Abstract
Tyrosine kinase 2 (TYK2) belongs to the Janus kinase (JAK) family of tyrosine kinases, which transmit signals from activated cytokine receptors. GWAS have consistently implicated TYK2 in psoriasis susceptibility. We performed an in-depth association analysis of TYK2 using GWAS and resequencing data. Strong genetic association of three nonsynonymous variants in the exonic regions of the TYK2 gene (rs34536443, rs12720356, and rs2304256) were found. rs12720356 encoding I684S is predicted to be deleterious based on its location in the pseudokinase domain. We analyzed PBMCs from 29 individuals representing the haplotypes containing each of the significantly associated signals. STAT4 phosphorylation was evaluated by phospho-flow cytometry after CD3/CD28 activation of cells followed by IL-12 stimulation. Individuals carrying the protective I684S variant manifested significantly reduced p-STAT4 levels in CD4 + CD25 + CD45RO+ (mean Stimulation Index (S.I.) 48.08, n = 10) and CD8 + CD25 + CD45RO + cells (S.I. 55.71, n = 10), compared to controls homozygous for the ancestral haplotype (S.I. 68.19, n = 10 (p = 0.002) and 76.76 n = 10 (p = 0.0008) respectively). Reduced p-STAT4 levels were also observed in skin-homing, cutaneous lymphocyte associated antigen (CLA)-positive CD4 and CD8 cells from I684S carriers. No significant changes in p-STAT4 for the psoriasis-associated variant rs34536443 was found. These data establish the functional significance of the TYK2 I684S variant in psoriasis susceptibility.
Introduction
Psoriasis is an immunologically mediated inflammatory disease that affects about 1–2% of the population of the Western world1. Psoriasis is characterized by intense hyperproliferation and disturbed maturation of the epidermal cells, inflammatory infiltrates of the dermis and epidermis, and extensive new blood vessel formation2. Genetic results support the role of a dysregulated immune system through pathways that involve cells of both innate and acquired immunity3. Genome-wide association studies (GWAS) have identified 63 independent susceptibility loci in European-origin individuals, most of which appear to be regulatory in nature4. The major challenge of the post-GWAS era is the identification of causal variants and genes, which may be more straightforward for coding than for regulatory variation.
Tyrosine kinase 2 (TYK2) belongs to the Janus kinase (JAK) family of tyrosine kinases, which transmit signals from activated cytokine receptors5. GWAS identified TYK2 as a psoriasis susceptibility gene in 20106, this finding has since then been replicated by others, including ourselves7,8. In addition, the locus exhibits large effect size, and is one of the few psoriasis susceptibility regions whose strongest genetic signals involve coding variants. Genetic variation in TYK2 has also been associated with several other autoimmune diseases, including multiple sclerosis, systemic sclerosis, Type 1 diabetes, Crohn’s disease, ulcerative colitis, and systemic lupus erythematosus9–17. TYK2 associates with the cytoplasmic domain of cytokine receptors to transmit cytokine signals by phosphorylating receptor subunits18,19. In a recent study, selective inhibitors for TYK2 were used to determine the specific contribution of TYK2 catalytic activity to cytokine responses, using a receptor-distal phospho-STAT readout20. TYK2 catalytic activity was found to be required for signaling events downstream of IL-12 and IL-23, but did not contribute significantly to signaling downstream of IFN-α, IL-6, IL-10, and IL-22, which instead depended on JAK1 catalytic activity20. Interestingly, JAK2 contributed to IL-23, but not to IL-12 signaling, suggesting that IL-12 might be the most appropriate stimulus for the evaluation of TYK2 coding variation. In this study, we therefore selected IL-12 as a stimulus for the evaluation of functional effects of genetic variation in TYK2.
Results and Discussion
The identification of three nonsynonymous variants in TYK2 using GWAS and resequencing data
Using Plink version 1.0721 to perform conditional haplotype-based association analysis of the TYK2 genomic region (Supplementary Information and Table S1), we found strong independent genetic associations between psoriasis and three nonsynonymous variants in exonic regions of the TYK2 gene (rs34536443 G > C, rs12720356 A > C, and rs2304256 C > A) (Fig. 1). We found that the minor (i.e., derived) allele for each SNP was psoriasis-protective, with the ancestral haplotype conferring the highest risk. Notably, among haplotypes with at least 1% frequency in the underlying population, the protective alleles at the 1st (rs34536443) and 2nd (rs12720356) positions occur only with the protective allele at the 3rd position (rs2304256) (Fig. 1). SNP rs34536443 encoding TYK2 P1104A is located in the kinase domain, is conserved across 20 mammalian species, predicted to be strongly damaging by Polyphen22 and was found to alter TYK2 phosphorylation in response to IFN-β23. SNP rs12720356 encoding TYK2 I684S is located in the pseudokinase domain, is also conserved across 20 mammalian species, predicted to be strongly damaging, and was found to be catalytically-impaired yet signaling-competent in the context of interferon (IFN)-α activation, likely due to its ability to exert a heterodimeric scaffolding function with other JAKs24. SNP rs2304256 encoding V362F resides in the FERM domain involved in cytokine receptor binding, is not predicted to be damaging, and is not conserved across 20 mammalian species or across other JAK family members25.
Figure 1.
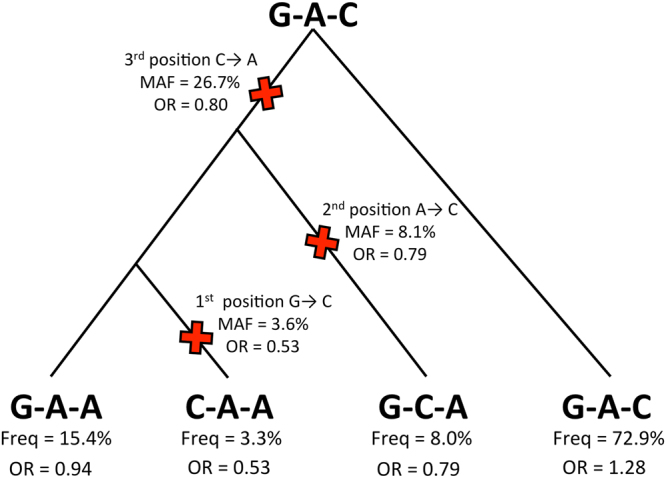
TYK2 variants and haplotypes analyzed in this study. The figure shows the most parsimonious ancestry for the four most common TYK2 haplotypes (frequency > 0.5%). The TYK2 gene tree is rooted at the ancestral G-A-C haplotype with red “X”’s indicating the most parsimonious history of mutation events giving rise to the three independently associated psoriasis SNPs rs34536443 G > C, rs12720356 A > C, and rs2304256 C > A. The ancestral allele at each SNP was determined using the UCSC Genome Browser and in each case agreed with the major allele. The three SNPs are presented in chromosomal (p → q) order on the plus strand, which corresponds to C-terminal to N-terminal order in the TYK2 gene. The minor/protective allele frequency (MAF) and single marker odds ratio (OR) are shown for each SNP. Frequencies and odds ratios are also shown for each haplotype.
The TYK2 I684S variant impairs IL-12 stimulated pSTAT4 response
Based on the aforementioned pharmacological evidence for specificity of TYK2 signaling downstream of the IL-12 receptor20, we used phospho-flow cytometry to measure IL-12-stimulated T-cell STAT4 phosphorylation in CD3/CD28-activated peripheral blood mononuclear cells (PBMC). In preliminary experiments, we determined the optimal time and dose of IL-12 stimulation (Figure S1) as well as CD3/CD28 activation (Figure S2). As shown in Figure S3, IL-12 receptors are induced in response to CD3/CD28 activation in purified T-cell populations, to a very similar extent to that seen in PBMC. We studied individuals representing three rs34536443-rs12720356-rs2304256 haplotypes: GAC (ancestral, n = 10), CAA, n = 9, and GCA, n = 10. Characteristics of the study population are shown in Table S2. In our study, the CAA and GCA haplotypes were each paired with the ancestral GAC haplotype. The GAA haplotype, whose component SNP rs2304256 C > A is not highly conserved or predicted to be damaging, was not evaluated.
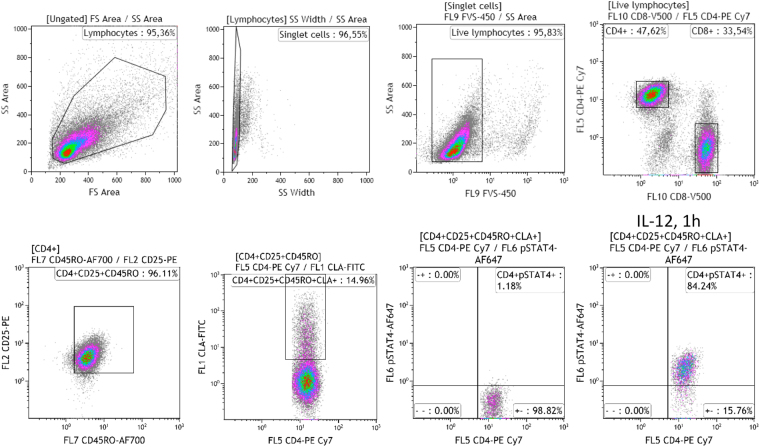
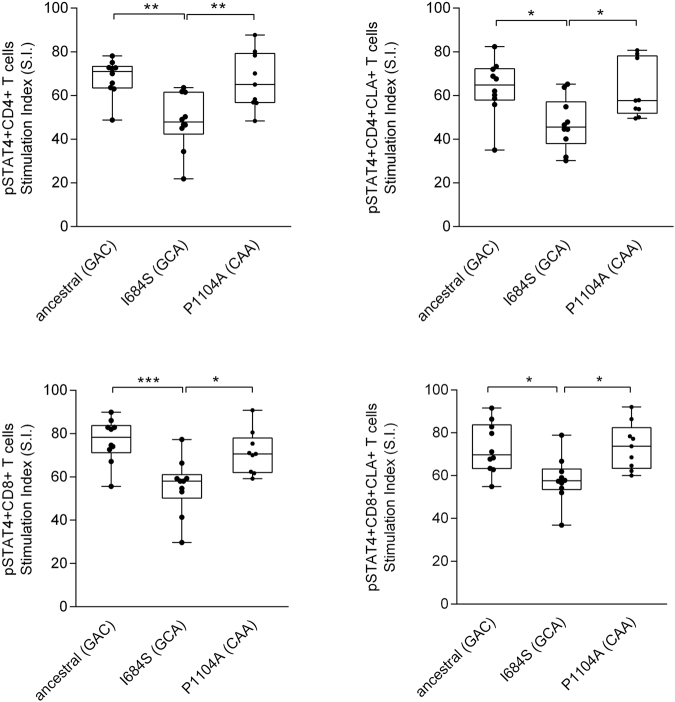
The gating strategy is shown in Fig. 2. We found that individuals carrying one copy of the GCA haplotype encoding the protective I684S variant (genotype GCA/GAC) had significantly reduced p-STAT4 levels in CD4+ and CD8+, activated (CD25+) memory (CD45RO+) T-cells, compared to individuals homozygous for the ancestral GAC haplotype (Fig. 3). Similar reductions in p-STAT4 were also observed in skin-homing, cutaneous lymphocyte associated antigen (CLA)-positive CD4 and CD8 cells from carriers of the protective I684S variant (Fig. 3). In contrast, we found no significant changes in p-STAT4 for individuals of the CAA/GAC haplotype harboring the psoriasis-associated variant rs34536443 encoding P1104A in the kinase domain, relative to homozygotes for the ancestral haplotype GAC (Fig. 3). We obtained the same result after restricting the analysis to controls (individuals without psoriasis, Figure S4).
Figure 2.
Gating strategy for phospho-flow cytometry of p-STAT4. Lymphocytes were gated on a forward (FS) and side scatter (SS) density plot and on a SS area versus SS width density plot to remove doublet cells. A live/dead stain (FVS-450) was used to exclude dead lymphocytes. Live, activated, memory, skin-homing CD4+ or CD8+ T lymphocytes were selected based on the expression of CD25, CD45RO, and cutaneous lymphocyte antigen (CLA). pSTAT4 expression was detected in cells activated by anti-CD3/CD28 and stimulated with or without 50 ng/ml IL-12 for 1 h.
Figure 3.
Functional effects of genetic variants in TYK2 on STAT4 phosphorylation in CD4+ and CD8+ T lymphocytes after stimulation with IL-12. PBMCs derived from individuals carrying the ancestral haplotype (n = 10) and from two haplotypes representing the genetic variants I684S (n = 10) and P1104A (n = 9), were cultured for 72 h with anti-CD3/CD28 beads, rested overnight, then stimulated with 50 ng/ml IL-12 for 1 h and analyzed for intracellular pSTAT4 expression by multicolor flow cytometry. Stimulation Index (S.I.) of pSTAT4 in CD25 + CD45RO + CD4+ and in lymphocyte antigen positive (CLA+) CD25 + CD45RO + CD4 + T-lymphocytes (upper panel). S.I. of pSTAT4 in CD25+CD45RO+CD8 and in CLA+CD25+CD45RO+CD8 T- lymphocytes (lower panel). Box plots: midline indicates median, box extends from 25th to 75th percentile, and whiskers indicate minimum to maximum. A p value < 0.05 was considered to be significant. *Indicates p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.001.
Because JAK-family tyrosine phosphorylation is regarded as an obligate downstream signal transduction response to cytokine stimulation, we also assessed TYK2 tyrosine phosphorylation by ELISA. The characteristics of the analyzed individuals are shown in Table S3. However, as shown in Figure S5, there were no significant differences in TYK2 phosphorylation across any of the three haplotypes.
TYK2 regulates IL-12 dependent gene expression
In order to ask whether any particular gene responses to IL-12 might be specific targets of TYK2 genetic variation, we carried out RNA-seq analysis of CD3/CD28-activated PBMC harvested after 1 and 4 h of IL-12 treatment. For these experiments, at least five individuals of each haplotype were selected for analysis (Table S4). Differential expression analysis across all three haplotypes as a function of IL-12 treatment identified 52 significantly (FDR <= 0.1 and fold change (FC) log2|FC| ≥ log2|1.5|) dysregulated genes at 1 h (43 up, 9 down, Table S5) and 233 dysregulated genes at 4 h (132 up, 101 down; Table S6). These dysregulated genes displayed significant overlap with a list of 28 IL-12 responsive genes identified from the literature by searching the NCI Biosystems database (4 and 6 genes at 1 h and 4 h, p = 1.05 × 10−7 and 1.09 × 10−7, respectively, Tables S5 and S6). Not unexpectedly, genes significantly up-regulated by 4 h of IL-12 treatment were most highly enriched for the Gene Ontology term “Regulation of interferon gamma mediated signaling pathway” (22.8-fold, FDR = 3.9 × 10−4, Table S7). Notably, after 4 hours of IL-12 treatment, the most significantly up-regulated gene was IL18RAP (average 2.37-fold, FDR p = 1.0 × 10−13), which also manifested a nominally significant effect of TYK2 genotype (ANOVA p = 7.2 × 10−3, Table S10). IL18RAP encodes an accessory subunit of the heterodimeric receptor for interleukin 18 (IL-18), a proinflammatory cytokine involved in inducing cell-mediated immunity. This protein enhances the IL-18-binding activity of the IL-18 receptor and plays a role in signaling by IL-18. IL18RAP is overexpressed 3.4-fold in lesional psoriatic vs. normal skin26, and mutations in this gene are associated with susceptibility to Crohn’s disease, celiac disease, and leprosy.
Regarding the functional effects of rs12720356 encoding TYK2 I684S, we have recently shown that the combined effects of IFN-γ and TNF contribute to increased vascular inflammation, a major cause of cardiovascular comorbidity in psoriasis27. While we were unable to demonstrate a significant effect of TYK2 genotype on IFN-γ mRNA expression in this relatively small RNA-seq study (Tables S9 and S10), our finding that IL-12 stimulation rapidly and significantly increases IFN-γ gene expression in anti-CD3/CD28-stimulated PBMC (Tables S5 and S6) has potential functional relevance to systemic inflammation in psoriasis. More generally, a Gene Ontology analysis of genes meeting an ANOVA p-value criterion of 0.05 for an effect of s12720356 genotype demonstrated significant (FDR ≤ 0.1) functional enrichment for “lymphocyte activation” and “T-cell activation” (Table S11). Finally, in support for functional relevance in the skin, we assessed the possible expression quantitative trait locus (eQTL) properties of rs12720356 encoding I684S on our RNA-seq-based gene expression dataset of skin samples26,28 (Table S12). The top hit in this eQTL analysis was GCSAM (germinal center associated signaling and motility, aka GCET2, Gene ID: 257144, eQTL p = 2.3 × 10−5), which manifests strongly biased expression not only in lymph nodes, but also in skin (ref. Entrez Gene). Collectively, these suggestive findings merit further analysis in a larger sample.
In this study, we selected individuals for functional studies based on 3-SNP haplotype-based genotypes instead of single-SNP genotypes. As shown in Fig. 1, rs34536443C is found on only one haplotype (CAA), whereas rs34536443G can belong to two different haplotypes—the ancestral GAC haplotype or the derived GAA haplotype—which carry different odds ratios in our study. More importantly, we utilized heterozygotes for the minor alleles at rs34536443 and rs12720356, rather than homozygotes. It is possible that by analyzing heterozygotes, we may have been able to capture an inhibitory effect of the TYK2 684S variant on its “wild-type” 684I counterpart, in the context of TYK2 homodimers bound to the IL-12 receptor. The existence of such homodimers in T-cells is strongly suggested by pharmacological evidence20. Our major findings complement those of Dendrou et al.29, who found a significant reduction in STAT4 phosphorylation in CD4 and CD8 memory T-cells in rs34536443 C/C vs. G/G homozygotes29. There were several differences between the two studies with regards to the experimental protocol, which might account for the differences in results. We stimulated whole PBMC, whereas Dendrou et al. stimulated purified CD3+ T-cells. We utilized CD3/CD28 beads for 72 hr followed by 1 hr of IL-12 stimulation, whereas Dendrou et al. activated with phytohemaglutinin for 72 hr followed by IL-2 for 24 hr prior to stimulation with IL-12 for 15 min. It is worth noting that in both our studies and those of Dendrou et al.29, hypofunctional TYK2 alleles, which are predicted to be functionally damaging to TYK2 signaling, are associated with a protective effect against psoriasis.
In conclusion, our results demonstrate a significant decrease in STAT4 phosphorylation in GCA/GAC heterozygotes (bearing one copy of the rare C allele at rs12720356 encoding TYK2 684 S in the pseudokinase domain), relative to either GAC/GAC homozygotes or CAA/GAC heterozygotes. This decrease in STAT4 phosphorylation was even detectable in skin-homing, cutaneous lymphocyte associated antigen (CLA)-positive CD4 and CD8 T-cells from carriers of the protective I684S variant. Taken together, these data support the functional significance of the TYK2 I684S variant in psoriasis susceptibility, and set the stage for further mechanistic investigation.
Methods
CD3/CD28 activation and IL-12 stimulation of PBMCs
Characteristics of the subjects chosen for analysis of STAT4 phosphorylation are shown in Table S2. Peripheral blood mononuclear cells (PBMC) were obtained by Ficoll-Hypaque centrifugation and cryopreserved in liquid nitrogen until analysis. Cryopreserved PBMCs were thawed, washed by centrifugation at 250 × g in complete medium, and activated with 25 µl per 106 cells anti-CD3/anti-CD28 beads (Gibco) for 3 days. Cells were seeded at a density of 1.0 × 106 cells/well (24 well plate) in RPMI 1640 supplemented with 1% L-glutamine, 1% penicillin and streptomycin, and 10% FBS. On the third day, beads were separated from cells by pipetting up and down five times followed by immediate magnetic capture of the beads. The activated PBMCs were rested overnight in fresh medium and then stimulated by addition of 50 ng/ml IL-12 (R&D Systems) for 1 h.
Flow cytometric analysis of phosphorylated STAT4
After washing, cells were stained with antibodies directed to cutaneous lymphocyte antigen (CLA)-FITC (BioLegend), the activation marker CD25-PE (BD Biosciences), CD3-PerCP-Cy5.5, and CD8-V500 (BD Biosciences), in PBS supplemented with 0.1% FBS and 2% pooled heat-inactivated (HI) human serum for the blocking of Fc-receptors. Cells were then treated with Fixation Buffer (BD Biosciences), and subsequently with Perm Buffer III (BD Biosciences) according to the manufacturer’s protocol. For intracellular detection of pSTAT4, cells were treated with titrated anti-pSTAT4 Alexa Fluor 647 (BD Biosciences) after fixation and permeabilization. Flow cytometry was performed on a Gallios analyser (Beckman Coulter) and data were analyzed using Kaluza® Flow Analysis Software, version 1.3 (Beckman Coulter). Lymphocytes were gated on a forward-(FS) and side scatter (SS) density plot and on a SS area versus SS width density plot to remove cell doublets. A live/dead stain (FVS-450) was used to exclude dead lymphocytes. Live, activated (CD25+) memory (CD45RO+) CD4+ or CD8+ T lymphocytes were selected based on the expression of CD25 and CD45RO, and skin-homing T-cells were further selected based on surface expression of cutaneous lymphoid antigen (CLA). The gating strategy is depicted in Fig. 2. The Stimulation Index (S.I.) was obtained by dividing the percentage of pSTAT4+ cells after IL-12 stimulation by the percentage of pSTAT4+ cells in unstimulated cells. The gate defining pSTAT4 positivity was set such that 1% of IL-12-unstimulated cells fell within the pSTAT4 gate.
Statistical analysis
We used two-way ANOVA followed by Holm-Sidak test for multiple comparisons for the analysis of pTYK2 measured by ELISA using GraphPad Prism 6. One-way ANOVA followed by Holm-Sidak test for multiple comparisons was used to evaluate the comparison of pSTAT4-upregulation after IL-12 stimulation in individuals carrying nonsynonymous variants or the ancestral haplotype, using GraphPad Prism 6. The results are shown as mean and the standard deviation (mean ± SD). A p value < 0.05 was considered to be significant.
Additional methodologic details are provided in the Supplementary Information.
Database submission
The RNA-seq data described in this work has been deposited in the Gene Expression Omnibus database (GEO accession number TBD).
Electronic supplementary material
Acknowledgements
The authors thank Sue J. Sohn, Ph.D. (Genentech) for her advice regarding the IL-12 stimulated STAT4 phosphorylation assay. This work was supported by awards from the National Institutes of Health (R01AR042742, R01AR050511, R01AR054966, R01AR063611, R01AR065183) to JTE, K01-AR072129 to LCT and K01-AR064765 to AJ, as well as a GAIN award from the Foundation for the National Institutes of Health. LCT is supported by awards from the Dermatology Foundation, the National Psoriasis Foundation, and the Arthritis National Research Foundation. JTE is supported by the Ann Arbor Veterans Affairs Hospital, and PES, AJ, SL, RPN, and JTE are supported by the Dawn and Dudley Holmes Foundation and the Babcock Memorial Trust. CE, CS and DV are supported by the Ingrid Asp Foundation, the Welander Foundation, the Swedish Psoriasis Association and the Medical Research Council.
Author Contributions
C.E. and J.T.E. conceived the study, C.E., C.S., S.L., D.V., R.P.N. and A.J. performed the experiments, M.Z., P.E.S. and L.C.T. performed the computational analysis, C.E., C.S., S.L., D.V., R.P.N., A.J., M.Z., P.E.S., L.C.T. and J.T.E. analyzed the data, all authors reviewed the manuscript
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25282-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gudjonsson, J. E. & Elder, J. T. Psoriasis: epidemiology. Clin Dermatol25, 535–546, 10.1016/j.clindermatol.2007.08.007 (2007). [DOI] [PubMed]
- 2.Kim J, Krueger JG. The immunopathogenesis of psoriasis. Dermatologic clinics. 2015;33:13–23. doi: 10.1016/j.det.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Lowes MA, Suarez-Farinas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–255. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsoi, L. C. et al. Large-scale meta-analysis identifies 18 novel psoriasis susceptibility loci. Nat Commun in press (2017).
- 5.Hirahara K, Schwartz D, Gadina M, Kanno Y, O’Shea JJ. Targeting cytokine signaling in autoimmunity: back to the future and beyond. Curr Opin Immunol. 2016;43:89–97. doi: 10.1016/j.coi.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Genetic Analysis of Psoriasis, C. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet42, 985–990, 10.1038/ng.694 (2010). [DOI] [PMC free article] [PubMed]
- 7.Ellinghaus D, et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am J Hum Genet. 2012;90:636–647. doi: 10.1016/j.ajhg.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang L, et al. Genetic association and interaction between the IRF5 and TYK2 genes and systemic lupus erythematosus in the Han Chinese population. Inflammation research: official journal of the European Histamine Research Society… [et al.] 2015;64:817–824. doi: 10.1007/s00011-015-0865-2. [DOI] [PubMed] [Google Scholar]
- 10.Sigurdsson S, et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. American journal of human genetics. 2005;76:528–537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mero IL, et al. A rare variant of the TYK2 gene is confirmed to be associated with multiple sclerosis. Eur J Hum Genet. 2010;18:502–504. doi: 10.1038/ejhg.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet42, 1118–1125, 10.1038/ng.717 (2010). [DOI] [PMC free article] [PubMed]
- 14.Cunninghame Graham DS, et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS genetics. 2011;7:e1002341. doi: 10.1371/journal.pgen.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagafuchi S, et al. TYK2 Promoter Variant and Diabetes Mellitus in the Japanese. EBioMedicine. 2015;2:744–749. doi: 10.1016/j.ebiom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Isac, E. et al. Influence of TYK2 in systemic sclerosis susceptibility: a new locus in the IL-12 pathway. Annals of the rheumatic diseases, 10.1136/annrheumdis-2015-208154 (2015). [DOI] [PMC free article] [PubMed]
- 17.Lee, Y. H. & Bae, S. C. Association between TYK2 polymorphisms and susceptibility to autoimmune rheumatic diseases: a meta-analysis. Lupus, 10.1177/0961203316638933 (2016). [DOI] [PubMed]
- 18.O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36:542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babon JJ, Lucet IS, Murphy JM, Nicola NA, Varghese LN. The molecular regulation of Janus kinase (JAK) activation. Biochem J. 2014;462:1–13. doi: 10.1042/BJ20140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sohn SJ, et al. A restricted role for TYK2 catalytic activity in human cytokine responses revealed by novel TYK2-selective inhibitors. J Immunol. 2013;191:2205–2216. doi: 10.4049/jimmunol.1202859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet81, 559–575, 10.1086/519795 (2007). [DOI] [PMC free article] [PubMed]
- 22.Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat Methods7, 248–249, 10.1038/nmeth0410-248 (2010). [DOI] [PMC free article] [PubMed]
- 23.Couturier, N. et al. Tyrosine kinase 2 variant influences T lymphocyte polarization and multiple sclerosis susceptibility. Brain134, 693-703, 10.1093/brain/awr010 (2011). [DOI] [PubMed]
- 24.Li Z, et al. Two rare disease-associated Tyk2 variants are catalytically impaired but signaling competent. J Immunol. 2013;190:2335–2344. doi: 10.4049/jimmunol.1203118. [DOI] [PubMed] [Google Scholar]
- 25.Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ. Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat Struct Mol Biol. 2014;21:443–448. doi: 10.1038/nsmb.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, et al. Transcriptome analysis of psoriasis in a large case-control Sample: RNA-Seq rovides insights into disease mechanisms. J Invest Dermatol. 2014;134:1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehta NN, et al. IFN-gamma and TNF-alpha synergism may provide a link between psoriasis and inflammatory atherogenesis. Sci Rep. 2017;7:13831. doi: 10.1038/s41598-017-14365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsoi LC, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dendrou CA, et al. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci Transl Med. 2016;8:363ra149. doi: 10.1126/scitranslmed.aag1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.