Abstract
Hair color is one of the most recognizable visual traits in European populations and is under strong genetic control. Here we report the results of a genome-wide association study meta-analysis of almost 300,000 participants of European descent. We identified 123 autosomal and one X-chromosome loci significantly associated with hair color; all but 13 are novel. Collectively, SNPs associated with hair color within these loci explain 34.6% of red hair, 24.8% of blond hair and 26.1% of black hair heritability in the study populations. These results confirm the polygenic nature of complex phenotypes and improve our understanding of melanin pigment metabolism in humans.
Human pigmentation refers to coloration of external tissues due to variations in quantity, ratio and distribution of the two main types of the pigment melanin: eumelanin and pheomelanin1. Most melanin is produced by melanosomes2,3, large organelles specialized in melanin synthesis and transportation located mainly in the epidermis, hair and iris as well as the central nervous system. Early humans had a darkly pigmented skin4,5 which protected against high Ultraviolet radiation (UVR) and its consequences such as skin cancer6 and folate depletion7. European and Asian populations evolved to lighter skin pigmentation8,9, as they migrated towards northern latitudes with lower UVR4. The lighter pigmentation maximizes UVR absorption needed to maintain adequate vitamin D levels. In Europeans, pigmentation of skin, hair and or eyes has characteristic geographic distributions because of natural selection10 and perhaps genetic drift; a role for sexual selection has been debated 11–13.
Hair color is one of the most prominent traits in humans. Twin studies suggest that up to 97% of variation in hair color may be explained by heritable factors14 and genome-wide association studies (GWAS) 15–20 have identified several chromosomal regions associated with hair color and related pigmentation traits21. Except for red hair, known variants have a relatively low predictive value22 and the heritability gap remains relatively large14 which suggests that many hair color genes, remain undiscovered.
Here we report the results of a meta-analysis of two GWAS carried out in two large discovery cohort studies: 157,653 research participants from the 23andMe, Inc. customer base18 and 133,238 individuals from the UK Biobank (UKBB). Participants in both studies self-reported the natural color of their hair in adulthood (Supplementary Figure 1 and Supplementary Methods). For the purpose of this work, each hair color category collected (black, dark brown, light brown, red and blond) was assigned numerical values ranging from lowest (blond) to highest (black). These codes were used as the outcome variable in linear regression based GWAS analyses. To minimize population admixture and stratification, the analyses were restricted to individuals of European ancestry (Supplementary Figures 2 and 3) and adjusted for the first ten principal components (PCs) of the genotype matrix, as well as age and sex.
The analyses confirmed a strong association between hair color and PCs, especially in the less ethnically homogeneous 23andMe dataset, which includes participants of more varied European origin, in line with the known North-South cline in hair color variation and other regional differences in hair color across Europe12 (Supplementary Table 1). The strongest associations in both groups were with sex (Table 1). Women were more likely to report blond (OR=1.20 and OR=1.29 in the 23andMe and UKBB participants, respectively), or red hair (OR=1.72 and OR=1.40, respectively) than any other color and three to five times less likely to report black hair (OR=0.35 and OR=0.20, respectively) compared to men.
Table 1.
Effect of sex on the hair color phenotypes in the 23andMe (N=157,653 independent participants) and UK Biobank (N=133,238 independent participants) cohorts
| 23andMe | Odds | Standard | 95% Confidence Interval | |
|---|---|---|---|---|
| Ratio | Error | low | upper | |
| Blond (all) | 1.202 | 0.024 | 1.174 | 1.230 |
| Red | 1.721 | 0.014 | 1.675 | 1.768 |
| Light Brown | 1.116 | 0.013 | 1.088 | 1.145 |
| Dark Brown | 0.663 | 0.011 | 0.650 | 0.677 |
| Black | 0.348 | 0.030 | 0.329 | 0.369 |
| UKBB | Odds | Standard | 95% Confidence Interval | |
|---|---|---|---|---|
| Ratio | Error | Low | upper | |
| Blond | 1.285 | 0.018 | 1.241 | 1.330 |
| Red | 1.395 | 0.026 | 1.325 | 1.469 |
| Light Brown | 1.101 | 0.011 | 1.077 | 1.125 |
| Dark Brown | 0.993 | 0.011 | 0.971 | 1.015 |
| Black | 0.195 | 0.033 | 0.182 | 0.208 |
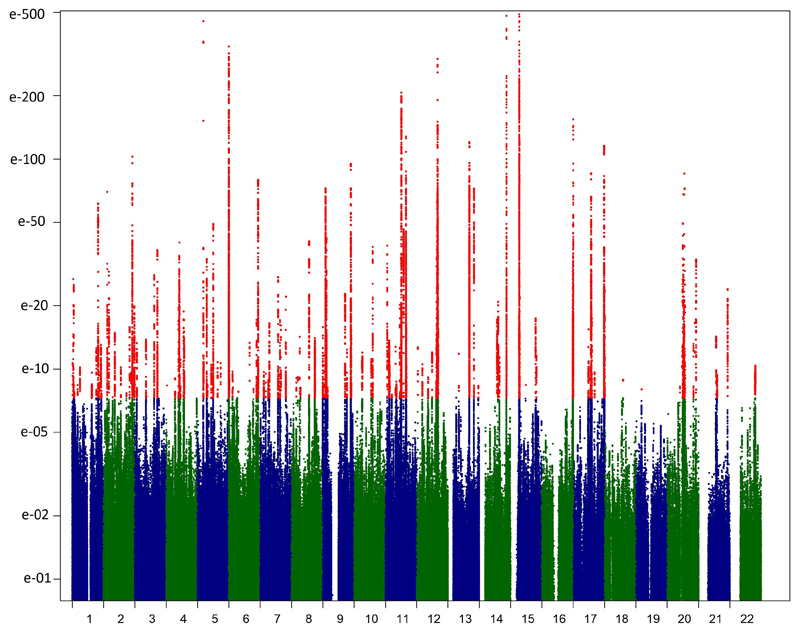
Genomic inflation factors 23 (λGC) from the 23andMe and the UKBB GWAS were 1.147 and 1.146, respectively, in line with expectations of high power to detect large polygenic effects in these large samples24 (Supplementary Figure 4). Meta-analyzed GWAS results reached conventional genome-wide significance (p<5x10-08) in many regions, primarily clustering around 123 distinct autosomal genomic SNPs and one X-chromosomal locus (Figure 1, Supplementary Table 2), mostly new (Table 2). In line with power expectations (Supplementary Figure 5), 75 of these regions were genome-wide significant in at least one of the two cohorts and always at least nominally significant (p<0.01) in the other.
Figure 1.
Manhattan plot of the inverse variance meta-analysis for association with hair color of the 23andMe and UKBB cohorts (meta-analysis N=290,891). The unadjusted significance of association (y-axis) for each SNP on different chromosomes is shown in alternating navy and green along the x-axis with polymorphisms reaching significance at GWAS level (p<5x10-08) depicted in red. The values on the y-axis were truncated at p=10-500.
Table 2. A selection of genes newly associated with hair color.
The selection was based on the strength of their effect, which is defined as the standardized linear regression coefficient. Results are given for the UK Biobank, 23andMe, their meta-analysis as well as the meta-analysis results from the VisiGen Consortium. These results were generated linear models and effect sizes (Beta) are given in SD units. The A, C, T and G under the “Reference Allele field” denote the nucleotide of the allele for which the effect size and allele frequencies are reported. Frequencies are given for the reference allele and are the average of observed frequencies in the 23andMe and UK Biobank. Associations with p-values of less than 10-100 are reported as “p<10-100”.
| UK Biobank | 23andMe | Meta-analysis | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Pos(Build37) | SNP ID | Ref. Allele | Freq. | Nearest Gene | N | Beta | SE | p-value | N | Beta | SE | p-value | Beta | SE | p-value |
| 1 | 8207579 | rs80293268 | G | 0.047 | SLC45A1 | 132221 | 0.194 | 0.009 | 1.54E-97 | 157651 | 0.157 | 0.009 | 1.29E-67 | 0.175 | 0.007 | <E-100 |
| 1 | 205181062 | rs2369633 | T | 0.089 | DSTYK | 132887 | -0.071 | 0.007 | 9.20E-26 | 157651 | -0.077 | 0.006 | 3.15E-38 | -0.075 | 0.005 | 3.44E-62 |
| 2 | 28613302 | rs71443018 | G | 0.039 | FOSL2 | 126428 | 0.133 | 0.01 | 2.14E-39 | 157651 | 0.148 | 0.012 | 4.18E-33 | 0.139 | 0.008 | 1.36E-70 |
| 9 | 126808006 | rs58979150 | T | 0.108 | LHX2 | 132883 | 0.089 | 0.006 | 1.03E-44 | 157651 | 0.083 | 0.005 | 9.93E-53 | 0.086 | 0.004 | 1.40E-95 |
| 13 | 78391757 | rs1279403 | T | 0.406 | EDNRB | 133238 | -0.086 | 0.004 | <E-100 | 157651 | -0.074 | 0.004 | 4.57E-95 | -0.08 | 0.003 | <E-100 |
| 15 | 48426484 | rs1426654 | G | 0.021 | SHC4 | 133238 | 0.188 | 0.069 | 0.006 | 157651 | 0.289 | 0.03 | 2.12E-21 | 0.273 | 0.028 | 1.24E-22 |
| 17 | 39551099 | rs117612447 | T | 0.029 | KRT31 | 133238 | 0.063 | 0.011 | 2.95E-08 | 157651 | 0.064 | 0.011 | 2.09E-09 | 0.063 | 0.008 | 3.29E-16 |
| 20 | 52661068 | rs73132911 | T | 0.046 | BCAS1 | 132836 | 0.089 | 0.009 | 6.78E-22 | 157651 | 0.046 | 0.008 | 2.54E-09 | 0.064 | 0.006 | 5.85E-27 |
Previously known pigmentation loci were all strongly associated in the meta-analysis results: HERC2 (rs12913832), IRF4 (rs12203592), MC1R (rs1805007), as well as others, showed some of the strongest statistical evidence for association ever published for human complex traits. Strong associations were found for genes whose mutations reportedly cause impairment of pigmentation such as Waardenburg (EDNRB, rs1279403, p<10-100; MITF, rs9823839, p<10-100), Hermansky-Pudlak (HPS5, rs201668750, p=4.68x10-11), Trichomegaly (FGF5 rs7681907, p=5.684x10-25) or Ablepharon-Macrostomia (TWIST2, rs11684254, p=1.233x10-20) Syndromes. Many polymorphisms significantly (p<5x10-08) associated with hair color in our meta-analysis had existing entries in the GWAS Catalog21. In previous publications, they were associated to several phenotypes, including most known pigmentation loci (Supplementary Table 3).
Among the associated loci, some of the strongest effects were observed for two solute carrier 45A family members (SLC45A1, rs80293268, p<10-100 and the SLC45A2 gene, rs16891982, p<10-100); polymorphisms near a third solute carrier gene were also significantly associated with the trait (rs60086398 upstream of SLC7A1, p=4.93x10-08). In addition, forkhead box family genes (FOXO6, rs3856254, p=4.0x10-09 and FOXE1, rs3021523, p=4.23x10-23) and sex determining region Y (SRY)-box genes (SOX5 rs9971729, p=8.8x10-17 and SOX6, rs1531903, p=9.1x10-16) were among those highlighted in our results. An additional locus, located on chromosome X, on the second intron of the collagen type IV alpha 6 gene was also significantly associated (COL46A, rs1266744, p=5.03x10-12). Chromosome Y information was not analyzed. Interestingly, given the observed strong association of hair color with sex, there was no particular difference in effect sizes observed for these loci among men and women in either cohort (Supplementary Table 4, Supplementary Figure 6); only one SNPs significantly associated with hair color in the meta-analysis showed significant (p=1.6x10-08) interaction with sex in the 23andMe (Supplementary Table 5), but much weaker interaction in the UK Biobank cohort (p=0.04). As reported before10, some hair color genes are subject to significant natural selection (Supplementary Table 6); SNPs associated with hair color in our meta-analysis, tended to have lower selection score centiles and higher than average evidence for natural selection within European populations (p=0.04) and compared to Africans (Supplementary Figure 7).
To further validate the results and to introduce a testing dataset, we collected GWAS summary statistics from 10 additional cohorts with 27,865 European participants from International Visible Trait Genetics (VisiGen) Consortium25 and meta-analyzed them. For 114 of the 123 autosomal loci highlighted by the discovery GWAS meta-analysis, the direction of the association was the same as observed in the meta-analysis; despite the lower statistical power of the replication due to smaller sample sizes, most leading SNP loci from the discovery meta-analysis (75 of the 123 autosomal regions) replicated at least at a nominal level and the same direction of association (p<0.05); for 35 of these loci the association was stronger even after correction for multiple testing (Supplementary Table 2).
Next, we assessed the potential relationship of the most associated polymorphisms and expression of the genes nearest to them. In line with most previous GWAS26, the majority of these polymorphic loci had eQTL effects in several tissues. The strongest associations were observed with transcript of the CBWD1 (rs478882, p=1.30x10-30), PPM1A (rs7154748, p=3.30x10-14) and RALY genes (rs6059655 being associated with ASIP gene expression, p=6.0x10-09) in sun-exposed skin tissues (Supplementary Table 7).
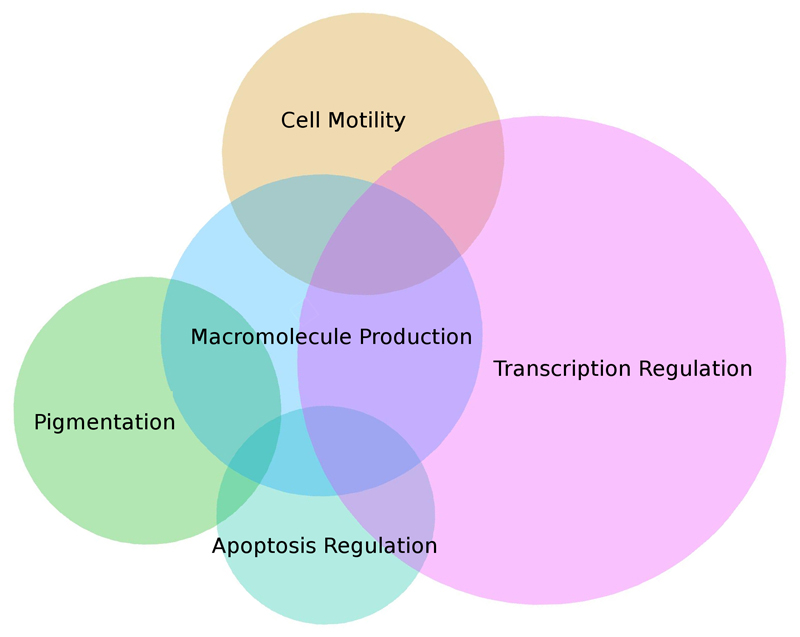
As expected, genes showing the strongest association in the meta-analysis were significantly enriched for several Gene Ontology entries, especially pigmentation, melanin biosynthetic and metabolic processes, etc. (Figure 2, Supplementary Table 8).
Figure 2.
Gene Ontology Biological Processes annotations for genes adjacent to the SNPs showing the strongest associations with hair color via GWAS meta-analysis in the 23andMe and UKBB cohorts.
A conditional analysis of the discovery cohorts identified 258 SNPs independently associated with hair color (Supplementary table 9). These SNPs explain overall 20.68% of the hair color heritability (using ordinal categories) and 34.58% (SE=3.64%) of the population liability scale27 heritability for red hair (vs. any other color, assuming population prevalence is as in the UKBB at 0.047), 24.80% for blond hair (SE=2.49%, assuming a prevalence of 0.11) and 26.12% (SE=3.11%) of the black hair heritability (prevalence 0.046, Table 3).
Table 3.
Phenotypic variance explained by the identified autosomal loci significantly associated with hair color. The current estimates are given as the ratio of the genetic variance, V(G), over the phenotypic variance (Vp) and scaled over the population prevalence, V(G)/Vp_L, (estimated in the UKBB cohort, N=133,238), on the right. The estimates of genetic variance explained by known SNPs prior to this study were taken from previous publications. The phenotypes in this table were compared with all other hair colors. Since 80% of the participants reported some shade of brown hair color (dark or light), the heritabilities for these two phenotypes were considered baseline and were not calculated.
| Current heritability estimates | Previous estimates | ||||||
|---|---|---|---|---|---|---|---|
| Phenotype | V(G)/Vp | SE | V(G)/Vp_L | SE | Prevalence | V(G)/Vp | SE |
| Blond | 0.094 | 0.009 | 0.248 | 0.025 | 0.113 | 0.058 | 0.022 |
| Red | 0.074 | 0.008 | 0.346 | 0.036 | 0.046 | 0.069 | 0.069 |
| Black | 0.056 | 0.007 | 0.261 | 0.031 | 0.047 | 0.005 | 0.005 |
Finally, we modelled hair color prediction in two cohorts (QIMR N=7,283 and RS N=7,724) using the 258 independently associated SNPs from the discovery GWAS meta-analysis (Supplementary Table 9) together with previously reported SNP predictors for hair color from the HIrisPlex System28. We split the data into model building (80%) and validation (20%) sets to assure that marker discovery, model building and model validation were independently executed. In both cohorts, prediction accuracies were high for black (QIMR AUC=0.91, RS AUC=0.81) and red (0.87 and 0.84) hair colors, but lower for blond (0.79 and 0.74) and brown (0.76 and 0.64; Supplementary Table 10, Supplementary Figure 8). Using the same datasets, these new models outperformed the previous HIrisPlex model22 (QIMR/RS black 0.82 vs 0.77, red 0.87 vs. 0.83, blond 0.67 vs. 0.65, brown 0.66 vs. 0.57, Supplementary Table 10).
Our work identified over a hundred new genetic loci involved in hair pigmentation in Europeans and raises interesting questions. First, the observation of higher prevalence of lighter hair colors among women (Supplementary Figure 9), follows previous findings based on objective quantitative measurement of hair color29,30 suggesting that sex is truly associated with hair color, independent of socially driven self-reporting bias. Second, although hair pigmentation spans a spectrum from very bright (blond) to very dark (black), the genetic mechanisms don’t always follow this linear scale, as red hair color often has unique predisposing genetic factors 16,17. However, our results explain even higher portions of heritability than before14 for all hair colors and not just for the extremes of the light-dark hair color spectrum. Third, hair color is a trait that follows special distribution patterns of distribution, therefore is prone to issues of population structure bias that may be controlled in several ways. A comparison of different methodologies (Supplementary Figure 10) shows that our approach is roughly equivalent with others. Fourth, annotation of genetic regions based on physical distances and association probability most likely underestimates the number of regions involved in hair pigmentation. For example, the involvement of OCA2 and HERC2 genes in human pigmentation is not simply due to linkage disequilibrium31, yet because of their proximity, both loci in our study were assigned to the same association region. This would, however, not affect the conditional analysis at a marker level, which discriminates separate effects.
In conclusion, this large GWAS meta-analysis has improved our knowledge on the genetic controls of human hair and pigmentation by bringing the number of known genes into the hundreds. The newly identified genetic loci explain substantial portions of the hair color phenotypic variability and will guide future research into better understanding the functional mechanisms linking these genes to pigmentation variation. Our findings are also useful in the future for both the better molecular understanding of human pigmentation including their DNA-based prediction as relevant in forensic and anthropological applications, and the diseases that result from biological impairment of pigmentation including the development of treatment strategies.
Online Methods
Data Availability
This work used data from two primary sources. The original datasets can be accessed as follows: For UK Biobank data, through the UK Biobank Access management, as specified here: http://www.ukbiobank.ac.uk/register-apply/. The hair color data accession codes are 1747.0.0, 1747.1.0 and 1747.2.0. The participants age UK Biobank accession code is 21022, for sex 31.0.0 and the pre-computed principal components used here 22009.0.1 through 22009.0.10.
For the 23andMe participants requests for summary statistics access can be made at https://researchers.23andme.org/collaborations. There are no accession codes available.
For the TwinsUK datasets access can be asked through http://www.twinsuk.ac.uk/data-access/ and access to the secondary source of data through the corresponding authors.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 12052.
The ALSPAC work is supported by a Medical Research Council program grant (MC_UU_12013/4 to D.M.E). The UK Medical Research Council and the Wellcome Trust (Grant refs: 092731 and 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. D.M. Evans is supported by an Australian Research Council Future Fellowship (FT130101709). This publication is the work of the authors and DME will serve as guarantor for the contents of this paper. ALSPAC GWAS data was generated by Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe.
The ERF Study was supported by the joint grant from the Netherlands Organization for Scientific Research (NWO, 91203014), the Center of Medical Systems Biology (CMSB), Hersenstichting Nederland, Internationale Stichting Alzheimer Onderzoek (ISAO), Alzheimer Association project number 04516, Hersenstichting Nederland project number 12F04(2).76, and the Interuniversity Attraction Poles (IUAP) program. As a part of EUROSPAN (European Special Populations Research Network) ERF was supported by European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947) and also received funding from the European Community's Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the programme "Quality of Life and Management of the Living Resources" of 5th Framework Programme (no. QLG2-CT-2002-01254). High-throughput analysis of the ERF data was supported by joint grant from Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043).
The INGI research was supported by funds from Compagnia di San Paolo, Torino, Italy; Fondazione Cariplo, Italy and Ministry of Health, Ricerca Finalizzata 2008 and CCM 2010 and Telethon, Italy. Aditional support was provided by the Italian Ministry of Health (RF 2010 to PG), FVG Region, and Fondo Trieste.
The NTR study was supported by multiple grants from the Netherlands Organization for Scientific Research (NWO: 016-115-035, 463-06-001, 451-04-034), ZonMW (31160008, 911-09-032); Institute for Health and Care Research (EMGO+); Biomolecular Resources Research Infrastructure (BBMRI-NL, 184.021.007), European Research Council (ERC-230374). Genotyping was made possible by grants from NWO/SPI 56-464-14192, Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health, Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls (USA) and the National Institutes of Health (NIH R01 HD042157-01A1, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995). B.D. Lin is supported by a PhD grant (201206180099) from the China Scholarship Council.
QIMR funding was provided by the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498), the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016, DP0343921), the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254), and the U.S. National Institutes of Health (NIH grants AA07535, AA10248, AA13320, AA13321, AA13326, AA14041, MH66206). Statistical analyses were carried out on the Genetic Cluster Computer, which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003). S.E.M and D.L.D. are supported by the National Health and Medical Research Council (NHMRC) Fellowship Scheme.
The 20-year follow-up of Generation 2 of the Raine Study was funded by Australian National Health and Medical Research Council (NHMRC) project grant 1021105, Lions Eye Institute, the Australian Foundation for the Prevention of Blindness and the Ophthalmic Research Institute of Australia. SY is supported by NHMRC Early Career Fellowship (CJ Martin - Overseas Biomedical Fellowship).
The Rotterdam Study is supported by the Netherlands Organization of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) project nr. 050-060-810. The Rotterdam Study is supported by the Erasmus MC and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research (NWO); the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE) the Netherlands Genomics Initiative (NGI); the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sport; the European Commission (DG XII); and the Municipality of Rotterdam. The generation and management of GWAS genotype data for the Rotterdam Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC.
The TwinsUK study was funded by the Wellcome Trust (105022/Z/14/Z); European Community’s Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research (NIHR)- funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London. SNP Genotyping was performed by the Wellcome Trust Sanger Institute and National Eye Institute via NIH/CIDR.
Footnotes
Author Contribution
PGH, AV, FL jointly wrote the manuscript, coordinated meta-analyses and prediction modelling; NAF, DME, VB, AV, GH, GM, SMR, DLD, GZ, SDG, SEM, BDL, GW, JJH, DV, GG, IG, CS, APC, MB, DT, MC, AR, SY, AWH, YC, CZ, AGU, MAH, TN, MF, DAH each conducted part of the analyses described in this work; GDS, PG, CMvD, MAI, DAM, DIB, NGM, MF contributed populations samples and data used for analyses; MK and TDS jointly coordinated the work and participated in manuscript preparation.
Competing Financial Interests
NF and DAH are employees of the 23andMe Inc. consumer genetics company.
References
- 1.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–50. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 2.Randhawa M, et al. Evidence for the ectopic synthesis of melanin in human adipose tissue. FASEB J. 2009;23:835–43. doi: 10.1096/fj.08-116327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sturm RA, Teasdale RD, Box NF. Human pigmentation genes: identification, structure and consequences of polymorphic variation. Gene. 2001;277:49–62. doi: 10.1016/s0378-1119(01)00694-1. [DOI] [PubMed] [Google Scholar]
- 4.Jablonski NG, Chaplin G. The evolution of human skin coloration. J Hum Evol. 2000;39:57–106. doi: 10.1006/jhev.2000.0403. [DOI] [PubMed] [Google Scholar]
- 5.Jablonski NG, Chaplin G. Colloquium paper: human skin pigmentation as an adaptation to UV radiation. Proc Natl Acad Sci U S A. 2010;107(Suppl 2):8962–8. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greaves M. Was skin cancer a selective force for black pigmentation in early hominin evolution? Proc Biol Sci. 2014;281:20132955. doi: 10.1098/rspb.2013.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branda RF, Eaton JW. Skin color and nutrient photolysis: an evolutionary hypothesis. Science. 1978;201:625–6. doi: 10.1126/science.675247. [DOI] [PubMed] [Google Scholar]
- 8.Norton HL, et al. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 2007;24:710–22. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- 9.Wilde S, et al. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc Natl Acad Sci U S A. 2014;111:4832–7. doi: 10.1073/pnas.1316513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field Y, et al. Detection of human adaptation during the past 2000 years. Science. 2016 doi: 10.1126/science.aag0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki K. Sexual selection as a cause of human skin colour variation: Darwin's hypothesis revisited. Ann Hum Biol. 2002;29:589–608. doi: 10.1080/0301446021000019144. [DOI] [PubMed] [Google Scholar]
- 12.Frost P. European hair and eye color - A case of frequency-dependent sexual selection? Evolution and Human Behavior. 2006;27:85–103. [Google Scholar]
- 13.Madrigal L, Kelly W. Human skin-color sexual dimorphism: a test of the sexual selection hypothesis. Am J Phys Anthropol. 2007;132:470–82. doi: 10.1002/ajpa.20453. [DOI] [PubMed] [Google Scholar]
- 14.Lin BD, et al. Heritability and Genome-Wide Association Studies for Hair Color in a Dutch Twin Family Based Sample. Genes (Basel) 2015;6:559–76. doi: 10.3390/genes6030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 16.Han J, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sulem P, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40:835–7. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson N, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenny EE, et al. Melanesian blond hair is caused by an amino acid change in TYRP1. Science. 2012;336:554. doi: 10.1126/science.1217849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M, et al. Genome-wide association studies identify several new loci associated with pigmentation traits and skin cancer risk in European Americans. Hum Mol Genet. 2013;22:2948–59. doi: 10.1093/hmg/ddt142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burdett T, et al. The NHGRI-EBI Catalog of published genome-wide association studies. [Accessed May 8th, 2016]; Vol. Version 1.0. Available at: www.ebi.ac.uk/gwas.
- 22.Walsh S, et al. Developmental validation of the HIrisPlex system: DNA-based eye and hair colour prediction for forensic and anthropological usage. Forensic Sci Int Genet. 2014;9:150–61. doi: 10.1016/j.fsigen.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–12. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, et al. Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Hum Genet. 2015;134:823–35. doi: 10.1007/s00439-015-1559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh S, et al. The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. Forensic Sci Int Genet. 2013;7:98–115. doi: 10.1016/j.fsigen.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Mengel-From J, Wong TH, Morling N, Rees JL, Jackson IJ. Genetic determinants of hair and eye colours in the Scottish and Danish populations. BMC Genet. 2009;10:88. doi: 10.1186/1471-2156-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shekar SN, et al. Spectrophotometric methods for quantifying pigmentation in human hair-influence of MC1R genotype and environment. Photochem Photobiol. 2008;84:719–26. doi: 10.1111/j.1751-1097.2007.00237.x. [DOI] [PubMed] [Google Scholar]
- 31.Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22:446–55. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This work used data from two primary sources. The original datasets can be accessed as follows: For UK Biobank data, through the UK Biobank Access management, as specified here: http://www.ukbiobank.ac.uk/register-apply/. The hair color data accession codes are 1747.0.0, 1747.1.0 and 1747.2.0. The participants age UK Biobank accession code is 21022, for sex 31.0.0 and the pre-computed principal components used here 22009.0.1 through 22009.0.10.
For the 23andMe participants requests for summary statistics access can be made at https://researchers.23andme.org/collaborations. There are no accession codes available.
For the TwinsUK datasets access can be asked through http://www.twinsuk.ac.uk/data-access/ and access to the secondary source of data through the corresponding authors.