Abstract
The variation and taxonomic diversity among mammalian gut microbiomes raises several questions about the factors that contribute to the rates and patterns of change in these microbial communities. By comparing the microbiome compositions of 112 species representing 14 mammalian orders, we assessed how host and ecological factors contribute to microbiome diversification. Except in rare cases, the same bacterial phyla predominate in mammalian gut microbiomes, and there has been some convergence of microbiome compositions according to dietary category across all mammalians lineages except Chiropterans (bats), which possess high proportions of Proteobacteria and tend to be most similar to one another regardless of diet. At lower taxonomic ranks (families, genera, 97% OTUs), bacteria are more likely to be associated with a particular mammalian lineage than with a particular dietary category, resulting in a strong phylogenetic signal in the degree to which microbiomes diverge. Despite different physiologies, the gut microbiomes of several mammalian lineages have diverged at roughly the same rate over the past 75 million years; however, the gut microbiomes of Cetartiodactyla (ruminants, whales, hippopotami) have evolved much faster and those of Chiropterans much slower. Contrary to expectations, the number of dietary transitions within a lineage does not influence rates of microbiome divergence, but instead, some of the most dramatic changes are associated with the loss of bacterial taxa, such as those accompanying the transition from terrestrial to marine lifestyles and the evolution of hominids.
Keywords: molecular evolution, microbiome, evolutionary rates, phylosymbiosis
Introduction
There is tremendous variation in the composition of the gut microbiomes of mammalian species—and even among individuals of the same species. Both genetic and environmental factors, such as sex, geography, diet, and disease-state, contribute to differences in microbial community composition among individuals (Degnan et al. 2012; Bonder et al. 2016; Gilbert et al. 2016). Across a broad taxonomic range, the gut microbiome often appears adapted to assist in metabolic processes related to host diet (Muegge et al. 2011a; Delsuc et al. 2014b; Sanders et al. 2015a). For example, in contrast to dolphins, baleen whales harbor bacteria that specialize in degrading chitin, a prominent component of the invertebrate exoskeletons that comprise their diet (Sanders et al. 2015a). Even at the level of individuals, changes in diet or feeding regimes have been shown to alter microbial community compositions in both humans and mice (Wu et al. 2011; David et al. 2014; Carmody et al. 2015).
The gut environment is subject to a constant influx of microbial colonizers, and yet, mammalian species harbor distinct microbiomes and can be readily differentiated based on their resident microbes. Among very recently diverged species with similar diets, and even in cases where co-occurring species participate in microbial transfer, it is possible to partition host species based on their microbiomes (Moeller et al. 2013; Song et al. 2013; Amato et al. 2015). And while it has repeatedly been shown that variation in environmental factors, such as diet and lifestyle, contribute to differences in the microbiome composition in human populations (De Filippo et al. 2010a; O’Keefe et al. 2015; Rampelli et al. 2015), genetic factors have also been shown to play a role. Genome-wide studies indicate that individuals of different ethnicities experience varying levels of selection on genetic sites associated with microbiome composition (Blekhman et al. 2015). Furthermore, genetic differences among individuals are associated with the abundance of certain microbial taxa (Goodrich et al. 2014, 2016a; Davenport et al. 2015)
Despite the high level of host- and species-specificity of gut microbiomes, and the potential for rapid change in response to diet and environment, it was recently reported that microbiome composition diverges at a relatively constant rate over evolutionary timescales (Moeller et al. 2014). Comparisons of samples from great apes revealed that species accumulate differences in microbiome composition in a clocklike manner, but that humans show accelerated divergence due to the loss of microbial diversity arising from changes in lifestyle and diet. To explore how microbiomes diverge across different taxonomic groups, we analyzed the microbiome compositions of over 100 mammalian species, and assessed the taxonomic and dietary factors contributing to microbiome diversification. Among major mammalian orders, there is substantial variation in rates of microbiome divergence, with two of the most intensely sampled orders, Cetartiodactyla and Primates, presenting opposite trends. Microbiome divergence in the Cetartiodactyla lineage is associated with changes in diet and environment, whereas Primates steadily accumulate differences in their microbiome composition regardless of diet.
Materials and Methods
Data acquisition and taxonomic sampling
Sequence data used to assess gut microbial diversity were obtained for an initial total of 116 species representing 14 orders of mammals, as available from the QIITA website (qiita.ucsd.edu) and other sources (Table S1). Because DNA extraction methods can affect the recovery of certain bacterial taxa (Yuan et al. 2012), we limited analyses to studies that applied a bead-beating approach to obtain DNA from fecal samples (Table S2). Due to the variation in the numbers of individuals sampled from each species (which ranged from one to several hundred individuals), we randomly selected five individuals from each of the species datasets for which larger numbers of samples were available. In the two instances where a particular species was surveyed in two separate studies where more than five conspecifics were sampled, we included individuals from both studies (Table S2). Because several of the samples from insectivorous species showed signs of contamination from soil bacteria (Delsuc et al. 2014), we included only those samples that passed a source-tracking threshold set by the authors. To determine how well samples of this size represent total microbial diversity, we performed a subsampling procedure on the datasets of Pan paniscus and Gorilla gorilla (Figure S1).
Sequence quality filtering and preprocessing
Sequence files were visualized for quality in FastQC, and sequence reads were trimmed and filtered with Prinseq (Andrews & others 2010; Schmieder & Edwards 2011). Only reads with average quality scores >25 over a 20-nucleotide window were retained. For studies that did not release FASTQ data, the available FASTA files were merged with the quality-filtered FASTQ files. Reads were sorted according the particular 16S rRNA region they overlapped (V2 or V4). For each of the region-specific datasets, reads were aligned in mothur (Schloss et al. 2009) to the SILVA reference database (Quast et al. 2012) and trimmed at uniform positions (V2: 200 nt ending at position 6333; V4: 100 nt beginning at position 13862). Because sequence datasets varied in length, these positions were selected to maximize the number of sequences overlapping a uniform region. Sequences whose lengths fell short of 75% of either of the defined regions were removed.
Microbial community analysis
Estimates of diversity within and among taxonomic ranks were calculated on the V2 and V4 regions separately. Sequences were clustered de novo into 97% OTUs with Vsearch (Rognes et al. 2016). Sequences aligning with less than 70% identity to the SILVA reference database (Quast et al. 2012) and those deemed chimeric by UCHIME (Edgar et al. 2011) were removed. Further processing and analysis were performed with QIIME (Caporaso et al. 2010), such that OTUs represented by a single read as well as those that could not be assigned to Bacteria or Archaea (including unassigned, chloroplast, and eukaryotic sequences) were removed. Samples that did not rarefy to the threshold number of sequences (V2: 1500 sequences; V4: 5000 sequences) were also omitted, reducing the number of species included in our analyses to 112 (Table S2). Due to the differences in sequencing platforms and variation in sampling depth of published studies, rarefaction depths were chosen to maximize number and diversity of hosts sampled. Because the number of individuals per species ranged from 1 to 5, reads from all individuals of a species were collapsed into a single pool. We rarefied the collapsed single pool 10 times and averaged OTU counts across the rarefactions to generate a representative sample for each species.
Comparing microbial diversity by host lineage and diet
Host diet and body mass information were obtained from published sources (Table S1). Host species were consolidated into four dietary categories: herbivore, omnivore, predatory carnivore (i.e., eating vertebrate prey) and invertivorous carnivore. This last dietary category includes baleen whales, myrmecophagous mammals, and some bat species, as their diet is principally invertebrate-based. In contrast, insectivores that consume a wide range of plants, such as the nine-banded and pink-fairy armadillos, were designated as omnivores.
The relationship between body mass and microbial diversity was analyzed with Kendall rank correlation tests. Microbial diversity was indexed by both a taxonomic metric (total number of OTUs) and a phylogenetic metric [Faith’s Phylogenetic Diversity (Faith 1992)]. To investigate the phylogenetic independence of these trends, we conducted a PGLS analysis using a Brownian motion model of evolution (Felsenstein, 1985). We performed Kendall rank correlation tests among taxa assigned to the various dietary categories. To determine the relative influence of small-bodied organisms on the relationship between body mass and microbial diversity, we repeated the analysis after excluding species weighing more than one kilogram. Furthermore, this analysis was also implemented within selected host lineages—termed the “K-T lineages”—that all originated around the time of the K-T mass extinction and that contained more than five sampled species. In the V4 dataset, five lineages (Carnivora, Cetartiodactyla, Chiroptera, Primates, and Xenarthra), and in the V2 dataset, three lineages (Carnivora, Cetartiodactyla, and Primates) met these criteria.
To compare the relative influence of host dietary category and K-T lineage, we performed a perMANOVA analysis on unweighted uniFrac distances using 999 permutations. We compared the relative abundances of bacteria and archaea at various taxonomic ranks, ranging from phyla to 97% OTUs (roughly equivalent to species), using Kruskal-Wallis tests, applying a Bonferroni correction for multiple testing (Supplemental Data). We determined microbial lineages that were significantly enriched in species assigned to a particular dietary category or host phylogenetic lineage. We also calculated the core microbiome (microbial lineages present in over 95% of the species) across all mammals as well as individually for each mammalian order, for each K-T lineage, and for taxa within each of the dietary categories. Beta-diversity was visualized with PCoA plots ordinated by weighted and unweighted UniFrac distances on each of the region-specific datasets.
Host phylogenies
Divergence times for the major mammalian lineages and radiations were obtained from published sources (Table S3). Genera were assumed to be monophyletic, such that intergeneric divergence times could be applied to all species within a given genus. In those few instances where published phylogenies showed incongruencies in the branching order of closely related clades, we considered the node as an unresolved polytomy.
Rates of microbiome divergence
Average rates of microbiome divergence within each of the K-T lineages were based OTU dissimilarities among species’ microbiomes in relation to the corresponding species’ divergence times. To test whether the microbiome divergence rates among K-T lineages were consistent among different dissimilarity metrics, we calculated the microbiome divergence rate applying both phylogenetic metrics (unweighted and weighted UniFrac) and non-phylogenetic metrics (Bray-Curtis dissimilarity and percentage of shared OTUs) of dissimilarity. Phylogenetic metrics give more weight to the gains and losses of divergent microbial taxa than to those taxa that just pass the 97% OTU threshold. Moreover, these metrics offer an additional perspective on microbiomes dissimilarity in that some account for the relative abundance of microbial taxa whereas others are based only on the presence/absence of microbial taxa. The percentage of shared OTUs was calculated by dividing the number of shared OTUs by the total number of OTUs between two taxa, with the percentage of unique OTUs is equal to 100 minus the fraction of shared OTUs.
We calculated microbiome divergence rates for all examined species in the V2 and V4 datasets and within each of the K-T lineages. We did not include intraspecies comparisons of microbiome composition because these data were not available for all species. We also determined the microbiome divergence rate within selected sublineages that had a common ancestor (MRCA) within the last 30 million years (for example, the Cetartiodactyla lineage as well as its monophyletic sublineages Cetacea and Ruminantia). Analyzing sublineages within a clade allows recognition of any deviations in microbiome divergence rates. To evaluate the influence of host phylogeny on microbiome dissimilarity, we performed Mantel tests between matrices of host divergence times and microbiome dissimilarities with 10,000 permutations.
To examine the effect of host dietary changes on rates of microbiome divergence, we sorted all pairwise comparisons among hosts into subcategories denoting whether their diets were the same or differed. In addition to the six possible transitions among the four dietary categories, we distinguished whether species are of the same dietary category due to common ancestry or dietary convergence, resulting in eight additional subcategories. (To distinguish dietary convergence from common ancestry, we examined the phylogenetic relationships of hosts and determined whether all species descended from the common ancestor of the focal species have the same diet.) For this analysis, we pooled all pairwise-difference values for the V2 and V4 regions and in cases where a particular species-pair was compared for both the V2 and V4 regions, the mean dissimilarity was calculated. For each of the 14 diet-change subcategories, the relationship between divergence time and dissimilarity was analyzed with Kendall rank correlation tests. To compare rates of microbiome divergence among host lineages and dietary change subcategories, we generated 95% confidence intervals by bootstrapping 5,000 replicates for both the slope and y-intercept in a linear regression of microbiome dissimilarity and divergence time.
Results
Body size, diet, and gut microbial diversity
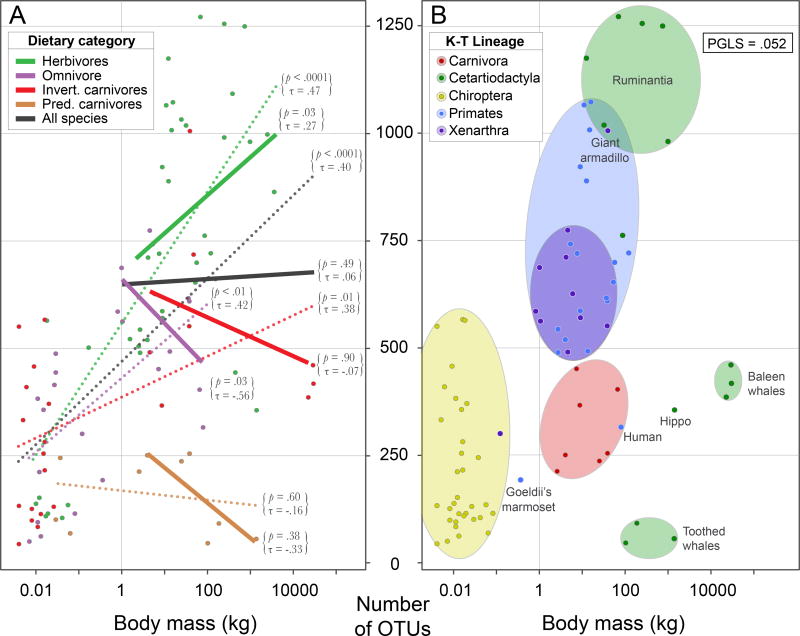
Microbial diversity within gut microbiomes is reported as being higher in larger hosts (Godon et al. 2016). This trend manifests only when all taxa are considered, and it is not evident in each of the individual dietary categories or when the smallest organisms (those weighing less than one kilogram) are excluded (Figure 1a). Furthermore, this trend also depends on the taxonomic scale at which the association is examined (Figure 1b). Focusing initially on variation detected in the V4 region rarefied to depths of 5000 reads per individual (since this maximizes the number and taxonomic breadth of species being compared), we detect a positive association between body mass and microbial diversity (PGLS, p = .05). After separating taxa into different dietary categories, herbivores exhibit the largest gain in microbial diversity with body size, increasing, on average, at about twice the rate of omnivores and carnivorous invertivores (Figure 1a).
Figure 1. Association between body mass and gut microbial diversity.
Microbial diversity is expressed as the number of 97% OTUs present in each mammalian host (colored-coded dots) surveyed for sequence variation in the V4 region of 16S rDNA, rarefied to 5000 reads. (A) Host species sorted to dietary category. Solid lines correspond to the regression for species greater than 1 kg, and dotted lines correspond to the regression for all species, in a particular dietary category. After removing hosts <1 kg, only herbivores display a significant positive association (assessed by Kendall rank correlation test) between microbial diversity and body mass. (B) Host species sorted taxonomically. Color-coded ellipses enclose those host species assigned to one of the five K-T lineages. Note that there is no significant relationship between microbial diversity and body mass when hosts are sorted taxonomically and that this is particularly pronounced for Cetartiodactyla, in which large marine mammals have among the lowest OTU diversity.
Among carnivores, the association between body mass and gut microbial diversity is restricted to invertivores, and there is no significant relationship in predatory carnivores. We examined the relationship between body mass and gut microbiome diversity in additional ways to determine the strength of the association in different taxonomic or dietary groups, and to identify factors that might contribute to the trend. Herbivores, which showed the strongest trend, typically have the highest gut microbial diversity (although the giant armadillo, an insectivore, exhibits the same level of microbial diversity as that of large herbivorous ruminants), whereas predatory carnivores harbor the lowest gut microbial diversity, with larger hosts having roughly the same, or sometimes lower, numbers of OTUs than smaller hosts.
Notably, the gut microbiomes of all sampled mammals weighing less than 100 grams, regardless to which dietary category they were assigned, contain low numbers of 97% OTUs (Figure 1a). Excluding these small-bodied hosts from the analysis reveals that only herbivores—not omnivores or either invertivorous or predatory carnivores—exhibit a significant increase in microbial diversity with body mass, as indexed by total number of OTUs (Figure 1a) or by Faith’s Phylogenetic Diversity (Figure S2).
Furthermore, confining analyses to the K-T lineages (i.e., the five mammalian orders originating at the K-T boundary that each contained more than five sampled species), we find that none exhibits a significant positive relationship between body mass and microbial diversity (Figure 1b) and that some display a negative association. The decrease in microbial diversity with body mass in several of these K-T lineages can be traced to dietary differences among the constituent taxa. For example, within the Cetartiodactyla, carnivorous marine Cetaceans (toothed and baleen whales) have the largest body masses and gut sizes, but harbor significantly lower gut microbial diversity than do ruminants.
Aside from differences in microbial diversity that are attributable to body size and diet, several other features emerged from these comparisons. For example, marine-dwelling herbivores harbor less microbial diversity than land-dwelling herbivores of comparable size—a trend observed for different mammalian groups in several geographic locations (such as manatees and hippos). Among primates, humans, which were already known to possess the lowest microbial diversity among the great apes, maintain about twice the gut microbial diversity of the Goeldi's monkey, which has a diet predominated by fungi (Porter et al. 2007). When microbial diversity is assessed by Faith’s Phylogenetic Diversity, the trends observed for the V4 datasets are almost identical to those observed when microbial diversity is assessed by the number of OTUs (Figure S2). Many of the associations reported above were also observed in the V2 dataset, but fewer reach statistical significance due to the smaller number of sampled species (Figure S3).
The influence of diet and phylogeny on microbiome contents
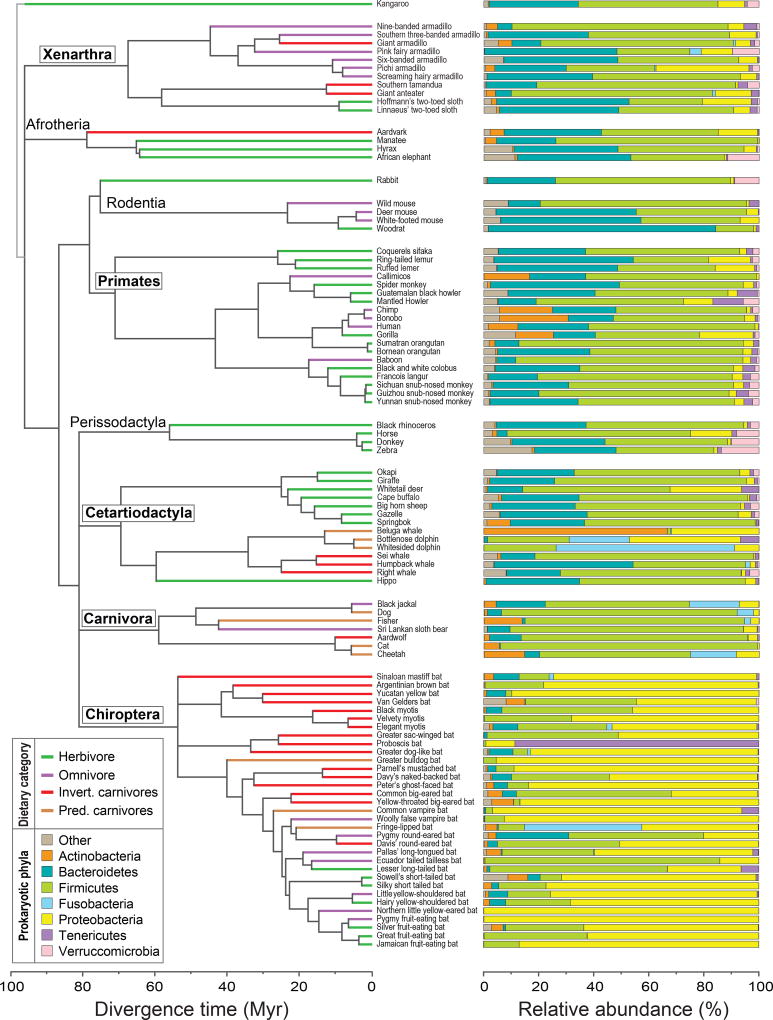
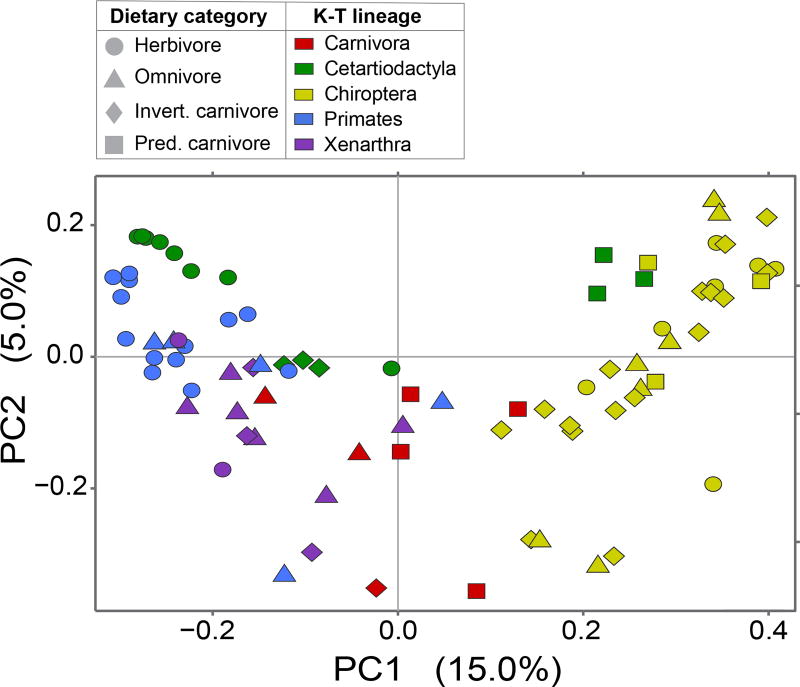
Both the breadth of taxonomic diversity and the relative abundances of bacterial lineages vary substantially with host taxonomy and diet. Four bacterial phyla—Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria—constitute the core mammalian microbiome (i.e., present in over 95% of all samples). Among the K-T lineages, Chiropterans are enriched in Proteobacteria and are the most compositionally distinct, whereas Bacteroidetes and Firmicutes predominate in the other lineages (Figure 2). Accordingly, when species are ordinated based on OTU composition, Chiropterans generally group together (yellow symbols in Figure 3), whereas other taxa assort according to both host phylogeny and dietary category. Excluding Chiropterans from the analysis, the PCoA plot separates herbivores, which are enriched in Bacteroidetes and other less abundant phyla (such as Planctomycetes and Verrucomicrobia) from primary carnivores, with omnivores and invertivores situated intermediate to the two (Figure 3). Although the V2 dataset was not as extensive in its coverage of host species, it also separates herbivorous from carnivorous species (with the exception of the herbivorous panda), with omnivorous species intermediate to these two clusters (Figure S6). Notably, predatory mammals form a distinct cluster due to the high relative abundance of Fusobacteria, as previously reported (Roggenbuck et al. 2014; Nelson et al. 2013).
Figure 2. Phylogenetic relationships and gut microbiome compositions of mammalian hosts.
Names of the five K-T lineages are boxed. Terminal branches are color-coded according to host dietary category. Taxonomic classification of gut microbes is at the level of phylum. Branching orders and divergence times obtained from sources listed in Table S3.
Figure 3. Principal coordinates plot showing similarity in gut microbiome composition among hosts.
Ordination based on unweighted Unifrac distances among 97% OTUs. Geometric shapes denote host dietary categories, with host lineages color-coded.
Within the four phyla that constitute their core microbiome, all surveyed mammals possess two classes of Firmicutes (Bacilli and Clostridia) and two classes of Proteobacteria (Beta- and Gamma-) (Table S4). Although a single order, Clostridiales, is part of the mammalian core gut microbiome, within this order, herbivores and carnivores harbor different families (Ruminococcaceae and Peptostreptococcaceae, respectively) (Table S5). Herbivores harbor a greater microbial diversity at almost every taxonomic level.
Based on a perMANOVA analysis, phylogenetic lineage has a larger relative effect on microbiome composition (Psuedo-F 4,83 = 5.90, p < .001) than does host dietary category (Pseudo-F3,83 = 2.81, p < .001), as evident by the observation that the relative abundances of several microbial lineages differ among K-T lineages but not among dietary categories (Tables S6 and S7). At lower taxonomic ranks, the majority of bacterial lineages are more likely to be associated with a particular K-T lineage rather than with a particular dietary category, even though non-Chiropteran species still sort according to diet when clustered using a phylogenetic method (Figure 3). Approximately 10% (40) of archaeal and bacterial families vary significantly in their abundances according to host lineage (Table S6), whereas only 2% (8) families vary significantly according to host dietary group (Table S7). K-T lineages also vary dramatically in the number of bacterial lineages that constitute their core microbiomes: Cetartiodactyla species share just a single family (Clostridiaceae), whereas Xenarthran species share 15 families from five phyla as part of their core microbiome (Table S4).
Host divergence affects microbiome dissimilarity
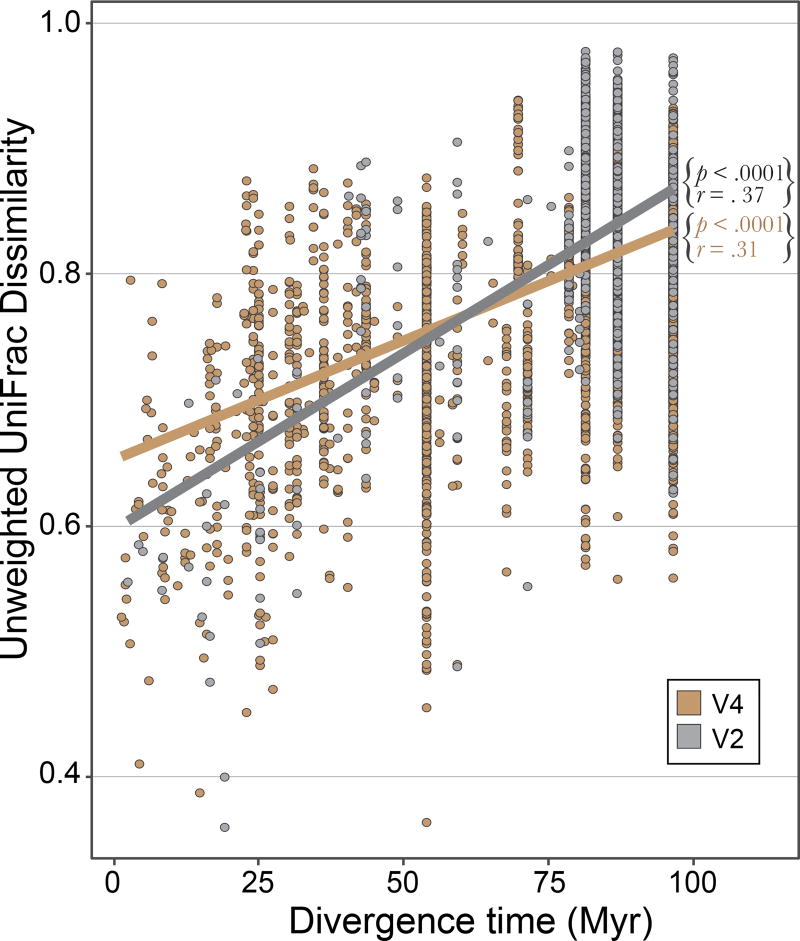
Although divergent host species assigned to the same dietary category share several bacterial taxa, there is a strong phylogenetic signal in the degree to which microbiomes diverge, such that more closely related species generally exhibit more similar communities of bacteria (Figure 4). Based on unweighted UniFrac dissimilarities, approximately 1.5% of shared phylogenetic branch-length between gut microbiomes is lost every 10 million years (Table S8). Based on a non-phylogenetic metric, this amounts to a loss of about 2% of shared OTUs over the same interval of time (Figure S7, Table S8). This relationship is confounded by the fact the closely related organisms often have more similar lifestyles, diets and niches; however, the trend persists over long evolutionary timescales. There is a significant positive relationship between divergence time and microbiome dissimilarity spanning 100 million years of mammalian evolution—an association maintained despite numerous instances of dietary convergence among species from different lineages and of closely related species occupying very different environments. Although the V2 and V4 regions furnish different amounts of variation, when considering all mammals, both datasets yield roughly the same absolute rate of microbiome divergence (Figure 4).
Figure 4. Rate of microbiome divergence across mammals.
Dissimilarities among species calculated from unweighted Unifrac distances of 97% OTUs for both the V2 (silver) and V4 (gold) datasets. Strengths of correlations assessed by Mantel tests with 10,000 permutations.
The relationship between microbiome dissimilarity and host divergence plateaus and is best fit by a polynomial second-order function (Voung’s closeness test, p < .0001). To establish the date of this saturation point, we calculated the intersection between the random expectation of dissimilarity between any two species and the regression model. All four dissimilarity metrics reached a saturation point between 65 and 75 million years ago (Figure S7). Therefore, K-T lineages represent an appropriate timescale to make meaningful comparisons among host lineages because the majority share a common ancestor that diverged around this time. Among the K-T lineages, Chiroptera and Cetartiodactyla display rates of microbiome divergence that differ significantly from the mammalian average both in terms of percentage of shared OTUs and percentage of unweighted UniFrac distance lost per 10 million years (Table 1) as well as the other surveyed dissimilarity metrics (Table S8). The Cetartiodactyla microbiomes diverge at roughly three times the average rate, whereas Chiropteran microbiomes diverged at rates that were significantly slower. For Carnivora, the rate of microbiome divergence could only be assessed with the V2 dataset, for which a greater range of species was sampled, and the resulting rate was similar to the average (Table S9). Additionally, three mammalian lineages, Perissodactyla, Rodentia, Afrotheria (which were not considered K-T lineages due to their more limited sampling) show similar patterns of divergence (Figure S8).
Table 1.
Rates of microbiome divergence within K-T lineages.
| Host lineagea | Number of host species |
Percentage of unweighted UniFrac distance lost per 10 Myrc |
Percentage of shared OTUs lost per 10 Myrc |
Number of shared OTUs lost per 10 Myr |
Dietary categoriesc |
Number of dietary transitionsd |
Number of possible transitionse |
|---|---|---|---|---|---|---|---|
| Cetartiodactylab | 14 | 4.88 (4.20, 5.52) | 5.95 (4.95, 6.87) | 41 | H,IC,PC | 2 | 26 |
| Primates | 19 | 1.31 (0.801, 1.83) | 2,11 (1.56, 2.62) | 14 | H,O | 3 | 36 |
| Xenarthra | 11 | 0.74 (0.024, 1.47) | 1.21 (0.47, 2.08) | 8 | H,O,IC | 3 | 20 |
| Carnivora | 7 | 0.0612 (−3.18, 0.99) | 1.06 (−3.37, 2.87) | 3 | O,IC,PC | 3 | 12 |
| Chiropterab | 33 | 0.363 (0.18, 0.91) | 0.76 (0.31, 1.17) | 2 | H,O,IC,PC | 10 | 61 |
| Average | 84 | 1.52 (1.19, 1.86) | 2.16 (1.85, 2.47) | 10 | H,O,IC,PC | 21 | 155 |
Analyses confined to K-T lineages, which are those lineages with >5 sampled species.
Highlighted rows indicate those lineages whose divergence rates differ significantly from the average.
Rates of microbiome divergence were inferred from changes in microbiome compositions in species assigned to a particular K-T lineage and calculated by unweighted phylogenetic and taxonomic beta diversity metrics. 95% confidence intervals (in parentheses) were generated from bootstrapped replicates for linear slope correlations.
H, herbivore; O, omnivore; IC, invertivorous carnivore; PC, predatory carnivore
Dietary transitions inferred by parsimony based on current host diet and phylogenetic relationships
Number of possible dietary transitions calculated as the sum of terminal and internal branches for a K-T lineage phylogeny
Effects of dietary change on rates of microbiome divergence
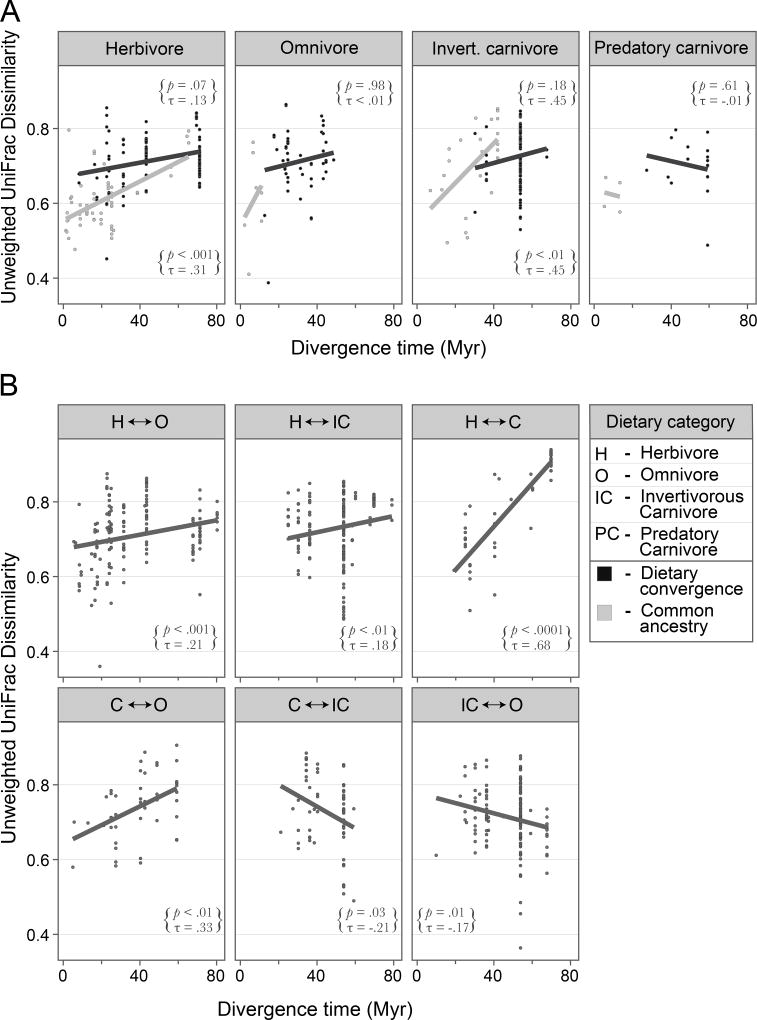
To determine the effect of dietary transitions on the rate of microbiome divergence, we first asked whether related species of the same dietary category displayed different degrees of microbiome dissimilarity depending on whether their diet arose through shared ancestry or by convergence. Overall, comparisons among species that evolved the same diet independently had higher microbiome dissimilarity than those species that maintained the same diet since descending from a common ancestor (p < .0001), a result that was apparent for hosts in all dietary categories (Figure 5a). One potentially confounding factor is that species that have undergone dietary convergence are more distantly related than those species having the same diet due to ancestry, so we limited our analyses to taxa where there were no biases in divergence times and obtained the identical result (p < .0001).
Figure 5. Effect of dietary transition on rates of microbiome divergence.
Dietary transitions inferred by parsimony from the phylogeny in Figure 2. Rates are based on comparisons among species that diverged < 80 million years ago. Microbiome dissimilarities among species were calculated from unweighted Unifrac distances of 97% OTUs. Strengths of associations were assessed with a Kendall rank correlation test. (A) Rates of microbiome divergence among species that have the same diet due to either dietary convergence (upper p and τ values) or common ancestry (lower p and τ values). Note that neither omnivores nor predatory carnivores with the same diet due to common ancestry had sufficient comparisons to perform statistical tests. (B) Rates of microbiome divergence for different types of dietary transition.
To determine whether particular types of dietary transitions are associated with enhanced levels of microbiome dissimilarities, we analyzed 14 categories of dietary transitions within the K-T lineages (thereby excluding comparisons among species that diverged over 75 million years ago). We observed positive relationships between host divergence time and microbiome dissimilarity in those cases where species have the same diet due to ancestry, but not in cases where species acquired the same diet independently (Figure 5a). The transition between carnivory and herbivory produces the steepest rates of microbiome divergence, and for two of dietary transitions—i.e., the transition between carnivorous invertivore and omnivore, and between carnivorous invertivore and predatory carnivore—there were negative associations between divergence time and microbiome dissimilarity (Figure 5b). However, the number of observations limited the statistical power to compare divergence rates across dietary categories.
How constant is the rate of microbiome divergence?
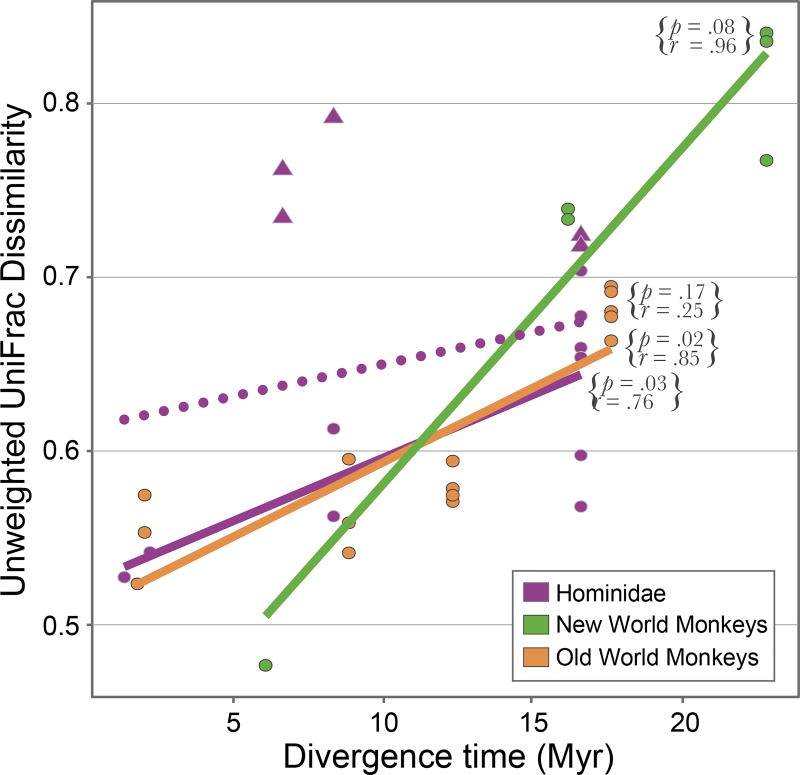
To examine whether microbiome divergence occurs in periodic bursts or at more uniform rates, we analyzed subclades within those K-T lineages in which multiple sister taxa were sampled and whose representative species span different divergence intervals. Within Primates, both the Old World Monkey (OWM) lineage and the New World Monkey (NWM) lineage display linear, but highly accelerated, rates of microbiome evolution over the past 20 million years (Figure 6). When humans are excluded, the rate of microbiome divergence in Hominidae is very similar to the rate observed in OWM, with species’ microbiomes losing 8% of their shared phylogenetic branch length and 10% of their shared OTUs per 10 million years. Rates of microbiome divergence in each of these primate families are 10 times faster compared to the rate observed across all Primates, again supporting the view that the extent of microbiome divergence reaches a saturation point.
Figure 6. Similar rates of microbiome divergence in primate sublineages.
Relationship between host divergence time and microbiome dissimilarity among three lineages of Primates: Hominidae, Old World monkeys (OWM), and New World monkeys (NWM). Circles represent comparisons between nonhuman apes, and triangles represent comparisons between humans and other hominid species. The dotted line represents the relationship within Hominidae when humans, whose gut microbiomes are known to evolve at accelerated rates (Moeller et al. 2014), are included. The strength of the correlation was assessed using a Mantel test with the complete set of possible permutations.
In contrast to Primates, in which changes in the microbiome occur at a relatively constant rate, changes to the Cetartiodactyla microbiome occur with periodic bursts in response to changes in diet and environment. When Cetartiodacytla are categorized by diet, there is no significant trend between divergence time and microbiome dissimilarity. However, we observe significant differences in microbiome dissimilarity among species that share the same diet versus those with different diets.
Analytical Constraints
Because this study derives data from multiple sources, several factors, such as differences in sampling (wild vs. captive individuals; taxonomic coverage of host species) and/or sequencing protocol could potentially affect estimates of microbiome diversity and comparisons across samples. The available datasets did not allow direct comparisons of wild and captive individuals of the same species; however, we observed no significant difference in the microbiome divergence rates of captive or wild individuals (Figure S9). Similarly, neither the sequencing method nor the particular 16S rRNA region sequenced had a significant effect on microbiome composition (Table S10). However, DNA extraction method yielded a significant effect on microbiome composition, but its effect was slight relative to biological factors, such as diet and K-T lineage.
Discussion
We amassed microbiome data from a phylogenetically diverse cohort of over 100 mammalian host species to analyze the pace of evolution in microbial community composition and the factors that contribute to gut microbiome divergence. Previous studies have shown that there is a broad phylogenetic signal in the divergence of microbiomes but that other factors, most notably diet and body mass, influence the diversity and composition of gut microbial communities (Ley et al. 2008; Ochman et al. 2010; Muegge et al. 2011a; Phillips et al. 2012; Delsuc et al. 2014b; Sanders et al. 2015a; Godon et al. 2016; Groussin et al. 2017; Hale et al. 2017). With this expanded sampling, we sought to determine the background rate at which microbiome changes accrue and the host features that accelerate or confine this rate. Overall, as taxa diverge, their microbiomes diverge as well—at a rate of approximately 2% of shared 97% OTUs or 1.5% of shared phylogenetic branch length lost every 10 million years. This rate of change is common to taxonomic groups that differ in their habitats and geographic ranges as well as in their feeding habits and the degree of heterogeneity of dietary-types within a lineage, and we use it as a baseline for comparing how mammalian microbiomes change over evolutionary timescales.
Because it is necessary to determine when comparisons of microbiome divergence are still meaningful, we plotted the degree of microbiome dissimilarity based on a variety of metrics against host divergence times. For each of the dissimilarity metrics, this saturation point, i.e., the point at which microbiome divergence asymptotes and no longer becomes more dissimilar with time, occurs between 65 and 75 million years ago. For those metrics based on the presence or absence of microbial lineages (i.e., unweighted UniFrac and % shared OTUs), rare microbial taxa have a greater impact on the microbiome dissimilarity between host species. These unweighted metrics saturate at slightly later dates and explain a greater amount of variation than metrics based on the relative abundances of taxa (Figure S7).
Microbiome composition can fluctuate in response to dietary and environmental changes. Seasonal availability of food and modifications to the diet of individuals have been shown to influence microbiome composition in several mammalian species (Turnbaugh et al. 2009; Kohl & Dearing 2014; Davenport et al. 2014; Amato et al. 2014; Degnan et al. 2012; Sun et al. 2016; Gomez et al. 2016). In humans, adopting a high-protein diet increases the abundance of bile-tolerant bacteria, whereas a plant-based diet enriches microbial taxa that specialize in carbohydrate fermentation (David et al. 2014; O’Keefe et al. 2015), paralleling the differences observed in gut microbiomes of hunter gathers vs. humans in industrialized societies (Rampelli et al. 2015; Morton et al. 2015a).
Despite the potential for large shifts in the microbiome composition of individuals and the diversity within populations due to diet, mammalian species can be distinguished from one another based on their microbiome compositions (Nelson et al. 2013; Delsuc et al. 2014b; Moeller et al. 2014; Sanders et al. 2015a). Even though captivity can decrease microbial diversity (McKenzie et al. 2017) and “humanize” the composition of gut microbiomes (Clayton et al. 2016), the degree of similarity among the microbiomes of captive primate species remained largely concordant with host phylogeny, suggesting that species retain their lineage-specific microbial taxa during compositional shifts.
Although there is a pervasive phylogenetic signal in the microbiome composition of mammals, we detected several factors that alter rates of microbiome divergence. First is body size—the body masses of mammals surveyed span six orders of magnitude, and the K-T lineages at the extremes showed the most disparate rates of microbiome evolution. Cetartiodactyla, which include the most massive terrestrial and marine mammals (ruminants, hippopotami, whales), diverged significantly faster than the baseline rate, whereas Chiropterans (bats), the smallest hosts, diverged significantly slower. Chiropterans harbor the lowest microbial diversity, while Cetartiodactyla include several species that possess among the highest gut microbial diversity.
It was previously reported that microbial community diversity increases with host mass (Godon et al. 2016). It is noteworthy that the sampling by Godon et al., who initially reported the broad trend between body mass and gut microbial diversity, was biased towards herbivorous hosts. By sampling across a range of dietary categories, we find that larger hosts do not universally harbor more microbial diversity, and that the positive association between body mass and microbiome diversity is largely limited to herbivores. Omnivores and carnivores have simpler intestinal tracts and, unlike herbivores, their gut capacity does not scale linearly with body mass (Clauss et al. 2013; Karasov & Douglas 2013). This suggests that gut capacity, which depends both on gut physiology as well as host size, is a more accurate measure of microbial diversity than body mass alone. This is supported by that fact the smallest hosts, regardless of their dietary category, harbor about the same levels of microbial diversity, whereas the largest hosts show considerable variation (ranging from 50 OTUs in carnivores to 1250 OTUs in herbivores), indicating that diet has a greater effect on microbial diversity and microbiome divergence in larger hosts. And although it has been suggested that larger, species-rich communities are less subject to turnover and change (Nemergut et al. 2013), we find there is no significant difference in the rates of microbiome divergence for different diet categories, even though herbivores tend to have much higher alpha diversity.
Contrary to what might be anticipated, the incidence of transitions between dietary categories within a lineage does not significantly alter its overall rate of microbiome divergence. Evaluating the number of actual to potential dietary transitions (i.e., the number of dietary transitions vs. the number of internal + terminal branches) within each of the K-T lineages, we find no acceleration in the rates of microbiome divergence related to the number of dietary changes (Table 1, Table S8). Chiropterans were assigned to all four dietary categories and display many dietary transitions but maintain the most slowly evolving microbiomes. This suggests that even after a dietary transition, gut physiology of the host restricts the composition of the gut microbiome. For instance, the giant panda switched to a strictly herbivorous diet roughly 2 million years ago but maintains carnivore-like microbiome that reflects the constraints of a simple gastrointestinal tract with rapid transit time typical of its carnivorous/omnivorous relatives (Zhu et al. 2011; Xue et al. 2015). Additionally, Sanders et al. suggested that difference between microbiome composition of baleen whales and terrestrial invertivores is reflective of the pre-adapted artiodactyl multi-chambered stomach. We find that the degree of microbiome divergence between two sister lineages that experienced a dietary transition is related to the age of the transition, showing the accumulation of microbiome changes is associated with phylogenetic distance.
A final factor found to influence rates of microbiome divergence is host lifestyle. Although host size and diet can confound the analysis of ecological factors, there are several cases where marine mammals displayed lower levels of microbial diversity (Figure 1a) and accelerated rates of microbiome divergence in comparison to terrestrial mammals with the same diet. For example, the same dietary transition (e.g., herbivory and invertivorous carnivory) occurred in both the Cetartiodactyla and Xenarthra lineages. Yet, when such changes are associated with a transition from terrestrial to marine environments, the rates of microbiome divergence are augmented due to the loss of taxa (Table 1). Similarly, in primates, which over their 75-million year history have sustained the most constant rate of microbiome divergence among mammals, there has been an acceleration in the human lineage due to the dramatic loss of bacteria at all taxonomic levels (Moeller et al. 2014). When compared to other mammalian lineages, primates have more parental care and sociality (Clutton-Brock 1991; Storey & Ziegler 2016), which can both increase the potential for transmission and safeguard against loss of bacterial taxa (Tung et al. 2015; Moeller et al. 2016). Despite the high levels of social behavior in humans, the reduction of microbial diversity and accelerated divergence of human microbiomes have been ascribed to changes in lifestyle, including dietary features, improved sanitation and antibiotic usage (Blaser & Falkow 2009; De Filippo et al. 2010b; Yatsunenko et al. 2012; Morton et al. 2015b).
As previously shown, the microbiome compositions of mammalian species assort according to both diet and phylogeny (Ley et al. 2008; Ochman et al. 2010; Muegge et al. 2011b; Delsuc et al. 2014a; Sanders et al. 2015b). Host species with the same diet often converge on similar microbiome contents at high taxonomic ranks, whereas host phylogeny is associated with bacteria at lower taxonomic ranks (Groussin et al. 2017). This broad pattern is evident for the majority of K-T lineages: we find that diet serves as a good predictor of the relative abundance of some of the core bacterial phyla, such as Bacteroidetes, Firmicutes, and Proteobacteria, but at the level of bacterial family and genus, the number of taxa associated with a particular phylogenetic lineage outweighs the number associated with diet (Table S6, Table S7). Note, however, that this pattern does not hold true for Chiropterans, which cluster apart from other mammals regardless of host diet (Figure 3). Although tests based on differential abundance could potentially result in some false positives (Gloor et al. 2016; Weiss et al. 2017), the overall trend—that host phylogeny is associated with the diversity of microbes at lower taxonomic ranks and diet associated with the diversity of microbes at higher taxonomic ranks—would not be affected.
Our finding that Chiropteran microbiomes do not display either a strong dietary or phylogenetic signal conflicts with a previous study that reported a distinction between bats with plant and animal-based diets (Carrillo-Araujo et al. 2015). Additionally, there is an earlier report that the pattern of divergence among microbiomes of insectivorous bats was congruent with host phylogeny (Phillips et al. 2012). Because these studies sampled microbiomes from either intestinal mucosa or dissected segments of the gut—neither of which is not directly comparable to fecal samples—we did not include their data in our analyses. And in neither case was the reported pattern apparent in our more comprehensive dataset. Chiropterans have a gut physiology that reflects their volant lifestyle, and like bats, the microbiomes of small passerine birds do not assort by diet (Hird et al. 2015). Bats and small birds have evolved convergent adaptations to increase paracellular absorption during the digestive process (Caviedes-Vidal et al. 2007; Price et al. 2015), suggesting that such digestive features may limit microbial diversity.
By analyzing a diverse set of mammalian species in a phylogenetic context, it was possible to identify several trends influencing microbiome composition that previously went unrecognized. Although mammals exhibit a robust phylogenetic signal dating back at least 75 million years, such that a large component of microbiome divergence between taxa is time dependent. The rate at which microbiomes diverge is influenced by host size, gut physiology and environment but is relatively insensitive to diet and the number (or type) of dietary transitions within a lineage. The most anomalous order is the Chiroptera, where gut physiology has constrained the evolution of microbiome composition despite the numerous dietary changes that accompanied rapid speciation. At the opposite end of the spectrum, rates of microbiome divergence have greatly accelerated in Cetartiodactyls, accompanying the transition from terrestrial to marine environments, and in Hominids, during the evolution of the human lineage.
Supplementary Material
Acknowledgments
We thank Kim Hammond for assistance with preparation of the figures, and Louis-Marie Bobay for helpful discussions during data analysis and manuscript preparation. This work was supported by grants from the National Institutes of Health (R35 GM118038 to H. Ochman) and the National Science Foundation (Graduate Research Fellowship 2016226761 to A. Nishida).
Footnotes
Data accessibility
All 16S rRNA sequencing data are publically available from cited sources (Table S1).
Author contributions
A.H.N. and H.O. conceived and designed study; A.H.N. analyzed data; A.H.N. and H.O. wrote the manuscript.
References
- Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, Garber PA. The role of gut microbes in satisfying the nutritional demands of adult and juvenile wild, black howler monkeys (Alouatta pigra) American Journal of Physical Anthropology. 2014;155:652–664. doi: 10.1002/ajpa.22621. [DOI] [PubMed] [Google Scholar]
- Amato KR, Yeoman CJ, Cerda G, Danzy Cramer J, Berg Miller ME, Gomez A, Leigh SR. Variable responses of human and non-human primate gut microbiomes to a Western diet. Microbiome. 2015;3:53. doi: 10.1186/s40168-015-0120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 Available from https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nature Reviews Microbiology. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, Clark AG. Host genetic variation impacts microbiome composition across human body sites. Genome Biology. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Zhernakova DV. The effect of host genetics on the gut microbiome. Nature Genetics. 2016;48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host & Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Araujo M, Taş N, Alcántara-Hernández RJ, Gaona O, Schondube JE, Medellín RA, Falcón LI. Phyllostomid bat microbiome composition is associated to host phylogeny and feeding strategies. Frontiers in Microbiology. 2015;6:447. doi: 10.3389/fmicb.2015.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss M, Steuer P, Müller DWH, Codron D, Hummel J. Herbivory and body size: allometries of diet quality and gastrointestinal physiology, and implications for herbivore ecology and dinosaur gigantism. PLoS One. 2013;8:e68714. doi: 10.1371/journal.pone.0068714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, Knights D. Captivity humanizes the primate microbiome. Proceedings of the National Academy of Sciences. 2016;113:10376–10381. doi: 10.1073/pnas.1521835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock TH. The Evolution of Parental Care. Princeton, NJ: Princeton University Press; 1991. [Google Scholar]
- Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, Gilad Y. Genome-wide association studies of the human gut microbiota. PLoS One. 2015;10:e0140301. doi: 10.1371/journal.pone.0140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport ER, Mizrahi-Man O, Michelini K, Barreiro LB, Ober C, Gilad Y. Seasonal variation in human gut microbiome composition. PLoS One. 2014;9:e90731. doi: 10.1371/journal.pone.0090731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Biddinger SB. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML, Ochman H. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proceedings of the National Academy of Sciences. 2012;109:13034–13039. doi: 10.1073/pnas.1110994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, González A, Knight R. Convergence of gut microbiomes in myrmecophagous mammals. Molecular Ecology. 2014;23:1301–1317. doi: 10.1111/mec.12501. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992;61:1–10. [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. The American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Knight R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- Godon JJ, Arulazhagan P, Steyer JP, Hamelin J. Vertebrate bacterial gut diversity: size also matters. BMC Ecology. 2016;16:12. doi: 10.1186/s12898-016-0071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A, Rothman JM, Petrzelkova K, Yeoman CJ, Vlckova K, Umaña JD, Leigh SR. Temporal variation selects for diet–microbe co-metabolic traits in the gut of Gorilla spp. The ISME Journal. 2016;10:514–526. doi: 10.1038/ismej.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Ley RE. Genetic determinants of the gut microbiome in UK twins. Cell Host & Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Spector TD. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Wu JR, Pawlowsky-Glahn V, Egozcue JJ. It's all relative: analyzing microbiome data as compositions. Annals of epidemiology. 2016;26:322–329. doi: 10.1016/j.annepidem.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, Alm EJ. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nature Communications. 2017;8:14319. doi: 10.1038/ncomms14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale VL, Tan CL, Niu K, Yang Y, Knight R, Zhang Q, Amato KR. Diet versus phylogeny: a comparison of gut microbiota in captive colobine monkey species. Microbial Ecology. 2017:1–13. doi: 10.1007/s00248-017-1041-8. [DOI] [PubMed] [Google Scholar]
- Hird SM, Sánchez C, Carstens BC, Brumfield RT. Comparative gut microbiota of 59 neotropical bird species. Frontiers in Microbiology. 2015;6:1403. doi: 10.3389/fmicb.2015.01403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov WH, Douglas AE. Comparative digestive physiology. Comprehensive Physiology. 2013;3:741–783. doi: 10.1002/cphy.c110054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl KD, Dearing MD. Wild-caught rodents retain a majority of their natural gut microbiota upon entrance into captivity. Environmental Microbiology Reports. 2014;6:191–195. doi: 10.1111/1758-2229.12118. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie VJ, Song SJ, Delsuc F, Prest TL, Oliverio AM, Korpita TM, Knight R. The effects of captivity on the mammalian gut microbiome. Integrative and Comparative Biology. 2017;486:222–227. doi: 10.1093/icb/icx090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AHA, Foerster S, Wilson ML, Pusey AE, Hahn BH, Ochman H. Social behavior shapes the chimpanzee pan-microbiome. Science Advances. 2016;2:e1500997. doi: 10.1126/sciadv.1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, Ochman H. Rapid changes in the gut microbiome during human evolution. Proceedings of the National Academy of Sciences. 2014;111:16431–16435. doi: 10.1073/pnas.1419136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Peeters M, Ndjango J-B, Li Y, Hahn BH, Ochman H. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Research. 2013;23:1715–1720. doi: 10.1101/gr.154773.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton ER, Lynch J, Froment A, Lafosse S, Heyer E, Przeworski M, Ségurel L. Variation in rural African gut microbiota is strongly correlated with colonization by entamoeba and subsistence. PLoS Genetics. 2015;11:e1005658. doi: 10.1371/journal.pgen.1005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson TM, Rogers TL, Carlini AR, Brown MV. Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environmental Microbiology. 2013;15:1132–1145. doi: 10.1111/1462-2920.12022. [DOI] [PubMed] [Google Scholar]
- Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, Ferrenberg S. Patterns and processes of microbial community assembly. Microbiology and Molecular Biology Reviews. 2013;77:342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe SJD, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Vipperla K. Fat, fibre and cancer risk in African Americans and rural Africans. Nature Communications. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Worobey M, Kuo C-H, Ndjango J-BN, Peeters M, Hahn BH, Hugenholtz P. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biology. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CD, Phelan G, Dowd SE, McDonuough MM, Ferguson AW, Delton Hanson J, Baker RJ. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Molecular Ecology. 2012;21:2617–2627. doi: 10.1111/j.1365-294X.2012.05568.x. [DOI] [PubMed] [Google Scholar]
- Porter LM, Sterr SM, Garber PA. Habitat use and ranging behavior of Callimico goeldii. International Journal of Primatology. 2007;28:1035–1058. [Google Scholar]
- Price ER, Brun A, Caviedes-Vidal E, Karasov WH. Digestive adaptations of aerial lifestyles. Physiology. 2015;30:69–78. doi: 10.1152/physiol.00020.2014. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C, Candela M. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Current Biology. 2015;25:1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- Roggenbuck M, Schnell IB, Blom N, Bælum J, Bertelsen MF, Sicheritz-Pontén T, Hansen LH. The microbiome of New World vultures. Nature Communications. 2014;5:5498. doi: 10.1038/ncomms6498. [DOI] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JG, Beichman AC, Roman J, Scott JJ, Emerson D, McCarthy JJ, Girguis PR. Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nature Communications. 2015;6:8285. doi: 10.1038/ncomms9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Sahl JW. Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Gordon JI. Cohabiting family members share microbiota with one another and with their dogs. eLife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey AE, Ziegler TE. Primate paternal care: interactions between biology and social experience. Hormones and Behavior. 2016;77:260–271. doi: 10.1016/j.yhbeh.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Wang X, Bernstein S, Huffman MA, Xia D-P, Gu Z, Li J. Marked variation between winter and spring gut microbiota in free-ranging Tibetan Macaques (Macaca thibetana) Scientific Reports. 2016;6:26035. doi: 10.1038/srep26035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J, Barreiro LB, Burns MB, Grenier J-C, Lynch J, Grieneisen LE, Archie EA. Social networks predict gut microbiome composition in wild baboons. eLife. 2015;4:e05224. doi: 10.7554/eLife.05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science Translational Medicine. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Hyde ER. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Zhang W, Wang L, Hou R, Zhang M, Fei L, Zhang Z. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. mBio. 2015;6:1–12. doi: 10.1128/mBio.00022-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PloS One. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wu Q, Dai J, Zhang S, Wei F. Evidence of cellulose metabolism by the giant panda gut microbiome. Proceedings of the National Academy of Sciences. 2011;108:17714–17719. doi: 10.1073/pnas.1017956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.