Abstract
A MADS family gene, OsMADS6, was isolated from a rice (Oryza sativa L.) young flower cDNA library using OsAMDS1 as a probe. With this clone, various MADS box genes that encode for protein-to-protein interaction partners of the OsMADS6 protein were isolated by the yeast two-hybrid screening method. On the basis of sequence homology, OsMADS6 and the selected partners can be classified in the APETALA1/AGAMOUS-LIKE9 (AP1/AGL9) family. One of the interaction partners, OsMADS14, was selected for further study. Both genes began expression at early stages of flower development, and their expression was extended into the later stages. In mature flowers the OsMADS6 transcript was detectable in lodicules and also weakly in sterile lemmas and carpels, whereas the OsMADS14 transcript was detectable in sterile lemmas, paleas/lemmas, stamens, and carpels. Using the yeast two-hybrid system, we demonstrated that the region containing of the 109th to 137th amino acid residues of OsMADS6 is indispensable in the interaction with OsMADS14. Site-directed mutation analysis revealed that the four periodical leucine residues within the region are essential for this interaction. Furthermore, it was shown that the 14 amino acid residues located immediately downstream of the K domain enhance the interaction, and that the two leucine residues within this region play an important role in that enhancement.
The vegetative-to-reproductive transition is an important event in a plant's development. Upon initiation of the reproductive phase, the development of the floral meristem is initiated by floral meristem identity genes such as LFY (LEAFY) and AP1 (APETALA1) in Arabidopsis (Mandel et al., 1992; Weigel et al., 1992; Hempel et al., 1997). At a later step, the fate of floral organ primordia is specified by three classes of homeotic genes (for review, see Weigel and Meyerowitz, 1994): AP1, AG (AGAMOUS), PI (PISTILATA), and AP3 in Arabidopsis (Yanofsky et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994), and SQUA (SQUAMOSA), PLE (PLENA), GLO (GLOBOSA), and DEF (DEFICIENS) in snapdragon (Sommer et al., 1990; Huijser et al., 1992; Tröbner et al., 1992; Bradley et al., 1993). These homeotic genes belong to the MADS-domain protein gene family. MADS box genes encode a family of highly conserved transcription factors that participate in signal transduction and developmental control in plants, animals, yeast, and fungi.
In addition to the three classes of organ identity genes in plants, there are a large number of other MADS box genes whose function is less well defined. In Arabidopsis at least 17 AGL (AG-LIKE) genes have been isolated (Ma et al., 1991; Mandel and Yanofsky, 1995; Rounsley et al., 1995). It was revealed that some of these genes are floral organ specific and appear to be involved in controlling floral organ initiation and development. For example, AGL2 and AGL4 are first expressed very early in flower development (after the floral meristem has emerged from the inflorescence meristem but before any of the organ primordia emerge), suggesting that AGL2 and AGL4 play important roles in the intermediate step between inflorescence initiation and floral organ initiation (Flanagan and Ma, 1994; Savidge et al., 1995). In addition to flower development, several MADS box genes are involved in the control of ovule and seed development, vegetative growth, root development, embryogenesis, or symbiotic induction (Mandel et al., 1994; Angenent et al., 1995; Heard and Dunn, 1995; Flanagan et al., 1996; Perry et al., 1996; Buchner and Boutin, 1998; Zhang and Forde, 1998).
Plant MADS box proteins consist of a MADS box domain, an I region, a K domain, and a C-terminal region. The conserved MADS box domain is required for sequence-specific DNA binding and dimerization (Mizukami et al., 1996; Riechmann et al., 1996a, 1996b; West et al., 1998). The MADS box domains bind to the consensus DNA sequence, the CArG motif (Huang et al., 1993, 1995; Tilly et al., 1998). In AP3 and PI the MADS box and the I region are needed for nuclear localization of the proteins (McGonigle et al., 1996). The K domain is the second conserved region, carrying 65 to 70 amino acid residues and located in the middle of the MADS box proteins. The K domain was named due to its structural resemblance to the coiled-coil domain of keratin, and has been suggested to be involved in protein-to-protein interactions (Ma et al., 1991; Pnueli et al., 1991; Theissen et al., 1995). The C-terminal region is rich in acidic amino acids, which are characteristic of transactivation domains. Using the yeast two-hybrid system, we recently demonstrated that OsMADS16, a rice (Oryza sativa L.) AP3 homolog, contains a transcription activation domain in the C-terminal region of the protein (Moon et al., 1999).
It has been demonstrated that the K domain is required for interactions between MADS box proteins (Davies et al., 1996; Fan et al., 1997). In experiments using the yeast two-hybrid system, GLO and DEF, the B class proteins of snapdragon, specifically selected each other as a partner in the protein-to-protein interaction, and the K domain played an important role in the interaction (Davies et al., 1996). Furthermore, in studies using the same system, AG interacted with AGL2, AGL4, AGL6, and AGL9 (Fan et al., 1997). In that study it was demonstrated that the K domain is necessary in the protein-to-protein interaction. These results indicate that MADS box proteins cooperate with other MADS box proteins in a K-domain-mediated interaction to carry out their functions.
Almost all of our knowledge about the interaction between MADS box proteins has been obtained from the two dicots Arabidopsis and snapdragon. Although many MADS box proteins were isolated from monocots, including maize, sorghum, orchid, and rice (Lu et al., 1993; Schmidt et al., 1993; Chung et al., 1995; Kang et al., 1995, 1997; Mena et al., 1995; Montag et al., 1995; Greco et al., 1997; Kang and An, 1997), the interaction between the MADS box proteins has not been elucidated. We previously reported the isolation of OsMADS1, a rice MADS box gene that exhibited the highest homology with AGL2 (Chung et al., 1994). It was demonstrated that ectopic expression of the OsMADS1 gene in homologous and heterologous plants resulted in early flowering, suggesting that the rice MADS gene is involved in flower induction (Chung et al., 1994). In the present study, we report the isolation of a rice AP1/AGL9 family gene, OsMADS6, by screening a young flower cDNA library from rice using OsMADS1 as a probe. With this gene we isolated the protein-to-protein interaction partners by the yeast two-hybrid system. We also investigated the motif responsible for the interaction between MADS box proteins.
MATERIALS AND METHODS
Library Screening and Sequence Analysis
The expression cDNA libraries were constructed from mRNA isolated from young rice (Oryza sativa L.) panicles (less than 2 cm height) using Uni-ZAP XR and HybriZAP vectors (Stratagene; Moon et al., 1999). Total cDNAs of a phagemid form were obtained by the mass in vivo excision method. Hybridization was performed with 105 plaques using a labeled probe prepared from the OsMADS1 coding region. The cDNA clones were rescued by in vivo excision using a helper phage (ExAssist, Stratagene). Double-stranded DNA was used as a template for DNA sequence analysis following the manufacturer's instructions (Thermo Sequnase cycle sequencing kit, Amersham). Amino acid sequence homology was compared using the BLASTX alignment program (Altschul et al., 1997).
Plasmid Construction
The binding domain vector pBDGAL4 and the activation domain vector pADGAL4 were purchased from Stratagene. The sequences containing a portion of OsMADS1, OsMADS3, OsMADS4, OsMADS5, OsMADS6, OsMADS7, OsMADS8, OsMADS14, OsMADS15, OsMADS17, and OsMADS18 were generated by PCR. DNA sequences encoding the following amino acid residues were amplified: 85 to 257 in OsMADS1 (OsMADS1-KC), 87 to 236 in OsMADS3 (OsMADS3-KC), 84 to 210 in OsMADS4 (OsMADS4-KC), 89 to 225 in OsMADS5 (OsMADS5-KC), 1 to 90 in OsMADS6 (OsMADS6-MI), 1 to 170 in OsMADS6 (OsMADS6-MIKC14), 86 to 156 in OsMADS6 (OsMADS6-K), 86 to 170 in OsMADS6 (OsMADS6-KC14), 86 to 250 in OsMADS6 (OsMADS6-KC), 90 to 249 in OsMADS7 (OsMADS7-KC), 90 to 248 in OsMADS8 (OsMADS8-KC), 1 to 90 in OsMADS14 (OsMADS14-MI), 1 to 172 in OsMADS14 (OsMADS14-MIKC14), 90 to 172 in OsMADS14 (OsMADS14-KC14), 90 to 246 in OsMADS14 (OsMADS14-KC), and the K domain and C region of OsMADS15 (OsMADS15-KC), OsMADS17 (OsMADS17-KC), and OsMADS18 (OsMADS18-KC). Regions containing amino acids 86 to 110 (OsMADS6-KI), 109 to 137 (OsMADS6-KII), 138 to 170 (OsMADS6-KIII), and 109 to 170 (OsMADS6-KII+KIII) of OsMADS6 were also amplified by PCR.
Replacement mutants in the K box region of OsMADS6 were generated by PCR as follows: Leu-110 with Ser and Leu-118 with Arg in the KII fragment (OsMADS6-KIIS110R118); Leu-126 with Arg and Leu-134 with Arg in the KII fragment (OsMADS6-KIIR126R134); Leu-110 with Ser and Leu-118 with Arg in the KII and KIII fragments (OsMADS6-KIIS110R118+KIII); Leu-159 with Arg and Leu-166 with Arg in the KII and KIII fragments (OsMADS6-KII+KIIIR159R 166); and Leu-110 with Ser, Leu-118 with Arg, Leu-159 with Arg, and Leu-166 with Arg of the KII and KIII fragments (OsMADS6-KIIS110R118 +KIIIR159R 166). For all constructs, the 5′ EcoRI site and the 3′ SalI site were introduced by PCR. The PCR profile used was 1 min at 95°C, 1 min at 57°C, and 1.5 min at 72°C for a total of 40 cycles. The PCR products were digested with EcoRI and SalI, ligated to pBDGAL4 or pADGAL4, and transformed into appropriate hosts. The sequences of all inserts were determined to confirm the proper fusion of the constructs.
Yeast Two-Hybrid Screening
The yeast (Saccharomyces cereviseae) strain YRG-2 (Matα, ura3-52, his3-200, ade2-101, lys2-801, trp1-901, leu2-3, 112, gal4-542, gal80-538, LYS::UASGAL1-TATAGAL1-HIS3, URA3::UASGAL4 17mers(x3)-TATACYC1-lacZ), was purchased from Stratagene. YRG-2 was transformed with pBD/OsMADS6-KC14, the binding domain plasmid containing the K domain and 14 amino acid residues of the C-terminal region of OsMADS6, using a modified lithium acetate method (Gietz et al., 1992). The transformants were tested for imidazoleglycerol-phosphate dehydratase (HIS3) reporter gene expression and found not to express this gene, as indicated by the absence of growth on a medium lacking His.
The strain was transformed with 100 μg of plasmid DNA of the HybriZAP (Stratagene) cDNA library, along with 3 mg of salmon-sperm carrier DNA, using the lithium acetate method. The transformants were plated on SD (synthetic dropout) medium lacking Trp, Leu, and His, and containing 1 mm 3-aminotriazole (SD-Trp-Leu-His+3-AT; Kaiser et al., 1994). Approximately 1.4 × 106 transformants were obtained, as estimated based on the number of transformants grown on the SD-Trp-Leu plate. The 59 colonies that grew on the SD-Trp-Leu-His+3-AT plates after 5 d were subsequently transferred onto the filter paper on the SD-Trp-Leu-His plate and incubated for 1 d. The β-galactosidase activity was measured by filter assay according to the method of Breeden and Nasmyth (1985). The colonies that turned blue in less than 6 h were selected for isolation of DNA, and were then retransferred into the YRG-2 yeast strain containing pBDGAL4 or pBD/OsMADS6-KC14. Plasmid DNA was recovered from yeast according to the method of Hoffman and Winston (1987) and transformed into Escherichia coli strain XL-1-Blue (Stratagene) by electroporation.
Isolation of the 5′ Region of the OsMADS14 cDNA
The 5′ region of the OsMADS14 cDNA was isolated by PCR using the T3 primer (5′-AATTAACCCTCACTAAAGGG-3′) and the OsMADS14 gene-specific primer (5′-ATGGACTCGAGCATTAGTTGG-3′) located within the K domain. The template was total cDNA in vivo excised from the Uni-ZAP (Stratagene) cDNA library. The amplified fragment was cloned into pBlueScript II KS(+) (Stratagene).
DNA and RNA Blot Analyses
Genomic DNA was prepared from 2-weak-old rice seedlings according to the protocol of Shure et al. (1983). DNAs (10 μg) were digested with EcoRI, HindIII, or PstI, subjected to electrophoresis in a 0.8% (w/v) agarose gel, and then blotted onto a Hybond-N+ filter (Amersham). The filter was prehybridized, hybridized, and washed according to the method of Moon et al. (1999).
Total RNA was isolated (TRI reagent, Molecular Research Center, Cincinnati) from young flowers with a panicle size of 1 to 5 cm, flowers at the early vacuolated pollen stage, flowers at the late vacuolated pollen stage, leaves, roots, lodicules, sterile lemmas, paleas/lemmas, stamens, and carpels. Total RNA (20 μg) was fractionated on a 1.3% (w/v) agarose gel as described previously (Sambrook et al., 1989). After RNA transfer onto a nylon membrane, the blots were prehybridized, hybridized, and washed according to the method of Moon et al. (1999).
The OsMADS6-specific probe (296-bp fragment between nucleotides 748 and 1043) and the OsMADS14-specific probe (626-bp fragment between nucleotides 730 and 1355) were labeled by the random priming method (Amersham).
Quantitative Assay of β-Galactosidase Activity
Mid to late exponential-phase yeast cells were collected and resuspended in Z buffer (Miller, 1972). The cells were assayed for β-galactosidase activity as described by Miller (1972) using O-nitrophenyl β-d-galactopyranoside as a substrate. The activity unit was calculated using the formula: 1,000 × A420/(A600 × assay time in minutes × assay volume in milliliters).
Construction of Phylogenetic Trees
Alignment of conceptual amino acid sequences was made using the Jotun Hein method in the program MegAlign of the DNASTAR (Madison, WI) phylogenetic package. Phylogenetic trees were constructed by comparing 170 amino acid sequences comprising the MADS box, the I region, and the K domain.
RESULTS
Isolation of a Rice cDNA Clone Encoding a MADS Box Protein
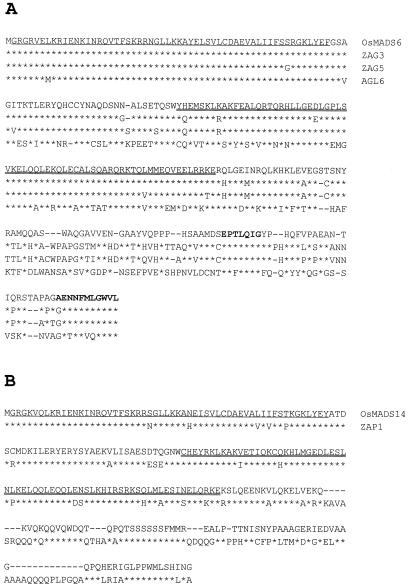
A cDNA clone was isolated by screening the Uni-ZAP cDNA library that was prepared from rice floral primordia using the OsMADS1 cDNA as a probe (Chung et al., 1994). This clone was designated OsMADS6. DNA sequence analysis showed that this cDNA clone is 1,043 nucleotides long and encodes a putative protein of 250 amino acid residues (calculated Mr = 28,400; accession no. U78782). The MADS box domain of the cDNA clone is located between the 2nd and 57th amino acids of the protein (Fig. 1A). This region is the most conserved region, as observed from other MADS box proteins. The second conserved domain, the K box, is located between the residues 91 and 156.
Figure 1.
Deduced amino acid sequence comparisons of MADS box proteins. A, Alignment of the amino acid sequences of OsMADS6, ZAG3, ZAG5, and AGL6. The two conserved motifs of the C region are indicated in bold. B, Alignment of the amino acid sequences of OsMADS14 and ZAP1, an AP1 homolog of maize. The MADS box regions are underlined and the K domains are double underlined. Asterisks indicate identical amino acid residues. Dashes indicate gaps introduced to maximize alignments.
The OsMADS6 protein contains two variable regions: the I region between the MADS and K boxes and the C-terminal region downstream of the K box (Purugganan et al., 1995). The C-terminal region of OsMADS6 has two short motifs, EPTLQIG and AENNFMLGWVL, which are partially conserved in ZAG3, ZAG5, AGL2, AGL4, and AGL9 (Fig. 1A). Based on amino acid sequence similarity of the entire coding region, OsMADS6 can be grouped into the AP1/AGL9 family (Purugganan et al., 1995), which includes AGL2 and AGL4 of Arabidopsis (Ma et al., 1991), ZAG3 and ZAG5, the maize homolog of AG (Mena et al., 1995), floral binding protein 2 (FBP2) of petunia (Angenent et al., 1994), TM5 of tomato (Pnueli et al., 1994), and OsMADS1, OsMADS5, OsMADS7, and OsMADS8 of rice (Chung et al., 1994; Kang and An, 1997; Kang et al., 1997). Among these genes, OsMADS6 was the most homologous to ZAG3 (84%) and ZAG5 (82%).
Two-Hybrid Screening
We conducted yeast two-hybrid screening to identify proteins that interact with OsMADS6. We initially made a fusion between the GAL4 binding domain and the OsMADS6 protein containing the K domain and the C-terminal region. This fusion molecule (pBD/OsMADS6-KC) by itself was able to activate the HIS3 and β-galactosidase (LacZ) selectable reporter genes, which were under the control of the GAL1 and GAL4 upstream activating sequences, respectively. This indicates that the K domain and the C-terminal region carries an activation domain. It was recently observed that other MADS box proteins also carry an activation domain in the C-terminal regions (Moon et al., 1999). Therefore, we made another molecule that was identical to pBD/OsMADS6-KC except that most of the C-terminal end was deleted, leaving only the 14 amino acid residues of the C region located immediately downstream of the K region. This plasmid, pBD/OsMADS6-KC14, was introduced into the yeast strain YRG-2, and the transformants were tested for activation of the HIS3 selectable marker. The transformants did not grow on a medium lacking His, demonstrating that the fragment containing the K domain and 14 amino acid residues of the C region of OsMADS6 does not contain an activator domain.
We therefore proceeded to introduce the cDNA expression library constructed from the mRNA of young rice panicles into the YRG-2 yeast strain containing pBD/OsMADS6-KC14. A total of 1.4 × 106 transformants was screened for their ability to grow on a medium lacking His. This initial screening identified 59 colonies, which were subsequently tested for activation of the LacZ gene. These experiments resulted in the identification of 45 colonies that activated both HIS3 and LacZ. Plasmid DNAs were prepared from these colonies and retransferred into the YRG-2 strain to confirm whether the activation is indeed due to the presence of the fusion protein. We observed that 39 plasmids were able to activate the LacZ gene only in the presence of pBD/OsMADS6-KC14. Sequence determination of these clones revealed that 38 plasmids contained an ORF that exhibited a significant homology to MADS box proteins (Table I). The remaining plasmid had some homology to MADS genes, but was significantly different from typical plant MADS box genes. This clone was not studied further.
Table I.
The MADS proteins isolated by yeast two-hybrid screening using OsMADS6 as bait
| Gene Name | No. of Isolated Clone(s) | Homologous Gene | Reference |
|---|---|---|---|
| OsMADS1 | 6 | AGL2 | Chung et al. (1994) |
| OsMADS5 | 2 | AGL2 | Kang and An (1997) |
| OsMADS7 | 3 | AGL2 | Kang et al. (1997) |
| OsMADS14 | 12 | ZAP1 | This study |
| OsMADS15 | 13 | ZAP1 | This study |
| OsMADS17 | 1 | ZAG3 | This study |
| OsMADS18 | 1 | ZAP1 | This study |
Eleven of the clones encoded for previously identified MADS box proteins; of these, six clones belong to OsMADS1 (Chung et al., 1994), two to OsMADS5 (Kang and An, 1997), and three to OsMADS7 (Kang et al., 1997). The remaining plasmids encode for MADS box proteins not previously reported. Twelve of these were partial clones of an identical MADS box gene, although the 5′ ends were different from each other (Fig. 2). This gene was designated OsMADS14. Thirteen clones encoded for an identical protein of another MADS box protein. The gene for these clones was designated OsMADS15. Both the OsMADS14 and OsMADS15 proteins were highly homologous to ZAP1. Among the remaining two clones, one clone, OsMADS17, showed a high similarity with ZAG3 and the last clone, OsMADS18, was the most homologous to ZAP1.
Figure 2.
The nucleotide sequence and the deduced amino acid sequence of OsMADS14. The MADS box region is underlined, and the K domain is double underlined. The 10 repeats of the GGA sequence in the 5′-UTR are indicated in bold type. The primer sequence used in isolation of the 5′ region of the gene is underlined. Arrowheads and numbers below the amino acid sequence indicate positions of the first amino acid of the fusion proteins selected by the yeast two-hybrid screening and the number of selected clones with the same first amino acid, respectively. The XhoI site used for generation of the gene-specific probe (accession no. AF058697) is indicated in bold type.
Isolation of the OsMADS14 cDNA Clone Containing an Entire ORF
All of the twelve cDNA clones of OsMADS14 selected by the two-hybrid screening were partial, lacking the 5′ region that encodes for the N-terminal end of the protein (Fig. 2). The 5′ region was isolated by PCR using a vector primer and the cDNA specific primer. A cDNA clone of 1,355 bp, containing the entire ORF, was generated by connecting the 5′ region to the cDNA clone obtained from the two-hybrid screening. It contains a 287-bp 5′-UTR and an ORF of 246 amino acid residues (calculated Mr = 28,500; accession no. AF058697).
The 5′ UTR of OsMADS14 cDNA contains 10 repeats of the GGA sequence (Fig. 2), and such repeat sequences were previously observed from other rice MADS-box genes (Chung et al., 1994; Kang and An, 1997; Kang et al., 1997). The OsMADS14 protein contains a MADS box domain which consists of 56 conserved amino acids present in the N-terminal region of all of the MADS transcription factors (Figs. 1B and 2). The K box domain, a region considered to participate in the protein-to-protein interaction, is also present between amino acid residues 91 and 158. Amino acid sequence comparison revealed that OsMADS14 was 72.4% homologous to ZAP1, an AP1 homolog of maize.
Expression Patterns of OsMADS6 and OsMADS14
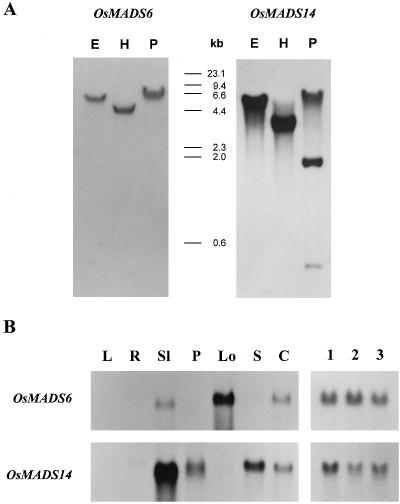
It has been well established that there are a large number of MADS box genes in the rice genome (Chung et al., 1994). Therefore, it was necessary to identify the region that does not cross hybridize with other MADS box genes by genomic DNA blot analyses. It was observed that the 300-bp PstI–EcoRI fragment located at the C-terminal region of OsMADS6 hybridized to a single DNA fragment (Fig. 3A). Likewise, the 630-bp XhoI fragment of OsMADS14 was shown to be a gene-specific region. In genomic DNA analysis of OsMADS14, three PstI fragments were hybridized with the probe (Fig. 3A), due to the presence of two PstI sites in the region used for the probe.
Figure 3.
Genomic DNA blot analysis and RNA blot analysis of OsMADS6 and OsMADS14. A, Southern blot analysis of OsMADS6 and OsMADS14. The rice genomic DNA was digested with EcoRI (E), HindIII (H), or PstI (P). The numbers indicate the size (in kb) of the DNA markers. B, RNA blot analysis of OsMADS6 and OsMADS14. Ethidium bromide staining of 25S and 17S rRNAs demonstrated equal amounts of RNA loading (data not shown). Lane L, Leaves; lane R, roots; lane Sl, sterile lemmas; lane P, paleas/lemmas; lane Lo, lodicules; lane S, stamens; lane C, carpels; lane 1, young flowers with a panicle size of 1 to 5 cm; lane 2, flowers at the early vacuolated pollen stage; lane 3, flowers at the late vacuolated pollen stage.
RNA blot analyses were conducted using the gene-specific probes. The results showed that the OsMADS6 transcript was detectable primarily in lodicules and also weakly in sterile lemmas and carpels of flowers at the late vacuolated pollen stage (Fig. 3B). However, the transcript was not detectable in stamens, paleas/lemmas, and vegetative organs. The spatial expression pattern of OsMADS14 was different from that of OsMADS6. Transcripts of this clone were detectable primarily in sterile lemmas and also weakly in paleas/lemmas, stamens, and carpels (Fig. 3B). However, the OsMADS14 transcripts were not detected in lodicules and vegetative organs. During flower development the OsMADS6 and OsMADS14 genes were expressed at an early stage, and their expression was extended into later stages of flower development (Fig. 3B).
Interaction between OsMADS6 and Other MADS Box Proteins
The yeast two-hybrid screening resulted in identification of seven types of MADS box proteins that interact with OsMADS6. To confirm these results, we investigated the protein-to-protein interaction between OsMADS6 and other rice MADS box proteins. The C-terminal half containing the K domain and C region of OsMADS1, OsMADS3, OsMADS4, OsMADS5, OsMADS6, OsMADS7, OsMADS8, OsMADS14, OsMADS15, OsMADS17, and OsMADS18 was fused to the activation domain of GAL4 using the pADGAL4 vector. These plasmids were introduced into the yeast strain YRG-2 containing the binding domain plasmid pBD/OsMADS6-KC14. The colonies that grew on a medium lacking Leu and Trp were examined for β-galactosidase activity. The results in Table II show that the KC regions of OsMADS1, OsMADS5, OsMADS7, OsMADS8, OsMADS14, OsMADS15, OsMADS17, and OsMADS18 were able to activate the LacZ gene. However, the KC regions of OsMADS3, OsMADS4, and OsMADS6 did not activate the reporter gene. Western blot analyses showed that the β-galactosidase protein level was proportional to the enzyme activity (data not shown).
Table II.
Quantitative assay of protein-to-protein interaction between OsMADS6 and rice MADS proteins
| Activation Domain Plasmid | β-Galactosidase Activitya
|
|
|---|---|---|
| pBD/OsMADS6-KC14b | pBDGAL4c | |
| pAD/OsMADS1-KC | 88.16 ± 1.357 | 0.04 ± 0.011 |
| pAD/OsMADS3-KC | 0.10 ± 0.014 | 0.04 ± 0.013 |
| pAD/OsMADS4-KC | 0.06 ± 0.012 | 0.04 ± 0.013 |
| pAD/OsMADS5-KC | 67.99 ± 2.096 | 0.05 ± 0.014 |
| pAD/OsMADS6-KC | 0.29 ± 0.023 | 0.03 ± 0.012 |
| pAD/OsMADS7-KC | 39.04 ± 3.417 | 0.04 ± 0.013 |
| pAD/OsMADS8-KC | 17.91 ± 0.631 | 0.04 ± 0.011 |
| pAD/OsMADS14-KC | 84.66 ± 4.389 | 0.02 ± 0.006 |
| pAD/OsMADS15-KC | 44.83 ± 1.937 | 0.02 ± 0.006 |
| pAD/OsMADS17-KC | 39.58 ± 3.619 | 0.04 ± 0.011 |
| pAD/OsMADS18-KC | 52.32 ± 2.175 | 0.04 ± 0.013 |
| None | 0.09 ± 0.032 | 0.04 ± 0.013 |
β-Galactosidase activity unit = 1,000 × A420/[A600 × reaction time (min) × volume of culture (mL)].
GAL4 DNA binding domain fusion protein.
pBDGAL4 is used as a negative control.
To confirm the lack of interaction, the region containing the K domain and 14 amino acids of the C-terminal region of the OsMADS3 and OsMAS4 proteins were fused to the binding domain vector pBDGAL4. Most of the C-terminal regions of OsMADS3 and OsMADS4 were not included in the construction to avoid a potential activator domain. Introduction of these plasmids into the YRG-2 strain containing pAD/OSMADS6-KC did not activate the LacZ gene (data not shown). These results showed that OsMADS6 interacts with OsMADS1, 5, 7, 8, 14, 15, 17, and 18, members of the AP1/AGL9 family, but not to the B and C class of MADS box proteins, OsMADS4 and OsMADS3; it also failed to interact with OsMADS6 itself.
Identification of the Protein-to-Protein Interaction Motif
The motif responsible for the protein-to-protein interaction between OsMADS6 and OsMADS14 was investigated using the yeast two-hybrid system. The MADS box domain and the I region (MI), the K domain and 14 amino acid residues of the C-region (KC14), the MI and KC14 region (MIKC14), and the K domain (K) of OsMADS6 were connected to the activation domain and the binding domain of GAL4. The K domain and C-terminal region (KC) of OsMADS6 was fused to the activation domain of GAL4. Similarly, the MI region, the MIKC14 region, and the KC14 region of OsMADS14 were connected to the activation domain and the binding domain of GAL4, and the KC region of OsMADS14 to the activation domain. The transformants that grew on a medium lacking Leu and Trp were examined for activation of the LacZ gene by a β-galactosidase activity analysis (Table III). When pAD/OsMADS6-K and pBD/OsMADS14-KC14 were introduced into YRG-2, the LacZ gene was activated. Similarly, pBD/OsMADS6-K and pAD/OsMADS14-KC activated the reporter gene expression. Moreover, OsMADS6-MIKC14 activated the LacZ gene in the presence of OsMADS14-MIKC14. However, the C region by itself containing amino acids 171 to 250 of OsMADS6 did not activate the LacZ gene in the presence of OsMADS14-KC (data not shown). Furthermore, OsMADS6-MI and OsMADS14-MI did not activate the LacZ gene (Table III). These results suggest that the K box is primarily responsible for heterodimerization between OsMADS6 and OsMADS14. Interestingly, including the 14 amino acid residues immediately downstream of the K box enhanced the enzyme activity by 5- or 20-fold (Table III), suggesting that the 14 residues stabilized or enhanced the interaction in the yeast two-hybrid system.
Table III.
Investigation of the motif responsible for protein-to-protein interaction between OsMADS6 and OsMADS14
| Activation Domain Plasmid | Binding Domain Plasmid | β-Galactosidase Activitya |
|---|---|---|
| pAD/OsMADS14-MIKC14 | pBD/OsMADS6-MIKC14 | 10.21 ± 1.007 |
| pAD/OsMADS14-MIKC14 | pBD/OsMADS6-MI | 0.09 ± 0.011 |
| pAD/OsMADS14-MI | pBD/OsMADS6-MIKC14 | 0.06 ± 0.007 |
| pAD/OsMADS14-MI | pBD/OsMADS6-MI | 0.07 ± 0.032 |
| pAD/OsMADS14-KC | pBD/OsMADS6-K | 13.22 ± 1.310 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KC14 | 62.12 ± 7.583 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KI | 0.14 ± 0.015 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KII | 18.21 ± 3.224 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KIII | 0.16 ± 0.023 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KII+KIII | 53.40 ± 2.824 |
| pAD/OsMADS6-MIKC14 | pBD/OsMADS14-MIKC14 | 8.14 ± 0.075 |
| pAD/OsMADS6-MI | pBD/OsMADS14-MIKC14 | 0.08 ± 0.010 |
| pAD/OsMADS6-MIKC14 | pBD/OsMADS14-MI | 0.06 ± 0.011 |
| pAD/OsMADS6-MI | pBD/OsMADS14-MI | 0.05 ± 0.008 |
| pAD/OsMADS6-K | pBD/OsMADS14-KC14 | 3.21 ± 1.024 |
| pAD/OsMADS6-KC14 | pBD/OsMADS14-KC14 | 61.23 ± 7.972 |
| pAD/OsMADS6-KC | pBD/OsMADS14-KC14 | 59.14 ± 4.927 |
| pAD/OsMADS6-KI | pBD/OsMADS14-KC14 | 0.04 ± 0.007 |
| pAD/OsMADS6-KII | pBD/OsMADS14-KC14 | 0.06 ± 0.032 |
| pAD/OsMADS6-KIII | pBD/OsMADS14-KC14 | 0.06 ± 0.016 |
| pAD/OsMADS6-KII+KIII | pBD/OsMADS14-KC14 | 17.43 ± 2.972 |
β-Galactosidase activity unit = 1,000 × A420/[A600 × reaction time (min) × volume of culture (mL)].
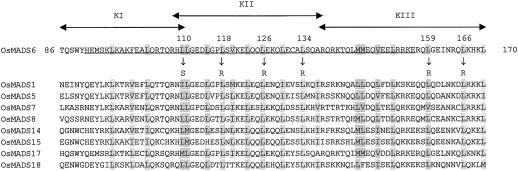
To narrow down the motif responsible for the protein-to-protein interaction, the KC14 region of OsMADS6 was divided into three regions; KI (86th to 110th amino acid), KII (109th to 137th amino acid), and KIII (138th to 170th amino acid) (Fig. 4). The three regions were connected into the binding domain of GAL4 and introduced into the yeast strain YRG-2 containing pAD/OsMADS14-KC. The results in Table III show that only the KII region activated the LacZ gene, suggesting that this region plays an important role in the protein-to-protein interaction. The experiment also indicated that the KIII region alone did not bind to OsMADS14, but enhanced the interaction between these proteins. When pAD/OsMADS14-KC14 was used as the activation domain plasmid instead of pAD/OsMADS14-KC, similar results were observed, although the β-galactosidase activities generally decreased (data not shown). Also, when those regions of OsMADS6 were fused into the activation domain and OsMADS14 was connected to the binding domain, similar results were obtained, except that pAD/OsMADS6-KII did not activate the LacZ gene in the presence of pBD/OsMADS14-KC14 (Table III). None of the activation domains or binding domain plasmids used in these experiments activated the LacZ gene by itself (data not shown).
Figure 4.
Amino acid sequence alignment of the region containing the K domain of OsMADS6 with those of OsMADS1, OsMADS5, OsMADS7, OsMADS8, OsMADS14, OsMADS15, OsMADS17, and OsMADS18. The region of OsMADS6 was divided into three regions: the KI region (amino acids 86–110), the KII region (amino acids 109–137), and the KIII region (amino acids 138–170). The entire K domain of OsMADS6 is underlined. The replaced amino acids are indicated below each Leu with arrows. The conserved hydrophobic residues, such as Leu, Ile, Val, and Met, are shaded. The numbers indicate the positions of mutagenized Leu residues and the first and last amino acids of the K region elucidated in this study.
Identification of the Amino Acid Residues Responsible for the Protein-to-Protein Interaction
Because it was determined that the KII and KIII regions containing amino acids 109 to 170 of OsMADS6 play an important role in the protein-to-protein interaction between OsMADS6 and OsMADS14, we investigated the amino acid residues responsible for the interaction. The KII and KIII regions of OsMADS6 have periodical and conserved Leu residues (Fig. 4). It has been previously suggested that such hydrophobic repeats may be involved in protein-to-protein interactions (Ma et al., 1991; Pnueli et al., 1991; Theissen et al., 1995). Therefore, we made the following five mutant fragments: OsMADS6-KIIS110R118: replacement of Leu-110 residue with Ser and Leu-118 with Arg in the KII fragment; OsMADS6-KIIR126R134: replacement of Leu-126 and Leu-134 Leus with Arg residues in the KII fragment; OsMADS6-KIIS110R118+KIII: replacement of Leu-110 with Ser and Leu-118 with Arg in the KII+KIII fragment; OsMADS6-KII+KIIIR159R166: replacement of Leu-159 and Leu-166 with Arg residues in the KII+KIII fragment; and OsMADS6-KIIS110R118+KIIIR159R166: replacement of Leu-110 with Ser and Leu-118, Leu-159, and Leu-166 Leus with Arg residues in the KII+KIII fragment.
These mutant fragments were connected to the binding domain of GAL4 and transferred into the YRG-2 strain containing the activation domain plasmid pAD/OsMADS14-KC. The results in Table IV show that mutations in Leu-110 and Leu-118 diminished the interacting ability of the KII fragment. Similarly, mutations of Leu-126 and Leu-134 also significantly affected the activity. These results suggest that the four periodical Leus in the KII region are necessary for interaction between the K box regions. When the mutations at Leu-110 and Leu-118 were introduced into the KII+KIII fragment, the enzyme activity was reduced, but still retained a significant level of activity. However, when mutations were introduced into both KII and KIII regions by replacing Leu-110, Leu-118, Leu-159, and Leu-166, the activity was almost completely diminished. These results, together with the results shown in Table III, indicate that the KIII region alone is not sufficient for protein interaction, but is able to enhance the interaction between MADS box proteins.
Table IV.
Investigation of the amino acid residues responsible for the interaction between OsMADS6 and OsMADS14 by site-directed mutagenesis of amino acids in the K domain and 14 amino acids of the C region of OsMADS6
| Activation Domain Plasmid | Binding Domain Plasmid | β-Galactosidase Activitya |
|---|---|---|
| pAD/OsMADS14-KC | pBD/OsMADS6-KII | 10.52 ± 2.460 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KIIS110R118 | 0.17 ± 0.016 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KIIR126R134 | 0.05 ± 0.015 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KII+KIII | 50.63 ± 1.983 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KIIS110R118+KIII | 29.33 ± 2.660 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KIIS110R118+KIIIR159R166 | 0.07 ± 0.012 |
| pAD/OsMADS14-KC | pBD/OsMADS6-KII+KIIIR159R166 | 0.07 ± 0.016 |
| pAD/OsMADS6-KII | pBD/OsMADS14-KC14 | 0.09 ± 0.027 |
| pAD/OsMADS6-KIIS110R118 | pBD/OsMADS14-KC14 | 0.08 ± 0.032 |
| pAD/OsMADS6-KIIR126R134 | pBD/OsMADS14-KC14 | 0.08 ± 0.016 |
| pAD/OsMADS6-KII+KIII | pBD/OsMADS14-KC14 | 13.91 ± 0.905 |
| pAD/OsMADS6-KIIS110R118+KIII | pBD/OsMADS14-KC14 | 3.11 ± 0.247 |
| pAD/OsMADS6-KIIS110R118+KIIIR159R166 | pBD/OsMADS14-KC14 | 0.09 ± 0.007 |
| pAD/OsMADS6-KII+KIIIR159R166 | pBD/OsMADS14-KC14 | 0.09 ± 0.013 |
β-Galactosidase activity unit = 1,000 × A420/[A600 × reaction time (min) × volume of culture (mL)].
When the mutant fragments were used in the construction of activation domain plasmids and their interaction ability in the yeast strain carrying the binding domain plasmid pBD/OsMADS14-KC14 was tested, similar results were obtained except that pAD/OsMADS6-KII and pBD/OsMADS14-KC14 did not activate the LacZ gene (Table IV). Taken together, these results suggest that the Leu residues in the KII region are important for the protein interaction and the Leu residues in the KIII region are involved in enhancing the interaction.
DISCUSSION
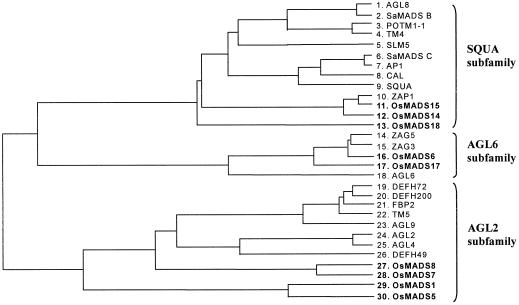
We have isolated the OsMADS6 gene by screening a young rice flower library using OsMADS1 as a probe. On the basis of deduced amino acid sequences, the OsMADS6 gene can be classified into the AGL6 subfamily (Theissen et al., 1996), which belongs to the AP1/AGL9 family (Purugganan et al., 1995) (Fig. 5). In addition, we have isolated various MADS box genes that encode for protein-to-protein interaction partners of OsMADS6 by the yeast two-hybrid screening method.
Figure 5.
Phylogenetic tree showing the relationship among AP1/AGL9 family proteins. Rice MADS box proteins are indicated in bold type. The horizontal branches are proportional to the number of base substitutions. 1, 7, 8, 18, 23, 24, and 25, Arabidopsis; 9, 19, 20, and 26, snapdragon; 10, 14, and 15, maize; 11, 12, 13, 16, 17, 27, 28, 29, and 30, rice; 21, petunia; 3, potato; 4 and 22, tomato; 5, Silene latifolia; 2 and 6, Sinapis alba.
It was observed that the region containing the K domain and the C-terminal region of OsMADS6 had transcription activation ability in yeast. The region responsible for the activation appears to be located in the C-terminal region, since deletion of the region eliminated the activation ability. Recently, we reported that OsMADS16, a rice AP3 homolog, contains a motif responsible for the transcription activation ability in the C-terminal region (Moon et al., 1999). We also observed the transcription activation ability of other rice MADS box proteins such as OsMADS1, OsMADS3, OsMADS5, OsMADS7, OsMADS8, and OsMADS14 (unpublished data), and found that one of the roles of the C-terminal region of MADS box proteins is transcription activation.
Almost all of the genes isolated by two-hybrid screening using OsMADS6 corresponded to MADS box proteins. This suggests that MADS box proteins efficiently interact with each other in yeast. The isolated clones belonged to seven MADS genes that can be classified into three groups in the AP1/AGL9 family (Table I; Fig. 5). The OsMADS1, OsMADS5, and OsMADS7 genes belong to the AGL2 subfamily, the OsMADS17 gene belongs to the AGL6 subfamily, and the OsMADS14, OsMADS15, and OsMADS18 genes belong to the SQUA subfamily (Theissen et al., 1996). In addition to these seven MADS box proteins, OsMADS6 also interacted with OsMADS8, which is also a member of the AGL2 subfamily (Kang et al., 1997). However, OsMADS6 did not interact with OsMADS3 and OsMADS4, rice homologs of AG and PI, respectively (Table II). In addition, OsMADS6 failed to interact with itself.
In studies using the yeast two-hybrid system, it was previously reported that AG, a C-class protein in Arabidopsis, interacted with AP1/AGL9 family proteins such as AGL2, AGL4, AGL6, and AGL9, most of which are expressed at the early flowering stage (Fan et al., 1997). Moreover, MADS box proteins expressed at the early (SQUA), intermediate (DEFH200 and DEFH72), and late (DEFH49) stages of flower development were also identified as interaction partners of PLE, a C-class MADS box protein in snapdragon, by the two-hybrid screening method (Davies et al., 1996). Sequence similarity and expression patterns showed that DEFH200 and DEFH72 were very similar to FBP2 and TM5, and DEFH49 was very similar to AGL2. These results indicate that C-class proteins and AP1/AGL9 family proteins interact with each other in Arabidopsis and snapdragon. However, we demonstrated that in rice the AP1/AGL9 family proteins interact with each other within the family, but do not interact with OsMADS3, a C-class protein.
The temporal expression patterns of the rice AP1/AGL9 family genes, such as OsMADS1, OsMADS5, OsMADS6, OsMADS7, OsMADS8, and OsMADS14, were similar (Chung et al., 1994; Kang and An, 1997; Kang et al., 1997). These six MADS box genes were expressed at the early stage of the flower development and their expressions were extended into later stages of flower development. However, the spatial expression patterns of these MADS box genes were different. In mature flowers the OsMADS1 transcript was present in paleas/lemmas and carpels (Chung et al., 1994), OsMADS5 in anthers and weakly in carpels (Kang and An, 1997), OsMADS7 and OsMADS8 in carpels and weakly in anthers (Kang et al., 1997), OsMADS14 in sterile lemmas, paleas/lemmas, stamens, and carpels, and OsMADS6 in lodicules, sterile lemmas, and carpels (Fig. 3B). It is likely that an AP1/AGL9 protein cooperates with another AP1/AGL9 protein coexisting in specific cell types at a particular developmental stage.
Ectopic expression of OsMADS6 and OsMADS14 in rice exhibited extreme early flowering and dwarfism, which were also observed in rice plants ectopically expressing OsMADS1, OsMADS5, OsMADS7, and OsMADS8 (unpublished results). However, the early flowering and dwarfism in transgenic plants of those four MADS genes was not as extreme as in those of OsMADS6 and OsMADS14, indicating that OsMADS6 and OsMADS14 might regulate a very early stage of flower development. Further in vivo studies are now being undertaken to identify whether a heterodimer between OsMADS6 and its interaction partners participates in these developmental processes.
It is known that MADS box proteins interact with each other to form dimers (Davies et al., 1996; Fan et al., 1997). In the present study, motifs responsible for the interaction between OsMADS14 and OsMADS6 proteins were investigated using the yeast two-hybrid screening method. Our results showed that the K domain plays a very important role in the protein-to-protein interaction. In Arabidopsis, it was previously reported that AG forms a dimer with various AGL proteins via K-domain-mediated interaction (Fan et al., 1997). It was also reported that the K domain was essential for protein-to-protein interaction between GLO and DEF (Davies et al., 1996).
It has also been suggested that the AG and AGL proteins contain two helices in the K domain, which may participate in the protein-to-protein interaction (Ma et al., 1991). The KI and KII regions of OsMADS6 contain one helix each. In the present study, we demonstrated that the KII region carrying the second helix plays a more important role in the interaction. The site-directed mutation analysis revealed that Leu-126 and Leu-134 of the KII region play an important role in the interaction. These amino acids are the hydrophobic residues suggested to cluster on one face of the putative helices (Theissen et al., 1995). Therefore, it can be proposed that the putative helix structure plays an important role in the K-domain-mediated interaction between OsMADS14 and OsMADS6.
In Arabidopsis, it was reported that the C region of the AG and AGL proteins enhanced the K-domain-mediated interaction between these proteins (Fan et al., 1997). In the present study, we showed that the 14 amino acid residues located immediately downstream of the K domain enhanced the interaction between OsMADS6 and OsMADS14. In Arabidopsis, ag-4 mutants that result from deletions of 12 or 14 amino acids downstream of the K domain retain a partial AG activity (Sieburth et al., 1995). Therefore, it appears that the region is not essential for function, but is necessary for full activity, probably by enhancing the interaction between AG and its partner. The deleted amino acids correspond to the residues between amino acids 142 and 153 or 142 and 155 of OsMADS6, which are located in the KIII region.
We demonstrated by site-directed mutagenesis experiments that Leu-110, Leu-118, Leu-126, and Leu-134 in the KII region and at Leu-159 and Leu-166 residues in the KIII region of OsMADS6 play a very important role in the protein interaction. These periodical hydrophobic residues are conserved in OsMADS6 and its interaction partners such as OsMADS1, OsMADS5, OsMADS7, OsMADS8, OsMADS14, OsMADS15, OsMADS17, and OsMADS18 (Fig. 4), supporting the idea that these amino acids are important for the function of MADS box proteins.
ACKNOWLEDGMENTS
We wish to thank Sung Key Jang for valuable suggestions during yeast two-hybrid experiments, Dong-Hoon Jeong, Jongmin Nam, and Jinwon Lee for DNA sequencing, and Chahm An for critical reading of the manuscript.
Footnotes
This work was supported in part by grants from the Korean Science and Engineering Foundation (no. 96–0401–06–01–3) and from the Korea Research Foundation (no. 1998–019–D00090).
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang J, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, van Dijken A, van Went JL, Dons HJM, van Tunen AJ. A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell. 1995;7:1569–1582. doi: 10.1105/tpc.7.10.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, Weiss D, van Tunen AJ. Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. Plant J. 1994;5:33–44. doi: 10.1046/j.1365-313x.1994.5010033.x. [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Buchner P, Boutin J-P. A MADS box transcription factor of the AP1/AGL9 subfamily is also expressed in the seed coat of pea (Pisum sativum) during development. Plant Mol Biol. 1998;38:1253–1255. doi: 10.1023/a:1006008212200. [DOI] [PubMed] [Google Scholar]
- Chung Y-Y, Kim S-R, Finkel D, Yanofsky MF, An G. Early flowering and reduced apical dominance result from ectopic expression of a rice MADS box gene. Plant Mol Biol. 1994;26:657–665. doi: 10.1007/BF00013751. [DOI] [PubMed] [Google Scholar]
- Chung Y-Y, Kim S-R, Kang H-G, Noh Y-S, Park MC, Finkel D, An G. Characterization of two rice MADS box genes homologous to GLOBOSA. Plant Sci. 1995;109:45–56. [Google Scholar]
- Davies B, Egea-Cortines M, de Andrade Silva E, Saedler H, Sommer H. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 1996;15:4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Fan H-Y, Hu Y, Tudor M, Ma H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 1997;12:999–1010. doi: 10.1046/j.1365-313x.1997.12050999.x. [DOI] [PubMed] [Google Scholar]
- Flanagan CA, Hu Y, Ma H. Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant J. 1996;10:343–353. doi: 10.1046/j.1365-313x.1996.10020343.x. [DOI] [PubMed] [Google Scholar]
- Flanagan CA, Ma H. Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol Biol. 1994;26:581–595. doi: 10.1007/BF00013745. [DOI] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Greco R, Stagi L, Colombo L, Angenent GC, Sari-Gorla M, Pe ME. MADS box genes expressed in developing inflorescences of rice and sorghum. Mol Gen Genet. 1997;253:615–623. doi: 10.1007/s004380050364. [DOI] [PubMed] [Google Scholar]
- Heard J, Dunn K. Symbiotic induction of a MADS-box gene during development of alfalfa root nodules. Proc Natl Acad Sci USA. 1995;92:5273–5277. doi: 10.1073/pnas.92.12.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF. Floral determination and expression of floral regulatory genes in Arabidopsis. Development. 1997;124:3845–3853. doi: 10.1242/dev.124.19.3845. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Huang H, Mizukami Y, Hu Y, Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993;21:4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tudor M, Weiss CA, Hu Y, Ma H. The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol Biol. 1995;28:549–567. doi: 10.1007/BF00020401. [DOI] [PubMed] [Google Scholar]
- Huijser PW, Klein J, Lonnig W-E, Meijer H, Saedler H, Sommer H. Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 1992;11:1239–1249. doi: 10.1002/j.1460-2075.1992.tb05168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brochman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics, A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kang H-G, An G. Isolation and characterization of a rice MADS box gene belonging to the AGL2 gene family. Mol Cells. 1997;7:45–51. [PubMed] [Google Scholar]
- Kang H-G, Jang S, Chung J-E, Cho Y-G, An G. Characterization of two rice MADS box genes that control flowering time. Mol Cell. 1997;7:559–566. [PubMed] [Google Scholar]
- Kang H-G, Noh Y-S, Chung Y-Y, Costa MA, An K, An G. Phenotypic alterations of petal and sepal by ectopic expression of a rice MADS box gene in tobacco. Plant Mol Biol. 1995;29:1–10. doi: 10.1007/BF00019114. [DOI] [PubMed] [Google Scholar]
- Lu Z-X, Wu M, Loh C-S, Yeong CY, Goh C-J. Nucleotide sequence of a flower-specific MADS box cDNA clone from orchid. Plant Mol Biol. 1993;23:901–904. doi: 10.1007/BF00021545. [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991;5:484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Lutziger I, Kuhlemeier C. A ubiquitously expressed MADS-box gene from Nicotiana tabacum. Plant Mol Biol. 1994;25:319–321. doi: 10.1007/BF00023247. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF. The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell. 1995;7:1763–1771. doi: 10.1105/tpc.7.11.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle B, Bouhidel K, Irish VF. Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 1996;10:1812–1821. doi: 10.1101/gad.10.14.1812. [DOI] [PubMed] [Google Scholar]
- Mena M, Mandel MA, Lerner DR, Yanofsky MF, Schmidt RJ. A characterization of the MADS-box gene family in maize. Plant J. 1995;8:845–854. doi: 10.1046/j.1365-313x.1995.8060845.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Mizukami Y, Huang H, Tudor M, Hu Y, Ma H. Functional domains of the floral regulator AGAMOUS: characterization of the DNA binding domain and analysis of dominant negative mutations. Plant Cell. 1996;8:831–845. doi: 10.1105/tpc.8.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag K, Salamini F, Thompson RD. ZEMa, a member of a novel group of MADS box genes, is alternatively spliced in maize endosperm. Nucleic Acids Res. 1995;23:2168–2177. doi: 10.1093/nar/23.12.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon Y-H, Jung J-Y, Kang H-G, An G. Identification of a rice APETALA3 homolog by yeast two-hybrid screening. Plant Mol Biol. 1999;40:167–177. doi: 10.1023/a:1026429922616. [DOI] [PubMed] [Google Scholar]
- Perry SE, Nichols KW, Fernandez DE. The MADS domain protein AGL15 localizes to the nucleus during early stages of seed development. Plant Cell. 1996;8:1977–1989. doi: 10.1105/tpc.8.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer Z, Lifschitz E. The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic gene from Antirrhinum and Arabidopsis. Plant J. 1991;1:255–266. [PubMed] [Google Scholar]
- Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell. 1994;6:163–173. doi: 10.1105/tpc.6.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics. 1995;140:345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA. 1996a;93:4793–4798. doi: 10.1073/pnas.93.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996b;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell. 1995;7:1259–1269. doi: 10.1105/tpc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Savidge B, Rounsley SD, Yanofsky MF. Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell. 1995;7:721–733. doi: 10.1105/tpc.7.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Velt B, Mandel MA, Mena M, Hake S, Yanofsky MF. Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell. 1993;5:729–737. doi: 10.1105/tpc.5.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M, Wessler S, Federoff N. Molecular identification and isolation of the waxy locus in maize. Cell. 1983;35:225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Running MP, Meyerowitz EM. Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell. 1995;7:1249–1258. doi: 10.1105/tpc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H, Beltran J-P, Huijser PW, Pape H, Lonnig W-E, Saedler H, Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Kim JT, Saedler H. Classification and phylogeny of the MADS-box gene families in the morphological evolution of eukaryotes. J Mol Evol. 1996;43:484–516. doi: 10.1007/BF02337521. [DOI] [PubMed] [Google Scholar]
- Theissen G, Strater T, Fisher A, Saedler H. Structural characterization, chromosomal localization and phylogenetic evaluation of two pairs of AGAMOUS-like MADS-box genes from maize. Gene. 1995;156:155–166. doi: 10.1016/0378-1119(95)00020-7. [DOI] [PubMed] [Google Scholar]
- Tilly JJ, Allen DW, Jack T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development. 1998;125:1647–1657. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lonnig W-E, Saedler H, Sommer H, Schwarz-Sommer Z. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]