SUMMARY
The glycogen synthase kinase-3 (GSK3) family kinases are central cellular regulators highly conserved in all eukaryotes. In Arabidopsis, the GSK3-like kinase BIN2 phosphorylates a range of proteins to control broad developmental processes, and BIN2 is degraded through unknown mechanism upon receptor kinase-mediated brassinosteroid (BR) signaling. Here we identify KIB1 as an F-box E3 ubiquitin ligase that promotes the degradation of BIN2 while blocking its substrate access. Loss-of-function mutations of KIB1 and its homologs abolished BR-induced BIN2 degradation and caused severe BR-insensitive phenotypes. KIB1 directly interacted with BIN2 in a BR-dependent manner and promoted BIN2 ubiquitination in vitro. Expression of an F-box-truncated KIB1 caused BIN2 accumulation but dephosphorylation of its substrate BZR1 and activation of BR responses, because KIB1 blocked BIN2 binding to BZR1. Our study demonstrates that KIB1 plays an essential role in BR signaling by inhibiting BIN2 through dual mechanisms of blocking substrate access and promoting degradation.
eTOC Blurb
Zhu et al. identifies KIB1 as an F-box E3 ubiquitin ligase required for brassinosteroid signal transduction, and further reveals a dual mechanism of KIB1-mediating GSK3 inactivation: blocking substrate access by the same binding event that leads subsequently to its ubiquitination and proteasomal degradation.
INTRODUCTION
As sessile organisms, plants need exceptionally high levels of developmental plasticity, which requires sophisticated signaling mechanisms. Plant growth is controlled by integrating complex environmental signals with endogenous hormones (Chaiwanon et al., 2016). Among these, brassinosteroid (BR) is a major growth-promoting hormone that integrates information about internal nutrient/energy status with other hormonal and environmental signals (Chaiwanon et al., 2016; Zhang et al., 2016). BR activation of the BRI1 receptor kinase initiates a phosphorylation cascade that leads to inhibition of the GSK3-like kinase BIN2 (Kim and Wang, 2010; Zhu et al., 2013). BIN2 phosphorylates and regulates other kinases and a number of key transcription factors, thereby playing a central role in BR regulation of gene expression and specific cellular and developmental processes (Li and Nam, 2002; Youn and Kim, 2015).
Among the BIN2 regulated transcription factors, BRISSINOZOLE-RESISTANT 1 (BZR1) family of transcription factors play a major role in BR promotion of plant growth, as dominant mutations that cause constitutive activation of BZR1 or its homolog BZR2 (also named BES1) suppress most of the growth defects of the BR deficient and insensitive mutants (Wang et al., 2002; Yin et al., 2002). BZR1 binds to thousands of BR-responsive promoters (Sun et al., 2010), and regulates their expression by interacting with other transcription regulators (Oh et al., 2014a; Oh et al., 2014b). BIN2 phosphorylation inhibits BZR1 nuclear localization and DNA-binding activity (Gampala et al., 2007; Vert and Chory, 2006), whereas upon BR inactivation of BIN2, protein phosphatase 2A (PP2A) dephosphorylates BZR1 to enable BR responsive gene expression (Sun et al., 2010; Tang et al., 2011; Wang et al., 2012). The accumulation of BZR1 and activity of BIN2 are also regulated by sugar signaling mediated by the Target of Rapamycin (TOR) pathway, which allows nutrient/energy signaling to modulate steroid-dependent growth (Xiong et al., 2016; Zhang et al., 2016). In addition to BZR1, BIN2 also phosphorylates the PHYTOCHROME INTERACTING FACTOR 4 (PIF4), AUXIN RESPONSE FACTOR 2 (ARF2), and CESTA transcription factors to regulate cell elongation (Bernardo-Garcia et al., 2014; Khan et al., 2014; Vert et al., 2008), the AUXIN RESPONSE FACTOR 7 (ARF7) to modulate lateral root development (Cho et al., 2014), and the ENHANCER OF GLABRA 3 (EGL3) and TRANSPARENT TESTA GLABRA1 (TTG1) factors to regulate root hair formation (Cheng et al., 2014). Further, BIN2 phosphorylates components of a mitogen activated protein (MAP) kinase pathway to mediate BR regulation of stomata development (Gudesblat et al., 2012; Khan et al., 2013; Kim et al., 2012), and phosphorylates the SnRK2 kinases to modulate plant responses to the stress hormone abscisic acid (Cai et al., 2014). These observations indicate that BIN2 plays a central role in cellular and developmental regulation, and therefore regulation of BIN2 level is crucial for growth and stress adaptation in plants.
Despite BIN2’s central function and extensive studies of the BR signaling pathway, the mechanisms by which BR restrains cellular BIN2 level are not fully understood. Upon BR binding and activation (Hothorn et al., 2011; She et al., 2011), BRI1 phosphorylates the BR-SIGNALING KINASE1 (BSK1) and the CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1) kinase (Kim et al., 2011; Tang et al., 2008), which in turn phosphorylate and activate the BRI1 SUPPRESSOR 1 (BSU1) phosphatase (Kim et al., 2011; Kim et al., 2009). BSU1 dephosphorylates BIN2 at Tyr200 to reduce its kinase activity (Kim et al., 2009). There is evidence that BR signaling involves proteasome-mediated BIN2 degradation (Peng et al., 2008), and interestingly glucocorticoid also promotes GSK3 degradation in mammalian cells (Dominguez and Green, 2000; Failor et al., 2007). However, the protein factors and molecular mechanisms for regulating GSK3 degradation have remained unknown.
In this study, we report the identification of an E3 ubiquitin ligase for GSK3s as an essential component of the BR signaling pathway in Arabidopsis. Genetic analysis demonstrates that KIB1 is essential for BR signaling, and specifically for BR-induced BIN2 degradation. Biochemical analyses revealed that KIB1 not only mediates BIN2 ubiquitination but also blocks its substrate access. Our study demonstrates that BR signaling induces KIB1-BIN2 interaction, which directly prevents BIN2 interacting with its substrate and subsequently causes BIN2 ubiquitination and degradation.
RESULTS
Loss of function kib1 mutation suppresses bzr1-1D and increases BZR1 phosphorylation
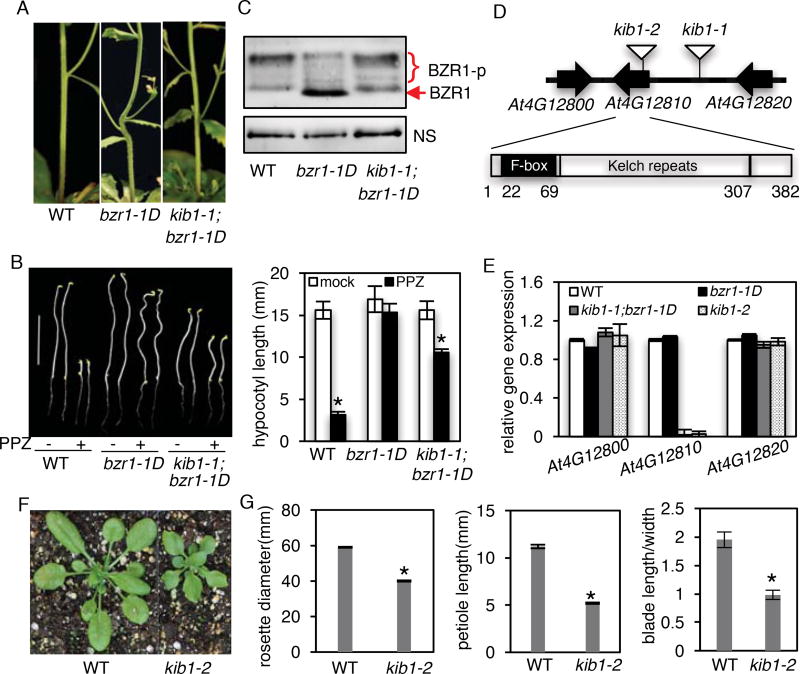
The dominant gain-of-function bzr1-1D mutation (containing P234L substitution) enhances BZR1’s interaction with PP2A and hence its dephosphorylation (Tang et al., 2011), causing constitutive BR-response phenotypes, including BR-independent hypocotyl elongation and stem kink due to organ-fusion (Gendron et al., 2012; Wang et al., 2002) (Figure 1A). Genetic mutations that enhance BIN2 kinase activity or increase BIN2 protein level are expected to increase phosphorylation of bzr1-1D and suppress the bzr1-1D phenotypes. We thus screened for T-DNA-mutagenized suppressors of bzr1-1D (Fan et al., 2012), and identified a mutant that showed no kink at the stem-branch junctions (Figure 1A). We named the mutation kink suppressed in bzr1-1D (kib1-1). The kib1-1 mutation also suppressed the other phenotypes of bzr1-1D, including the resistance to BR biosynthesis inhibitor propiconazole (PPZ) (Figure 1B) and late-flowering phenotypes (Figure S1A and S1B). The suppression of multiple phenotypes of bzr1-1D suggests that kib1-1 mutation affects the protein level or activity of bzr1-1D. Indeed, immunoblot analysis showed that the kib1-1;bzr1-1D double mutant accumulated more phosphorylated BZR1 and less unphosphorylated BZR1 protein than the bzr1-1D single mutant (Figure 1C). The results suggest that the kib1-1 mutation increased BZR1 phosphorylation.
Figure 1. The kib1 mutation suppresses bzr1-D phenotype by increasing the phosphorylation of BZR1.
(A) The kib1 mutation suppresses the stem bending phenotype of bzr1-1D. Stems of 8-week-old plants of wild-type (WT), bzr1-1D, and kib1-1;bzr1-1D are shown.
(B) kib1-1;bzr1-1D is less resistant to propiconazole (PPZ) than bzr1-1D. Left panel: seedlings of the indicated genotypes grown on the medium with or without 2 µM PPZ in the dark for 5 days. Scale bar is 10 mm; right panel: the quantitation of hypocotyl lengths in the indicated genotypes.
(C) Immunoblot analysis of phosphorylated (BZR1-p) and de-phosphorylated BZR1 in seven-day-old light-grown seedlings of the indicated genotypes using an anti-BZR1 antibody. Nonspecific bands (NS) show protein loadings.
(D) Diagrams of the T-DNA insertion sites in kib1-1 and kib1–2 mutants (upper panel) and a gene model of KIB1 (lower panel).
(E) qRT-PCR analysis of the expression level of KIB1 and its neighbor genes in the indicated genotypes. The data are shown as means of three biological repeats ± SD.
(F) Plant morphology of WT and kib1–2 grown in the soil for 3 weeks.
(G) Quantification of the rosette diameters, petiole lengths, ration between blade length and blade width of WT and kib1–2 plants. The rosette diameters are measured from three-week-old plants grown in a long-day condition. Petiole lengths, blade lengths and blade widths were determined for the 6th leaves.
The data in (B) and (G) are shown as means ± SD, n=15. The asterisks indicate significant differences by Student’s t test (*p < 0.05) when compared to wild-type. Also see Figure S1, S2 and S3.
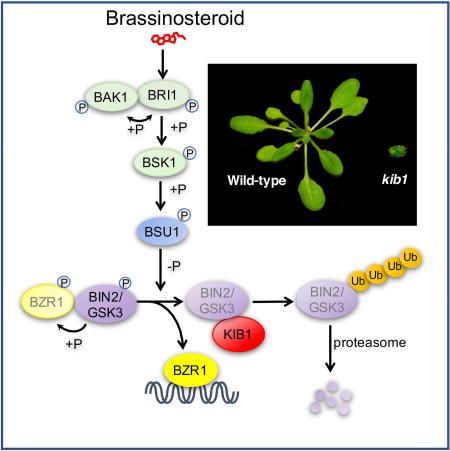
We next investigated the nature of the kib1-1 mutation. Using thermal asymmetric interlaced PCR(Liu et al., 1995), we identified a T-DNA flanking sequence in the promoter of AT4G12810 in the kib1-1;bzr1-1D mutant (Figure 1D). Quantitative RT-PCR analysis showed that the expression level of AT4G12810 was greatly reduced in kib1-1;bzr1-1D, whereas the expression levels of its neighboring genes AT4G12800 and AT4G12820 were not affected (Figure 1E). To confirm that loss of AT4G12810 expression is responsible for the kib1-1 phenotype, we obtained another T-DNA insertion mutant kib1–2 (SAIL_1230_F06), in which a T-DNA was inserted in an exon of AT4G12810 (Figure 1D). Quantitative RT-PCR analysis detected no expression of AT4G12810 in kib1–2 (Figure 1E). We crossed kib1–2 to the bzr1-1D mutant, and kib1–2;bzr1-1D double mutant had straight stems (Figure S1C), indicating that, similar to kib1-1, kib1–2 suppresses the stem kink phenotype of bzr1-1D. Homozygous kib1–2 mutants showed shorter hypocotyls compared to wild-type seedlings grown on the medium (Figure S1D and S1E). Expressing KIB1-YFP under the KIB1 native promoter in kib1–2 mutant restored the mutant hypocotyl phenotype (Figure S1D and S1E). kib1–2 displayed rosette leaves with shorter petioles and wider but shorter leaf blades when grown in the soil (Figure 1F and 1G), thus resembling weak BR deficient mutants. These results confirm that loss of AT4G12810 function is responsible for the bzr1-1D-suppression phenotypes of kib1-1 and kib1–2, and thus we named AT4G12810 gene KIB1.
KIB1 encodes a 382-amino acid protein that is a member of the F-box family protein, and KIB1 has three closest homologous genes that we named KIB2, KIB3 and KIB4 (Figure S2A). KIB1 and its homologs contain a putative F-box domain at the N-terminal region and three kelch repeats at C-terminal region (Figure 1D, Figure S2B). The expression pattern of KIB1 was examined using KIB1 promoter fused with GUS reporter gene. GUS stain results showed KIB1 expression broadly in various tissues, including young seedlings, leaves, stems, flower buds and flowers (Figure S3A). A KIB1-YFP (yellow fluorescent protein) fusion protein expressed from KIB1 promoter (KIB1::KIB1–YFP) in transgenic plants showed a strong signal in the nucleolus and weaker signal in the cytoplasm (Figure S3B).
KIB1 is an essential component for BR signaling
To further analyze the function of KIB1, we transformed wild-type Arabidopsis with T-DNA constructs that overexpressed KIB1 fused to the Myc tag (KIB1-Myc) or the yellow fluorescence protein (KIB1-YFP) from the constitutive 35S promoter. With both constructs, we obtained more than 30 transgenic plants that had larger rosette leaves with longer petioles compared to wild type (Figure 2A, Figure S4A and S4B), and three transgenic plants that were extreme dwarfs resembling the BR receptor mutant bri1 (Figure 2A, 2B and 2C, Figure S4C). Immunoblotting analysis detected KIB1-Myc overexpression in the larger plants but not in the dwarf plants (Figure 2A). Quantitative RT-PCR analysis showed that the dwarf plants had greatly reduced expression of KIB1 and its three closest homologous genes KIB2, KIB3 and KIB4 (Figure 2B). These results show that the larger plants were KIB1-overexpression (KIB1-ox) lines, and the dwarf plants were KIB1-cosuppression (kib1-cs) lines. The KIB1-ox seedlings displayed longer hypocotyls compared to wild-type (Figure S4D and S4E), while the hypocotyl lengths of kib1-cs seedlings were severely short (Figure S4D and S4E). The KIB1-ox lines were insensitive to the BR biosynthesis inhibitor PPZ, similar to bzr1-1D and BIN2 loss-of function triple mutant bin2-3bil1bil3 (Figure 2D, Figure S4F and S4G). Overexpression of KIB1-YFP in the bzr1-1D mutant background severely enhanced the stem-bending phenotype of bzr1-1D (Figure 2E). These results demonstrate that overexpression of KIB1 activates the BR signal transduction pathway.
Figure 2. KIB1 plays a positive role in the BR signal transduction pathway.
(A) Plant morphology and KIB1-Myc protein levels of three-week-old WT and two independent KIB1-Myc transgenic Arabidopsis (#9 and #16). Tubulin protein levels show the protein loadings.
(B) qRT-PCR analysis of the expression level of KIB1 and its homologs in WT and KIB1-Myc (#16) co-suppression line (kib1-cs). The data are shown as means of three biological repeats ± SD.
(C) Four-week-old plants of KIB1-Myc co-suppression lines (#1, #16 and #25), WT and bri1–116 mutant. Inset shows close-up view.
(D) KIB1 overexpression line is insensitive to PPZ. WT and KIB1-ox transgenic plants were grown on the medium with (+) or without (−) 2 µM PPZ in the dark for 7 days. Scale bar is 10 mm.
(E) A transgenic plant overexpressing KIB1 in the bzr1-1D mutant shows severe stem bending phenotype.
Also see Figure S4.
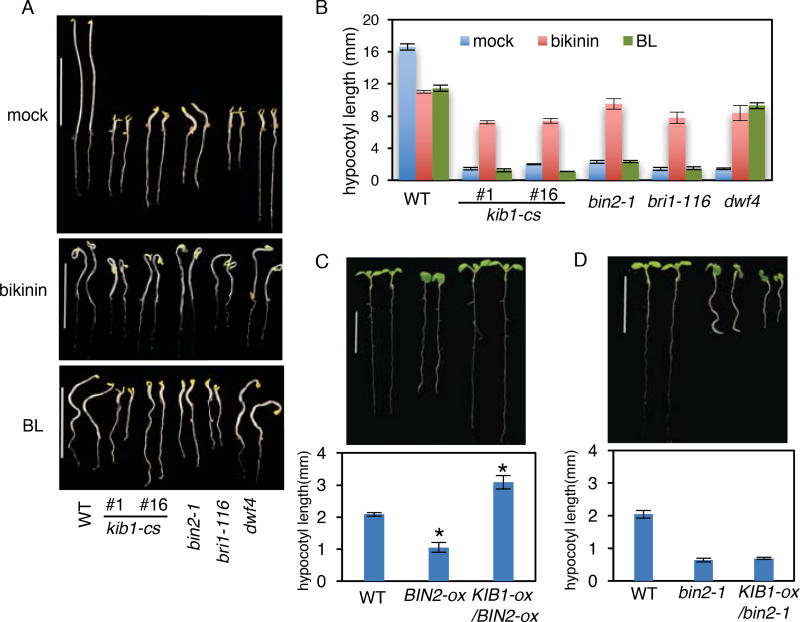
To determine the step of BR signal transduction that KIB1 acts on, we compared the kib1-cs plants with BR biosynthetic and signaling mutants for their morphology and responses to BR and bikinin, a chemical inhibitor of GSK3 kinases (De Rybel et al., 2009). The kib1-cs plants, grown under the light or in the dark, had phenotypes nearly identical to the BR insensitive mutants bri1–116 and bin2-1 or the BR-deficient mutant dwf4 (Figure 2C, 3A and Figure S4C–S4G). The dwf4 mutant, which is deficient in BR synthesize, was rescued by brassinolide (BL, the most active form of BR), whereas bri1–116 and bin2-1, in which BIN2 is not inactivated by BL (Kim et al., 2009), were only rescued by bikinin but not by BL (Figure 3A and 3B). Similar to bri1–116 and bin2-1, the kib1-cs seedlings were insensitive to BL, but were rescued by bikinin (Figure 3A and 3B), suggesting that kib1-cs is defective in BR inactivation of BIN2. The results demonstrate that KIB1, together with its close homologs, is required for BR signal transduction upstream of BIN2. Consistent with this notion, KIB1 overexpression suppressed the dwarf phenotypes of transgenic plants overexpressing wild-type BIN2(Kim et al., 2009) (Figure 3C), but did not suppress the strong dwarf phenotypes of the homozygous dominant bin2-1 mutant (Figure 3D, Figure S5), possibly because KIB1 inhibits wild-type BIN2 but not the mutant bin2-1, which contains E263K substitution (Peng et al., 2008).
Figure 3. KIB1 acts upstream of BIN2 in the BR signaling pathway.
(A–B) The kib1-cs mutant is rescued by bikinin treatment, but not BL treatment. Morphology (A) and hypocotyl length measurement (B) of the indicated genotype seedlings grown on the medium containing bikinin (30 µM), BL (1 µ M) or mock solution in the dark for 5 days.
(C–D) Overexpression of KIB1 partially rescues the BIN2-ox transgenic Arabidopsis, but not the bin2-1 mutant. Morphology (upper panel) and hypocotyl length measurement (lower panel) of seven-day-old light-grown seedlings of the indicated genotypes.
The data in (B), (C) and (D) are shown as means ± SD, n=15. The asterisks indicate significant differences by Student’s t test (*p < 0.05) when compared to wild-type.
Also see Figure S5.
KIB1 promotes BR-induced BIN2 degradation
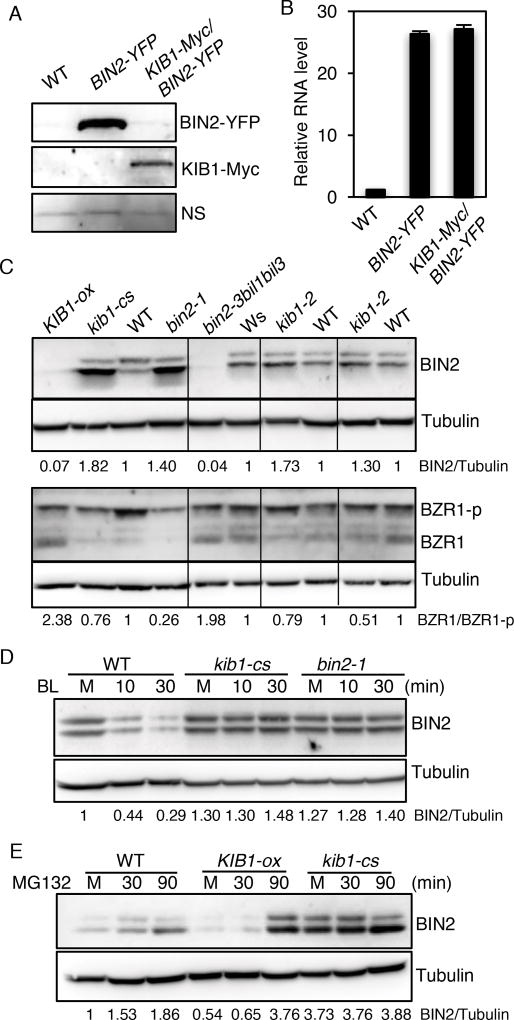
KIB1 contains a putative F-box motif (Figure 1D, Figure S2B), a structure found in many E3 ubiquitin ligases (Cardozo and Pagano, 2004; Gagne et al., 2002). To test whether KIB1 functions as an E3 ubiquitin ligase that promotes BIN2 degradation, we analyzed the effects of overexpression and loss of function of KIB1 on the level of BIN2 protein. Immunoblot analysis showed that overexpression of KIB1-Myc decreased the BIN2-YFP protein level dramatically in transgenic Arabidopsis (Figure 4A), whereas quantitative RT-PCR analysis showed no obvious effect on BIN2-YFP RNA level (Figure 4B). Furthermore, we found that the endogenous BIN2 protein level was decreased in the KIB1-ox line (Figure 4C), which correlated with increased levels of unphosphorylated form of BZR1 (Figure 4C), similar to that observed in the bin2-3bil1bil2 triple mutant (Figure 4C). In contrast, the kib1-cs plants showed an increased level of BIN2 and enhanced BZR1 phosphorylation, similar to the gain-of-function mutant bin2-1 (Figure 4C). These results demonstrate that KIB1 promotes BIN2 degradation, leading to subsequent accumulation of unphosphorylated BZR1.
Figure 4. KIB1 is required for BR-induced BIN2 degradation.
(A) Overexpression of KIB1 decreased BIN2 protein level. Immunoblot shows BIN2-YPF and KIB1-Myc protein levels in the seven-day-old BIN2-YFP and KIB1-Myc/BIN2-YFP transgenic Arabidopsis using the anti-GFP or anti-Myc antibody. Nonspecific bands (NS) show the protein loadings.
(B) Quantitative RT-PCR analysis of the expression level of BIN2 in the plants from (A). The gene expression levels were normalized to PP2A and presented as values relative to that of WT. The data are shown as means of three biological repeats ± SD.
(C) Immunoblot analyses of BIN2, phosphorylated (BZR1-p) and de-phosphorylated BZR1(BZR1) levels of ten-day-old seedlings of the indicated genetic backgrounds using the anti-BIN2, anti-BZR1 or anti-Tubulin anti-body. The numbers below the blots indicate the relative ratios of the signal intensity between BIN2 and Tubulin bands (BIN2/Tubulin) or the ratios of the signal intensity between de-phosphorylated BZR1 and phosphorylated BZR1(BZR1/BZR1-p) bands. The ratios were normalized to WT control for each set of experiments.
(D–E) KIB1 is required for BR-induced BIN2 degradation by the proteasome. (D), ten-day-old seedlings of WT, kib1-cs and bin2-1 grown on the medium with 2 µM PPZ under the light were treated with mock solution for 30 minutes (M) or with BL (100 nM) for 10 and 30 minutes. (E), seven-day-old seedlings of WT, KIB1-ox and kib1-cs grown under the light were treated with mock solution (M, 30 min) or with 10 µM MG132 (30 or 90 min). The immunoblots were probed with the anti-BIN2 antibody or anti-Tubulin antibody. The numbers below the blots indicate the relative ratios of the signal intensity between BIN2 and tubulin bands (BIN2/Tubulin).
Previous studies showed that BR induces BIN2 degradation by the proteasome (Peng et al., 2008). We therefore further tested whether KIB1 is required for BR-induced BIN2 degradation. Consistent with the previous reports (Peng et al., 2008), we observed BR-induced degradation of BIN2 in wild-type plants (Figure 4D). However, BR had very little effect on BIN2 level in the kib1-cs plants, similar to the bin2-1 dominant mutant (Figure 4D). By contrast, the KIB1-ox plants had a lower level of BIN2 than wild-type plants (Figure 4E). Treatment with the proteasome inhibitor MG132 increased BIN2 levels in the wilt-type and KIB1-ox plants but not in kib1-cs (Figure 4E). These results indicate that KIB1 is required for BR-induced BIN2 degradation by the proteasome.
KIB1 is an E3 ubiquitin ligase for GSK3s
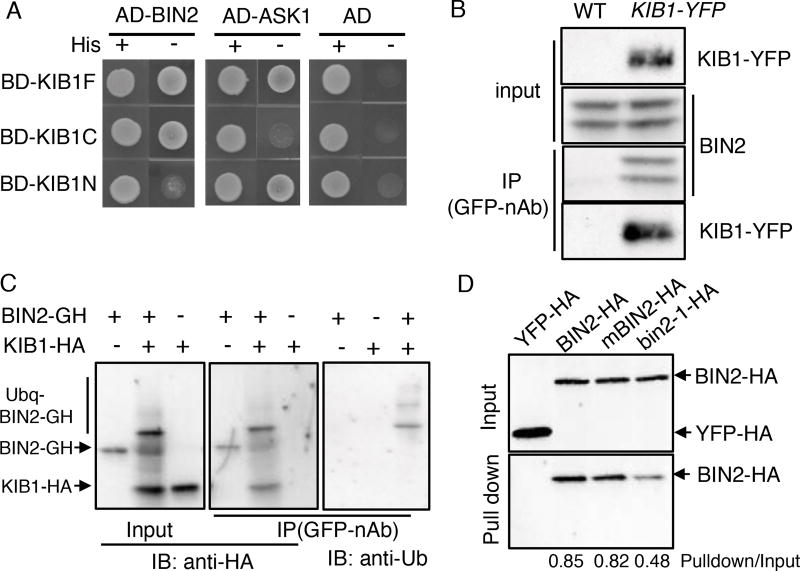
We next tested whether KIB1 interacts with BIN2. Yeast two-hybrid assays showed that BIN2 interacted with both the full-length KIB1 (KIB1F) and its C-terminal region containing three kelch repeats (amino acid 71–382, KIB1C), but not the N-terminal F-box domain (amino acid 1–70, KIB1N) (Figure 5A, Figure S6A and S6B). In contrast, the N-terminal F-box domain of KIB1 interacted with the Arabidopsis SKP1-like1 (ASK1)(Gagne et al., 2002)(Figure. 5A), which is consistent with the F-box motif being known to interact with the SKP1 core component to form the SKP1-Cullin-F-box (SCF) E3 ubiquitin ligase complex (Cardozo and Pagano, 2004). In addition, KIB1C interacted with several other Arabidopsis GSK3 family members (AtASK11, 12, 13, 22 and 23) (Figure S6A), and these Arabidopsis GSK3s have been shown to interact with BZR1 in yeast two-hybrid assays and to play redundant roles with BIN2 in BR signaling (Kim et al., 2009). Similarly, KIB2 also interacted with BIN2 in yeast (Figure S6C). In co-immunoprecipitation assays, BIN2 was detected after GFP-nAb immunoprecipitation using extracts from KIB1::KIB1-YFP transgenic plants but not from wild-type plants (Figure 5B), indicating that KIB1 associates with BIN2 in vivo. To determine if KIB1 is a E3 ligase for BIN2, we performed in vitro ubiquitination assays, and the results showed that KIB1 indeed can mediate BIN2 ubiquitination (Figure 5C).
Figure 5. KIB1 interacts with BIN2 and promotes BIN2 ubiquitination.
(A) Yeast-two-hybrid assay shows the interaction between the fragments of KIB1 and BIN2 or ASK1. KIB1F, full-length KIB1; KIB1N, amino acid 1–70; KIBC, amino acid 71–382.
(B) Co-immunoprecipitation assay shows KIB1 is associated with BIN2 in vivo. Total proteins extracted from WT or KIB1::KIB1-YFP transgenic plants were immunoprecipitated with the GFP-nAb, and then probed with the anti-GFP or anti-BIN2 antibody.
(C) In vitro ubiquitination assays show KIB1 promotes BIN2 ubiquitination. BIN2-GFP-HA (BIN2-GH) was incubated with or without KIB1-HA for the ubiquitination assay reactions. The BIN2-GFP-HA protein was immunoprecipitated using the GFP-nAb, and then analyzed by immunoblots using the anti-HA and anti-Ub antibody separately.
(D) In vitro pull-down assays show KIB1 binds to BIN2. BIN2, BIN2Y200F (mBIN2) and bin2-1 fused with HA tags were pulled down by KIB1-YFP-HA immobilized on the GFP-nAb magnetic agarose beads and immunoblotted using an anti-HA antibody. YFP-HA was used as a negative control. The numbers below the blots indicate the ratio of the relative density between pull-down and input signals (Pulldown/Input).
Also see Figure S6.
Previous studies showed that phosphorylation of Tyr200 is required for full BIN2 kinase activity (Kim et al., 2009), and that the bin2-1 mutation (E263K) blocks BR-induced BIN2 degradation (Peng et al., 2008). We thus tested whether Y200F and E263K substitutions affect BIN2 interaction with KIB1. KIB1-Myc pulled down BIN2 in vitro (Figure 5D). While the Y200F mutation showed no obvious effect on KIB1-BIN2 interaction, the bin2-1 mutation reduced the KIB1-BIN2 interaction (Figure 5D). Similarly, in yeast, KIB1 and its homolog KIB2 interacted strongly with the wild-type BIN2, but only weakly with bin2-1 (Figure S6B and S6C). The reduced KIB1 interaction with bin2-1 is consistent with the dwarf phenotype of KIB1-ox/bin2-1 (Figure 3D, Figure S5), which suggests that KIB1 is unable to inactivate bin2-1. Together the results provide genetic evidence supporting the requirement of interaction with KIB1 for BR-induced BIN2 degradation.
KIB1 binding blocks BIN2 access to its substrate BZR1
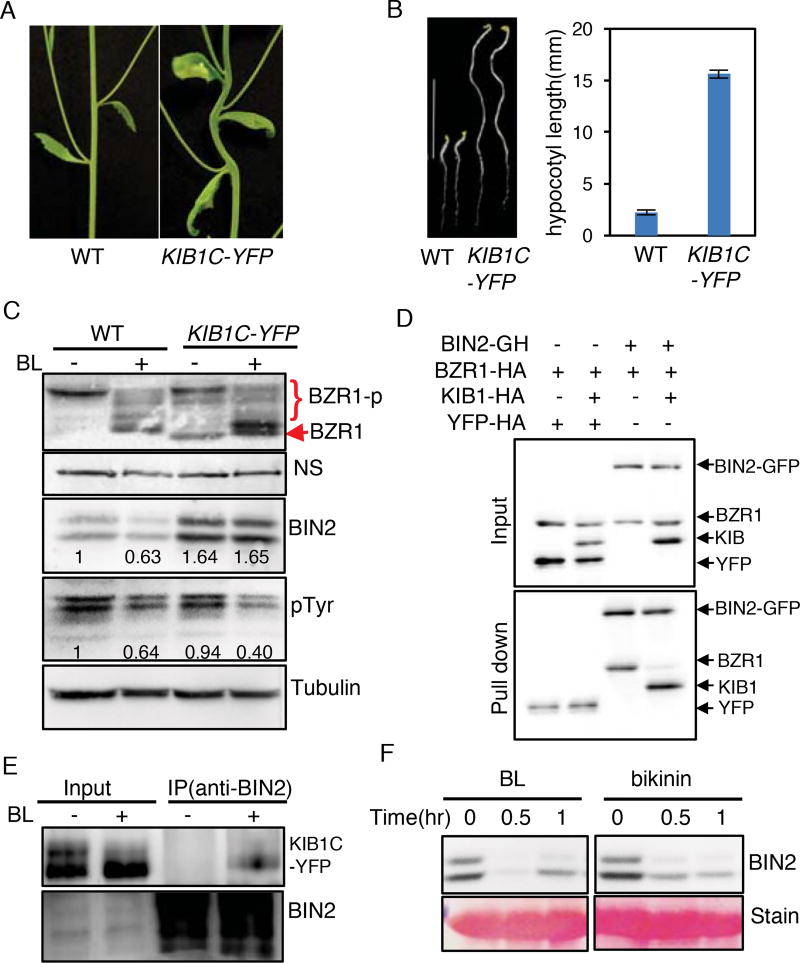
Deletion of F-box sequences has been shown to create a dominant-negative form of E3 ligase that interacts with substrate but is unable to mediate its ubiquitination, thus causing substrate accumulation (Yaron et al., 1998). We overexpressed an F-box-truncated KIB1 (KIB1C) fused with YFP in Arabidopsis (KIB1C-YFP). Surprisingly, the transgenic seedlings showed constitutive BR-response phenotypes such as stem kinks (Figure 6A), insensitivity to BR biosynthesis inhibitor PPZ (Figure 6B) and large and curly leaves (Figure S7A), similar to bzr1-1D and plants overexpressing full-length KIB1 (Figure 1 and 2). The KIB1C-YFP plants also displayed reduced lateral root development (Figure S7B and S7C), similar to the phenotypes observed in the bin2/gsk3 loss-of-function mutants (Cho et al., 2014). Consistent with the growth phenotype, the KIB1C-YFP plants accumulated increased levels of dephosphorylated BZR1 compared to wild type both before and after BR treatment (Figure 6C). However, in contrast to the KIB1-ox plants in which BIN2 is degraded, these KIB1C-YFP plants accumulated a slightly increased level of BIN2 without BL treatment, and did not show obvious BIN2 degradation upon BL treatment in contrast to the wild type plants (Figure 6C). Thus, it appears that the F-box-truncated KIB1 caused BIN2 accumulation through a dominant negative effect but somehow decreased the phosphorylation of its substrate BZR1 (Figure 6C). One possibility is that KIB1 binding blocks BIN2 access to its substrate BZR1. Indeed, in vitro pull-down assays showed that BIN2 bound to BZR1 in the absence of KIB1, but bound to only KIB1 not BZR1 when both KIB1 and BZR1 were present (Figure 6D), indicating that KIB1 prevents BIN2 interaction with BZR1.
Figure 6. KIB1-BIN2 interaction blocks BIN2 accessing its substrate BZR1.
(A) KIB1C-YFP transgenic Arabidopsis grown under the light for 5 weeks shows stem bending.
(B) KIB1C-YFP transgenic Arabidopsis shows constitutive BR-response phenotypes. Left panel: seven-day-old seedlings of WT and KIB1C-YFP grown in the dark on the medium with or without 2 µM PPZ. Scale bar is 10 mm; right panel: the quantitation of hypocotyl lengths in the indicated genotypes. The data are shown as means ± SD, n=15.
(C) Immunoblot analyses of BZR1, BIN2 and phospho-GSK3 using the anti-BZR1, anti-BIN2, anti-pTyr279/Tyr216 (pTyr) or anti-Tubulin antibody from ten-day-old seedlings of WT and KIB1C-YFP grown on the medium containing 2 µM PPZ (−) or 100 nM BL (+). Nonspecific bands (NS) and Tubulin protein level show the equal loadings. The numbers inside the images indicate the relative ratios of the signal intensity between BIN2 or pTyr and tubulin bands. The ratio of WT was set to 1.
(D) In vitro pull down assay shows KIB1 blocks BIN2 binding to BZR1. The mixtures of the indicated proteins BZR1-HA, KIB1-HA or YFP-HA were pulled down by BIN2-GFP-HA immobilized on the GFP-nAb magnetic agarose beads, and analyzed by immunoblots using an anti-HA antibody. YFP-HA was used as a negative control.
(E) Co-immunoprecipitation assay shows BR enhances BIN2-KIB1C interaction. Protein extracts from seven-day-old light-grown KIBC-YFP seedlings grown on the medium containing 2 µM PPZ treated with 100 nM BL or mock solution for 10 minutes were immunoprecipitated using an anti-BIN2 antibody, and analyzed by immunoblots using an anti-GFP antibody.
(F) Inhibiting kinase activity of BIN2 by bikinin causes BIN2 degradation. Wild-type seedlings were grown on the medium containing 2 µM PPZ under the light for 10 days, and then treated with 100 nM BL or 30 µM bikinin for the indicated time. Proteins were immunoblotted using an anti-BIN2 antibody. Ponceau S staining bands show the protein loadings.
Also see Figure S7.
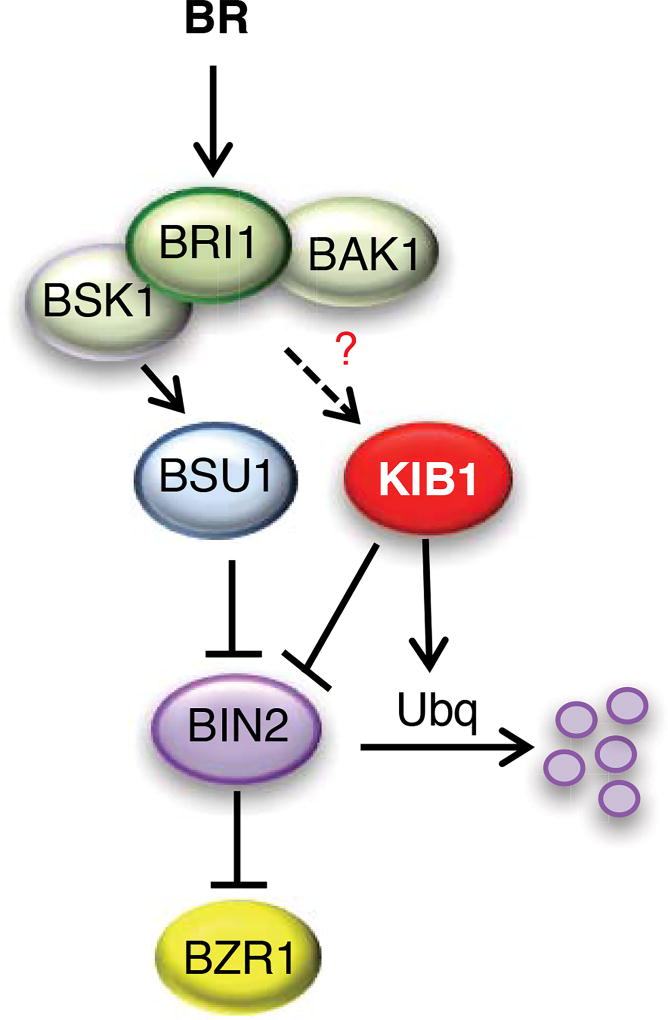
Consistent with the previous reports that BR signaling causes BIN2 degradation and BSU1-mediated dephosphorylation of BIN2 at pTyr200 (Kim et al., 2009; Peng et al., 2008), BL treatment of wild type plants decreased the levels of BIN2 detected by both anti-BIN2 and anti-pTyr200-GSK3 antibodies (anti-pTyr, corresponding to Tyr279 and Tyr216 of human GSK3α/β) (Figure 6C). In contrast, BL treatment of the KIB1C-YFP plants had no obvious effect on BIN2 protein level, but caused obvious dephosphorylation of BIN2 at pTyr200 (Figure 6C), suggesting that KIB1C-YFP abolished BIN2 degradation but did not interfere with BR-induced dephosphorylation of BIN2 at pTyr200. BL treatment also caused a mobility shift of KIB1C-YFP in immunoblot that can be mimicked by in vitro phosphatase treatment (Figure 6E and Figure S7D and S7E), indicating that BR induces KIB1 dephosphorylation. Further, co-immunoprecipitation assays showed that BR enhanced BIN2-KIB1C interaction (Figure 6E). These results suggest that the BR-induced dephosphorylation of pTyr200 is likely an upstream event that leads to BIN2 inactivation, KIB1 dephosphorylation, and BIN2-KIB1 interaction. Indeed, similar to BR, bikinin also caused BIN2 degradation (Figure 6F), suggesting that inhibiting BIN2 kinase activity is sufficient for triggering its degradation. Together our results support a model that upon BR signaling BSU1-mediated dephosphorylation and partial inactivation of BIN2 leads to recruitment of KIB1, which blocks BIN2 interaction with its substrates and further promotes BIN2 ubiquitination and degradation, ensuring robust signaling outputs (Figure 7).
Figure 7. A model for KIB1 mediating BR-induced BIN2 inactivation and degradation.
BR activates the BRI1-BAK1 receptor kinase complex, triggering sequential phosphorylation of the BSK1 kinase and BSU1 phosphatase; BSU1 dephosphorylates BIN2 to reduce its kinase activity, which, through an unknown mechanism, leads to BIN2 binding by KIB1. KIB1 binding both excludes BIN2 association with its substrate BZR1 and facilitates the ubiquitination, leading to degradation, of BIN2. It remains unclear whether upstream BR signaling also modifies KIB1, such as by dephosphorylation, to enhance its interaction with BIN2.
DISCUSSION
Our study demonstrates that the KIB1 family F-box proteins are essential components of the BR signaling pathway, and they positively regulate BR signaling by both promoting ubiquitination/degradation and blocking substrate phosphorylation of GSK3 kinases. Previous studies demonstrated that GSK3 inactivation by BR signaling involves both BSU1-mediated dephosphorylation and proteasome-mediated BIN2 degradation (Kim et al., 2009; Peng et al., 2008). However, the functional importance and underlying mechanism of BIN2 degradation have remained unclear. Here our genetic evidence indicates that BR promotion of plant growth requires BIN2 degradation and inactivation mediated by KIB1. Further, we find that KIB1 binding with BIN2 not only mediates BR-induced ubiquitination and degradation of BIN2 but also directly blocks BIN2 access to its substrate BZR1. As such, our study reveals an essential step of BR signal transduction and uncovers dual mechanisms of GSK3 inhibition by its E3 ligase KIB1 through blocking substrate docking while promoting degradation. Together, the studies illustrate a model for tight regulation of GSK3/BIN2 by BR-signaling through a combination of BSU1-mediated dephosphorylation and KIB1-mediated inhibition and degradation (Figure 7).
The functions of KIB1 are supported by strong genetic and biochemical evidence. Both in vitro and in vivo assays demonstrate that KIB1 directly interacts with BIN2 to promote BIN2 ubiquitination and subsequent degradation by the proteasome. Further, the interaction is enhanced by BR treatment in vivo, consistent with the role of KIB1 in BR promoting BIN2 degradation. The severe BR-insensitive dwarf phenotypes, BR-insensitive accumulation of BIN2, and increased phosphorylation of BZR1, observed in the kib1-cs plants clearly demonstrate the essential role for the KIB1 family proteins in BIN2 inactivation/degradation and BR signaling. The stronger phenotypes of kib1-cs, in which four homologous genes are suppressed, than the kib1 single mutants, indicate that KIB1 family members redundantly fulfill an essential role in mediating BR signaling. Consistent with this notion, yeast two-hybrid assays showed that both KIB1 and its homolog KIB2 interact with BIN2, while KIB1 interacts with all six GSK3s that interact with BZR1 (Kim et al., 2009) (Figure S6). These results indicate that KIB1 and its homologs play similar roles in regulating GSK3s. Analysis of additional single and higher-order knockout mutants will be required to determine the degree of overlap between roles played by different KIB family members.
Regulation of cellular GSK3 level is likely to influence broadly GSK3-regulated processes. Further, KIB1 binding may block BIN2 access to specific substrates or non-selectively block all its substrates. The KIB1C-YFP plants displayed reduced lateral root development (Figure S7B and S7C), which is regulated through BIN2 phosphorylation of the auxin response factor 7 (ARF7) (Cho et al., 2014), suggesting that KIB1 also blocks BIN2 phosphorylation of ARF7. Therefore, KIB1 and its homologs may redundantly function as master regulators of GSK3s, impacting broadly GSK3-regulated processes in plants. Whether different KIB1 family members play distinct roles in different signaling and developmental context requires further genetic and molecular dissection.
Overexpression of an F-box-truncated KIB1, which binds BIN2 but lacks E3 ubiquitin ligase activity, caused BIN2 accumulation as expected but surprisingly resulted in phenotypes that are consistent with BIN2 inactivation, including decreased levels of BZR1 phosphorylation. A previous study has shown that BIN2 phosphorylation of its substrate BZR1 is mediated by a direct kinase-substrate docking mechanisms (Peng et al., 2010). The in vitro binding competition assays showed that KIB1-BIN2 binding prevents BIN2-BZR1 interaction, revealing KIB1 inhibits BIN2 by blocking substrate docking. Our study thus uncovers a mechanism of dual actions of E3 ubiquitin ligase: blocking substrate access while promoting ubiquitination/degradation. Such a dual mechanism of inactivation presumably provides rapid and efficient inhibition of BIN2, although the relative contributions of the “blocking” and “ubiquitination” activities in vivo remain unknown. Similar dual mechanisms may have been evolved broadly for other E3 ligase-substrate pairs but have been overlooked in research.
We have previously shown that BR-induced phospho-relay from BRI1, BSK1/CDG1, to BSU1 triggers BSU1 dephosphorylation of BIN2 at pTyr200 (Kim et al., 2011; Kim et al., 2009). However, mutating Tyr200 to Phe (Y200F) reduced but did not fully abolish BIN2 kinase activity based on in vitro kinase assays and the phenotypes of overexpression transgenic plants (Kim et al., 2009). This is consistent with additional mechanisms, such as KIB1-mediated inhibition and degradation, being required for effective BIN2 inactivation. The observations that BR induces pTyr200 dephosphorylation and BIN2-KIB1C interaction in the KIB1C-YFP plants suggest that BSU1-mediated dephosphorylation and inhibition of BIN2 kinase promotes its interaction with KIB1. Consistent with this notion is the observation that direct inactivation of BIN2 by its chemical inhibitor bikinin causes BIN2 degradation (Figure 6F). Since bikinin inhibits BIN2 as an ATP competitor (De Rybel et al., 2009), the results suggest that inhibiting BIN2 kinase activity is sufficient for triggering its degradation. While the Y200F mutation of BIN2 showed no obvious effect on KIB1 binding in vitro and in yeast (Figure 5D and Figure S6B, S6C), there is evidence that BIN2 is dephosphorylated at multiple residues upon BR treatment (Kim et al., 2009), and thus dephosphorylation at additional auto-phosphorylated residues other than Tyr200 may affect KIB1 binding. Alternatively, BIN2 phosphorylation of KIB1 might inhibit its E3 ubiquitin ligase function and thus stabilize BIN2. Our results support a model that upon upstream BR signaling, an initial partial and/or reversible inactivation of BIN2 by BSU1-mediated dephosphorylation leads to further inhibition and eventual degradation mediated by KIB1, which are required for effective de-repression of BIN2 substrates such as BZR1.
It is interesting to note that glucocorticoid induces GSK3 ubiquitination/degradation in mammalian cells, and this involves phosphorylation of GSK3 by Glucocorticoid-Induced Kinase (Sgk) and Akt kinases (Failor et al., 2007). In addition, a GSK3-binding protein (GBP) was shown to inhibit substrate phosphorylation and promote GSK3 depletion in Xenopus embryo patterning (Dominguez and Green, 2000; Farr et al., 2000). However, no E3 ubiquitin ligase that promotes GSK3 degradation in animals has been reported. While GSK3β has been shown to be ubiquitinated by the E3 ligase TNF Receptor Associated Factor 6 (TRAF6) in the Toll-like receptor 3 (TLR3)-mediated pro-inflammatory signaling pathway, such ubiquitination promotes assembly of functional signaling complex but has no effect on GSK3 degradation (Ko et al., 2015). In the Wnt signaling pathway, GSK3β is mono-ubiquitinated by ubiquitin E3 ligase β-TrCP, which suppresses recruitment of their substrate β-catenin without affecting GSK3β degradation (Gao et al., 2014). While the mechanisms for regulating GSK3 degradation remain obscure in animal systems, the KIB1-mediated regulation of BIN2 in the BR signaling pathway provides an example of how cell surface signaling regulates GSK3’s stability as well as activity in plants.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author, Zhi-Yong Wang (zywang24stanford.edu).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
All the Arabidopsis thaliana plants used in this study were in Col-0 ecotype background, except the bin2-1 bil2 bil3 mutant which is in Ws ecotype background. The plants were grown in greenhouses with a 16-h light/8-h dark cycle at 22–24°C for general growth and seed harvesting. For seedlings grown on the medium in Petri dishes, the sterilized seeds were grown on 1/2 Murashige and Skoog (MS) medium containing 1% sucrose and supplemented with 0.7% phytoagar. Plates were placed in a growth chamber under the constant light condition at 22°C.
METHOD DETAIL
T-DNA Insertion Mutagenesis and bzr1-1D Suppressor Screen
The bzr1-1D mutant plants were transformed with the pSKI015 plasmid (Weigel et al., 2000) using the floral dipping method (Clough and Bent, 1998). T2 seeds were harvested from each group of 20 T1 plants that showed typical bzr1-1D phenotypes (i. e. stem-bending phenotype). About 200 T2 seeds of each pool (average 10 T2 seeds from each T1 line) were grown in the soil at 22–24°C in a green house, and the plants were screened for the lack of the stem-bending phenotype of bzr1-1D. The kib1-1 mutant was isolated after screening T2 seeds from about 2000 T1 lines. The T-DNA flanking sequence was recovered by thermal asymmetric interlaced PCR (TAIL-PCR)(Liu et al., 1995).
Plasmids Construction and Plant Transformation
To generate KIB1-Myc, the full-length KIB1 coding sequence was cloned into the gateway compatible p1390-4Myc-His vector. To generate KIB1-YFP and KIB1C-YFP transgenic lines, the full-length KIB1 coding sequence or F-box-truncated KIB1(KIB1C) sequence with start codon was cloned into the gateway compatible pX-YFP vector. To generate KIB1::KIB1-YFP, 1.5 kb of KIB1 promoter sequence plus the full-length KIB1 genomic sequence was cloned into the gateway compatible pEGTW vector. To generate KIB1-GUS, 1.5 kb of KIB1 promoter sequence was cloned into the gateway compatible pMDC164 vector. Gene specific primers are listed in Key Resource Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-Myc(9B11) | Cell Signaling Technology | #2276;RRID:AB_2148465 |
|
| ||
| Mouse monoclonal anti-GFP (JL-8) | Clontech | 632381; RRID: AB_2313808 |
|
| ||
| Rat monoclonal anti-Tubulin (YL1/2) | abcam | Ab6;RRID:AB_343284 |
|
| ||
| Rat monoclonal anti-HA | Roche | 11867423001; RRID:AB_390919 |
|
| ||
| Mouse monoclonal anti-Ubiquitin (P4D1) | Cell signaling | 3936; RRID:AB_331292 |
|
| ||
| Rabbit polyclonal anti-BZR1 | This paper | homemade |
|
| ||
| Rabbit polyclonal anti-BIN2 | This paper | Homemade |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| Brassinolide (BL) | Wako | 635-00811 |
|
| ||
| Propiconazole (PPZ) | Control Solutions | N/A |
|
| ||
| Bikinin | ChemBridge | 5122035 |
|
| ||
| lambda protein phosphatase | NEB | P0753 |
|
| ||
| MG132 | Sigma-Aldrich | C2211 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| GFP-nAb Magnetic Agarose Kit | Allele Biotech | ABP-NABGFPXK20 |
|
| ||
| Auto-ubiquitinylation kit | Enzo life science | BML-UW0970-0001 |
|
| ||
| Protein A/G plus agarose | Pierce | 20423 |
|
| ||
| TnT quick couple transcription/translation system | Promega | L1170 |
|
| ||
| Spectrum plant total RNA kit | Sigma-Aldrich | STRN250 |
|
| ||
| M-MLV reverse transcriptase | ThermoFisher | 28025013 |
|
| ||
| Experimental Models: Organisms/Strains | ||
|
| ||
| Arabidopsis: Col-0, Ws | ABRC | |
|
| ||
| Arabidopsis: bzr1-1D | Wang et al., 2002 | N/A |
|
| ||
| Arabidopsis bin2-1, bin2-3bil1bil3 |
Li and Nam.,
2002; Yan et al., 2009 |
N/A |
|
| ||
| Arabidopsis: dwf4 | Choe et al., 1998 | N/A |
|
| ||
| Arabidopsis: bri1–116 | Wang et al., 2001 | N/A |
|
| ||
| Arabidopsis: BIN2-ox | Kim et al., 2010 | N/A |
|
| ||
| Arabidopsis: KIB1-ox, kib1-cs, kib1-1,
KIB1C-ox,KIB1-YFP/bzr1-1D, KIB1-ox/BIN2-ox, KIB1-ox/bin2-1, KIB1::KIB1-YFP, KIB1-GUS |
This paper | N/A |
|
| ||
| Arabidopsis: kib1–2 | ABRC | SAIL_1230_F06 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers for KIB1qRT-PCR | ||
| F: ATTCGATACTTGCCGTGGAC | This paper | Customer order |
| R: GGATGAGCCATGGAGTTGTT | ||
|
| ||
| Primers for KIB2qRT-PCR | ||
| F: TCCAGCTGCTTGAGAGTTGA | This paper | Customer order |
| R: ACAGGCTGCTTTGAATCGTC | ||
|
| ||
| Primers for KIB3qRT-PCR | ||
| F: GGTTTTTCCCCACTTTTGGT | This paper | Customer order |
| R: CGTCACTGGTTTTGTCCCTT | ||
|
| ||
| Primers for KIB4qRT-PCR | ||
| F: GGAGTCGTTGTTTGGCAAGT | This paper | Customer order |
| R: AACCACAGTCACCGGTTCTC | ||
|
| ||
| Primers for AT3G03726qRT-PCR | ||
| F: CAACGATTTCCACGTTTTGA | This paper | Customer order |
| R: CCACAAAACCGCAGTGTCTA | ||
|
| ||
| Primers for AT4G12800qRT-PCR | ||
| F: TCCGACAAGACAACATTCCA | This paper | Customer order |
| R: GGTACGGTATCCAGGGAGGT | ||
|
| ||
| Primers for BIN2qRT-PCR | ||
| F:TCTGCTGGTTGTGTTCTTGC | This paper | Customer order |
| R: AAGATCTTGTGCCAGGGATG | ||
|
| ||
| Primers for PP2AqRT-PCR | ||
| F: TATCGGATGACGATTCTTCGTGCAG | This paper | Customer order |
| R: GCTTGGTCGACTATCGGAATGAGAG | ||
|
| ||
| Primers for KIB1 promoter cloning | ||
| F:CACCTCTCCATCTAACACACTAAGCACA | This paper | Customer order |
| R: GGCCAAGTAAAGACCTAGAAACAT | ||
|
| ||
| Primers for full length KIB1 cloning | ||
| F: CACCATGACACATAAGAAGCAGAAGAAG | This paper | Customer order |
| R: TTAAAGCAACCATTTTGAAGTA | ||
|
| ||
| Primer for KIB1N cloning | ||
| R: ACTATGTAGGTTTTGTTTTCATGAGG | This paper | Customer order |
|
| ||
| Primer for KIB1C cloning | ||
| F:CACCATGAAGAGACCTCGGGTTTGATTTAGTTA | This paper | Customer order |
|
| ||
| Primers for full length KIB2 cloning | ||
| F: CACCATGGCGCCTCTCAACTCTCA | This paper | Customer order |
| R: AAGCAACCATTTTCCACCAA | ||
|
| ||
| Primers for full length ASK1 cloning | ||
| F: CACCATGTCTGCGAAGAAGATTGTGTTGAA | This paper | Customer order |
| R: TTCAAAAGCCCATTGGTTCTCTCTGCG | ||
|
| ||
| Recombinant DNA | ||
|
| ||
| KIB1::KIB1-YFP | This paper | N/A |
|
| ||
| 35S::KIB1-YFP | This paper | N/A |
|
| ||
| 35S::KIB1-Mys | This paper | N/A |
|
| ||
| 35s::KIB1C-YFP | This paper | N/A |
|
| ||
| KIB1-GUS | This paper | N/A |
|
| ||
| BD-KIB1F, BD-KIB1C, BD-KIB1N, AD-KIB1, AD-KIB2, AD-ASK1 | This paper | N/A |
|
| ||
| AD-BIN2, AD-bin2-1, AD-mBIN2, BD-AtASK11, BD-AtASK12, BD-AtASK13, BD-AtASK22, BD-AtASK23 | Kim et al., 2009 | N/A |
|
| ||
| AD-BIN2-GFP | This paper | N/A |
|
| ||
| AD-KIB1-YFP | This paper | N/A |
|
| ||
| pXDGATcy86 | Ding et al., 2007 | N/A |
|
| ||
| pGADT7-GW | Lu et al., 2010 | N/A |
|
| ||
| pMDC164 | Curtis et al., 2003 | N/A |
|
| ||
| Software and Algorithms | ||
|
| ||
| Clustal Omega | Sievers et al., 2011 | http://www.clustal.org/omega/ RRID:SCR_001591 |
|
| ||
| ImageJ | Schneider et al., 2012 | https://imagej.net/ImageJ RRID:SCR_003070 |
|
| ||
| Fiji | Schindelin et al., 2012 | https://imagej.net/Fiji RRID:SCR_002285 |
|
| ||
| PHYLIP | Felsenstein, 1989 | http://evolution.genetics .washington.edu/phylip/ RRID:SCR_006244 |
The resulting vectors were transformed into Arabidopsis plants by floral dip (Clough and Bent, 1998). KIB1-Myc, KIB1-YFP, KIB1C-YFP and KIB1-GUS constructs were transformed to the wild-type Col-0. KIB1::KIB1-YFP construct was transformed to wild-type Col-0 or kib1–2 mutant.
KIB1-YFP/BIN2-Myc and KIB1-Myc/BIN2-YFP transgenic lines were generated by crossing KIB1-YFP with BIN2-Myc transgenic Arabidopsis or crossing KIB1-Myc with BIN2-YFP transgenic Arabidopsis.
Total RNA Extraction and Quantitative RT–PCR Analysis
Total RNA was extracted from seedlings or specific tissues using the Spectrum Plant Total RNA kit. M-MLV reverse transcriptase was used to synthesize cDNA from RNA. Quantitative real-time PCR (qRT-PCR) was performed using LightCycler® 480 (Roche) and the Bioline SYBR green master mix. Gene expression levels were normalized to that of PP2A and are shown relative to the expression levels in wild type. Gene specific primers are listed in Key Resource Table.
Co-immunoprecipitation (co-IP) Assays
Plant proteins were extracted using IP buffer (50 mM Tris-Cl pH7.5, 1mM EDTA, 75mM NaCl, 0.1% Triton X-100, 5% Glycerol, 1mM PMSF, 1× Protease Inhibitor). After centrifugation at 20,000g for 10 minutes at 4°C, the supernatant was incubated for 1 hour with the GFP-nAb magnetic agarose beads. The beads were then washed for three times with IP buffer. The eluted samples were analyzed by immunoblots using the indicated antibodies.
Yeast-two-hybrid Assay
The various fragments of KIB1 cDNA, full-length KIB2 and ASK1 were subcloned into the gateway compatible pGCADT7 or pXDGATcy86 vector. The constructs were co-transformed into yeast AH109 cells. Yeast clones were grown on the synthetic dropout medium with histidine (+His) or without histidine (−His) containing 3-amino-1, 2, 4-triazole (3-AT, 1mM).
Protein pull-down Assays
KIB1-YFP-HA, BIN2-HA, mBIN2-HA, bin2-1-HA or YFP-HA proteins were synthesized by TNT T7 quick coupled in vitro transcription/translation system with the vectors AD-KIB1-YFP, AD-BIN2, AD-bin2-1, and AD-mBIN2. For the pull-down assays using KIB1-YFP-HA as bait, KIB1-YFP-HA proteins were pre-incubated with GFP-nAb magnetic agarose for 1 hour. After removing unbound proteins, BIN2-HA, mBIN2-HA, bin2-1-HA or YFP-HA proteins were incubated with KIB1-YFP-HA immobilized on GFP-nAb magnetic agarose beads for 1 hour in binding buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl) at 4°C, and then the beads were washed by the washing buffer (10 mM Tris-HCl pH 7.5, 500 mM NaCl) for three times. The pulled-down proteins were analyzed by immunoblot using an anti-HA antibody.
In Vitro Ubiquitination Analysis
BIN2-GFP-HA and KIB1-HA were synthesized using TnT T7 in vitro transcription/translation system with the vectors AD-BIN2-GFP and AD-KIB1. BIN2-GFP-HA with or without KIB1-HA was incubated in the Ub E3 ligase buffer (50 mM Tris pH 7.6), 0.6 mM DTT, 2 mM Mg-ATP, 1.5 ng/µl E1, 10 ng/µl E2 (UbcH5a), 1 µg/µl ubiquitin, 1 µM histidine-purified recombinant Cullin-1, ASK1 and Rbx1 for 1 hour at 30°C. The ubiquitination products were mixed with the same volume of 2× sample loading buffer were heated at 95 °C for 5min and separated by SDS-PAGE and immunoblotted using the anti-HA or anti-Ub antibody. The BIN2-GFP-HA protein was immunoprecipitated from the ubiquitination reaction mixtures using the GFP-nAb magnetic agarose beads. After 1 hour of incubation at 4°C, the beads were washed for three times with wash buffer (10mM Tris-HCl pH 7.5, 500mM NaCl). The immunoprecipitates were eluted using 2× sample loading buffer. Samples were heated at 95 °C for 5min and separated by SDS-PAGE and analyzed using anti-HA or anti-Ub antibody.
QUANTIFICATION AND STATISTICAL ANALYSIS
Root and hypocotyl lengths were measured from images using ImageJ software. Band intensity quantification of protein signals from western blot was performed using the ImageJ.
Statistical significance of root and hypocotyl lengths and gene expression were examined by Student’s t test (*p < 0.05).
Supplementary Material
HIGHLIGHTS.
KIB1 is an essential positive regulator in brassionosteroid signaling.
KIB1 mediates BR-induced ubiquitination and degradation of GSK3 kinase BIN2.
KIB1 binding to BIN2 prevents BIN2-substrate interaction and promotes BIN2 degradation.
Acknowledgments
We thank Dr. Matthew Scott for helpful comments on the manuscript. This study was supported by a grant from NIH (R01GM066258) to Z-Y. W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J-Y.Z., D.C., and Z-Y.W. conceived the projects. J-Y.Z., Y.L., D.C., H.Y., E.O., Y.B., and S.Z. performed the experiments. J-Y.Z. and Z-Y.W. wrote the manuscript.
References
- Bernardo-Garcia S, de Lucas M, Martinez C, Espinosa-Ruiz A, Daviere JM, Prat S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes & development. 2014;28:1681–1694. doi: 10.1101/gad.243675.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Liu J, Wang H, Yang C, Chen Y, Li Y, Pan S, Dong R, Tang G, Barajas-Lopez Jde D, et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:9651–9656. doi: 10.1073/pnas.1316717111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nature Reviews Molecular Cell Biology. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Wang W, Zhu JY, Oh E, Wang ZY. Information Integration and Communication in Plant Growth Regulation. Cell. 2016;164:1257–1268. doi: 10.1016/j.cell.2016.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhu W, Chen Y, Ito S, Asami T, Wang X. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife. 2014;3:e02525. doi: 10.7554/eLife.02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ, et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nature Cell Biology. 2014;16:66–76. doi: 10.1038/ncb2893. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Vert G, Rozhon W, Mayerhofer J, Peelman F, Coutuer S, Denayer T, Jansen L, Nguyen L, et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chemistry & Biology. 2009;16:594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez I, Green JB. Dorsal downregulation of GSK3beta by a non-Wnt-like mechanism is an early molecular consequence of cortical rotation in early Xenopus embryos. Development. 2000;127:861–868. doi: 10.1242/dev.127.4.861. [DOI] [PubMed] [Google Scholar]
- Failor KL, Desyatnikov Y, Finger LA, Firestone GL. Glucocorticoid-induced degradation of glycogen synthase kinase-3 protein is triggered by serum- and glucocorticoid-induced protein kinase and Akt signaling and controls beta-catenin dynamics and tight junction formation in mammary epithelial tumor cells. Molecular Endocrinology. 2007;21:2403–2415. doi: 10.1210/me.2007-0143. [DOI] [PubMed] [Google Scholar]
- Fan XY, Sun Y, Cao DM, Bai MY, Luo XM, Yang HJ, Wei CQ, Zhu SW, Chong K, Wang ZY. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol Plant. 2012;5:591–600. doi: 10.1093/mp/sss041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr GH, 3rd, Ferkey DM, Yost C, Pierce SB, Weaver C, Kimelman D. Interaction among GSK-3, GBP, axin, and APC in Xenopus axis specification. J Cell Biol. 2000;148:691–702. doi: 10.1083/jcb.148.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Chen G, Romero G, Moschos S, Xu X, Hu J. Induction of Gsk3beta-beta-TrCP interaction is required for late phase stabilization of beta-catenin in canonical Wnt signaling. J Biol Chem. 2014;289:7099–7108. doi: 10.1074/jbc.M113.532606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Liu JS, Fan M, Bai MY, Wenkel S, Springer PS, Barton MK, Wang ZY. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci USA. 2012;109:21152–21157. doi: 10.1073/pnas.1210799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat GE, Schneider-Pizon J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, Boeren S, Zhiponova M, de Vries S, Jonak C, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nature Cell Biology. 2012;14:548–554. doi: 10.1038/ncb2471. [DOI] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature. 2011;474:467–471. doi: 10.1038/nature10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Rozhon W, Bigeard J, Pflieger D, Husar S, Pitzschke A, Teige M, Jonak C, Hirt H, Poppenberger B. Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J Biol Chem. 2013;288:7519–7527. doi: 10.1074/jbc.M112.384453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Rozhon W, Unterholzner SJ, Chen T, Eremina M, Wurzinger B, Bachmair A, Teige M, Sieberer T, Isono E, et al. Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nature Communications. 2014;5:4687. doi: 10.1038/ncomms5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Guan S, Burlingame AL, Wang Z-Y. The CDG1 Kinase Mediates Brassinosteroid Signal Transduction from BRI1 Receptor Kinase to BSU1 Phosphatase and GSK3-like Kinase BIN2. Molecular Cell. 2011;43:561–571. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Guan S, Sun Y, Deng Z, Tang W, Shang J, Sun Y, Burlingame AL, Wang Z-Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Bio. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482:419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- Ko R, Park JH, Ha H, Choi Y, Lee SY. Glycogen synthase kinase 3beta ubiquitination by TRAF6 regulates TLR3-mediated pro-inflammatory cytokine production. Nature Communications. 2015;6:6765. doi: 10.1038/ncomms7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife. 2014a;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Ryu H, Hwang I, Wang ZY. TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nature Communications. 2014b;5:4140. doi: 10.1038/ncomms5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P, Yan Z, Zhu Y, Li J. Regulation of the Arabidopsis GSK3-like Kinase BRASSINOSTEROID-INSENSITIVE 2 through Proteasome-Mediated Protein Degradation. Mol Plant. 2008;1:338–346. doi: 10.1093/mp/ssn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P, Zhao J, Zhu Y, Asami T, Li J. A direct docking mechanism for a plant GSK3-like kinase to phosphorylate its substrates. J Biol Chem. 2010;285:24646–24653. doi: 10.1074/jbc.M110.142547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Yang M, Wang ZY, Chai J. Structural insight into brassinosteroid perception by BRI1. Nature. 2011;474:472–476. doi: 10.1038/nature10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan X-Y, Cao D-M, He K, Tang W, Zhu J-Y, He J-X, Bai M-Y, Zhu S, Oh E, et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim T-W, Zhou H-W, Deng Z, Gampala SS, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Bio. 2011;13:124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Zhang R, Meng Z, Deng K, Que Y, Zhuo F, Feng L, Guo S, Datla R, Ren M. Brassinosteriod Insensitive 2 (BIN2) acts as a downstream effector of the Target of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. The New Phytologist. 2016;213:233–249. doi: 10.1111/nph.14118. [DOI] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Youn JH, Kim TW. Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol Plant. 2015;8:552–565. doi: 10.1016/j.molp.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhu JY, Roh J, Marchive C, Kim SK, Meyer C, Sun Y, Wang W, Wang ZY. TOR Signaling Promotes Accumulation of BZR1 to Balance Growth with Carbon Availability in Arabidopsis. Current Biology. 2016;26:1854–1860. doi: 10.1016/j.cub.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Sae-Seaw J, Wang ZY. Brassinosteroid signalling. Development. 2013;140:1615–1620. doi: 10.1242/dev.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.