Abstract
Approximately 80% of Neisseria gonorrhoeae and 17.5% of Neisseria meningitidis clinical isolates carry a ~59 kb genomic island known as the gonococcal genetic island (GGI). About half of the GGI consists of genes encoding a type IV secretion system (T4SS), and most of these genes are clustered in a ~28 kb region at one end of the GGI. Two additional genes (parA and parB) are found at the other end of the island. The remainder of the GGI consists mostly of hypothetical proteins, with several being identified as DNA binding or processing proteins. The T4SS genes show similarity to those of the F-plasmid family of conjugation systems, with similarity in gene order and a low but significant level of sequence identity for the encoded proteins. However, several GGI-encoded proteins are unique from the F-plasmid system, such as AtlA, Yag, and TraA. Interestingly, the gonococcal T4SS does not act as a conjugation system. Instead, this T4SS secretes ssDNA into the extracellular milieu, where it can serve to transform highly competent Neisseria species, thereby increasing the transfer of genetic information. Although many of the T4SS proteins are expressed at low levels, this system has been implicated in several cellular processes. The secreted ssDNA is involved in the initial stages of biofilm formation, and the presence of the T4SS enables TonB-independent intracellular survival of N. gonorrhoeae strains during infection of cervical cells. Other GGI-like T4SS have been identified in several other α-, β- and γ-Proteobacteria, but the function of these GGI-like T4SSs is unknown. Remarkably, the presence of the GGI is related to resistance to several antibiotics. Here we describe our current knowledge about the GGI and its unique ssDNA-secreting T4SS.
1 The gonococcal genetic island (GGI)
Neisseria gonorrhoeae is the etiological agent of the disease gonorrhea, which according to estimates of the World Health Organization infects over 100 million people annually (World Health Organization 2012). In the majority of cases, gonorrhea is an uncomplicated mucosal infection, but if left untreated gonorrhea can cause severe complications like pelvic inflammatory disease, infertility, neonatal conjunctivitis, septic arthritis, blindness and disseminated gonococcal infections (Unemo 2015; Hill et al. 2016). The type IV secretion system (T4SS) of Neisseria gonorrhoeae is the only known T4SS that secretes ssDNA into the extracellular milieu (Dillard and Seifert 2001; Hamilton et al. 2005). The secreted ssDNA serves to transform highly competent Neisseria species and is involved in the initial stages of biofilm formation (Dillard and Seifert 2001; Hamilton and Dillard 2006; Zweig et al. 2014). The T4SS is encoded on the 59-kb gonococcal genetic island (GGI), a genetic island found in approximately 80% of N. gonorrhoeae and 17.5% of N. meningitidis clinical isolates (Dillard and Seifert 2001; Hamilton et al. 2005; Snyder et al. 2005; Woodhams et al. 2012) (GenBank accession #CP003909.1). The GC-content of the GGI, at 44%, is significantly lower than the GC-content of the gonococcal chromosome with 51% (Hamilton et al. 2005). Its DNA sequence varies in dinucleotide frequencies and contains a deviant number of DNA-uptake sequences (DUS) in comparison to the rest of the chromosome (Hamilton et al. 2005; Spencer-Smith et al. 2016). The DUS is a 10 or 12 bp sequence that is specific to a subset of species in the Neisseriaceae and occurs throughout their genomes (Goodman and Scocca 1988; Ambur et al. 2007). DNA containing this sequence is preferentially taken up during natural transformation, thus favoring uptake of DNA from closely related species over foreign DNA (Goodman and Scocca 1988). On the gonococcal chromosome, DUS occur approximately once every 1.1 kb, whereas only 6 DUS are present on the 59-kb GGI (Hamilton et al. 2005) (Figure 13.1). Thus, the GGI is most likely horizontally acquired. Both sides of the GGI are flanked by direct repeat sequences, a consensus difA site and an imperfect difB site that harbors 4 mismatches compared to the consensus site (Hamilton et al. 2005). dif sites, found in the replication terminus region of most sequenced bacterial genomes, are recognized by the recombinase XerCD to resolve plasmid or chromosome dimers during replication (Blakely et al. 1993; Carnoy and Roten 2009; Castillo et al. 2017). Moreover, XerCD-mediated recombination was found to be exploited by some mobile genetic elements like the cholera-toxin encoding bacteriophage CTXφ that integrates via XerCD into the Vibrio cholerae chromosome (Huber and Waldor 2002). A XerCD-mediated integration and excision of the GGI was previously reported (Hamilton et al. 2005; Domínguez et al. 2011). Under laboratory conditions, excision of the GGI occurred once every 106 cells during growth for 18 hours. Replacement of difB by another consensus difA site increased its excision rate to once every 103 cells (Domínguez et al. 2011). Recently, it was shown that excision of the GGI follows a tightly controlled interaction between XerCD and FtsK, where XerD binding to difB might favor a particular conformation of XerD that causes the excision of the GGI to occur only rarely (Fournes et al. 2016).
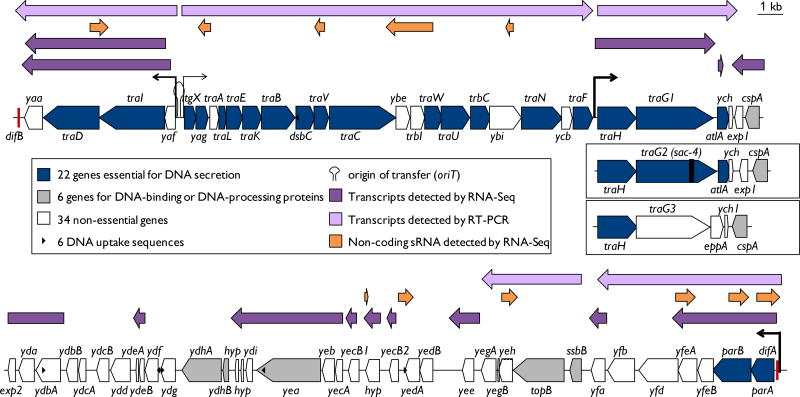
Figure 13.1. Map of the gonococcal genetic island (GGI).
The 22 genes essential to DNA secretion (blue) are encoded on four operons. Note that the transcript containing ydbA is continuous with the transcript containing cspA. dif sites are shown in red. Transcripts were detected using reverse transcriptase PCR (Pachulec et al. 2014, Jain et al. 2012, light purple) and RNA-seq (Remmele et al. 2014, dark purple). A number of sRNAs have also been identified in this region (Remmele et al. 2014, orange). Expression from these operons is variable, as represented by the thickness of the promoters. The three traG alleles are shown in parallel to one another. The GGI in strain MS11 was originally thought to be 57 kb in length, but re-sequencing identified a repeat at yecB and an additional ORF making the complete GGI 59 kb (accession #CP003909.1).
Close homologs of the GGI have also been identified in Neisseria meningitidis. The GGIs in N. meningitidis vary in size and carry a number of different mutations compared to the GGI from N. gonorrhoeae (Woodhams et al. 2012). The GGIs of some meningococcal strains accumulated up to eight mismatches within difB and exhibit deleterious mutations likely rendering their T4SS inactive (Domínguez et al. 2011; Woodhams et al. 2012). It was hypothesized that these GGIs degraded over time but that the high number of mutations within difB prevented loss of the GGI in past generations (Domínguez et al. 2011). The meningococcal strain 01/241471 carries four mutations in difB and mutations in genes that were shown to be essential for ssDNA secretion in N. gonorrhoeae (Woodhams et al. 2012). Interestingly, excision of the GGI could be observed in this strain, albeit at a frequency even lower than in N. gonorrhoeae. Nevertheless, some meningococcal strains harbor GGI variants that seem to encode an intact T4SS. However, the meningococcal T4SS seems not to secrete ssDNA nor does it confer Ton-independent intracellular survival as its gonococcal counterpart (discussed below). No connection could be observed between the presence of the T4SS and the infection process (Woodhams et al. 2012).
The GGI of N. gonorrhoeae strain MS11 comprises at least 65 ORFs of which 19 ORFs show similarity to genes of the E. coli F-plasmid T4SS, with a low but significant level of sequence similarity for encoded proteins and a conserved gene order (Hamilton et al. 2005) (Figure 13.1). The majority of those genes are encoded within the first 27.5 kb of the GGI, the region between yaa and ych, from which many genes were shown to be essential for ssDNA secretion (Hamilton et al. 2001, 2005; Pachulec et al. 2014). The origin of transfer (oriT) was identified between the yaf and ltgX genes (Salgado-Pabón et al. 2007). The exp1-difB region encodes at least 38 ORFs of which several show homology to DNA processing and modifying proteins, including SsbB, TopB, Yea, and CspA. However, most of the genes encode proteins with an unknown function, and only parA and parB encoded within the exp1-difB region are essential for ssDNA secretion (Hamilton et al. 2005; Pachulec et al. 2014).
2 Related type IV secretion systems
2.1 The T4SS encoded in the GGI shows homology to the F-plasmid encoded T4SS
The yaa-ych region encodes mostly proteins that show homology with T4SS proteins (Hamilton et al. 2005; Pachulec et al. 2014). The ltgX-ych region encodes mainly proteins involved in the formation and stabilization of the mating pair complex and proteins involved in the assembly of the pilus (Hamilton et al. 2005). Although the GGI-encoded T4SS does not act in conjugation and does not lead to mating pairs, we will use this terminology for comparison to homologous systems. The genes coding for 13 proteins in this region (TraL, TraE, TraK, TraB, TraV, TraC, TraW, TraU, TrbC, TraN, TraF, TraH and TraG) are ordered in the same orientation as in the E. coli F-plasmid (Hamilton et al. 2005). The gene encoding the peptidoglycan transglycosylase LtgX is oriented in a different direction, but represents a functional homolog of Orf169 of the F-plasmid encoding a lytic transglycosylase (Hamilton et al. 2005; Kohler et al. 2007). Phylogenetic analysis shows that most of these proteins are associated with mating pair formation (MPF) complexes of the MPFF family (Pachulec 2010). Several proteins encoded within the ltgX-ych region, e.g. Yag, DsbC, Ybe, Ybi, Ycb, AtlA and Ych are not present in the F-plasmid. Close homologs of the TraA pilin and of TrbI, the protein that circularizes pilin subunits are not found in the F-plasmid, but are found in T4SSs that contain an MPF complex that belongs to the MPFT family (Eisenbrandt et al. 1999; Hamilton et al. 2005; Jain et al. 2012). A functional analysis has shown that neither TraA and TrbI, nor Ybe, Ybi, Ycb, or Ych are essential for ssDNA secretion, but all proteins with homology to proteins encoded on the F-plasmid are essential (Hamilton et al. 2001, 2005; Pachulec et al. 2014). Remarkably, although not found in the F-plasmid, AtlA, Yag, and DsbC encoded in this region are required for DNA secretion (Hamilton et al. 2001, 2005; Pachulec et al. 2014). The yaf-yaa region encodes the relaxase TraI and the coupling protein TraD together with the small hypothetical proteins Yaf and Yaa (Pachulec et al. 2014). TraI belongs to the MOBH family of relaxases, and in phylogenetic analyses, the coupling protein TraD also clusters together with other coupling proteins that are always encoded closely to a relaxase of the MOBH family (Garcillán-Barcia et al. 2009; Pachulec et al. 2014). Furthermore, synteny is conserved for these relaxases and coupling proteins, as well as Yaa, a small hypothetical protein with 4 putative transmembrane domains (Pachulec et al. 2014). The fourth protein in this operon, Yaf, shows only little homology to other proteins (Hamilton et al. 2005; Pachulec et al. 2014). Thus, the T4SS encoded in the GGI is composed of a mating pair formation complex related to the MPFF family, a mobilization region related to the MOBH family, and contains a pilin and pilin circularization machinery most often found associated with the MPFT family (Hamilton et al. 2005; Garcillán-Barcia et al. 2009; Pachulec 2010; Pachulec et al. 2014).
2.2 GGI-like T4SS are widely spread
By searching the available genomes on the absynte website (http://archaea.u-psud.fr/absynte/), several regions were identified which encode T4SS systems showing homology and synteny of their MPF complex components, their pilin and their pilin circularization machinery to the T4SS encoded on the GGI. Here we refer to these systems as GGI-like T4SSs (Pachulec et al. 2014). Most of the organisms containing a GGI-like T4SS belong to the group of β-proteobacteria. GGI-like T4SSs were also identified in α- and γ-proteobacteria where most of these T4SSs are encoded on the chromosome. Alcaligenes denitrificans, Acidovorax JS42 and N. aromaticivorans contained both a chromosomally encoded and a plasmid encoded GGI-like T4SS (Pachulec et al. 2014).
In close proximity of the GGI-like T4SS genes, relaxases and coupling protein genes could be identified. Several systems contained a relaxase of the MOBH family (TraI), with a related coupling protein (TraD) and a small membrane protein (Yaa). Other systems were identified where the GGI-like MPF proteins were found together with a relaxase and a coupling protein linked to the MOBF family (Pachulec et al. 2014). This observation suggests that relaxases of the MOBH and MOBF families both function together with GGI-like MPFF systems. The parA-exp1 region was only found associated with the GGI-like T4SS of Neisseriaceae, suggesting that in the other systems the ParA/ParB system is not important for DNA transport (Pachulec et al. 2014). Moreover, no specific site of insertion into the chromosome could be determined for other GGI-like sequences. Remarkably, genes encoding proteins with homology to the Exp1 protein are present in most of the GGI-like T4SSs. Exp1 is a small protein with a signal sequence, which was identified in a mini-TnphoA screen for gonococcal proteins that were exported out of the cytoplasm (Boyle-Vavra and Seifert 1995; Dillard and Seifert 1997). An exp1 knock-out mutant was still able to secrete ssDNA, suggesting that the gene is not involved in the secretion of DNA (Pachulec et al. 2014).
3 DNA secretion provides donor DNA for natural transformation
The N. gonorrhoeae T4SS secretes ssDNA into the extracellular space in a contact independent manner (Dillard and Seifert 2001; Hamilton et al. 2005). No protein effectors or dsDNA have been identified as a substrate of this system. The secreted DNA is hypothesized to be cleaved by the relaxase TraI at the oriT, a region of the GGI located between the divergent yaf and ltgX promoters (Salgado-Pabón et al. 2007). ssDNA is then shuttled across both the inner and outer membrane and into the extracellular space, where it has been shown to be 5’ protected and susceptible only to 3’ ssDNA-specific DNases (Salgado-Pabón et al. 2007). Neisseria spp. are naturally transformable; N. gonorrhoeae is able to take up and recombine DNA into its chromosome during all stages of growth (Sparling 1966). When co-cultured, the ssDNA output by donor strains of N. gonorrhoeae containing a functional T4SS can be taken up and chromosomally incorporated by competent recipient cells (Dillard and Seifert 2001; Hamilton et al. 2001). High-frequency DNA release and transformation of recipient cells is T4SS-dependent; GGI+ donor strains yield up to 500-fold more transformants than donors with mutated T4SS genes that are deficient in DNA secretion (Dillard and Seifert 2001; Hamilton et al. 2001; Pachulec et al. 2014). This process is also dependent on natural transformation. A pilT mutant, which lacks the ability to retract its type IV pili and therefore is unable to take up DNA, could not act as a recipient (Kohler et al. 2013).
4 Secreted DNA affects biofilm formation
In addition to the role of secreted DNA in the transfer of genetic information, other roles, such as acting as a nutrient source or as an important structural component of biofilms have been suggested (For review see: Vorkapic et al. 2016). At the onset of biofilm formation, the initially planktonic bacteria attach reversibly to the surface and start to produce an extracellular matrix of polymeric substances, which results in their irreversible attachment to the surface (Dunne 2002). These polymeric substances can consist of exopolysaccharides, secreted proteins, membrane vesicles or extracellular DNA. For many bacteria, including N. gonorrhoeae, it was demonstrated that extracellular DNA is an important component of the biofilm (Steichen et al. 2011; Zweig et al. 2014; Vorkapic et al. 2016). Growth of N. gonorrhoeae MS11 biofilms in a medium containing Exonuclease I, which specifically degrades ssDNA, delayed biofilm formation in its initial phases (Zweig et al. 2014). Strongly delayed biofilm formation was also observed in a ΔtraB strain (MS11ΔtraB), which is impaired in ssDNA secretion in comparison with its parental strain (Pachulec et al. 2014; Zweig et al. 2014). Restoration of the ΔtraB strain with traB (MS11ΔtraB::traB) resulted in biofilm formation comparable to the MS11 parental strain. This result demonstrates that the ssDNA secreted by the T4SS plays an important role in the initial phases of biofilm formation (Zweig et al. 2014).
5 The T4SS functions in tonB-independent iron uptake
N. gonorrhoeae must acquire iron from its environment to survive. During infection of the iron-deplete female lower genital tract, this is accomplished by upregulating expression of genes related to iron scavenging from host transferrin and lactoferrin (e.g. tbp, lbp, fbp) and other iron-regulated genes (West and Sparling 1985; McClure et al. 2015). These scavenging proteins rely on TonB, as demonstrated during intracellular infection and replication within ME180 cervical epithelial cells (Hagen and Cornelissen 2006). However, Zola et al. reported that an N. gonorrhoeae mutant lacking the tonB gene is able to survive intracellularly during infection of ME180 cervical cells, provided that it has a GGI with intact structural components of the T4SS (Zola et al. 2010). This phenotype is independent of the ability of N. gonorrhoeae to actively take up or secrete DNA, as tested by assaying intracellular growth of gonococcal strains lacking pilT or the non-structural GGI genes parA, traD, and traI (Zola et al. 2010). The mechanism of iron uptake for these mutants is unknown, as is the role of the T4SS in this process. However, the implication that the T4SS machinery can affect bacterial survival independent of DNA secretion opens an intriguing series of new questions for future investigation.
6 Presence of the GGI correlates with antibiotic resistance
N. gonorrhoeae elaborates different surface-exposed components such as type IV pili and opacity proteins which frequently undergo phase- and antigenic variation allowing for efficient evasion of the host immune system (Hill et al. 2016). Moreover, the ability to rapidly exchange DNA, mutations and a high recombination rate promote spread of resistances against various antimicrobials, which is a major health concern. The only option left for first-line monotherapy of gonorrhea is the use of third-generation cephalosporins, although treatment failures have already been reported (Unemo 2015). Recently, first-line dual-therapy of uncomplicated gonorrhea infections has been introduced in several high-income countries, and has been successful thus far (Unemo 2015).
In a 2016 study, a significant positive correlation between the presence of the GGI and resistance to a variety of common antimicrobials was found (Harrison et al. 2016). Investigators mined whole genome sequencing data for antimicrobial resistance (AMR) determinants from 289 gonococcal isolates from which MIC (minimal inhibitory concentration) values against several antimicrobials were available. With the mined data, genotypic AMR datasets were generated comprising predictions about the resistances of strains to certain antimicrobials, which were then compared to the reported MIC values, if available. For instance, cefixime resistance was predicted and found in 56% of the GGI+ strains, but only in 4% of the GGI− strains. 74% of the GGI+ strains were predicted to be resistant to Ciprofloxacin compared to 33% of the GGI− strains. The mechanisms governing this association are unknown, however possibilities include increased horizontal gene transfer due to the presence of the T4SS, activity of the putative YdbB-YdcA toxin-antitoxin system, or competition somewhere during the evolutionary history of N. gonorrhoeae between plasmids and the GGI prior to its chromosomal insertion that could have altered resistance phenotypes (Harrison et al. 2016).
7 Characterized components of the T4SS apparatus
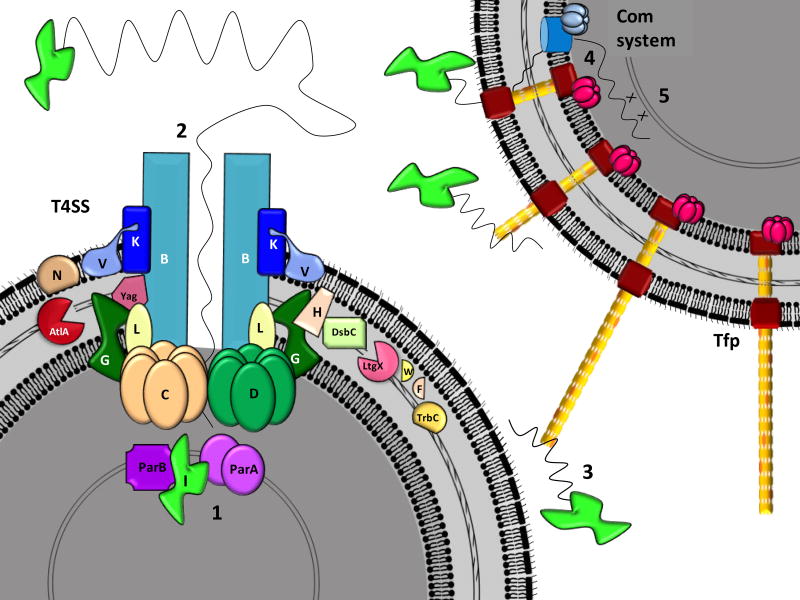
Several components encoded within the GGI have been characterized. Here we describe what is currently known about these components (Figure 13.2).
Figure 13.2. Model of horizontal gene transfer as facilitated by the gonococcal T4SS.
The core outer membrane complex of the gonococcal T4SS is made of TraK, TraB, and TraV. Insertion into the membrane is thought to depend on the cutting of the peptidoglycan layer by AtlA, and periplasmic proteins such as Yag, TraW, TraF, TraH, TrbC, and LtgX are likely involved in assembly of the apparatus. Transport of DNA and incorporation into the recipient cell is thought to occur as follows: (1) TraI nicks the chromosome at the oriT and is localized to the secretion system with the help of the partitioning proteins ParA and ParB. (2) The TraI-ssDNA complex is transported out of the cell, facilitated by the coupling protein TraD and the ATPase TraC. (3) Once outside the cell, the ssDNA may bind ComP on the type IV pilus of a neighboring cell (shown as orange, where the major pilin protein is shown as yellow), (4) Retraction of the pilus brings the DNA into the recipient periplasm, where it is then transported across the inner membrane by the competence (Com) system. (5) Finally, the DNA is incorporated into the genome by homologous recombination.
7.1 TraI
TraI of N. gonorrhoeae belongs to the MobH-family of relaxases, which consists of relaxases often found on large plasmids, genetic islands and integrative conjugative elements (Garcillán-Barcia et al. 2009). The available information on MobH relaxases is scarce, since no MobH relaxase has been analyzed biochemically (Garcillán-Barcia et al. 2009). The general architecture of MobH-relaxases is an N-terminal relaxase domain, followed by a middle part of unknown function that varies in length among proteins of this family, and a C-terminal TraI_C_2 domain (formerly known as DUF1528) (Garcillán-Barcia et al. 2009). A unique feature of MobH relaxases is the presence of two different motifs predicted to be involved in coordination of divalent metal ions. The first motif is reminiscent of the 3-histidine motif of MobF relaxases which, due to its different sequence arrangement, was termed “alternative 3-histidine motif” (Garcillán-Barcia et al. 2009). A second motif is the HD-hydrolase domain, present in metal-dependent phosphohydrolases (Aravind and Koonin 1998; Salgado-Pabón et al. 2007; Garcillán-Barcia et al. 2009). Surprisingly, amino acid substitutions in TraI revealed that neither the 3-histidine motif nor the HD-hydrolase motif had an influence on ssDNA secretion, which leaves their physiological role unclear (Salgado-Pabón et al. 2007). Moreover, it was shown that Y93, one of three conserved tyrosines in TraI, is essential for DNA secretion suggesting a role in the DNA transfer process. Since the secreted DNA in N. gonorrhoeae was shown to be 5’ protected from DNase treatment, it is assumed that TraI remains bound to the DNA (Salgado-Pabón et al. 2007). Gonococcal TraI and a small subset of closely related relaxases contain a hydrophobic N-terminus, which suggests its association to the cellular membrane (Salgado-Pabón et al. 2007). Secretion assays showed that the hydrophobic N-terminus is essential for ssDNA secretion, but its function is not yet known (Salgado-Pabón et al. 2007).
7.2 TraA and TrbI
Pilin proteins assemble into the pilus fiber, which facilitates attachment of donor and recipient cells in conjugation systems and might also form a conduit for substrate secretion (Babic et al. 2008; Cabezón et al. 2015; Costa et al. 2016). The GGI encodes TraA, a pilin homolog and TrbI, which shows homology to the serine protease involved in the circularization of the pilin subunit of the RP4 plasmid (Eisenbrandt et al. 1999, 2000; Hamilton et al. 2005). Sequence comparison of the DNA-secreting MS11 strain with other N. gonorrhoeae GGI+ strains showed that the sequence of the traA gene of MS11 comprises a frameshift mutation resulting in a truncation of the last 14 amino acids of TraA (Jain et al. 2011). Only when the full length gonococcal TraA was expressed in Escherichia coli, it was processed to a 68-amino acid long circular peptide exclusively by the leader peptidase LepB and TrbI. First, TraA is co-translationally inserted into the inner membrane, which is followed by cleavage of the N-terminal signal sequence by LepB. Then, TrbI cleaves three residues at the C-terminus resulting in a covalent intermediate of TrbI with TraA, which eventually leads to the circularization of the TraA pilin (Jain et al. 2011). Mutagenesis studies of TrbI showed that the conserved Lys-93 and Asp-155 residues are essential. However, mutagenesis of the putative catalytic Ser-52 did not influence circularization. Mutagenesis of other serine residues could not identify a catalytic serine, indicating that TrbI either contains redundant catalytic serine residues or does not employ the serine-lysine dyad mechanism seen in other systems (Jain et al. 2011). Deletion of neither traA nor trbI in MS11 affected DNA secretion (Pachulec et al. 2014). This shows that the DNA-secreting MS11 strain expresses a defective pilin subunit, which is not processed and most likely is not assembled into a pilus, and shows that neither the pilin protein nor pilus assembly is required for DNA secretion into the medium.
7.3 SsbB
The single-stranded DNA binding protein SsbB is encoded within a genetic cluster present in several genetic islands of different proteobacteria (Jain et al. 2012). This cluster encodes, next to SsbB, the ParA and ParB partitioning proteins, the TopB topoisomerase and four conserved hypothetical proteins. Deletion of ssbB did not affect DNA secretion nor marker transfer, suggesting no function for SsbB in DNA secretion (Jain et al. 2012). Since the cluster is conserved, it might play a role in maintenance of the genetic islands. Biochemical characterization of SsbB showed that it forms stable tetramers that bind ssDNA with high affinity. The minimal ssDNA binding frame of an SsbB monomer is 15 nucleotides, whereas it is 70 nucleotides for two SsbB tetramers (Jain et al. 2012).
7.4 TraK, TraV, TraB: The core complex
The core complex of the gonococcal T4SS is comprised of TraB, TraK, and TraV (homologs of VirB10, VirB9, and VirB7 of plasmid pKM101, respectively) (Chandran et al. 2009; Ramsey et al. 2014). These gonococcal proteins have limited sequence homology to their F-plasmid counterparts (between 23% and 29%) but exhibit robust similarity in their interactions and cellular localization. Both TraK and TraB have been shown to localize to the outer membrane, and TraK has been shown to interact with TraV by bacterial two-hybrid assay (Ramsey et al. 2014). These findings are in alignment with those of the F-plasmid system as well as the VirB/D4 system of Agrobacterium tumefaciens (Chandran et al. 2009).
Native expression levels of gonococcal TraK and TraB are low, such that they are undetectable by western blot. Thin-section electron microscopy and immunogold labeling of TraK further corroborated this finding, revealing that TraK could only be observed in a subset of cells (Ramsey et al. 2014). Due to the low expression levels, a more detailed structural characterization of the core complex is challenging.
7.5 AtlA, LtgX, EppA, and Yag – peptidoglycan-related proteins
AtlA is a homolog of bacteriophage lambda endolysin (gpR), an enzyme that destroys the E. coli cell wall during the lytic stage of lambda infection (Dillard and Seifert 1997). Purified AtlA has a lytic transglycosylase activity and degrades peptidoglycan (Kohler et al. 2007). T4SSs often utilize a lytic transglycosylase, and it is assumed that these enzymes serve to make a localized break in the peptidoglycan to allow assembly of the T4SS apparatus (Dijkstra and Keck 1996; Baron et al. 1997). This is in accordance with the finding that AtlA is required for DNA secretion by N. gonorrhoeae (Dillard and Seifert 2001).
Some gonococcal strains lack atlA and instead carry the gene eppA. EppA is an M23-family metallopeptidase capable of degrading peptidoglycan. The purified enzyme was demonstrated to digest peptidoglycan by cutting peptide crosslinks (Kohler et al. 2013). Interestingly, strains carrying eppA instead of atlA do not secrete DNA (Kohler et al. 2013).
The E. coli F-plasmid carries a gene for a lytic transglycosylase encoded by orf169 (Manwaring et al. 1999). N. gonorrhoeae encodes a homolog of Orf169 named LtgX. Deletion of ltgX results in loss of DNA secretion (Kohler et al. 2007). It is not clear why gonococci would need two peptidoglycan-degrading enzymes for DNA secretion.
The gene just downstream of ltgX is yag (Hamilton et al. 2005). Yag has an OmpA-like domain that has been found in other proteins to bind to peptidoglycan. An in-frame deletion of yag resulted in a loss of DNA secretion, and this phenotype could be complemented by expression of yag from a distant locus on the chromosome (Pachulec et al. 2014). Thus, we predict that Yag binds to peptidoglycan at the site of the T4SS’s passage through the cell wall, and might bind the apparatus to the wall or otherwise facilitate T4SS assembly.
7.6 ParA and ParB
parA and parB encode a pair of partitioning proteins and are essential for T4SS-dependent DNA secretion in N. gonorrhoeae (Hamilton et al. 2005; Pachulec et al. 2014). Gonococcal ParA is a member of the ParA/MinD ATPase family within the P-loop GTPase superfamily, a superfamily of proteins characterized by a deviant Walker A box containing two conserved lysine residues (Lutkenhaus 2012; Roberts et al. 2012). A point mutation in the Walker A box of gonococcal ParA diminishes DNA secretion, demonstrating dependence on the ATPase function of ParA for function of the T4SS (Hamilton et al. 2005). ParA proteins play an important role in spatial organization of plasmids, chromosomes, and protein complexes in many other systems and employ two distinct approaches to do so (Atmakuri et al. 2007; Lutkenhaus 2012; Roberts et al. 2012). One mechanism, employed by the MinCDE system in E. coli, involves oscillation of protein(s) to identify the middle of the cell. The other mechanism, termed the “landmark mechanism”, uses a protein that is anchored to a specific location within the cell as a guide by which all the other proteins and the substrate of interest are positioned in relation (Lutkenhaus 2012).
In Agrobacterium tumefaciens, which possesses a VirB/D4 T4SS that injects tumor inducing T-DNA into the cells of its plant host, the partitioning proteins VirC1 (ParA homolog) and VirC2 (ParB homolog) have been shown to form a complex and direct the localization of the relaxosome and T-DNA complexes to the cellular poles where the T4SS machinery is located. It is thought that this system utilizes the landmark mechanism wherein VirC1 and VirC2, along with VirD1, form a complex at the pole and then recruit the relaxase VirD2 and substrate DNA to initiate transfer (Atmakuri et al. 2007).
Evidence of ParA proteins working to direct localization and segregation beyond plasmid DNA is accumulating; VirC1 activity affects the nucleoprotein complex VirD2-DNA, and “orphan” ParA proteins that lack an identified cognate ParB have been shown to act in segregating protein complexes (Atmakuri et al. 2007; Roberts et al. 2012). It is reasonable to think that gonococcal ParAB may be working similarly, with ParA interacting either nonspecifically with DNA or binding specific sequence(s) and ParB activating ParA ATPase activity to both localize the DNA and interact with the relaxase TraI to initiate transfer of the substrate through the T4SS (Lutkenhaus 2012).
7.7 TraG
traG is encoded on the third operon of the GGI, in an operon with traH proximal to the promoter followed by traG and atlA, all of which are essential for T4SS-dependent DNA secretion (Dillard and Seifert 2001; Hamilton et al. 2001; Kohler et al. 2013; Pachulec et al. 2014). It is transcribed at comparable levels to the parA operon, and both of these have higher transcript levels than the yaf and ltgX operons (Pachulec et al. 2014; Ramsey et al. 2015). The traH operon is especially interesting because this region of the GGI exhibits variation between gonococcal strains (Dillard and Seifert 2001; Hamilton et al. 2001; Salgado-Pabón 2008; Kohler et al. 2013). Firstly, three “classes” of the traH operon have been identified (Dillard and Seifert 2001). There are strains that carry the traG allele termed traG1 alongside atlA, which encodes a peptidoglycan lytic transglycosylase (Dillard and Seifert 1997; Kohler et al. 2007). These strains are able to secrete DNA, as shown with strain MS11 (Hamilton et al. 2001; Kohler et al. 2013). Other strains have been identified that contain an allele of traG with an extra sequence termed sac-4. This allele, named traG2, is also found accompanied by atlA, and these strains are capable of DNA secretion (Dillard and Seifert 2001; Kohler et al. 2013). Some groups reported that the sac-4 sequence may be associated with serum resistance in N. gonorrhoeae (Mcshan et al. 1987; Nowicki et al. 1997), however more recent studies found no relationship between serum resistance phenotypes and the presence of the sac-4 allele (Dillard and Seifert 2001; Chen et al. 2008). Interestingly, both atlA and the sac-4 allele of traG are significantly overrepresented in disseminated gonococcal infection isolates compared to local infections isolates (Dillard and Seifert 2001). A third variant, traG3, has been found that maintains the conserved N-terminus of the traG gene, but is shorter than traG1 and lacks atlA. In place of atlA these strains possess eppA, followed by a variant of ych, named ych1, and they lack the gene often found adjacent to ych, exp1. These traG3 strains are unable to secrete DNA (Dillard and Seifert 2001; Hamilton et al. 2001; Kohler et al. 2013). Additional variation has been identified in the exp1-cspA-exp2 region of the GGI (Salgado-Pabón 2008). These genes are not essential for DNA secretion, and the source and consequences of these variations are presently unknown (Pachulec et al. 2014).
In gonococci, TraG localizes to the inner membrane, and it has five transmembrane domains, putting the N-terminus in the periplasm and the C-terminus in the cytosol (Kohler et al. 2013). Functionally similar T4SS inner membrane proteins include A. tumefaciens VirB6 and F-plasmid TraG (Kohler et al. 2013). TraGF is hypothesized to form part of the transmembrane channel that allows substrate passage into the recipient cell (Firth and Skurray 1992; Frost et al. 1994; Audette et al. 2007). The inability of gonococcal TraG3 to function for DNA secretion suggests that TraGGGI might have more than a structural role and might affect substrate specificity, possibly by interacting with the coupling protein or the relaxosome. In the F-plasmid system, there is evidence that TraGF interacts with proteins in the recipient cell periplasm during conjugation, which would be consistent with the hypothesis that TraGGGI may interact with other T4SS proteins to affect T4SS function (Audette et al. 2007).
8 Transcriptional regulation
8.1 Operon structure
Analysis of the mRNA transcripts of GGI genes focused mainly on the regions that contain genes involved in DNA secretion. These analyses identified at least 5 different operons: the yaf-yaa operon, the ltgX-traF operon, the traH-ych operon, the ssbB-yegA operon and the parA-yfa operon (Hamilton et al. 2001, 2005; Kohler et al. 2007; Salgado-Pabón et al. 2007; Jain et al. 2012; Pachulec et al. 2014) (Figure 13.1). The yaa-yaf operon contains genes for the relaxase TraI and for the coupling protein TraD. The operon is divergently transcribed from the ltgX-traF operon leaving an intergenic region between yaf and ltgX of 630 bp which comprises the promoters for yaf and ltgX as well as the oriT (Salgado-Pabón et al. 2007; Pachulec et al. 2014; Ramsey et al. 2015). traD, traI and yaa are conserved among GGI-like T4SS, with the exception of yaf which is, apart from N. gonorrhoeae, only found in the GGI-like T4SS of N. meningitidis and N. bacilliformis (Pachulec et al. 2014). Mutation of yaf had no effect on DNA secretion, whereas deletion of yaa resulted in a 7-fold increase of secreted DNA. Interestingly, mutation of yaa results in an increase of transcript levels of the genes traI, traD, ltgX, traH and parA, representing all four GGI operons containing genes essential for DNA secretion. However, the mechanism by which mutation of yaa increases the secreted DNA levels remains unknown (Pachulec et al. 2014). The ltgX-traF operon contains 14 of the essential 21 genes, of which the majority encode for structural components of the T4SS (Salgado-Pabón et al. 2007; Pachulec et al. 2014). The traH-ych operon encodes four genes including the variable traG and a gene for the lytic transglycosylase AtlA, which was not found in other GGI-like T4SS (Dillard and Seifert 2001; Pachulec et al. 2014). In the genetic region close to the difB site, only the parA and parB genes are essential for DNA secretion (Pachulec et al. 2014). They are located at the beginning of the parA-yfa operon. This is followed by another operon from ssbB to yegA (Jain et al. 2012). This parA-yegA operon structure seems in part conserved; it is often found at the borders of large genetic islands like the PAGI-3(SG), PAGI-2(C) and clc-like genetic islands found in Pseudomonas aeruginosa and other organisms (Larbig et al. 2002).
8.2 Transcript levels and sRNAs
Targeted qRT-PCR studies have provided a transcriptional profile of the GGI, specifically for the four operons containing genes for DNA secretion (Salgado-Pabón et al. 2010; Pachulec et al. 2014; Ramsey et al. 2015). The Pyaf operon, comprised of yaf, traI, traD, and yaa, produces low transcript levels as determined by qRT-PCR of the traI and traD genes (Salgado-Pabón et al. 2010; Pachulec et al. 2014; Ramsey et al. 2015). Overexpression of this operon using a strong ermC promoter increased DNA secretion above wild type levels, but only in non-piliated gonococci (Salgado-Pabón et al. 2010). The PltgX promoter has extremely low expression levels as measured by ltgX and traK transcript levels (Pachulec et al. 2014; Ramsey et al. 2015). These transcript levels have been consistently reported to be lower than those of the Pyaf operon, and traK levels are shown to be significantly lower than those of traI (Pachulec et al. 2014; Ramsey et al. 2015). Alteration of both Pyaf and PltgX to consensus promoter sequences increased transcription and protein levels from these promoters, as measured by qRT-PCR of traI and traK mRNA and western blotting for TraI and TraK proteins (Ramsey et al. 2015). The gonococcal strain with these consensus promoters exhibited variable DNA secretion with a non-significant trend of increased DNA secretion over the wild type strain (Ramsey et al. 2015). Transcript levels for all three genes under control of PtraH have been reported to be significantly higher than those of traI and traK (representing the yaf and ltgX operons), with traH having the most robust expression, followed by atlA, and finally traG (Ramsey et al. 2015). This result may indicate that the traH promoter is stronger than Pyaf and PltgX, although not all transcription initiation events are able to transcribe the entire operon. In addition to the PtraH promoter, another promoter has been reported immediately upstream of atlA, which would explain its increased transcript levels over traG (Remmele et al. 2014). The final operon of the GGI that contains genes essential to DNA secretion depends upon the parA promoter (Hamilton et al. 2005; Pachulec et al. 2014). Expression of this operon is more robust than that of the Pyaf and PltgX operons, and transcripts are at least as abundant as those on the traH operon, if not more so (Pachulec et al. 2014; Ramsey et al. 2015).
The N. gonorrhoeae transcriptome has also been assessed by Remmele et al. for the GGI+ strain MS11 using differential RNA-Seq methods. This study mapped three of the four transcripts for T4SS genes but did not detect a transcript for the ltgX operon, the latter was likely below the detection limit in this study. Furthermore, in the RNA-Seq study, seven antisense transcripts were detected among the T4SS genes (Figure 13.1). Three of these sRNAs are internal to coding sequences (traV, ybi, and parB), three overlap the beginnings of genes (traD, traA, parA) and one overlaps with ybe, trbI, and traW. The functions of these antisense transcripts are not known, but it is reasonable to assume that they may decrease transcript stability for the T4SS genes or decrease T4SS protein translation (Remmele et al. 2014).
9 Translational regulation
Regulation of the T4SS at the translational level has been identified by Ramsey et al. for the traH transcript of the GGI (Ramsey et al. 2015). The 5’UTR of this transcript contains two stem loops (SL1 and SL2) which, when folded, the stem loop proximal to traH (SL2) occludes its RBS and start codon, thereby preventing ribosome binding and translation of TraH, TraG, and possibly AtlA. Mutation to prevent stem loop formation increases protein levels upwards of 8×104-fold compared to the wild type sequence. Furthermore, the 5’UTR also has the capacity to form one alternative stem loop (SL3) using a portion of both SL1 and SL2 that does not occlude the RBS or start codon of traH. Genetic modification of the 5’UTR to make the formation of SL3 favorable also increased protein levels from this operon on the order of 104. These findings suggest that an RNA switch mechanism regulates translation for the traH operon-encoded transcript in which formation of SL1 and SL2 is favorable and translation of these proteins is “OFF” by default. When the formation of SL3 becomes favorable, perhaps by the binding of an unidentified factor to the upstream leg of SL1, translation is effectively turned “ON”. While manipulating this regulatory mechanism increased protein levels, it did not significantly alter DNA secretion by the T4SS when tested in combination with the consensus promoter mutations increasing Pyaf and PltgX transcription as discussed in section 7.2 (Ramsey et al. 2015). There may be additional translational regulation of the T4SS. A sequence found in SL1 is also present on the antisense strand at the ltgX translational start site, suggesting that proteins encoded on the big operon may also be subject to translational control.
10 Concluding Remarks
10.1 Expression of the T4SS
Although the GGI-encoded T4SS has dramatic effects on co-culture transformation in vitro, the numbers of T4SS apparatuses observed on N. gonorrhoeae cells by microscopy and the amounts of T4SS proteins detected on western blots are very low (Hamilton et al. 2001; Ramsey et al. 2014). Thus, future studies of the T4SS will either have to rely on genetic methods or will require knowledge that allows for increased production of the T4SS. Recent identification of factors controlling translation represents progress in understanding the regulation of the system (Ramsey et al. 2015). The sequence conserved at the beginning of ltgX and in stem-loop 1 (SL1) of traH suggest that an unknown factor may control translation of some proteins on both of these transcripts (Ramsey et al. 2015). It is possible that identification of this factor will lead to a method for increasing T4SS production.
Although understanding T4SS expression would lead to advances in understanding the T4SS in multiple areas, it is possible that forcing expression of the T4SS genes would allow for more functional studies. Initial attempts at producing a constitutive expression strain did not lead to significantly more secretion, and the over-expressed proteins examined were subject to degradation in the periplasm (Ramsey et al. 2015). However, it is possible that multiple proteins in addition to TraH and TraG are also translationally regulated, and expression constructs that facilitate translation of these proteins may lead to more T4SS expression.
Progress is being made in identifying regulatory factors as well as environmental conditions that increase T4SS expression. Initial studies suggest that amounts of iron and other metals affect expression of traD, and the transcriptional regulators Fur and FarR both affect transcript levels (Dillard 2014).
10.2 Unique components of the GGI and F-plasmid like systems
T4SSs related to the E. coli F-plasmid have a number of components that do not have homologs in the A. tumefaciens VirB/D4 system or other highly characterized systems including TraU, TraW, TrbC, TraN, TraF, TraH, and the TraG C-terminal regions (Lawley et al. 2003). These F-plasmid specific proteins are of interest as well as the proteins that differ between the gonococcal system and F-plasmid. In the N. gonorrhoeae T4SS, very tight translational regulation of TraH and TraG was found, but it is not clear why these particular components are a regulation target (Ramsey et al. 2015). A number of GGI encoded proteins are different from F-plasmid (Hamilton et al. 2005). Examination of these proteins and their interactions may reveal how gonococci are able to secrete DNA directly into the medium instead of into a target cell. Noteworthy examples include the need for two peptidoglycanases in gonococci (AtlA and LtgX), as well as the presence of two proteins with similarity to TraN (Ybi and TraN) (Hamilton et al. 2005; Kohler et al. 2007). Furthermore, the relaxosome components are not conserved and may explain important differences that allow gonococcal DNA secretion.
Variant forms of the GGI raise a number of questions about T4SS function. The versions of the T4SS that carry eppA instead of atlA have not been found to secrete DNA, suggesting that they perform a different function (Woodhams et al. 2012; Kohler et al. 2013). Do these versions of the apparatus secrete protein effectors? Do they transfer DNA by conjugation? Since the T4SS has been found to be important during intracellular growth, examination of possible phenotypes in that milieu may be fruitful (Zola et al. 2010).
10.3 Effects on horizontal gene transfer
The abundance of N. gonorrhoeae genome sequences and the ability to cheaply sequence additional strains allows for an examination of the effects of the T4SS on recombination in the gonococcal population. The mechanism of T4SS-mediated gene transfer may result in some regions of the chromosome being transferred more than others or GGI+ strains more effectively spreading their genes to other strains or species. The identification of antibiotic resistance associated with the genomic presence of the GGI suggests that the T4SS does have measureable effects on gonococcal populations (Harrison et al. 2016). Experiments to identify the mechanisms underlying the increased likelihood of GGI+ strains to exhibit antimicrobial resistance may give the most insight into the medically relevant role of the T4SS in N. gonorrhoeae infection.
Acknowledgments
This work was supported in part by funding from the National Institutes of Health (NIH) grant R01AI047958.
Contributor Information
Melanie M. Callaghan, Department of Medical Microbiology and Immunology, University of Wisconsin-Madison, 1550 Linden Dr., Madison, WI 53706, USA
Jan-Hendrik Heilers, Institut für Biologie II – Mikrobiologie, Albert-Ludwigs-Universität Freiburg, Schänzlestraße 1, 79104 Freiburg.
Chris van der Does, Institut für Biologie II – Mikrobiologie, Albert-Ludwigs-Universität Freiburg, Schänzlestraße 1, 79104 Freiburg.
Joseph P. Dillard, Department of Medical Microbiology and Immunology, University of Wisconsin-Madison, 1550 Linden Dr., Madison, WI 53706, USA
References
- Ambur OH, Frye SA, Tønjum T. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J Bacteriol. 2007;189(5):2077–2085. doi: 10.1128/JB.01408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23(12):469–472. doi: 10.1016/S0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 2007;26(10):2540–2551. doi: 10.1038/sj.emboj.7601696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audette GF, Manchak J, Beatty P, Klimke WA, Frost LS. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology. 2007;153(2):442–451. doi: 10.1099/mic.0.2006/001917-0. [DOI] [PubMed] [Google Scholar]
- Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319(5869):1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- Baron C, Llosa M, Zhou S, Zambryski PC. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1*. J Bacteriol. 1997;179(4):1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely G, May G, McCulloch R, Arciszewska LK, Burke M, Lovett ST, Sherratt DJ. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993;75(2):351–361. doi: 10.1016/0092-8674(93)80076-Q. [DOI] [PubMed] [Google Scholar]
- Boyle-Vavra S, Seifert HS. Shuttle mutagenesis: A mini-transposon for producing PhoA fusions with exported proteins in Neisseria gonorrhoeae. Gene. 1995;155(1):101–106. doi: 10.1016/0378-1119(94)00890-5. [DOI] [PubMed] [Google Scholar]
- Cabezón E, Ripoll-Rozada J, Peña A, de la Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. 2015;39(1):81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- Carnoy C, Roten CA. The dif/Xer recombination systems in Proteobacteria. PLoS ONE. 2009;4(9):e6531. doi: 10.1371/journal.pone.0006531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo F, Benmohamed A, Szatmari G. Xer site specific recombination: Double and single recombinase systems. Front Microbiol. 2017;8(453) doi: 10.3389/fmicb.2017.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462(7276):1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wu Z, Chen R, Shuai J, Xu L, Yu Y, Tu Y. Study on the classification of gonococcal island of different genotypes and effects of sac-4 gene on serum resistance of Neisseria gonorrhoeae. Chinese J Epidemiol. 2008;29(5):482–485. [PubMed] [Google Scholar]
- Costa TRD, Ilangovan A, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, Waksman G. Structure of the bacterial sex F pilus reveals an assembly of a stoichiometric protein-phospholipid complex. Cell. 2016;166(6):1436–1444.e10. doi: 10.1016/j.cell.2016.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra AJ, Keck W. Peptidoglycan as a barrier to transenvelope transport. J Bacteriol. 1996;178(19):5555–5562. doi: 10.1007/b112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard JP. Regulation of the gonococcal type IV secretion system involves two transcriptional repressors, two proteases, and an RNA switch. Asheville, North Carolina, USA: 2014. [Google Scholar]
- Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41(1):263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- Dillard JP, Seifert HS. A peptidoglycan hydrolase similar to bacteriophage endolysins acts as an autolysin in Neisseria gonorrhoeae. Mol Microbiol. 1997;25(5):893–901. doi: 10.1111/j.1365-2958.1997.mmi522.x. [DOI] [PubMed] [Google Scholar]
- Domínguez NM, Hackett KT, Dillard JP. XerCD-mediated site-specific recombination leads to loss of the 57-kilobase gonococcal genetic island. J Bacteriol. 2011;193(2):377–388. doi: 10.1128/JB.00948-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne WM. Bacterial Adhesion: Seen Any Good Biofilms Lately? Clin Microbiol Rev. 2002;15(2):155–166. doi: 10.1128/CMR.15.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbrandt R, Kalkum M, Lai EM, Lurz R, Kado CL, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274(32):22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- Eisenbrandt R, Kalkum M, Lurz R, Lanka E. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J Bacteriol. 2000;182(23):6751–6761. doi: 10.1128/JB.182.23.6751-6761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth N, Skurray R. Characterization of the F plasmid bifunctional conjugation gene, traG. MGG Mol Gen Genet. 1992;232(1):145–153. doi: 10.1007/BF00299147. [DOI] [PubMed] [Google Scholar]
- Fournes F, Crozat E, Pages C, Tardin C, Salomé L, Cornet F, Rosseau P. FtsK translocation permits discrimination between an endogenous and an imported Xer/dif recombination complex. Proc Natl Acad Sci. 2016;113(28):7882–7887. doi: 10.1073/pnas.1523178113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Ippen-Ihler K, Skurray RA. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58(2):162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcillán-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- Goodman SD, Scocca JJ. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85(18):6982–6. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen TA, Cornelissen CN. Neisseria gonorrhoeae requires expression of TonB and the putative transporter TdfF to replicate within cervical epithelial cells. Mol Microbiol. 2006;62(4):1144–1157. doi: 10.1111/j.1365-2958.2006.05429.x. [DOI] [PubMed] [Google Scholar]
- Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: From DNA donation to homologous recombination. Mol Microbiol. 2006;59(2):376–385. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- Hamilton HL, Domínguez NM, Schwartz KJ, et al. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55(6):1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- Hamilton HL, Schwartz KJ, Dillard JP. Insertion-duplication mutagenesis of Neisseria: Use in characterization of DNA transfer genes in the gonococcal genetic island. J Bacteriol. 2001;183(16):4718–4726. doi: 10.1128/JB.183.16.4718-4726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OB, Clemence M, Dillard JP, Tang CM, Trees D, Grad YH, Maiden MCJ. Genomic analyses of Neisseria gonorrhoeae reveal an association of the gonococcal genetic island with antimicrobial resistance. J Infect. 2016;73(6):578–587. doi: 10.1016/j.jinf.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SA, Masters TL, Wachter J. Gonorrhea – an evolving disease of the new millennium. Microb Cell. 2016;3(9):371–389. doi: 10.15698/mic2016.09.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417(417):656–659. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- Jain S, Kahnt J, van der Does C. Processing and maturation of the pilin of the type IV secretion system encoded within the gonococcal genetic island. J Biol Chem. 2011;286(51):43601–43610. doi: 10.1074/jbc.M111.264028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Zweig M, Peeters E, Seiwering K, Hackett KH, Dillard JP, van der Does C. Characterization of the single stranded DNA binding protein SsbB encoded in the gonoccocal genetic island. PLoS ONE. 2012;7(4):e35285. doi: 10.1371/journal.pone.0035285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler PL, Chan YA, Hackett KT, Turner N, Hamilton HL, Cloud-Hansen C, Dillard JP. Mating pair formation homologue TraG is a variable membrane protein essential for contact-independent type IV secretion of chromosomal DNA by Neisseria gonorrhoeae. J Bacteriol. 2013;195(8):1666–1679. doi: 10.1128/JB.02098-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler PL, Hamilton HL, Cloud-Hansen K, Dillard JP. AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J Bacteriol. 2007;189(15):5421–5428. doi: 10.1128/JB.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larbig KD, Christmann A, Johann A, Klockgether J, Hartsch T, Merkl R, Wiehlmann L, Fritz HJ, Tümmler B. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J Bacteriol. 2002;184(23):6665–6680. doi: 10.1128/JB.184.23.6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224(1):1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. The ParA/MinD family puts things in their place. Trends Microbiol. 2012;20(9):411–418. doi: 10.1016/j.tim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manwaring NP, Skurray Ra, Firth N. Nucleotide sequence of the F plasmid leading region. Plasmid. 1999;41:219–225. doi: 10.1006/plas.1999.1390. [DOI] [PubMed] [Google Scholar]
- McClure R, Nudel K, Massari P, Tjaden B, Su X, Rice PA, Genco CA. The gonococcal transcriptome during infection of the lower genital tract in women. PLoS ONE. 2015;10(8):e0133982. doi: 10.1371/journal.pone.0133982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcshan WM, Williams RP, Hull RA. A recombinant molecule from a disseminating strain of Neisseria gonorrhoeae that confers serum bactericidal resistance. Infect Immun. 1987;55(12):3017–3022. doi: 10.1128/iai.55.12.3017-3022.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki S, Ram P, Pham T, Goluszko P, Morse S, Anderson GD, Nowicki B. Pelvic inflammatory disease isolates of Neisseria gonorrhoeae are distinguished by C1q-dependent virulence for newborn rats and by the sac-4 region. Infect Immun. 1997;65:2094–2099. [Google Scholar]
- Pachulec E. Dissertation. University of Groningen; 2010. The type IV secretion systems of Neisseria gonorrhoeae. [Google Scholar]
- Pachulec E, Siewering K, Bender T, Heller EM, Salgado-Pabón W, Schmoller SK, Woodhams KL, Dillard JP, van der Does C. Functional Analysis of the Gonococcal Genetic Island of Neisseria gonorrhoeae. PLoS ONE. 2014;9(10):e109613. doi: 10.1371/journal.pone.0109613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey ME, Bender T, Klimowicz AK, Hackett KT, Yamamoto A, Jolicoeur A, Callaghan MM, Wassarman KM, van der Does C, Dillard JP. Targeted mutagenesis of intergenic regions in the Neisseria gonorrhoeae gonococcal genetic island reveals multiple regulatory mechanisms controlling type IV secretion. Mol Microbiol. 2015;97(6):1168–1185. doi: 10.1111/mmi.13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey ME, Hackett KT, Bender T, Kotha C, van der Does C, Dillard JP. TraK and TraB are conserved outer membrane proteins of the Neisseria gonorrhoeae type IV secretion system and are expressed at low levels in wild-type cells. J Bacteriol. 2014;196(16):2954–2968. doi: 10.1128/JB.01825-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmele CW, Xian Y, Albrecht M, Faulstich M, Fraunholz M, Heinrichs E, Dittrich MT, Müller T, Reinhardt R, Rudel T. Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nucleic Acids Res. 2014;42(16):10579–10595. doi: 10.1093/nar/gku762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MAJ, Wadhams GH, Hadfield KA, Tickner S, Armitage JP. ParA-like protein uses nonspecific chromosomal DNA binding to partition protein complexes. Proc Natl Acad Sci. 2012;109(17):6698–6703. doi: 10.1073/pnas.1114000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Pabón W. Dissertation. University of Wisconsin-Madison; 2008. Neisseria gonorrhoeae processing of chromosomal DNA for direct secretion via a type IV secretion system: Requirement of a novel relaxase homologue. [Google Scholar]
- Salgado-Pabón W, Du Y, Hackett KT, Lyons KM, Arvidson CG, Dillard JP. Increased expression of the type IV secretion system in piliated Neisseria gonorrhoeae variants. J Bacteriol. 2010;192(7):1912–1920. doi: 10.1128/JB.01357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Pabón W, Jain S, Turner N, van der Does C, Dillard JP. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol Microbiol. 2007;66(4):930–947. doi: 10.1111/j.1365-2958.2007.05966.x.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder LAS, Jarvis SA, Saunders NJ. Complete and variant forms of the “gonococcal genetic island” in Neisseria meningitidis. Microbiology. 2005;151(12):4005–4013. doi: 10.1099/mic.0.27925-0. [DOI] [PubMed] [Google Scholar]
- Sparling PF. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966;92(5):1364–71. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer-Smith R, Roberts S, Gurung N, Snyder LA. DNA uptake sequences in Neisseria gonorrhoeae as intrinsic transcriptional terminators and markers of horizontal gene transfer. Microb Genomics. 2016;2(8):1–11. doi: 10.1099/mgen.0.000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen CT, Cho C, Shao JQ, Apicella MA. The Neisseria gonorrhoeae biofilm matrix contains DNA, and an endogenous nuclease controls its incorporation. Infect Immun. 2011;79(4):1504–1511. doi: 10.1128/IAI.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M. Current and future antimicrobial treatment of gonorrhoea – the rapidly evolving Neisseria gonorrhoeae continues to challenge. BMC Infect Dis. 2015;15(364):1–15. doi: 10.1186/s12879-015-1029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorkapic D, Pressler K, Schild S. Multifaceted roles of extracellular DNA in bacterial physiology. Curr Genet. 2016;62:71–79. doi: 10.1007/s00294-015-0514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SEH, Sparling PF. Response of Neisseria gonorrhoeae to iron limitation: alterations in expression of membrane proteins without apparent siderophore production. Infect Immun. 1985;47(2):388–94. doi: 10.1128/iai.47.2.388-394.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhams KL, Benet ZL, Blonsky SE, Hackett KT, Dillard JP. Prevalence and detailed mapping of the gonococcal genetic island in Neisseria meningitidis. J Bacteriol. 2012;194(9):2275–2285. doi: 10.1128/JB.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae 2012 [Google Scholar]
- Zola TA, Strange HR, Dominguez NM, Dillard JP, Cornelissen CN. Type IV secretion machinery promotes ton-independent intracellular survival of Neisseria gonorrhoeae within cervical epithelial cells. Infect Immun. 2010;78(6):2429–2437. doi: 10.1128/IAI.00228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweig M, Schork S, Koerdt A, Siewering K, Sternberg C, Thormann K, Albers SV, Molin S, van der Does C. Secreted single-stranded DNA is involved in the initial phase of biofilm formation by Neisseria gonorrhoeae. Environ Microbiol. 2014;16(4):1040–1052. doi: 10.1111/1462-2920.12291. [DOI] [PubMed] [Google Scholar]