Abstract
The H3K4 demethylase KDM5B is amplified and overexpressed in luminal breast cancer, suggesting it might constitute a potential cancer therapy target. Here we characterize, in breast cancer cells, the molecular effects of a recently developed small-molecule inhibitor of the KDM5 family of proteins (KDM5i), either alone or in combination with the DNA demethylating agent 5-aza-2′-deoxycytidine (DAC). KDM5i treatment alone increased expression of a small number of genes, whereas combined treatment with DAC enhanced the effects of the latter for increasing expression of hundreds of DAC-responsive genes. ChIP-seq studies revealed that KDM5i resulted in the broadening of existing H3K4me3 peaks. Furthermore, cells treated with the drug combination exhibited increased promoter and gene body H3K4me3 occupancy at DAC-responsive genes compared to DAC alone. Importantly, treatment with either DAC or DAC+KDM5i induced a dramatic increase in H3K27ac at enhancers with an associated significant increase in target gene expression, suggesting a previously unappreciated effect of DAC on transcriptional regulation. KDM5i synergized with DAC to reduce the viability of luminal breast cancer cells in in-vitro assays. Our study provides the first look into the molecular effects of a novel KDM5i compound and suggests that combinatorial inhibition along with DAC represents a new area to explore in translational epigenetics.
Introduction
Post-translational modification of histone tails serves a critical role in regulating chromatin structure and gene regulation. Canonically, methylation of histone H3 lysine 4 (H3K4) has been associated with transcriptional activation including enrichment of monomethylation of H3K4 (H3K4me1) at primed and active enhancers and dimethylation and trimethylation (H3K4me2 and H3K4me3, respectively) at active gene promoter regions (1). H3K4 methylation is dynamically regulated by histone methyltransferases and demethylases (KDM’s) with the latter including the flavin adenine dinucleotide (FAD) dependent polyamine oxidases LSD1 and LSD2, and the Jumonji (JmJc) domain containing 2-oxoglutarate (2-OG) dependent oxygenases KDM5 (Jarid1) family (2,3). Whereas FAD-dependent KDM’s can only remove H3K4me1 and H3K4me2, the four homologous KDM5 proteins (KDM5A-D) also catalyze the removal of H3K4me3 (3–5). Importantly, KDM5A and KDM5B are overexpressed or amplified in a number of human cancers and have been described as critical regulators of tumorigenesis (4,6), making these proteins ideal candidates for targeted inhibition with small molecules.
Although several specific small molecule inhibitors have been developed for the FAD-dependent KDMs, inhibitors specific to JmJc domain containing KDM’s have only recently begun to emerge. A majority of these latter compounds competitively inhibit 2-OG binding in the JmJc domain (7,8). However, since this active site is strongly conserved in more than 30 distinct demethylases, many of these inhibitors lack specificity for a given family member (9,10). Recently, a pan-KDM5 inhibitor, CPI-455, that specifically targets the active site of this family was developed and shown to have 200-fold selectivity for KDM5A over the closely related KDM4C, and more than 500 fold selectivity against other JmJc domain containing proteins (11). Although this compound was unsuitable for in-vivo studies due to low bioavailability, Vinogradova et al. demonstrated that this compound could specifically increase H3K4 methylation in a number of cancer cell lines in-vitro (11). Importantly, the molecular and epigenomic effects of this compound, such as gene expression changes and H3K4 localization patterns, have yet to be elucidated.
Here, we characterize the molecular and epigenomic effects of CPI-455 alone, and in combination with the well-characterized DNA methyltransferase (DNMT) inhibitor, 5-aza-2′-deoxycytidine (DAC). DAC is a cytosine analog that irreversibly binds, inhibits the catalytic activity of, and degrades the three active DNMTs, thereby resulting in the passive loss of DNA methylation throughout the genome (12). This drug, and its congener, 5-azacytidine (AZA), induces re-expression of silenced genes with abnormal promoter, CpG island (CpGi) DNA methylation in cancer, some being important tumor suppressor genes (12–14). These drugs are FDA approved for treatment of the pre-leukemic syndrome, myelodysplastic syndrome (15). Importantly, these actions of DAC and AZA are accompanied, at promoters of the above genes, by reductions of the repressive histone modification H3K9me2 (16–18) and increases in the active marks H3K4me2 and H3K4me3 (16,19,20). We have investigated whether increases in levels of H3K4me3 mediated by KDM5 inhibition (KDM5i) might further reorganize chromatin architecture, thereby enhancing the effects of DAC when these compounds are administered together. We outline, through analyses of genome wide expression & DNA methylation patterns, global H3K4me3 occupancy, and enhancer activities, how combination of the above drugs augments the transcriptional activating effects of DAC at hundreds of loci. These dynamics are associated with increases in H3K4me3 occupancy in targeted gene promoters and gene bodies. Furthermore, we observed a dramatic increase in enhancer activity following DAC treatment that was associated with significantly increased expression of target genes with unmethylated promoters, suggesting a previously unappreciated role for DAC. Finally, KDM5i and DAC synergistically inhibited the growth of three different luminal breast cancer cell lines in in-vitro assays. Our study suggests that DAC+KDM5i is worthy of further development and research as a novel paradigm in translational epigenetics.
Materials and Methods
KDM5 Inhibitors
Two KDM5 inhibitor compounds, CPI-203 and CPI-455 (11), were obtained from Constellation Pharmaceuticals. Both compounds were dissolved in dimethyl sulfoxide (DMSO).
Cell Lines
MCF-7, T-47D, and MCF10A cell lines were purchased from the American Type Culture Collection (ATCC). EFM-19 was obtained from Dr. Cynthia Zahnow at The Johns Hopkins University School of Medicine. All cell lines were negative for mycoplasma contamination as determined by testing with Lonza Mycoalert Mycoplasma Detection Kit (#LT07-418). Cell line authentication was not conducted. All experiments were conducted within 2 months of thawing frozen stocks. MCF-7 was cultured in MEM with 10% fetal bovine serum (FBS), T-47D and EFM-19 were grown in RPMI 1640 with 10% FBS, and MCF10A was cultured with MEGM Bulletkit (Lonza #CC-3150) with 100ng/mL cholera toxin. All cell lines were incubated at 37°C, 5% CO2 atmosphere.
Drug Treatments
Due to low bioavailabilty, for KDM5i treatment in breast cancer cell lines, cells were cultured with base media plus 2% FBS. Drug vehicle (DMSO) was used as a treatment “mock” control. For combination treatments, cells were incubated with DAC (Sigma Aldrich) or vehicle control (PBS) in base media plus 10% FBS for 72 hours; drug media was replaced every 24 hours. Next, DAC media was replaced with KDM5i or vehicle control (DMSO) containing media plus 2% FBS. KDM5 inhibitor dose was a constant ratio of 150:1 relative to DAC. For MCF10A, since no serum is added to the base medium, KDM5i was administered in complete medium. For viability studies, media was replaced with fresh drug media at days 3 and 7 of KDM5i treatment. Viability was measured at day 10 for cancer cell lines or day 7 for MCF10A of KDM5i treatment with Promega Cell Titer Aqueous Non-Radioactive Cell Proliferation Assay (MTS) (#G541). Data shown represents mean and standard error from three independent replicates. Isobolograms were generated in CompuSyn software.
Western Blots and Antibodies
Protein extraction, quantitation, and western blotting were performed as previously described (21). For western blotting, primary antibodies included: anti-trimethyl H3K4 (Cell Signaling #9751S), anti-dimethyl H3K4 (Millipore #07-030), anti-monomethyl H3K4 (Abcam #ab8895), anti-trimethyl H3K27 (Millipore #07-449), anti-dimethyl H3K27 (Upstate #07-452), anti-dimethyl H3K9 (Millipore #07-212), anti-dimethyl H3K36 (Upstate #07-274), and anti-H3 (Abcam #ab1791). Secondary antibodies included: anti-Mouse IgG-HRP (GE #NA931V) and anti-Rabbit IgG-HRP (GE #NA934V). ChIP antibodies included: anti-trimethyl H3K4 (Millipore #07-473) and anti-acetyl H3K27 (abcam #ab4729).
Breast Cancer Cell Line Screen
Cell lines and screening platform was conducted as per Haverty et al. (22). Cell viability was assessed in an 8-day assay with compound added on day 2 and 5.
Histone Purification and Mass Spectrometry
Core histones were purified, prepared for mass spectrometry, and analyzed as previously described (11,23).
Global Expression Analysis
MCF-7 cells were treated with as described above and harvested after 3 days of exposure to KDM5i. Whole genome expression analysis of three independent replicates was assayed on the Agilent Human 4×44k v2 expression array as previously described (24,25). Rank ordered gene set enrichment analysis (GSEA) and PANTHER Pathway gene ontology analysis were performed as previously described (26,27).
Quantitative PCR
Local expression analysis was performed as previously described by our group (24). Primers utilized are listed in Supplementary Table 1.
DNA Methylation Arrays
Genomic DNA was extracted according to the protocol described above was extracted and hybridized to the Illumina Infinium HumanMethylation 450 array according to manufacturer’s instructions. Methylation analysis was performed as previously described (28).
ChIP-Seq
MCF-7 cells were treated with DAC, KDM5i, or both as described above and harvested on day 3 of KDM5i treatment. For ChIP and ChIP-Seq, DNA was isolated as described and sequenced on Illumina Hi-Seq as previously described (21), and mapped to hg19 in Bowtie2. Next, since KDM5i increases H3K4me3 levels, we performed quantile normalization to control for sequencing variation between samples. We measured sequencing depth in adjacent 25bp bins with bamCoverage in deepTools2 (29). Differences in read counts between samples were normalized with normalizeQuantiles in the Limma R package. ChIP samples were normalized to respective inputs with bigwigCompare in deepTools2. Peaks were called using MACS2 bdgcallpeak in the Galaxy platform (cutoff= 2.0, min size= 1500, max gap=300) (30,31). Differential ChIP-seq enrichment was measured using a custom formula. First we measured differential read count per bin (Diff. read count/bin) with the following formula: (Normalized Reads Counts per bin of Treatment ChIP-Normalized Reads Counts per bin of Treatment Input) - (Normalized Reads Counts per bin on Control ChIP - Normalized Reads Counts per bin of Control Input). To filter out background noise, all bins with Diff. read count/bin smaller than the median of all bins were assigned a value of 0. Nearby (max gap=100) positive or negative bins were grouped together, creating an “observed differential area”. To find areas with statistically significant differential enrichment, we then calculated an “expected differential area” value as an area with the same size as the observed differential area but assigned the median Diff. read count/bin value for each bin. Chi-square statistics where used to compare the reads counts for the observed differential area vs. expected differential area. Finally, we filtered out all differentially enriched regions <500 bp in size and with a Bonferoni corrected p-value >0.01. All customized algorithms are available on github.com/Baylin-Easwaran-Labs.
Enhancer Analysis
MCF-7 enhancers were identified via ChromHMM as previously described (32). To control for the effect of promoter H3K4me3 or H3K27ac signal, all enhancers within 5kb of any TSS were removed. Super-enhancers were identified with a custom algorithm similar to ROSE (33) (github.com/Baylin-Easwaran-Labs) to accommodate quantile-normalized files described above. Target genes were identified with RNA Pol II ChIA-PET data for interactions of enhancer regions with gene promoters. Interactions observed in less than 2 of 4 replicates were discarded. Enhancers with differential ChIP enrichment were defined as any enhancer with a significant observed differential area as described above.
Flow Cytometry
For apoptosis assays, 2×105 cells were harvested at the indicated times and then stained with Annexin V-FITC and Propidium Iodide according to manufacturer’s protocol. For cell cycle analysis, 2×105 cells were harvested at the indicated times and stained with Propidium Iodide and RNase A containing buffer according to manufacturer’s protocol. Cells were analyzed on a FACSCalibur flow cytometer with FlowJo software.
Data Sets
MCF-7 KDM5B ChIP-seq and MCF-7 ChromHMM enhancer data was obtained from GEO (GSE46073 and GSE57498, respectively). MCF-7 RNA Pol II ChIA-PET data was obtained from ENCODE (wgENCODEH001430). All sequencing and microarray data generated from this study have been submitted to NCBI GEO under accession number GSE97484.
Results
KDM5i increases global H3K4me3 levels
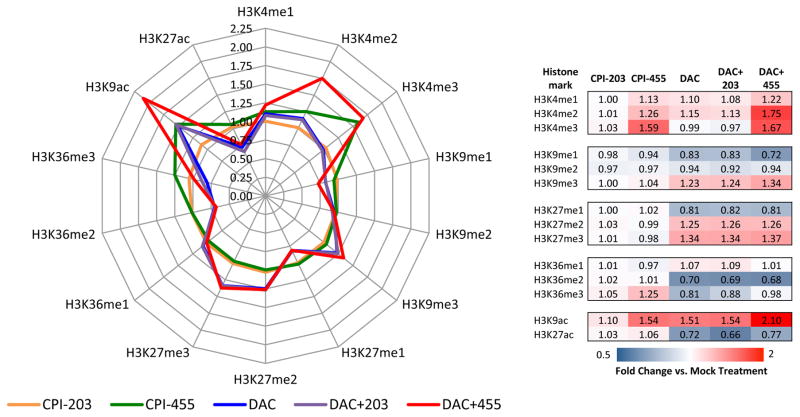
Recent studies (4,6) have highlighted the importance of KDM5B in breast cancer oncogenesis. Accordingly, we screened 36 breast cancer cell lines for sensitivity to CPI-455 (Supp. Table 2). To determine if KDM5i could potentiate the effect of DAC, we chose to investigate the biochemical, molecular, and epigenomic effects of this treatment paradigm in a cell line with relatively modest sensitivity to KDM5i, MCF-7. Exposure to CPI-455, but not the inactive control compound CPI-203, for 72 hours, increased overall cellular H3K4me2 and H3K4me3 levels (Fig. 1) as assayed by quantitative mass spectrometry. This KDM5i mediated increase in H3K4 methylation is similar in magnitude to those of previous studies using siRNA-mediated knockdown of KDM5B (6) or a distinct KDM5 inhibitor (7,34). Interestingly, increases in H3K9ac were also detected, but the mechanism driving these gains is unclear. As determined by immunoblot, H3K4me3 increased in a dose-dependent fashion in response to CPI-455 (Supp. Fig. 1A), but no changes in H3K4me2 could be detected by this method, potentially due to lack of antibody sensitivity to the subtle quantitative change in total abundance of this modification. CPI-455 appears to specifically affect the methylation status of H3K4, as no significant changes in other H3 modifications were detected (Fig. 1; Supp. Fig. 1B). No effect on any of the tested H3 methylation marks was detected after exposure to the inactive compound, CPI-203 (Fig. 1; Supp. Fig. 1).
Figure 1. Changes in histone modification levels following KDM5i alone or in combination with DAC.
Enrichment of various H3 modifications were analyzed by mass spectrometry. Enrichment fold change is shown relative to treatment mock. Left- radar plot of fold enrichment. Right- Table heatmap of enrichment fold change.
Combination treatment with DAC enhanced the effects of KDM5i, as marked increases in H3K4me2 and slight increases in H3K4me3 were detected (Fig. 1). Although DAC alone led to increases in abundance of the repressive histone marks H3K27me3 and H3K9me3, no further changes were mediated by the addition of CPI-455. Interestingly, DAC dramatically affected histone H3 acetylation. H3K9ac increased in response to DAC, with further increases observed after exposure to both compounds, while marked decreases in H3K27ac were also observed (Fig. 1). Validation with immunoblot demonstrated that combination treatment predominantly affected H3K4me3 levels, but lacked the quantitative sensitivity to detect DAC mediated changes in the histone methylation marks tested (Supp. Fig. 1C). Nevertheless, CPI-455 administered alone or in combination with DAC, produces specific, global increases in methylation of H3K4.
KDM5i enhances the effects of DAC on gene expression
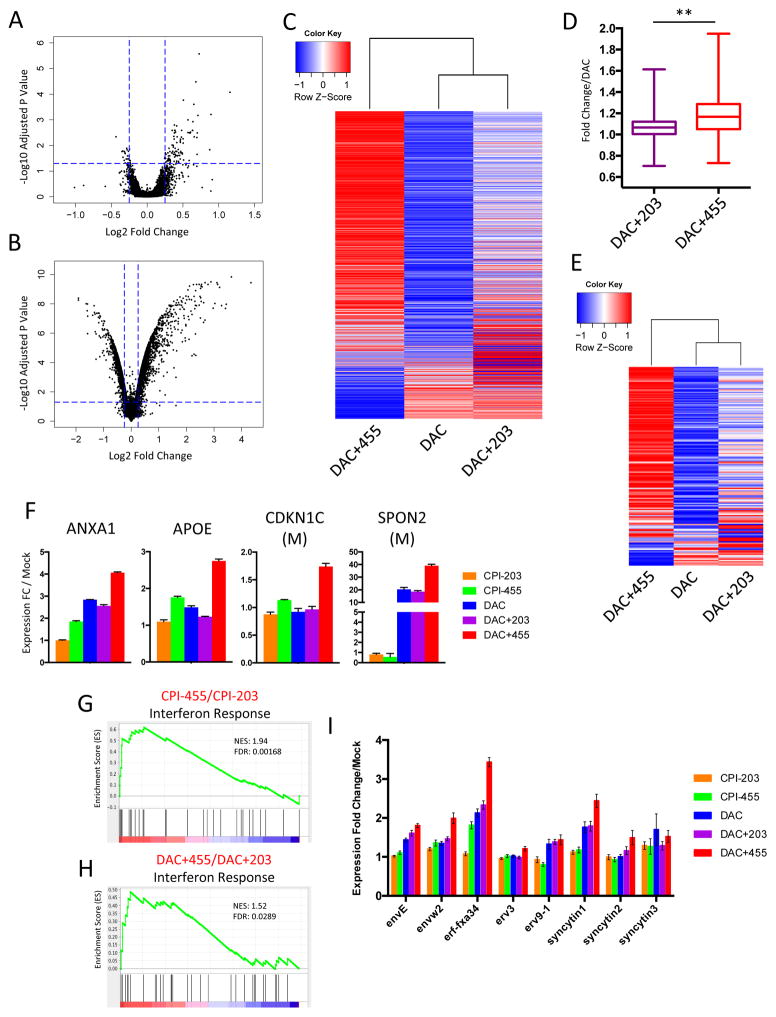
Given that CPI-455 mediated a marked increase in global H3K4 methylation, what are the ramifications for gene expression? Surprisingly, as assessed by Agilent 4×44k v2 microarray, despite our observation that CPI-455 induced global increases in H3K4me2 and H3K4me3 levels (Fig. 1), following 72 hours of exposure to CPI-455, only 94 genes were significantly (adj. p <.05) upregulated and 23 genes were significantly down-regulated relative to cells exposed to CPI-203 (Fig. 2A). Among these upregulated genes, we observed significant enrichment for transcripts upregulated by KDM5B RNAi (Supp. Fig. 2A). Next, we hypothesized that since DAC results in genome-wide decreases in DNA methylation (12) and reorganization of chromatin (16–18), KDM5i might facilitate a further increase in expression of previously silenced genes by driving up levels of the active H3K4me3 mark at selected gene promoters. Indeed, of the approximately 800 genes most upregulated by DAC treatment alone (Fig. 2B), most had further expression increases with subsequently added CPI-455 administration versus the control compound, CPI-203 (Fig. 2C,D; Supp. Fig. 2B–D). Furthermore, when assessing only genes with hypermethylated, CpGi promoters (Supp. Fig. 2E), nearly all were further upregulated by the addition of CPI-455, but not CPI-203 (Fig. 2E). Similar patterns were observed upon validation of the microarray data by qRT-PCR at several loci (Fig. 2F).
Figure 2. Addition of CPI-455 to DAC treatment results in increased expression of DAC regulated genes.
A. Expression changes after exposure to CPI-455 as compared to CPI-203. B. Expression changes after exposure to DAC as compared to mock. C. Expression changes (relative to mock) at all DAC upregulated genes. D. Cumulative fold expression change for genes in (C) relative to DAC for DAC+203 & DAC+455 (**=p<.001, Kolmogorov-Smirnov test). E. Expression changes (relative to mock) at all methylated CpGi DAC upregulated genes. F. Validation of microarray data at panel of 4 genes. Expression for each gene was measured by qRT-PCR. M indicates that the gene has hypermethylated, CpG island containing promoter. G&H. GSEA analysis for Interferon Response gene set (from Li et al. (26)) in either CPI-455 relative to CPI-203 (G) or DAC+455 relative to DAC+203 (H). I. Expression of 8 ERV elements as measured by qRT-PCR.
Importantly, KDM5i enhances the transcriptional activation effects of DAC for genes involved in critical homeostatic and immunomodulatory functions. Our group (24,25,35,36) and others (37) have previously shown that DNA demethylating drugs induce a viral defense response which drives inflammation, interferon response, and chemokine signaling via upregulation of endogenous retroviral elements (ERV’s). Although KDM5i alone significantly increased the expression of relatively few genes, these immunomodulatory pathways, as well as key developmental pathways, were significantly upregulated (Fig. 2G; Supp. Fig. 3A,B). With the addition of CPI-455 to DAC treatment, these pathways, as well as cell death and apoptosis pathways, were significantly further upregulated (Fig. 2H; Supp. Fig. 3A,C,D). Critically, in combination treated cells, this increased viral defense response was associated with distinct upregulation of a number of ERV’s relative to DAC alone (Fig. 2I). These data highlight the concept that epigenetic therapy, including KDM5i, may be able to improve the efficacy of immunotherapy (38).
KDM5i does not affect DNA methylation
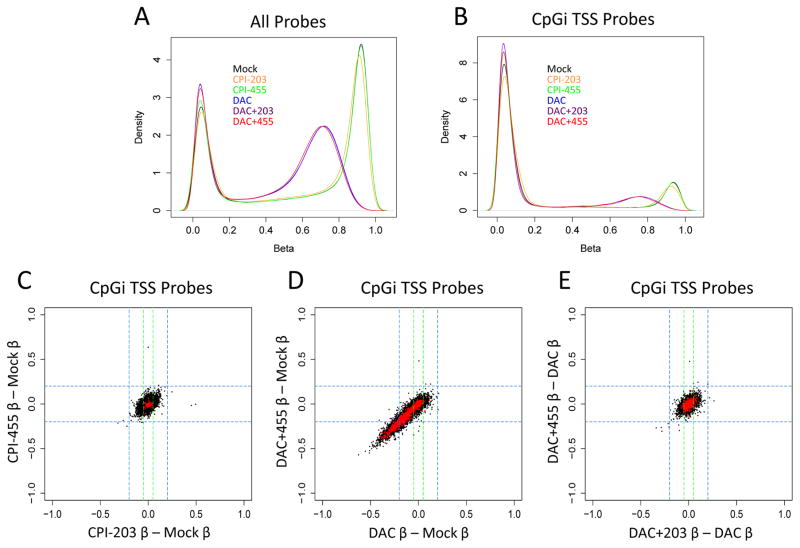
To rule out the possibility that the above increases in gene expression produced by KDM5i are not the result of decreases in DNA methylation, we measured genome-wide methylation levels with the Illumina HumanMethylation450 BeadChip. As expected, DAC, but not CPI-203 or CPI-455, treatment resulted in a substantial drop in total DNA methylation levels both globally, (Fig. 3A) and at CpGi promoters (Fig. 3B). Similarly, adding KDM5i to DAC treatment did not significantly increase the genome-wide DNA de-methylation levels induced by the latter drug (Fig. 3A,B). These same patterns are seen at individual promoters of differentially expressed genes as well; CPI-455 alone did not induce significant changes at either genes differentially expressed by the drug, or any other loci tested (Fig. 3C). DAC and DAC+455, on the other hand, induced significant demethylation at a number of differentially expressed genes (Fig. 3D), but with no additional effects to those seen with DAC alone (Fig. 3E). These findings indicate that KDM5i, either alone or in combination with DAC, has no effect on DNA methylation.
Figure 3. Combination treatment does not result in further DNA demethylation relative to DAC alone.
A&B. Density plot of beta values (an estimate of methylation levels using ratio of methylated probe intensity and overall intensity) for MCF-7 cells exposed to DAC +/− KDM5i using either all probes (A) or only CpGi probes near TSS (B). C. Delta beta values for all CpGi TSS probes (in black) in CPI-455 (y-axis) and CPI-203 (x-axis) relative to mock. Probes corresponding to CPI-455 upregulated genes highlighted in red. D. Delta beta values for all CpGi TSS probes (in black) in DAC+455 (y-axis) and DAC (x-axis) relative to mock. Probes corresponding to DAC upregulated genes highlighted in red. E. Delta beta values for all CpGi TSS probes (in black) in DAC+455 (y-axis) and DAC+203 (x-axis) relative to DAC. Probes corresponding to DAC upregulated genes highlighted in red.
KDM5i expands existing H3K4me3 peaks
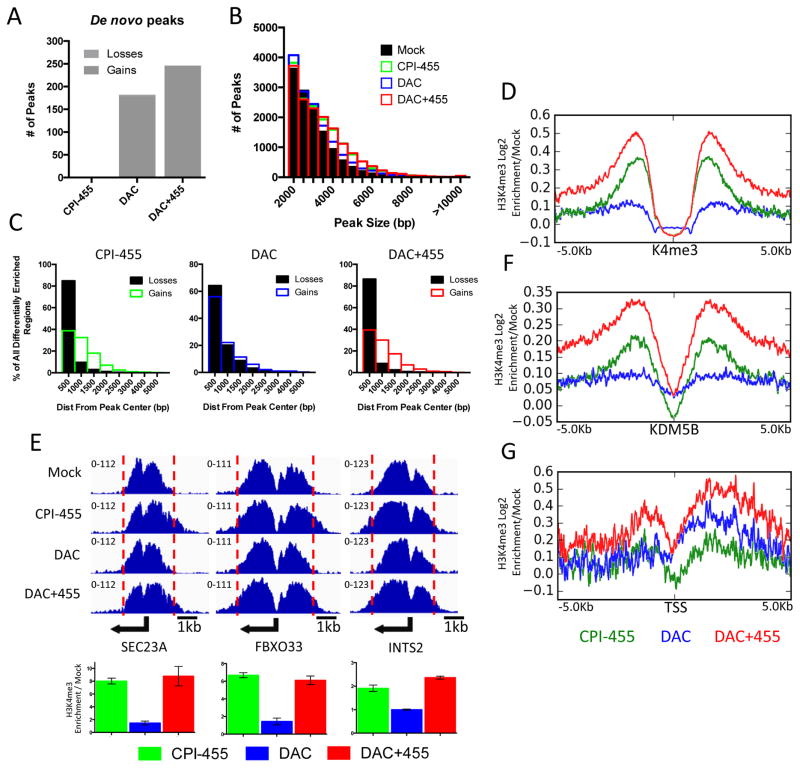
Although it has been shown that CPI-455 increases global H3K4me3 levels (Fig. 1; (11)), we now define, via ChIP-seq analyses, where this occurs throughout the genome in MCF-7 cells and that a subset of KDM5i mediated increases in this active transcription mark may drive the increased expression of DAC responsive genes. KDM5i treatment alone did not establish novel de novo H3K4me3 peaks whereas DAC or DAC+455 treatment resulted in 178 or 245 novel peaks, respectively (Fig. 4A, Supp. Fig. 4A,B). However, consistent with the global increase in H3K4me3 levels, KDM5i treatment alone appears to mediate an expansion of existing H3K4me3 peaks (Fig. 4B), suggesting that KDM5 proteins may play an essential role in delineating the boundaries for regions of H3K4me3 enrichment. Loss of catalytic activity via KDM5i minimizes these boundaries, thereby resulting in the subsequent spreading of the H3K4me3 peak into neighboring regions. In this regard, the increases in size of H3K4me3 peaks after KDM5i alone results in a gain in signal strength in the flanking regions of pre-existing peaks that is often coupled with decreased H3K4me3 signal near the center of these peaks (Fig. 4C,D; Supp. Fig. 4C,D). This pattern was verified at individual promoters by local ChIP-qPCR assays (Fig. 4E). Finally, we found that in KDM5i treated cells, the spreading of H3K4me3 signals extends beyond regions occupied by KDM5B in MCF-7 cells (Fig. 4F).
Figure 4. Treatment with CPI-455 results in spreading of H3K4 trimethylation.
A. Total number of de novo gained or lost H3K4me3 peaks after exposure to CPI-455, DAC, or both. B. Frequency distribution of peak size in each treatment group. C. Bar plot depicting distance of differentially enriched regions from center of aligned peak after treatment with CPI-455 (left panel), DAC (middle panel), or both (right panel). D. Tag density plot of H3K4me3 gains relative to mock at all basally marked H3K4me3 regions in MCF-7. E. H3K4me3 ChIP-seq genome view for 3 genes (top), with qPCR validation (bottom). Error bars indicate SE of three biological replicates. F&G. Tag density plot of H3K4me3 gains relative to mock at KDM5B marked regions in MCF-7 (F), and promoters of all genes upregulated by DAC (G).
What are the consequences of the above KDM5i mediated changes in H3K4me3 occupancies with respect to increases in expression of genes induced by DAC? Compared to DAC alone, we observed a substantial increase in H3K4me3 within the promoters and bodies of genes in cells treated with both DAC and KDM5i (Fig. 4G). KDM5i alone resulted in minimal changes in H3K4me3 levels at these loci (Fig. 4G). Importantly, no changes in H3K4me3 levels were observed at genes with either unaffected or decreased expression following the addition of KDM5i to DAC treated cells (Supp. Fig. 4E). These data suggest that KDM5i mediated increases in H3K4me3 promoter occupancy and this may help drive the increased expression of a subset of DAC responsive genes.
DAC increases enhancer activity
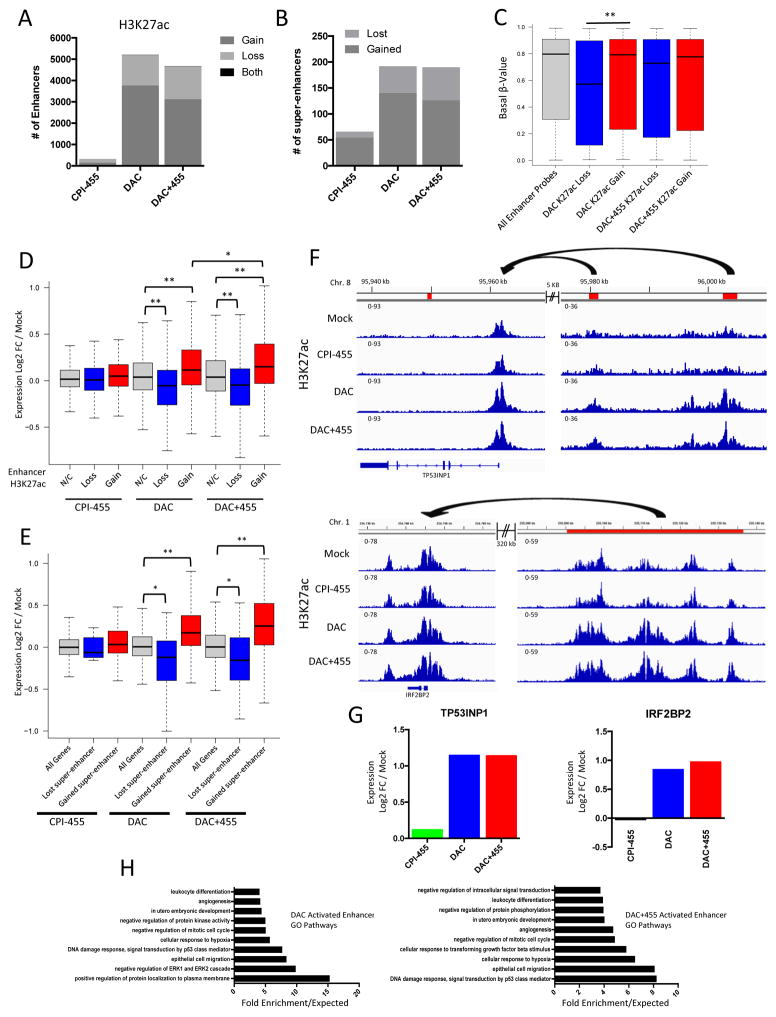
Our genomics approach has uncovered novel ramifications for our drug effects on enhancer and super-enhancer regions which may be closely linked to the observed effects on gene expression patterns. A previous study (39) found that loss of KDM5B in mouse embryonic stem cells (mESC’s) resulted in a misregulation of H3K4me3 and a subsequent decrease in H3K27ac at enhancers, implying a loss of activity. Furthermore, DAC has recently been suggested to have an antagonistic effect on super-enhancer activity in a number of cancer cell lines, though the effect at regular enhancers remains unclear (40). We analyzed H3K4me3 and H3K27ac localization at candidate enhancers (as identified by ChromHMM) in MCF-7 via ChIP-seq (32,41). Although DAC alone had a relatively small effect on enhancer H3K4me3, KDM5i treatment, either alone or in combination with DAC, resulted in significant gains in H3K4me3 levels (Supp. Fig. 5A). Unlike the pattern observed in a previous study (39), minimal changes in enhancer H3K27ac levels were observed after KDM5i alone (Fig. 5A). In contrast to its effects on global levels of H3K27ac, DAC treatment, either alone or with KDM5i, resulted in a dramatic enrichment of H3K27ac with significant gains in this mark at thousands of enhancers, though some losses of this mark from these regions were observed as well (Fig. 5A; Supp. Fig. 5B,C). Changes in enhancer H3K27ac enrichment were predominantly mutually exclusive of gains or losses in H3K4me3 (Supp. Fig. 5D,E). Furthermore, DAC alone or in combination with KDM5i led to the establishment of many novel super-enhancer domains (Fig. 5B; Supp. Fig. 5F,G). These data suggest an inverse relationship between DNA methylation and H3K27ac; compared to enhancers with H3K27ac losses, gains in this mark occurred at enhancers with higher levels of basal DNA methylation in MCF-7 (Fig. 5C) and accompanied losses of DNA methylation at these loci following DAC treatment (Supp. Fig. 5H).
Figure 5. DAC treatment increases enhancer activity.
A. Number of enhancers with H3K27ac gains, losses, or both after exposure to CPI-455, DAC, or DAC+455. B. Number of super-enhancers gained or lost after each treatment as compared to treatment mock. C. Basal methylation levels of enhancers with significant differential H3K27ac enrichment after DAC or DAC+455 (** = p<.01, Kolmogorov-Smirnov test). D. Effects of enhancer H3K27ac gains or losses on target gene expression (non-CpGi and unmethylated CpGi genes only) after exposure to CPI-455, DAC, or DAC+455. Enhancer/promoter interactions determined via Encode RNA Pol II ChIA-PET in MCF-7 (* = p<.05, ** = p<.01, unpaired t-test with Welch’s correction). E. Effects of gained or lost super-enhancers on target gene expression (non-CpGi and unmethylated CpGi genes only) after exposure to CPI-455, DAC, or DAC+455. Super-enhancer/promoter interactions determined via Encode RNA Pol II ChIA-PET in MCF-7 (* = p<.05, ** = p<.01, unpaired t-test with Welch’s correction). F. Genome views of H3K27ac enrichment after Mock, CPI-455, DAC, or DAC+455 treatment at TP53INP1 with upstream enhancers (top) and IRF2BP2 with upstream super-enhancer (bottom). Enhancers or super-enhancers indicated by red boxes, RNA Pol II ChIA-PET validated interaction indicated by arrows. G. Expression changes for TP53INP1 and IRF2BP2 after treatment with CPI-455, DAC, or DAC+455. H. Panther GO overrepresentation analysis of upregulated (log2FC/mock ≥0.25) gene targets of enhancers that gain H3K27ac after DAC (left) or DAC+455 (right) treatment; shown are the top 10 most overrepresented pathways following each drug treatment.
Using ENCODE RNA Pol II ChIA-PET data to stringently identify candidate gene promoter targets of enhancers, we find potentially significant effects for enhancer function alterations induced by our drug treatments. To control for the DNA demethylating effects of DAC, we excluded all genes containing promoters with hypermethylated CpG islands. First, the minimal changes in enhancer H3K4me3 or H3K27ac induced by KDM5i were not associated with significant changes in target gene expression (Fig. 5D, Supp. Fig. 5I). Second, after treatment with DAC or DAC plus KDM5i, H3K27ac gains at standard and super-enhancers were associated with significant increases in expression of target genes (Fig. 5D,E). Targeted genes include TP53INP1 (Fig. 5F,G), a tumor suppressor gene for which loss of function hinders p53 activity and causes increased aggressiveness of breast and pancreatic cancers (42,43) and IRF2BP2 (Fig. 5F,G), a target gene of p53 that mediates cell growth arrest during cell stress and genotoxic insults (44). These types of changes are reflected in alterations to key signaling pathways and biological processes relevant to the anti-tumorigenic effects of DAC (36,45) including: upregulation of pathways related to immune response, cellular differentiation, and development as well as down-regulation of cell cycling control and kinase signaling (Fig. 5H). Finally, losses in enhancer H3K27ac after these treatments were associated with a significant decrease in expression of target genes (Fig. 5D). These findings indicate that although the effects of KDM5i on enhancer activity are minimal, DAC significantly induces changes in enhancer activity by dramatically altering H3K27ac localization at these loci, thereby elucidating a previously unappreciated action of this compound that may significantly contribute to the anti-tumorigenic effects of this drug.
DAC+KDM5i synergistically inhibit cell growth
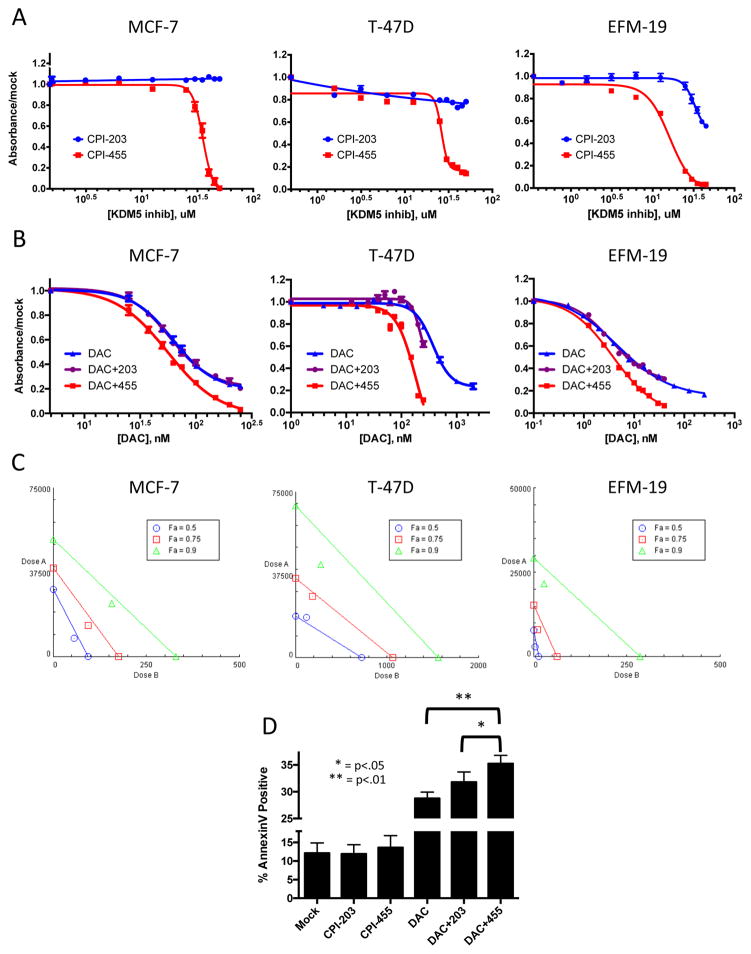
Given the above effects for CPI-455, alone and in combination with DAC, to increase apoptosis signaling, it is important to determine the effects of our drug combination on cell viability. We found that there are significant effects on the behavior of cancer cell lines that occur concomitantly with all of the molecular changes characterized in the preceding sections. Only very high doses (>20 uM) of CPI-455 alone affected cell viability (Fig. 6A) in the three luminal breast cancer cell lines, MCF-7, T-47D, and EFM-19. The control drug, CPI-203, had no effect except at very high doses in EFM-19. Lower CPI-455 doses, which increase H3K4me3 levels, had no significant effects on viability (Fig. 6A; Supp. Fig. 1). When combined with DAC, treatment with CPI-455, but not control drug CPI-203, led to significant decreases in viability in the three luminal cancer cell lines (Fig. 6B). These interactions were pharmacologically synergistic at all tested effect levels in MCF-7 and EFM-19, and at higher effect levels in T-47D (Fig. 6C). This synergistic inhibition of growth was not associated with changes in cell cycle (Supp. Fig. 6A), but rather appears to be mediated by increased induction of apoptosis in DAC plus CPI-455 treated cells versus those treated with DAC alone or DAC plus CPI-203 (Fig. 6D). Interestingly, the non-tumorigenic, but immortalized mammary epithelial cell line, MCF10A, was far more sensitive to CPI-455 than the above cancer cell lines (Supp. Tab. 2; Supp. Fig. 6B), but the mechanism driving this sensitivity is unclear. However, as opposed to the results for the cancer cell lines, no synergistic interaction was detected (Supp. Fig. 6C,D).
Figure 6. CPI-455 and DAC synergize to inhibit cell growth in 3 luminal breast cancer cell lines.
A&B. Viability dose response curves for 10 day exposure to CPI-203 or CPI-455 alone (A), or combination treatment with 3 days DAC (dosed at 1:150 ratio with KDM5i) followed by 10 days KDM5i (B) in MCF-7, T-47D, and EFM-19. C. Isobolograms generated from combination viability data in MCF-7, T-47D, and EFM-19 in (B). D. Annexin V binding in MCF-7 as determined by flow cytometry.
Discussion
The pan-KDM5 inhibitor, CPI-455, alone and in combination with DAC, is found to increase global levels of H3K4me3 and leads to the expansion of existing H3K4me3 peaks throughout the genome (Fig. 4). Importantly, the expansions occur at genomic regions where KDM5B is bound (Fig. 4F), indicating that this protein may be responsible for maintaining some of the borders of transcriptionally active regions. Key aspects of our findings thus fit those of others including data for another small molecule inhibitor of KDM5 (KDM5-C70) (7,34) which suggested a functional role for KDM5 proteins in regulating H3K4 methylation dynamics. Similarly, knockdown of KMD5B in MCF-7 led to subtle global increases in the H3K4me3/H3K4me2 ratio, and in mESC’s, KDM5B knockdown led to the spreading of H3K4me3 peaks into gene bodies (6,39). All of these above findings, including our present work, indicate that CPI-455 specifically and potently inhibits KDM5 proteins, to account for the alterations in H3K4me3 levels we now report.
Although CPI-455 alone leads to significant gains in H3K4me3 at thousands of gene promoters, only modest changes are induced for global gene expression patterns. Knockdown of KDM5B resulted in a similar dynamic (6). Importantly though, we observed significant enrichment for genes upregulated by CPI-455 alone and genes upregulated by KDM5B RNAi (6) (Supp. Fig. 2A), indicating that these specific target genes are regulated by the catalytic activity of this protein. As discussed by Yamamoto et al., since KDM5B commonly localizes near the promoters of actively transcribed genes, our data further support the hypothesis that this family of proteins are not master regulators of transcription, but rather function in fine-tuning transcription of target loci. Furthermore, although H3K4me3 is associated with active transcription, isolated gains in this mark are insufficient to increase expression at most genes. We propose that a small subset of genes may be directly regulated for transcription by KDM5 catalytic activity, but at most genes, H3K4me3 gains must coincide with changes in other epigenetic marks as well as regulatory input from signal transduction for gene reactivation to occur.
One of the most important findings in our study is that the above dynamics for CPI-455 may enhance the transcriptional activating effects of DAC. At most genes with upregulated expression after DAC, the addition of KDM5i led to significant expression increases (Fig. 2) including at ERV’s and genes in AZA-inducible immunomodulatory pathways (Fig. 2; Supp. Fig. 3). Our group recently demonstrated that DNMT inhibitors leads to the reactivation of ERV’s, induction of the interferon response, and subsequent sensitization to immune checkpoint therapy in a pre-clinical melanoma model (36). These data indicate that combination therapy with DAC and KDM5i might further sensitize to immune checkpoint blockade, and further studies are merited to verify translational potential of such a combination.
We found that genes with increased expression following the addition of CPI-455 to DAC treated cells were associated with increased promoter and gene body H3K4me3 occupancy after combination drug treatment (Fig. 2, Fig. 4G). Since CPI-455 alone produced neither expression of associated genes nor H3K4me3 increases at these loci, the increased H3K4me3 appears to be specifically mediated by the combinatorial actions of these two compounds together to further increase the transcriptional activity of these genes. As discussed above, we propose that focal increases in H3K4me3 are insufficient to drive expression increases independent of other epigenetic changes. Following DAC treatment, DNA methylation and repressive chromatin marks are lost from promoters (12,16). Although further study is needed, these data suggest that combinatorial administration of DAC and KDM5i further remodels the cancer epigenome, thereby facilitating greater changes to genome wide expression patterns than either drug alone.
The findings that DAC potentially exerts a profound effect on enhancer activity represents a major and previously underappreciated role for this compound in transcriptional regulation. Previous studies have suggested a potential role for KDM5B and DNA methylation in regulating enhancer and super-enhancer activity. In contrast to a previous study of the loss of KDM5B in mESC’s via RNAi (39), we now find that KDM5i treatment alone led to minimal changes in H3K27ac enrichment at enhancers (Fig. 5). This likely reflects the inherent differences in epigenome plasticity between these distinct cell types. Conversely, DAC robustly alters enhancer H3K27ac enrichment. Despite decreasing global H3K27ac levels (Fig. 1), DAC mediated significant gains in this activating mark at thousands of putative enhancers and established over one hundred novel super-enhancers (Fig. 5; Supp. Fig. 5). These perturbations were linked to significant changes in candidate target gene expression (Fig. 5). Among these upregulated enhancer targets was a significant overrepresentation for genes involved in pathways relevant to the anti-tumorigenic effects of DAC (Fig. 5H), suggesting that enhancer activation may be a critical mechanism of action for this drug.
The action of DAC on enhancer elements might partially explain the counterintuitive observation that a number of unmethylated CpG island containing promoter genes are re-expressed following DAC treatment (Supp. Fig. 2E), although epistatic pathway interactions likely contribute to this observation as well. To our knowledge, only one study regarding the role of DAC at distal regulatory elements has been published (40). The authors found that DNMT inhibition led to decreased activity of super-enhancers in MCF-7. Although we too observed a loss of some of these elements following DAC treatment, the predominant effect of this compound was the gain of super-enhancers (Fig. 5; Supp. Fig. 5). Given the profound increase in H3K27ac we observed at thousands of enhancers, our data suggest that DAC treatment dramatically reorganizes enhancer chromatin, potentially leading to increased activity at thousands of these elements. Gains in H3K27ac were biased to enhancers with high levels of DNA methylation, suggesting that, as other groups have proposed (46–48), DNA methylation is inversely correlated with enhancer activity. Pharmacological removal of DNA methylation with DAC relieves this repression, likely allowing the recruitment of activating complexes and subsequent activation of these regulatory elements. Furthermore, future studies should investigate the ability of DAC to establish de novo enhancer elements.
Our results indicate that the combinatorial action of DAC and KDM5i has a small, but significant effect on cellular viability, which may be a key phenotypic correlate of the molecular changes defined throughout our study. Whereas KDM5i alone did not reduce viability at doses which mediated global H3K4me3 increases (Fig. 1; Fig. 4; Fig. 6), we found that when combined with DAC, these compounds could synergistically reduce net cell growth (Fig. 6). This reduction appears to be driven by increased apoptosis. Although the exact mechanism remains unclear, our observation of increased activation of apoptosis and death receptor signaling pathways in cells treated with both compounds (Supp. Fig. 3D) further supports this hypothesis and suggests that the KDM5i mediated expression increases of DAC responsive genes plays a critical role in mediating this phenotype. Future studies with more potent KDM5 inhibitors should address if this synergistic interaction can be further enhanced as well as validate the in-vivo efficacy of this treatment paradigm.
In the context of another well-studied epigenetic drug combination, our group (35,49) and others (50) have previously demonstrated that DNMT and HDAC inhibition synergistically reactivate silenced genes and reduce cancer cell viability. Similarly, here we show that KDM5i in combination with DAC enhances the reactivation of hypermethylated genes and synergistically inhibits cell growth, though neither effect was as potent as the DNMT and HDAC inhibitor combination. These data suggest that histone acetylation exerts a more dominant effect on transcriptional regulation and, as discussed above, KDM5 proteins and H3K4 methylation likely function to fine-tune gene expression. Nevertheless, our findings suggest that combined inhibition of DNMT’s and KDM5 proteins represents an exciting new avenue of investigation in translational epigenetics.
Supplementary Material
Acknowledgments
These studies were supported by research grants from the Memorial Sloan-Kettering Cancer Center sub-grant through the Department of Defense Breast Cancer Research Program (Award No. W81XWH-13-1-0199) (S.B. Baylin), The Hodson Trust (S.B. Baylin), The Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (S.B. Baylin), and the Evelyn Grollman Glick Young Scholar Award (H. Easwaran).
We thank the Genentech cell line screening facility for the breast cancer cell line screen of CPI-455 and CPI-203 and Kathy Bender for manuscript preparation and submission.
Footnotes
Conflict of Interest Statement: C.W., T.K.C., D.A., and M.C. are employees of Genentech Inc. and may hold Roche stock. P.T. is an employee and shareholder of Constellation Pharmaceuticals Inc.
References
- 1.Calo E, Wysocka J. Modification of Enhancer Chromatin: What, How, and Why? Mol Cell. 2013:825–37. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black JC, Van Rechem C, Whetstine JR. Mol Cell [Internet] Vol. 48. Elsevier Inc; 2012. Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact; pp. 491–507. Available from: http://dx.doi.org/10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, et al. The Retinoblastoma Binding Protein RBP2 Is an H3K4 Demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, et al. PLU-1 Is an H3K4 Demethylase Involved in Transcriptional Repression and Breast Cancer Cell Proliferation. Mol Cell. 2007;25:801–12. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Christensen J, Agger K, Cloos PAC, Pasini D, Rose S, Sennels L, et al. RBP2 Belongs to a Family of Demethylases, Specific for Tri-and Dimethylated Lysine 4 on Histone 3. Cell. 2007;128:1063–76. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto S, Wu Z, Russnes HG, Takagi S, Peluffo G, Vaske C, et al. Cancer Cell [Internet] Vol. 25. Elsevier Inc; 2014. JARID1B is a luminal lineage-driving oncogene in breast cancer; pp. 762–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol [Internet]. Nature Research. 2016;12:539–45. doi: 10.1038/nchembio.2087. [cited 2016 Aug 22] [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen PB, Staller P. Epigenomics [Internet] Vol. 6. Future Medicine Ltd; London, UK: 2014. The KDM5 family of histone demethylases as targets in oncology drug discovery; pp. 277–86. [cited 2016 Aug 18] [DOI] [PubMed] [Google Scholar]
- 9.Heinemann B, Nielsen JM, Hudlebusch HR, Lees MJ, Larsen DV, Boesen T, et al. Inhibition of demethylases by GSK-J1/J4. Nature [Internet] Nature Research. 2014;514:E1–2. doi: 10.1038/nature13688. [cited 2016 Aug 29] [DOI] [PubMed] [Google Scholar]
- 10.Sayegh J, Cao J, Zou MR, Morales A, Blair LP, Norcia M, et al. Identification of small molecule inhibitors of Jumonji AT-rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high throughput screen. J Biol Chem [Internet] American Society for Biochemistry and Molecular Biology. 2013;288:9408–17. doi: 10.1074/jbc.M112.419861. [cited 2016 Aug 29] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vinogradova M, Gehling VS, Gustafson A, Arora S, Tindell CA, Wilson C, et al. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat Chem Biol [Internet] Nature Research. 2016;12:531–8. doi: 10.1038/nchembio.2085. [cited 2016 Aug 18] [DOI] [PubMed] [Google Scholar]
- 12.Baylin SB, Jones PA. Nat Rev Cancer [Internet] Vol. 11. Nature Publishing Group; 2011. A decade of exploring the cancer epigenome - biological and translational implications; pp. 726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman JGBS. Gene silencing in cancer in association with promoter hypermethylation. N Eng J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The Epigenomics of Cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egger G, Liang G, Aparicio A, Jones PA. Nature [Internet] Vol. 429. Nature Publishing Group; 2004. Epigenetics in human disease and prospects for epigenetic therapy; pp. 457–63. [cited 2016 Aug 29] [DOI] [PubMed] [Google Scholar]
- 16.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Cancer Res [Internet] Vol. 66. American Association for Cancer Research; 2006. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state; pp. 3541–9. [cited 2016 Aug 29] [DOI] [PubMed] [Google Scholar]
- 17.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JCY, Liang G, et al. Cancer Res [Internet] Vol. 62. American Association for Cancer Research; 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine; pp. 6456–61. [cited 2016 Aug 29] [PubMed] [Google Scholar]
- 18.Komashko VM, Farnham PJ. 5-Azacytidine Treatment Reorganizes Genomic Histone Modification Patterns. Epigenetics. 2010;5:229–40. doi: 10.4161/epi.5.3.11409. [DOI] [PubMed] [Google Scholar]
- 19.McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, et al. Cancer Res [Internet] Vol. 68. American Association for Cancer Research; 2008. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells; pp. 5753–9. [cited 2016 Aug 29] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fahrner JA, Eguchi S, Herman JG, Baylin SB. Cancer Res [Internet] Vol. 62. American Association for Cancer Research; 2002. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer; pp. 7213–8. [cited 2016 Aug 29] [PubMed] [Google Scholar]
- 21.O’Hagan HM, Wang W, Sen S, DeStefano Shields C, Lee SS, Zhang YW, et al. Cancer Cell [Internet] Vol. 20. Elsevier Inc; 2011. Oxidative Damage Targets Complexes Containing DNA Methyltransferases, SIRT1, and Polycomb Members to Promoter CpG Islands; pp. 606–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haverty PM, Lin E, Tan J, Yu Y, Lam B, Lianoglou S, et al. Nature [Internet] Vol. 533. Nature Publishing Group; 2016. Analysis Reproducible pharmacogenomic profiling of cancer cell line panels; pp. 333–7.7. [DOI] [PubMed] [Google Scholar]
- 23.Maile TM, Izrael-tomasevic A, Cheung T, Guler GD, Tindell C, Masselot A, et al. Mass Spectrometric Quantification of Histone Post-translational Modifications by a Hybrid Chemical Labeling Method. Mol Cell Proteomics. 2015;14:1148–58. doi: 10.1074/mcp.O114.046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Chiappinelli KB, Guzzetta Aa, Easwaran H, Yen R-WC, Vatapalli R, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget [Internet] 2014;5:587–98. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrangle J, Wang W, Koch A, Easwaran H, Mohammad HP, Vendetti F, et al. Alterations of immune response of Non-Small Cell Lung Cancer with Azacytidine. Oncotarget [Internet] 2013;4:2067–79. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Proc Natl Acad Sci U S A. Vol. 102. United States: 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles; pp. 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. Nucleic Acids Res. Vol. 44. England: 2016. PANTHER version 10: expanded protein families and functions, and analysis tools; pp. D336–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang YW, Wang Z, Xie W, Cai Y, Xia L, Easwaran H, et al. Mol Cell [Internet] Vol. 65. Elsevier Inc; 2017. Acetylation Enhances TET2 Function in Protecting against Abnormal DNA Methylation during Oxidative Stress; pp. 323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. Nucleic Acids Res. Vol. 42. England: 2014. deepTools: a flexible platform for exploring deep-sequencing data; pp. W187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, et al. Nucleic Acids Res. Vol. 44. England: 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update; pp. W3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu T, Meyer CA, Eeckhoute JJ, Johnson DS, Bernstein BE, et al. Genome Biol [Internet] Vol. 9. England: 2008. Model-based Analysis of ChIP-Seq (MACS) p. R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taberlay PC, Statham AL, Kelly TK, Clark SJ, Jones PA. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 2014;24:1421–32. doi: 10.1101/gr.163485.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Cell [Internet] Vol. 153. Elsevier Inc; 2013. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes; pp. 307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Galen P, Viny AD, Ram O, Ryan RJH, Cotton MJ, Donohue L, et al. A Multiplexed System for Quantitative Comparisons of Chromatin Landscapes. Mol Cell. 2016;61:170–80. doi: 10.1016/j.molcel.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields C, Wenzel A, Hicks J, Ballew M, Stone M, Tran PT, Zahnow CA, Hellmann MD, Strissel PL, Strick RBS. A combination epigenetic therapy ties blocking MYC to reversing immune evasion and treating lung cancer. Cell. 2017;171:1284–1300. doi: 10.1016/j.cell.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–86. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, et al. Cell [Internet] Vol. 162. Elsevier Inc; 2015. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts; pp. 961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiappinelli KB, Zahnow CA, Ahuja N, Bylin SB. Combining epigenetic and immunotherapy to combat cancer. Cancer Res. 2016:1683–9. doi: 10.1158/0008-5472.CAN-15-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kidder BL, Hu G, Zhao K. KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol [Internet] 2014;15:R32. doi: 10.1186/gb-2014-15-2-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlet J, Duymich CE, Lay FD, Mundbjerg K, Dalsgaard Sørensen K, Liang G, et al. Mol Cell [Internet] Vol. 62. Elsevier Inc; 2016. Bivalent Regions of Cytosine Methylation and H3K27 Acetylation Suggest an Active Role for DNA Methylation at Enhancers; pp. 422–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst J, Kellis M. Nat Methods [Internet] Vol. 9. Nature Publishing Group; 2012. ChromHMM: automating chromatin-state discovery and characterization; pp. 215–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gironella M, Seux M, Xie M-J, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A [Internet] 2007;104:16170–5. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C-M, Zhao J, Deng H-Y. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J Biomed Sci [Internet] Journal of Biomedical Science. 2013;20:79. doi: 10.1186/1423-0127-20-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koeppel M, Van Heeringen SJ, Smeenk L, Navis AC, Janssen-Megens EM, Lohrum M. The novel p53 target gene IRF2BP2 participates in cell survival during the p53 stress response. Nucleic Acids Res. 2009;37:322–35. doi: 10.1093/nar/gkn940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, et al. Cancer Cell [Internet] Vol. 21. Elsevier Inc; 2012. Transient Low Doses of DNA-Demethylating Agents Exert Durable Antitumor Effects on Hematological and Epithelial Tumor Cells; pp. 430–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature [Internet] 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 47.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. Nature [Internet] Vol. 489. Nature Publishing Group; 2012. The accessible chromatin landscape of the human genome; pp. 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu W, Briones V, Lister R, McIntosh C, Han Y, Lee EY, et al. CG hypomethylation in Lsh−/− mouse embryonic fibroblasts is associated with de novo H3K4me1 formation and altered cellular plasticity. Proc Natl Acad Sci U S A [Internet] 2014;111:5890–5. doi: 10.1073/pnas.1320945111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 50.Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. 2002;2:485–95. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.