Abstract
LapA RNAs, proteins, and activities increased in response to systemin, methyl jasmonate, abscisic acid (ABA), ethylene, water deficit, and salinity in tomato (Lycopersicon esculentum). Salicylic acid inhibited wound-induced increases of LapA RNAs. Experiments using the ABA-deficient flacca mutant indicated that ABA was essential for wound and systemin induction of LapA, and ABA and systemin acted synergistically to induce LapA gene expression. In contrast, pin2 (proteinase inhibitor 2) was not dependent on exogenous ABA. Whereas both LapA and le4 (L. esculentum dehydrin) were up-regulated by increases in ABA, salinity, and water deficit, only LapA was regulated by octadecanoid pathway signals. Comparison of LapA expression with that of the PR-1 (pathogenesis-related 1) and GluB (basic β-1,3-glucanase) genes indicated that these PR protein genes were modulated by a systemin-independent jasmonic acid-signaling pathway. These studies showed that at least four signaling pathways were utilized during tomato wound and defense responses. Analysis of the expression of a LapA1:GUS gene in transgenic plants indicated that the LapA1 promoter was active during floral and fruit development and was used during vegetative growth only in response to wounding, Pseudomonas syringae pv tomato infection, or wound signals. This comprehensive understanding of the regulation of LapA genes indicated that this regulatory program is distinct from the wound-induced pin2, ABA-responsive le4, and PR protein genes.
Plants respond quickly to pathogen and herbivore attacks by activating wound- and defense-response genes (Bowles, 1990; Dixon et al., 1994; Yang et al., 1996). Tomato (Lycopersicon esculentum) wound/defense-response genes are often expressed both locally and systemically (Enkerli et al., 1993; Pautot et al., 1993; Bergey et al., 1996). The signals that mediate systemic responses must be transmitted rapidly throughout the plant and may involve cell-to-cell signaling. Putative systemic signals include ethylene (Ecker and Davis, 1987; O'Donnell et al., 1996), SA (Malamy et al., 1990), ABA (Peña-Cortés et al., 1989), JA (Farmer and Ryan, 1990), and systemin (Pearce et al., 1991), as well as electrical and hydraulic signals (Wildon et al., 1992; Malone et al., 1994; Herde et al., 1996; Stankovic and Davies, 1998).
Multiple signal transduction pathways interact to activate or suppress wound- and defense-response genes in the Solanaceae. Wound-response genes, such as the pin (proteinase inhibitor) genes are activated by systemin and octadecanoid pathway products such as JA (Farmer and Ryan, 1990, 1992; Pearce et al., 1991; Peña-Cortés et al., 1995). Systemin acts locally and systemically to induce synthesis of JA, which induces the expression of wound-response genes (Narváez-Vásquez et al., 1995). ABA has also been reported as a local and systemic signal for the induction of pin genes in potato and tomato (Peña-Cortés et al., 1989, 1991, 1995). However, the role of ABA in the induction of wound-response genes in tomato has remained controversial (Schaller and Ryan, 1995; Birkenmeier and Ryan, 1998).
Several tomato wound-response genes are negatively regulated by SA, which acts at multiple steps in the octadecanoid signaling pathway (Doherty et al., 1988; Li et al., 1992; Peña-Cortés et al., 1993; Doares et al., 1995; O'Donnell et al., 1996). This contrasts to SA induction of the PR (pathogenesis-related) protein genes (van Kan et al., 1995). The SA and octadecanoid signaling pathways are reciprocally regulated by a wound mitogen-activated protein kinase (Seo et al., 1995) and a small GTP-binding protein (Sano et al., 1994). This cross-talk may aid in separating early responses to wounding that accompany pathogen or pest attack from long-term responses, such as PR gene expression and the development of SAR. The interactions of ethylene with the wound- and SA-signaling pathways are not completely understood. Ethylene is important for the development of necrotic symptoms that accompany pathogen invasion, but is not essential for the development of SAR (Bent et al., 1992; Lawton et al., 1994; Lund et al., 1998). Several PR transcripts accumulate in response to ethylene or ethephon treatments (Ecker and Davis, 1987; Raz and Fluhr, 1993; van Kan et al., 1995), while ethylene treatments do not induce some wound-response genes (Ryan, 1974; Kernan and Thornberg, 1989). Recent data have suggested that ethylene and JA interact to induce wound-response genes (Xu et al., 1994; O'Donnell et al., 1996).
In addition to responding to wound and defense signals, the expression of each wound- and defense-response gene is modulated during development. While some defense-response genes are silent throughout all of vegetative development and are solely induced in response to stress, other defense- and wound-response genes are expressed in specific vegetative or reproductive organs (Peña-Cortés et al., 1991; Titarenko et al., 1997). Many wound- and defense-response genes are expressed only in floral buds (Peña-Cortés et al., 1991) or in a subset of the floral organs (Lotan et al., 1989; Cote et al., 1991; Uknes et al., 1993; Constabel and Brisson, 1995), while other genes are expressed in all mature floral organs (Peña-Cortés et al., 1991). Finally, the developmental programming may be distinct in different species. For example, the pin2 genes of potato are only expressed in tubers and developing floral buds, while the pin2 genes of tomato are expressed in all mature floral organs (Peña-Cortés et al., 1991).
In tomato, LapA (Leu aminopeptidase) transcripts, proteins, and activities increase locally and systemically in response to wounding (Pautot et al., 1993; Gu et al., 1996b). Two tomato genes, LapA1 and LapA2, encode the 55-kD subunits of this exopeptidase (Gu et al., 1996a). The Lap genes of potato and Arabidopsis are regulated differently than the tomato LapA genes, since the Arabidopsis Lap gene is constitutively expressed (Bartling and Nosek, 1994) and potato Lap RNAs do not accumulate systemically after wounding (Hildmann et al., 1992), nor are they detected in response to pathogens (Herbers et al., 1994).
Given the fact that LapA transcripts and proteins are abundant after wounding, pathogen invasion, and insect infestation, LAP-A may play an important role in the tomato defense response (Pautot et al., 1993). For this reason, it was important to develop a comprehensive understanding of LapA expression (at the RNA, protein, and activity level) in response to wound/defense-response signals (including ethylene, SA, JA, ABA, and systemin) and during abiotic stress. Comparisons of LapA gene expression patterns relative to patterns of expression for the wound-response gene pin2, the ABA-response gene le4, and three PR protein genes (PR-1, PR-4, and GluB [basic β-1,3-glucanase]) in ABA-deficient or ABA-producing lines demonstrated that each gene responded to wound and defense signals in a distinct manner. These data indicated that at least four signaling pathways are required to modulate wound/defense gene expression in tomato plants. Finally, to understand the organ specificity of LapA responses to wound signals, transgenic tomato and tobacco (Nicotiana tabacum) plants expressing a chimeric LapA1:GUS gene were analyzed.
MATERIALS AND METHODS
Plant Growth, Tissue Harvest, and Storage
Tomato (Lycopersicon esculentum cv Peto 238R, cv Ailsa Craig flacca, and cv Ailsa Craig) plants were grown in soil (University of California mix III) in growth chambers with 16-h (30°C)/8-h (20°C) light/dark cycles. Plants were watered daily and supplemented with 14:14:14 fertilizer (Osmocote, Scotts-Sierra, Maysville, OH). Immediately after treatments, leaves were excised, placed directly into liquid nitrogen, and stored at −80°C until use.
Treatments with Wound- and Defense-Response Molecules
Shoots from 3- to 4-week-old Peto 238R tomato plants were excised 5 cm above the soil for the ABA, MeJA, and ethylene treatments. Shoots from 3-week-old Peto 238R plants were excised below the third leaf from the plant shoot apex for the systemin and SA treatments. The 24-h 10 μm MeJA treatment and control have been described previously (Gu et al., 1996a). For ABA treatments, shoots were placed in flasks filled with 100 μm ABA (pH 6.0) or water (control) for 24 h. For ethylene treatments, excised shoots were placed in flasks filled with water and incubated in airtight glass desiccators containing a ripe banana and apple or without fruit (control) for 24 h. Ethylene levels typically rose to 29 ppm (D.P. Puthoff and L.L. Walling, unpublished results).
The systemin treatment was a modification of that of Pearce et al. (1993). Excised shoots were placed in a microfuge tubes with 90 μL of 15 mm sodium phosphate buffer (pH 6.5) or 90 μL of 15 mm sodium phosphate buffer with 1 pmol of systemin. After 10 min, shoots were transferred to a flask filled with water for 12 h. Systemin was kindly provided by Dr. C.A. Ryan (Washington State University, Pullman). For SA treatments, shoots were placed in flasks filled with distilled water (control) or 0.1 to 0.5 mm SA (Sigma) for 24 h. The maximum concentration of SA tolerated by tomato shoots without inducing physical damage was 0.5 mm.
Five- to six-week-old flacca plants were excised below the third leaf from the shoot apex. Four-week-old Ailsa Craig plants were at a similar stage in development. Shoots were treated with systemin (above) or wounded (Pautot et al., 1991), and were subsequently placed in water with or without 100 μm ABA and incubated in closed desiccators. Systemin-treated and wounded leaves were collected 12 and 24 h later, respectively.
Water Deficit and Salinity Treatments
After 3 weeks of growth (described above), water was withheld from Peto 238R plants and leaves were harvested at 4.5 d (at the initial signs of wilting), 5 d, and 5.5 d later. Control plants were watered once per day and leaves were harvested at the same time as the plants experiencing 5.5 d of water deficit. For salinity treatments, Peto 238R plants were watered with 300 mL of 300 mm NaCl, 400 mm NaCl, or water (control) for 3 d, and leaves were harvested.
Construction of a LapA1:GUS Fusion Gene
A 970-bp HindIII/DdeI fragment from the λLapA1 genomic subclone pLapA1-EH (W.S. Chao, V. Pautot, F.M. Holzer, and L.L. Walling, unpublished data), was end-filled using Klenow enzyme and cloned into the filled-in BamHI site of pBI101 (CLONTECH). Site-directed mutagenesis was used to remove residual vector sequences and restore the integrity of the LapA1 5′-UTR (Chao, 1996). pLapA1:GUS was transformed into Agrobacterium tumefaciens (LBA4404 or EHA105), and transformants were confirmed by minilysates (Gelvin and Schilperoort, 1988; Birnboim and Doly, 1979).
Tobacco and Tomato Transformation
The tomato lines UC82b (Sunseeds Genetics, Hollister, CA) and VF36 (provided by Dr. S. McCormick, U.S. Department of Agriculture/Agricultural Research Service, Albany, CA) plants and tobacco (Nicotiana tabacum cv Xanthi) plants were used in the transformation experiments. LapA1:GUS transgenic plants were regenerated from tomato cotyledons and tobacco leaf discs using a modification of protocols described by Fillatti et al. (1987) and McCormick (1991). Details were described in Chao (1996). Fifteen independent tomato lines and 12 independent tobacco lines were characterized. DNA blots with HindIII/EcoRI-digested genomic DNAs (10 μg/lane) from T0 plants and reconstruction lanes with pLapA1:GUS were used to determine transgene copy number (Walling et al., 1988). The expression of the LapA1:GUS gene in T1 and T0 plants was confirmed by wounding of cotyledon segments and GUS histochemical staining. Transgenic tomato and tobacco plants expressing 35S:GUS (pBI121, CLONTECH) were also made.
GUS Activity Assays
The expression of the chimeric LapA1:GUS gene was monitored using histochemical and fluorometric assays for GUS activity (Jefferson, 1987). To reduce endogenous GUS activity, 20% methanol (v/v) was added to the assay buffers (Kosugi et al., 1990). Fluorescence was measured using a mini fluorometer (TKO 100, Hoefer Scientific Instruments, San Francisco). Protein concentrations were determined using a bicinchoninic acid protein assay reagent (Pierce). To reduce interference caused by β-mercaptoethanol, samples were preincubated with an equal volume of 0.1 m iodoacetamide in 0.1 mm Tris-HCl (pH 8) at 37°C for 20 min (Hill and Straka, 1988).
Wounding, MeJA Treatment, and Infection of LapA1:GUS Plants
T1 (LapA1:GUS) and UC82b tomato plants with six to eight leaves and T1 (LapA1:GUS) and Xanthi tobacco plants with five to seven leaves were used. Leaves of four to six individual plants per transgenic line were wounded (Pautot et al., 1991) or served as controls. Leaves were harvested into liquid nitrogen 24 h later. Intact 7- to 10-d-old seedlings were treated with MeJA by submerging roots in 10 μm MeJA/0.002% ethanol or 0.002% ethanol (control).
T1 LapA1:GUS and UC82b plants with six to eight leaves were used for the infection studies. Three to four upper leaves were used. Half of the leaflets on a leaf served as the mock-infected control and were gently swabbed with water using cotton-tipped applicators. The remaining leaflets were inoculated with a Pseudomonas syringae pv tomato suspension (3 × 108 cfu/mL) using cotton swabs (Pautot et al., 1991). Leaflets were harvested 24 h later.
RNA Blot Analyses
RNA blots and washes were performed as described previously (Pautot et al., 1991). Blots were exposed to film (Hyper-MP, Amersham) at −80°C with an intensifying screen (DuPont) for 24 h unless indicated otherwise. Autoradiographic signals were quantitated using a phosphor imager (Molecular Dynamics). Probes were labeled using [α-32P]dCTP by nick translation. Transcript sizes were determined by running an RNA ladder (GIBCO-BRL) in parallel lanes. The pLe4 cDNA was described previously (Cohen et al., 1991; Kahn et al., 1993). The GluB, PR-1, and PR-4 cDNA clones from tomato have been described previously (van Kan et al., 1992, 1995), and were kindly provided by Dr. P.J.G.M. de Wit (Wageningen Agricultural University, Wageningen, Netherlands). The tomato pin2 clone pT2-47 (Graham et al., 1985) and the LapA cDNA clone pDR57 (Pautot et al., 1993) have also been described previously.
Total Protein Extraction, Fractionation, and Immunoblot Analyses
Total leaf proteins were extracted and fractionated by two-dimensional PAGE as described by Wang et al. (1992). Electro-transfer and immunoblot procedures were described in Gu et al. (1996b). A 1:500 dilution of the LAP-A polyclonal antiserum and the preimmune serum were used (Gu et al., 1996b).
Aminopeptidase Activity Assay
Native proteins were extracted from leaves of treated and control plants (Gu et al., 1996b). Protein concentrations were determined by a modified Bradford method (Ramagli and Rodriguez, 1985). The assays were performed in triplicate in 96-well microplates with 2 μg of protein and 250 μL of assay solution (1 mm l-Leu-p-nitroanilide [Sigma], 50 mm Tris-HCl, pH 8.0, and 0.5 mm MnCl2). After 30 min, the amount of p-nitroaniline generated was measured spectrophotometrically at A405 using a microplate reader (E-Max, Molecular Devices, Menlo Park, CA).
In Situ Hybridizations
Floral buds (10-mm) were harvested, fixed, and imbedded in methacrylate as described by Kronenberger et al. (1993). Five-millimeter transverse sections of tomato buds were made. Sections were hybridized to a digoxigenin-labeled antisense or sense LapA1 RNAs. Digoxigenin-labeled RNAs were synthesized using T3 or T7 RNA polymerase (GIBCO-BRL) and pBS-LapA1 according to the manufacturer's instructions (Boehringer Mannheim). pBS-LapA1 has a 1.6-kb EcoRI/XbaI fragment from pDR57 inserted into the EcoRI/XbaI sites of pBS-KS+ (Pautot et al., 1993).
RESULTS
LapA Was Induced by Wound Signals: Systemin, ABA, MeJA, and Ethylene
To understand the impact of wound signals on LapA RNA levels, tomato plants were treated with MeJA, systemin, ABA, or ethylene. RNA blots were hybridized with probes for LapA1 and genes that respond to one or more of these signals (le4, PR-1, PR-4, and GluB). Le4 encodes a dehydrin-like protein and is induced by exogenous ABA and water deficit (Cohen et al., 1991; Kahn et al., 1993). PR-1, PR-4, and GluB RNAs and proteins accumulate in response to SA or ethephon (an ethylene-releasing compound) (Christ and Mösinger, 1989; van Kan et al., 1995; Tonero et al., 1997). PR-1 and PR-4 encode extracellular proteins. PR-1 has antifungal activity but its mechanism of action is not known (Niderman et al., 1995). PR-4 is similar to Win and hevein proteins; the role of PR-4 in defense has yet to be elucidated (Linthorst et al., 1991). GluB encodes an intracellular, basic β-1,3-glucanase whose activity can hydrolyze pathogen cell walls (van Kan et al., 1992, 1995).
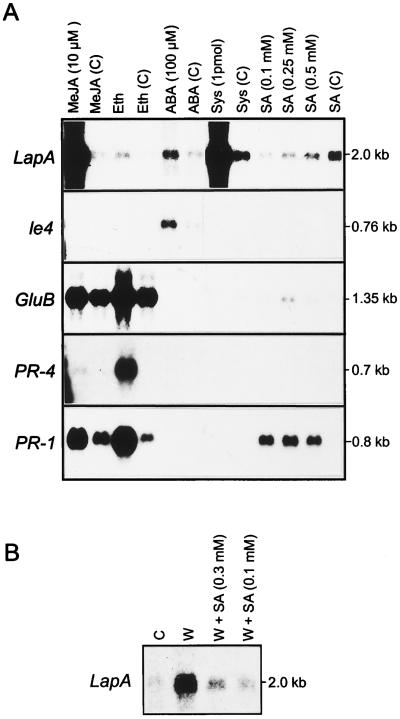
LapA was strongly induced by MeJA and systemin (Fig. 1A). A 57-fold increase in LapA transcripts occurred in leaves after 24 h of exposure to 10 μm MeJA. LapA RNAs were 2.5-fold more abundant in systemin-treated plants than in MeJA-treated plants. Treatment of shoots with 100 μm ABA increased LapA RNA levels 2-fold, which is similar to the increase measured for the well-characterized ABA- and water-deficit-response gene le4 (Cohen et al., 1991). This may be a minimal estimate of LapA induction in response to ABA, since larger increases in le4 transcripts were observed in an excised leaf assay (Cohen et al., 1991). In contrast to LapA, le4 RNA levels did not increase in response to systemin, MeJA, or ethylene treatments.
Figure 1.
RNA blot analyses of plants treated with wound- and defense-response signal molecules. A, Tomato plants treated with 10 μm MeJA, 29 ppm ethylene (Eth), 100 μm ABA, 1.0 pmol of systemin (Sys), or 0.1, 0.25, or 0.5 mm SA. For each treatment, the corresponding control is shown as a C in parentheses. B, Tomato plants were wounded and incubated in the absence (lane W) or presence of 0.1 mm or 0.3 mm SA (lanes W + SA). Total RNAs were extracted from treated and healthy control (lane C) leaves. The RNA blots were hybridized with 32P-labeled LapA, le4, GluB, PR-4, or PR-1 probes. The RNA sizes are indicated in kb. Data in A and B are from representative experiments. Photographs presented are optimized for visualization of weak autoradiographic signals. Hybridization signals were quantitated using a phosphor imager.
Relative to the control, there was a small increase in LapA RNAs (2-fold) in plants exposed to ethylene (Fig. 1A). After ethylene treatment, PR-1, PR-4, and GluB transcripts increased 14-, 16-, and 3-fold, respectively. The response of the tomato PR-1, PR-4, and GluB genes to pathogens, ethylene, and SA is well established (Christ and Mösinger, 1989; van Kan et al., 1995; Tonero et al., 1997), but less is known about their responses to wound signals. PR-1, PR-4, and GluB RNAs were unchanged after ABA or systemin treatments (Fig. 1A). While PR-4 transcripts did not accumulate in response to MeJA, MeJA caused both GluB and PR-1 RNAs to accumulate relative to the control plants. GluB and PR-1 RNA levels were elevated in the MeJA and ethylene controls relative to controls from other treatments (i.e. ABA, systemin, or SA). This may be due to the fact that the MeJA and ethylene treatments were done in a closed environment and a volatile signal may have accumulated to induce GluB and PR-1. It is clear that LapA, le4, and PR-4 transcript levels were not modulated by this additional signal(s).
During the course of these studies, we noted that the position of the incision and the age of the seedling used in the excised shoot assay was important (see Methods). When 3-week-old plants were used in this assay (systemin and SA treatments), LapA RNAs were detected in controls. This is in contrast to the extremely low to undetectable levels of LapA RNAs in leaves from excised 4-week-old shoots (MeJA, ethylene, and ABA treatments) or from intact plants (Pautot et al., 1993). It is clear that the developmental state must influence the tomato response to shoot excision. Several other studies have indicated that plant age may influence wound signaling (Wolfson and Murdock, 1990; Alarcon and Malone, 1995).
LapA RNA Levels Decreased in Response to Exogenous SA
Using the excised shoot assay, PR-4 transcripts were not detected after any of the SA treatments (Fig. 1A). In contrast, PR-1 and GluB transcripts increased after 0.1 and 0.25 mm SA treatments, respectively. The steady-state levels of PR-1 and GluB RNAs were distinct, suggesting differences in either transcriptional or posttranscriptional regulation. SA inhibited the accumulation of LapA transcripts, since control leaves had higher levels of LapA transcripts than SA-treated leaves (Fig. 1A). When wounding was followed by 0.1 or 0.3 mm SA treatments, LapA transcript levels were significantly reduced relative to wounded plants (Fig. 1B).
ABA Was Required for Wound-Induced Activation of LapA
To determine if endogenous ABA was required for wound and systemin induction of LapA, the expression of LapA was examined in the ABA-deficient flacca mutant and the ABA-proficient Ailsa Craig lines. Shoots were treated with systemin or were wounded, and subsequently incubated in water or 100 μm ABA (Fig. 2A). High levels of LapA and pin2 transcripts and low levels of le4 RNAs were detected after wounding in cv Ailsa Craig. In flacca plants, the le4 and LapA transcripts were undetectable in healthy or wounded leaves. Low levels of pin2 transcripts were consistently detected in healthy flacca leaves and, after wounding, the levels of pin2 RNAs increased 2-fold (Fig. 2A).
Figure 2.
RNA blot analysis of LapA gene expression in ABA-deficient (flacca) and control (Ailsa Craig) plants and in response to salinity and water deficit. A, Plants were mechanically wounded (W) or treated with 1 pmol of systemin (Sys), and excised shoots were subsequently incubated in water with or without 100 μm ABA. Ailsa Craig leaves were mechanically wounded (Ailsa, W). Shoots of healthy flacca plants were incubated in water (H) or ABA (H + ABA). Shoots of wounded flacca plants were incubated in water (W) or ABA (W + ABA). Flacca shoots were treated with systemin (Sys), systemin + ABA (Sys +ABA), or incubated with 15 mm phosphate buffer (Sys [C]). The blots were exposed to film for 48 h. B, Tomato plants were treated with 100 μm ABA, 300 mm NaCl, or 400 mm NaCl, or were not watered (water deficit) for 4.5, 5.0, or 5.5 d. Total RNA was extracted from treated and control (C) leaves. The RNA blots were hybridized with 32P-labeled LapA, le4, GluB, PR-4, or PR-1 probes. The autoradiographic signals of each band were quantitated using a phosphor imager.
Treatment of flacca shoots with 100 μm ABA caused LapA RNAs to rise 5-fold (Fig. 2A). When flacca was wounded and ABA treated, LapA RNAs increased 7-fold. ABA supplementation of healthy or wounded flacca plants caused pin2 RNAs to increase only 2- to 3-fold. Le4 RNA levels also increased when healthy flacca leaves were treated with ABA; wounding did not further increase le4 transcript abundance.
To examine if ABA has a role in systemin signal transduction, flacca shoots were treated with 15 mm phosphate buffer (control), systemin, or systemin plus 100 μm ABA (Fig. 2A). In buffer-treated flacca shoots, LapA or le4 RNAs were undetectable and low levels of pin2 RNAs were observed. After systemin treatment, LapA RNA increased 2-fold in flacca leaves. In contrast, a 50-fold induction of LapA transcripts was detected when flacca shoots were treated with both systemin and ABA. These levels were comparable to LapA levels in wounded cv Ailsa Craig leaves, and suggest that ABA and systemin act synergistically. These data indicated that ABA was critical for maximal accumulation of LapA transcripts in response to systemin and that pin2 was regulated in a different manner. pin2 RNAs increased 9-fold in flacca leaves in response to systemin and, when applied simultaneously, systemin and ABA increased pin2 transcripts 18-fold. Le4 transcripts did not increase in response to systemin (Figs. 1A and 2A). The level of le4 RNAs in leaves of flacca treated with both systemin and ABA was actually lower than that observed with ABA alone.
LapA Is Induced during Water Deficit and Salinity Stress
ABA is not only an important signal in the wound response of tomato, but it is also an important component in abiotic stresses such as water deficit and salinity (Bray, 1993; Chandler and Robertson, 1994). Therefore, we measured changes in LapA RNA levels during water deficit. LapA RNA levels increased during water deficit and reached maximal levels in plants stressed for 5 d (Fig. 2B); Le4 served as positive control (Cohen et al., 1991; Kahn et al., 1993). Le4 transcripts were present at higher levels than LapA RNAs and accumulated throughout the entire stress period. By d 5, le4 RNA levels had increased 53-fold.
To determine if LapA RNAs accumulated in response to salinity, tomato plants were watered with 300 or 400 mm NaCl for 3 d. LapA RNAs increased 4- to 6-fold in response to salinity treatments (Fig. 2B), whereas a more dramatic increase (22-fold) in le4 RNA levels was observed. None of the PR gene transcripts accumulated in response to water deficit or salinity stress, which is consistent with the observation that exogenous ABA treatments did not induce PR-1, PR-4, or GluB gene expression (Fig. 1A).
LAP-A Proteins and Activities Were Elevated after Stress Treatments
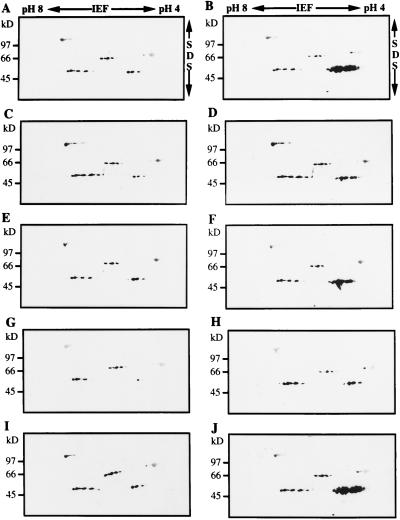
To determine if there was a coordinate induction of LapA RNAs and proteins, total proteins were extracted from leaves that were subjected to water deficit or treated with MeJA, systemin, ABA, or NaCl. Immunoblots showed that four classes of LAP-related proteins and one class of non-LAP protein were resolved (Gu et al., 1996b). The 90-kD proteins were not related to LAP, since they were recognized by preimmune serum (Gu et al., 1996b). The 66- and 77-kD LAP-like polypeptides and 55-kD LAP proteins with neutral pIs (LAP-N) were detected in all control and treated tomato leaf samples (Fig. 3). Only the 55-kD LAP-A polypeptides (with acidic pIs) were induced after stress treatments (Fig. 3, B, D, F, H, and J). LAP-A proteins were most abundant in MeJA- and systemin-treated leaves (Fig. 3, B and J), which is consistent with RNA blot analyses (Figs. 1A and 2B). While the levels of LapA RNA varied in the control plants (Figs. 1A and 2B), the LAP-A protein levels varied only slightly (Fig. 3, A, C, E, G, and I).
Figure 3.
Immunoblots of proteins that accumulated in response to wound signals and abiotic stress. Total proteins (80 μg) were fractionated by two-dimensional PAGE. The gels were electroblotted onto nitrocellulose and the blots were incubated with a 1:500 dilution of the LAP-A polyclonal antiserum. The pH range for IEF and molecular mass markers (in kD) are indicated. The 55-kD LAP-A proteins had a pI range of 5.6 to 5.9 (Gu et al., 1996b). A, Control plants for MeJA treatment. B, Plants treated with 10 μm MeJA for 12 h. C, Control plants for ABA treatment. D, Plants treated with 100 μm ABA for 12 h. E, Control plants for 5-d water deficit treatment. F, Plants 5 d after water was withheld. G, Control plants for 300 mm NaCl treatment. H, Plants 3 d after 300 mm NaCl treatment. I, Control plants for systemin treatment. J, Plants 12 h after treatment with 1 pmol of systemin.
To determine if LAP-A protein levels is correlated with LAP activities, aminopeptidase activity assays were performed. Relative aminopeptidase activities increased in leaves of MeJA-, systemin-, ABA-, water-deficit-, and NaCl-treated plants (Fig. 4). The large increases in aminopeptidase activities noted in the MeJA- and systemin-treated samples paralleled the large increases in LapA mRNAs and proteins (Fig. 1A). Since the activity assays were performed on total soluble leaf protein extracts, a direct correlation between the amount of LAP-A proteins and LAP activities could not be made. The observed changes in aminopeptidase activities may have been due to increases in LAP-A and/or changes in the activities or levels of additional tomato leaf aminopeptidases (Gu et al., 1996b; Walling and Gu, 1996).
Figure 4.
Relative aminopeptidase activities. Native proteins were extracted from control (C) leaves and from leaves treated with 10 μm MeJA, 100 μm ABA, 5 d of water deficit, 300 mm NaCl, or 1 pmol of systemin (Sys). Aminopeptidase activity was measured in triplicate spectrophotometrically at A405 by the release of p-nitroaniline. Aminopeptidase activity was calculated as the A405 per milligram of protein. Relative aminopeptidase activities and sds are shown; the highest aminopeptidase levels were detected in the systemin-treated leaves (6.2 A405/μg protein); this value was set at 100%. Each analysis was replicated two times.
There was substantial variation in the aminopeptidase levels detected in the five controls for these studies. This may have been due to the fact that the treatment regimes varied. Four-week-old plants in soil were used for the water deficit and salinity studies and the aminopeptidase activities were similar in their controls. Three-week-old (systemin)- or 4-week-old (ABA and MeJA) excised shoots were used for the other treatments. The impact of seedling age on the LapA RNA levels detected in controls was noted (Fig. 1A). Finally, although the ages of the seedlings in the ABA and the MeJA treatments were the same, the ABA- and MeJA-treated plants were incubated in open and closed environments, respectively.
The LapA1 Promoter Was Activated by Wound Signals and P. syringae pv tomato
LapA genes are primarily controlled at the transcriptional level (W.S. Chao, V. Pautot, F.M. Holzer, and L.L. Walling, unpublished data). Therefore, a LapA1:GUS fusion was used to investigate LapA1 promoter activity in response to pathogens, to wound signals, and during development. The response of the LapA1 promoter to wounding was characterized using 15 independent LapA1:GUS transgenic tomato lines (Table I). No GUS activity was detected in wounded or nonwounded leaves from UC82b control plants. Basal GUS activity levels in nonwounded leaves from the transgenic tomato lines varied (12–1,191 nmol 4-methylumbelliferone min−1 mg−1 protein). After wounding, GUS activity increased in all LapA1:GUS transgenic tomato lines except the U55 line. Increases in GUS activity levels after wounding was variable and ranged from 1.3-fold (line V13) to 40-fold (line V14). Wound induction of the LapA1 promoter was also noted in the 12 independent LapA1:GUS transgenic tobacco lines characterized (Chao, 1996). In general, wound induction was less dramatic, ranging from 2- to 8-fold; however, one transgenic LapA1:GUS tobacco line exhibited a 72-fold induction (data not shown).
Table I.
Fluorometric analysis of GUS activity in transgenic tomato lines in response to wounding
| Transgenic Line | GUS Activity
|
Ratio (W/H) | |
|---|---|---|---|
| Wounded (W) | Healthy (H) | ||
| nmol 4-MU min−1 mg−1 protein | |||
| UC82b | 0 | 0 | – |
| V13 | 1,074 | 853 | 1.3 |
| V14 | 3,373 | 84 | 40.2 |
| V15 | 2,257 | 154 | 14.7 |
| U17 | 454 | 43 | 10.6 |
| U26 | 634 | 144 | 4.4 |
| U33 | 1,072 | 88 | 12.2 |
| U38 | 1,639 | 443 | 3.7 |
| U48 | 222 | 22 | 10.1 |
| U49 | 2,075 | 256 | 4.2 |
| U55 | 722 | 1,191 | 0.6 |
| U63 | 170 | 71 | 2.4 |
| U69 | 109 | 12 | 9.1 |
| U78 | 3,750 | 407 | 9.2 |
| U83 | 694 | 404 | 1.7 |
| U93 | 2,467 | 90 | 27.4 |
Fifteen independent LapA1:GUS transgenic lines were analyzed. Transgenic lines are designated to indicate their parentage: UC82b (U) or VFNT (V). All lines had one to two copies of the LapA1:GUS transgene, except U38, which had five copies. GUS activity was measured in leaf extracts. Four to six GUS-positive T1 plants per line were mechanically wounded or served as healthy controls. Leaves were harvested 24 h later and pooled for each treatment. GUS and protein levels were determined as described in “Materials and Methods.”
GUS activity increased significantly in six transgenic tomato lines 24 h after P. syringae pv tomato inoculation (Table II). Nontransformed UC82b leaves had low to undetectable levels of GUS activity in infected and mock-infected leaves, respectively. The levels of GUS activity in mock-infected LapA1:GUS transgenic tomato plants were variable, but were similar to basal levels in untreated leaves (Table I). Line U49 showed the most dramatic increase in GUS activity (174-fold) in response to P. syringae pv tomato infection. These data were well correlated with the wounding results (Table I).
Table II.
Fluorometric analysis of GUS activity in transgenic tomato lines in response to P. syringae pv tomato infection
| Transgenic Line | GUS Activity
|
Ratio (I/M) | |
|---|---|---|---|
| Infected (I) | Mock-infected (M) | ||
| nmol 4-MU min−1 mg−1 protein | |||
| UC82b | 16 | 0 | – |
| V14 | 5,328 | 806 | 6.6 |
| V15 | 1,274 | 239 | 5.3 |
| U38 | 6,703 | 192 | 34.9 |
| U49 | 12,509 | 72 | 173.7 |
| U78 | 10,509 | 129 | 82.9 |
| U93 | 8,196 | 66 | 124.2 |
GUS activity in leaf extracts from six LapA1:GUS transgenic lines was measured 24 h after P. syringae pv tomato inoculation or mock infection. GUS and protein levels were determined as described in Methods.
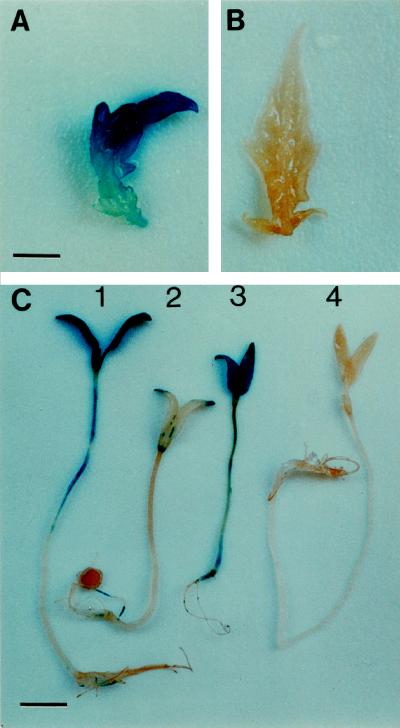
Histochemical staining for GUS activity showed that, like the control line UC82b, LapA1:GUS seedlings did not display significant GUS staining of cotyledons, hypocotyls, roots, or primary leaves (Fig. 5, B and C). Occasionally, GUS staining was detected at random sites on the LapA1:GUS seedlings, and this was correlated with sites of inadvertent mechanical wounding. 35S:GUS seedlings served as a positive control, and uniform GUS staining in the cotyledons, hypocotyls, and roots was detected (Fig. 5C). When treated with MeJA, the LapA1:GUS seedlings showed strong GUS staining in the aerial portions of the transgenic tomato plants: primary leaves (Fig. 5A), cotyledons, and hypocotyls (Fig. 5C). Cotyledons showed the highest level of GUS staining and the most apical portion of the hypocotyl in most seedlings also exhibited strong GUS staining. GUS staining was rarely detected in roots of JA-treated or control plants.
Figure 5.
LapA1 promoter activity in response to MeJA and during seedling development. A and B, Primary leaf from 10-d-old LapA1:GUS (line U49) (A) and UC82b control seedlings (B) treated with 10 μm MeJA. Bar = 3.1 mm. C, Seven-day-old LapA1:GUS (U49) seedling treated with 10 μm MeJA (1), 7-d-old LapA1:GUS seedling (U49) treated with 0.02% EtOH (2), 7-d-old 35S:GUS seedling (3), and 7-d-old UC82b seedling (4). Bar = 8.6 mm.
The LAPA1 Promoter Was Active in Flowers and Fruit
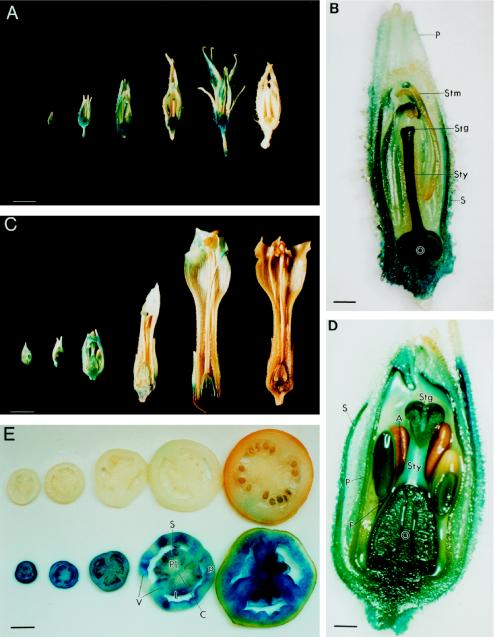
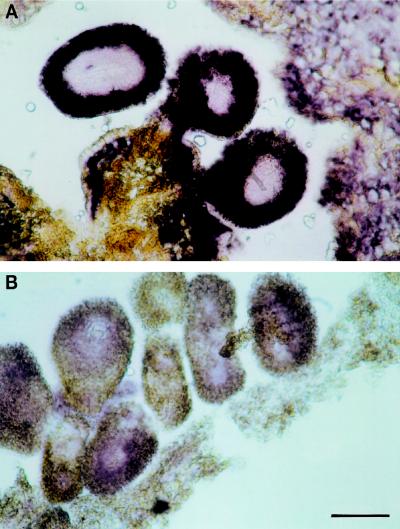
Examination of the LapA1:GUS transgene expression in both tomato and tobacco indicated that the LapA1 promoter was active in reproductive organs and in developing tomato fruit (Fig. 6). Strong GUS staining was consistently observed in the stamens, ovaries, stigma, and styles of 2-mm buds to fully opened flowers (1.6-cm) from LapA1:GUS transgenic tomatoes (Fig. 6, A and B). GUS staining was more uniform in petals and sepals of younger buds (2 mm) than in older buds (0.8 cm and larger) or petals of fully opened flowers (Fig. 6A). This is well correlated with the accumulation of LapA RNAs during tomato flower development (V. Pautot, F.M. Holzer, J. Chaufaux, and L.L. Walling, unpublished data; Milligan and Gasser, 1995). In situ hybridizations with a LapA antisense RNA probe showed that LapA transcripts were the most abundant in the integument of the ovaries and in the placental region (Fig. 7). LapA RNAs were also detected at lower levels throughout other floral organs (V. Pautot, F.M. Holzer, J. Chaufaux, and L.L. Walling, unpublished data), and LAP-A proteins were detected in all floral organs of open flowers (C.J. Tu, F.M. Holzer, and L.L. Walling, data not shown). Similar results were obtained when transgenic LapA1:GUS tobacco lines were examined (Fig. 6, C and D). All LapA1:GUS flowers (0.5–5 cm) had GUS activity in sepals, petals, stamens, pistils, and sepal trichomes. In tobacco, pistils, ovaries, and stigmas exhibited the strongest GUS staining throughout floral development (Fig. 6, C and D).
Figure 6.
LapA1 promoter activity during flower and fruit development. A, Flowers of LapA1:GUS tomato plants (U49) were excised, cut in half, and infiltrated with GUS histochemical substrate. Flowers displayed are 0.3, 0.8, 1.2, 1.6, and 1.6 cm in length (left to right). A 1.5-cm UC82b flower is presented at the right end of the panel. Variation in GUS activity in tomato and tobacco styles was due to the fact that GUS activity was not readily detected unless the style was bisected. Bar = 4.3 mm. B, Eleven-fold enlargement of the 1.2-cm LapA1:GUS tomato flower. Bar = 0.92 mm. S, Sepal; P, petal; O, ovary; Sty, style; Stg, stigma; Stm, stamen. C, Flowers of LapA1:GUS tobacco plants (line X2; Chao, 1996) were excised, cut in half, and infiltrated with GUS histochemical substrate. Flowers displayed are 0.5, 0.5, 1.0, 3.0, and 5.0 cm in length (left to right). A 5-cm N. tabacum cv Xanthi flower is present at the right end of the panel. Bar = 6.3 mm. D, Fourteen-fold enlargement of the 1.0-cm LapA1:GUS tobacco flower (line X2). Bar =0.71 mm. S, Sepal; P, petal; O, ovary; Sty, style; Stg, stigma; A, anther; F, filament. E, Top, Fruit from control UC82b; bottom, fruits of LapA1:GUS (U49) ranging in size from 0.5 mm to 7 cm incubated with GUS histochemical substrate. Bar = 1.25 cm. P, Pericarp; L, locular tissue; S, seed; Pl, placental tissue; V, vascular bundle; C, collumella.
Figure 7.
In situ hybridization of an antisense LapA RNA with a tomato floral bud. A methacrylate-imbedded transverse section of a 10-mm floral bud of tomato was hybridized with digoxigenin-labeled antisense (A) and sense (B) LapA RNAs. Bar = 88 μm.
The LapA1 promoter was active in all stages of tomato fruit development in the different LapA1:GUS transgenic lines (Fig. 6E). The pericarp generally had the highest levels of GUS staining, but staining was also seen in locular tissue, seeds, placental tissue, vascular bundles, and collumella. In most cases, GUS staining was most uniform in the earlier stages of fruit development (data not shown). While all fruit exhibited GUS staining, the degree of GUS staining was variable, and approximately 10% of the fruit had lower levels of GUS activity (data not shown).
DISCUSSION
Multiple Signal Transduction Pathways Regulate Wound- and Defense-Response Genes
The LapA RNAs and proteins accumulate in response to wounding and P. syringae pv tomato infection in tomato (Pautot et al., 1993; Gu et al., 1996b). Therefore, it was important to determine if LapA genes were regulated by the octadecanoid- or SA-dependent defense-response pathways or if LapA utilized one of the more recently identified JA-independent (Titarenko et al., 1997) or SA-independent signal transduction pathways (Penninckx et al., 1995; Pieterse et al., 1996). To this end, tomato plants were treated with wound/defense signals and assessed for levels of LapA and three PR gene transcripts. These studies indicated that at least four signaling pathways were important for the expression of wound- and defense-response genes in tomato (Fig. 8). Similar to pin2, LapA was modulated by MeJA and systemin, signals associated with the octadecanoid-signaling pathway (Fig. 1; Schaller and Ryan, 1995). Consistent with the role of SA in blocking the octadecanoid signaling pathway (Peña-Cortés et al., 1993; Doares et al., 1995; O'Donnell et al., 1996), wound induction of LapA was suppressed by SA. LapA was not strongly induced by exogenous ethylene (Fig. 1). However, if it were similar to pin genes, LapA would require JA for maximal activation by ethylene (O'Donnell et al., 1996).
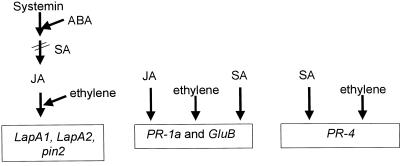
Figure 8.
Schematic diagram showing the four independent signal transduction pathways that activate wound and defense genes in tomato. The PR genes (PR-1, PR-4, and GluB) utilize two signaling pathways: a SA-dependent and an ethylene-dependent pathway. PR-1 and GluB utilize a third pathway, a systemin-independent, JA-dependent pathway, which may be analogous to the SA-independent pathway used to induce defensins in Arabidopsis (Penninckx et al., 1995). The fourth signaling pathway is the octadecanoid pathway, which in tomato utilizes systemin, ABA, JA, and ethylene to activate the wound-response genes (LapA1, LapA2, and pin2) and SA to down-regulate the pathway. There is evidence for a fifth signaling pathway in Arabidopsis. A JA-independent mechanism for wound-response gene activation was recently described, but tomato genes activated by this pathway have yet to be identified (Titarenko et al., 1997).
PR-4 may be regulated by two signaling pathways that were not utilized by LapA: an ethylene-dependent and a SA-dependent pathway. PR-4 was strongly induced by exogenous ethylene (Fig. 1; van Kan et al., 1995) and therefore utilized an ethylene signaling mechanism distinct from that used by LapA or pin genes. The excised-shoot assay utilized in these studies did not detect increases in PR-4 transcripts in response to 0.5 mm SA. However, excised-leaf assays have demonstrated that SA is an important regulator of PR-4 gene expression (van Kan et al., 1995). The SA signaling pathway has been elegantly elucidated in Arabidopsis (Dangl et al., 1996; Ryals et al., 1996; Yang et al., 1996), and is assumed to function in a similar manner in the Solanaceae.
A more complex circuitry was used to modulate expression of the tomato PR-1 and GluB genes. PR-1 and GluB RNAs accumulated in response to exogenous SA and ethylene, and similar observations were made by other investigators (Christ and Mösinger, 1989; van Kan et al., 1995; Tonero et al., 1997). The ethylene and SA signal transduction pathways utilized by PR-1, GluB, and PR-4 were probably the same. However, while SA increased the levels of both PR-1 and GluB RNAs, the steady-state levels of these RNAs were distinct. These data indicate that there are substantial differences in transcriptional and/or posttranscriptional controls that modulate these genes in response to SA.
Finally, PR-1 and GluB genes were also regulated by a signaling pathway that was activated by JA but not by systemin. At present it is not known why the JA generated after systemin treatments was insufficient for PR-1 and GluB transcript accumulation. It is possible that the systemin-induced JA was present in a subset of tomato leaf cells that were not competent for PR-1 and GluB gene expression; similar theories regarding JA compartmentalization have been proposed by Harms et al. (1995). Alternatively, systemin may have induced an inhibitor to interfere with JA induction of PR-1 and GluB expression, which would be consistent with the reciprocal regulation of the oxylipin and SA signal transduction pathways (Seo et al., 1995). At the present time, it is not known how the SA-independent and JA-dependent signaling pathways identified in Arabidopsis or tobacco relate to the pathways being elucidated in tomato (Penninckx et al., 1995; Pieterse et al., 1996; Vidal et al., 1997). However, it is clear from studies in Arabidopsis that some genes (such as defensins) can be induced by exogenous JA and ethylene but not by SA (Penninckx et al., 1995). These data indicate the independence of these signaling mechanisms. The fact that Arabidopsis defensins are not wound induced suggests that they may utilize the JA and ethylene signaling pathways that are similar to those used by the tomato PR-4, PR-1, and GluB and are distinct from JA-dependent or -independent wound responses (Titarenko et al., 1997).
ABA Is Essential for Wound Induction of LapA
The role of ABA in the modulation of wound-response gene expression in tomato remains controversial. Peña-Cortés et al. (1989, 1991, 1995) have concluded that ABA is essential for pin2 gene expression and acts early in the octadecanoid pathway. On the other hand, other studies have concluded that ABA does not have a primary role in oxylipin signal transduction pathway (Schaller and Ryan, 1995; Birkenmeier and Ryan, 1998). These discrepancies suggested to us that the role of ABA in wound-response gene induction needed to be re-evaluated, since significant differences in plant genotypes and treatments were present in the previous studies. The studies reported here with ABA-producing lines indicated that PR gene expression was not influenced by exogenous ABA. Our data support the idea that the systemin-independent, JA-responsive mechanism for PR-1 and GluB gene expression is distinct from the JA-signaling mechanisms utilized by LapA and pin2 genes.
Examination of LapA, pin2, and le4 transcript levels in the ABA-deficient line flacca indicated that there were significant differences in the role of ABA in the regulation of each of these genes. First, pin2 RNAs accumulated in nonwounded and wounded flacca leaves, whereas LapA RNAs and le4 RNAs were undetectable. Second, the impact of exogenous ABA on le4 and LapA gene expression was accentuated in flacca plants relative to ABA-producing plants. Third, ABA was critical for maximal accumulation of LapA transcripts in response to systemin, and ABA and systemin appeared to act synergistically to modulate LapA RNA levels. In contrast, pin2 transcript accumulation was not dependent on exogenous ABA for systemin induction. Finally, while ABA promoted le4 transcript accumulation, le4 did not respond to the signals of the octadecanoid pathway.
Collectively, these data indicate that although both LapA and pin2 genes utilized the octadecanoid signaling pathway, they responded differentially to ABA. LapA responses to ABA were more similar to those of le4 than to those of pin2. Wound induction of LapA was either dependent on ABA or required ABA levels that exceeded the residual levels in the flacca line. These data may indicate that pin2 gene expression was more sensitive to the residual levels of ABA in flacca plants (Neill and Horgan, 1985) than were le4 (Cohen and Bray, 1990) and LapA. Alternatively, the basal pin2 transcript levels detected in flacca plants were reflective of ABA-independent expression. Data from Peña-Cortés and colleagues (1989, 1996) support the idea that pin2 expression in nonwounded flacca leaves was due to residual ABA levels. Using an excised leaf assay, the sitiens mutant (which accumulates less ABA than flacca) exhibited no increase in pin2 RNAs, while the ABA-producing control showed a marked increase in pin2 mRNAs. This interpretation is also supported by Carrera and Prat (1998), who showed that transgenic tomato plants expressing the mutant abi1 allele from Arabidopsis, which blocks the ABA signal transduction cascade, prevents the accumulation of pin2 and LapA transcripts in response to ABA.
Comparison of the data presented here and those from Peña-Cortés et al. (1989, 1996), Carrera and Prat (1998), and Birkenmeirer and Ryan (1998) showed that the results obtained from excised shoot versus excised leaf assays are different. First, using the excised leaf assay, pin2 mRNAs are not detected in healthy, ABA-proficient plants (Peña-Cortés et al., 1989, 1996; Carrera and Prat, 1998). Excised shoot assays routinely detect pin2 transcripts (Birkenmeirer and Ryan, 1998; Fig. 1). Second, exogenous ABA caused larger increases in pin2 RNA levels in excised leaves than in excised shoots. This is consistent with the difference in le4 expression noted in our studies and in previous studies that utilized detached leaf assays (Cohen et al., 1991).
LapA Genes Are Induced during Abiotic Stresses That Are Accompanied by ABA Accumulation
Endogenous ABA levels increase when plants are exposed to a saline environment (Downton and Loveys, 1981; Walker and Dumbroff, 1981), water deficit (Zeevaart and Creelman, 1988), or low-temperature stress (Chen et al., 1983). Like the ABA- and water-deficit-response gene le4 (Cohen and Bray, 1990; Cohen et al., 1991; Kahn et al., 1993), LapA RNAs increased in response to water deficit and increases in salinity. However, differences in the le4 and LapA responses were noted. le4 RNAs increased more dramatically and persisted for a longer period of time than the LapA transcripts. It is also important that while tomato LapA RNAs increased during water-deficit stress, water deficit did not change potato Lap transcript levels (Hildmann et al., 1992). Several barley JIP (jasmonate-induced protein) genes are also water-deficit, ABA, and JA-induced; however, these genes are not induced by salinity stress, suggesting differences in stress signaling pathways in barley and tomato (Reinbothe et al., 1992).
It is not clear if the signal transduction pathways used for expression of LapA genes in response to water deficit and salinity stress were the same and corresponded to the octadecanoid pathway, or if they represent different signal transduction pathways. Clearly, le4, which is not modulated by systemin or MeJA, must utilize a signal transduction pathway distinct from the octadecanoid pathway. However, it is possible that there is cross-talk between the octadecanoid and abiotic-stress signal transduction pathways to coordinate LapA gene responses. Alternatively, LapA RNA induction by abiotic stress might solely utilize the wound-response octadecanoid pathway, since increases in JA have been measured for several abiotic stresses (Creelman and Mullet, 1995). It is also possible that the active oxygen species that accumulate during water deficit (Davies and Mansfield, 1983; Inzé and van Montagu, 1995) might activate the octadecanoid signaling pathway by causing lipid peroxidation and lipoxygenase production (Keppler and Novacky, 1989; Ádám et al., 1989). Rises in ABA could further activate the octadecanoid pathway to ultimately increase JA levels and activate LapA gene expression.
The LapA1 Promoter Is Responsive to Wound and Developmental Signals
Analysis of transgenic tomato and tobacco plants expressing a chimeric LapA1:GUS transgene demonstrated that the LapA1 promoter sequences that responded to wound and developmental signals were located within the 1st kb of the LapA1 5′-flanking sequences. Similar results have been reported for the expression of the tomato LapA1 gene in potato (Ruíz-Rivero and Prat, 1998) and the tomato LapA2 gene in Arabidopsis (A. El Amrani and V. Pautot, unpublished data). The LapA1 promoter was not active in vegetative organs unless tissues were wounded, P. syringae pv tomato infected, or treated with a wound signal such as MeJA.
The lack of LapA1 promoter activity in cotyledons after germination indicated that the LAP-A protein does not have a role in the mobilization of storage protein reserves (Walling and Gu, 1996). Aminopeptidases with properties similar to LAP-A have been characterized from kidney bean and barley seeds (Sopanen and Mikola, 1975; Mikkonen, 1992). The data presented here indicate that the kidney bean and barley seed LAPs are likely to be analogs of the constitutively expressed LAP-N of tomato and Arabidopsis LAP (Bartling and Nosek, 1994; Gu et al., 1996b; C.J. Tu and L.L. Walling, unpublished data).
Compared with other wound- and defense-response genes (Lotan et al., 1989; Cote et al., 1991; Uknes et al., 1993; Constabel and Brisson, 1995), the LapA1 promoter has a unique developmental specificity. In transgenic tomato, the LapA1 promoter was active in all floral organs, which is similar to the activity seen for tomato pin2 RNA accumulation (Peña-Cortés et al., 1991). However, unlike pin2 genes, LapA genes were active throughout all of fruit development. Other wound-response genes (i.e. the wound-induced ACC synthase gene and a pin1-like gene) are expressed only during the ripening phase of fruit development (Margossian et al., 1988; Li et al., 1992), when endogenous ethylene levels rise. Recently, Ruíz-Rivero and Prat (1998) reported an analysis of the tomato LapA1 promoter in transgenic potato, and their results differed from the data reported here. They did not detect LapA1 promoter activity in stigmas, styles, or ovaries, although expression in other floral organs was reported. At present, it is not known what signals are responsible for the activation of the LapA1 promoter in tomato flowers and fruit. However, the signaling mechanisms appear to be different in potato and tomato (Peña-Cortés et al., 1991; Ruíz-Rivero and Prat, 1998). The availability of tomato mutants that impact biosynthesis or perception of ABA, JA, and ethylene may aid in resolving their roles in developmental programming of LapA gene expression.
Plants utilize intricate systems for the expression of defense genes during floral and fruit development. The overlapping patterns of expression of the vast array of defense- and wound-response genes may ensure production of viable seeds. LAP-A proteins may play a defensive (or protective) role in tomato flowers and fruit by protecting gametes from damage by insect or pathogen attack. While the role of protease inhibitors in the control of insect predation is established and highly publicized (Johnson et al., 1989; Xu et al., 1996), a few studies have shown that proteases are important in plant defense. For example, one study showed that a Cys endoprotease confers resistance to maize against fall armyworm (Jiang et al., 1995). It is possible that exopeptidases such as LAP-A or the tomato wound-induced carboxypeptidases may have important roles in plant defense (Pautot et al., 1993; Mehta et al., 1996; Walling and Gu, 1996).
In animals, exopeptidases are important in the activation and inactivation of bioactive peptides and regulation of protein half-lives (Taylor, 1996; Varshavsky, 1996; Bradshaw et al., 1998). In a similar manner, LAP-A may serve to modulate levels or activities of regulatory proteins or peptides. Alternatively, LAP-A may facilitate turnover of proteins that are damaged due to reactive oxygen species generated during wounding, or may hydrolyze proteins to supply the pool of amino acids to support the substantial changes in protein synthesis associated with wounding. Current studies using antisense plants and plants overexpressing LapA are in progress. These studies will help to resolve the roles of LAP-As during floral and fruit development and during wounding and defense responses.
ACKNOWLEDGMENTS
We would like to thank members of the Walling laboratory for helpful discussions; David Puthoff for ethylene measurements; Fran Holzer for aid in figure modifications; Dr. Timothy Close (Department of Botany and Plant Sciences, University of California, Riverside) for reading earlier versions of this manuscript and for the use of his microplate reader; Dr. Richard Whitkus (Department of Botany and Plant Sciences, University of California, Riverside) for the use of his fluorometer; Dr. Jan Oakes (Calgene, Davis, CA) for her training of W.S.C. in tomato transformation; and Dr. Jocelyne Kronenberger (Laboratoire de Biologie Cellulaire, Institut National de la Recherche Agronomique) for her aid with in situ hybridizations.
Abbreviations:
- JA
jasmonic acid
- MeJA
methyl jasmonate
- SA
salicylic acid: SAR, systemic acquired resistance
Footnotes
This research was supported by a National Science Foundation grant (no. IBN–9318260) to L.L.W. W.S.C. was partially supported by the National Science Foundation training grant (no. GER–5355042).
LITERATURE CITED
- Ádám A, Farkas T, Somlya G, Hevesi M, Kiraly Z. Consequence of O2− generation during a bacterially induced hypersensitive reaction in tobacco: deterioration of membrane lipids. Physiol Mol Plant Pathol. 1989;34:13–26. [Google Scholar]
- Alarcon JJ, Malone M. The influence of plant age on wound induction of proteinase inhibitors in tomato. Physiol Plant. 1995;95:423–427. [Google Scholar]
- Bartling D, Nosek J. Molecular and immunological characterization of leucine aminopeptidase in Arabidopsis thaliana: a new antibody suggests a semi-constitutive regulation of a phylogenetically old enzyme. Plant Sci. 1994;9:199–209. [Google Scholar]
- Bent AF, Innes RW, Ecker JR, Staskawicz BJ. Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant-Microbe Interact. 1992;5:371–378. doi: 10.1094/mpmi-5-372. [DOI] [PubMed] [Google Scholar]
- Bergey DR, Howe GA, Ryan CA. Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proc Natl Acad Sci USA. 1996;93:12053–12058. doi: 10.1073/pnas.93.22.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier GF, Ryan CA. Wound signaling in tomato plants: evidence that ABA is not a primary signal for defense gene activation. Plant Physiol. 1998;117:687–693. doi: 10.1104/pp.117.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D. Defense-related proteins in higher plants. Annu Rev Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- Bradshaw RA, Brickey WW, Walker KW. N-terminal processing: the methionine aminopeptidase and N-alpha-acetyl transferase families. Trends Biochem Sci. 1998;23:263–267. doi: 10.1016/s0968-0004(98)01227-4. [DOI] [PubMed] [Google Scholar]
- Bray EA. Molecular responses to water deficit. Plant Physiol. 1993;103:1035–1040. doi: 10.1104/pp.103.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Prat S. Expression of the Arabidopsis abi1-1 mutant allele inhibits proteinase inhibitor wound-induction in tomato. Plant J. 1998;15:765–771. doi: 10.1046/j.1365-313x.1998.00261.x. [DOI] [PubMed] [Google Scholar]
- Chandler PM, Robertson M. Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:113–141. [Google Scholar]
- Chao WS (1996) Characterization of wound-induced leucine aminopeptidase genes (LapA1 and LapA2) in tomato and their expression in response to environmental and developmental cues. PhD Thesis. University of California, Riverside
- Chen H-H, Li PH, Brenner ML. Involvement of abscisic acid in potato cold acclimation. Plant Physiol. 1983;71:362–365. doi: 10.1104/pp.71.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ U, Mösinger E. Pathogenesis-related proteins of tomato 1. Induction by Phytophthora infestans and other biotic and abiotic inducers and correlations with resistance. Physiol Mol Plant Path. 1989;35:53–65. [Google Scholar]
- Cohen A, Bray EA. Characterization of three mRNAs that accumulate in wilted tomato leaves in response to elevated levels of endogenous abscisic acid. Planta. 1990;182:27–33. doi: 10.1007/BF00239979. [DOI] [PubMed] [Google Scholar]
- Cohen A, Plant Á, Moses MS, Bray EA. Organ-specific and environmentally regulated expression of two abscisic acid-induced genes of tomato. Plant Physiol. 1991;97:1367–1374. doi: 10.1104/pp.97.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constabel CP, Brisson N. Stigma- and vascular-specific expression of the PR-10a gene of potato: a novel pattern of expression of a pathogenesis-related gene. Mol Plant-Microbe Interact. 1995;8:104–113. [Google Scholar]
- Cote F, Cutt JR, Asselin A, Klessig DF. Pathogenesis-related acidic β-1,3-glucanase genes of tobacco are regulated by both stress and developmental signals. Mol Plant-Microbe Interact. 1991;4:173–181. doi: 10.1094/mpmi-4-173. [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc Natl Acad Sci USA. 1995;92:4114–4119. doi: 10.1073/pnas.92.10.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Dietrich RA, Richberg MH. Death don't have no mercy: cell death programs in plant-microbe interactions. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WJ, Mansfield TA (1983) The role of abscisic acid in drought avoidance. In FT Addicott, ed, Abscisic Acid. Praeger, New York, pp 237–268
- Dixon RA, Harrison MJ, Lamb CJ. Early events in the activation of plant defense responses. Annu Rev Phytopath. 1994;32:479–501. [Google Scholar]
- Doares SH, Narvaez-Vasquez J, Conconi A, Ryan CA. Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 1995;108:1741–1746. doi: 10.1104/pp.108.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty HM, Selvendran RR, Bowles DJ. The wound response of tomato plants can be inhibited by aspirin and related hydroxy-benzoic acids. Physiol Mol Plant Pathol. 1988;33:377–384. [Google Scholar]
- Downton WJS, Loveys BR. Abscisic acid content and osmotic relations of salt-stressed grapevine leaves. Aust J Plant Physiol. 1981;8:443–453. [Google Scholar]
- Ecker J, Davis RW. Plant defense genes are regulated by ethylene. Proc Natl Acad Sci USA. 1987;84:5202–5206. doi: 10.1073/pnas.84.15.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkerli J, Gisi U, Mösinger E. Systemic acquired resistance to Phytophthora infestans in tomato and the role of pathogenesis-related proteins. Physiol Mol Plant Path. 1993;43:161–171. [Google Scholar]
- Farmer EE, Ryan CA. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci USA. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell. 1992;4:129–134. doi: 10.1105/tpc.4.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti JJ, Kiser J, Ronald R, Comai L. Efficient transfer of glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Biotechniques. 1987;5:726–730. [Google Scholar]
- Gelvin SB, Schilperoort RA (1988) Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp A3–A4
- Graham JS, Pearce G, Merryweather J, Titani K, Ericsson L, Ryan CA. Wound-induced proteinase inhibitors from tomato leaves: the cDNA-deduced primary sequence of pre-inhibitor II. J Biol Chem. 1985;260:6561–6564. [PubMed] [Google Scholar]
- Gu Y-Q, Chao WS, Walling LL. Localization and post-translational processing of the wound-induced leucine aminopeptidase proteins of tomato. J Biol Chem. 1996a;271:25880–25887. doi: 10.1074/jbc.271.42.25880. [DOI] [PubMed] [Google Scholar]
- Gu Y-Q, Pautot V, Holzer FM, Walling LL. A complex array of proteins related to the multimeric leucine aminopeptidase of tomato. Plant Physiol. 1996b;110:1257–1266. doi: 10.1104/pp.110.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms K, Atzorn R, Brash A, Kühn H, Wasternack C, Willmitzer L, Peña-Cortés H. Expression of a flax allene oxide synthase cDNA leads to increased endogenous jasmonic acid (JA) levels in transgenic potato but not to a corresponding activation of JA-responding genes. Plant Cell. 1995;7:1645–1654. doi: 10.1105/tpc.7.10.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K, Prat S, Willmitzer L. Functional analysis of a leucine aminopeptidase from Solanum tuberosum L. Planta. 1994;194:230–240. [PubMed] [Google Scholar]
- Herde O, Atzorn R, Fisahn J, Wasternack C, Willmitzer L, Peña-Cortés H. Localized wounding by heat initiates the accumulation of proteinase inhibitor II in abscisic acid-deficient plant by triggering jasmonic acid biosynthesis. Plant Physiol. 1996;112:853–860. doi: 10.1104/pp.112.2.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildmann T, Ebneth M, Peña-Cortés H, Sánchez-Serrano J, Willmitzer L, Prat S. General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell. 1992;4:1157–1170. doi: 10.1105/tpc.4.9.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill HD, Straka JG. Protein determination using bicinchoninic acid in the presence of sulfhydryl reagents. Anal Biochem. 1988;170:203–208. doi: 10.1016/0003-2697(88)90109-1. [DOI] [PubMed] [Google Scholar]
- Inzé D, van Montagu MV. Oxidative stress in plants. Curr Opin Biotech. 1995;6:153–158. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Jiang B, Siregar U, Willeford KO, Luthe DS, Williams WP. Association of a 33-kilodalton cysteine proteinase found in corn callus with the inhibition of fall armyworm larval growth. Plant Physiol. 1995;108:1631–1640. doi: 10.1104/pp.108.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Narvrez J, An GH, Ryan CA. Expression of proteinase inhibitor-I and inhibitor-II in transgenic tobacco plants: effects on natural defense against Manduca sexta larvae. Proc Natl Acad Sci USA. 1989;86:9871–9875. doi: 10.1073/pnas.86.24.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn TL, Fender SE, Bray EA, O'Connell MA. Plant Physiol. 1993;103:597–605. doi: 10.1104/pp.103.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler LD, Novacky A. The initiation of membrane lipid peroxidation during bacteria-induced hypersensitive reaction. Physiol Mol Plant Pathol. 1989;30:233–245. [Google Scholar]
- Kernan A, Thornberg RW. Auxin levels regulate the expression of a wound-inducible proteinase inhibitor II-chloramphenicol acetyl transferase gene fusion in vitro and in vivo. Plant Physiol. 1989;91:73–78. doi: 10.1104/pp.91.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y. An improved assay for β-glucuronidase in transformed cells: methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Sci. 1990;70:133–140. [Google Scholar]
- Kronenberger J, Desprez T, Höfte H, Caboche M, Traas J. A methacrylate embedding procedure developed for immunolocalization in plant tissues is also compatible with in situ hybridization. Cell Biol Int. 1993;17:1013–1021. [Google Scholar]
- Lawton KA, Potter SL, Uknes S, Ryals J. Acquired resistance signal transduction in Arabidopsis is ethylene independent. Plant Cell. 1994;6:581–588. doi: 10.1105/tpc.6.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Parsons BL, Liu D, Mattoo AK. Accumulation of wound-inducible ACC synthase transcript in tomato fruit is inhibited by salicylic acid and polyamines. Plant Mol Biol. 1992;18:477–487. doi: 10.1007/BF00040664. [DOI] [PubMed] [Google Scholar]
- Linthorst HJM, Danhash N, Brederode FT, Vankan JAL, Dewit P, Bol JF. Tobacco and tomato PR proteins homologous to Win and pro-hevein lack the hevein domain. Mol Plant-Microbe Interact. 1991;4:586–592. doi: 10.1094/mpmi-4-586. [DOI] [PubMed] [Google Scholar]
- Lotan T, Ori N, Fluhr R. Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell. 1989;1:881–887. doi: 10.1105/tpc.1.9.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund ST, Stall RE, Klee HJ. Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell. 1998;10:371–382. doi: 10.1105/tpc.10.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I. Salicylic acid, a likely endogenous signal in the resistance response of tobacco to viral infection. Science. 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- Malone M, Alarcon J-J, Palumbo L (1994) An hydraulic interpretation of rapid, long-distance wound signaling in the tomato. Planta 193: 181–185
- Margossian LJ, Federman AD, Giovannoni JJ, Fischer RL. Ethylene-regulated expression of a tomato fruit ripening gene encoding a proteinase inhibitor I with a glutamic residue at the reactive site. Proc Natl Acad Sci USA. 1988;85:8012–8016. doi: 10.1073/pnas.85.21.8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S (1991) Transformation of tomato with Agrobacterium tumefaciens. In Lindsey K, ed, Plant Tissue Culture Manual: Fundamentals and Applications, Vol B6. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–9
- Mehta RA, Warmbardt RD, Mattoo AK. Tomato fruit carboxypeptidase: properties, induction upon wounding, and immunocytochemical localization. Plant Physiol. 1996;110:883–892. doi: 10.1104/pp.110.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkonen A. Purification and characterization of leucine aminopeptidase from kidney bean cotyledons. Physiol Plant. 1992;84:393–398. [Google Scholar]
- Milligan SB, Gasser CS. Nature and regulation of pistil-expressed genes in tomato. Plant Mol Biol. 1995;28:691–711. doi: 10.1007/BF00021194. [DOI] [PubMed] [Google Scholar]
- Narváez-Vásquez J, Pearce G, Orozco-Cárdenas ML, Franceschi VR, Ryan CA. Autoradiographic and biochemical evidence for the systemic translocation of systemin in tomato plants. Planta. 1995;195:593–600. [Google Scholar]
- Neill SJ, Horgan R. Abscisic acid production and water relations in wilty tomato mutants subjected to water deficiency. J Exp Bot. 1985;36:1222–1231. [Google Scholar]
- Niderman T, Genetet I, Bruyere T, Gees R, Stintzi A, Legrand M, Fritig B, Mosinger E. Pathogenesis-related PR–1 proteins are antifungal: isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995;108:17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ. Ethylene as a signal mediating the wound response of tomato plants. Science. 1996;274:1914–1918. doi: 10.1126/science.274.5294.1914. [DOI] [PubMed] [Google Scholar]
- Pautot V, Holzer FM, Reisch B, Walling LL. Leucine aminopeptidase: an inducible component of the defense response in Lycopersicon esculentum (tomato) Proc Natl Acad Sci USA. 1993;90:9906–9910. doi: 10.1073/pnas.90.21.9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautot V, Holzer FM, Walling LL. Differential expression of tomato proteinase inhibitor I and II genes during bacterial pathogen invasion and wounding. Mol Plant-Microbe Interact. 1991;4:284–292. doi: 10.1094/mpmi-4-284. [DOI] [PubMed] [Google Scholar]
- Pearce G, Johnson S, Ryan CA. Structure-activity of deleted and substituted systemin, an 18-amino acid polypeptide inducer of plant defensive genes. J Biol Chem. 1993;268:212–216. [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253:895–898. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- Peña-Cortés H, Albrecht T, Prat S, Weiler EW, Willmitzer L. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta. 1993;191:123–128. [Google Scholar]
- Peña-Cortés H, Fisahn J, Willmitzer L. Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc Natl Acad Sci USA. 1995;92:4106–4113. doi: 10.1073/pnas.92.10.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés H, Prat S, Atzorn R, Wasternack C, Willmitzer L. Abscisic acid-deficient plants do not accumulate proteinase inhibitor II following systemin treatment. Planta. 1996;198:447–451. [Google Scholar]
- Peña-Cortés H, Sánchez-Serrano JJ, Mertens R, Willmitzer L, Prat S. Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato. Proc Natl Acad Sci USA. 1989;86:9851–9855. doi: 10.1073/pnas.86.24.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés H, Willmitzer L, Sánchez-Serrano JJ. Abscisic acid mediates wound induction but not developmental-specific expression of the proteinase inhibitor II gene family. Plant Cell. 1991;3:963–972. doi: 10.1105/tpc.3.9.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblanx GW, Buchala A, Métraux J-P, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1995;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, Hoffland E, van Pelt JA, van Loon LC. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell. 1996;8:1225–1237. doi: 10.1105/tpc.8.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagli LS, Rodriguez LV. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis. 1985;6:559–563. [Google Scholar]
- Raz V, Fluhr R. Ethylene is transduced via protein phosphorylation events in plants. Plant Cell. 1993;5:523–530. doi: 10.1105/tpc.5.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Lehmann J, Parthier B. Differential accumulation of methyl jasmonate-induced mRNAs in response to abscisic acid and desiccation in barley (Hordeum vulgare) Physiol Plant. 1992;86:49–56. [Google Scholar]
- Ruíz-Rivero O, Prat S. A −308 deletion of the tomato LAP promoters is able to direct flower-specific and MeJA-induced expression in transgenic plants. Plant Mol Biol. 1998;36:639–648. doi: 10.1023/a:1005980028203. [DOI] [PubMed] [Google Scholar]
- Ryals J, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA. Assay and biochemical properties of the proteinase inhibitor inducing factor, a wound hormone. Plant Physiol. 1974;54:328–332. doi: 10.1104/pp.54.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Seo S, Orudgev E, Youssefian S, Ishizuka K, Ohashi Y. Expression of the gene for a small GTP biding protein in transgenic tobacco elevates endogenous cytokinin levels, abnormally induces salicylic acid in response to wounding, and increases resistance to tobacco mosaic virus infection. Proc Natl Acad Sci USA. 1994;91:10556–10560. doi: 10.1073/pnas.91.22.10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Ryan CA. Systemin: a polypeptide defense signal in plants. BioEssays. 1995;18:27–33. doi: 10.1002/bies.950180108. [DOI] [PubMed] [Google Scholar]
- Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y. Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science. 1995;270:1988–1992. doi: 10.1126/science.270.5244.1988. [DOI] [PubMed] [Google Scholar]
- Sopanen T, Mikola J. Purification and partial characterization of barley leucine aminopeptidase. Plant Physiol. 1975;55:809–814. doi: 10.1104/pp.55.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankovic B, Davies E. The wound response in tomato involves rapid growth and electrical responses, systemically up-regulated transcription of proteinase inhibitor and calmodulin and down-regulated translation. Plant Cell Physiol. 1998;39:268–274. [Google Scholar]
- Taylor A. Aminopeptidases. Georgetown, TX: R.G. Landes; 1996. [Google Scholar]
- Titarenko E, Rojo E, Leó J, Sánchez-Serrano JJ. Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 1997;115:817–826. doi: 10.1104/pp.115.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Gdea J, Conejero V, Vera P. Two PR1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Mol Plant-Microbe Interact. 1997;10:624–634. doi: 10.1094/MPMI.1997.10.5.624. [DOI] [PubMed] [Google Scholar]
- Uknes S, Dincher S, Friedrich L, Negrotto D, Williams S, Thompson-Taylor H, Potter S, Ward E, Ryals J. Regulation of pathogenesis-related protein-1a gene expression in tobacco. Plant Cell. 1993;5:159–169. doi: 10.1105/tpc.5.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan JAL, Cozijnsen T, Danhash N, de Wit PJGM. Induction of tomato stress protein RNAs by ethephon, 2,6-dichloroisonicotinic acid and salicylate. Plant Mol Biol. 1995;27:1205–1213. doi: 10.1007/BF00020894. [DOI] [PubMed] [Google Scholar]
- van Kan JAL, Joosten MHAJ, Wagemakers CAM, van den Berg-Velthuis GCM, de Wit PJGM. Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Mol Biol. 1992;20:513–517. doi: 10.1007/BF00040610. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S, de Léon IP, Denecke J, Palva ET. Salicyclic acid and the plant pathogen Erwinia carotovora induce defense genes via antagonistic pathways. Plant J. 1997;11:115–123. [Google Scholar]
- Walker MA, Dumbroff EB. Effect of salt stress on abscisic acid and cytokinin levels in tomato. Z Pflanzenphysiol. 1981;101:461–470. [Google Scholar]
- Walling LL, Chang YC, Demmin DS, Holzer FM. Isolation, characterization and evolutionary relatedness of three members from the soybean multigene family encoding chlorophyll a/b binding proteins. Nucleic Acids Res. 1988;16:10477–10492. doi: 10.1093/nar/16.22.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LL, Gu Y-Q. Plant aminopeptidases: occurrence function and characterization. In: Taylor A, editor. The Aminopeptidases. Georgetown, TX: RG Landes; 1996. pp. 174–219. [Google Scholar]
- Wang C-S, Walling LL, Eckard KJ, Lord EM. Patterns of protein accumulation in developing anthers of Lilium longiflorum correlate with histological events. Am J Bot. 1992;79:118–127. [Google Scholar]
- Wildon DC, Thain JF, Minchin PEH, Gubb IR, Reilly AJ, Skipper YD, Doherty HM, O'Donnell PJ, Bowles DJ. Electrical signaling and systemic proteinase inhibitor in the wounded plant. Nature. 1992;360:62–64. [Google Scholar]
- Wolfson JL, Murdock LL. Growth of Manduca sexta on wounded tomato: role of induced proteinase inhibitors. Entomol Exp Appl. 1990;54:257–264. [Google Scholar]
- Xu D, Xue Q, McElroy D, Mawal Y, Hilder VA, Wu R. Constitutive expression of a cowpea trypsin inhibitor gene, CpTi, in transgenic rice plants confers resistance to two major rice insect pests. Mol Breed. 1996;2:167–173. [Google Scholar]
- Xu Y, Chang P-FL, Liu D, Narasimhan ML, Raghothama KG, Hasagawa PM, Bressan RA. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Shah I, Klessig DF. Signal perception and transduction in plant defense responses. Genes Dev. 1996;11:1621–1639. doi: 10.1101/gad.11.13.1621. [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:439–473. [Google Scholar]