Abstract
When investigating Mendelian disease using exome or genome sequencing, distinguishing disease-causing genetic variants from the multitude of candidate variants is a complex, multidimensional task. Many prioritization tools and online interpretation resources exist, and professional organizations have offered clinical guidelines for review and return of prioritization results. In this Review, we describe the strengths and weaknesses of widely used computational approaches, explain their roles in the diagnostic and discovery process and discuss how they can inform (and misinform) expert reviewers. We place variant prioritization in the wider context of gene prioritization, burden testing and genotype–phenotype association, and we discuss opportunities and challenges introduced by whole-genome sequencing.
Despite the power of DNA sequencing for genetic discovery1 and the many computational tools and online resources available, fewer than 50% of Mendelian disorders are resolved after sequencing affected families2. The reasons are manifold: there are new disease phenotypes, new genes for known diseases and many unknown disease-causing variants still await discovery. To understand the scope of the challenge, consider that the genetic causes underlying more than 3,000 known Mendelian disorders remain unknown2. Variant prioritization is central to every Mendelian disease discovery and diagnosis effort. Put simply, it is the process of determining which variants identified in the course of genetic testing, whole-exome sequencing (WES) and whole-genome sequencing (WGS) are most likely to damage gene function and underlie the disease phenotype.
With the advent of WES and WGS, variant prioritization has grown ever more critical to discovery and diagnosis. It has also grown more complicated because of the sheer number of variants. Every individual’s genome contains millions of variants, many of which will never have been seen before3,4. The identification of the one or two variants responsible for a patient’s Mendelian disease is a classic ‘needle in the haystack’ problem5. Whereas early tools addressed this complexity by using the scant means at their disposal for prioritization — phylogenetic conservation and protein structures — the latest tools combine these data with other information such as population allele frequencies, functional genomics data and other genome annotations. Some even use the predictions of other tools to inform their own. The scope of prioritization has also been widened. Some tools have expanded their scope beyond single nucleotide variants (SNVs) to prioritize more complex forms of variation such as insertions, deletions and structural variants, and others provide the means to prioritize variants in non-coding regions. These approaches lead to greater accuracy, but they can also complicate interpretation; therefore, understanding how these approaches work is essential for those engaged in genome-based diagnostic activities.
Although variant prioritization is central to Mendelian disease discovery and diagnosis, it is only part of a bigger picture that includes gene prioritization. Gene prioritization tools use information such as variant allele frequencies, genotype frequencies, inheritance models, family histories and patient phenotypes to identify and prioritize likely damaged genes associated with a phenotype, as opposed to simply identifying potentially damaging variants. Although this may seem a subtle distinction, it is, in fact, a fundamental difference from the perspective of the underlying algorithms. Many gene prioritization tools use an approach called burden testing — a key concept that is increasingly central to WES- and WGS-driven discovery and diagnostic efforts6–11.
The manner in which the results of variant and gene prioritization tools are delivered to users is also changing. The last several years have seen a proliferation of decision support frameworks for variant interpretation12–15. These interactive, often web-based, platforms are a big step forwards from simple command line-based analyses. Within these interactive environments, variant and gene prioritization scores are only one component of a dynamic, multifactorial approach to discovery and diagnosis that uses population-scale variation resources, such as the Exome Aggregation Consortium (ExAC)4, the genome Aggregation Database (gnomAD; see Further information), the 1000 Genomes Project3, disease genotype–phenotype associations such as Online Mendelian Inheritance in Man (OMIM)16 and ClinVar17, and workflows based on guidelines established by the American College of Medical Genetics and the Association for Clinical Genetic Science of the United Kingdom15.
Despite all of these advances, attributing disease causation to prioritized variants remains an inexact process. No phrase better summarizes the current state of affairs than ‘variant of uncertain significance’ (VUS). The key to understanding this phrase is to grasp that a variant that damages a gene is not necessarily damaging to an individual’s health (BOX 1). Understanding the cascading steps underlying variant and gene prioritization, how prioritization scores are combined with adjunct information such as phenotype and family history, and how they are judged to be medically significant are the subjects of this Review.
Box 1. Damaging does not mean pathogenic.
Variant prioritization tools such as SIFT (Sorts Intolerant From Tolerant) and PolyPhen2 (polymorphism phenotyping version 2) use the terms ‘damaging’ and ‘tolerated’ to describe whether a variant is predicted to affect protein function or be functionally neutral, respectively. We emphasize that the term damaging should never be logically equated with causal for a disease phenotype, because a variant that damages a gene is not necessary damaging to an individual’s health.
The term ‘pathogenic’ has become widely used to describe a damaging variant that is (potentially) disease-causing. This is straightforward for dominant Mendelian disorders for which pathogenic variants typically cause the disease phenotype but more complex for recessive disorders for which both copies of the gene must harbour variants for pathogenicity (see the figure). Consider a variant producing a stop codon, p.Arg510Ter, in hexosaminidase subunit-α (HEXA), which is a gene that is implicated in Tay–Sachs disease. Obviously, this variant changes the transcript in which it resides: the resulting protein is probably nonfunctional due to truncation and may be subject to nonsense-mediated decay. However, this does not mean that it will necessarily be pathogenic to the individual, as many Mendelian diseases such as Tay–Sachs disease, are recessive. Cystic fibrosis is another well-known example, for which the genomes of approximately 1 in 20 healthy Western Europeans contain a damaging variant in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. As the disease is recessive, there are no negative health consequences to carriers of damaging variants. For recessive diseases, two copies of the pathogenic variant must be present, or it must be in trans to another pathogenic variant, as a so-called compound heterozygote (see the figure).
The association of damaging variants with pathogenicity has other pitfalls as well. A variant elsewhere in the genome may introduce a seemingly minor and conservative amino acid substitution that may nonetheless damage the patient’s health, thereby causing a dominant Mendelian disease. For example, the semi-conservative amino acid-changing variant p.Arg143Gln in the gap junction protein-β2 (GJB2) gene is implicated with non-syndromic hearing loss. This variant has been shown in functional studies to encode a protein with impaired function and curated by multiple laboratories in the ClinVar database to be pathogenic.
In a study from 2010, variants implicated in cystic fibrosis and related disorders were assessed using three prediction tools107. This study shed light on the differences between predictions and causative alleles. For example, the CFTR variant p.Arg75Gln is predicted to be damaging because it alters a highly conserved position in the protein, but the phenotypic effect is mild. The converse was shown by p.Val520Phe, a deleterious mutation at a non-conserved position in the CFTR protein. In another example, the truncating breast cancer type 2 susceptibility protein (BRCA2) variant p.Tyr791Phe is seemingly damaging — it causes the loss of the 93 C-terminal amino acids of the protein implicated in hereditary breast cancer, but does not cause the disease phenotype (see ClinVar database where it is curated as benign by multiple laboratories and an expert panel). BRCA2 provides another example of the complex relationship between damaging and pathogenic variants. Damaging BRCA2 alleles are typically classified as pathogenic, but they are not immediately disease-causing; instead, they increase cancer risk over a lifetime.

Describing variants
Variant annotation
We define a genetic variant (or, for brevity, ‘variant’) as a specific allele at a particular locus. Although variant discovery is outside the scope of this article, reviews are available elsewhere18–21. The first step of variant prioritization is annotation, which is the process of describing the nature and the effect of the DNA alterations produced by a variant. With this goal in mind, the variant call format (VCF)22 has standardized the reporting of genetic variation observed in a cohort and formalized a syntax with which to describe annotations that are vital to variant prioritization. Variant annotation tools such as the Variant Effect Predictor (VEP)23, the Variant Annotation, Analysis and Search Tool (VAAST) suite’s Variant Annotation Tool (VAT)24 and single nucleotide polymorphism effect (SnpEff)25 relate variants to annotated gene models in order to determine their location and effect on a transcript. For example, an SNV may result in a missense codon that alters the translated amino acid or it may result in a stop codon that terminates translation prematurely. Most variant prioritization tools use controlled vocabularies for variant annotation because of the scale of the data and to maximize reproducibility and interoper-ability between tools. The Sequence Ontology26 (SO) provides a widely used terminology for variant annotation, describing a variant in terms of the ‘sequence alteration’ it causes. Examples of sequence alterations include insertions, deletions and substitutions. Once the variant alteration has been described, the next step is to describe its effect.
Variant effects
A variant’s effect describes how it changes the annotated reference sequence features that contain it. Examples include a missense variant (SO:0001583), which induces an amino acid change, or a splice donor variant (SO:0001575), in which the alteration disrupts the dinucleotide at the 5 end of an intron27. The Sequence Ontology can also describe changes caused by more exotic forms of alterations, including structural variants that may introduce effects such as transcript ablation (that is, a deletion of a sequence encoding a transcript) and transcript amplification (that is, a duplication of a sequence encoding a transcript). The Sequence Ontology variant effect terms have been created in collaboration with Ensembl, and many variant annotation tools23,24,25 have adopted them. The common terminology that the Sequence Ontology provides for describing variant effects enables the comparison of annotations across tools, and Sequence Ontology terms are used by most genetic variant databases, such as ClinVar, dbVar, dbSNP and Ensembl Variation17,28–30.
Complications
Gene models describe the intron–exon structure of a gene’s transcripts and, for protein-coding genes, their start and stop codons. Variant annotations are wholly dependent on the gene models within which they reside. However, gene models are often incomplete and change over time31. Moreover, the number of human genes is still unknown32,33, and the precise structure of many genes is still being debated. GenBank and Ensembl both provide reference gene models for the human genome. In general, Ensembl tries to be inclusive, whereas GenBank is more conservative, requiring more peer-reviewed evidence for its gene models. At the time of writing, Ensembl contains 26,998 protein-coding genes and 81,787 mRNA transcripts, whereas GenBank’s RefSeq collection has 21,104 protein-coding genes and 34,799 mRNA transcripts. Even when both data sets have a model for a gene, its exon coordinates, transcript numbers, and start and stop codons often vary. Thus, a variant may lie in a coding exon in one provider’s gene model but reside in the intron or even an intergenic region in the other model.
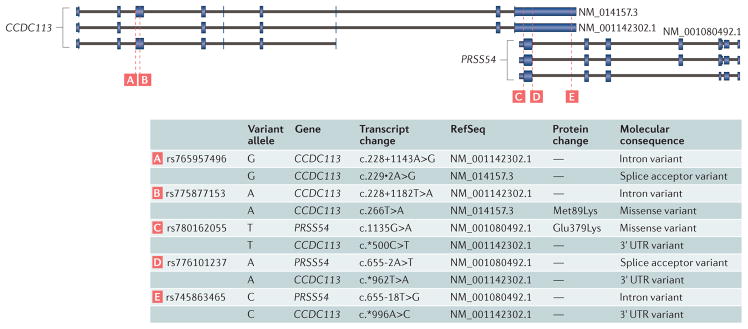
Alternative splicing further complicates variant annotation, because the effect of a variant can vary on a transcript-by-transcript basis. For example, it may occur in an intron of one transcript but within an exon of another (FIG. 1). A common strategy to deal with this complication is to annotate the variant based on the transcript (or transcripts) with the most severe effect. The rationale for this approach is to avoid missing potentially causal variants (false negatives) at the expense of enriching for false positives that could be eliminated through other means of prioritization (for example, population allele frequency) and manual inspection.
Figure 1. A demonstration of the multiple possible effects of a single variant across transcripts and genes.
The complexity of genomic annotation adds to the complexity of variant annotation. In this example, two genes, coiled-coil domain-containing 113 (CCDC113) and protease serine 54 (PRSS54) overlap on different strands of the genome, and both have multiple observed transcripts. Variants intersecting this extent of the genome show different effects depending on the gene and the transcript inspected. For example, the rs780162055 variant from the single nucleotide polymorphism database (dbSNP) is a missense variant with a protein effect for PRSS54 and a 3′ untranslated region (3′ UTR) variant for CCDC113. This proliferation of effects has data management implications for variant interpretation.
Prioritizing variants
Identifying the genetic cause of a Mendelian disease requires the systematic prioritization of the one or two causative variants from among the thousands or millions of variants identified in a typical exome or genome, respectively. The simplest imaginable approach is to use Sequence Ontology terms to quickly prioritize variants in an ad hoc manner under the assumption that, for example, a variant creating a premature stop codon is typically more damaging than a missense variant. However, this is a poor approach because the average human harbours hundreds of putative loss-of-function alleles in both heterozygous and homozygous states4,22,34,35 (TABLE 1). Such a simplistic filtering approach is also ill-advised because a stop codon in a poorly conserved gene may be more tolerated than a missense variant in another highly conserved gene. Furthermore, synonymous changes (those that do not alter the amino acid encoded) have been implicated in human diseases by affecting splicing36 and mRNA stability37, and by altering protein conformation38.
Table 1.
Median number of protein-coding variants and effects among world super-populations*
| Super-population code | Synonymous (het; hom alt) | Missense (het; hom alt) | Frameshift (het; hom alt) | Stop gain (het; hom alt) | Start lost (het; hom alt) | Splice donor (het; hom alt) | Splice acceptor (het; hom alt) | ||
|---|---|---|---|---|---|---|---|---|---|
| Total | SIFT Del | PP Del | |||||||
| EUR | 6961; 4317 | 7220; 4452 | 116; 55 | 116; 38 | 151; 146 | 93; 35 | 61; 52 | 184; 99 | 114; 72 |
| AFR | 9296; 4673 | 9347; 4820 | 163; 56 | 156; 31 | 196; 150 | 123; 32 | 78; 51 | 231; 116 | 150; 80 |
| AMR | 7257; 4314 | 7449; 4479 | 121; 56 | 121; 38 | 154; 145 | 96; 34 | 62; 50 | 187; 101 | 117; 76 |
| SAS | 7180; 4397 | 7366; 4550 | 123; 56 | 121; 39 | 159; 148 | 93; 36 | 68; 49 | 186; 103 | 117; 78 |
| EAS | 6502; 4759 | 6802; 4908 | 105; 66 | 113; 45 | 143; 149 | 89; 38 | 62; 54 | 171; 112 | 115; 86 |
AFR, individuals of African descent; AMR, individuals of admixed descent from the Americas; EAS, individuals of East-Asian descent; EUR, individuals of European descent; PP Del, PolyPhen2 predicted the missense variant to be deleterious; SAS, individuals of South-Asian descent; SIFT Del, SIFT predicted the missense variant to be deleterious.
We measured the average number of heterozygous (het) and homozygous alternate (hom alt) genotype counts among the 2,504 individuals sequenced by the 1000 Genomes Project. All genetic variants affecting genes were annotated with the Variant Effect Predictor and categorized by their most deleterious predicted effect.
Identifying pathogenic variants given the vast candidate pool of benign variants in a human exome or genome is a fundamentally challenging problem that has given rise to diverse variant prioritization tools. Traditional approaches use conservation and protein structure to predict the consequence of a missense change on protein function. More powerful techniques39,40 have recently been developed that widen the scope of prioritization (that is, not just missense changes) and improve accuracy. These performance gains are achieved by integrating population allele frequency, and gene conservation and constraint into prioritization calculations. In the following paragraphs, we explain how these data are used for variant prioritization. TABLE 2 provides an overview of the tools that use them, their scope of application and which information sources they use.
Table 2.
Commonly used software for assessing variant impact
| Tool | Category | Coding (missense only) | Indel | Non- coding | Method summary |
|---|---|---|---|---|---|
| SIFT112 | Missense prediction | Y (Y) | N | N | The degree of protein sequence conservation is used to predict the impact of a missense variant |
| PolyPhen2 (REF. 113) | Missense prediction | Y (Y) | N | N | Uses protein sequence and structure to predict the impact of a missense variant |
| FATHMM114 | Missense prediction | Y (Y) | N | N | Uses protein sequence homology identified with HMMER3 (REF. 115) to predict the impact of a missense variant |
| PROVEAN116 | Missense and indel prediction | Y (N) | Y | N | The degree of protein sequence conservation is used to predict the impact of an amino acid change or an indel |
| REVEL117 | Missense prediction (ensemble method) | Y (Y) | N | N | Incorporates 18 individual scores from 13 different tools to produce an ensemble ‘pathogenicity’ score for missense variants |
| PhastCons118 | Sequence conservation | Y (N) | Y | Y | Uses multiple sequence alignments from diverse species to identify conserved elements |
| PhyloP92,118 | Sequence conservation | Y (N) | Y | Y | Uses multiple sequence alignments from diverse species to assign per-base P values of conservation |
| GERP++93 | Sequence conservation | Y (N) | Y | Y | Measures sequence conservation in the human genome through alignments to 43 other vertebrate genomes |
| MutationTaster2 (REF. 119) | Multi-data integration | Y (N) | Y | Y (intronic) | Integrates sequence conservation, as well as data from the 1000 Genomes Project, ENCODE90 and ClinVar, to predict the consequence of variants within a gene model |
| VAAST6,24,66 | Multi-data integration | Y (N) | Y | Y | Integrates variant frequency data with phylogenetic conservation for variant prioritization and burden testing |
| CADD39 | Multi-data integration | Y (N) | Y (short indels) | Y | Integration of conservation metrics, functional data (for example, DNase I hypersensitivity and transcription factor binding) and scores such as SIFT and PolyPhen2 to predict the deleteriousness of nucleotide or short indel change in the genome |
| FitCons40 | Multi-data integration | Y (N) | Y (short indels) | Y | Integrates functional genomic data from the ENCODE project to cluster genomic regions and to predict the probability of a fitness consequence based on sequence conservation and the degree of regional polymorphism in the human genome |
CADD, Combined Annotation-Dependent Depletion; ENCODE, Encyclopedia of DNA Elements; FATHMM, Functional Analysis Through Hidden Markov Models; FitCons, fitness consequence; GERP, Genomic Evolutionary Rate Profiling; HMMER3, a tool based on a hidden Markov Model (HMM) for searching sequence databases for homologues of protein or DNA sequences; indel, small insertion or deletion; PolyPhen2, polymorphism phenotyping version 2; PROVEAN, Protein Variation Effect Analyzer; REVEL, rare exome variant ensemble learner; SIFT, Sorts Intolerant From Tolerant; VAAST, Variant Annotation, Analysis and Search Tool.
Conservation
As missense variants are the most commonly observed non-synonymous alteration in a typical exome (TABLE 1), a long-standing variant prioritization strategy is to use phylogenetic conservation to distinguish damaging from tolerated missense variants. The degree of conservation is ascertained by aligning human protein sequences to homologous protein sequences from other organisms. The rationale behind this is simple: the more conserved a column is within the multiple alignment, the more damaging an amino acid-changing variant at that position will be. The corollary is that the less conserved the column, the more likely it will be tolerated. These assumptions are generally correct but not always. Just because a variant is predicted to be damaging by tools, such as Sorts Intolerant From Tolerant (SIFT)41, does not mean that it is pathogenic (BOX 1).
Users should also bear in mind that conservation-based approaches to prioritization suffer from two systematic limitations. First, although most human proteins are at least partially conserved across vertebrates, they frequently contain one or more poorly or non-conserved regions. Although many known disease-causing alleles reside in such regions, conservation-based approaches often fail to identify them as deleterious. An alternative approach used by polymorphism phenotyping version 2 (PolyPhen2) is to use protein structure information for improved accuracy, especially in less well-conserved regions, but the gains are modest42. A second major limitation is that phylogenetic conservation provides poor means for determining the impact of stop codons and frameshift-inducing variants. This is because the protein sequences from other organisms used to make the multiple alignments do not contain them. Instead, stop codon and frameshift-inducing variants are either not prioritized at all or assigned maximally damaging scores by default. One might be tempted to assume that such variants are necessarily damaging, but the truth is much more complex. It is now recognized that some proteins are tolerant to stop codons and frameshifts (especially when they occur near the protein’s carboxyl terminus)34. Moreover, in loci such as the ABO blood group gene43, a significant proportion of the human population has inherited at least one frameshifting variant. In many cases, even though these highly damaging alleles destroy protein function, they seem to have little (if any) impact on health, even when homozygous44. Obviously, other approaches beyond sequence conservation are needed for prioritization of stop codons and frameshifts. The advent of WES and WGS has also placed additional demands on prioritization tools regarding accuracy. High false-positive rates can dramatically lengthen the time required for manual review of potential disease-causing variants. SIFT and PolyPhen2, for example, predict on average between 154 and 219 deleterious changes, respectively, in a typical human exome from a healthy individual (TABLE 1); hence, the majority of these ‘deleterious’ changes are unlikely to be pathogenic. Given these facts, it is no wonder that variant prioritization tools have sought to improve accuracy by incorporating additional sources of information such as population allele frequency and gene constraint.
Population allele frequency
Although inter-species conservation has proved to be a useful, though imperfect, tool for variant prioritization, recent catalogues of genetic variation within the human population provide powerful and complementary means for prioritization (BOX 2). Large-scale genome and exome sequencing efforts such as the 1000 Genomes Project3,45–47, the US National Heart, Lung and Blood Institute (NHLBI) Exome Sequencing Project48 and, more recently, the ExAC4 and gnomAD (see Further Information) projects, have catalogued protein-coding variation observed among the exomes of more than 60,000 individuals. The use of these resources cannot be overstated, as they provide an exquisitely detailed map of the landscape of human genetic variation from the common to the incredibly rare. Indeed, the ExAC consortium showed the extent of coding variation that exists in the human exome, observing an average of 1 coding variant every 8 base pairs. Further still, more than half of the 9 million variants uncovered were so rare that they were observed only once, as a heterozygote in a single individual (that is, an allele frequency of 1 out of 121,412 chromosomes on average among all subpopulations measured). Recognizing the power of these resources to establish an a priori expectation for a variant’s relevance to disease, variant prioritization tools such as VAAST24 and ANNOVAR49 use these allele frequencies to prioritize rare variants.
Box 2. Variant interpretation resources.
Genomic data repositories
Multiple catalogues of observed variants assembled from cohorts of thousands of genomes and/or exomes are now available. These resources are absolutely crucial adjuncts to the variant interpretation process.
The 1000 Genomes Project
The 1000 Genomes Project3 sequenced 2,504 individuals using whole-genome sequencing (WGS) to catalogue variants and their frequencies genome-wide in 26 different population groups. The individuals in this study are self-declared as healthy and no further phenotype data were collected.
The NHLBI Exome Sequencing Project
The US National Heart, Lung and Blood Institute (NHLBI) Exome Sequencing Project sequenced the exomes of ~6,500 individuals with phenotypes pertinent to heart, lung and blood disorders and provided the first glimpse into the extent of extremely rare protein-coding variation that exists in the human population.
ExAC
The Exome Aggregation Consortium (ExAC)4 is an aggregation of 60,706 exomes, the goal of which is to provide a deep catalogue of protein-coding variation for both population studies and for the clinical interpretation of variants. ExAC represents 6 broad populations and 14 disease cohorts, although individuals with severe paediatric phenotypes were removed.
gnomAD
The genome Aggregation Database (gnomAD) is the successor to ExAC and, at the time of writing, comprises genetic variation observed from 123,136 whole-exome sequencing (WES) and 15,496 WGS data sets collected from unrelated individuals.
Data-sharing initiatives
Aside from centralized repositories of genomic data, there are also many efforts underway to address the urgent need for data sharing across institutions and borders. Data sharing can range from the simple (for example, discovery of a previously observed variant) to the more complex, in which parties endeavour to match patient genotypes, phenotypes and ancestries in an effort to corroborate a potential Mendelian disease discovery.
The GA4GH Beacon Project
The Global Alliance for Genomics and Health (GA4GH) Beacon Project108 allows researchers to search for a particular variant across a host of individual hospital and research facilities using the same interface.
Geno2MP and MyGene2
Similarly to the GA4GH Beacon Project, Genotype to Mendelian Phenotype (Geno2MP) is a service that houses anonymized and aggregated data that enable phenotypic querying. MyGene2 allows researchers and clinicians to identify and contact other researchers, clinicians or families who have shared both raw data and summary information about the same rare condition or candidate.
Databases of variant–disease and gene–disease associations
Comprehensive variant–phenotype databases
ClinVar17 and the Human Gene Mutation Database (HGMD)109,110 catalogue ever-increasing connections between variants and disease. ClinVar, as part of the larger ClinGen Resource88, is an open archive of variants, with clinical phenotypes, evidence and the interpreted clinical significance. Submitted variants are classified by type of submitter, number of agreeing submissions and the variant interpretation guidelines used. A key strength of this archive is the aggregation of data from multiple clinical laboratories, providing a growing record of support for each interpretation, in which the provenance for each interpretation is maintained. A benefit of this aggregation process is that disagreements about the significance of variants are collated and reported17. This is the first time such conflicts have been openly used as a tool to improve global understanding of the clinical significance of genetic variants.
Locus-specific databases
Many genes have established historical connections to disease and interpreting a variant that falls into one of these genes may be supported by evidence collected in genetic databases. The most established of these resources is Online Mendelian Inheritance in Man (OMIM)16, which covers more than 15,000 genes with literature-based curation. It also provides phenotypic terminology in the form of more than 5,000 condition names. Similarly, Orphanet111, a European rare disease network, also distributes sets of rare conditions and lists of associated genes.
Genes of uncertain significance
Genes that have not previously been associated with a disease or have limited evidence for association are often termed ‘genes of uncertain significance’ (GUS). Variants in this class of gene are assigned research status according to the American College of Medical Genetics (ACMG) and Association for Clinical Genetic Science (ACGS) guidelines and they are not often reported back to patients. PanelApp is a crowdsourcing tool that has been developed via the Genomics England project that supports the development of evidence-based gene panels to encourage standardization across genetic tests offered by different sites. It is hoped that through curation of evidence (currently unreportable) GUS will transition to clinical grade.
Population stratification
Although allele frequencies are a powerful tool for variant prioritization, population stratification can confound the fundamental assumption that rare variants are, a priori, more likely to be damaging than common ones. In many cases, the average frequency of a variant allele across populations is markedly lower than the maximum allele frequencies observed within individual subpopulations. For example, a variant that is very rare in individuals of European ancestry may be far more common in those of African ancestry, or vice versa (FIG. 2a). Further complicating interpretation, many ethnicities are still underrepresented in publicly available genetic variation resources. Indeed, population stratification is a particular problem for Central American individuals of mixed ancestry50, whose genomes often contain rare variants of presumably Native American origin. These alleles superficially meet the requirement of being rare in all populations sampled so far and are therefore often predicted to be relevant to a rare disease phenotype. However, the fact that genomes of admixed Central Americans have not been sequenced as extensively as individuals of European ancestry increases the likelihood that the allele is, in fact, relatively common in Central Americans.
Figure 2. Population stratification and regional constraint within a gene are critical to variant interpretation.
a | For a particular variant, although the overall allele frequency may be low enough to be a plausible candidate with respect to a disease phenotype, the allele frequency is often substantially higher in specific subpopulations, thereby casting doubt on its relevance to rare disease phenotypes. In the example shown (source: http://gnomad.broadinstitute.org/variant/1-216172299-C-G), the rs79444516 variant of usherin (USH2A) is low in European populations but considerably higher in African populations. b | Constraint (that is, tolerance to genetic variation) can vary dramatically from region to region in a given gene. In this example, potassium voltage-gated channel subfamily Q member 2 (KCNQ2) shows higher constraint in the functionally important ion transport domain, as indicated by the scarcity of missense and loss-of-function (LOF) variants, relative to regions of lower functional importance in the same gene.
An established maxim of human genetics is that alleles causing Mendelian diseases do not discriminate: they should be rare in all ancestries. Therefore, allele frequencies among diverse ancestries should always be examined when prioritizing candidate variants for Mendelian disease. It is important to note that diseases such as cystic fibrosis and sickle cell anaemia represent well-known exceptions to this maxim and reflect situations in which the causal polymorphisms are under balancing selection, which keeps these alleles at higher frequency, because they confer protection (when in a heterozygous state) from illnesses such as cholera51 and malaria52, respectively.
Population-scale variant catalogues from diverse ancestries enable increased scrutiny of variants for which rare disease association was ascertained from small sample sizes or from single-ancestry cohorts (for example, European descent). A recent analysis showed that many reportedly pathogenic variants in ClinVar have markedly higher allele frequencies than predicted by the disease prevalence, suggesting that they may reflect spurious associations53. The majority of these discrepancies are observed for ClinVar ‘zero star submissions’, emphasizing the need for ClinVar users to understand submission guidelines and classification procedures (BOX 2). Similarly, resources such as ExAC have been used to refute the implication of new variants in rare diseases on the basis of the overly high frequency of the implicated allele in healthy individuals53,54.
Gene constraint
Using population-scale measurements of variant density and allele frequencies, multiple groups have developed statistical models of gene-wide constraint that model the tolerance of a gene to amino acid-changing or loss-of-function (LOF) variation relative to all other genes in the human genome. Such tools are essentially ranking genes on the strength of purifying selection. For example, the Residual Variation Intolerance Score (RVIS) uses ~6,500 exomes from the NHLBI Exome Sequencing Project and a linear model comparing the number of common functional variants observed in a gene against the total number of variants observed in the gene55. Genes with significantly more common functional variants than expected are inferred to have low constraint, whereas constrained genes have less common functional variation than expected. More recently, ExAC used 60,706 exomes to measure the probability of loss-of-function intolerance (pLI) for each gene in the human genome. Building on previous work56, modelling the expected number of de novo mutations per gene, pLI compares the observed and expected numbers of LOF variants to derive a probability that each gene is intolerant of LOF mutations4. The closer the pLI is to 1, the more intolerant to variation the gene is predicted to be. Gene-wide measures of constraint are effectively assuming a dominant model of inheritance; for example, the most LOF-intolerant genes (that is, pLI >0.9) encompass the majority of known severe haploinsufficient human disease genes. However, such measures have limited use for recessive disease genes and pLI is by no means a perfect predictor of the disease association of a gene4. This is especially true for adult-onset hereditary cancer, in which the disease often manifests after reproduction (for example, the pLI for BRCA1, BRCA2 and ATM is 0.0, despite the occurrence of well-known pathogenic variants in these genes).
A logical improvement on gene-wide constraint measures is the calculation of regional constraint along the gene. The rationale for regional measures is simple: genic ‘regions’ (that is, exons or portions of an exon) that encode crucial domains or subunits of a protein will be under stronger purifying selection than other regions of the protein. For example, the ExAC data set shows high constraint within the ion transport domain of sodium and potassium channel genes underlying both seizure and heart disorders (for example, early infantile epileptic encephalopathy and long QT syndrome), whereas other regions in these genes show far less constraint (FIG. 2b). Therefore, although gene-wide constraint measures are informative, prioritizing candidate variants on the basis of regional constraint is more nuanced and reduces both false-negative and false-positive predictions.
Caveats
Those engaged in variant and gene prioritization should also bear in mind that no two genes are alike. Large genes (for example, titin (TTN), filaggrin (FLG) and usherin (USH2A)) are more likely to harbour a possibly deleterious variant by chance, simply because they are comprised of more nucleotides. Furthermore, genes that are members of large, paralogous gene families (for example, mucins, keratins and olfactory receptors) are also likely to harbour false-positive variants that are unrelated to a disease phenotype owing to problems with the exome capture and sequence mapping57 process. However, simply ignoring variants in such genes is ill-advised: some TTN mutations, for example, cause autosomal recessive and dominant cardiomyopathies58, as well as various muscular dystrophies59,60. Similarly, mutations in keratins underlie several disorders of the skin and appendages61,62. For these problematic genes, burden test-based approaches (see below) can prove especially efficacious.
Burden testing
A critical distinguishing feature of gene prioritization tools is whether or not they use a burden test. Burden tests aggregate the variants observed at a given locus within one or more probands to calculate a sum or burden score. These scores are then used to prioritize genes rather than variants: the greater the burden, the more likely the gene is to be damaged. Many different scoring methods exist, but one commonality is the use of variant frequency information, so that common variants contribute less burden than rare ones. Most burden testing software tools also evaluate potentially damaging genotypes in the context of other genotypes observed at the same locus in a control population. This controls for gene-specific effects, so that the burden scores of larger, highly variable genes, such as TTN, do not always rise to the top of the candidate list.
Burden tests were originally proposed as a means to identify common rather than rare diseases, but the efficacy of the approach for Mendelian disease discovery is now making it popular for these applications as well63. There are several types of burden test64 and many tools are available for carrying out these analyses; examples include KBAC9, SKAT-O65, VT8 and VAAST6.
The burden-testing process is easiest to understand for dominant diseases. Consider, for example, a proband with a dominant Mendelian disease who has a de novo missense variant located in a particular gene. The effect of the variant is to change a tryptophan to a cysteine: a non-conservative amino acid change. Imagine that the variant is predicted to be maximally damaging by SIFT because it lies at a highly conserved position on the protein. Moreover, the de novo variant is novel; that is, it has never been observed in population-scale variant catalogues (BOX 2). All things considered, this variant would seem to be an excellent candidate for disease causation. Now imagine that 50% of all healthy individuals have some other equally damaging missense variant or even a more severe frameshifting variant at some other location in the gene; does it still seem so certain that the de novo variant is pathogenic? Logically, the hypothesis that the proband’s highly damaging de novo variant is disease-causing is now far less certain in light of these facts. Burden-testing tools automate this interpretive process.
One key feature of burden tests is their ability to score the diverse combinations of different types of variants that comprise genotypes using a single scoring scheme. VAAST, for example, can score and rank recessive genotypes that are combinations of missense, frameshifting and splice site-damaging variants using a single scoring scheme that also includes amino acid substitution scores, variant population frequencies and phylogenic conservation24,66. This means that the burden score for a proband with a damaging splice variant on one chromosome and a missense variant on the other can be compared with another individual whose genotype is comprised of a frameshift-inducing variant in trans to a missense variant. This is a computationally complex task, but it has considerable utility, as burden tests provide a means to rank genotypes for gene prioritization purposes and to explore the distribution of burden at a given locus for a population. One can then speak of a proband having a burden at a candidate disease locus that exceeds 95% or 99.9% of all observed genotypes at that locus in the general population.
Burden tests are also well suited to large case– control and family-based studies. Traditional genome-wide association study (GWAS) tests, which proceed variant by variant, lose power as more and more variants are included in the analysis because ever-increasing multiple testing corrections are required. By contrast, burden tests scale much better to large WES and WGS data sets because the multiple test correction is the number of genes (a constant), no matter how many variants and individuals are included in the study.
Burden testing is also extensible to family-based analyses. Parent–child trios, for example, can be used to ‘Mendelize’ variants and to phase the data to ensure that only combinations of variants that are consistently inherited are considered in the calculations, thereby improving accuracy. Pedigree-VAAST (pVAAST)67, for example, can use multigenerational pedigrees in its calculations so that the final burden scores reflect co-segregation of variants and phenotypes across the pedigree.
Burden tests open new vistas for gene-based prioritization as well as diagnostic and discovery applications, especially for large case–control analyses, recessive diseases and family-based studies for which data complexity exceeds human interpretive capacities. Moreover, the tests can also be embedded into larger software frameworks to allow inclusion of important adjunct data in the calculations, such as the penetrance of the variant or genotype, disease prevalence and mode of inheritance (see below). For these reasons, burden tests are becoming widely used for gene prioritization, especially within decision support frameworks.
Relevance to disease
Clinical interpretation of variants and genotypes necessitates integration of diverse data types. For example, variants are often interpreted in the context of disease prevalence and mode of inheritance (see also BOX 2). The culmination of this aggregation of information is manual assessment of prioritized variants and genes using community-agreed-upon guidelines. Some of the data modalities most relevant to this assessment process are described below.
Penetrance, prevalence and mode of inheritance
Penetrance refers to the probability that having a pathogenic variant or genotype will result in disease. Penetrance is easiest to understand in the context of dominant and de novo variants. A dominant variant is said to be completely penetrant when every individual with the variant has the disease and every individual without the variant is unaffected. Reality is of course more complex. The impact of a variant may be delayed. Individuals with Huntington disease who have a completely penetrant variant are phenotypically normal as children, but develop the disease in their adult life68. A variant may also be incompletely penetrant: this term is less precise and is often used to describe variants that only produce disease in, for example, half of carriers. Such an allele is said to be 50% penetrant. Many of the variants responsible for familial cancers show incomplete penetrance. The term variable expressivity is used to describe variants that cause mild symptoms in some carriers and more severe ones in others. Neurofibromatosis type 1 has multiple variably expressed phenotypes. Incomplete penetrance and variable expressivity complicate interpretation because observing the variant in an unaffected individual does not necessarily mean it is not pathogenic.
Knowledge of the population prevalence of a disease provides a powerful means to exclude some candidate disease-causing variants from further consideration. A useful rule of thumb is that, for a dominant disease, the product of its population prevalence and its fractional penetrance is an upper bound for the population frequency of any candidate disease-causing variant. Consider the case for a dominant Mendelian disease that occurs with a population prevalence of 1 in 10,000 individuals. If we assume 50% penetrance, then by this rule of thumb, any variant (population stratification issues aside) having a frequency greater than 1 in 5,000 in the general population is a poor candidate. For recessive diseases, the situation is more complex. In this case, it is the population genotype frequency, rather than individual variant frequencies, that must be less than the population disease prevalence. In the case of simple recessive diseases, the population genotype frequency is the square of variant frequency. This means that a recessive disease-causing variant can be relatively frequent in the population, which greatly increases the number of potential candidates.
A further complication for prioritization is that many recessive disease cases result from compound heterozygous genotypes. These are recessive genotypes in which both the maternal and paternal copies of a gene harbour a damaging variant, but these variants are distinct and occur at different positions in the maternal and paternal copies of the gene (BOX 1). In this case, the population genotype frequency is obtained by multiplying the constituent variant frequencies and, all things being equal, this value should be less than the observed incidence of the Mendelian disease. The complication in this case is that once the scope of prioritization has been extended to include compound heterozygous genotypes, every possible combination of rare variants in every locus must be considered in burden calculations. This requirement greatly increases the complexity of prioritization tasks, requiring specialized algorithms, as discussed below. Another complication is that the variants need to be in trans to one another, one located on the maternal chromosome, the other on the paternal. One easy way to determine this is to sequence the proband’s parents: doing so makes it possible to restrict the search to combinations of variants in which one is inherited from the mother and the other from the father. Family studies can further reduce the search space by focusing on heterozygote pairs that are observed in affected siblings but absent in unaffected siblings.
Taking all of these factors into account — conservation, constraint, mode of inheritance, variant and genotype population frequencies and penetrance — the task of assessing the thousands or even millions of variants in a typical WES or WGS requires an automated process. Variant prioritization tools that make a prediction in isolation for each variant are ill-suited to this problem. This is the domain of integrative gene prioritization tools. Existing tools use a variety of strategies. Genome Mining (GEMINI)69, seqr (see Further Information), Variant Association Tools70 and ANNOVAR49, for example, use filtering approaches that prioritize variants only if they follow a specified mode of inheritance, are predicted to affect protein sequence or function and have a population frequency below a specified threshold. By contrast, tools such as VAAST and SKAT-O use a probabilistic approach that uses background population frequencies to determine genotype frequencies and combines these with mode of inheritance and penetrance to identify damaged genes and disease-causing alleles using a burden test. Some go even further: pVAAST67, for example, can use multigenerational pedigrees of sequenced relatives, following the segregation of every variant in the family and correlating it with disease status. This results in greater power for family-based studies.
Phenotype
Intuitively, a proband’s phenotype has a crucial role in every gene and variant prioritization analysis. Mendelian diseases characteristically manifest themselves as recurring collections of stereotypical symptoms that together define a disease phenotype or condition. Unfortunately, many different conditions produce overlapping constellations of symptoms, hence the need for genome-sequence-based precision medicine.
If the disease were known, diagnosis would be simple; however, what are available before diagnosis are clinical symptoms and the results of diagnostic tests. Consider the case for a patient with medium-chain acyl co-enzyme A dehydrogenase (ACADM) deficiency (MCADD; see FIG. 3). Pre-diagnostic symptoms might include qualitative descriptions such as lethargy, seizures and hepatomegaly. Quantitative diagnostic results (that is, clinical measurements) might include controlled vocabulary-based descriptions of clinical tests and observed values such as those provided by Logical Observation Identifiers Names and Codes (LOINC), for example, an abnormal serum acylcarnitine profile. A genetic test yielding variant in the ACADM medium-chain-specific acyl-CoA dehydrogenase gene, with these clinical features, would make it possible to distinguish the disorder as MCADD from a series of related metabolic conditions.
Figure 3. Phenotypes are described across a spectrum of granularity, and different terminologies are used to define these features.

In this example, medium-chain acyl co-enzyme A dehydrogenase (ACADM) is used to show this granularity. At the broadest level, it is associated with the condition medium-chain acyl co-enzyme A dehydrogenase deficiency (MCADD), a metabolic disorder that is classified in databases such as Online Mendelian Inheritance in Man (OMIM) and Orphanet. Clinical terminologies such as Snomed and MedGen may also be used to categorize the condition. A condition is generally composed of multiple clinical features (such as lethargy) that describe the observable phenotypes. The Human Phenotype Ontology (HPO) is a widely used terminology that describes these features organized by the body system they manifest in. A key product of the HPO is the annotation of phenotype-to-gene and phenotype-to-condition files that are used in many downstream prioritization tools. At the most fine-grained level, the molecular phenotype of the patient is defined by the clinical measurements such as the concentration of urine organic acids. The most widely used terminology for these measurements are provided by Logical Observation Identifiers Names and Codes (LOINC), a universal code system for clinical data. A patient may be identified early in life as a result of newborn screening — by detecting unusual ratios of metabolites — or may be detected later in life as a result of experiencing one or more clinical features. These different levels of phenotypes are used to guide the patient towards the most appropriate test and to guide the prioritization of the genes and associated variants in the genetic analysis.
The Human Phenotype Ontology (HPO)71 provides hierarchical sets of disease names and clinical features (symptoms) for describing medical conditions and, crucially, the HPO also provides associations between symptoms and known disease genes. The disease–gene catalogue OMIM16 associates conditions and genes. Machine-readable phenotype descriptions, such as those produced using the online resources Phenotips72 and PhenoDB73, use HPO and OMIM terminology to produce standardized phenotype descriptions. Several tools exist for combining phenotype descriptions with variant and gene prioritization results (see REF. 74 for a review) to elevate rankings of potential candidates in variant prioritization. These tools vary from ontology-based semantic similarity methods to more complex machine-learning techniques75–80.
Phenotype analysis tools such as the Phenotype-Driven Variant Ontological Re-ranking tool (Phevor)80 and Phenolyzer78 can evaluate qualitative HPO-based phenotype descriptions such as ‘lethargy, seizures and hepatomegaly’ and use the broader structure of the HPO and its gene–symptom linkages in order to associate genes with proband phenotypes. They then combine this information with variant and gene prioritization results. They can even discover new gene–disease associations80. Another tool, Phenotypic Interpretation of Variants in Exomes (PHIVE)81, uses a different approach: it is a variant filtering tool that uses a combination of variant frequency, predicted deleteriousness of the allele and a semantic similarity-based phenotypic relevance score that uses model organism annotation to rank exonic variants.
Phenotype reprioritization tools straddle both realms of clinical application and disease–gene discovery. To simplify clinical analysis, Phenotypic Interpretation of Exomes (PhenIX)74 solely reports on known disease genes. Other tools can be used in both situations. For example, Phevor80 uses the knowledge collected in related ontologies such as the Gene Ontology (GO) to suggest new gene–disease associations.
Variant interpretation
Variant interpretation refers to the process of drawing direct connections from individual variants to disease phenotypes, and this process is central to both clinical reporting of results and incidental findings, and to research endeavours that include variant discovery and return of results. As a variant can be damaging to gene function but not disease-causing (BOX 1), candidates identified by variant or gene prioritization tools must be evaluated for causation. As a result of its complexity and impact on patient diagnosis and treatment, this process remains largely one of expert interpretation and literature review. As the complexity and amount of available genetic data have increased, interpretation has faced new challenges, and the need for standardized guidelines has become apparent. This fact is demonstrated by the 2012 CLARITY Challenge82, in which multiple groups interpreted the exomes of three parent–child trios, yielding inconsistent findings among the resulting variant reports.
As a result of these challenges, interpretation guidelines have been developed in Europe and the United States to standardize interpretation workflows so that decisions are made in a consistent manner. The UK Association for Clinical Genetic Science (ACGS)83 updated guidelines in 2013 that described a narrative list of the lines of evidence and necessity of this evidence to be used in variant interpretation. The American College of Medical Genetics (ACMG) has issued consensus guidelines distilled from community input. The ACMG guidelines provide a terminology to define clinical significance, a scheme for ranking evidence used to make variant–disease assertions and a set of rules for combining the evidence for a case84. In a recent announcement, the ACGS and British Society for Medical Genetics have recommended following the ACMG consensus guidelines, further consolidating a standardized clinical approach83,85.
The scope of these guidelines is strictly for the interpretation of variants suspected to be implicated in Mendelian disorders, and both organizations agreed on the need to standardize the description of variants using: Human Genome Variation Society (HGVS) nomenclature86, Human Genome Organisation (HUGO) gene identifiers87, named reference sequences with versioning, and five grades of clinical significance. Evidence is ranked into four classes: supporting, moderate, strong and very strong. The outcome is an assertion of either benign, likely benign, VUS, likely pathogenic or pathogenic. Many of the criteria are subjective; for example, quantification of co-segregation of the variant with the phenotype ranges from ‘supporting’ to ‘strong’ evidence, and there is a criterion for a variant being previously reported by a ‘reputable source’. Moreover, both guidelines acknowledge the many caveats involved in interpretation, such as variants that fall in the last exon and null variants that prove to be benign in heterozygous form.
It is widely acknowledged that these guidelines are general and that there is a pressing need to establish specific procedures for different genes and diseases. In response, clinical-domain working groups have been established, such as those administered via the ClinGen Resource from the US National Human Genome Research Institute (NHGRI)88. Their purpose is to extend these recommendations per gene or gene panel to accommodate any specific caveats that may exist84 (see also ClinGen Clinical Domains in Further information).
Clinical and research interpretation diverge whenever a variant of interest falls in a gene of uncertain significance (GUS)84. These are the genes for which there is no documented association with the disease or the phenotype. The guidelines are clear that these variants may only be clinically reported as VUS until further evidence is collected; for example, the discovery of additional individuals with similar phenotype and deleterious variants in the gene89. Research evidence is then collated in collaborative resources such as ClinVar and PanelApp (BOX 2).
Current challenges and emerging solutions
Mendelian disease research and diagnosis have been greatly empowered by the sequencing of exomes and targeted gene panels. Despite being effective, research laboratories are slowly transitioning to WGS because of its greater power to discover all forms of potentially causal variation. Clinical diagnostic laboratories are likely to follow this trend once costs decline sufficiently for insurance providers to reimburse WES- and WGS-based tests. However, aside from cost barriers, there are substantial analytical barriers to the systematic prioritization of the millions of genetic variants that are uncovered via WGS.
Non-coding variants
As the protein-coding exome represents less than 2% of the genome, most additional variants revealed through WGS lie in non-coding regions. The Encyclopedia of DNA Elements (ENCODE) project has emphasized that as much as 80%90 of the non-protein-coding portion of the genome is associated with biochemical ‘function’. Although the precise percentage and the definition of ‘function’ is debated91, it is clear that many non-coding regions, such as promoters, enhancers and splice sites, are crucial to gene function. More generally, non-coding nucleotide conservation can be used to prioritize non-coding variants in much the same way that SIFT uses protein-based alignments. Several such tools exist: some use conservation information directly, whereas others use conservation scores provided by third-party tools, such as phyloP92 and Genomic Evolutionary Rate Profiling (GERP)++93 (see TABLE 2 for details). However, it should be noted that non-coding variant prioritization tools are less accurate than their protein-coding counterparts. Although many new approaches are being developed39,94,95, there is simply insufficient understanding of the regulatory machinery encrypted in non-coding DNA to prioritize non-coding variants with similar accuracy to that of coding variants94.
Synonymous exonic variants
Synonymous exonic variations are scored and prioritized by several existing tools (TABLE 2). There are multiple mechanisms whereby these variants can cause disease, such as altering the fidelity of splicing or microRNA (miRNA) binding, affecting mRNA stability or altering translation dynamics. Software and other validation strategies are reviewed by Hunt et al.96.
Structural variants
Structural variants encompass both copy number variants (CNVs), such as deletions and duplications, and balanced rearrangements, such as inversions and reciprocal translocations. Although there are far fewer structural variants (that is, between ~5,000 and ~10,000) in a typical human genome47 than SNVs and small insertions and deletions (indels), their potential for phenotypic impact is disproportionately large because they can disrupt multiple genes, create gene fusions, ablate regulatory elements and alter gene dosage. Owing to a variety of technical reasons, structural variants remain the most difficult form of variation to detect, and WGS — as opposed to WES — is preferred for these analyses because of the greatly increased discovery power and resolution71. Despite the cost and complexity of WGS data, structural variant detection clearly improves the diagnostic yield for Mendelian disorders compared with WES97. For example, Wu et al. recently found that 11% of congenital scoliosis cases are explained by compound heterozygotes comprised of SNVs and large deletions in T box 6 (TBX6)98. Furthermore, Burn–McKeown syndrome was also found to be caused by compound heterozygous inheritance (see BOX 1) of a promoter deletion and an SNV in thioredoxin-like 4A (TXNL4A)99. More generally, balanced chromosomal abnormalities have been shown to underlie congenital abnormalities by disrupting topologically associating domains (TADs) in loci that are known to cause developmental disorders100. Long-read sequencing technologies were recently used to implicate a deletion of the first exon of protein kinase cAMP-dependent type I regulatory subunit-a (PRKAR1A) in autosomal dominant Carney complex101, and WGS studies of autism spectrum disorder estimated that between 1 in 5 and 1 in 20 individuals harbour a de novo structural mutation, further strengthening the argument for comprehensive WGS-based structural variant analysis in Mendelian disorders102.
Unfortunately, interpretation of structural variants observed in a family is complicated by the fact that population-scale variant databases such as ExAC are not yet available for structural variants. Consequently, it is very difficult to assess whether a structural variant of interest (for example, a deletion of multiple coding exons) is likely to be pathogenic on the basis of its allele frequency among diverse ancestries. However, it is clear that resources for these activities will eventually emerge from large projects such as the Genomics England 100,000 Genome Project (UK100K), the NHLBI Trans-Omics for Precision Medicine (TOPMed) programme and the NHGRI Centers for Common Disease Genomics, which are each focused on WGS of tens of thousands of individuals.
Graph-like genome representations
With the thirty-eighth major release of the human genome, the Genome Reference Consortium has transitioned away from traditional linear genome representations that represent little sequence or structural diversity. Instead, this and future versions of the human genome attempt to represent multiple alternative sequence ‘paths’ for loci that have high levels of nucleotide diversity or structural complexity103. Although ‘graph-like’ genome representations improve the representation of the true diversity of the human genome, it is imperative that SNV, indel and structural variant discovery methods account for these changes in order to maximize discovery accuracy by distinguishing truly paralogous loci from alternative sequence paths represented in the genome assembly for the same locus. Some software advances have been made in this regard104, but widespread adoption of current and future genome assemblies will require continued algorithm development.
Better decision support tools
Many analysis pipelines apply ‘linear’ approaches in which the primary goal is to identify a subset of variants that meet a series of evermore restrictive, hard-coded filters. Examples include enforcing an inheritance model, maximum allele frequency, genotype burden, variant effect and so forth. The obvious drawback to such approaches is the potential for false negatives when variants fail to meet filtering criteria, with no easy means to recover them for further consideration in light of other information. This pitfall of linear approaches is driving the creation of more integrative approaches using decision support tools. These tools are not so much pipelines as browser-based interpretation environments. Decision support tools enable more flexible, interactive analyses and generally provide easier means to analyse variant data in the context of external resources such as ExAC, OMIM and ClinVar, which greatly empower clinical decision-making. Academic examples include iobio14 and Variation Viewer105. Commercial tools increasingly have an important role in this domain, partly because of the complexity and cost of developing such software. Examples include Congenica’s Sapientia, WuXi’s NextCode, (see Further Information) and Fabric Genomics’ Opal platform12. These platforms offer customizable workflows and web-based user interfaces that facilitate expert review and interpretation of results. They also deal with a host of practical issues such as data security and privacy. These features make them ideal for clinical diagnosis, but the current generation of these applications still lacks the functionality that is necessary for larger case–control analyses for which data aggregation across multiple probands is essential for discovery.
Conclusions
Connecting variants to disease is a complex, multistep process. Its early steps are highly automated, but the final, most critical aspects are not. Instead, they rely on expert review and human interpretation. In this sense, the process resembles many of today’s big-data analysis activities, but it is further complicated by its clinical nature. Wrong answers can be devastating to patient health and family planning. For example, in a recent example of incorrect variant interpretation, members of a family received a diagnosis of long QT syndrome and an inappropriate course of treatment106.
Variant and gene prioritization scores are useful starting points for discovery and diagnosis of rare Mendelian diseases, but they are merely that: starting points. Those charged with their review and interpretation need to understand the computational workflows and the strengths and weaknesses of the many software tools that constitute them.
Prioritization scores should never be naively conflated with pathogenicity (BOX 1). Instead, it is crucial that they are considered in the context of genotype, disease prevalence, family history and phenotype. Variant interpretation resources such as ClinVar and ExAC are proving to be essential to this process. Many tools are now available to help with this integration process, and complex decision support environments are increasingly being used. Standardization of the interpretation process is also clearly desirable. The guidelines offered by the ACMG and ACGS are important steps forward in this regard. These are growing more sophisticated, and their granularity is improving to allow ever more gene- and disease-specific interpretation workflows. Nevertheless, those engaged in precision genomic medicine should bear in mind that VUS will remain one of the most frequently used terms in the precision medicine diagnostic vocabulary for some time to come.
Acknowledgments
The authors thank J. Chong for insightful discussions about the challenges of rare disease research at the University of Washington Center for Mendelian Genomics Workshop. This Review was supported by US National Institute of Health awards to A.Q. (NIH R01HG006693, NIH U24CA209999), K.E (NIH U41HG006834 (subcontract), NIH U01HG007437 (subcontract), NIH R01HG008628) and M.Y. (NIH R01GM104390, NIH UM1HL128711, NIH U01HL131698 and NSF IOS-1561337).
Glossary
- Mendelian disorders
Diseases or conditions that result from mutation at a genomic locus and are inherited according to Mendel’s laws
- Variant prioritization
The process of ranking the variants observed in an individual genome on the basis of factors such as the predicted consequence of each variant and the observed frequency in a population
- Population allele frequencies
The proportion of chromosomes within a population that carry a particular change at a given locus
- Gene prioritization
The process of associating a gene with a disease phenotype; this strategy is often used during variant prioritization
- Burden testing
A gene prioritization approach that scores, ranks and prioritizes genes based on genotypes rather than on single variants. The observed (or for some methods, the theoretical) distribution of burden scores within the wider population is often used to rank a proband’s genotype score. Many burden tests can also incorporate adjunct information into their calculations such as phylogenetic conservation, mode of inheritance and variant frequency data. Unlike variant prioritization tools, burden tests require access to genotype data for their calculations
- Decision support frameworks
Interactive, dynamic tools to guide medical decision-making by displaying and integrating patient data
- Nonsense-mediated decay (NMD)
A conserved eukaryotic pathway, the role of which is to detect and eliminate the translation of mRNAs that have premature stop codons
- Variant of uncertain significance (VUS)
Also known as variant of unknown significance. The canonical definition of a VUS is a variant in a disease-associated gene, the specific effect of which is unknown or uncertain. More generally, VUS can also be applied to variants in genes that lack direct disease association but are plausible given the biological function of the resulting protein
- Controlled vocabularies
Sets of agreed upon terms and definitions
- Exome
Generally, the portion of the genome that is translated into proteins
- Population stratification
The difference in allele frequencies across subpopulations
- Balancing selection
Under balancing selection, multiple alleles exist in a population when natural selection favours heterozygous genotypes
- Disease prevalence
The number of cases of a disease that are present in a population at a given point in time
- Purifying selection
Under purifying selection, deleterious alleles are selectively removed from a population
- Functional variants
Variants that alter gene function or expression
- Probands
The proband is the initial person of study in a genetics investigation. In the case of a family trio, the proband is usually the affected child
- De novo variant
A spontaneous mutation in a proband that is missing from the parents
- Phase
For a single variant, phase involves the determination of the parental chromosome on which a variant allele exists. When a proband and both parents have been sequenced, this can be directly determined for ‘informative sites’ where the allele transmission is unambiguous (for example, the proband is heterozygous A/G, the father is homozygous A/A, and the mother heterozygous A/G; in this case the G allele was clearly transmitted from the mother). More generally, phasing refers to the assignment of alleles from multiple variant sites to parental haplotypes
- Population genotype frequency
The proportion of individuals with a particular genotype at a given locus
- Incidental findings
In whole-exome sequencing (WES) or whole-genome sequencing (WGS), pathogenic and likely pathogenic variants in genes that are not relevant to the initial reason for sequencing may be found and reported back to the patient. These variants may relate to rare disease, disease risk, pharmacogenetic response, and status relating to prenatal screening
- Return of results
The process of returning findings from a research study, or incidental findings from a genetic test, back to the participant or patient
- Compound heterozygous inheritance
The situation in which a proband receives a damaging but different allele in the same gene, from each parent. Both copies of the gene are affected
- Topologically associating domains (TADs)
TADs are genomic regions in which loci have a higher probability of physical interaction
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
References
- 1.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 2.Chong JX, et al. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. This review summarizes findings from the study of more than 8,000 families with Mendelian disease phenotypes by the Centers for Mendelian Genomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. By sequencing the genomes of more than 2,500 individuals from diverse world ancestries, this study provides the first genome-wide map of both common and rare human genetic variation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. The ExAC-integrated exome sequencing data from 60,706 individuals provides an invaluable reference data set of genetic variation in protein-coding genes. Assessing variant allele frequencies in ExAC facilitates the interpretation of candidate variants observed in Mendelian disease families. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy B, et al. Using VAAST to identify disease-associated variants in next-generation sequencing data. Curr Protoc Hum Genet. 2014;81:6.14.1, 6.14.25. doi: 10.1002/0471142905.hg0614s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu MC, et al. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 2010;86:929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price AL, et al. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coonrod EM, Margraf RL, Russell A, Voelkerding KV, Reese MG. Clinical analysis of genome next-generation sequencing data using the Omicia platform. Expert Rev Mol Diagn. 2013;13:529–540. doi: 10.1586/14737159.2013.811907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doig KD, et al. PathOS: a decision support system for reporting high throughput sequencing of cancers in clinical diagnostic laboratories. Genome Med. 2017;9:38. doi: 10.1186/s13073-017-0427-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller CA, Qiao Y, DiSera T, D’Astous B, Marth GT. bam.iobio: a web-based, real-time, sequence alignment file inspector. Nat Methods. 2014;11:1189. doi: 10.1038/nmeth.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandeweyer G, Van Laer L, Loeys B, Van den Bulcke T, Kooy RF. VariantDB: a flexible annotation and filtering portal for next generation sequencing data. Genome Med. 2014;6:74. doi: 10.1186/s13073-014-0074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landrum MJ, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. ClinVar is an important repository for collating and understanding genome variant interpretation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zook JM, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32:246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 22.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaren W, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yandell M, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eilbeck K, et al. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. The Sequence Ontology is a project that initiated developing standardized terminologies for genomic sequence features and became widely used in both genome annotation and more recently in variant annotation. It is a key vocabulary used by tools that assign consequences to variants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cunningham F, Moore B, Ruiz-Schultz N, Ritchie GR, Eilbeck K. Improving the Sequence Ontology terminology for genomic variant annotation. J Biomed Semantics. 2015;6:32. doi: 10.1186/s13326-015-0030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aken BL, et al. Ensembl 2017. Nucleic Acids Res. 2017;45:D635–D642. doi: 10.1093/nar/gkw1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lappalainen I, et al. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41:D936–D941. doi: 10.1093/nar/gks1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eilbeck K, Moore B, Holt C, Yandell M. Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics. 2009;10:67. doi: 10.1186/1471-2105-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pertea M, Salzberg SL. Between a chicken and a grape: estimating the number of human genes. Genome Biol. 2010;11:206. doi: 10.1186/gb-2010-11-5-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ezkurdia I, et al. Multiple evidence strands suggest that there may be as few as 19 000 human protein-coding genes. Hum Mol Genet. 2014;23:5866–5878. doi: 10.1093/hmg/ddu309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. Through careful examination of LOF variants in 185 individuals, this study predicted that a typical human harbours roughly ~100 potential LOF variants in their genome, highlighting the challenge of isolating the one or two causal variants underlying a Mendelian disease phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saleheen D, et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. This manuscript studies individuals harbouring homozygous LOF variants in a population with a high rate of consanguinity, revealing more than 1,000 genes that were predicted to be completely knocked out in at least one individual studied. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheikh TI, Mittal K, Willis MJ, Vincent JB. A synonymous change, p. Gly16Gly in MECP2 Exon 1, causes a cryptic splice event in a Rett syndrome patient. Orphanet J Rare Dis. 2013;8:108. doi: 10.1186/1750-1172-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nackley AG, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 38.Kimchi-Sarfaty C, et al. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 39.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. This manuscript describes the Combined Annotation-Dependent Depletion (CADD) score, which integrates diverse genome annotations into a classifier to assess the relative deleteriousness of variants genome-wide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gulko B, Hubisz MJ, Gronau I, Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat Genet. 2015;47:276–283. doi: 10.1038/ng.3196. By integrating high-throughput functional data from the ENCODE project, the fitCons method estimates the probability of whether any genome-wide point mutation will result in a fitness consequence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yip SP. Sequence variation at the human ABO locus. Ann Hum Genet. 2002;66:1–27. doi: 10.1017/S0003480001008995. [DOI] [PubMed] [Google Scholar]
- 44.Kaiser VB, et al. Homozygous loss-of-function variants in European cosmopolitan and isolate populations. Hum Mol Genet. 2015;24:5464–5474. doi: 10.1093/hmg/ddv272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1, 092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. This study provides the first genome-wide map of all common forms of structural variation from thousands of human genomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kidd JM, et al. Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am J Hum Genet. 2012;91:660–671. doi: 10.1016/j.ajhg.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gabriel SE, Brigman KN, Koller BH, Boucher RC, Stutts MJ. Cystic fibrosis heterozygote resistance to cholera toxin in the cystic fibrosis mouse model. Science. 1994;266:107–109. doi: 10.1126/science.7524148. [DOI] [PubMed] [Google Scholar]
- 52.Hedrick PW. Population genetics of malaria resistance in humans. Heredity. 2011;107:283–304. doi: 10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah N, et al. Identification of misclassified ClinVar variants using disease population prevalence. 2016 doi: 10.1016/j.ajhg.2018.02.019. Preprint at bioRxiv http://dx.doi.org/10.1101/075416. [DOI] [PMC free article] [PubMed]
- 54.Minikel EV, MacArthur DG. Publicly available data provide evidence against NR1H3 R415Q Causing multiple sclerosis. Neuron. 2016;92:336–338. doi: 10.1016/j.neuron.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. The authors use genetic variation from 6,515 exomes in the NHLBI Exome Sequencing Project to develop the Residual Variation Intolerance Score (RVIS), which ranks genes by their intolerance to ‘functional’ (that is, missense or LOF) variation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shyr C, et al. FLAGS, frequently mutated genes in public exomes. BMC Med Genomics. 2014;7:64. doi: 10.1186/s12920-014-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nigro V, Savarese M. Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta Myol. 2014;33:1–12. [PMC free article] [PubMed] [Google Scholar]
- 60.Hackman P, et al. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am J Hum Genet. 2002;71:492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ang-Tiu CU, Nicolas MEO. Ichthyosis bullosa of Siemens. J Dermatol Case Rep. 2012;6:78–81. doi: 10.3315/jdcr.2012.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chamcheu JC, et al. Keratin gene mutations in disorders of human skin and its appendages. Arch Biochem Biophys. 2011;508:123–137. doi: 10.1016/j.abb.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Auer PL, Lettre G. Rare variant association studies: considerations, challenges and opportunities. Genome Med. 2015;7:16. doi: 10.1186/s13073-015-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee S, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu H, et al. VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol. 2013;37:622–634. doi: 10.1002/gepi.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu H, et al. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat Biotechnol. 2014;32:663–669. doi: 10.1038/nbt.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 69.Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang GT, Peng B, Leal SM. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am J Hum Genet. 2014;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Köhler S, et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 2017;45:D865–D876. doi: 10.1093/nar/gkw1039. The Human Phenotype Ontology provides a systematic description of clinical features and is annotated to both genes and diseases, making it an invaluable resource for variant prioritization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Girdea M, et al. PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat. 2013;34:1057–1065. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 73.Hamosh A, et al. PhenoDB: a new web-based tool for the collection, storage, and analysis of phenotypic features. Hum Mutat. 2013;34:566–571. doi: 10.1002/humu.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smedley D, Robinson PN. Phenotype-driven strategies for exome prioritization of human Mendelian disease genes. Genome Med. 2015;7:81. doi: 10.1186/s13073-015-0199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smedley D, et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat Protoc. 2015;10:2004–2015. doi: 10.1038/nprot.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Javed A, Agrawal S, Ng PC. Phen-Gen: combining phenotype and genotype to analyze rare disorders. Nat Methods. 2014;11:935–937. doi: 10.1038/nmeth.3046. [DOI] [PubMed] [Google Scholar]
- 77.Sifrim A, et al. eXtasy: variant prioritization by genomic data fusion. Nat Methods. 2013;10:1083–1084. doi: 10.1038/nmeth.2656. [DOI] [PubMed] [Google Scholar]
- 78.Yang H, Robinson PN, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods. 2015;12:841–843. doi: 10.1038/nmeth.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.James RA, et al. A visual and curatorial approach to clinical variant prioritization and disease gene discovery in genome-wide diagnostics. Genome Med. 2016;8:13. doi: 10.1186/s13073-016-0261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singleton MV, et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet. 2014;94:599–610. doi: 10.1016/j.ajhg.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robinson PN, et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014;24:340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brownstein CA, et al. An international effort towards developing standards for best practices in analysis, interpretation and reporting of clinical genome sequencing results in the CLARITY Challenge. Genome Biol. 2014;15:R53. doi: 10.1186/gb-2014-15-3-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wallis Y, et al. Practice guidelines for the evaluation of pathogenicity and the reporting of sequence variants in clinical molecular genetics. ACGS. 2013 http://www.acgs.uk.com/media/774853/evaluation_and_reporting_of_sequence_variants_bpgs_june_2013_-_finalpdf.pdf.
- 84.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. This paper provides the methodology with which to use the various lines of evidence for consistent variant interpretation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Association for Clinical Genetic Science. Consensus statement on adoption of American College of Medical Genetics and Genomics (ACMG) guidelines for sequence variant classification and interpretation. ACGS. 2016 http://www.acgs.uk.com/media/1032817/acgs_consensus_statement_on_adoption_of_acmg_guidelines__1_.pdf.
- 86.den Dunnen JT, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 87.Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rehm HL, et al. ClinGen — the Clinical Genome Resource. N Engl J Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.MacArthur DG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ponting CP, Hardison RC. What fraction of the human genome is functional? Genome Res. 2011;21:1769–1776. doi: 10.1101/gr.116814.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smedley D, et al. A whole-genome analysis framework for effective identification of pathogenic regulatory variants in Mendelian disease. Am J Hum Genet. 2016;99:595–606. doi: 10.1016/j.ajhg.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang YF, Gulko B, Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet. 2017;49:618–624. doi: 10.1038/ng.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hunt RC, Simhadri VL, Iandoli M, Sauna ZE, Kimchi-Sarfaty C. Exposing synonymous mutations. Trends Genet. 2014;30:308–321. doi: 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 97.Willig LK, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med. 2015;3:377–387. doi: 10.1016/S2213-2600(15)00139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu N, et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wieczorek D, et al. Compound heterozygosity of low-frequency promoter deletions and rare loss-of-function mutations in TXNL4A causes Burn–McKeown syndrome. Am J Hum Genet. 2014;95:698–707. doi: 10.1016/j.ajhg.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Redin C, et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet. 2017;49:36–45. doi: 10.1038/ng.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Merker J, et al. Long-read whole genome sequencing identifies causal structural variation in a Mendelian disease. Genet Med. 2017 doi: 10.1038/gim.2017.86. http://dx.doi.org/10.1038/gim.2017.86. [DOI] [PMC free article] [PubMed]
- 102.Brandler WM, et al. Frequency and complexity of de novo structural mutation in autism. Am J Hum Genet. 2016;98:667–679. doi: 10.1016/j.ajhg.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Church DM, et al. Extending reference assembly models. Genome Biol. 2015;16:13. doi: 10.1186/s13059-015-0587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jäger M, et al. Alternate-locus aware variant calling in whole genome sequencing. Genome Med. 2016;8:130. doi: 10.1186/s13073-016-0383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harrison SM, et al. Using ClinVar as a resource to support variant interpretation. Curr Protoc Hum Genet. 2016;89:8.16.1–8.16.23. doi: 10.1002/0471142905.hg0816s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ackerman JP, et al. The promise and peril of precision medicine: phenotyping still matters most. Mayo Clin Proc. 2016;91:1606–1616. doi: 10.1016/j.mayocp.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dorfman R, et al. Do common in silico tools predict the clinical consequences of amino-acid substitutions in the CFTR gene? Clin Genet. 2010;77:464–473. doi: 10.1111/j.1399-0004.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 108.Global Alliance for Genomics and Health. GENOMICS. A federated ecosystem for sharing genomic clinical data. Science. 2016;352:1278–1280. doi: 10.1126/science.aaf6162. [DOI] [PubMed] [Google Scholar]
- 109.Krawczak M, et al. Human gene mutation database-a biomedical information and research resource. Hum Mutat. 2000;15:45–51. doi: 10.1002/(SICI)1098-1004(200001)15:1<45::AID-HUMU10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 110.Samuels ME, Rouleau GA. The case for locus-specific databases. Nat Rev Genet. 2011;12:378–379. doi: 10.1038/nrg3011. [DOI] [PubMed] [Google Scholar]
- 111.Rath A, et al. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 112.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 113.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013:7.20.1, 7.20.41. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shihab HA, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ioannidis NM, et al. REVEL: an Ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]