaging multiple genomes is contentious, but the same effect could also be achieved by the frequently-observed association of individual filaments into higher-order cord-like structures (Beale et al., 2014; Bialas et al., 2014; Bruce et al., 2010; Elton et al., 2013; Morgan et al., 1956; Muraki et al., 2007; Muraki et al., 2004; Nishimura et al., 1990; Simpson-Holley et al., 2002). Physically associating multiple genomes, in a single particle or a cluster of particles, is unlikely to be advantageous in the high-multiplicity infections that characterise laboratory growth and (presumably) foci of infection within the host. However, it would be expected to increase the efficiency of low-multiplicity infections during the spread of viruses within and between hosts, given that virions typically lack at least one functional gene segment (Brooke et al., 2013; Heldt et al., 2015). Redundant copies of the genome may also provide some resistance to ultraviolet inactivation during between-host passage (Smirnov et al., 1991). Packaging additional genomes which lack a full complement of functional segments even raises the intriguing possibility of variable gene dosage for each segment within the context of a high-multiplicity infection (Brooke et al., 2014).
Secondly, it has recently been noted that influenza genomes can pass directly between cells though an actin-dependent, NA-independent process without packaging into virions (Mori et al., 2011; Roberts et al., 2015). Although this has not been demonstrated for filamentous influenza viruses, it is plausible that filaments could enhance cell-associated spread and provide an advantage in natural infections, for example by evading mucociliary clearance. However, direct transmission of a filamentous influenza virus has not been demonstrated.
Filaments May be an Adaptation to Spread Through Mucus
Influenza virions bind to sialic acid, which is cleaved by the viral NA. This neuraminidase activity is required during the initiation and within-host spread of an infection, to prevent virions being sequestered by sialic acid moieties on the mucins in respiratory mucus, as well as on the surfaces of infected cells when new virions are produced (Chlanda et al., 2015; Cohen et al., 2013; Matrosovich et al., 2004; Yang et al., 2014). Filaments appear to have an elevated NA activity, which would potentially enhance infectivity in natural hosts (Campbell et al., 2014a; Campbell et al., 2014b; Seladi-Schulman et al., 2014). Surprisingly, this activity is increased by mutations in M1 that allow filament formation, even when NA itself is unaltered (Campbell et al., 2014a; Campbell et al., 2014b; Seladi-Schulman et al., 2014). It is unclear whether this is due to the clustering of NA at the tips of filaments (Calder et al., 2010; Chlanda et al., 2015) or to the abundance of NA in filaments, which has not been determined.
There are only limited data available on the relative stability of filaments and spherical virions within mucus. Filaments do appear to be fragile during laboratory manipulations, but despite this they appear to be no more susceptible to heat inactivation than spherical strains (Beale et al., 2014; Seladi-Schulman et al., 2014). They can be bound by antibodies (Chu et al., 1949), but they appear to be no more susceptible to neutralisation than spherical virions in in vitro assays (Seladi-Schulman et al., 2014). Conversely, it has been suggested that non-infectious filaments may serve as an immune decoy by sequestering IgA antibodies away from smaller particles during infection (Vijayakrishnan et al., 2013).
Finally, as well as being a barrier to infection and within-host spread, mucus is ultimately the vehicle for between-host spread. Longer filaments would be able to extend through the low-viscosity periciliary layer, which coats the airway epithelium to a depth of around 7 μm (Button et al., 2012; Fahy & Dickey, 2010), potentially providing more efficient access to the gel-like airway mucus beyond and increasing the likelihood of their respiratory transmission. Respiratory transmission of influenza genomes in ferrets occurs most efficiently in large (> 4 μm) droplets of mucus. While the same distribution between droplet sizes is observed for both filamentous and spherical strains, it is notable that these droplets are large enough to contain intact filaments (Lakdawala et al., 2011).
Conclusion
Influenza viruses naturally exhibit a range of morphologies from small spherical particles to extremely long filamentous structures. It appears that filament production may be a common feature of orthomyxoviruses as it has been observed in influenza A, B and C viruses as well as in thogotovirus and infectious salmon anaemia virus. It is also notable that a number of unrelated respiratory viruses can form filamentous virions, including respiratory syncytial virus and certain paramyxoviruses (Compans et al., 1966; Liljeroos et al., 2013; Shaikh et al., 2012; Yao & Compans, 2000), although other respiratory viruses form exclusively spherical virions.
For influenza viruses filament formation is a heritable trait which is selected for in natural transmission. Despite this, long filaments have often been neglected in laboratory studies, and the reason for their production remains uncertain. Recent technical advances have allowed the structure of filaments to be analysed in detail and the conditions which select for them to be studied in a laboratory setting. This has strengthened the case for filaments as a class of well-formed viral structures which provide functional benefits during natural influenza infections. What these benefits are though remains uncertain, and in clarifying them we will need to fundamentally revise our models of how influenza virus genomes are transmitted outside of a laboratory setting.
Acknowledgements
We thank Dr Jeremy Rossman (University of Kent) for helpful comments on a draft of this manuscript. BD is funded by a Wellcome Trust studentship [105399/Z/14/Z]; EF is funded by an MRC programme grant [MR/K000241/1]; SV, DB and ECH are funded by core MRC funding to the MRC-University of Glasgow Centre for Virus Research and ECH is funded by an MRC Career Development Award [MR/N008618/1]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The final version of record is available at http://jgv.microbiologyresearch.org/content/journal/jgv/10.1099/jgv.0.000535#tab2
References
- Ada GL, Perry BT. Properties of the nucleic acid of the Ryan strain of filamentous influenza virus. Journal of general microbiology. 1958;19:40–54. doi: 10.1099/00221287-19-1-40. [DOI] [PubMed] [Google Scholar]
- Ada GL, Perry BT, Abbot A. Biological and physical properties of the Ryan strain of filamentous influenza virus. J Gen Microbiol. 1958;19:23–39. doi: 10.1099/00221287-19-1-23. [DOI] [PubMed] [Google Scholar]
- Ada GL, Perry BT, Edney M. Infectivity of influenza virus filaments. Nature. 1957;180:1134. doi: 10.1038/1801134a0. [DOI] [PubMed] [Google Scholar]
- Al-Mubarak F, Daly J, Christie D, Fountain D, Dunham SP. Identification of morphological differences between avian influenza A viruses grown in chicken and duck cells. Virus research. 2015 doi: 10.1016/j.virusres.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Almeida JD, Waterson AP. A morphological comparison of Bittner and influenza viruses. The Journal of hygiene. 1967a;65:467–474. doi: 10.1017/s0022172400046003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida JD, Waterson AP. Some observations on the envelope of an influenza virus. J Gen Microbiol. 1967b;46:107–110. doi: 10.1099/00221287-46-1-107. [DOI] [PubMed] [Google Scholar]
- Archetti I. Appearances associated with filamentous forms of influenza viruses. Archives of Virology. 1955;6:29–35. doi: 10.1007/BF01242050. [DOI] [PubMed] [Google Scholar]
- Basu A, Chadha M, Potdar V, Ganti K, Gangodkar S. Electron Tomography Imaging of the Pandemic H1N1 2009 Influenza Virus. Journal of Advanced Microscopy Research. 2012;7:7–13. [Google Scholar]
- Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe. 2014;15:239–247. doi: 10.1016/j.chom.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac DR, Melito PL, Devarennes SL, Hiebert SL, Rabb MJ, Lamboo LL, Jones SM, Booth TF. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PloS one. 2012;7:e29608. doi: 10.1371/journal.pone.0029608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas KM, Bussey KA, Stone RL, Takimoto T. Specific nucleoprotein residues affect influenza virus morphology. Journal of virology. 2014;88:2227–2234. doi: 10.1128/JVI.03354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas KM, Desmet EA, Takimoto T. Specific residues in the 2009 H1N1 swine-origin influenza matrix protein influence virion morphology and efficiency of viral spread in vitro. PloS one. 2012;7:e50595. doi: 10.1371/journal.pone.0050595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourmakina SV, Garcia-Sastre A. Reverse genetics studies on the filamentous morphology of influenza A virus. The Journal of general virology. 2003;84:517–527. doi: 10.1099/vir.0.18803-0. [DOI] [PubMed] [Google Scholar]
- Brooke CB, Ince WL, Wei J, Bennink JR, Yewdell JW. Influenza A virus nucleoprotein selectively decreases neuraminidase gene-segment packaging while enhancing viral fitness and transmissibility. Proc Natl Acad Sci U S A. 2014;111:16854–16859. doi: 10.1073/pnas.1415396111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke CB, Ince WL, Wrammert J, Ahmed R, Wilson PC, Bennink JR, Yewdell JW. Most influenza a virions fail to express at least one essential viral protein. J Virol. 2013;87:3155–3162. doi: 10.1128/JVI.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Digard P, Stuart AD. The Rab11 pathway is required for influenza A virus budding and filament formation. Journal of virology. 2010;84:5848–5859. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh LM, Calder LJ, Skehel JJ, Steinhauer DA. Influenza a viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes. Journal of virology. 2005;79:1262–1270. doi: 10.1128/JVI.79.2.1262-1270.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnet FM, Lind PE. Studies on filamentary forms of influenza virus with special reference to the use of dark-ground-microscopy. Archiv fur die gesamte Virusforschung. 1957;7:413–428. doi: 10.1007/BF01241959. [DOI] [PubMed] [Google Scholar]
- Button B, Cai LH, Ehre C, Kesimer M, Hill DB, Sheehan JK, Boucher RC, Rubinstein M. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science. 2012;337:937–941. doi: 10.1126/science.1223012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Danzy S, Kyriakis CS, Deymier MJ, Lowen AC, Steel J. The M segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to A/Puerto Rico/8/1934-based reassortant viruses. Journal of virology. 2014a;88:3802–3814. doi: 10.1128/JVI.03607-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Kyriakis CS, Marshall N, Suppiah S, Seladi-Schulman J, Danzy S, Lowen AC, Steel J. Residue 41 of the Eurasian avian-like swine influenza a virus matrix protein modulates virion filament length and efficiency of contact transmission. J Virol. 2014b;88:7569–7577. doi: 10.1128/JVI.00119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. Journal of virology. 2008;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. Journal of virology. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlanda P, Schraidt O, Kummer S, Riches J, Oberwinkler H, Prinz S, Krausslich HG, Briggs JA. Structural analysis of the roles of influenza A virus membrane-associated proteins in assembly and morphology. Journal of virology. 2015 doi: 10.1128/JVI.00592-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choppin PW. On the emergence of influenza virus filaments from host cells. Virology. 1963;21:278–281. doi: 10.1016/0042-6822(63)90273-3. [DOI] [PubMed] [Google Scholar]
- Choppin PW, Murphy JS, Tamm I. Studies of two kinds of virus particles which comprise influenza A2 virus strains. III. Morphological characteristics: independence to morphological and functional traits. J Exp Med. 1960;112:945–952. doi: 10.1084/jem.112.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choppin PW, Tamm I. Studies of Two Kinds of Virus Particles which Comprise Influenza A2 Virus Strains : I. Characterization of Stable Homogeneous Substrains in Reactions with Specific Antibody, Mucoprotein Inhibitors, and Erythrocytes. The Journal of experimental medicine. 1960;112:895–920. doi: 10.1084/jem.112.5.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CM, Dawson IM, Elford WJ. Filamentous forms associated with newly isolated influenza virus. The Lancet. 1949;253:602–603. doi: 10.1016/s0140-6736(49)91699-2. [DOI] [PubMed] [Google Scholar]
- Cohen M, Zhang X-Q, Senaati HP, Chen H-W, Varki NM, Schooley RT, Gagneux P. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J. 2013;10:321. doi: 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans RW, Holmes KV, Dales S, Choppin PW. An electron microscopic study of moderate and virulent virus-cell interactions of the parainfluenza virus SV5. Virology. 1966;30:411–426. doi: 10.1016/0042-6822(66)90119-x. [DOI] [PubMed] [Google Scholar]
- Cox JC, Hampson AW, Hamilton RC. An immunofluorescence study of influenza virus filament formation. Arch Virol. 1980;63:275–284. doi: 10.1007/BF01315033. [DOI] [PubMed] [Google Scholar]
- Crane M, Hyatt A. Viruses of fish: an overview of significant pathogens. Viruses. 2011;3:2025–2046. doi: 10.3390/v3112025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald HB, Isaacs A. Some properties of influenza virus filaments shown by electron microscopic particle counts. J Gen Microbiol. 1954;11:325–331. doi: 10.1099/00221287-11-2-325. [DOI] [PubMed] [Google Scholar]
- Elleman CJ, Barclay WS. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology. 2004;321:144–153. doi: 10.1016/j.virol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Elton D, Bruce EA, Bryant N, Wise HM, MacRae S, Rash A, Smith N, Turnbull ML, Medcalf L, Daly JM. The genetics of virus particle shape in equine influenza A virus. Influenza and other respiratory viruses. 2013;7:81–89. doi: 10.1111/irv.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami M, Enami K. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. Journal of virology. 1996;70:6653–6657. doi: 10.1128/jvi.70.10.6653-6657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy JV, Dickey BF. Airway mucus function and dysfunction. New England Journal of Medicine. 2010;363:2233–2247. doi: 10.1056/NEJMra0910061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavazzi C, Yver M, Isel C, Smyth RP, Rosa-Calatrava M, Lina B, Moules V, Marquet R. A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc Natl Acad Sci U S A. 2013;110:16604–16609. doi: 10.1073/pnas.1314419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proceedings of the Royal Society of London Series B, Biological sciences. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Forouhar F, Qiu S, Sha B, Luo M. The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology. 2001;289:34–44. doi: 10.1006/viro.2001.1119. [DOI] [PubMed] [Google Scholar]
- Hayase Y, Uno F, Nii S. Ultrahigh-resolution scanning electron microscopy of MDCK cells infected with influenza viruses. Journal of electron microscopy. 1995;44:281–288. [PubMed] [Google Scholar]
- Heldt FS, Kupke SY, Dorl S, Reichl U, Frensing T. Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection. Nat Commun. 2015;6:8938. doi: 10.1038/ncomms9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EC, Charles PD, Hester SS, Thomas B, Trudgian D, Martinez-Alonso M, Fodor E. Conserved and host-specific features of influenza virion architecture. Nat Commun. 2014;5:4816. doi: 10.1038/ncomms5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J Virol. 2008;82:11869–11879. doi: 10.1128/JVI.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EC, Fodor E. Transport of the influenza virus genome from nucleus to nucleus. Viruses. 2013;5:2424–2446. doi: 10.3390/v5102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Leser GP, Zhang J, Lamb RA. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. The EMBO journal. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibenge FS, Garate ON, Johnson G, Arriagada R, Kibenge MJ, Wadowska D. Isolation and identification of infectious salmon anaemia virus (ISAV) from Coho salmon in Chile. Diseases of aquatic organisms. 2001;45:9–18. doi: 10.3354/dao045009. [DOI] [PubMed] [Google Scholar]
- Kilbourne ED, Murphy JS. Genetic studies of influenza viruses. I. Viral morphology and growth capacity as exchangeable genetic traits. Rapid in ovo adaptation of early passage Asian strain isolates by combination with PR8. The Journal of experimental medicine. 1960;111:387–406. doi: 10.1084/jem.111.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren CWR, Nylund A. Morphology and morphogenesis of infectious salmon anaemia virus replicating in the endothelium of Atlantic salmon Salmo salar. Diseases of aquatic organisms. 1997;29:99–109. [Google Scholar]
- Kosoy OI, Lambert AJ, Hawkinson DJ, Pastula DM, Goldsmith CS, Hunt DC, Staples JE. Novel thogotovirus associated with febrile illness and death, United States, 2014. Emerging infectious diseases. 2015;21:760–764. doi: 10.3201/eid2105.150150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala SS, Lamirande EW, Suguitan AL, Jr, Wang W, Santos CP, Vogel L, Matsuoka Y, Lindsley WG, Jin H, Subbarao K. Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS pathogens. 2011;7:e1002443. doi: 10.1371/journal.ppat.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G, Narayan O, Rouse BT, Ferguson AE, Connell MC. A new influenza A virus infection in turkeys II. A highly pathogenic variant, a/turkey/ontario 772/66. The Canadian veterinary journalLa revue veterinaire canadienne. 1968;9:151–160. [PMC free article] [PubMed] [Google Scholar]
- Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci U S A. 2013;110:11133–11138. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Muller J, Ye Z. Association of influenza virus matrix protein with ribonucleoproteins may control viral growth and morphology. Virology. 2002;304:89–96. doi: 10.1006/viro.2002.1669. [DOI] [PubMed] [Google Scholar]
- Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitnaul LJ, Castrucci MR, Murti KG, Kawaoka Y. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. Journal of virology. 1996;70:873–879. doi: 10.1128/jvi.70.2.873-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Rose HM, Moore DH. Structure and development of viruses observed in the electron microscope. III. Influenza virus. J Exp Med. 1956;104:171–182. doi: 10.1084/jem.104.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Haruyama T, Nagata K. Tamiflu-resistant but HA-mediated cell-to-cell transmission through apical membranes of cell-associated influenza viruses. PloS one. 2011;6:e28178. doi: 10.1371/journal.pone.0028178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley VM, Wyckoff RWG. Electron micrography of the virus of influenza. Nature. 1946;157:1160. doi: 10.1038/157263a0. [DOI] [PubMed] [Google Scholar]
- Muraki Y, Murata T, Takashita E, Matsuzaki Y, Sugawara K, Hongo S. A mutation on influenza C virus M1 protein affects virion morphology by altering the membrane affinity of the protein. Journal of virology. 2007;81:8766–8773. doi: 10.1128/JVI.00075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki Y, Washioka H, Sugawara K, Matsuzaki Y, Takashita E, Hongo S. Identification of an amino acid residue on influenza C virus M1 protein responsible for formation of the cord-like structures of the virus. J Gen Virol. 2004;85:1885–1893. doi: 10.1099/vir.0.79937-0. [DOI] [PubMed] [Google Scholar]
- Murti KG, Webster RG. Distribution of hemagglutinin and neuraminidase on influenza virions as revealed by immunoelectron microscopy. Virology. 1986;149:36–43. doi: 10.1016/0042-6822(86)90084-x. [DOI] [PubMed] [Google Scholar]
- Nakajima N, Hata S, Sato Y, Tobiume M, Katano H, Kaneko K, Nagata N, Kataoka M, Ainai A, Hasegawa H. The first autopsy case of pandemic influenza (A/H1N1pdm) virus infection in Japan: detection of a high copy number of the virus in type II alveolar epithelial cells by pathological and virological examination. Jpn J Infect Dis. 2010;63:67–71. [PubMed] [Google Scholar]
- Nishimura H, Hara M, Sugawara K, Kitame F, Takiguchi K, Umetsu Y, Tonosaki A, Nakamura K. Characterization of the cord-like structures emerging from the surface of influenza C virus-infected cells. Virology. 1990;179:179–188. doi: 10.1016/0042-6822(90)90287-2. [DOI] [PubMed] [Google Scholar]
- Noda T. Native morphology of influenza virions. Frontiers in microbiology. 2011;2:269. doi: 10.3389/fmicb.2011.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J Gen Virol. 2007;88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KL, Manicassamy B, Lamb RA. Influenza a virus uses intercellular connections to spread to neighboring cells. Journal of virology. 2015;89:1537–1549. doi: 10.1128/JVI.03306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PC, Lamb RA, Compans RW. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology. 1998;240:127–137. doi: 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. Journal of virology. 2010a;84:5078–5088. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010b;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Leser GP, Lamb RA. Filamentous influenza virus enters cells via macropinocytosis. Journal of virology. 2012;86:10950–10960. doi: 10.1128/JVI.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok RW, Wrigley NG, Calder LJ, Cusack S, Wharton SA, Brown EB, Skehel JJ. Electron microscopy of the low pH structure of influenza virus haemagglutinin. EMBO J. 1986;5:41–49. doi: 10.1002/j.1460-2075.1986.tb04175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seladi-Schulman J, Campbell PJ, Suppiah S, Steel J, Lowen AC. Filament-Producing Mutants of Influenza A/Puerto Rico/8/1934 (H1N1) Virus Have Higher Neuraminidase Activities than the Spherical Wild-Type. PloS one. 2014;9:e112462. doi: 10.1371/journal.pone.0112462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seladi-Schulman J, Steel J, Lowen AC. Spherical influenza viruses have a fitness advantage in embryonated eggs, while filament-producing strains are selected in vivo. Journal of virology. 2013;87:13343–13353. doi: 10.1128/JVI.02004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh FY, Utley TJ, Craven RE, Rogers MC, Lapierre LA, Goldenring JR, Crowe JE., Jr Respiratory syncytial virus assembles into structured filamentous virion particles independently of host cytoskeleton and related proteins. PloS one. 2012;7:e40826. doi: 10.1371/journal.pone.0040826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. Cellular proteins in influenza virus particles. PLoS pathogens. 2008;4:e1000085. doi: 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge KF, Zhou NN, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti KG, Norwood M, et al. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- Sieczkarski SB, Whittaker GR. Characterization of the host cell entry of filamentous influenza virus. Archives of Virology. 2005;150:1783–1796. doi: 10.1007/s00705-005-0558-1. [DOI] [PubMed] [Google Scholar]
- Simpson-Holley M, Ellis D, Fisher D, Elton D, McCauley J, Digard P. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology. 2002;301:212–225. doi: 10.1006/viro.2002.1595. [DOI] [PubMed] [Google Scholar]
- Smirnov YuA, Kuznetsova MA, Kaverin NV. The genetic aspects of influenza virus filamentous particle formation. Arch Virol. 1991;118:279–284. doi: 10.1007/BF01314038. [DOI] [PubMed] [Google Scholar]
- Stevenson JP, Biddle F. Pleomorphism of influenza virus particles under the electron microscope. Nature. 1966;212:619–621. doi: 10.1038/212619a0. [DOI] [PubMed] [Google Scholar]
- Sugita Y, Noda T, Sagara H, Kawaoka Y. Ultracentrifugation deforms unfixed influenza A virions. The Journal of general virology. 2011;92:2485–2493. doi: 10.1099/vir.0.036715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine RC, Isaacs A. The structure of influenza virus filaments and spheres. J Gen Microbiol. 1957;16:195–204. doi: 10.1099/00221287-16-1-195. [DOI] [PubMed] [Google Scholar]
- Vijayakrishnan S, Loney C, Jackson D, Suphamungmee W, Rixon FJ, Bhella D. Cryotomography of budding influenza A virus reveals filaments with diverse morphologies that mostly do not bear a genome at their distal end. PLoS pathogens. 2013;9:e1003413. doi: 10.1371/journal.ppat.1003413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AC. Specific changes in the M1 protein during adaptation of influenza virus to mouse. Archives of Virology. 1995;140:383–389. doi: 10.1007/BF01309872. [DOI] [PubMed] [Google Scholar]
- Wasilewski S, Calder LJ, Grant T, Rosenthal PB. Distribution of surface glycoproteins on influenza A virus determined by electron cryotomography. Vaccine. 2012;30:7368–7373. doi: 10.1016/j.vaccine.2012.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS pathogens. 2012;8:e1002998. doi: 10.1371/journal.ppat.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrigley NG. Electron microscopy of influenza virus. British medical bulletin. 1979;35:35–38. doi: 10.1093/oxfordjournals.bmb.a071539. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Danev R, Nishiyama K, Sugawara K, Nagayama K. Zernike phase contrast electron microscopy of ice-embedded influenza A virus. Journal of structural biology. 2008;162:271–276. doi: 10.1016/j.jsb.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Yang X, Steukers L, Forier K, Xiong R, Braeckmans K, Van Reeth K, Nauwynck H. A beneficiary role for neuraminidase in influenza virus penetration through the respiratory mucus. PLoS One. 2014;9:e110026. doi: 10.1371/journal.pone.0110026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Compans RW. Filamentous particle formation by human parainfluenza virus type 2. The Journal of general virology. 2000;81:1305–1312. doi: 10.1099/0022-1317-81-5-1305. [DOI] [PubMed] [Google Scholar]
- Zebedee SL, Lamb RA. Growth restriction of influenza A virus by M2 protein antibody is genetically linked to the M1 protein. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:1061–1065. doi: 10.1073/pnas.86.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Pekosz A, Lamb RA. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. Journal of virology. 2000;74:4634–4644. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]