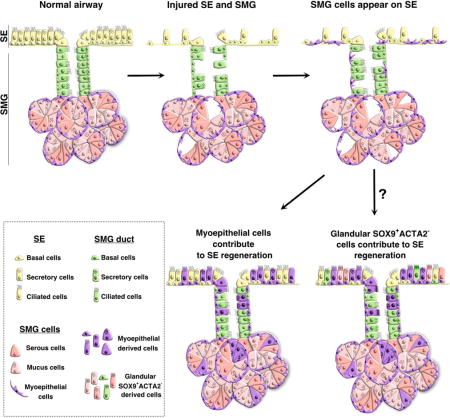
Summary
Cells demonstrate plasticity following injury, but the extent of this phenomenon and the cellular mechanisms involved remain underexplored. Using single cell RNA sequencing (scRNA-seq) and lineage tracing, we uncover that myoepithelial cells (MECs) of the submucosal glands (SMGs) proliferate and migrate to repopulate the airway surface epithelium (SE) in multiple injury models. Specifically, SMG-derived cells display multipotency and contribute to basal and luminal cell types of the SMGs and SE. Ex vivo expanded MECs have the potential to repopulate and differentiate into SE cells when grafted onto denuded airway scaffolds. Significantly, we find that SMG-like cells appear on the SE of both extra- and intra-lobular airways of large animal lungs following severe injury. We find that the transcription factor SOX9 is necessary for MEC plasticity in airway regeneration. Since SMGs are abundant and present deep within airways, they may serve as a reserve cell source for enhancing human airway regeneration.
Keywords: Cellular plasticity, Multipotent stem cells, reserve stem cells, Airway regeneration, Single cell RNA sequencing, Myoepithelial cells
eTOC blurb
Airway submucosal glands are reservoirs of mucous and anti-microbial products that provide defense against microbes. Here, Tata and colleagues uncover a hidden reserve multipotent stem cell population in SMGs that can proliferate, migrate, and transdifferentiate to repair surface epithelium following injury through a SOX9 dependent transcriptional program.
Introduction
Tissue homeostasis is fueled by resident stem/progenitor cells or by self-duplication of differentiated cells (Hogan et al., 2014; Visvader and Clevers, 2016). In adult tissues, stem/progenitor cells are located in specialized microenvironments tailored to accommodate the tissue needs (Hsu et al., 2011; Pardo-Saganta et al., 2015a; Scadden, 2014). In most tissues, stem cells and committed mature cells are thought to be irreversible and lineage restricted under homeostatic conditions but recent studies indicate that cells can be experimentally coaxed to exhibit plasticity (Donati et al., 2017; van Es et al., 2012; Rompolas et al., 2013; Stange et al., 2013; Tata and Rajagopal, 2016; Tata et al., 2013; Van Keymeulen et al., 2011). Specifically, in severe injury, committed cells revert to a stem cell state or cells from one region can migrate and display characteristics of an adjacent tissue to quickly replace damaged cells (Blanpain and Fuchs, 2014; Tetteh et al., 2015). For example, lineage tracing studies in skin epidermis revealed that cells from hair follicles and sebaceous glands can migrate and regenerate interfollicular epidermis following injury (Donati et al., 2017; Rompolas et al., 2013). In the small intestine, LGR5+ cells serve as stem cells and normally self-renew and generate all cell types of the villi (Barker et al., 2007). However, recent studies found that cells at the +4 position in the crypt, marked by Bmi1+/Tert+/HopX+, have the ability to generate LGR5+ cells following severe injury to crypts. Such cells have been proposed to serve as “reserve” stem cells for their ability to generate tissue specific resident stem cells after injury. Taken together, these and other results suggest that tissues harbor reserve or facultative stem cells that can act when resident stem cells are lost (Tetteh et al., 2015). Such plasticity mechanisms offer therapeutic avenues for enhancing regeneration, particularly following severe injuries that damage resident stem cells. However, the cellular sources and the mechanisms associated with such plasticity are poorly understood (Blanpain and Fuchs, 2014; Merrell and Stanger, 2016; Rajagopal and Stanger, 2016; Tata and Rajagopal, 2017).
Epithelial tissues such as the airway luminal SE require a rapid response to injury to prevent secondary damage and loss of barrier function (Hogan et al., 2014; Kotton and Morrisey, 2014). Airway epithelium consists of two broad tissue domains: the pseudostratified SE and SMGs. In the SE, lineage tracing analysis has identified multiple region-specific stem/progenitor cells along the proximo-distal axis of the airway. In the mouse trachea and human extra and intralobar airways, the epithelium consists of 3 major cell types: basal stem cells, and columnar luminal secretory and multiciliated cells. The basal cells self-renew and generate secretory club cells and multi-ciliated cells in homeostasis and regeneration (Borthwick et al., 2001; Cole et al., 2010; Franks et al., 2008; Ghosh et al., 2011; Hogan et al., 2014; Hong et al., 2004a, 2004b; Liu et al., 1994; Pardo-Saganta et al., 2015b; Rock et al., 2009). In the SMGs, the epithelial populations consist of distal acini and the proximal collecting ducts that connect them to the SE. Collecting ducts are morphologically similar to the SE and consist of basal and luminal cells while the distal SMGs contain luminal serous and mucous cells and basally located contractile MECs (Crystal et al., 2008; Franks et al., 2008; Hegab et al., 2010, 2011, 2012; Liu and Engelhardt, 2008; Wansleeben et al., 2014). In the human, SMGs are present throughout the cartilaginous airways. In mice, SMGs are primarily located in the proximal part of the trachea spanning the submucosal region between the larynx and cartilage-1 but are occasionally found up to cartilage-3, depending on the mouse strain, size, and age (Borthwick et al., 1999; Liu and Engelhardt, 2008).
At homeostasis, both SE and SMG cells are highly quiescent and display low turnover rates. Following injury, SE basal stem cells and SMG cells quickly respond to proliferate and generate both basal and luminal cells in the respective tissue domains. Intriguingly, a recent cell lineage tracing study, using a Krt14-crePR driver line, has suggested that SMG duct cells contribute to SE following hypoxia induced airway injury (Hegab et al., 2011). However, KRT14 is also expressed in a subset of SE basal cells. Therefore, additional studies using a lineage-tracing approach that is specific to SMG cells are required to definitively establish whether SMG cells can generate SE cells. Furthermore, unique markers that distinguish SMG epithelial cells from SE cells are needed to establish whether the cells from these two compartments interconvert and exhibit plasticity in homeostasis and after injury.
Here, using single cell RNA sequencing followed by stringent lineage tracing and functional assays, we demonstrate that MECs of the SMGs act as reserve stem cells that can proliferate, migrate and repopulate both basal and luminal compartments of the SE and SMGs following multiple injuries, including toxic gas exposure or viral infection. We find that SMG-derived MECs and their progeny lose the characteristics of SMGs and display features of SE cells. Using large animal injury models, we demonstrate that SMG-like cells emerge in pig extra-lobular as well as intra-lobular airway SE following severe injury. Using SOX9 loss of function mouse models, we also demonstrate that this transcription factor (TF) is necessary for MEC proliferation and migration to the SE after injury. Such conservation in cellular plasticity between small and large animal models suggests that SMGs embedded deep within the human airway submucosa likewise harbor a reserve stem cell population that can significantly contribute to repair and regeneration.
Results
Single cell RNA-seq identifies cells expressing SMG markers in SE following Naphthalene induced injury
Recent studies using lineage tracing analysis coupled with injury repair models have identified distinct region-specific cell populations that participate in airway regeneration (Hogan et al., 2014; Tata and Rajagopal, 2017). However, the cellular identities and cell states associated with regenerating tissues are poorly understood. To study airway injury-repair processes, we performed RNA-seq analysis on single cells isolated following naphthalene-induced injury. Naphthalene (naph) has been widely used to study airway injury repair mechanisms. While it is generally considered that secretory cells are the primary target of naphthalene, it has been noted that ciliated cells are also lost to some extent in this injury model (Buckpitt et al., 1995; Cole et al., 2010; Stripp et al., 1995; Van Winkle et al., 1995). Under our experimental conditions, most of the luminal SE cells, not just club cells, were lost within 24 h in the naphthalene treated airways but not in oil treated controls. Overall, about 76% ±1.74 (mean ± SEM) (n=3) of the total (DAPI+) cells were lost in the tracheal SE on day-1 post injury (Figure S1A-B). Immunohistochemistry (IHC) for KRT5 and KRT8, markers of basal and luminal cells respectively, revealed both of these cell types were lost, leaving behind small patches of basal cells (Figure S1C). Our attempts to isolate and recover a significant number of viable SE cells prior to day-5 for scRNA-seq analysis were unsuccessful. We therefore chose day-5 trachea to enzymatically dissociate and collect SE cells from naphthalene treated animals for scRNA-seq analysis. Quantification of total cell number and expression of basal (KRT5) and luminal (KRT8) cell markers indicated that most of the SE cells were recovered by day-5 (Figure S1). To avoid any artifacts in gene expression signatures we proceeded to Drop-seq based scRNA-seq without performing FACS (Figure 1A).
Figure 1. Single cell RNA-seq reveals a population of gland-like cells in the airway SE following naphthalene injury.
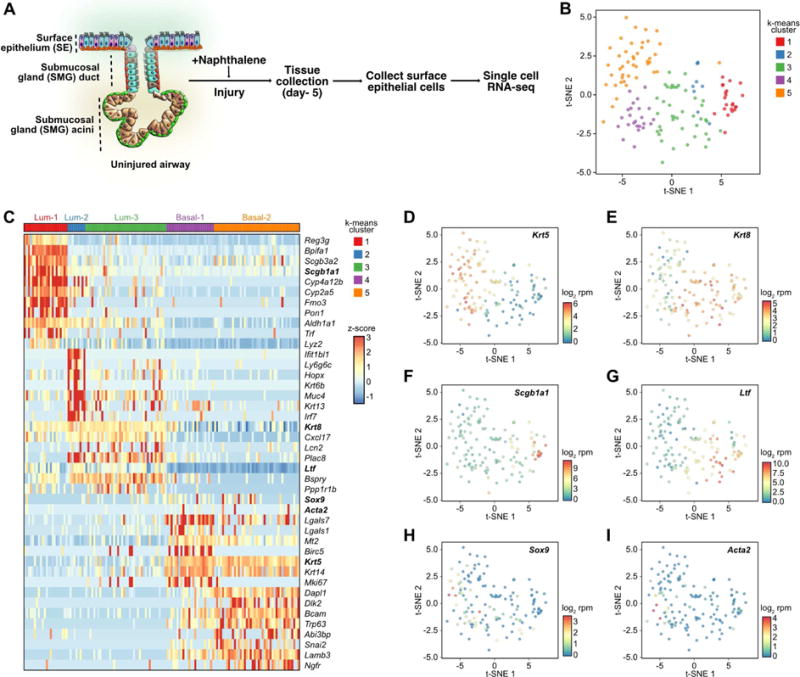
(A) Schematic of experimental design and single cell library preparation.
(B) t-SNE visualization of the single airway epithelial cells after naphthalene injury (n = 139 cells). Cells are color-coded by k-means clusters.
(C) Heat map of genes associated with the 5 clusters (Lum-1, Lum-2, Lum-3, Basal-1, and Basal-2). Values shown in terms of z-scores. Gene names in bold are individually featured in subsequent panels.
(D-I) t-SNE visualization of single airway epithelial cells after naphthalene injury (n = 139 cells). Cells are color coded by normalized expression of the indicated genes: (D) Krt5, (E) Krt8, (F) Scgb1a1, (G) Ltf, (lactoferrin) (H) Sox9, and (I) Acta2.
See also Figure S1.
After excluding non-epithelial cells, the final scRNA-seq dataset was comprised of 139 cells. Across all the single cells analyzed, an average of 22,149 ± 1019 (mean ± SEM) unique reads mapped to the mouse transcriptome. 1489 ± 30.73 (mean ± SEM) unique genes were found to be expressed (log2 reads per million mapped, rpm ≥ 1) in individual cells. We performed k-means clustering and visualized the data by t-distributed stochastic neighbor embedding (t-SNE) to reveal potential underlying structure in the data (Maaten, 2014). Five distinct clusters were identified across the 139 single cells (Figure 1B). Differential expression analysis revealed the key genes associated with each of the 5 clusters (Figure 1C). Clusters 1, 2, and 3 were determined to be different subtypes of luminal cells on the basis of high Krt8 expression, a universal marker of luminal epithelial cells in diverse columnar epithelial tissues (Figure 1E). Of the three luminal subtypes, Lum-1 most closely resembled normal airway secretory cells, as these cells were marked by Scgb1a1, Lyz, and Scgb3a2 expression (Figure 1F). These cells also highly expressed cytochrome p450 genes including Cyp4a12b and Cyp2a5, further indicating that these are secretory cells of the airways. Lum-2 cells were characterized by expression of inflammation-related genes Ifit1bl1 and Irf7. On the other hand, Lum-3 cells were not only defined by Ppp1r1b, Bspry, and Lcn2 expression, but also expressed Ltf (Figure 1G), which encodes the lactoferrin protein characteristic of serous cells of the mouse SMGs (Figure 1G). Interestingly, 5/41 (12.2%) of Lum-3 cells expressed Sox9, which encodes a transcription factor known to be highly and specifically expressed in developing SMGs (Figure 1H) and other glandular tissues (Anderson et al., 2017). Based on their distinct expression profiles, Lum-3 cells do not appear to correspond to a luminal cell type that is normally present in the uninjured airway epithelium.
Clusters 4 and 5 expressed high levels of basal cell markers including Krt5, Krt14, and Trp63 (also known as Tp63) (Figure 1D) and were therefore termed Basal-1 and Basal-2. Basal-1 expressed high levels of Lgals7, Lgals1, Birc5, as well as Mki67, which is a marker of proliferation. In contrast, Basal-2 was defined by Snai2, Lamb3, Ngfr, Dlk2, and Dapl1. Intriguingly, a fraction of the total basal cell population expressed Sox9 (17/67 cells or 25.4%) (Figure 1H) and Acta2 (also known as alpha-smooth muscle actin) (7/67 cells, or 10.4%) (Figure 1I). Of note, SOX9 and ACTA2 protein are not normally expressed in the SE but are known to be expressed in developing SMGs (SOX9) or in myoepithelial cells of the SMGs (ACTA2). Collectively, scRNA-seq revealed the presence of subpopulations of cells expressing canonical SMG markers in both the luminal and basal compartments of the airway SE 5 days following naphthalene-induced injury.
Airway luminal SE harbors cells expressing SMG cell markers following naphthalene-induced injury
To validate the scRNA-seq findings, we administered naphthalene or corn oil to mice and collected trachea on day-1, 3, and 5 for whole mount and section IHC. In controls KRT5 expression was observed in basal cells of the SE and SMGs, while ACTA2 was clearly confined to SMG myoepithelial cells (Figure 2A and B). Notably, SOX9 was expressed in both luminal and basal layers of the SMG cells but not in the SE, indicating that SOX9 serves a unique pan-SMG marker distinguishing SMGs from SE cells. In naphthalene-treated airways, significant changes were seen following injury. In particular, we observed stellate shaped cells co-expressing ACTA2, TP63 and KRT14, markers characteristic of myoepithelial cells of the SMGs, in regions around the gland pits where SMG ducts are connected to the SE (Figure 2A and Figure S1D). Significantly, we also observed SOX9 in SE cells of the naphthalene administered airways but not in corn oil treated controls. To temporally map these changes, we performed IHC analysis for ACTA2 in combination with SOX9 and Ki67, a marker of proliferating cells, at different times following injury. We then counted the ACTA2, SOX9, and Ki67 expressing cells on the SE on day-1, 3, and 5 in different regions along the proximo-distal axis. On day-3 post injury most of the markers are found between cartilage 0-3 (14.1% ± 2.74 ACTA2+ cells in naph (n=3) vs 0.06% ± 0.06 ACTA2+ cells in controls (n=3); 57.08% ± 4.07 SOX9+ cells in naph (n=4) vs 0.19% ± 0.12 SOX9+ cells in controls (n=3); 74.77% ± 4.49 Ki67+ cells in naph (n=4) vs 7.53% ± 0.63 Ki67+ cells in controls (n=3), (mean ± SEM)). At later times, the distribution of ACTA2+ and SOX9+ cells extended to cartilage 3-6 and 6-9, suggesting that the SMG-like cells migrate from proximal to distal regions of the airways. Interestingly, a sizeable number of the ACTA2+ (76.87% ± 3.94) and SOX9+ (77.32% ± 3.69) cells also expressed Ki67, indicating that SMG-like cells on the SE are highly proliferative (Figure 2E). Consistent with previous studies, naphthalene also caused significant damage to the SMG tissue, as shown by loss of mucous and serous cells (Liu and Engelhardt, 2008) (Figure S1D). Interestingly, on day-1, we noticed more proliferating cells in SMGs than in SE (Figure S1D). Taken together, our data suggest that cells expressing SMG cell markers first appear on the SE following naphthalene-induced airway injury and that these cells are highly proliferative and migratory.
Figure 2. Airway luminal SE harbors cells expressing SMG cell markers following naphthalene-induced injury.
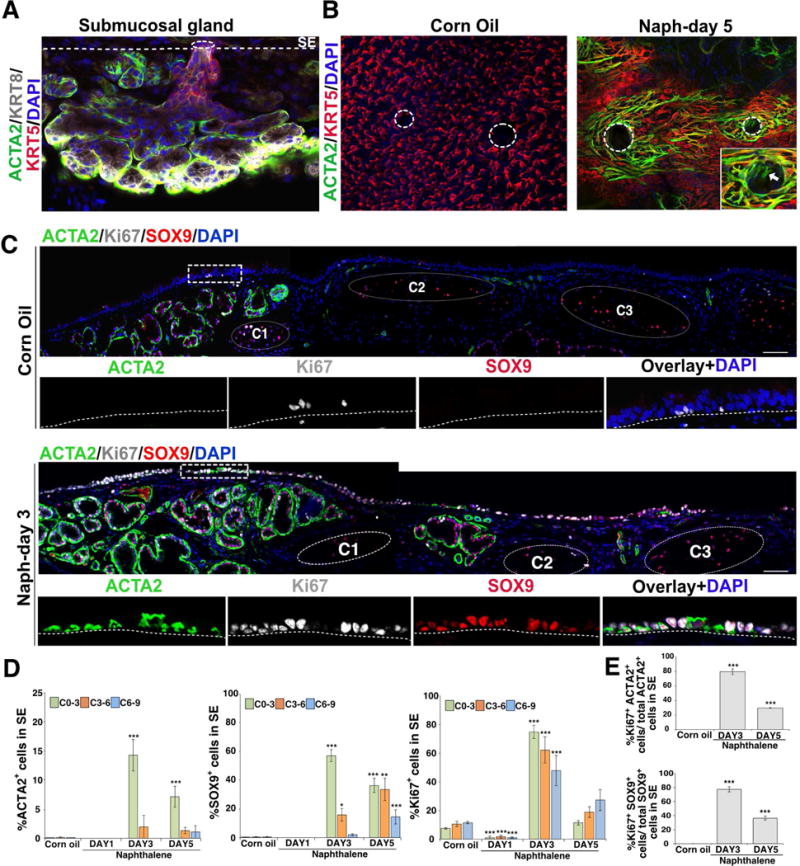
(A) Whole mount IHC of SMG for myoepithelial cell marker ACTA2 (green), basal cell marker KRT5 (red), luminal cell marker KRT8 (grey), and nuclear DAPI (blue). The dotted line indicates border between SMG and SE. Scale bar: 50μm.
(B) Whole mount IHC staining of tracheal SE in corn oil or naphthalene (naph) treated mice at day-5 after injury. ACTA2 (green), KRT5 (red), DAPI (blue). Dotted circles indicate gland pore opening. Higher magnification image of the gland pore is shown in inset right bottom. Scale bars: 50μm, n=3.
(C) Large panels: Tracheal sections from naphthalene exposed (day-3) or control (corn oil) mice stained for ACTA2 (green), SOX9 (red) and Ki67 proliferation marker (grey). Small panels: Single markers shown for regions delineated by dotted box. Dotted circles indicate cartilage rings marked as C1-C3. Scale bar: 50μm.
(D) Quantification of ACTA2+, SOX9+ and Ki67+ cells among total DAPI+ cells in SE in control and naph injured mice. Data are shown as mean ± SEM (n≥3 mice; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001). C0-3, C3-6, and C6-9 refers to SE spanning cartilage rings between 0-3 or cartilage 3-6 or cartilage 6-9, respectively.
(E) Quantification of ACTA2+, SOX9+ co-expressing Ki67+ cells in SE of control and day-3 and day-5 post naph injury. Data shown as mean ± SEM (n=3; ***p ≤ 0.001). p-values were determined compared to controls.
See also Figure S1 and Figure S2.
SMG-like cells appear on SE in multiple injury models
Different classes of injury models such as Sulphur dioxide (SO2) and chlorine-inhalation, influenza infection, and hypoxia, have been used to study repair mechanisms in trachea where SE basal stem cells were thought to serve as progenitor cells to replenish lost epithelium (Rock et al., 2011; Tata et al., 2013). To test whether the plasticity mechanisms observed in naphthalene-induced injury model occur in other injury models, we subjected wild type C57BL/6 mice to SO2 or mouse adapted H1N1 (PR8) infection-induced injury, both known to cause severe damage to luminal cells of the SE within 48 h post exposure, leaving behind few KRT5+ basal cells (Figure S2A). We collected tissues on day-5 post injury and performed IHC for SOX9 and ACTA2 (Figure S2) and found that SMG-like cells appeared in SE of SO2 and PR8 infected mice. However, fewer SOX9+ ACTA2+ cells were observed in SE of SO2 and PR8 infected airways compared to naphthalene injured SE. Of note, we observed a significant difference in the degree of injury in SO2 and PR8 infected airway SE compared to naphthalene injury models (Figure S2B-C). To determine whether a correlation exists between the number of SMG-like cells and degree of injury, we performed a similar quantification analysis as for naphthalene injury. We observed significantly fewer SMG cell markers on airway SE on day-3 post SO2 (1.19%± 0.62 SOX9+ cells and 0.09%± 0.09 ACTA2+ cells; n=3) or PR8 -induced (3.05%± 1.12 SOX9+ and 1.31% ± 0.80 ACTA2+ cells; n=3) injury compared to naph injury (57.08% ± 4.07 SOX9+ and 14.1% ± 2.74 ACTA2+ cells; n=3, (mean ± SEM)) (Figure 2D and Figure S2D-I). This data suggests that the degree of damage influences the extent of cell plasticity in different injury models (O’Koren et al., 2013). Nevertheless, a significant number of ACTA2+ (37.36% ± 8.96 in SO2 and 42.33% ± 5.76 in PR8 infection) and SOX9+ (44.17% ± 10.44 in SO2 and 47.54% ± 5.20 in PR8 infection) cells co-expressed Ki67, indicating that the SMG-like cells which emerge onto the SE are again highly proliferative (Figure S2F and I). Taken together, our data suggests that SMG-derived cells contribute to SE in multiple injury models.
SMG derived Sox9-lineage labeled cells repair SE and contribute to both luminal and basal cells following injury
To test the hypothesis that cells of the SMG contribute to SE repair, we bred a mouse line, Sox9-creER;R26-LSL-tdTomato (hereafter referred to as Sox9-tdt), in which tamoxifen administration specifically and permanently induces the expression of a red fluorescent protein, tdTomato (tdt), in Sox9-expressing cells with 93% efficiency. We observed efficient labeling of both basal and luminal cells of the SMG ducts and acini but not the SE (Figure 3A-B, and Figure S3A). Of note, Sox9-tdt lineage labeled cells do not contribute to SE even after a 10-month chase, indicating that SMGs do not normally contribute to SE lineages at homeostasis (Figure S3B). Thus, the Sox9-tdt model faithfully marks SMG cells and allows us to follow the fate of these cells following injury.
Figure 3. SMG derived Sox9-lineage labeled cells repair SE and contribute to both luminal and basal cells following injury.
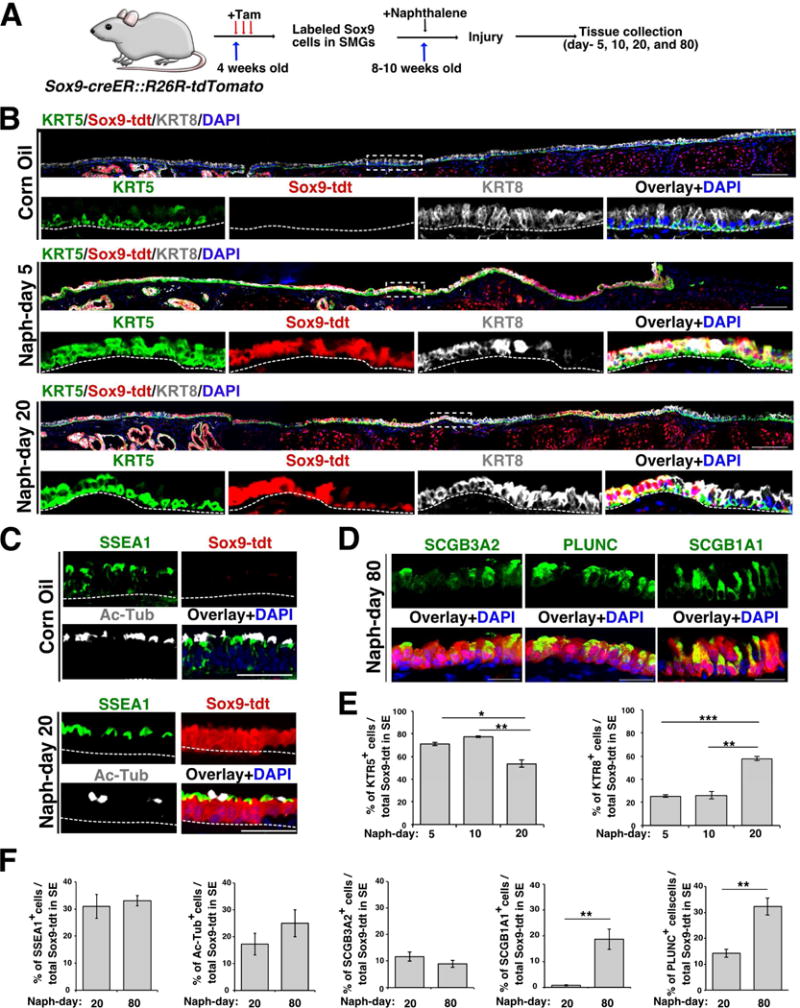
(A) Schematic of Sox9-tdt lineage trace, naphthalene injury and tissue collection.
(B) Co-staining of KRT5 (green) and KRT8 (grey) on tracheal sections collected from control and injured mice (day-5 and 20). Scale bar: 50μm. Dotted box indicates region shown at high magnification in panels below. The dotted line separates SE and mesenchyme. Scale bar: 50μm.
(C) Tracheal sections from Sox9-tdt naphthalene injured (day-20) or control mice stained for SSEA1 – secretory cell marker (green), AcTub (acetylated tubulin) – ciliated cell marker. Sox9-tdt (red) indicates Sox9-lineage labeled cells, nuclear DAPI (blue). Scale bar: 50μm.
(D) Staining of secretory cell markers: PLUNC, SCGB1A1 and SCGB3A2 (green) on day-80 naphthalene injured Sox9-tdt (red) mice. Nuclear DAPI (blue). Scale bar: 20μm.
(E) Quantification of KRT5+ and KRT8+ cells among total Sox9-tdt+ cells in SE on day-5, 10, and 20 post naphthalene injury. Data are shown as mean ± SEM (n=3; *p ≤ 0.05; ** p ≤ 0.01; ***p ≤ 0.001).
(F) Quantification of SSEA1+, Ac-Tub+, SCGB3A2+, SCGB1A1+, PLUNC+ cells among total Sox9-tdt+ cells in SE on day-20, and 80 post naphthalene injury. Data are shown as mean ± SEM (day 20: n=3, day 80: n=4 for all markers except SCGB1A1 (n=3); *p ≤0.05; **p ≤ 0.01; ***p ≤ 0.001).
See also Figure S3.
To prevent any residual tamoxifen from acting in response to injury, we administered tamoxifen at least 4-6 weeks prior to naphthalene-induced injury and tissues were collected on day-5, 10, and 20 post injury for lineage tracing analysis. We identified large patches of lineage labeled Sox9-tdt cells in the SE in injured airways but not in control animals (Figure 3B). Quantification of tdt+ cells in the SE showed 67.7% of the total SE cells were derived from Sox9-tdt lineage labeled SMG cells by day-20 following injury. We found that the lineage labeled cells extend more distally over time (up to cartilage-4 on day-5 and cartilage-9 on day-20) on SE, consistent with Sox9-tdt cells being a migratory population (Figure 3B). Interestingly, lineage labeled SMG-derived tdt+ cells express markers of basal (KRT5) and luminal cells (KRT8) and contribute to acetylated tubulin expressing (Ac-Tub+) ciliated cells and SSEA1+ secretory cells (Figure 3B-D and Figure S3C). However, we did not find SCGB1A1 expressing Sox9-tdt cells on day-20 post naphthalene injury. To test whether club cells require longer to express other markers (SCGB1A1, SCGB3A2, and BPIFA1 (also known as PLUNC)) we isolated airways from Sox9-tdt mice on day-80 post naphthalene injury. Marker analysis revealed Sox9-tdt cells expressing mature club cell markers (Figure 3D) on day-80 SE. Significantly, the proportion of Sox9-tdt+ KRT5+ expressing cells gradually declines over time (70.8%±1.5 on day-5 vs 77.6%±0.8 on day-10 vs 53.7%±3.4 on day-20, (mean ± SEM)) while the proportion of KRT8+ lineage labeled cells increases (25.5%±1.2 on day-5 vs 25.9%±3.1 on day-10 (n=3) vs 57.8%±1.7 on day-20 (n=3)) (mean ± SEM) (Figure 3E). We also observed a similar trend in the number of SCGB1A1 (1.29% ±0.23 on day 20 (n=3) vs 22.94% ±3.95 on day-80 (n=3)), PLUNC (14.28% ±1.52 on day-20 (n=3) vs 32.29% ±3.31 on day-80 (n=4)) and Ac-Tub (17.26% ±4.04 on day-20 (n=3) vs 25.01% ±5.07 on day-80 (n=4), (mean ± SEM)) expressing Sox9-tdt cells on the airway SE (Figure 3F). A model emerges in which Sox9-lineage labeled cells are initially KRT5+ in the SMGs and proliferate, migrate, differentiate, and mature into SE luminal cells following injury (Figure 3). One of the characteristic functions of airway SE is to establish a polarized barrier, with distinct apical junctional complexes. We therefore performed IHC for apical and basolateral polarity proteins such as ZO1 and CDH1. Confocal microscopy revealed SMG-derived Sox9-tdt lineage traced cells expressing ZO1 with a pattern similar to that seen in controls. Consistent with recent reports, we observed apical localization of ZO1 in early stages of regeneration where the epithelium is still composed of a single cell layer of cuboidal cells (Gao et al., 2015). The barrier therefore appears to be maintained at all times analyzed (Figure S3D). Together, our data demonstrates that SMG-derived Sox9-lineage labeled cells repair and contribute to SE basal and luminal cells and establish airway barrier following injury (Figure 3 and Figure S3).
SMG-derived Acta2-lineage labeled MECs repair the SE and contribute to both basal and luminal cells following injury
To test whether the Sox9-tdt cells that emerge from SMG onto the SE following naphthalene-induced injury are derived from ACTA2+ MECs, we bred Acta2-creER; R26R-LSL-tdTomato mice (Acta2-tdt). Following tamoxifen administration, we observed tdt expression in 75% of the ACTA2 and TP63-expressing cells of the acini of SMGs but not in SE or in SMG ducts (Figure 4 and Figure S4). We then performed lineage tracing in control and injured animals and harvesting trachea on day-5, day-12, day-20, and day-60 post injury for IHC (Figure 4). We observed large patches of tdt+ cells in SE of the naphthalene injured airways but not in controls (Figure 4B) and 42.6% of the total SE cells were derived from Acta2-tdt lineage labeled SMG cells on day-20 post injury. Moreover, the distribution of Acta2-tdt cells moved distally from day-5 to day-12 post injury (Figure 4B and data not shown). Similar to Sox9-tdt lineage, the Acta2-tdt lineage contributed to both basal and luminal cells and progressed from the KRT5+ basal cell state (78.2% ± 5.3 on day-5 vs 63.9% ± 4.8 on day-12 vs 53.48% ± 1.36 on day-20; n=3, (mean ± SEM)) to the KRT8+ luminal state (21.5% ± 5.4 on day-5 vs 36.6% ± 2.2 on day-12 vs 46.51% ± 1.36 on day-20; n=3, (mean ± SEM)) (Figure 4D). In addition, Acta2-tdt cells also expressed acetylated Tubulin and SSEA1, markers of ciliated and secretory cells, respectively, on day-20 post injury (Figure 4C and D). Again, we observed SCGB1A1 expression in Acta2-tdt-derived SE cells only at late stages (Figure 4E). Taken together, our data demonstrate that ACTA2+ MECs contribute to repair and regenerate SE following injury (Figure 4).
Figure 4. SMG-derived Acta2-lineage labeled MECs repair the SE and contribute to both basal and luminal cells following injury.
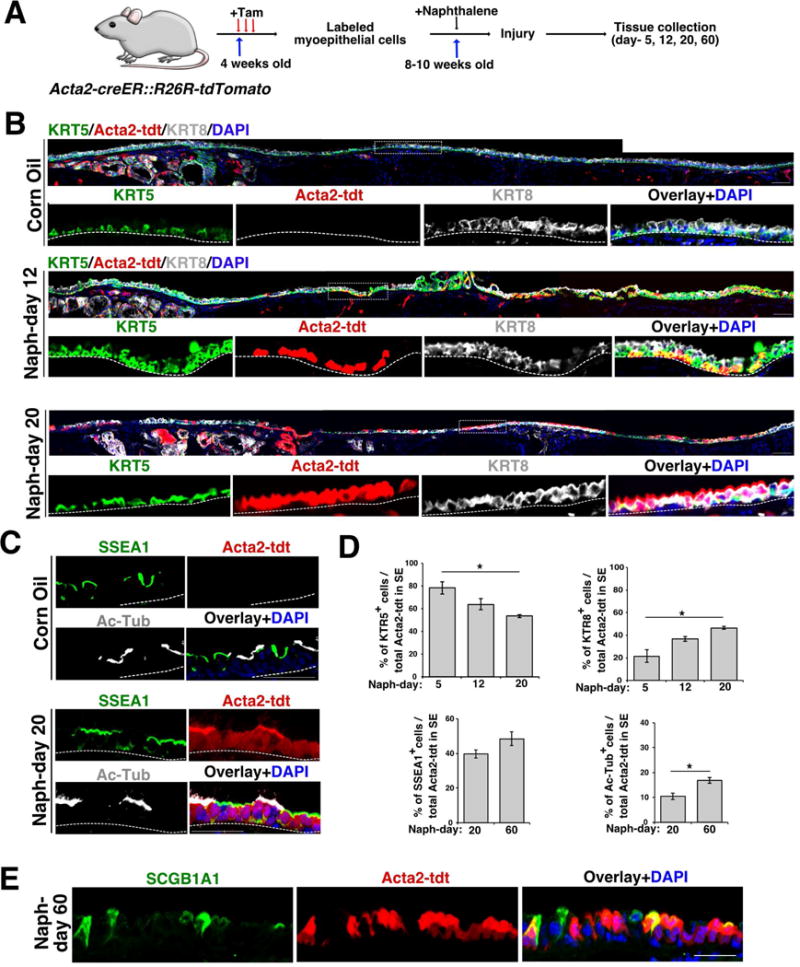
(A) Schematic of experimental design to label and trace Acta2-expressing myoepithelial cells in naphthalene-induced injury model.
(B) Tracheal sections from Acta2-tdt naphthalene injured (day-12 and 20) or control mice stained with KRT5 (green) and KRT8 (grey). Acta2-tdt lineage traced (red) and nuclear DAPI (blue). Scale bar: 50μm. n=3. Dashed line box indicates region of higher magnification shown in lower panel. The dashed line separates the SE and mesenchyme.
(C) Co-staining of SSEA1 (green) and Ac-Tub (grey) in naphthalene (day-20) injured and control Acta2 lineage traced (red) mice. DAPI was used to stain the nuclei (blue). Scale bar: 50μm. The dashed line separates the SE and mesenchyme.
(D) Quantification of KRT5+, KRT8+ (naph day-5, 12, and 20), SSEA1+ and Ac-Tub+ (naph day-20, and 60) cells among total Acta2-tdt+ cells in SE in controls and naphthalene injured mice. Data shown as mean ± SEM (n=3; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001).
(E) IHC for SCGB1A1 (green) on Acta2-tdt (red) airways on day-60 post naphthalene injury. Nuclear DAPI (blue). Scale bar: 20μm.
See also Figure S4.
SMG-derived Acta2-lineage labeled MECs contribute to both basal and luminal cells of the SMGs repair in vivo following injury
Distinct stem/progenitor cells are thought to contribute to distinct compartments in tissue homeostasis and regeneration in other epithelial tissues such as mammary glands (Van Keymeulen et al., 2011). Currently, it is not known whether ACTA2+ basal MECs of the SMGs have the ability to contribute to luminal cells of the SMGs. To test this, we utilized our naphthalene-induced injury model which causes significant damage to luminal cells of the SMGs. In control mice, we found no contribution of Acta2-tdt-lineage labeled cells to luminal cells (Figure S4). By contrast, a significant number of luminal cells were derived from the Acta2-tdt lineage in naphthalene treated airways (18.99% ± 2.65 KRT8+ Acta2-tdt cells in naph vs 0 KRT8+ Acta2-tdt cells in control mice (mean ± SEM); n=3) (Figure S4). Further analysis indicated that MEC-derived tdt+ luminal cells co-expressed LTF (48.53% ± 7.41) or MUC5B (21.92% ± 6.53) ((mean ± SEM); n=3), indicating that MEC-derived cells contribute to both serous and mucous cells, respectively (Figure S4E). Taken together, our data demonstrate that MECs contribute to basal and luminal cells of the SMGs and SEs of trachea (Figure 4 and Figure S4).
SMG-derived MECs display SE basal cell characteristics ex vivo
Our in vivo lineage tracing experiments indicate that MECs of the SMGs can proliferate, migrate and generate SE cells following injury. During this transition, the number of ACTA2+ cells sharply declines from day-3 to day-5 while the total number of cells increases on the SE indicating that MECs transition from an ACTA2+ to ACTA2− state characteristic of the basal cells of the SE. To study this phenomenon, we isolated MECs from SMGs of Acta2-tdt mice and cultured them ex vivo. The isolated cells replicated and formed small colonies in vitro by day-5. IHC for markers of MECs and basal cells on day-3 and day-5 cultures revealed that Acta2-tdt cells lost the expression of ACTA2 by day-5 while continuing to express the basal cell markers, KRT5, TP63, and KRT14 (Figure 5A-B and Figure S5A). We also observed the expression of AQP3, NGFR, and ITGA6, other markers of basal cells at late times (Figure S5B). During this transition, MEC-derived Acta2-lineage labeled cells changed their shape from a stellate to cobblestone morphology (Figure 5A-B). These data suggest that MECs can proliferate and change to SE basal cell-like cells in ex vivo cultures.
Figure 5. MEC-derived SE basal-like cells proliferate, engraft and generate basal and luminal cell types in ex vivo engraftment models.
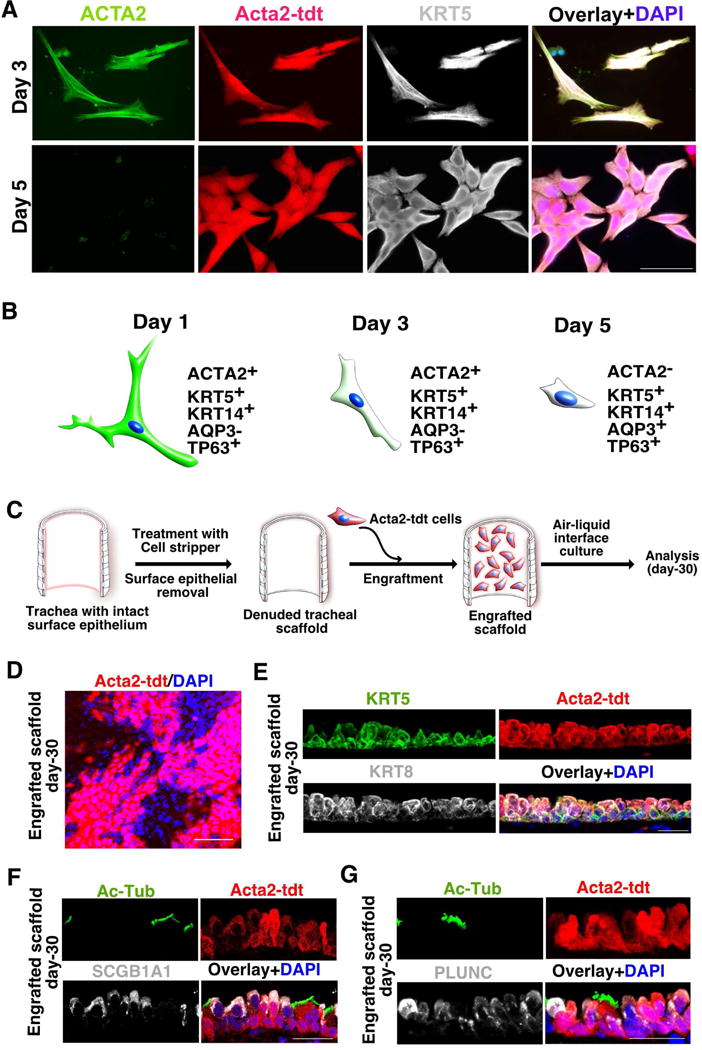
(A) Myoepithelial cells of SMG display SE basal cell characteristics in culture. Cultured cells on day-3 and day-5 co-stained with ACTA2 (green) and KRT5 (grey). Acta2-tdt (red) and nuclear DAPI (blue). Scale bar: 50μm.
(B) Schematic depicting the gradual loss of ACTA2 expression in SMG-derived MECs in culture.
(C) Schematic representation of engraftment of cultured MECs on a denuded tracheal scaffold. Isolated, cultured myoepithelial cells were seeded on denuded trachea followed by air-liquid interface culture and analysis.
(D) Whole mount IHC analysis for tdt-expressing engrafted cells on tracheal scaffold. Scale bar: 50μm.
(E) Co-staining of KRT5 (green) and KRT8 (grey) on trachea engraftment sections. Acta2-tdt (red) and nuclear DAPI (blue). n=3. Scale bar: 20μm
(F) IHC for Ac-Tub (green) and SCGB1A1 (grey) on tissue sections from Acta2-tdt (red) expressing cells engrafted on tracheal scaffold. Nuclear DAPI (blue). Scale bar: 20μm
(G) Sections from engrafted scaffolds expressing Acta2-tdt (red) co-stained with Ac-Tub (green) and PLUNC (grey). Nuclear DAPI (blue). Scale bar: 20μm
See also Figure S5.
MEC-derived SE basal-like cells proliferate, engraft and generate basal and luminal cell types in ex vivo engraftment models
Our lineage tracing and ex vivo experiments suggest that MECs convert into SE basal-like cells. To test the potential of cultured MEC-derived basal cells, we utilized an ex vivo tracheal regeneration model. We generated tracheal scaffolds by removing the SE using a non-enzymatic method. Whole mount IHC found that most of the SE basal and luminal cells were stripped away, leaving the denuded scaffold intact (Figure S5C). We then engrafted cultured Acta2-tdt MEC-derived basal-like cells onto the scaffolds and cultured them for 30 days in air-liquid-interface culture (Figure 5C). Whole mount imaging revealed patches of lineage labeled cells on the scaffold surface (n = 7 out of 12 scaffolds engrafted) suggesting that engrafted cells can proliferate and appropriately organize into epithelial sheets (Figure 5D). Subsequent section IHC showed that the engrafted cells generated both basal (KRT5+) and luminal (KRT8+) cells, including acetylated tubulin expressing ciliated cells as well as SCGB1A1 and PLUNC expressing secretory cells (Figure 5E-G). Thus, SMG-derived MECs have the ability to proliferate and generate SE cells ex vivo (Figure 5C-G).
SMG-like cells appear in SE of porcine proximal and distal lobular airway SE following severe injury
The distribution and abundance of SMGs varies between species. Unlike mouse airways, SMGs are present throughout cartilaginous airways in large mammals, including humans (Choi et al., 2000; Judge et al., 2014). Based on our current findings, we hypothesized that SMGs in large mammals have a greater ability to regenerate airway SE throughout the extra-lobular (i.e., trachea and bronchi) and intra-lobular airways (i.e., bronchioles). To test this hypothesis, we utilized a porcine airway injury model in which animals are exposed to chlorine gas at a concentration of 170±8ppm for 30 min to induce severe damage to the airways and alveoli (Gunnarsson et al., 1998; Wang et al., 2005). Filtered air exposed pigs served as controls (Figure 6A). We then isolated airway and lung tissues on day-2 post injury for marker analysis. Histological analysis of large airways (trachea and bronchi) indicated that SE cells are sloughed-off within 48 h of chlorine exposure but remain intact in control animals (Figure 6 and Figure S6A). Similar to the mouse naphthalene-induced injury model, SOX9 expression was observed in the SE of chlorine exposed porcine airways but not in controls where SOX9 is strictly confined to the SMGs (Figure 6B). Interestingly, the distribution of SOX9+ cells in the SE of the bronchioles was not uniform but tightly correlated with regions that had severe damage (Figure 6B). Most of these SOX9+ cells also co-expressed Ki67, a marker for proliferating cells, indicating that SOX9+ cells are highly proliferative in these damaged tissues (Figure S6B). In many cases patches of SOX9+ cells were seen in the SE extending from SMG acini and ducts. In contrast to mouse airway injury models, we did not find ACTA2+ cells on porcine SE after injury. To test whether porcine airway MECs have the ability to generate SE basal cells, we isolated, dissociated, and cultured SMGs from healthy porcine trachea similar to mouse MECs. We found small colonies of cells arising by day-3 and by day-5, 75% of the colonies were expressing MEC markers, ACTA2 and KRT5 and proliferation marker, Ki67. On day-9, KRT5 expression was maintained, but ACTA2 was no longer observed (Figure 6C). These data suggest that isolated porcine MECs have the ability to replicate and generate SE basal-like cells in ex vivo cultures. Our data suggests that SMG-derived cells act as reserve stem/progenitor cells throughout the airways in large animals and not just in the mouse trachea.
Figure 6. SMG-like cells appear in SE of porcine proximal and distal lobular airway SE following severe injury.
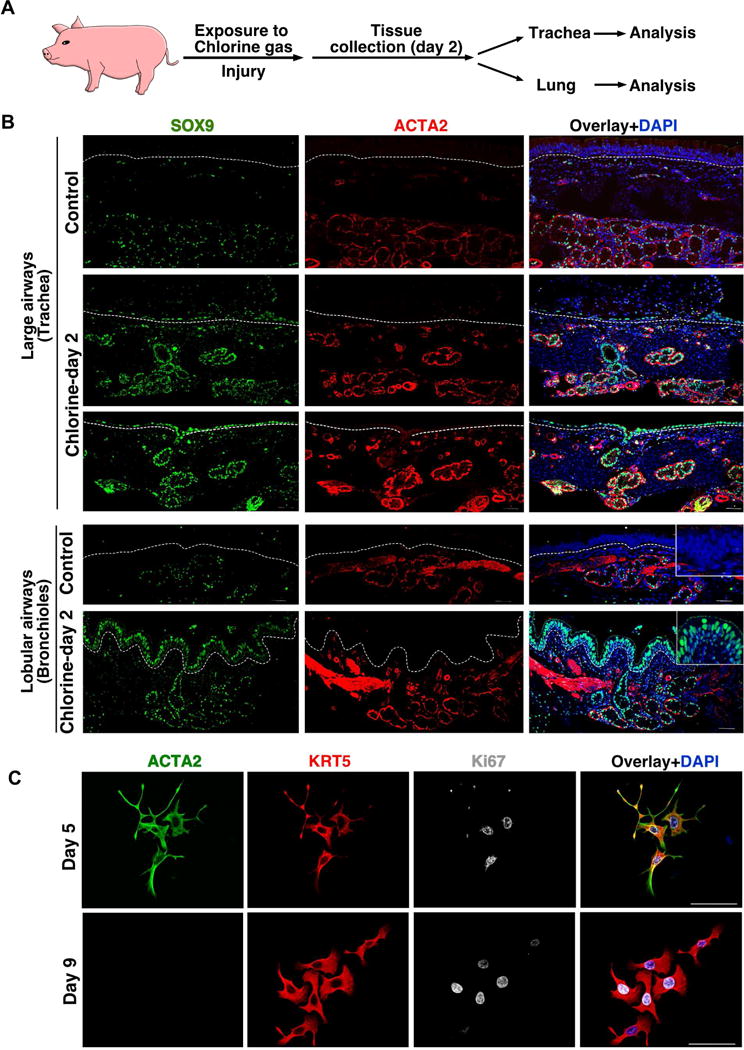
(A) Schematic representation of experimental design. Pigs were exposed to 170ppm chlorine gas for 30 min followed by collection of trachea or lung tissue on day-2 post injury.
(B) Co-IHC for indicated markers on tracheal sections (upper panels) or lobular airways (lower panel) from control or chlorine gas exposed pigs. The dashed line separates the SE and mesenchyme. Inset in the upper panel shows the higher magnification images. Scale bar: 50μm.
(C). MECs isolated from pig SMG display SE basal cell characteristics in culture. Cultured cells were fixed on day 5 or day 9 and co-stained with ACTA2 (green), KRT5 (red), Ki67 (grey) and nuclear DAPI (blue). Scale bar: 50μm.
See also Figure S6.
SOX9 dependent mechanisms are important for the contribution of MECs to SE repair after naphthalene-induced injury
To understand the molecular mechanisms underlying the proliferation and migration of MECs in response to injury, we analyzed our scRNA-seq data for genes differentially expressed between homeostatic SMGs and SMG cells that appeared on the SE. We focused our analysis on MECs (EpCAM+Acta2+) as this population was one of the major contributor to SE repair. We obtained 25 EpCAM+Acta2+ SE cells from injured airways and 36 EpCAM+Acta2+ cells from homeostatic SMGs (Figure S7A). Further analysis identified 92 genes that were significantly different between these populations; gene ontology analysis identified genes involved in cell cycle, translation, cell migration, regulators of cytoskeleton, wound healing, Inflammation, P53 signaling, Wnt signaling, and TNF/NF-kB signaling (Figure S7B). Many of the differentially enriched genes were also known targets of transcription factors, including MYC, SOX9, FOXA1, TP63, SMAD4, and KLF4 (Figure S7B). Interestingly, many of the SOX9 targets were highly enriched for cell migration (Fam60a, Ecm1, Iqgap1, Myh9, Itgb4, Tacstd2, Lgals3, Anxa1) (Babbin et al., 2007; Noritake et al., 2005; Simionescu-Bankston et al., 2013; Vicente-Manzanares et al., 2009; Xiong et al., 2012; Zhu et al., 2017) (Figure 7A). Based on this data, we hypothesized that SOX9 plays an essential role in MECs proliferation and migration to the SE following injury. To test this hypothesis, we generated Krt5-creER;Sox9f/f mice (Krt5-SOX9-LOF) which deletes SOX9 specifically in Krt5-expressing basal cell/myoepithelial cells upon tamoxifen administration (Figure 7B). We administered 3 doses of tamoxifen to Krt5-SOX9-LOF mice and control SOX9 f/f mice followed by naphthalene treatment. IHC for ACTA2 and SOX9 indicated loss of SOX9 specifically in MECs with 74.97% ± 3.34 (mean ± SEM); n=3) deletion efficiency (Figure 7C) within SMG acini. On day-4 post naphthalene injury, we observed significantly fewer DAPI+ cells on the SE compared to controls as well as significantly fewer SOX9 (5.73% ± 4.78 in Krt5-SOX9-LOF vs 32.49% ± 0.26 in controls, n=3), Ki67 (34.28% ± 4.73 in Krt5-SOX9-LOF vs 49.04% ± 2.33 in controls, n=3 and ACTA2 (3.71% ± 3.05 in Krt5-SOX9-LOF vs 11.74% ± 2.47 in controls, n=3) (mean ± SEM expressing cells on the SE indicating that significantly fewer MECs proliferated and migrated to the SE after loss of Sox9 in MECs (Figure 7E-F). These data support a model in which SOX9 is essential for MECs proliferation and their migration to the airway SE after naph-induced injury, potentially through regulating a set of down-stream target genes involved in this process.
Figure 7. SOX9 dependent mechanisms are important for the contribution of MECs to SE repair after naphthalene-induced injury.
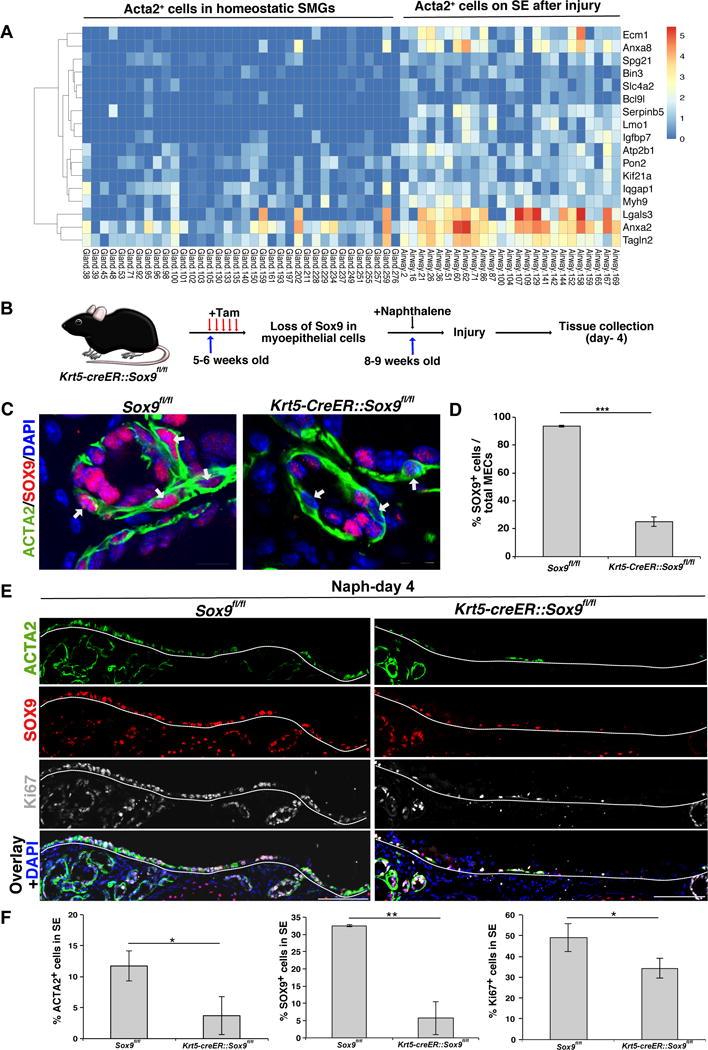
(A) Heatmap data from scRNA-seq analysis indicates potential SOX9 target genes that are differentially expressed in Acta2+ cells in homeostatic SMGs and Acta2+ cells isolated from SE following naphthalene-induced injury.
(B) Schematic representation of experimental design to delete Sox9 in myoepithelial cells followed by naphthalene administration to induce airway injury.
(C) Co-IHC for ACTA2 (green), SOX9 (red), and nuclear DAPI (blue) on tracheal sections. Arrows indicates nuclei of ACTA2+ myoepithelial cells. Scale bar: 20μm.
(D) Quantification of SOX9-expressing cells among myoepithelial cells of Krt5-SOX9-LOF and Sox9f/f mice. Data shown as mean ± SEM (n=3; ***p ≤ 0.001).
(E) IHC for ACTA2 (green), SOX9 (red), Ki67 (grey), DAPI (blue) on tracheal longitudinal sections collected from Krt5-SOX9-LOF and Sox9f/f mice on day-4 post naphthalene injury. The dashed line separates the SE cell layer from the mesenchyme. Scale bars: 50μm.
(F) Quantification of ACTA2+, SOX9+ and Ki67+ cells among total DAPI+ cells in SE spanning cartilage rings between 0-3 in naphthalene injured mice. Data shown are mean ± SEM (n=3; *p ≤ 0.05; **p ≤ 0.01).
See also Figure S7.
Discussion
Airway SMGs are highly abundant in human lungs where they function as mucous producing units, undergoing significant hypertrophy in conditions such as CF, asthma and COPD (Jeffery, 1991). Here we demonstrate that MECs within the SMGs also serve as progenitors, migrating long distances to regenerate the SE following severe injury. Intriguingly, the plasticity we observed is proportional to the degree of injury and occurred in multiple forms of injury models (Figure S2). However, it is possible that many factors including inflammation and other systemic factors may also influence the degree of plasticity in different forms of injury models. Previous studies using Krt14-promoter driven lineage tracing suggest that SMG duct cells contribute to SE repair following hypoxic injury. However, KRT14 is expressed in SE basal cells making it difficult to determine the contribution of SMG duct cells to SE repair. ScRNA-seq allowed us to identify SMG-like cells on SE following injury and revealed that SOX9 was specifically expressed in adult SMGs but not in SE. Using myoepithelial cell specific lineage tracing models, we demonstrate that SMG-derived MECs contribute to the SE repair. Accounting for lineage labeling differences, 67.7% of SE cells were derived from Sox9+ cells while 42.6% were derived from MECs. The different relative contribution of Sox9-tdt and Acta2-tdt cells suggests that cells other than MECs (SOX9+ACTA2−) within the SMGs also contribute to SE repair. Indeed previous studies suggested SMG duct cells contribute to SE repair (Hegab et al., 2012). Alternatively, SMG luminal cells may also contribute to the SE repair. Likewise, Mammary gland MECs are shown to act as lineage restricted mammary stem cells (Prater et al., 2014). Future studies are needed to uncover whether there are any subsets of MECs that possess higher “stemness” as such subsets were recently demonstrated to play a major role in alveoli regeneration (Nabhan et al., 2018; Zacharias et al., 2018).
Our data support a model in which, MECs migrate to the SE, lose their myoepithelial identity (defined by ACTA2 expression and morphology) and acquire SE basal cell characteristics following injury. The transition from MEC to airway basal cell may be driven by micro-environmental factors, such as the basement membrane or residual SE cells, or cell-autonomous mechanisms. Indeed, Sox9- and Acta2-lineage derived cells did not express mature club cell marker (SCGB1A1) until several weeks after injury, indicating that some cell-autonomous mechanisms likely constrain the differentiation of SSEA1+ immature club cells into mature club cells.
Recent studies have shown that MECs contribute to both basal and luminal lineages and act as multipotent progenitor cells during SMG development (Anderson et al., 2017). Interestingly, we find that MECs in the adult SMGs constrain their lineage potential at homeostasis but exhibit plasticity under adverse conditions. Such characteristics may thus qualify them to function as reserve multipotent stem cells (Blanpain and Fuchs, 2014; Merrell and Stanger, 2016; Tata and Rajagopal, 2016; Visvader and Clevers, 2016). Although this question remains an active area of investigation, it appears that while lineage restricted stem progenitor cells individually maintain the mammary basal and luminal compartments at homeostasis, but a subpopulation either intrinsically possess or acquire multipotency in transplantation assays or during pregnancy and lactation to contribute to all lineages (Rios et al., 2014; Van Keymeulen et al., 2011).
In contrast to SE, the lung SMGs are embedded deep within airway tissues, analogous to the intestinal crypts and gastric pits in the intestine and stomach, respectively. Such a location presumably protects the SMGs from normal wear and tear as well as airborne pathogens and toxic agents and enables them to serve as an important and overlooked source of reserve stem cells (Tetteh et al., 2015; Visvader and Clevers, 2016). Indeed, our large animal model suggests that the SMG cells contribute significantly to both extra- and intra-pulmonary airway regeneration (Figure 6). SMGs are present throughout cartilaginous airways of the human lungs and likely play a critical role in endogenous regeneration after infection and injury. Although we observed many SOX9+ cells on the SE of porcine injured airways, these cells did not express ACTA2 like the mouse. SMGs are abundant in porcine airways, so perhaps duct cells alone are sufficient to repair SE. Alternatively, MECs may downregulate and eventually lose the expression of ACTA2 en route to SE. Indeed, our experiments in mice suggest that MECs quickly lose ACTA2 expression when they migrate to the SE or when placed in culture (Figure 4 and 5). Nevertheless, isolated porcine MECs displayed SE basal-like cell characteristics in cultures suggesting that porcine MECs have the ability to generate SE basal cells (Figure 6).
Our scRNA-seq analysis not only identified SMG-like cells on the SE after injury, but also revealed differentially expressed genes in ACTA2+ MECs in response to injury. Many genes showed enrichment for different biological processes including cell cycle and cell migration. Some genes were previously shown to be regulated by the transcription factor SOX9 in multiple tissues, including hair follicle stem cells (Adam et al., 2015). Although SOX9 was expressed in both homeostatic MECs and MECs following injury, it remains to be tested how and whether SOX9 regulates different downstream target genes in different cellular contexts. Indeed, it has been demonstrated that SOX9 binds on different target gene regulatory elements in different cellular contexts in hair follicles and therefore recognized as a pioneer transcription factor (Adam et al., 2015).
Important questions for the future will be to determine whether Sox9+ SMG-derived SE persist in the airways, perhaps altering airway response to environmental agents or whether SMG glands continue to replenish the SE following repeated injuries. This research motivates future work to elucidate how endogenous repair mechanisms can be enhanced therapeutically and provide important new insights into human respiratory health and disease.
CONTACT FOR REAGENTS AND RESOURCE SHARING
Further information and requests for resource/reagents should be directed to and will be fulfilled by the lead contact Purushothama Rao Tata (purushothamarao.tata@duke.edu)
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Animal Studies
All laboratory mice were maintained under standard husbandry and housing conditions approved by the Duke University Institutional Animal Care and Use Committee. All animals were handled in accordance with the NIH and AAALAC guidelines for humane care and use of laboratory animals. Mice used for experiments were not involved in previous procedures and were drug or test naive. Mice were kept in a SPF breeding area under specified pathogen free conditions. Cages, bedding, food and water were autoclaved. Animals were maintained on the same 12 h light/dark cycle and monitored daily by caretakers and researchers. Health monitoring did not reveal any infections in the past 18 months. Male and female C57BL/6J, CD-1, and transgenic mice aged 6-12 weeks were used for experiments with similar aged mice for both control and treatment groups. Sox9-creERT2 (Soeda et al., 2010); (kindly provided by Henry Kronenberg, MGH, Boston) were bred with R26R-tdTomato mice (Jackson Laboratory 007914) to generate Sox9-creERT2::R26R-tdTomato males, which were then crossed with C57Bl/6 or CD-1 to generate experimental animals. Acta2-creERT2 (kindly provided by Hogan lab, Duke University; (Wendling et al., 2009)) females were bred with R26R-tdTomato (homozygous) males to obtain Acta2-creERT2-R26R-tdTomato and maintained on a C57Bl/6 background. To label Sox9 or Acta2-expressing cells, 3 doses of tamoxifen were administered intraperitoneally (2mg/20gms body weight) to induce cre recombinase mediated excision of a stop codon and subsequent expression of tdTomato. Mice were allowed to recover for 4 weeks between tamoxifen treatment and naphthalene injury. Krt5-creERT2 (Jackson Laboratory 029155) were bred with Sox9f/f (Jackson Laboratory 013106) to generate Krt5-creERT2;Sox9f/f males, which were then crossed to Sox9f/f females to generate experimental animals. Krt5-CreERT2::Sox9f/f and Sox9f/f littermates were maintained on a C57Bl/6j background. Krt5-creERT2;Sox9f/f and Sox9f/f mice were given five doses of tamoxifen (2mg/20gms body weight) followed by an interval of 3 weeks prior to naphthalene administration.
Domestic, juvenile American Yorkshire pigs (30-40 kg) of both sexes were obtained from a certified specific pathogen free breeder (Looper Farms, Granite Falls, NC). The animals were housed for an acclimation period in an adjoining facility for a minimum of 24 h prior to all studies. Animals were pre-medicated with ketamine (22 mg/kg IM) and acepromazine (1.1 mg/kg IM). Following pre-medication, propofol was administered as a single bolus (5-10 mg/kg IV) to facilitate tracheal intubation, and then continuously (5-10 mg/kg/hr IV) during mechanical ventilation. All animals were volume-control ventilated (AVEA, CareFusion Inc., Yorba Linda, CA) with a tidal volume of 7 ml/kg, a respiratory rate of 25 breaths per minute, and PEEP set to 5 cm H2O. Initial FiO2 was 0.21 and adjusted as needed to maintain SpO2 ≥ 90%. No further changes to the ventilator settings were made for the duration of experiments. Following chlorine or filtered air exposure, propofol was halted and replaced with an isotonic crystalloid solution (4 ml/kg/hr IV). Animals were weaned from mechanical ventilation until sustained spontaneous breathing was observed. Animals were returned to their enclosure and closely observed until upright. Animals were then monitored for 48 h for signs of respiratory distress, with heart rate and oxygenation saturation measured during voluntary feeding. After 48 h, animals were humanely euthanized while under deep anesthesia with potassium chloride (4-10 mmol/kg IV).
METHOD DETAILS
Chlorine Gas Exposure
Chlorine gas exposure and evacuation was performed according to a protocol approved by the Duke University Occupational & Environmental Safety Office. After animals were stably ventilated for 30 min to ensure appropriate sedation and analgesia, chlorine gas exposure was initiated by opening a valve on the inspiratory limb of the ventilator circuit. Compressed chlorine gas from a calibrated cylinder (Airgas National Welders, Durham, NC) was delivered via a 2-stage regulator through an adjustable flowmeter (Cole-Parmer) for 30 min at a concentration of 170 ± 8 ppm, with intermittent monitoring of the chlorine gas concentration (PortaSens II, Analytical Technology Inc., Collegeville, PA) to ensure consistent levels of exposure. Residual chlorine gas was scavenged under 100% vacuum of the exhaled breath via the unidirectional expiratory line of the ventilator circuit through a series of wet-scrubbing containment flasks containing 10% NaOH. As an additional safeguard, the animals and the ventilator circuit were placed under a negative pressure chamber with colorimetric indicators (Morphix Technologies) for the detection of leaked chlorine gas. These experiments are performed on two independent pigs.
Naphthalene injury model
Naphthalene administration was performed following standard procedures as described previously (Hong et al., 2001). Briefly, a single dose of naphthalene (dissolved in corn oil) was administered intraperitoneally at 275 mg/kg body weight to induce injury. Naphthalene is toxic and causes skin and eye irritation. So, care must be taken while preparing the reagent. Groups of 3-4 mice were sacrificed on day 1, 3, 5, 10, 12, 20, 60, and 80 post naphthalene and trachea were dissected. Corn oil administered mice served as vehicle control. Freshly prepared naphthalene was administered between 7:00 A.M. - 8:00 A.M.. Inconsistent injury was observed if administered later in the day or using frozen aliquots of naphthalene. No blinding was used, and no data was excluded from the analysis.
SO2 and Influenza infection induced injury
SO2 exposure and influenza infection was performed as previously described (Tata et al., 2013). Briefly, mice were placed in a chambered cage which was placed in a transparent polystyrene chamber. Compressed SO2 gas from a calibrated cylinder (Airgas Inc.) was delivered via a 2-stage regulator through an adjustable flowmeter for 3h 40 min at a concentration of 500ppm of SO2, with intermittent monitoring of the SO2 gas concentration to ensure consistent levels of exposure. Control mice were exposed to filtered air under otherwise the same conditions. After exposure, mice were returned to their cages where they received atmospheric air. For influenza-induced injury experiments, mice were anesthetized using ketamine/xylazine. Influenza virus A/Puerto Rico/8/34 (PR8) (kindly provided by Dr. Nicholas Heaton, Duke University) dose was determined based on titration experiments. Mice were anesthetized and administered 103 PFU virus, which was determined to result in a LD value of 75, through intranasal inhalation as previously described (Tata et al., 2013). Mice health was monitored every day. Mice were sacrificed on day-1, 3, and 5 post exposure/infection, and the trachea were harvested for histological analysis. No blinding was used and no data was excluded from the analysis. These experiments are performed on three independent mice.
Tissue harvesting, sectioning, and H&E staining
Mouse trachea were harvested and fixed in 4% PFA in PBS for 30 min at RT and then washed in PBS followed by overnight incubation in 30% sucrose at 4°C. Trachea were then incubated with 1:1 mixture of 30% sucrose (in PBS)/O.C.T. for 30 min at 4°C, embedded in O.C.T. and cryosectioned (6μm). Pig trachea were harvested and fixed overnight in 10% normal buffered formalin at 4°C before paraffin embedding. 5 μm thick sections were used for histological analysis.
Mouse SMG dissection and dissociation
Trachea were dissected from tamoxifen administered Acta2-tdt mice and placed in DMEM+pen/strep and cut in half (around cartilage 5) and opened through the trachea lumen. Tracheal pieces were incubated in 16U/ml dispase (in DMEM+pen/strep) for 30 min at RT. Digestion was stopped using 5%FBS DMEM+pen/strep. SE was manually peeled off with forceps. Tracheal pieces were washed 3 times in DMEM+pen/step then SMG were manually dissected with forceps, washed once with PBS, spun down at 1000g for 2 min and incubated with collagenase II/Hyaluronidase enzyme mix (in DMEM+pen/strep) for 30 min at 37°C. Then Trypsin-EDTA was added to a final concentration of 0.1% and incubated for an additional 20 min. The enzymatic digestion was stopped with 5%FBS in DMEM+pen/strep. The suspension was filtered through a 40 M cell strainer and spun down at 1000g for 2 min. Cell pellet was reconstituted in in SAGM media containing pen/strep, 1 μM A83-01, 10 μM Y-27632, 1 μM DMH-1, 1 μM CHIR 99021 and cultured at 37°C, 5% CO 2. Media was changed every other day between day-1 to 4 and every day after day-4 (Mou et al., 2016).
Pig SMG dissection and dissociation
Porcine tracheas were obtained from local slaughterhouse, with permission from the North Carolina Department of Agriculture and Consumer Services. Fresh trachea were transported in ice cold DMEM. Tracheal pieces were dipped in 95% ethanol for 10 sec and placed in DMEM+pen/strep. SMG were manually dissected under microscope, washed once in PBS and incubated in solution containing: 110 mM NaCl, 5 mM KCl, 1 mM MgCl2 and 2 mM CaCl2, 20mM HEPES, 10 mg/mL of Bovine Albumin Fraction V, 200 U/mL Collagenase I, 200 U/mL collagenase II, 200 U/mL Hyaluronidase in DMEM for 1h at 37°C with gentle agitation. Samp les were spun at 500g for 2 min and cell pellet was digested for 30 min. Samples were spun down, washed once with PBS, and incubated in 0.1% Trypsin-EDTA/DMEM for 20 min. Enzymatic digestion was stopped by addition of 5% FBS/DMEM. The suspension was filtered through a 40 M cell strainer and spun down at 1000g for 2 min. Cells were reconstituted and cultured in SAGM media containing anti/anti, gentamycin, 1 μM A83-01, 10 μM Y-27632, 1 μM DMH-1, 1 μM CHIR 99021, 0.5% matrigel. Cells were cultured in triplicates and was repeated on three independent tracheal cells.
SMG cell culture and immunocytochemistry
Cells isolated from SMG were grown on coverslips. Cells were fixed in 4% PFA for 15 min, washed twice for 5 min in PBS, and permeabilized for 10 min in PBT (PBS + 0.1% Triton X-100). The samples were then incubated for 30 min in 1%BSA in PBT, incubated with primary antibodies at RT for 1h, washed three times for 5 min in PBT and incubated with secondary antibody for 1h at RT. Cells were washed and coverslips were mounted on slides using Fluor G reagent with DAPI. No blinding was used and no data was excluded from the analysis.
Cells were cultured in triplicates and was repeated on three independent tracheal cells.
SMG cell engraftment and scaffold culture
Trachea from C57Bl/6 mice were dissected, cut in half longitudinally and trimmed to eliminate the region that contains SMGs (larynx to cartilage 3). Tracheal pieces were washed once in PBS and incubated with Cellstripper (Corning) over night at 37°C with continuous rotation to denude the SE. Next day, tracheal pieces were washed once in 10% FBS/DMEM, twice in PBS and placed on soaked Gelfoam (Pfizer). SMG cells isolated from Acta2-tdt mice were expanded for 5 days prior to grafting on denuded tracheal pieces. Scaffolds were cultured in an air-liquid-interface in PneumaCult™-ALI media for 30 days prior to histological analysis. The data presented in the results section was obtained from 7 successfully transplanted scaffolds out of 12 scaffolds engrafted. Engraftment efficiency was determined based on tdtomato expression on the scaffold surface.
Section Immunohistochemistry
Antigen retrieval was performed using 10 mM sodium citrate buffer in antigen retrieval system (Electron Microscopy Science) or water bath (90°C f or 10 min). Cryosections (6 μm) were washed in PBS, permeabilized with 0.1% Triton X-100 in PBS (PBT), blocked in 1% BSA in PBT for 30 min at RT. Incubation with primary antibodies (see key resource table) was performed for 1 h at RT. Next, slides were washed 3×5 min in PBT, incubated with appropriate secondary antibodies in blocking buffer for 1h at RT, washed 3×5 min PBT and mounted using Fluor G reagent with DAPI. 5-7μm paraffin embedded pig tissue sections were dewaxed, rehydrated before antibody staining. Immunostaining was performed on at least 3 sections from each trachea and this was repeated on 3-5 mice.
Whole mount immunohistochemistry
Antigen retrieval was performed in 95°C (water bath) for 15 min. Next, tissues were washed with PBS, permeabilized, blocked in 1.5% BSA 0.3% Triton in PBS (1.5% BSA 0.3% PBT) for 45 min at RT and incubated over night with primary antibody in blocking solution at 37°C. Next, samples were washed 3×1h in PBT, blocked in 1.5% BSA 0.3% PBT for 45 min at RT, and incubated with secondary antibody and DAPI in blocking solution at 37°C for 45 min. After three washes in PBT tissue was mounted on slides. Immunostaining was performed at least on 3-5 mice trachea.
Image acquisition, processing and quantification
Images were captured using Olympus Confocal Microscope FV3000 using a 20×, 40× or 60× objective (all images except Figure 5A) or a Zeiss wide-field fluorescence microscope (Figure 5A). Cells were manually counted based on IHC markers and DAPI. Images were processed using Olympus CellSens application or ImageJ and figures were prepared using Affinity Designer.
Single cell capture and library preparation
Cell suspensions at a density of 1000 cells/μl in PBS + 0.04% BSA were prepared for single cell sequencing using the Chromium™ Single Cell 3′ Reagent Version 1 Kit and Chromium™ Controller (10× Genomics, CA, USA) as previously described. Briefly, 20,000 cells per reaction were loaded for GEM (Gel Bead-in-Emulsion) generation and barcoding. GEM-RT was performed using a Thermocycler. Post GEM-RT cleanup and cDNA amplification was performed to isolate and amplify cDNA for library construction. Libraries were constructed using the Chromium™ Single Cell 3′ Reagent Kit (10× Genomics, CA, USA) and sample was sequenced in a single lane using the Illumina Nextseq 500 in Rapid Run Mode using a paired end flow cell. Read1: 26 cycles, Index2: 8 cycles, and Read2: 96 cycles.
scRNA-seq analysis
Raw Illumina sequencing output were processed through the 10× Genomics Cell Ranger software suite using default settings. Reads were mapped to the mm10 transcriptome. Read counts were normalized to log2 reads per million mapped (rpm) and a cutoff of log2 rpm ≥ 1 was used as a cutoff to determine whether a given gene was expressed in a cell.
To ensure that epithelial cells were included in the dataset, we first included any cell that expressed Epcam, Cdh1, Krt5, Krt8, or Krt14. We then removed endothelial cells, blood cells and macrophages by filtering out those expressing Esam, H2-ab1, H2-aa, Hba-a2, Hbb-bt, or Cd68. 139 cells were retained for further analysis. Any genes with fewer than 10 reads across the dataset were also removed from further consideration.
K-means clustering was performed in R using the stats package, followed by t-SNE visualization with the Rtsne package. Differential expression analysis was performed using edgeR in a one-vs-others approach, with a cutoff of FDR < 0.05 and logFC ≥ 1. Select differentially expressed genes were chosen for visualization in the heat map, which was created by the NMF R package. Selection of Acta2+ cells was determined using a cutoff of log2 rpm≥1. Enrichr (amp.pharm.mssm.edu/Enrichr/) was used to perform gene ontology, pathway analysis, and genomic and proteomic interaction analysis (Kuleshov et al., 2016).
QUANTIFICATION AND STATISTICAL ANALYSIS
The number of biological and technical replicates and animals are indicated in figure legends and text. Both male and female mice were used and all tested animals were included in data analysis. Briefly, the gender and mice strains used are: a) For quantitative analysis of ACTA2, SOX9 and Ki67 markers in WT (C57BL/6j) mice we used-1 male and 2 females for controls, 3 females were used for naph-day 1, 3, and 5; b) For Acta2-tdt lineage tracing experiments-1 male and 2 females for controls, 3 females for day-5 naphthalene, 3 females for naph-day12, 1 male and 2 females for naph-day 20, 3 females for naph-day 60 (C57BJ/6j). c) For Sox9-Tdt lineage tracing experiments-1 male and 2 females for controls, 3 females for naph-day 5, 3 females for naph-day10 (mixed background), 3 females for naph-day 20, 1 male and 3 females for naph day 80 (C57BL/6j).
Sample size was not predetermined. Data are presented as means with standard error (SEM) to indicate the variation within each experiment. We used D’Agnostino and Pearson omnibus normality test available in Prism to test whether our data are normally distributed. All data except Figure 2D-E, 3F (SCGB1A1 only), Figure S2E-G (ACTA2 and SOX9 only) and I, Figure 7F (Sox9 only) and S4D showed Gaussian distribution. We used Mann Whitney statistical test for non-normal distributions. Statistical differences between samples were assessed with Student two-tailed t-test calculated in Excel. p-values below 0.05 are considered significant (***p ≤ 0.001, **p ≤ 0.01, *p ≤ 0.05).
DATA REPORTING AND SOFTWARE AVAILABILITY
All relevant data are available from authors. All the software used in this work are described in the relevant methods sections. scRNA-seq data has been deposited to GEO (GSE111598).
KEY RESOURCES TABLE
The table highlights the genetically modified organisms and strains, cell lines, reagents, software, and source data essential to reproduce results presented in the manuscript. Depending on the nature of the study, this may include standard laboratory materials (i.e., food chow for metabolism studies), but the Table is not meant to be comprehensive list of all materials and resources used (e.g., essential chemicals such as SDS, sucrose, or standard culture media don’t need to be listed in the Table). Items in the Table must also be reported in the Method Details section within the context of their use. The number of primers and RNA sequences that may be listed in the Table is restricted to no more than ten each. If there are more than ten primers or RNA sequences to report, please provide this information as a supplementary document and reference this file (e.g., See Table S1 for XX) in the Key Resources Table.
Please note that ALL references cited in the Key Resources Table must be included in the References list. Please report the information as follows:
REAGENT or RESOURCE: Provide full descriptive name of the item so that it can be identified and linked with its description in the manuscript (e.g., provide version number for software, host source for antibody, strain name). In the Experimental Models section, please include all models used in the paper and describe each line/strain as: model organism: name used for strain/line in paper: genotype. (i.e., Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J). In the Biological Samples section, please list all samples obtained from commercial sources or biological repositories. Please note that software mentioned in the Methods Details or Data and Software Availability section needs to be also included in the table. See the sample Table at the end of this document for examples of how to report reagents.
SOURCE: Report the company, manufacturer, or individual that provided the item or where the item can obtained (e.g., stock center or repository). For materials distributed by Addgene, please cite the article describing the plasmid and include “Addgene” as part of the identifier. If an item is from another lab, please include the name of the principal investigator and a citation if it has been previously published. If the material is being reported for the first time in the current paper, please indicate as “this paper.” For software, please provide the company name if it is commercially available or cite the paper in which it has been initially described.
-
IDENTIFIER: Include catalog numbers (entered in the column as “Cat#” followed by the number, e.g., Cat#3879S). Where available, please include unique entities such as RRIDs, Model Organism Database numbers, accession numbers, and PDB or CAS IDs. For antibodies, if applicable and available, please also include the lot number or clone identity. For software or data resources, please include the URL where the resource can be downloaded. Please ensure accuracy of the identifiers, as they are essential for generation of hyperlinks to external sources when available. Please see the Elsevier list of Data Repositories with automated bidirectional linking for details. When listing more than one identifier for the same item, use semicolons to separate them (e.g. Cat#3879S; RRID: AB_2255011). If an identifier is not available, please enter “N/A” in the column.
-
○
A NOTE ABOUT RRIDs: We highly recommend using RRIDs as the identifier (in particular for antibodies and organisms, but also for software tools and databases). For more details on how to obtain or generate an RRID for existing or newly generated resources, please visit the RII or search for RRIDs.
-
○
Please use the empty table that follows to organize the information in the sections defined by the subheading, skipping sections not relevant to your study. Please do not add subheadings. To add a row, place the cursor at the end of the row above where you would like to add the row, just outside the right border of the table. Then press the ENTER key to add the row. You do not need to delete empty rows. Each entry must be on a separate row; do not list multiple items in a single table cell. Please see the sample table at the end of this document for examples of how reagents should be cited.
TABLE FOR AUTHOR TO COMPLETE
Please upload the completed table as a separate document. Please do not add subheadings to the Key Resources Table. If you wish to make an entry that does not fall into one of the subheadings below, please contact your handling editor. (NOTE: For authors publishing in Current Biology, please note that references within the KRT should be in numbered style, rather than Harvard.)
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal to Aquaporin 3 | Abcam | Ab125219 |
| Rabbit Polyclonal anti-alpha SMA | Abcam | Ab5694; |
| Mouse Monoclonal anti-alpha SMA | Sigma-Aldrich | A2547 RRID:AB_476701 |
| Rat Monoclonal anti-KRT8 | DSHB | TROMA-I RRID:AB_531826 |
| Rabbit Polyclonal anti-KRT5 | Abcam | Ab53121 RRID:AB_869889 |
| Mouse Monoclonal anti-KRT14 | ThermoFisher | MS-115-P1; |
| Rabbit Polyclonal anti-Ki67 | Abcam | Ab15580; RRID:AB_443209 |
| Rabbit Polyclonal anti-P63 | Genetex | GTX102425 RRID:AB_1952344 |
| Rat Monoclonal anti-Ki67 | ThermoFisher | 14-5698-82 RRID:AB_10854564 |
| Mouse Monoclonal anti-TP63 | OriGene | TA802078 RRID:AB_2626201 |
| Rabbit Polyclonal anti-Sox9 | Millipore | Ab5535 RRID:AB_2239761 |
| Mouse Monoclonal anti-Sox9 | Abcam | Ab76997 RRID:AB_2194156 |
| Mouse Monoclonal anti-Acetylated tubulin | ProteinTech | 66200-1-Ig |
| Rat monoclonal anti-E-cadherin | Innovative Research | 13-1900 RRID:AB_86571 |
| Goat polyclonal CC10 | Santa Cruz | Sc-9772 RRID:AB_2238819 |
| Rabbit polyclonal Plunc | Bingle et al., 2010 | kindly provided by Dr. Colin Bingle (The University of Sheffield, U.K.) |
| Rabbit polyclonal NGFR | Abcam | Ab8875 RRID:AB_306828 |
| Rat monoclonal Integrin alpha 6 | R&D | MAB13501 RRID:AB_2128311 |
| Anti-UEA1 | Vector Laboratories | RL-1062 RRID:AB_2336769 |
| Mouse Anti-SSEA1 | DSHB | MC-480 RRID:AB_528475 |
| Rabbit Polyclonal anti-Lactotransferrin | Millipore | 07-685 RRID:AB_11211872 |
| Rabbit Polyclonal ZO-1 | Abcam | Ab216880 |
| Alexa Fluoro 488 Donkey anti-mouse IgG | Invitrogen | A21202 RRID:AB_141607 |
| Alexa Fluoro 594 Donkey anti-mouse IgG | Invitrogen | A21203 RRID:AB_141633 |
| Alexa Fluoro 647 Donkey anti-mouse IgG | Invitrogen | A31571 RRID:AB_162542 |
| Alexa Fluoro 488 Donkey anti-rabbit IgG | Invitrogen | A21206 RRID:AB_141708 |
| Alexa Fluoro 594 Donkey anti-rabbit IgG | Invitrogen | A21207 RRID:AB_141637 |
| Alexa Fluoro 647 Donkey anti-rabbit IgG | Invitrogen |
A31573 RRID:AB_2536183 |
| Alexa Fluoro 488 Donkey anti-goat IgG | Invitrogen | A11055 RRID:AB_142672 |
| Alexa Fluoro 594 Donkey anti-goat IgG | Invitrogen | A11058 RRID:AB_142540 |
| Alexa Fluoro 647 Donkey anti-goat IgG | Invitrogen | A21447 RRID:AB_141844 |
| Alexa Fluoro 488 Donkey anti-rat IgG | Invitrogen | A21208 RRID:AB_141709 |
| Alexa Fluoro 594 Donkey anti-rat IgG | Invitrogen | A21209 RRID:AB_2535795 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Y-27632 2HCl ROCK1 (p160ROCK inhibitor) | Selleckchem | S1049 |
| DMH-1 (BMP antagonist) | Tocris | 4126 |
| CHIR 99021 (Wnt agonist) | Tocris | 4423 |
| A 83-01 (TGF antagonist) | Tocris | 2939 |
| Small Airway Epithelial Cell Growth Medium (SAGM) BulletKit | Lonza | CC-3118 |
| PneumaCult™-ALI Medium | STEMCELL technologies | 05001 |
| Matrigel | Corning | 356230 |
| Dulbecco’s Modification of Eagle’s Medium (DMEM) | Corning | 10-013-CV |
| PBS | Genesee Scientific | 25-507 |
| MgCl2 | Sigma | M4880 |
| NaCl | Sigma | S9888 |
| KCl | Invitrogen | AM9640G |
| CaCl2 | Sigma | C5670 |
| HEPES | Gibco | 15630080 |
| Bovine Albumin Fraction V (7.5% solution) | Gibco | 15260037 |
| Cellstripper | Corning | 25-056-CI |
| Hyaluronidase from bovine testes type I-S | Sigma | H3506-500MG |
| Collagenase, Type II, powder | Gibco | 17101-015 |
| Collagenase, Type I, powder | Gibco | 17100017 |
| Trypsin-EDTA (0.25%), phenol red | Gibco | 25200056 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | 15140122 |
| Antibiotic-Antimycotic (100X) | Gibco | 15240062 |
| Gentamycin sulfate | Sigma | G1914 |
| Paraformaldehyde solution 4% in PBS | Santa Cruz | sc-281692 |
| Tamoxifen | Sigma | T5648-1G |
| Bovine Serum Albumin | Sigma-Aldrich | A7906 |
| O.C.T. Compound | Fisher Scientific | 23-730-571 |
| Citrate Buffer 10x | Sigma-Aldrich | C9999 |
| Sucrose | Sigma-Aldrich | S0389 |
| Triton X-100 | Sigma-Aldrich | X100 |
| Fluoromount-G™, with DAPI | Thermo Fisher Scientific | 00-4959 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Airway surface and submucosal gland scRNA-seq | This paper | GSE111598 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | The Jackson Laboratory | Jax # 000664 |
| CD-1 IGS | Charles river | CRL# 022 |
| Sox9tm1(cre/ERT2)Haak | Soeda et al., 2010 | N/A |
| Tg(Acta2-cre/ERT2)12Pcn | Hogan lab Wendling et al., 2009 |
N/A |
| KRT5tm1.1(cre/ERT2)Blh | The Jackson Laboratory | Jax# 029155 |
| B6.129S7-Sox9tm2Crm/J | The Jackson Laboratory | Jax# 013106 |
| B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | Jax# 007909 |
| American Yorkshire pigs | Looper Farms, Granite Falls, NC | |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| ImageJ | NIH | N/A |
| 10x Genomics Cell Ranger | 10X Genomics | N/A |
| R Studio | N/A | N/A |
| Python | N/A | |
| Enrichr | Kuleshov et al, 2016 | http://amp.pharm.mssm.edu/Enrichr/ |
| Other | ||
Supplementary Material
Highlights.
SMGs harbor reserve multipotent stem cells that can contribute to SE repair
MECs contribute to both basal and luminal cells of the SMGs and SE after injury
SMG-like cells found on porcine SE following severe airway injury
SOX9 dependent mechanisms drive MECs proliferation and migration after injury
Acknowledgments
We thank Brigid Hogan, Ken Poss, Don Fox, and Nellwyn Hagan for critically reading the manuscript. We thank Brigid Hogan for sharing Acta2-creER mice and Henry Kronenberg for Sox9-creERT2 mice and members of the Hogan and Barkauskas laboratories for discussions. We thank the DMPI 10X genomics core for their assistance with scRNA-seq and Brigid Hogan and Dr. Colin Bingle (The University of Sheffield, U.K.) for the PLUNC antibody. This research was supported by a career development award from NHLBI/NIH to P.R.T. (4R00HL127181) and funds from Regeneration NeXT Initiative at Duke University. P.R.T. is a Whitehead Scholar. Research reported in this publication is supported in part by the CounterACT Program through the National Institute of Environmental Health Sciences of the National Institute of Health under award Number U01ES017219 to M.D.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contribution: A.T. designed, conceived and performed the experiments and co-wrote the manuscript; R.D.C. performed computational analysis; Y.K., A.J.M., J.T., A.D. performed IHC and quantified the number of cells; T.J.M. performed and helped with the porcine injury experiments. M.D.G. supervised pig injury experiments. P.R.T. designed, conceived and supervised the work and co-wrote the manuscript. All authors reviewed and edited the manuscript. Correspondence and requests for materials should be addressed to P.R.T.
Declaration of interests:
The authors declare no competing interests.
References
- Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015 doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PJ, Lynch TJ, Engelhardt JF. Multipotent Myoepithelial Progenitor Cells Are Born Early during Airway Submucosal Gland Development. Am J Respir Cell Mol Biol. 2017;56:716–726. doi: 10.1165/rcmb.2016-0304OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbin BA, Parkos CA, Mandell KJ, Winfree LM, Laur O, Ivanov AI, Nusrat A. Annexin 2 regulates intestinal epithelial cell spreading and wound closure through Rho-related signaling. Am J Pathol. 2007;170:951–966. doi: 10.2353/ajpath.2007.060647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344:1242281. doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthwick DW, West JD, Keighren MA, Flockhart JH, Innes BA, Dorin JR. Murine submucosal glands are clonally derived and show a cystic fibrosis gene-dependent distribution pattern. Am J Respir Cell Mol Biol. 1999;20:1181–1189. doi: 10.1165/ajrcmb.20.6.3475. [DOI] [PubMed] [Google Scholar]
- Borthwick DW, Shahbazian M, Krantz QT, Dorin JR, Randell SH. Evidence for stem-cell niches in the tracheal epithelium. Am J Respir Cell Mol Biol. 2001;24:662–670. doi: 10.1165/ajrcmb.24.6.4217. [DOI] [PubMed] [Google Scholar]
- Buckpitt A, Chang AM, Weir A, Van Winkle L, Duan X, Philpot R, Plopper C. Relationship of cytochrome P450 activity to Clara cell cytotoxicity. IV. Metabolism of naphthalene and naphthalene oxide in microdissected airways from mice, rats, and hamsters. Mol Pharmacol. 1995;47:74–81. [PubMed] [Google Scholar]
- Choi HK, Finkbeiner WE, Widdicombe JH. A comparative study of mammalian tracheal mucous glands. J Anat. 2000;197(Pt 3):361–372. doi: 10.1046/j.1469-7580.2000.19730361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole BB, Smith RW, Jenkins KM, Graham BB, Reynolds PR, Reynolds SD. Tracheal Basal cells: a facultative progenitor cell pool. Am J Pathol. 2010;177:362–376. doi: 10.2353/ajpath.2010.090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal RG, Randell SH, Engelhardt JF, Voynow J, Sunday ME. Airway epithelial cells: current concepts and challenges. Proc Am Thorac Soc. 2008;5:772–777. doi: 10.1513/pats.200805-041HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, Rognoni E, Hiratsuka T, Liakath-Ali K, Hoste E, Kar G, Kayikci M, Russell R, Kretzschmar K, Mulder KW, et al. Wounding induces dedifferentiation of epidermal Gata6(+) cells and acquisition of stem cell properties. Nat Cell Biol. 2017;19:603–613. doi: 10.1038/ncb3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee ANY, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TJ, Colby TV, Travis WD, Tuder RM, Reynolds HY, Brody AR, Cardoso WV, Crystal RG, Drake CJ, Engelhardt J, et al. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc. 2008;5:763–766. doi: 10.1513/pats.200803-025HR. [DOI] [PubMed] [Google Scholar]
- Gao X, Bali AS, Randell SH, Hogan BLM. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J Cell Biol. 2015;211:669–682. doi: 10.1083/jcb.201506014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Helm KM, Smith RW, Giordanengo MS, Li B, Shen H, Reynolds SD. A single cell functions as a tissue-specific stem cell and the in vitro niche-forming cell. Am J Respir Cell Mol Biol. 2011;45:459–469. doi: 10.1165/rcmb.2010-0314OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnarsson M, Walther SM, Seidal T, Bloom GD, Lennquist S. Exposure to chlorine gas: effects on pulmonary function and morphology in anaesthetised and mechanically ventilated pigs. J Appl Toxicol JAT. 1998;18:249–255. doi: 10.1002/(sici)1099-1263(199807/08)18:4<249::aid-jat507>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Hartmann JF, Hutchison CF, Jewell ME. Pig bronchial mucous membrane: a model system for assessing respiratory mucus release in vitro. Exp Lung Res. 1984;6:59–70. doi: 10.3109/01902148409087895. [DOI] [PubMed] [Google Scholar]
- Hegab AE, Kubo H, Fujino N, Suzuki T, He M, Kato H, Yamaya M. Isolation and characterization of murine multipotent lung stem cells. Stem Cells Dev. 2010;19:523–536. doi: 10.1089/scd.2009.0287. [DOI] [PubMed] [Google Scholar]
- Hegab AE, Ha VL, Gilbert JL, Zhang KX, Malkoski SP, Chon AT, Darmawan DO, Bisht B, Ooi AT, Pellegrini M, et al. Novel stem/progenitor cell population from murine tracheal submucosal gland ducts with multipotent regenerative potential. Stem Cells Dayt Ohio. 2011;29:1283–1293. doi: 10.1002/stem.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegab AE, Nickerson DW, Ha VL, Darmawan DO, Gomperts BN. Repair and regeneration of tracheal surface epithelium and submucosal glands in a mouse model of hypoxic-ischemic injury. Respirol Carlton Vic. 2012;17:1101–1113. doi: 10.1111/j.1440-1843.2012.02204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BLM, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CCW, Niklason L, Calle E, Le A, Randell SH, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Giangreco A, Hurley CM, Stripp BR. Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol. 2001;24:671–681. doi: 10.1165/ajrcmb.24.6.4498. [DOI] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol. 2004a;164:577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol. 2004b;286:L643–649. doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery PK. Morphology of the airway wall in asthma and in chronic obstructive pulmonary disease. Am Rev Respir Dis. 1991;143:1152–1158. doi: 10.1164/ajrccm/143.5_Pt_1.1152. discussion 1161. [DOI] [PubMed] [Google Scholar]
- Judge EP, Hughes JML, Egan JJ, Maguire M, Molloy EL, O’Dea S. Anatomy and bronchoscopy of the porcine lung. A model for translational respiratory medicine. Am J Respir Cell Mol Biol. 2014;51:334–343. doi: 10.1165/rcmb.2013-0453TR. [DOI] [PubMed] [Google Scholar]
- Kotton DN, Morrisey EE. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med. 2014;20:822–832. doi: 10.1038/nm.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Engelhardt JF. The glandular stem/progenitor cell niche in airway development and repair. Proc Am Thorac Soc. 2008;5:682–688. doi: 10.1513/pats.200801-003AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Nettesheim P, Randell SH. Growth and differentiation of tracheal epithelial progenitor cells. Am J Physiol. 1994;266:L296–307. doi: 10.1152/ajplung.1994.266.3.L296. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Anderson PJ, Xie W, Crooke AK, Liu X, Tyler SR, Luo M, Kusner DM, Zhang Y, Neff T, et al. Wnt Signaling Regulates Airway Epithelial Stem Cells in Adult Murine Submucosal Glands. Stem Cells Dayt Ohio. 2016;34:2758–2771. doi: 10.1002/stem.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L. Accelerating t-SNE using Tree-Based Algorithms. J Mach Learn Res. 2014;15:3221–3245. [Google Scholar]
- Merrell AJ, Stanger BZ. Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat Rev Mol Cell Biol. 2016;17:413–425. doi: 10.1038/nrm.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, Zhang B, Solomon GM, Turner B, Bihler H, et al. Dual SMAD Signaling Inhibition Enables Long-Term Expansion of Diverse Epithelial Basal Cells. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan A, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018 doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noritake J, Watanabe T, Sato K, Wang S, Kaibuchi K. IQGAP1: a key regulator of adhesion and migration. J Cell Sci. 2005;118:2085–2092. doi: 10.1242/jcs.02379. [DOI] [PubMed] [Google Scholar]
- O’Koren EG, Hogan BLM, Gunn MD. Loss of basal cells precedes bronchiolitis obliterans-like pathological changes in a murine model of chlorine gas inhalation. Am J Respir Cell Mol Biol. 2013;49:788–797. doi: 10.1165/rcmb.2012-0369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A, Tata PR, Law BM, Saez B, Chow RDW, Prabhu M, Gridley T, Rajagopal J. Parent stem cells can serve as niches for their daughter cells. Nature. 2015a;523:597–601. doi: 10.1038/nature14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A, Law BM, Tata PR, Villoria J, Saez B, Mou H, Zhao R, Rajagopal J. Injury Induces Direct Lineage Segregation of Functionally Distinct Airway Basal Stem/Progenitor Cell Subpopulations. Cell Stem Cell. 2015b;16:184–197. doi: 10.1016/j.stem.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prater MD, Petit V, Alasdair Russell I, Giraddi RR, Shehata M, Menon S, Schulte R, Kalajzic I, Rath N, Olson MF, et al. Mammary stem cells have myoepithelial cell properties. Nat Cell Biol. 2014;16:942–950. 1–7. doi: 10.1038/ncb3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal J, Stanger BZ. Plasticity in the Adult: How Should the Waddington Diagram Be Applied to Regenerating Tissues? Dev Cell. 2016;36:133–137. doi: 10.1016/j.devcel.2015.12.021. [DOI] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BLM. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BLM. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8:639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013;502:513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden DT. Nice neighborhood: emerging concepts of the stem cell niche. Cell. 2014;157:41–50. doi: 10.1016/j.cell.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionescu-Bankston A, Leoni G, Wang Y, Pham PP, Ramalingam A, DuHadaway JB, Faundez V, Nusrat A, Prendergast GC, Pavlath GK. The N-BAR domain protein, Bin3, regulates Rac1- and Cdc42-dependent processes in myogenesis. Dev Biol. 2013;382:160–171. doi: 10.1016/j.ydbio.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda T, Deng JM, de Crombrugghe B, Behringer RR, Nakamura T, Akiyama H. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis. 2000. 2010;48:635–644. doi: 10.1002/dvg.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange DE, Koo BK, Huch M, Sibbel G, Basak O, Lyubimova A, Kujala P, Bartfeld S, Koster J, Geahlen JH, et al. Differentiated troy(+) chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripp BR, Maxson K, Mera R, Singh G. Plasticity of airway cell proliferation and gene expression after acute naphthalene injury. Am J Physiol. 1995;269:L791–799. doi: 10.1152/ajplung.1995.269.6.L791. [DOI] [PubMed] [Google Scholar]
- Tata PR, Rajagopal J. Cellular plasticity: 1712 to the present day. Curr Opin Cell Biol. 2016;43:46–54. doi: 10.1016/j.ceb.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Rajagopal J. Plasticity in the lung: making and breaking cell identity. Dev Camb Engl. 2017;144:755–766. doi: 10.1242/dev.143784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh PW, Farin HF, Clevers H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 2015;25:100–108. doi: 10.1016/j.tcb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Van Winkle LS, Buckpitt AR, Nishio SJ, Isaac JM, Plopper CG. Cellular response in naphthalene-induced Clara cell injury and bronchiolar epithelial repair in mice. Am J Physiol. 1995;269:L800–818. doi: 10.1152/ajplung.1995.269.6.L800. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Clevers H. Tissue-specific designs of stem cell hierarchies. Nat Cell Biol. 2016;18:349–355. doi: 10.1038/ncb3332. [DOI] [PubMed] [Google Scholar]
- Wang J, Winskog C, Edston E, Walther SM. Inhaled and intravenous corticosteroids both attenuate chlorine gas-induced lung injury in pigs. Acta Anaesthesiol Scand. 2005;49:183–190. doi: 10.1111/j.1399-6576.2004.00563.x. [DOI] [PubMed] [Google Scholar]
- Wansleeben C, Bowie E, Hotten DF, Yu YRA, Hogan BLM. Age-related changes in the cellular composition and epithelial organization of the mouse trachea. PloS One. 2014;9:e93496. doi: 10.1371/journal.pone.0093496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling O, Bornert JM, Chambon P, Metzger D. Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genes N Y N 2000. 2009;47:14–18. doi: 10.1002/dvg.20448. [DOI] [PubMed] [Google Scholar]
- Xiong GP, Zhang JX, Gu SP, Wu YB, Liu JF. Overexpression of ECM1 contributes to migration and invasion in cholangiocarcinoma cell. Neoplasma. 2012;59:409–415. doi: 10.4149/neo_2012_053. [DOI] [PubMed] [Google Scholar]
- Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, Morrisey EE. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Zhang X, Weichert-Leahey N, Dong Z, Zhang C, Lopez G, Tao T, He S, Wood AC, Oldridge D, et al. LMO1 Synergizes with MYCN to Promote Neuroblastoma Initiation and Metastasis. Cancer Cell. 2017;32:310–323.e5. doi: 10.1016/j.ccell.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


