Abstract
Antibodies are the key circulating molecules that have evolved to fight infection by the adaptive immune system of vertebrates. Typical antibodies of most species contain six complementarity determining regions (CDRs), where the third CDR of the heavy chain (CDR H3) has the greatest diversity, and often makes the most significant contact with antigen. Generally, the process of V(D)J recombination produces a vast repertoire of antibodies; multiple V, D, and J gene segments recombine with additional junctional diversity at the V-D and D-J joints, and additional combinatorial possibilities occur through heavy and light chain pairing. Despite these processes, the overall structure of the resulting antibody is largely conserved, and binding to antigen occurs predominantly through the CDR loops of the immunoglobulin V domains. Bovines have deviated from this general paradigm by having few VH regions and thus little germline combinatorial diversity, but their antibodies contain long CDR H3 regions, with substantial diversity generated through somatic hypermutation. A subset of the repertoire comprises antibodies with ultralong CDR H3s, which can reach over seventy amino acids in length. Structurally, these unusual antibodies form a β-ribbon ‘stalk’ and disulfide bonded ‘knob’ that protrude far from the antibody surface. These long CDR H3s allow cows to mount a particularly robust immune response when immunizied with viral antigens, particularly to broadly neutralizing epitopes on a stabilized HIV gp140 trimer, which has been a challenge for other species. The unusual genetics and structural biology of cows provide for a unique paradigm for creation of immune diversity, and could enable generation of antibodies against especially challenging targets and epitopes.
1. Overview of cow antibodies
The immunology of domesticated species has been inextricably linked to humans throughout history. Cows have a unique place in the history of modern immunology research by being central to the discovery of the first vaccine. Indeed, the word “vaccine” itself is derived from the Latin “vacca” (cow). In the 1760s, Edward Jenner realized that dairy workers seemed to be resistant to the deadly smallpox virus because they had been infected with cowpox, which causes only mild disease in humans. Jenner then began inoculating humans with sera from cowpox-infected cows, and found that he could prevent infection with smallpox (reviewed in (Baxby, 1999)). Despite the genetic divergence of cowpox and smallpox, conserved neutralizing epitopes could mediate an effective antibody response that could cross-neutralize these different viruses.
Interestingly, the discovery of immunologic tolerance also occurred in cattle; Ray D. Owen showed that dizygotic twin calves possess red blood cells of their twin, which were not self-immunoreactive (Owen, 1945). Similarly, Sir Peter Medawar’s initial work on skin transplantation showed that dizygotic bovine twins would not reject grafts from their sibling (Anderson et al., 1951). This latter work ultimately led to the series of classic experiments in mice and other species establishing key tolerance principles for which the Nobel Prize was awarded in 1960 (Billingham and Brent, 1956;Billingham, Brent and Medawar, 1953). Despite these important historical experiments, cows have not been deeply studied at the molecular immunological level compared to many other model organisms, such as mice, rabbits and macaques. Recent work, however, has shown that cow antibodies have unusually long CDR H3s with novel protruding ‘stalk’ and ‘knob’ structures (Figure 1) and underlying genetics (Wang et al., 2013). Importantly, cows can mount particularly robust responses against challenging viruses like HIV-1 (Sok et al., 2017), suggesting that these unusual antibodies may have particular utility in targeting certain antigenic epitopes and thereby warrant further investigation.
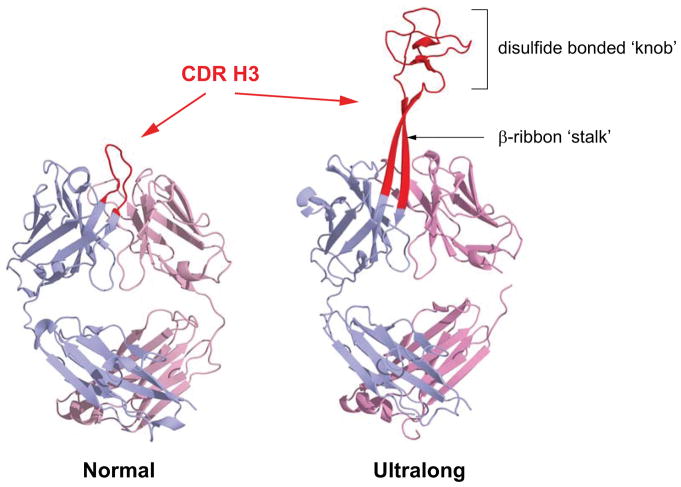
Figure 1. Comparison of ‘normal’ and ‘ultralong’ CDR H3s.
Structure of an antibody with a ‘normal’ CDR H3 length, Ofatumumab (left, PDB: 3giz) and an ultralong CDR H3 cow antibody, BLV1H12 (right, PDB: 4k3d). The CDR H3s are colored red with the heavy chain in light blue and light chain in grey. The β-ribbon ‘stalk’ and disulfide bonded ‘knob’ motifs are labelled.
An initial clue to the unusual antibodies of cows came from the immunogenetic study of Berens et al., where several immunoglobulin heavy chain sequences were analysed from fetal and adult lymphoid tissue, and one sequence termed F18M was found to have a CDR H3 of 51 amino acids (Berens, Wylie and Lopez, 1997). Interestingly, this transcript was derived from fetal tissue and, in hindsight, appears to have an unmutated (germline) V-D-J rearranged sequence. Shortly thereafter, several more exceptionally long CDR H3 sequences were studied in more detail by Kaushik and colleagues who were investigating Bovine Leukemia Virus (BLV) infected B-cells from cows. A total of 44 heterohybridomas from BLV infected B-cells were obtained and sequencing of the heavy chains revealed four unusually long CDR H3s (9% incidence), ranging from 56 to 61 amino acids. Further studies by several different groups found the ultralong sequences at a frequency of 1–22% of heavy chains, with some of the frequency variation possibly arising from different procedural methods, tissues analyzed, breeds and immune status of the cattle(Deiss et al., 2017;Liljavirta et al., 2014;Ma et al., 2016;Walther, Czerny and Diesterbeck, 2013;Wang et al., 2013). Cattle of different breeds all appear to have these unusually long CDR H3s, but at slightly different frequencies when examined side-by-side (Walther et al., 2016). Kaushik’s group identified multiple cysteine residues in CDR H3, found that they usually occurred in even numbers, and predicted that these may form unusual structures (Saini et al., 1999). Interestingly, these four sequences (termed BLV1H12, BLV5B8, BLV5D3, and BLV8C11), which were highly homologous outside of CDR H3, were predicted to utilize the same germline variable region gene (originally termed g1.110.20). Importantly, radiolabelled clones secreted properly formed IgM antibody, suggesting that the antibodies with long CDR H3s were functional (Saini et al., 1999). The antigens for these antibodies were unknown; however, some appeared polyspecific when tested on a panel of antigens (Saini et al., 1999). While originally identified in several IgM antibodies, ultralong CDR H3 sequences were ultimately identified in all other isotypes (Walther, Czerny and Diesterbeck, 2013;Wang et al., 2013).
2. Genetics of ultralong CDR H3 cow antibodies
a. Genomic organization of the IGH locus
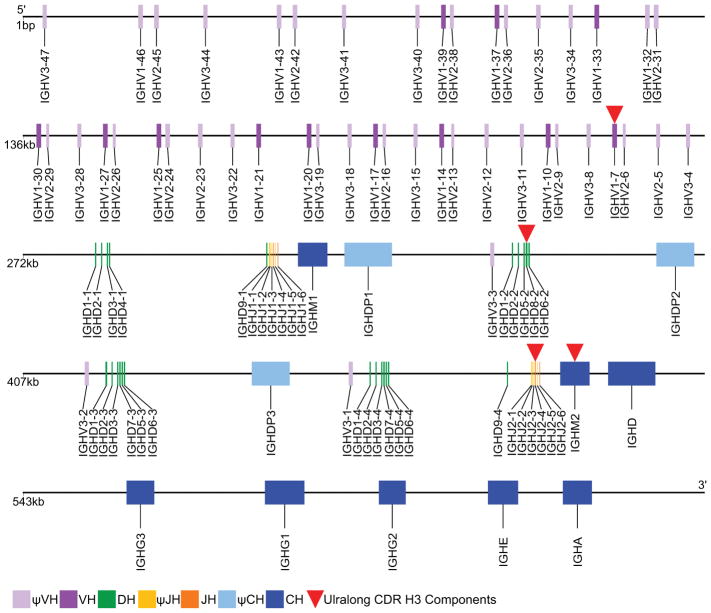
The immunoglobulin heavy chain of cattle was originally thought to be encoded in two distinct loci (Hayes and Petit, 1993;Hosseini et al., 2004;Tobin-Janzen and Womack, 1992), on autosomes BTA11 and BTA21. However, more recently, it has been shown that chromosome 11 only harbours a truncated μCH2, while the complete functional locus has been localized by fluorescent in situ hybridization to BTA21q24 (Ma et al., 2016). All five immunoglobulin heavy chain isotypes known to eutherian (i.e. placental) mammals are also expressed in cattle and encoded on chromosome 21: IgM (Mousavi et al., 1998), IgD (Zhao et al., 2002), IgG (Kacskovics and Butler, 1996;Rabbani et al., 1997;Symons, Clarkson and Beale, 1989), IgE (Mousavi, Rabbani and Hammarstrom, 1997), and IgA (Brown et al., 1997). There are three sub-isotypes of bovine IgG, and more interestingly two IgM (Figure 2). Both of the μ genes appear functional as they both must be inactivated to ablate B lymphopoeisis (Kuroiwa et al., 2009). One μCH (IGHM1) has a pseudogene δCH (psi-IGHD1) 3′ from it and is one of three δCH pseudogenes. The IgD (psi-IgD1) just 3′ of IGM1 has a frameshift in CH2 and thus has been considered nonfunctional. However, the frameshift is a single base insertion in a homopolymer run, which is notoriously difficult to resolve on any sequencing platform. The other two δ pseudogenes are missing a 5′ splice site, >200 bases of CH1, and a hinge exon in addition to large intronic deletions (ranging from 500bp-1kb). The second μCH (IGHM2) precedes a functional δCH (Ma et al., 2016). The functional consequence of two IgM is unclear; however, it is interesting that ultralong CDR H3 antibodies appear to exclusively utilize IgM2 (Ma et al., 2016). The constant genes are ordered 5′-IGHM-IGHD-IGHG3-IGHG1-IGHG2-IGHE-IGHA-3′, with the upstream functional IGHM2 and three pseudo IGHD genes interspersed between clusters of V-D-J elements (Figure 2).
Figure 2. Bovine IGH locus genomic organization.
Schematic of the orientation of coding elements of the cattle immunoglobulin heavy chain locus. Key at bottom left shows the purple variable (V) segments, green diversity (D), orange joining (J) and blue constant (C) genes. Pseudo V, J and C genes are paler tones. Elements used preferentially in the ultralong CDR H3 antibodies are marked by red triangles.
The bovine immunoglobulin heavy chain locus contains a paucity of functional variable gene segments compared to most vertebrates (Berens, Wylie and Lopez, 1997;Liljavirta et al., 2014) [although pig (Eguchi-Ogawa et al., 2010) and rabbit (DiPietro et al., 1992) also have a low variable gene count) (Jung et al., 2006)]. Only twelve potentially functional V gene segments exist in cows, all belonging to a single subgroup (IGHV1) (Lopez, Perez and Wylie, 1998;Niku et al., 2012;Saini, Hein and Kaushik, 1997), which is most closely homologous to the human IGHV4 subgroup. More pseudogenes exist in this and two other variable subgroups (IGHV2 and IGHV3) (Niku et al., 2012). As discussed below, this initial dearth of rearrangement possibilities is compensated for by substantial activation induced cytidine deaminase (AID)-mediated somatic hypermutation in the periphery in a second diversification (pre-antigen exposure) in the gut-associated lymphoid tissue of the Peyer’s patches. Four of 12 joining genes are functional at the bovine IGHJ locus. A total of 23 diversity gene segments, 16 of which appear to be functional, are found within four clusters of DH segments (Ma et al., 2016). Thus, although restricted in number and diversity, there are adequate canonical V(D)J segments for generation of a heavy chain repertoire at this locus, yet some segments (IGHV1-7, IGHD8-2, IGHJ2-4; marked by red in Figure 2) have evolved specifically for use in the ultralong CDR3 antibodies. The variable gene IGHV1-7 (by IMGT nomenclature, previously termed g1.110.20, IGHV10/34, IGHV1S3, IGHV1-1, and VHBUL), whose 3′ end contains a protracted block of nucleotides that encodes an extended F β-strand, is the most 3′ of the functional variable gene segments. This variable segment recombines with the exceptionally long IGHD8-2 (previously termed DH2 (Shojaei, Saini and Kaushik, 2003)) to encode the ‘stalk’ and ‘knob’ of the ultralong CDR H3 antibodies, as well as the descending strand that connects to the J segment encoded G β-strand, as described below.
b. Germline basis for ultralong CDR H3 antibody formation
The subset of antibodies with ultralong CDR H3 appears to be generated through a V-D-J recombination event that only utilizes one VH (IGHV1-7), one DH (IGHD8-2) and one JH (IGHJ2-4) (Deiss et al., 2017;Kaushik, Shojaei and Saini, 2002;Koti, Kataeva and Kaushik, 2008;Wang et al., 2013). While junctional diversity at the V-D and D-J joints in the shorter cow antibody gene repertoire is significant (Liljavirta et al., 2014), for ultralong CDR H3 antibody genes there appears to be little diversity at the D-J joint, and only limited diversity characterized by long insertions at the V-D joint. Koti et al. suggested that atypical nucleotide insertions termed conserved short nucleotide sequences (CSNS; primarily of adenine stretches) play a role in these V-D junctions (Koti, Kataeva and Kaushik, 2010). Of note, the structural features encoded at the V-D and D-J joints are the ascending and descending β-strands of the protruding stalk, respectively. Therefore, it is likely that only a subset of junctional residues can encode amino acids that maintain the β-strand structures. In this regard, a significant stretch of adenines in this V-D junctional diversity encodes a relatively conserved stretch of amino acids in the ascending strand. It is unclear whether these junctional residues are added by terminal deoxynucleotidyl transferase (TdT), and then the conserved residues are selected during B-cell development (Liljavirta et al., 2014), or whether atypical nucleotide insertional processes (Koti, Kataeva and Kaushik, 2010) occur to specifically add these residues during V-D recombination. In any event, there appears to be little recombinational diversity in antibodies with ultralong CDR H3 sequences, with only minor junctional diversity at the V-D junction. Compared to antibody genes from other species, this mechanism to produce functional variable regions is unusual in not enabling significant diversity through V(D)J recombination.
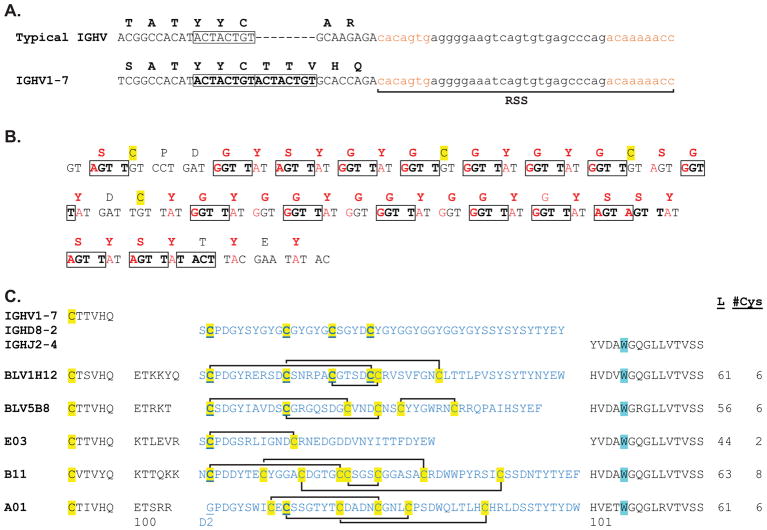
The IGHV1-7 gene is utilized in nearly every ultralong CDR H3 antibody analysed; however, it is also found rearranged to shorter DH segments in the shorter repertoire (Deiss et al., 2017). Interestingly, CDR H1 and CDR H2 of IGHV1-7 seem to retain a greater level of germline conservation than other VHs, which suggests they act as a scaffold for the ultralong CDR H3 microdomain, in lieu of accruing somatic hypermutations during affinity maturation. IGHV1-7 is unusual amongst all vertebrate VH regions in that it encodes three additional amino acids at the C-terminal region (Figure 3A). All immunoglobulin domains encode two conserved cysteines, which form a disulfide bond at the core of the immunoglobulin β-sandwich structure. The second cysteine is typically three amino acids from the C-terminus of the variable region encoded by the VH gene. In most vertebrates, an amino acid motif of CAR or CAK is readily recognized in this region. Notably, the positively charged arginine or lysine in CAR or CAK nearly always forms a salt bridge with a conserved aspartate encoded by the JH region, thus stabilizing the base of the CDR H3 loop. Ultralong CDR H3 antibodies do not have CAR/K but instead have a CTTVHQ motif encoded by IGHV1-7 that does not participate in a salt bridge, but instead initiates the ascending β-strand of the stalk, which protrudes from the antibody surface. Genetically, an eight basepair duplication at the 3′ end of the IGHV1-7 region shifts the reading frame, and encodes three additional amino acids (Figure 3A). As the recombination signal sequence (RSS) immediately follows the inserted sequence, the frame shift has no further consequences on the protein sequence; the residues after CTTVHQ will be then encoded by junctional residues and IGHD8-2. In addition to the germline eight bp duplication, IGHV1-7 also encodes amino acids in CDR H2, which interact with the ultralong CDR H3 stalk, and may play stabilizing roles. For example, AsnH50 in CDR H2 forms a hydrogen bond to GlnH97 in CDR H3. To summarize, bovine IGHV1-7 is unusual compared to other VH regions in vertebrates in having a germline frameshift insertion which lengthens the VH region and encodes a protruding β-ribbon, and additionally encodes potentially stabilizing residues that interact with the ultralong CDR H3.
Figure 3. Genetic basis for encoding ultralong CDR H3s.
(A) A germline insertion in germline IGHV1-7. The nucleotide and encoded amino acid sequence of the 3′ end of a typical vertebrate IGVH region is shown above and IGHV1-7 shown below. The coding region nucleotides are in uppercase letters. The recombination signal sequence (RSS) is shown in small letters, with the heptamer and nonamer in brown. IGHV1-7, which is used in ultralong CDR H3 antibodies, has an 8 base-pair duplication in the germline (boxed). This duplication shifts the reading frame and extends the amino acid sequence, allowing the initial portion of the ‘stalk’ β-strand to be encoded. (B) Nucleotide and amino acid sequence of the germline IGHD8-2 gene. Nucleotides which can be mutated to a residue that encodes a cysteine are shown in red, as are the corresponding amino acids. RGYW/WRCY mutational hotspots for AID are boxed. Note that a large fraction of codons of IGHD8-2 are mutational hotspots that can be mutated to cysteine. (C) Sequences of the five Fabs with ultralong CDR H3s whose structures have been determined. The germline regions of the IGHV1-7, IGHD8-2, and IGHJ2-4 are shown above, with the CDR H3 lengths and number of cysteines indicated to the right. The cysteines are highlighted in yellow, and the disulfide connectivities of the knob region are indicated. The cysteine and tryptophan, which define the boundaries of CDR H3, are highlighted in yellow and cyan, respectively. The amino acid numbers, using the system of Stanfield et al., are shown below (Stanfield, Wilson and Smider, 2016).
The IGHD8-2 region is particularly unusual because of its long length; it encodes 48 amino acids (Figure 3B) (Shojaei, Saini and Kaushik, 2003) that include the entire knob domain and a portion of the descending strand of the stalk. However, several other features are also apparent. A CPDG motif at the N-terminal end includes the first cysteine that is nearly always conserved in ultralong CDR H3 sequences (Figure 3). This cysteine forms a disulfide bond that is spatially conserved, despite the fact that its bonding partner is not conserved in the germline (Stanfield, Wilson and Smider, 2016). The CPDG motif also forms a β-turn, which leads into the first short β-strand that is conserved in the core of the knob domain (see below). The 3′ end of IGHD8-2 encodes several alternating tyrosines, which form stacking interactions at the C-terminus of CDR H3 and stabilize the descending strand of the stalk.
While we may infer that the knob encoded by the germline IGHD8-2 forms a distinct β-sheet core structure, as seen in five affinity-matured cow antibody structures (described below in Section 3), this motif is not obviously encoded by IGHD8-2. Internally IGHD8-2 encodes a repeating pattern of GYG amino acids. This repeating motif is also seen in other bovine DH regions (though of shorter lengths), and is also found in other proteins in nature including keratins and prisilkin, which function in the extracellular matrix. In this regard, a BLAST search using multiple GYG repeats did not yield any obvious results suggesting a structure for the germline DH; GYG-repeat containing proteins include a HET-domain containing protein (genebank:EIW77936.1), an ankyrin-repeat domain containing protein (genebank: EZA54628.1), keratins (e.g. genebank: KFQ43969.1), the RNA binding region of RNP-1 (genebank: APR83217.1), the glycine/tyrosine rich eggshell protein (genebank: ABF13207.1), and several predicted and so-called ‘hypothetical’ proteins. Thus, there currently is no significant insight into the role and importance of the GYG repeats in the germline structure of the knob. Although the small β-sheet has been seen in the crystallographic studies of five affinity-matured Fab fragments (Stanfield, Wilson and Smider, 2016), the amino acid identities of the β-strands in these five structures are diverse, and the residues encoding the three β-strands cannot be readily mapped to the germline IGHD8-2 gene. Thus, the structural features encoded by IGHD8-2 in the knob region Gly-Tyr-Gly repeats are not entirely clear. IGHD8-2 does encode four cysteines, so presumably two disulfide bonds can form in the germline knob. The IGHJ2-4 region appears to be used in all bovine heavy chains (shorter and ultralong), and encodes the short β-strand at the end of CDR H3. This region is similar in sequence and structure to JH regions of other vertebrates.
For ultralong CDR H3 antibodies, few germline light chains appear to be utilized in pairing with IGHV1-7 heavy chains. Adult bovines nearly exclusively use lambda light chains (Arun, Breuer and Hermanns, 1996;Pasman et al., 2010), and the germline V30 (previously termed Vλx1) was used in the four original ultralong CDR H3 antibodies found by the Kaushik group (Saini et al., 2003), as well as several of the recently described anti-g140 SOSIP trimer antibodies (Sok et al., 2017). The germline V30 light chain has structural features (see below) that may optimize its utility in ultralong CDR H3 antibodies. This gene is the most highly expressed VL in the fetal repertoire (Liljavirta et al., 2014), and maintains expression throughout adult life (Pasman, Merico and Kaushik, 2017). Interestingly, although lambda light chains predominate in adults, kappa light chains were identified in up to 22% of the neonatal repertoire (Pasman, Merico and Kaushik, 2017), although It is unclear whether these kappa light chains can pair with heavy chains containing ultralong CDR H3s. Interestingly, IGHV1-7 and IGHD8-2 are also highly expressed in neonatal calves, suggesting that ultralong CDR H3 antibodies may be a predominant component of the neonatal repertoire (Pasman, Merico and Kaushik, 2017). The genetic locus of the lambda light chain has been assembled and analysed in some detail; approximately 17 functional Vλ genes were identified with preferential expression of the vλ1 family (Pasman et al., 2010), and particularly V30 (Liljavirta et al., 2014); however, detailed analysis of light chain usage and VH-VL pairing in the context of an immune response and ultralong CDR H3 heavy chains has not yet been examined.
c. Somatic factors impacting diversity
In addition to encoding the CPDG β-turn motif and the descending β-strand of ultralong CDR H3s, the IGHD8-2 region has high mutagenic potential based on the number of RGYW (or in reverse, WRCY) hotspots (where R=A/G, W=A/T, Y=T/C). These sequences are substrates for the activation induced cytidine deaminase (AID), which is the key enzyme involved in somatic hypermutation of antibody genes (Muramatsu et al., 2000). Indeed, there are 19 hotspots in the 148 basepair IGHD8-2 region, covering nearly all of the protein coding sequence (Figure 3). Furthermore, the codon usage of the Gly-Tyr-Gly repeating motif is (i) biased towards comprising a hotspot as well as (ii) being optimized for cysteine mutations. The codon encoding glycine is GGT, and for tyrosine is TAT. GGT is the most underrepresented (i.e. the rarest) of the four codons encoding glycine in the bovine genome, yet is used nearly exclusively to encode multiple glycines in the IGHD8-2 germline gene. Similarly, TAT is rarer than TAC, the other codon that encodes tyrosine in the genome. However, the Gly-Tyr-Gly repeats, encoded by GGTTATGGT invariably include the GGTT hotspot for AID. Remarkably, both GGT and TAT codons can be mutated to cysteine with just one basepair change (both GGT and TAT to TGT). Thus, the IGHD8-2 gene appears to be ‘hard coded’ for potential mutation, through multiple hotspots, and specifically towards mutation to cysteine. Of note, other shorter bovine DH regions also have Gly-Tyr-Gly repeats and, although they do not have the remarkable length as IGHD8-2, they may also have hotspots and proclivity to mutate to cysteine. Thus, these genetic characteristics may be general features of bovine germline CDR H3s.
d. Sequence features of matured ultralong CDR H3s
Unlike humans or mice, several species appear to activate AID immediately following V(D)J recombination, and utilize somatic hypermutation to create diversity for the primary immunoglobulin repertoire (Liljavirta et al., 2013;Reynaud et al., 1995;Yasuda et al., 2006;Zhao, Jackson and Aitken, 2006). AID is highly expressed in bovine fetal spleen and Peyer’s patch (prior to antigen exposure), and serves to diversify the primary repertoire in cows (Liljavirta et al., 2013). While this mechanism may appear fundamentally different compared to other vertebrates who primarily develop their B-cells in the bone marrow, species like bovines primarily develop B-cells in gut-associated lymphoid tissue, which appears to be an analog of the chicken’s bursa of fabricius (Yasuda et al., 2006;Yasuda et al., 2002). Recently, however, evidence of AID activity was found in antigen naïve B-cells derived from the gut in well-controlled studies in transgenic mice (Yeap et al., 2015), suggesting that, in this compartment, AID-mediated diversification may indeed occur to diversify the primary repertoire in Mus musculus. Indeed, bovine ultralong CDR H3 sequences that are not fetally derived are very heavily mutated in their CDR H3 (Kaushik et al., 2009;Wang et al., 2013). Thus, a potential mechanism to reconcile these species differences is that the primary repertoire derived from gut B-cell development utilizes AID for primary diversification, whereas those B-cells derived from bone marrow development do not. The species differences, therefore, may be due to the relative proportion of the repertoire derived from these different compartments used for B-cell maturation. Of considerable note, the ability to mutate prior to antigen exposure dramatically increases the potential repertoire diversity, and has implications in vaccinology where germline/naïve versus mutated sequences in the repertoire may be important (Yeap et al., 2015).
In cows, the ability to generate further diversity in the primary repertoire through somatic hypermutation is very important – in the ultralong subset of antibodies, only single V, D, and J genes appear to produce the entire repertoire. Even in shorter CDR H3 antibodies, the homology between the remaining VH regions is so high that only limited diversity can be obtained through the use of alternative DH regions. Thus, it appears that, in cattle, a primary driver of antibody gene diversification is AID-mediated somatic mutation. When these ultralong CDR H3 antibody genes in the periphery are analyzed, they have unusual characteristics. First, several conserved non-germline encoded residues resulting from junctional diversity at the V-D joint are observed (Figure 3C and Figure 4). As mentioned, there are typically 5–6 non-templated amino acid residues in this region that encode the distal portion of the ascending β-strand of the stalk. Next, a nearly universally conserved cysteine in the knob is present. This cysteine, typically in the context of a CPDG motif, allowed for the derivation of a numbering system for bovine ultralong CDR H3 (Figure 3, C). Due to the exceptionally long length of these CDR H3 regions, standard numbering schemes like Kabat and Chothia (Al-Lazikani, Lesk and Chothia, 1997;Kabat and Wu, 1991) do not easily apply (Stanfield, Wilson and Smider, 2016). When mature sequences are aligned with this conserved cysteine (at position D2), other cysteines conserved with the germline IGHD8-2 region can be identified. Interestingly, it appears that, as one moves from the N to C-terminal direction in CDR H3, the conservation of the cysteines becomes increasingly less. Thus, the first germline-encoded cysteine is nearly completely conserved, whereas the fourth cysteine is the least conserved. Furthermore, presumably as a result of somatic hypermutation, non-germline encoded cysteines are clearly evident and can be found throughout the CDR H3 knob region. As mentioned above, the Gly-Tyr-Gly repeating motif is comprised of an AID hotspot, and each of these residues can be mutated to cysteine with only one nucleotide change.
Figure 4. Genetic components mapped onto the Fab structure of BLV5B8.
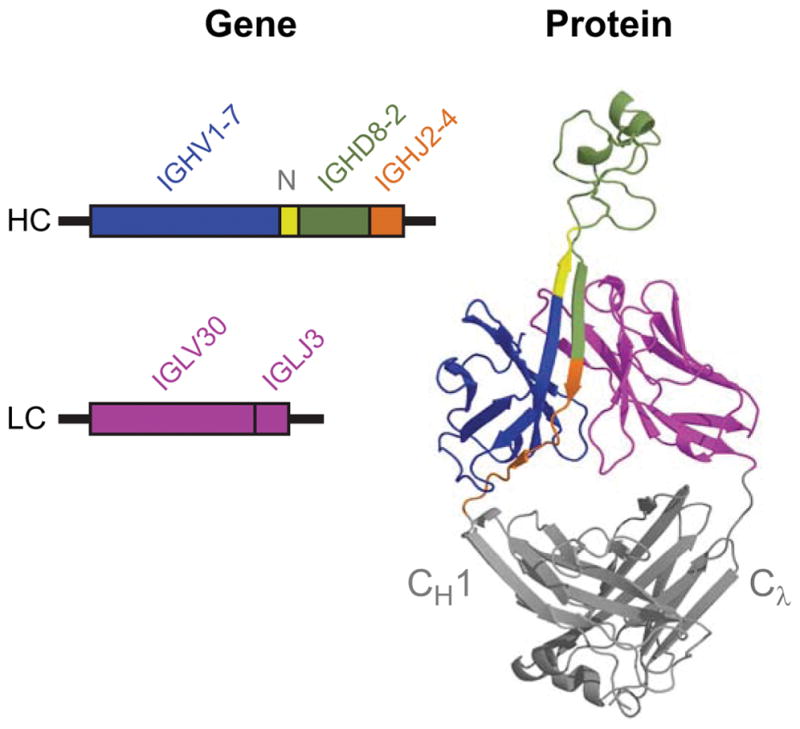
The rearranged genes (left) and their corresponding structural components (right) are color coded (blue, IGHV1-7; green, IGHD8-2; orange, IGHJ2-4, Pink, both IGLV30 and IGLJ3). In yellow is the area of V-D junction (labeled N) that encodes the ascending β-strand. This region is conserved and it is unclear whether it is derived from typical TdT catalyzed N-region insertions or other mechanisms like nucleotide capture. The CH1 and Cλ regions are indicated on the right.
Although there is not extensive junctional diversity in ultralong CDR H3 transcripts, there is considerable length variation within CDR H3. Given the potential structural constraints imposed by the ascending and descending strands of the β-ribbon stalk, which are each encoded at a recombinational junction, other sources of length diversity have been analyzed (Deiss et al., 2017). Surprisingly, deletions internal to IGHD8-2 (e.g. not at the V-D or D-J junctions) were identified in nearly 50% of recombined mature ultralong CDR H3 transcripts. These deletions range from 3–54 basepairs (1–18 codons) (Deiss et al., 2017). While AID is known to induce deletions, the number of nucleotides deleted, as well as the unusually high frequency of deletions, seem unique to bovine ultralong CDR H3 sequences. These deletions presumably resulted from AID-mediated processes, and could potentially be enhanced through the multiple repeats in IGHD8-2 that can allow DNA slippage events to occur. From the standpoint of a functional protein, and creation of diversity, deletions between cysteines potentially dramatically alter the surface loops on the knob, as well as the disulfide bonding patterns. Thus, in addition to SH providing an ability to alter amino acid content and mutation to and from cysteines, the feature of deleting residues can additionally alter protein secondary structure by changing loop length and disulfide bonds.
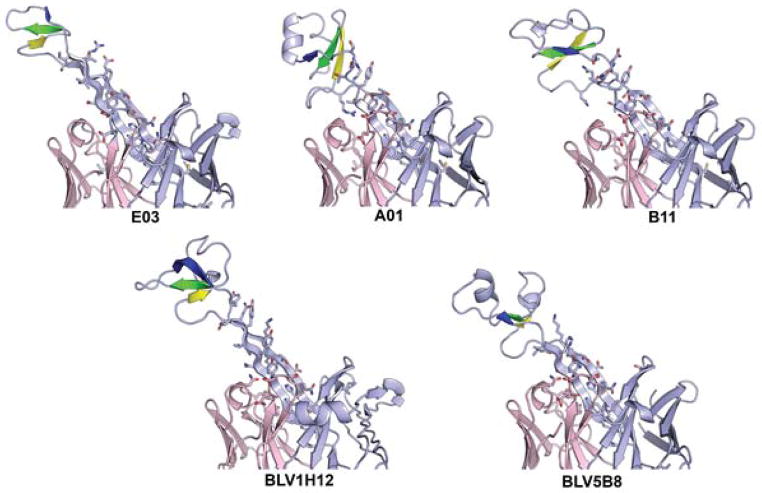
3. Structural biology of ultralong CDR H3 antibodies
Currently, five bovine Fab structures have been deposited to the Protein Data Bank (PDB), for Fabs BLV1H12 (4K3D, 1.85Å), BLV5B8 (4K3E, 2.2Å) (Wang et al., 2013), B11 (5IHU, 2.06Å), E03 (5IJV, 2.2Å), and A01 (5ILT, 2.0Å) (Stanfield, Wilson and Smider, 2016). These Fabs contain CDR H3 regions that range in length from 44 to 63 residues, and that contain anywhere from 1 to 4 disulfides (Table 1). While the long CDR H3s all adopt the stalk and knob architecture, the other five CDR loops have limited sequence diversity and have structures largely consistent with the canonical structures seen previously in human and murine Fabs (Al-Lazikani, Lesk and Chothia, 1997;Chothia and Lesk, 1987;Chothia et al., 1992), except for CDR L3 that has a bend in its tip to accommodate and likely support the C-terminal strand of the stalk. The five currently determined structures all contain the same λ light chain (germline V30).
Table 1.
Diverse structural features of ultralong CDR H3 Fabs
| Fab (PDB ID) | CDR H3 AA length | Protrusion distance (Å)a | No. Cys (disulfide patterns) | Stalk length (residues pairs)b |
|---|---|---|---|---|
| BLV1H12 (4K3D) | 61 | 41 | 6 (1–4, 2–6, 3–5) | 12 |
| BLV5B8 (4K3E) | 56 | 36 | 6 (1–3, 2–4, 5–6) | 12 |
| E03 (5IJV) | 44 | 37 | 2 (1–2) | 12 |
| B11 (5IHU) | 63 | 36 | 8 (1–4, 2–7, 3–8, 5–6) | 8 |
| A01 (5ILT) | 61 | 32 | 6 (1–4, 2–5, 3–6) | 8 |
Approximate distance from the tips of CDRs L1–L3, H1, H2 to the distal tip of the H3 knob
Length of hydrogen-bonded stalk domain in residues pairs (the number of amino acids forming the two-stranded β-ribbon divided by two)
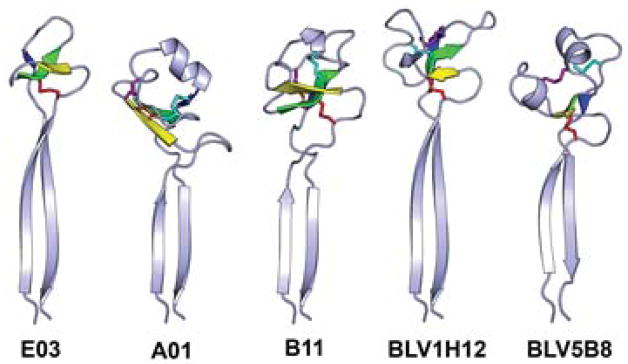
The knob domains all contain three short anti-parallel β-strands, connected by two loops of variable length and secondary structure (Figure 6). Each knob domain is initiated by a type-I β turn at the ‘CPDG’ sequence motif (C/G P/S D G/XD in the five current structures) encoded by the 5′ end of the DH gene. The D and G are the i+1 and i+2 residues in the β-turn. While this sequence motif is usually recognizable, there is no conserved sequence within the rest of the knob domains, and even the three β-strands vary in length and sequence. There is one spatially conserved disulfide from the first Cys residue in the CPDG motif to a Cys located on the second β-strand; however, the number of residues between this disulfide bonded pair can vary. Fab A02 lacks the highly conserved Cys in the CPDG motif, but nevertheless maintains the type I β-turn found in the other knob domains. The other disulfide bonds in the knob domain occur at different positions and with different disulfide connectivities (Table 1). The knob domains have limited structural homology with the small cyclic peptide Kalata B1 (PDB 1N1U, (Daly, Clark and Craik, 2003)), WW domain FE65 (PDB 2HO2, (Meiyappan, Birrane and Ladias, 2007)) and part of the tumor necrosis factor receptor superfamily member 13C (PDB 1OQE, (Liu et al., 2003)).
Figure 6. The stalk and knob domains of the five currently known bovine Fab ultralong CDR H3 regions.
The three strands in the core knob domain are colored yellow, green and blue and the disulfide bonds are shown as sticks. The orange disulfide is always formed from the first Cys in the CPDG motif to a Cys in the second core β-strand.
The β-ribbon stalk regions can also vary in length. E03 and BLV1H12 have long, regular β-ribbon stalks with 12 residues in each strand (e.g. 12 residue pairs (Table 1)), but A01, B11, and BLV5B8 have shorter β-ribbons of 8 residues on each side, with the B11 ribbon topped by short non-β linkers to the knob (Figure 6). The descending strands of the stalks generally contain stacking hydrophobic residues, likely adding stability to the stalk (Figure 7). Superposition of the Fab variable heavy chain domains shows that the stalk/knob units all extend at approximately the same angle from the Fabs, suggesting a fairly high degree of rigidity for the stalk. But the knobs can twist by as much as almost 60° with respect to their stalks, so that superposition of the stalks does not then superpose the knobs and vice versa. The combination of differing ribbon length and knob position results in different overall lengths for the stalk/knob units with respect to the antibody surface. The distance between the top of the knob domain to a plane roughly bisecting the tips of the other five CDR loops ranges between 32 to 41 Å (Table 1). Residue conservation is found at the N-terminal base of the stalk (C, T/V, T/S/I, V, H/Y, Q, E/K) and in the descending strand in the ladder of alternating aromatic residues. Along with several hydrogen bonds within the stalk regions, the surrounding CDR residues may play a role in the support and stabilization of the stalk domain, especially the descending strand of the stalk (Figure 7). The tip of CDR L3 (AEDSS, residues L92-95) stacks against a Tyr residue in the descending strand, 7 residues before the conserved TrpH103 (the CDR H3’s are too large for conventional Kabat numbering, so we will describe residue positions with respect to conserved residue TrpH103, as described in (Stanfield, Wilson and Smider, 2016)). The main chain of this Tyr also hydrogen bonds to AsnL30. TyrL32 stacks against a Glu/Asp found 6 residues before H103 and TyrL49 stacks against a His/Tyr found 4 residues upstream of H103. Thus, there are key residues both within CDR H3, and in other CDRs which appear to provide structural support to the unusual architecture of the heavy chain ultralong CDR H3.
Figure 7. The bovine stalk and surrounding CDR residues.
The Fab heavy and light chains are shown in light blue and pink, respectively The stalk regions all have very similar take-off angles from the body of the Fabs and appear to be structurally stable. Ladders of alternating aromatic residues may add to the stabilization of the stalks. The non-H3 CDR loops are well conserved and may also aid in support for the long stalk.
4. Biochemistry and potential function of ultralong CDR H3 antibodies
a. Antigen binding
Unlike any other vertebrates examined, cows are able to mount a rapid broadly neutralizing antibody response against the stable engineered HIV gp140 SOSIP Env trimer (Sok et al., 2017). Several cow monoclonal antibodies were isolated and their epitopes mapped to the CD4 binding site of the gp120 component of the Env glycoprotein. Of note, all of the antibodies reported in this study had ultralong CDR H3s of 60 or 61 amino acids. One antibody termed NC-Cow1 was able to neutralize 72% of the strains in a 117 strain panel at high potency. The likely mechanism for the neutralization of these antibodies is that their long CDR H3 is able to navigate through the glycans in the glycan shield surrounding the CD4 binding site to contact conserved the protein epitopes on the glycoprotein that interact with CD4 receptor. Indeed, electron microscopy was able to visualize the long CDR H3 binding to the CD4 binding site, without evidence of direct antigen contact by the other CDRs. In this regard, current evidence suggests that some of the other five CDRs, as opposed to contacting antigen, play a structural role through supporting the conformation of the long CDR3 loop at its base. In addition to the direct EM evidence with NC-Cow1, an antibody targeting bovine viral diarrheal virus (BVDV) was identified and removal of its knob domain completely abrogated binding to the virus (Wang et al., 2013). Alanine scanning mutagenesis of the entire CDR H3 was able to identify the key residues involved in binding. Although these studies do not rule out the possibility that some cow ultralong CDR H3 antibodies may be found that have binding contributions from the other CDRs, the fact that little diversity is seen in the other CDRs (Deiss et al., 2017), with only a single VH region and utilization of a limited number of light chains, which are often in nearly germline configuration, suggests that the main driver of antigen binding is the ultralong CDR H3. Further crystallographic information on a diversity of antigens with this unusual antibody class will shed more light on their binding and recognition mechanisms at the atomic level.
A key feature of any antibody is its affinity and specificity. While early experiments with the ultralong antibodies derived from BLV infected B-cells suggested that some ultralong CDR H3 antibodies had polyspecific binding properties (Saini et al., 1999), ELISAs and cell-based binding assays of the anti-HIV and anti-BVDV monoclonal antibodies show that these particular monoclonal antibodies bind with high specificity to their cognate antigens (Sok et al., 2017;Wang et al., 2013). Neutralization potency of antibodies targeting HIV BG505 SOSIP suggests their binding affinities are in the low nanomolar range. Indeed, surface plasmon resonance analysis showed subnanomolar Kds for NS-Cow1 and other anti-HIV cow antibodies (V. Smider and M. Vadnais, unpublished). Thus, the unusual antibodies with ultralong CDR H3s derived from cattle appear to have similar antigen binding characteristics in terms of specificity and affinity as other antibodies derived by affinity maturation from other vertebrate species.
b. Potential function and evolution
It is presently unclear why bovines have evolved the ultralong CDR H3 system, which differs dramatically from other vertebrate repertoires at both the genetic and structural levels. At least two germline genetic events occurred during evolution to enable creation of this system: (i) the in-frame 8 basepair duplication at the 3′ end of IGHV1-7, and (ii) the generation of the ultralong IGHD8-2 diversity gene. On one hand, it is possible that this system developed as an alternative mechanism to create structural diversity in the repertoire. In this vein, whereas multiple VH, DH, and JH segments can form enormous combinatorial diversity in some organisms, bovines, and perhaps other mammals, may have minimalized combinatorial diversity in favor of a mechanism whereby structural diversity is created prior to antigen exposure through AID-driven processes that alter loops and structures by varying the disulfide connectivities and amino-acid content of the knob domain. An alternative hypothesis, however, is that specific microorganisms drove evolution of this system. The digestive physiology of cows is distinct in utilizing a four-chambered stomach, with the rumen compartment acting as a large fermenter (80L for a full grown cow). In order to process cellulose and other feedstuff, the rumen contains a highly diverse and symbiotic mixture of both prokaryotic and eukaryotic microorganisms, particularly bacteria and protozoa (Bryant, 1959;Hungate, 1975;Hungate, Bryant and Mah, 1964;Mao et al., 2015;Sharpe, Latham and Reiter, 1969), of which the latter may present an unusual antigenic burden to which other vertebrates are not typically exposed. Indeed, cows must contain these microorganisms within the rumen compartment, as dissemination of these microorganisms via the blood or tissue can cause life-threatening disease(Miesner and Reppert, 2017;Ward and Ducharme, 1994;Zinicola et al., 2015). Serum from cattle is known to possess antibodies reactive to rumen microorganisms, suggesting that an immune response against their rumen antigens is common (Sharpe, Latham and Reiter, 1969).
An alternative hypothesis is that certain infections (as opposed to rumen symbionts) have driven development of the unusual architecture of bovine ultralong CDR H3s. As a species, cows are exposed to several retroviruses including bovine leukemia virus (BLV), which can cause substantial morbidity within a herd, bovine syncytial virus (BSV), and bovine immunodeficiency-like virus (BIV), which is analogous to HIV but only causes mild disease in bovines and does not lead to acquired immunodeficiency syndrome (AIDS) (Carpenter et al., 1992;Flaming et al., 1993;Gonda et al., 1990;Jacobs et al., 1992;Onuma et al., 1992). A recent observation in studying broadly neutralizing antibodies to certain human viruses was that these antibodies often had long CDR H3s; human antibodies typically average 12–15 residues, whereas in broadly neutralizing antibodies against HIV, the CDR H3 can be >30 amino acids (reviewed (Burton and Hangartner, 2016;Burton and Mascola, 2015)).
In light of the importance of these rare human antibodies, the HIV immunization experiments described above were performed. Four cows were immunized with a soluble, native-like gp140 Env trimer (BG505 SOSIP), and the neutralizing antibody response was impressive (Sok et al., 2017). Immunization of one cow led to a remarkable 96% serum neutralization breadth (of 117 cross-clade isolates), with isolation of the monoclonal antibody NS-Cow1 with an ultralong CDR H3 of 60 amino acids that could potently neutralize 72% of cross-clade isolates with an IC50 of 0.028 μg/ml. Broad neutralization properties developed rapidly; breadth (20%) could even be observed after a single prime and homologous boost. Such responses are unprecedented in animal immunization against HIV, and could make cows a novel model for evaluating vaccine candidates because of their unusual, and potentially optimized, repertoire for such types of antigen. Of note, cross-reactive immune responses between HIV and BIV were noted decades ago (Jacobs et al., 1992), as was crossreactivity of serum antibodies against different bovine retroviruses. Thus, a potential hypothesis for evolution of the unusual ultralong CDR H3 cow antibody repertoire is that it was selected to combat certain classes of viruses, and potentially retroviruses, as a biologically economic means to cross-react against several related and perhaps even poorly related strains.
5. Perspectives
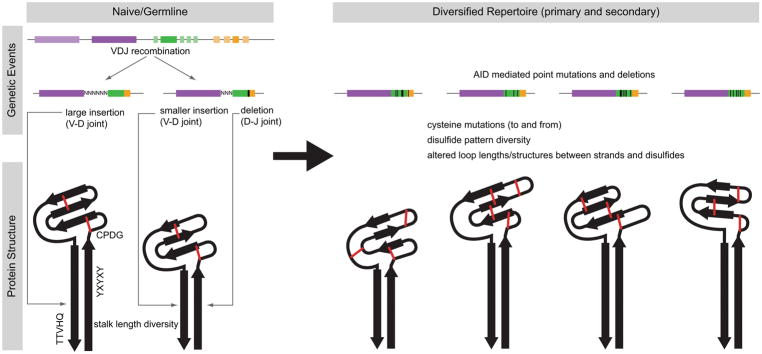
One of the most fundamental questions in the history of immunology was how a limited amount of genetic material in the genome could encode an apparently limitless array of antibodies in its repertoire to protect the host from the vast numbers of possible infections as well as cancer. The discovery of V(D)J recombination several decades ago (Hozumi and Tonegawa, 1976), and the resulting understanding of the combinatorial nature of antibody diversity development provided a fundamental and compelling answer to this question. Humans, for example, have a variety of heavy variable light and heavy chain germline genes that can recombine with different (D) and J regions. The ultralong CDR H3 antibody system of cows, however, appears to utilize a different mechanism to achieve similar ends (Figure 8). Only a single V-D-J recombination event leads to the precursor “naïve” antibody molecule. This naïve molecule has the motifs associated with ‘stalk’ and ‘knob’ formation with four germline-encoded cysteines that presumably form two disulfide bonds. The V-D and D-J junctions can provide limited junctional diversity (as occurs in other mammalian systems), which may alter the length of the ‘stalk’ domain. This single genetic unit then undergoes somatic hypermutation that can potentially ‘reshape’ the knob domain through (i) amino acid point mutations, (ii) mutations to and from cysteine, altering loop structures and disulfide connectivities, and (iii) nucleotide deletions which can further alter loop lengths, as well as disulfide bond patterns (Figure 8). While detailed molecular and cellular studies, including the tissue compartment(s) involved (such as bone marrow vs Peyer’s patch) or the temporal nature of these events, have not been performed, all of these events can be mediated by AID, which serves to shape the primary repertoire (before antigen exposure) in bovines. Thus, although cows perform V(D)J recombination, the primary driver of genetic and ultimately structural diversity may be somatic mutation by the AID enzyme.
Figure 8. Genetic and structural model for ultralong CDR H3 antibody repertoire formation.
One or more germline V-D-J recombination events using IGHV1-7 (dark purple), IGHD8-2 (dark green), and IGHJ2-4 (dark orange) is shown in the upper left. Junctional diversity, including insertions and deletions, can potentially alter the length of the stalk, as the V-D and D-J joints occur in regions encoding the ascending and descending strands, respectively. Presumably, two disulfide bonds can form in the knob of the naïve/germline antibody (shown in red). The primary repertoire is further diversified in the gut associated lymphoid tissue by AID-mediated processes (upper right). Nucleotide changes that alter amino acid content, particularly to cysteines, can change disulfide patterns and loops (red lines). Larger deletions (black rectangles) in the DH region can also potentially change loop lengths and disulfide patterns. The β-strands of the knob core and stalk domain are represented by black arrows.
From a structural view, it is intriguing that a single germline protein can apparently evolve, through mutation, into a myriad of different disulfide-bonded ‘knob’ minidomains. In this regard, it has not yet been determined whether the germline precursor antibody can adopt one or many conformations in its knob domain, or whether it possesses a defined (single) structure. Crystal structures of five affinity-matured Fab fragments show a conserved short β-sheet core; however, neither the amino acid sequences of the β-strands nor their linear positions are conserved across different knob sequences. Thus, although it seems likely that the β-sheet core should be encoded in the germline, the details regarding its formation are unclear. Thus, a paradox appears to exist; while this presumably important β-sheet feature is conserved between several affinity-matured knobs, the genetic basis for this conserved structure is not readily apparent in the germline IGHD8-2 DNA. In contrast, the underlying genetic basis for the other conserved structural motifs of the ultralong CDR H3, including the ascending and descending strands of the stalk, and β-turn of the knob, is readily identifiable in IGHV1-7 or IGHD8-2. The germline DH region encodes the β-turn, the 4 cysteines with intervening Gly-Tyr-Gly repeats, and the descending strand of the stalk. With regard to the cysteines, while we presume that two disulfide bonds form in the germline structure, there are actual several theoretical possibilities. These two disulfide bonds may adopt one of three different configurations (e.g. 1–2, 3–4; 1–3, 2–4; or 1–4, 2–3 patterns). Alternatively, it is possible that some cysteines are unpaired; there are six configurations of four cysteines where one disulfide bond forms but the remaining two cysteines are unpaired. A final possibility is that all four cysteines are unpaired, and that disulfides only form during the SH process, when the knob is fully folded or adopts an alternate conformation. In the context of these possibilities, the Gly-Tyr-Gly repeats are likely important, as either (i) the Gly-Tyr-Gly repeats can form a β-sheet core allowing at least the first highly conserved cysteine to form a disulfide bond, or (ii) perhaps less likely, the knob itself is partially stabilized through formation of the disulfide bonds and does not form a β-sheet core until the requisite mutations from AID occur within the Gly-Tyr-Gly regions. An intriguing question related to the first possibility (discussed below) is whether more than one β-sheet structure/disulfide combination could exist in the germline, which would enable further diversity in the structural repertoire.
The structure of the germline knob should give further insights into the generation of diversity in the ultralong CDR H3 repertoire. The germline knob encoded by the IGHD8-2 region is not obviously intrinsically disordered; in fact, predictive algorithms like IUPred or DisEMBL (www.iupred.enzim.hu, and www.dis.embl.de) score the germline DH as more likely to be highly ordered. Coupled with the likelihood of constraints on the N- and C-termini on the knob from the close proximity to the β-strands forming the stalk, as well as two probable disulfide bonds, this suggests that one or more stable structures of the germline knob may exist. Indeed, some germline CDRs have been reported to be able to form more than one conformation for the same sequence (Wedemayer et al., 1997;Yin et al., 2003). This ‘structural plasticity’ has been suggested to provide added diversity to the naïve repertoire, and explain the tendency for germline antibodies to have oligospecific or polyspecific antigen binding properties. Furthermore, germline CDR H3s have been shown to be flexible compared to their affinity-matured counterparts in certain cases that have been examined (Adhikary et al., 2015;Jimenez et al., 2004;Thielges et al., 2008;Zimmermann et al., 2006). It has been postulated that germline CDR amino acid content has been evolved for structural flexibility (Babor and Kortemme, 2009), and ‘induced fit’ of the antibody with antigen has been proposed (Rini, Schulze-Gahmen and Wilson, 1992;Stanfield et al., 2007). CDR H3s in all species, and their corresponding germline DH regions, are overrepresented by tyrosine and glycine (Babor and Kortemme, 2009;Birtalan et al., 2008;Fellouse, Wiesmann and Sidhu, 2004). Thus, if the knob region encoded by bovine IGHD8-2 behaves like other germline CDR H3s, it may have substantial flexibility. The presence of four cysteines, and the potential to form different disulfide patterns, could ‘lock in’ different knob loop patterns as part of the repertoire during somatic hypermutation.
The recent discovery of two IgM genes in cattle is interesting in that ultralong CDR H3 antibodies appear to utilize IgM2 (Ma et al., 2016). Each IGHM gene has upstream V, D, and J sequences, which appear to rearrange and splice to IGHM1 or IGHM2. Thus, while ultralong CDR H3 antibodies use IGHM2, other V-D-J rearrangements with shorter CDR H3s can also use this isotype. No mammalian species other than bovines have been reported to have more than one IGHM gene; however, crocodilians interestingly have three (Cheng et al., 2013). This duplicative arrangement of gene segments suggests that independent V-D-J events could occur in a single B-cell, resulting in expression of two heavy chains; however, this has not been investigated. Furthermore, both IGHM genes have μ switch regions, suggesting that a V-D-J recombination event upstream of IGHM1 could class switch to IGHM2. Indeed, Ma et al. determined that class switch recombination to IGHM2 from IGHM1 does occur in splenocytes. Of course, during an immune response, class switch recombination from either IGHM1 or IGHM2 to downstream γ, ε, or α constant region genes would also occur. Quantitative PCR of IGHM1 versus IGHM2 gene expression from a variety of lymphoid tissues at different developmental stages did not show any tissue-specific patterns for the two genes, although IGHM2 had slightly higher expression levels in all tissues examined (Ma et al., 2016). Thus, the evolutionary significance of two IGHM genes, and their relationship to the unusual ultralong CDR H3 antibodies of cattle remains a mystery.
In conclusion, the ultralong CDR H3 subset of bovine antibodies illuminates unexpected mechanisms to produce a structurally diverse immune repertoire. These antibodies contain CDR H3s that project out far from the typical antibody paratope using a β-ribbon stalk and a disulfide-bonded knob containing a small conserved β-sheet core. The knob loops and disulfide patterns are quite diverse in sequence and structure, and we are just beginning to understand the genesis of this diversity that appears to occur through heavy SH that includes significant deletions. The intrigue of these antibodies is heightened by cow immunization experiments with the Envelope glycoprotein of HIV, where potent broadly neutralizing antibodies with ultralong CDR H3s were rapidly induced- a finding so far not observed in any other species by immunization with the HIV Env antigen. The potential of cow antibodies to penetrate through glycan-coated antigens in viruses (as in HIV), or bind to cryptic or recessed epitopes, such as those in enzymes, pores, or channels, is likely to be the subject of further investigation, as antibodies to these types of epitopes could have important applications in research and medicine.
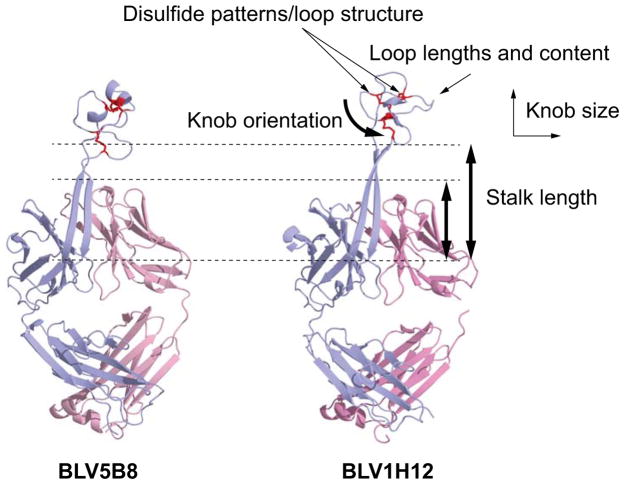
Figure 5. Structural diversity in the bovine ultralong CDR H3 repertoire.
These include diverse disulfide patterns, driven by cysteine mutations, different loop lengths and amino acid content formed through AID point mutations as well as diversity in deletions, knob orientation, size, and stalk length.
Acknowledgments
This work was supported by NIH grant R01 GM105826-01 to VVS, R21 AI120791 to VVS and MFC, NSF grant IOS-1656870 to MFC, and NIH UM1 AI100663 to IAW.
References
- Adhikary R, Yu W, Oda M, Walker RC, Chen T, Stanfield RL, Wilson IA, Zimmermann J, Romesberg FE. Adaptive mutations alter antibody structure and dynamics during affinity maturation. Biochemistry. 2015;54:2085. doi: 10.1021/bi501417q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol. 1997;273:927. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- Anderson D, Billingham RE, Lampkin GH, Medawar PB. The use of skin grafting to distinguish between monozygotic and dizygotic twins in cattle. Heredity. 1951;5:379. [Google Scholar]
- Arun SS, Breuer W, Hermanns W. Immunohistochemical examination of light-chain expression (lambda/kappa ratio) in canine, feline, equine, bovine and porcine plasma cells. Zentralbl Veterinarmed A. 1996;43:573. doi: 10.1111/j.1439-0442.1996.tb00489.x. [DOI] [PubMed] [Google Scholar]
- Babor M, Kortemme T. Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility. Proteins. 2009;75:846. doi: 10.1002/prot.22293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxby D. Edward Jenner’s inquiry; a bicentenary analysis. Vaccine. 1999;17:301. doi: 10.1016/s0264-410x(98)00207-2. [DOI] [PubMed] [Google Scholar]
- Berens SJ, Wylie DE, Lopez OJ. Use of a single VH family and long CDR3s in the variable region of cattle Ig heavy chains. Int Immunol. 1997;9:189. doi: 10.1093/intimm/9.1.189. [DOI] [PubMed] [Google Scholar]
- Billingham RE, Brent L. Acquired tolerance of foreign cells in newborn animals. Proc R Soc Lond B Biol Sci. 1956;146:78. doi: 10.1098/rspb.1956.0073. [DOI] [PubMed] [Google Scholar]
- Billingham RE, Brent L, Medawar PB. Actively acquired tolerance of foreign cells. Nature. 1953;172:603. doi: 10.1038/172603a0. [DOI] [PubMed] [Google Scholar]
- Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377:1518. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- Brown WR, Rabbani H, Butler JE, Hammarstrom L. Characterization of the bovine Cα gene. Immunology. 1997;91:1. doi: 10.1046/j.1365-2567.1997.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant MP. Bacterial species of the rumen. Bacteriol Rev. 1959;23:125. doi: 10.1128/br.23.3.125-153.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol. 2016;34:635. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16:571. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S, Miller LD, Alexandersen S, Whetstone CA, VanDerMaaten MJ, Viuff B, Wannemuehler Y, Miller JM, Roth JA. Characterization of early pathogenic effects after experimental infection of calves with bovine immunodeficiency-like virus. J Virol. 1992;66:1074. doi: 10.1128/jvi.66.2.1074-1083.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Gao Y, Wang T, Sun Y, Wei Z, Li L, Ren L, Guo Y, Hu X, Lu Y, Wang X, Liu G, Zhang C, Yu J, Pan-Hammarstrom Q, Hammarstrom L, Wu X, Li N, Zhao Y. Extensive diversification of IgH subclass-encoding genes and IgM subclass switching in crocodilians. Nat Commun. 2013;4:1337. doi: 10.1038/ncomms2317. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987;196:901. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM, Gherardi E, Tomlinson IM, Walter G, Marks JD, Llewelyn MB, Winter G. Structural repertoire of the human VH segments. J Mol Biol. 1992;227:799. doi: 10.1016/0022-2836(92)90224-8. [DOI] [PubMed] [Google Scholar]
- Daly NL, Clark RJ, Craik DJ. Disulfide folding pathways of cystine knot proteins. Tying the knot within the circular backbone of the cyclotides. J Biol Chem. 2003;278:6314. doi: 10.1074/jbc.M210492200. [DOI] [PubMed] [Google Scholar]
- Deiss TC, Vadnais ML, Wang F, Chen PL, Torkamani A, Mwangi W, Lefranc M-P, Criscitiello MF, Smider VV. Immunogenetic factors driving formation of ultralong VH CDR3 in Bos taurus antibodies. 2017 doi: 10.1038/cmi.2017.117. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPietro LA, Sethupathi P, Kingzette M, Zhai SK, Suter M, Knight KL. Limited repertoire of utilized VH gene segments in a VHa3-allotype-suppressed rabbit. Int Immunol. 1992;4:555. doi: 10.1093/intimm/4.5.555. [DOI] [PubMed] [Google Scholar]
- Eguchi-Ogawa T, Wertz N, Sun XZ, Piumi F, Uenishi H, Wells K, Chardon P, Tobin GJ, Butler JE. Antibody repertoire development in fetal and neonatal piglets. XI. The relationship of variable heavy chain gene usage and the genomic organization of the variable heavy chain locus. J Immunol. 2010;184:3734. doi: 10.4049/jimmunol.0903616. [DOI] [PubMed] [Google Scholar]
- Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: A dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci U S A. 2004;101:12467. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaming K, van der Maaten M, Whetstone C, Carpenter S, Frank D, Roth J. Effect of bovine immunodeficiency-like virus infection on immune function in experimentally infected cattle. Vet Immunol Immunopathol. 1993;36:91. doi: 10.1016/0165-2427(93)90100-i. [DOI] [PubMed] [Google Scholar]
- Gonda MA, Oberste MS, Garvey KJ, Pallansch LA, Battles JK, Pifat DY, Bess JW, Jr, Nagashima K. Development of the bovine immunodeficiency-like virus as a model of lentivirus disease. Dev Biol Stand. 1990;72:97. [PubMed] [Google Scholar]
- Hayes HC, Petit EJ. Mapping of the β-lactoglobulin gene and of an immunoglobulin m heavy chain-like sequence to homoeologous cattle, sheep, and goat chromosomes. Mamm Genome. 1993;4:207. doi: 10.1007/BF00417564. [DOI] [PubMed] [Google Scholar]
- Hosseini A, Campbell G, Prorocic M, Aitken R. Duplicated copies of the bovine JH locus contribute to the Ig repertoire. Int Immunol. 2004;16:843. doi: 10.1093/intimm/dxh085. [DOI] [PubMed] [Google Scholar]
- Hozumi N, Tonegawa S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc Natl Acad Sci U S A. 1976;73:3628. doi: 10.1073/pnas.73.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungate RE. The rumen microbial ecosystem. Annu Rev Ecol Syst. 1975;6:39. [Google Scholar]
- Hungate RE, Bryant MP, Mah RA. The rumen bacteria and protozoa. Annu Rev Microbiol. 1964;18:131. doi: 10.1146/annurev.mi.18.100164.001023. [DOI] [PubMed] [Google Scholar]
- Jacobs RM, Smith HE, Gregory B, Valli VE, Whetstone CA. Detection of multiple retroviral infections in cattle and cross-reactivity of bovine immunodeficiency-like virus and human immunodeficiency virus type 1 proteins using bovine and human sera in a western blot assay. Can J Vet Res. 1992;56:353. [PMC free article] [PubMed] [Google Scholar]
- Jimenez R, Salazar G, Yin J, Joo T, Romesberg FE. Protein dynamics and the immunological evolution of molecular recognition. Proc Natl Acad Sci U S A. 2004;101:3803. doi: 10.1073/pnas.0305745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kabat EA, Wu TT. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J Immunol. 1991;147:1709. [PubMed] [Google Scholar]
- Kacskovics I, Butler JE. The heterogeneity of bovine IgG2--VIII. The complete cDNA sequence of bovine IgG2a (a2) and an IgG1. Mol Immunol. 1996;33:189. doi: 10.1016/0161-5890(95)00107-7. [DOI] [PubMed] [Google Scholar]
- Kaushik A, Shojaei F, Saini SS. Novel insight into antibody diversification from cattle. Vet Immunol Immunopathol. 2002;87:347. doi: 10.1016/s0165-2427(02)00063-6. [DOI] [PubMed] [Google Scholar]
- Kaushik AK, Kehrli ME, Jr, Kurtz A, Ng S, Koti M, Shojaei F, Saini SS. Somatic hypermutations and isotype restricted exceptionally long CDR3H contribute to antibody diversification in cattle. Vet Immunol Immunopathol. 2009;127:106. doi: 10.1016/j.vetimm.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Koti M, Kataeva G, Kaushik A. Organization of DH-gene locus is distinct in cattle. Dev Biol. 2008;132:307. doi: 10.1159/000317176. [DOI] [PubMed] [Google Scholar]
- Koti M, Kataeva G, Kaushik AK. Novel atypical nucleotide insertions specifically at VH-DH junction generate exceptionally long CDR3H in cattle antibodies. Mol Immunol. 2010;47:2119. doi: 10.1016/j.molimm.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Kasinathan P, Sathiyaseelan T, Jiao JA, Matsushita H, Sathiyaseelan J, Wu H, Mellquist J, Hammitt M, Koster J, Kamoda S, Tachibana K, Ishida I, Robl JM. Antigen-specific human polyclonal antibodies from hyperimmunized cattle. Nat Biotechnol. 2009;27:173. doi: 10.1038/nbt.1521. [DOI] [PubMed] [Google Scholar]
- Liljavirta J, Ekman A, Knight JS, Pernthaner A, Iivanainen A, Niku M. Activation-induced cytidine deaminase (AID) is strongly expressed in the fetal bovine ileal peyer’s patch and spleen and is associated with expansion of the primary antibody repertoire in the absence of exogenous antigens. Mucosal Immunol. 2013;6:942. doi: 10.1038/mi.2012.132. [DOI] [PubMed] [Google Scholar]
- Liljavirta J, Niku M, Pessa-Morikawa T, Ekman A, Iivanainen A. Expansion of the preimmune antibody repertoire by junctional diversity in Bos taurus. PLoS One. 2014;9:e99808. doi: 10.1371/journal.pone.0099808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hong X, Kappler J, Jiang L, Zhang R, Xu L, Pan CH, Martin WE, Murphy RC, Shu HB, Dai S, Zhang G. Ligand-receptor binding revealed by the TNF family member Tall-1. Nature. 2003;423:49. doi: 10.1038/nature01543. [DOI] [PubMed] [Google Scholar]
- Lopez O, Perez C, Wylie D. A single VH family and long CDR3s are the targets for hypermutation in bovine immunoglobulin heavy chains. Immunol Rev. 1998;162:55. doi: 10.1111/j.1600-065x.1998.tb01429.x. [DOI] [PubMed] [Google Scholar]
- Ma L, Qin T, Chu D, Cheng X, Wang J, Wang X, Wang P, Han H, Ren L, Aitken R, Hammarstrom L, Li N, Zhao Y. Internal duplications of DH, JH, and C region genes create an unusual IgH gene locus in cattle. J Immunol. 2016;196:4358. doi: 10.4049/jimmunol.1600158. [DOI] [PubMed] [Google Scholar]
- Mao S, Zhang M, Liu J, Zhu W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci Rep. 2015;5:16116. doi: 10.1038/srep16116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiyappan M, Birrane G, Ladias JA. Structural basis for polyproline recognition by the FE65 WW domain. J Mol Biol. 2007;372:970. doi: 10.1016/j.jmb.2007.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesner MD, Reppert EJ. Diagnosis and treatment of hardware disease. Vet Clin North Am Food Anim Pract. 2017;33 doi: 10.1016/j.cvfa.2017.06.007. pii: S0749. [DOI] [PubMed] [Google Scholar]
- Mousavi M, Rabbani H, Hammarstrom L. Characterization of the bovine epsilon gene. Immunology. 1997;92:369. doi: 10.1046/j.1365-2567.1997.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi M, Rabbani H, Pilstrom L, Hammarstrom L. Characterization of the gene for the membrane and secretory form of the IgM heavy-chain constant region gene (Cμ) of the cow (Bos taurus) Immunology. 1998;93:581. doi: 10.1046/j.1365-2567.1998.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Niku M, Liljavirta J, Durkin K, Schroderus E, Iivanainen A. The bovine genomic DNA sequence data reveal three IGHV subgroups, only one of which is functionally expressed. Dev Comp Immunol. 2012;37:457. doi: 10.1016/j.dci.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Onuma M, Koomoto E, Furuyama H, Yasutomi Y, Taniyama H, Iwai H, Kawakami Y. Infection and dysfunction of monocytes induced by experimental inoculation of calves with bovine immunodeficiency-like virus. J Acquir Immune Defic Syndr. 1992;5:1009. [PubMed] [Google Scholar]
- Owen RD. Immunogenetic consequences of vascular anastomoses between bovine twins. Science. 1945;102:400. doi: 10.1126/science.102.2651.400. [DOI] [PubMed] [Google Scholar]
- Pasman Y, Merico D, Kaushik AK. Preferential expression of IGHV and IGHD encoding antibodies with exceptionally long CDR3H and a rapid global shift in transcriptome characterizes development of bovine neonatal immunity. Dev Comp Immunol. 2017;67:495. doi: 10.1016/j.dci.2016.08.020. [DOI] [PubMed] [Google Scholar]
- Pasman Y, Saini SS, Smith E, Kaushik AK. Organization and genomic complexity of bovine λ-light chain gene locus. Vet Immunol Immunopathol. 2010;135:306. doi: 10.1016/j.vetimm.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Rabbani H, Brown WR, Butler JE, Hammarstrom L. Polymorphism of the IGHG3 gene in cattle. Immunogenetics. 1997;46:326. doi: 10.1007/s002510050279. [DOI] [PubMed] [Google Scholar]
- Reynaud CA, Garcia C, Hein WR, Weill JC. Hypermutation generating the sheep immunoglobulin repertoire is an antigen-independent process. Cell. 1995;80:115. doi: 10.1016/0092-8674(95)90456-5. [DOI] [PubMed] [Google Scholar]
- Rini JM, Schulze-Gahmen U, Wilson IA. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992;255:959. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- Saini SS, Allore B, Jacobs RM, Kaushik A. Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur J Immunol. 1999;29:2420. doi: 10.1002/(SICI)1521-4141(199908)29:08<2420::AID-IMMU2420>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Saini SS, Farrugia W, Ramsland PA, Kaushik AK. Bovine IgM antibodies with exceptionally long complementarity-determining region 3 of the heavy chain share unique structural properties conferring restricted VH + Vλ pairings. Int Immunol. 2003;15:845. doi: 10.1093/intimm/dxg083. [DOI] [PubMed] [Google Scholar]
- Saini SS, Hein WR, Kaushik A. A single predominantly expressed polymorphic immunoglobulin VH gene family, related to mammalian group, I, clan, II, is identified in cattle. Mol Immunol. 1997;34:641. doi: 10.1016/s0161-5890(97)00055-2. [DOI] [PubMed] [Google Scholar]
- Sharpe ME, Latham MJ, Reiter B. The occurrence of natural antibodies to rumen bacteria. J Gen Microbiol. 1969;56:353. doi: 10.1099/00221287-56-3-353. [DOI] [PubMed] [Google Scholar]
- Shojaei F, Saini SS, Kaushik AK. Unusually long germline DH genes contribute to large sized CDR3H in bovine antibodies. Mol Immunol. 2003;40:61. doi: 10.1016/s0161-5890(03)00098-1. [DOI] [PubMed] [Google Scholar]
- Sok D, Le KM, Vadnais M, Saye-Francisco KL, Jardine JG, Torres JL, Berndsen ZT, Kong L, Stanfield R, Ruiz J, Ramos A, Liang CH, Chen PL, Criscitiello MF, Mwangi W, Wilson IA, Ward AB, Smider VV, Burton DR. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature. 2017;548:108. doi: 10.1038/nature23301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Dooley H, Verdino P, Flajnik MF, Wilson IA. Maturation of shark single-domain (IgNAR) antibodies: Evidence for induced-fit binding. J Mol Biol. 2007;367:358. doi: 10.1016/j.jmb.2006.12.045. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Wilson IA, Smider VV. Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies. Sci Immunol. 2016;1:aaf7962. doi: 10.1126/sciimmunol.aaf7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons DB, Clarkson CA, Beale D. Structure of bovine immunoglobulin constant region heavy chain γ1 and γ2 genes. Mol Immunol. 1989;26:841. doi: 10.1016/0161-5890(89)90140-5. [DOI] [PubMed] [Google Scholar]
- Thielges MC, Zimmermann J, Yu W, Oda M, Romesberg FE. Exploring the energy landscape of antibody-antigen complexes: protein dynamics, flexibility, and molecular recognition. Biochemistry. 2008;47:7237. doi: 10.1021/bi800374q. [DOI] [PubMed] [Google Scholar]
- Tobin-Janzen TC, Womack JE. Comparative mapping of IGHG1, IGHM, FES, and FOS in domestic cattle. Immunogenetics. 1992;36:157. doi: 10.1007/BF00661092. [DOI] [PubMed] [Google Scholar]
- Walther S, Czerny CP, Diesterbeck US. Exceptionally long CDR3H are not isotype restricted in bovine immunoglobulins. PLoS One. 2013;8:e64234. doi: 10.1371/journal.pone.0064234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther S, Tietze M, Czerny CP, Konig S, Diesterbeck US. Development of a bioinformatics framework for the detection of gene conversion and the analysis of combinatorial diversity in immunoglobulin heavy chains in four cattle breeds. PLoS One. 2016;11:e0164567. doi: 10.1371/journal.pone.0164567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, Wilson IA, Schultz PG, Smider VV. Reshaping antibody diversity. Cell. 2013;153:1379. doi: 10.1016/j.cell.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JL, Ducharme NG. Traumatic reticuloperitonitis in dairy cows. J Am Vet Med Assoc. 1994;204:874. [PubMed] [Google Scholar]
- Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;276:1665. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Jenne CN, Kennedy LJ, Reynolds JD. The sheep and cattle peyer’s patch as a site of B-cell development. Vet Res. 2006;37:401. doi: 10.1051/vetres:2006008. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Tanaka S, Arakawa H, Taura Y, Yokomizo Y, Ekino S. A comparative study of gut-associated lymphoid tissue in calf and chicken. Anat Rec. 2002;266:207. doi: 10.1002/ar.10062. [DOI] [PubMed] [Google Scholar]
- Yeap LS, Hwang JK, Du Z, Meyers RM, Meng FL, Jakubauskaite A, Liu M, Mani V, Neuberg D, Kepler TB, Wang JH, Alt FW. Sequence-intrinsic mechanisms that target AID mutational outcomes on antibody genes. Cell. 2015;163:1124. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Beuscher AEt, Andryski SE, Stevens RC, Schultz PG. Structural plasticity and the evolution of antibody affinity and specificity. J Mol Biol. 2003;330:651. doi: 10.1016/s0022-2836(03)00631-4. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Jackson SM, Aitken R. The bovine antibody repertoire. Dev Comp Immunol. 2006;30:175. doi: 10.1016/j.dci.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Kacskovics I, Pan Q, Liberles DA, Geli J, Davis SK, Rabbani H, Hammarstrom L. Artiodactyl IgD: The missing link. J Immunol. 2002;169:4408. doi: 10.4049/jimmunol.169.8.4408. [DOI] [PubMed] [Google Scholar]
- Zimmermann J, Oakman EL, Thorpe IF, Shi X, Abbyad P, Brooks CL, 3rd, Boxer SG, Romesberg FE. Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc Natl Acad Sci U S A. 2006;103:13722. doi: 10.1073/pnas.0603282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinicola M, Lima F, Lima S, Machado V, Gomez M, Dopfer D, Guard C, Bicalho R. Altered microbiomes in bovine digital dermatitis lesions, and the gut as a pathogen reservoir. PLoS One. 2015;10:e0120504. doi: 10.1371/journal.pone.0120504. [DOI] [PMC free article] [PubMed] [Google Scholar]