Abstract
The SQUAMOSA-promoter binding like (SPL) gene family encodes transcription factors that have been shown in many species to influence plant growth and development, but information about these genes in barley (Hordeum vulgare L.) is limited. This study identified 17 barley SPL genes, within eight distinct groups, that are orthologs of SPL genes described in Arabidopsis, wheat, and rice. Sixteen barley SPLs undergo alternative splicing. Seven SPLs contain a putative miR156 target site and the transcript levels of the miR156-targeted HvSPLs (HvSPL3, 13 and 23) were lower in vegetative than in reproductive phase but this was true also for some SPL genes such as HvSPL6 that were not regulated by miR156. Because SPL gene products regulate miR172, which is also involved in floral development, the expression of miR172 was studied. An antagonistic expression pattern of miR156 and miR172b during the vegetative and the reproductive phases signifies their apparent function in barley growth phase transition. Characterization of a barley mir172 mutant having an abnormal, indeterminate spikelet phenotype suggests the possible feedback role of AP2/miR172 module on HvSPL genes. This is the first comprehensive analysis of the miR156/SPL/miR172 axis in barley that provides a basis to elucidate their roles in various biological processes.
Introduction
Barley (Hordeum vulgare L.) is a widely cultivated cereal grain. Cereal inflorescences are known as spikes. Each spike is composed of multiple spikelets formed directly on the main axis1. During domestication, both the yield and the architecture of cereal plants have been modified. Plant architecture and grain yield are complex traits that are encoded by many genes and regulatory factors. In the current study, our main research target was to explore the involvement of SPLs in barley growth phase transition from vegetative to reproductive stage. Transcription factors play important roles in plant growth and development by inducing or suppressing the expression of their target genes. The SQUAMOSA promoter binding like (SPL) protein family is one of the plant specific transcription factor families and each member shares a highly conserved 76 amino acid long DNA binding domain known as the SBP domain2,3. The SBP domain consists of three functionally important motifs, including two zinc-binding sites, Cys–Cys–Cys–His (Zn-1) and Cys–Cys– His–Cys (Zn-2), and a nuclear localization signal (NLS) located at the C-terminus of the domain3,4.
The first SPL gene was identified in Antirrhinum majus, and it controls flowering by binding to the promoter of SQUAMOSA (SQUA)2. Subsequently, multiple SPL genes were identified in Arabidopsis thaliana5, green algae (Chlamydomonas)6,7, moss8, silver birch9, tomato10, rice11, maize12, soybean13, wheat14 and cotton15. Studies have identified 16 SPL genes in A. thaliana16,17, 19 in rice18, and 28 in Populus trichocarpa19.
Studies of A. thaliana have shown that SPL genes have diverse functions in plant growth and development. Constitutive expression of SPL3 produced very early flowering20. SPL8 affected reproductive development through the genes involved in GA (gibberellic acid) biosynthesis21,22. SPL2, SPL10 and SPL11 have been associated with shoot maturation23. SPL9 and SPL13 controlled shoot development in Arabidopsis24. SPL7 was identified as a central regulator of copper homeostasis25. The miR156-targeted SPL9 promoted sesquiterpene biosynthesis by binding to the promoter region of TPS2126 and it negatively regulated anthocyanin levels by modulating the expression of the MYB-bHLH-WD40 complex27.
SBP1 silencing in A. majus resulted in a late to non-flowering phenotype, and SBP1-mediated transition to flowering occurred due to the positive regulation of FUL/LFY meristem identity genes28. Paralogous SBP1, SBP2 and CNR genes differentially controlled leaf initiation and reproductive phase transition in petunia29. SPL genes in monocot plants also have been shown to affect important developmental processes. In rice, overexpression of SPL14 promoted panicle branching and higher grain yield30; SPL16 regulates grain size, shape, and quality31; SPL13 positively controlled grain weight, length, and thickness32; and the interaction of the SPL14 protein with human OTUB1 like deubiquitinase enhanced grain yield33. In bread wheat, it was found that the miR156-SPL module regulated bread wheat plant architecture by interacting with a strigolactone signalling repressor gene, DWARF5334. The maize SBP-box transcription factors unbranched2 and unbranched3 alters plant architecture and affect yield traits in maize35. Genetic modification of the miR156-SPL4 module controls aerial axillary bud formation, branching, biomass yield, and re-growth after cutting in switchgrass36.
MicroRNAs (miRNAs) are non-coding RNAs that can complementarily bind to target sites and repress expression via cleavage or repression of translation37. In Arabidopsis, 10 of the 16 SPL genes are targets of miR1565,20 and 11 of the 19 SPL genes in rice have been identified as a targets of miR15618. The miR156 complementary sites are present in the coding region or in the 3′ un-translated region (3′-UTR). In A. thaliana, two miRNAs, miR156 and miR172, regulated the juvenile to adult developmental phase change38; SPL9 and SPL10 promoted the expression of miR172b by binding to its promoter and acted independently of this and its target genes38; and the expression of miR156 was higher in the juvenile phase than in the adult phase, whereas the expression of miR172 was lower in the juvenile phase than in the adult phase38. The miR172 is known for the regulation of AP2-like transcription factors through transcript cleavage and translational repression in Arabidopsis39,40. Expression of miR172 promotes the vegetative phase change in maize by repressing an AP2-like gene Glossy1541. In barley, suppression of miR172 guided cleavage of AP2 mRNA produces cleistogamous flowering42 and affects spikelet determinacy43. Perturbed interaction between AP2 and miR172 leads to striking differences in the size and shape of the barley spike44. The characterization of SPL genes has not been conducted for barley as it has for Arabidopsis and rice. This is the first comprehensive study of SPL genes in barley and includes analysis of phylogeny, motif composition, gene structure, miRNA target site, alternative splicing events and spatio-temporal expression patterns. In addition, the expression patterns of SPL genes and of miR156 and miR172 from vegetative to reproductive phases revealed their possible functional relationships. The expression of AP2 and SPL genes in the spikes of mir172 mutants and its wild-type counterpart golden promise (GP) elucidated their involvement in spike development.
Results
Identification of SPL Genes in Barley
A total 17 putative SPL genes were identified in barley and were designated as HvSPL. In this study, HvSPLs were specified based on their similarity to wheat and rice orthologs (Table 1). Full length coding sequences of the HvSPLs ranged from 339 to 3393 bp (Table S1), and the deduced proteins ranged from 112 to 1130 amino acids (Table S2). The 17 HvSPL genes were unevenly distributed on chromosomes 1H, 2H, 3H, 5H, 6H, 7H and Un (Unnumbered) (Table 1).
Table 1.
Characteristics of identified SPL genes in H. vulgare.
| Genea | Gene symbolb | CDSc length (bp) | Domaind | Deduced protein (aa)e | Chf | Position on genomeg | Exonh |
|---|---|---|---|---|---|---|---|
| HvSPL1 | HORVU7Hr1G042370 | 2526 | SBP, DEXDC, ANK | 841 | 7H | 122553615–122558679 | 10 |
| HvSPL3 | HORVU6Hr1G019700 | 1524 | SBP | 507 | 6H | 53909817–53916886 | 5 |
| HvSPL6 | HORVU5Hr1G117190 | 2895 | SBP, DEXDC, ANK | 964 | 5H | 650559269–650565702 | 11 |
| HvSPL7 | HORVU2Hr1G097580 | 630 | SBP | 209 | 2H | 679640052–679642809 | 2 |
| HvSPL7A | HORVU2Hr1G097610 | 612 | SBP | 203 | 2H | 679689970–679692277 | 2 |
| HvSPL 8 | HORVU0Hr1G039150 | 1089 | SBP | 362 | Un | 248118980–248122768 | 3 |
| HvSPL9 | HORVU1Hr1G060770 | 978 | SBP | 325 | 1H | 441051831–441063137 | 1 |
| HvSPL11 | HORVU6Hr1G031450 | 981 | SBP | 326 | 6H | 133169150–133173306 | 4 |
| HvSPL13 | HORVU2Hr1G048280 | 588 | SBP | 195 | 2H | 269391017–269400061 | 3 |
| HvSPL15 | HORVU7Hr1G051400 | 3393 | SBP,DEXDC, ANK | 1130 | 7H | 192879289–192885430 | 10 |
| HvSPL16 | HORVU5Hr1G076380 | 1179 | SBP | 392 | 5H | 551053196–551057389 | 3 |
| HvSPL17 | HORVU5Hr1G073440 | 1251 | SBP | 416 | 5H | 539014598–539018210 | 3 |
| HvSPL18 | HORVU0Hr1G039170 | 1203 | SBP | 400 | Un | 248141188–248147114 | 3 |
| HvSPL 20 | HORVU7Hr1G110980 | 1350 | SBP | 449 | 7H | 631945657–631949862 | 3 |
| HvSPL21 | HORVU6Hr1G030490 | 1476 | SBP | 491 | 6H | 127986525–127990179 | 3 |
| HvSPL 22 | HORVU7Hr1G110950 | 339 | SBP | 112 | 7H | 631926981–631927880 | 1 |
| HvSPL23 | HORVU3Hr1G094730 | 1290 | SBP | 429 | 3H | 647368957–647372642 | 3 |
aName referred to H. vulgare SPLs in this work.
bGene accession number in database.
cLength of coding DNA sequence.
dDomain predicted by SMART tool.
eLength (number of amino acids).
fChromosome position of the HvSPL genes.
gLocation of HvSPL genes on barley genome.
hExon number in HvSPL genes.
Phylogenetic Relationship of SPL Genes in Barley, Arabidopsis, Rice, and Wheat
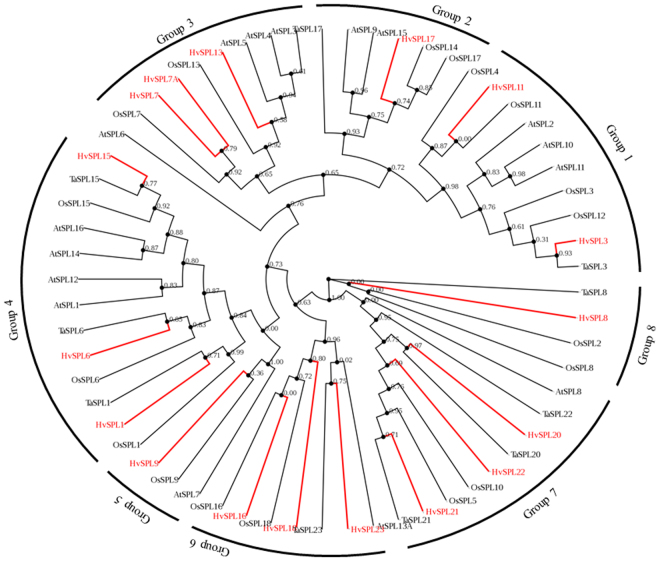
A phylogenetic tree was constructed using the maximum likelihood method based on 61 SBP domains sequences from barley, rice, wheat, and A. thaliana (Fig. 1; Table S4). The SPL proteins were assigned to one of eight groups with one Arabidopsis protein (AtSPL6) outlier. As expected, HvSPL proteins were more closely related to those of wheat and rice than to those of A. thaliana. HvSPLs were present in all groups. Maximum numbers of HvSPL genes (9) were found in the same clades where SPL orthologs from wheat were grouped. Other HvSPLs were found to be more closely related to rice SPL orthologs. Interestingly, HvSPL11 was orthologous to two rice SPL paralogs, SPL11 and SPL4 (Fig. 1). Rice SPL7 was orthologous to two barley SPL paralogs, SPL7 and SPL7A. Likewise, HvSPL17 was orthologs to OsSPL14 and OsSPL17 genes.
Figure 1.
Phylogenetic analysis of SPL proteins based on their SBP domain sequences. The maximum likelihood phylogenetic tree of SPL proteins from Arabidopsis thaliana (AtSPL), rice (OsSPL), wheat (TaSPL) and barley (HvSPL) using PhyML3.0. HvSPLs are shown in red color.
Structural Features and Conserved Motif Analysis of Barley SPL Genes
Genetic structural diversity may enable the evolution of multi-gene families. To gain further insight into the structure of HvSPL genes, we compared the predicted numbers, lengths, and arrangements of introns and exons. At least one intron was present in all HvSPL genes except HvSPL9, and the number of exons ranged from 1 to 11 (Fig. S1A). We searched for conserved motif sequences present inside of SBP domain and in predicted HvSPL full length proteins, and their compositions and diversities were analysed. Analysis of the conserved domains of HvSPL proteins showed that barley SBP domain has two zinc binding motifs (Cys3His and Cys2HisCys) that include eight conserved cysteine and histidine residues (Fig. S1B). A putative NLS motif was identified at the C-terminus of the Cys2HisCys motif. HvSPL1, 6 and 15 also contained DEXDC and ANK (ankyrin) domain (Table 1). Further investigation discovered ten motif sequences that are less distantly diverged among their groups and their combinations were analysed (Fig. S2). Motifs 1, 2, 7 and 8 are involved in the makeup of the Zn-2, Zn-1, NLS (nuclear localization signal) and joint peptide of SBP domain, but the identities of other motifs are unknown. All predicted HvSPL proteins contain conserved motif combinations within their phylogenetic group, suggesting that the variable amino acids outside of the SBP domains are responsible for the diversity of SPLs in A. thaliana, barley, wheat, and rice.
Cis-Regulatory Elements in HvSPLs Promoter Regions
Presence of cis-regulatory motifs was analysed using 1 Kb upstream genomic sequences of HvSPL genes. The identified cis-regulatory motifs were belonged to mainly four categories based on functional aptitude namely, light responsive elements, growth and development responsive elements, stress responsive and hormone responsive elements (Table 2). The 17 SPL genes were grouped according to phylogeny and HvSPL9 from Group 5, HvSPL17 from 2, and HvSPL18 from group 6 were found to contain plant_AP-2-like regulatory motif. The light responsive elements, G-Box, sp1, GA-motif, GAG-motif, I-box, GC-motif, ACE motif, MNF1 and Box 1 were enriched in most of the genes. In case of growth and development responsive elements, AC-II, CCGTCC-box, ATGCAAAT motif, GCN4_motif, O2-site, CCGTCC-box, Skn-1_motif, circadian and plant_AP-2-like etc. were more abundant. Further, stress responsive elements like ARE, A-box MBS, LTR and HSE were responsive to various abiotic stresses whereas, Box W1 and W-box were reported in biotic stresses. Numerous cis-regualtory elements responsive to hormones were identified such as ERE and ABRE were ethylene and abscisic acid responsive elements; TGA element and AuxRE-core were reported as auxin responsive; GARE-motif, P-box and TATC-box as gibberellin responsive and CGTCA and TGACG motifs are related to MeJa response.
Table 2.
Cis-regulatory elements predicted in promoter region of HvSPL genes.
| Gene | Light Response | Growth and Development | Stress Response | Hormone Response |
|---|---|---|---|---|
| HvSPL3 | Box II –like sequence, CATT-motif, GA-motif, GAG-motif,Sp1 | 5UTR Py-rich stretch, AC-II,CCGTCC-box, GCN4_motif, Skn-1_motif, circadian | A-box, ARE, HSE,MBS, TC-rich repeats, box E | CGTCA-motif, TCA-element, TGACG-motif |
| HvSPL11 | ATC-motif, BoxI, GAG-motif, GC-motif, I-box,MNF1,Sp1 | CAT-box, GCN4_motif, Skn-1_motif, circadian | ARE | CGTCA-motif, TCA-element, TGA-element, TGACG-motif |
| HvSPL17 | ATCC-motif, Box I, CATT-motif, G-Box, GA-motif, GAG-motif, Gap-box,Sp1 | HD-Zip 3,Skn-1_motif, plant_AP-2-like | ARE, HSE, TC-rich repeats | TGA-element |
| HvSPL13 | ACE,ATCT-motif, Box 4,GA-motif, LAMP-element | ATGCAAAT motif, GCN4_motif, O2-site | Box-W1, HSE, W box | CGTCA-motif, EIRE, TGACG-motif |
| HvSPL7 | ACE,CG-motif, G-box, GATA-motif, GC-motif, I-box,Sp1,box II | CCGTCC-box, RY-element, Skn-1_motif | A-box, LTR, box S | ABRE, CGTCA-motif, TATC-box, TGACG-motif |
| HvSPL7A | ATC-motif, ATCT-motif, G-Box, GA-motif, GAG-motif, GC-motif, Sp1,TCT-motif | Skn-1_motif | box S | ABRE |
| HvSPL15 | ACE,G-Box, GC-motif,Sp1 | AC-II,CAT-box, CCGTCC-box, O2-site,dOCT | A-box, ARE, MBS, TC-rich repeats | ABRE, motif Iib |
| HvSPL6 | ACE, GC-motif, MNF1, Pc-CMA2c, Sp1,TCT-motif | CAT-box, CCAAT-box, CCGTCC-box, OCT | A-box, Box-W1, TCCACCT-motif, W box | CGTCA-motif, TGA-element, TGACG-motif |
| HvSPL1 | ACE,AE-box, Box 4,G-box, GT1-motif,I-box,MRE,Sp1, TCT-motif | O2-site, Skn-1_motif, circadian | Box-W1, MBS, W box | AuxRE, TCA-element |
| HvSPL9 | GC-motif, L-box, MNF1, Sp1 | AC-I,AC-II, CAT-box, GCN4_motif,O2-site, Skn-1_motif, plant_AP-2-like | Not found | ABRE, CGTCA-motif, GARE-motif, TGACG-motif, motif IIb |
| HvSPL16 | CATT-motif, G-Box,GAG-motif, I-box, SpI | 5UTR Py-rich stretch, AC-I,CAT-box, Skn-1_motif | HSE, TCCACCT-motif | ABRE |
| HvSPL18 | G-Box,GAG-motif, GT1-motif, I-box,Sp1, TCCC-motif | CCGTCC-box, GCN4_motif, HD-Zip 1,HD-Zip 2, RY-element, circadian, plant_AP-2-like | A-box, ARE, TC-rich repeats | ABRE |
| HvSPL23 | ATCC-motif, G-box,GC-motif, MNF1,Sp1 | 5UTR Py-rich stretch, CCAAT-box, Skn-1_motif | Box-W1, LTR, TC-rich repeats, W box | GARE-motif, motif IIb |
| HvSPL21 | ACE, Box I, GT1-motif, TCCC-motif | ATGCAAAT motif, GCN4_motif, Skn-1_motif, TA-rich region | TC-rich repeats | CGTCA-motif, ERE, TCA-element, TGACG-motif |
| HvSPL 22 | Box II,G-Box, GC-motif, Sp1, TATCCAT/C-motif | CCAAT-box, CCGTCC-box, Skn-1_motif | A-box, Box-W1, HSE,W box | CGTCA-motif, TATC-box, TGA-element, TGACG-motif |
| HvSPL 20 | C-box,G-box, GC-motif,Sp1, chs-Unit 1 m1 | AC-II,CCGTCC-box, GCN4_motif, O2-site, Skn-1_motif | A-box, ARE, Box-W1, MBS, TCCACCT-motif, W box | ABRE, CGTCA-motif, GARE-motif, P-box,TGACG-motif |
| HvSPL 8 | G-Box, GATA-motif, chs-CMA2a | Circadian | Box-W1,MBS, W box | TCA-element |
MiR156 Family in H. vulgare and Their Target Site in HvSPL Genes
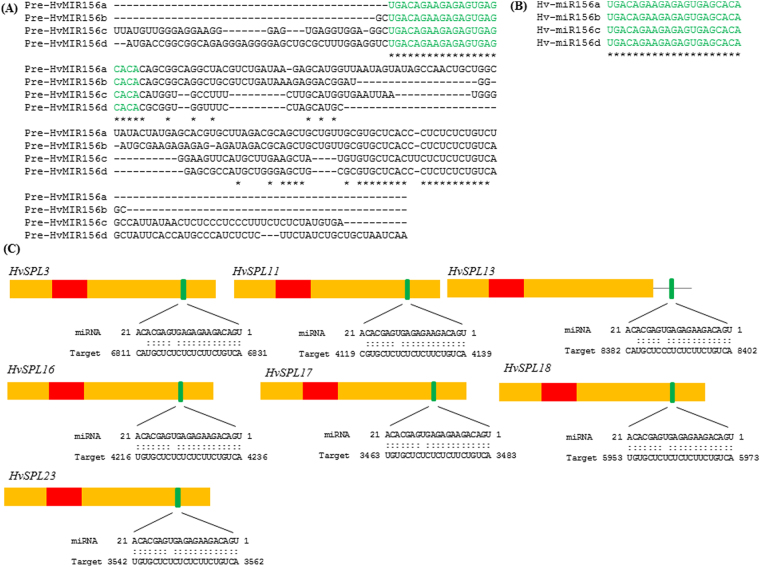
Previous studies have reported that a subset of SPL genes are regulated by miR156 in plant species including A. thaliana17, soybean13, rice18, poplar19, and maize45. Therefore, miR156 family in barley genome and its target site in HvSPLs was studied. Two putative members of miR156 family, Hv-miR156a (accession number MI0016449) and Hv-miR156b (accession number MI0030546) were identified for barley in the miRbase database (http://www.mirbase.org/cgibin/mirna_summary.pl?fam=MIPF0000008) and another two (Hv-miR156c and Hv-miR156d) were identified in the mirEX2.0 database (Fig. 2A, 2B). The mature miR156 sequences of all four members were identical, but divergence was observed in the precursor sequences which showed 71 to 87% homology. Putative miR156 binding sites were found for HvSPL3, HvSPL11, HvSPL16, HvSPL17, HvSPL18 and HvSPL23 in their coding regions and for HvSPL13 in the 3′UTR (Fig. 2C; Table S5), suggesting that regulation by miR156 is restricted to this subset of HvSPL genes.
Figure 2.
miR156 family members and their target site in Barley SPL genes. (A) Alignment of precursor sequences of four miR156 family members. (B) Alignment of mature sequences of four miR156 family members. Green colour denotes the mature sequence of miR156a/b/c and d. (C) miR156 target site in HvSPL3, 11, 13, 16, 17, 18 & 23 genes. Yellow box represent CDS, red box SBP domain and line 3′UTR. The miR156 target sites with the nucleotide positions of HvSPL transcripts are shown in green. RNA sequence of each complementary site from 5′ to 3′ and the predicted miRNA sequence from 3′ to 5′ are indicated.
Validation of Alternative Splicing Events in HvSPL Genes
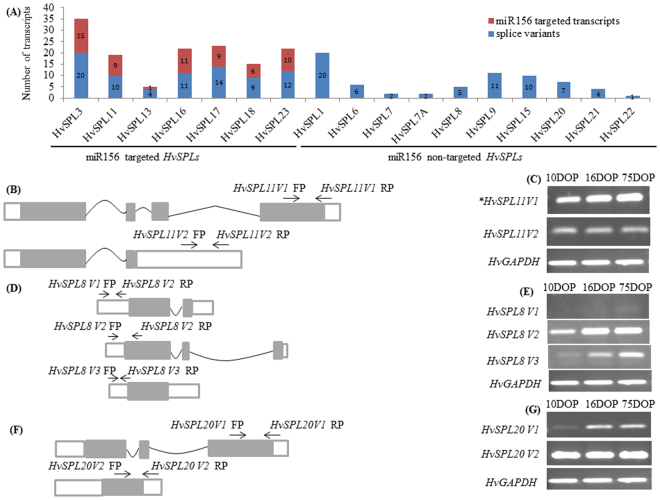
Multiple transcripts can be formed from one gene by selecting different splice sites during mRNA production. Therefore, various transcript isoforms may generate truncated proteins that could influence their stability levels, sub-cellular localizations, protein-protein interactions and other functions. The ensemble database predicted splice variants in 16 of 17 HvSPL genes, except HvSPL22 (Fig. 3A; Table S6). All HvSPLs with miR156 binding sites were predicted to produce splice variants (4 to 20 numbers). Similarly, miR156 non-targeted (that is, lacking a miR156 binding site) HvSPLs also generated splice variants (1 to 20 numbers) of varying length. HvSPL1 and 3 produced the highest numbers (20) of predicted splice variants. Interestingly, differences in the miR156 target site among splice variants were also observed. In case of HvSPL3 (20 splice variants), HvSPL11 (4 splice variants) and HvSPL17 (14 splice variants), only 15, 1 and 9 number of splice variants contained miR156 target site.
Figure 3.
Alternative splicing event in HvSPL genes and expression analysis. (A) Number of Splice variants in HvSPL genes and miR156 target site distribution. (B,C) The splice variants of barley SPL11 and their expression pattern in 10, 16 and 75 day old plant. The two variants (SPL11V1 and V2) expressed differentially. (D,E) Splice variant of barley SPL8 and their expression patterns. The three variants (SPL8V1, V2 and V3) of SPL8 expressed lower in juvenile and higher in reproductive phase. (F,G) Two splice variant (SPL20V1 and V2) of barley SPL20 and their expression patterns. *Asterisks denote the presence of the miR156 complementary sequence in the splice variants of barley SPL11 gene. The brown colour box represents coding region, black line denotes intron and the white rectangle denotes 5′ & 3′ UTR regions. The arrow shows the site of forward and reverse primers. For more clarity gel area showing relevant bands were cropped. The full-length gels are presented in Supplementary Fig. S3.
To distinguish the major and minor splice variants and their differential expression patterns, several HvSPLs (HvSPL8, HvSPL11, and HvSPL20) were selected at random for analysis by semi-quantitative RT-PCR in 10, 16, and 75 days old (vegetative to reproductive phase) barley plants (Fig. 3B–G). Splice variant 1 of HvSPL11 (HvSPL11 V1), which contained a miR156 target site, showed lower expression at vegetative and higher at the reproductive phase and was the major transcript (Fig. 3B,C). However, expression remained constant for variant 2 of HvSPL11 (SPL11 V2), which lacked a miR156 target site and was a minor transcript. Similarly, differential expression of splice variants of HvSPL8 and HvSPL20 was observed at different developmental stages (Fig. 5D–G). Expression of HvSPL8 V2 and HvSPL8 V3 was lower in the vegetative phase but higher in the reproductive phase; these were the major transcripts. Expression of HvSPL20 V1 was higher at the reproductive stage than in the vegetative stage. Expression of HvSPL20 V2 was constant in all stages and was the key transcript for HvSPL20. These results suggest that splice variants of HvSPLs are produced and that they may contribute to the diversity of encoded proteins with the potential to play important roles at various stages of plant growth and development.
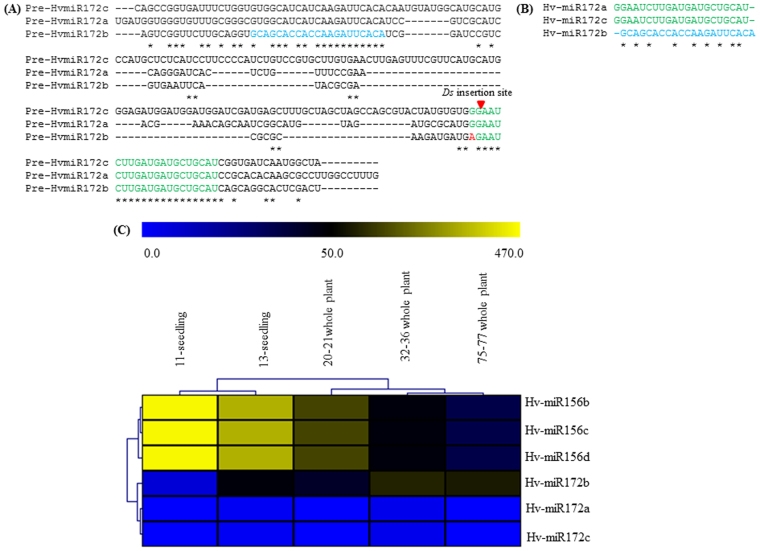
Figure 5.
Barley miR172 sequences and expression analysis of miR156 and miR172 family members. (A) Alignment of three miR172 (miR172a/b and c) precursor sequences. (B) Alignment of three miR172 mature sequences. Green and blue colours differentiate mature sequences of miR172a & c and miR172b respectively. (C) Heat map showing the expression profiling of Hv-miR156b/c/d and Hv-miR172a/b/c. Expression data was obtained from publically available database mirex2.0. Antagonistic expression pattern of miR156 and miR172b was observed during vegetative and reproductive phases of barley. The respective transcripts of miR156 and miR172 has been shown in RPM (reads per million). The red triangle indicates Ds insertion site in miR172c mature sequence.
Spatio-temporal Expression Pattern of HvSPL Genes
In the absence of HvSPL mutants, the expression patterns of various HvSPLs may provide clues about their potential functions. We examined the spatio-temporal expression patterns of HvSPLs in eight tissues (4-day-old embryos (EMB), roots (Roo), shoots (LEA), developing inflorescences (INF1; 5 mm) and (INF2; 1–1.5 cm), developing tillers internodes (NOD), developing grain 5 days post anthesis (DPA), developing grain with bracts removed at 5 DPA (CAR5) and 15 DPA (CAR15)) of barley via hierarchical clustering analysis (Fig. 4). Nine HvSPLs (HvSPL1, 3, 6, 11, 13, 15, 16, 17 and 23), including six that are targeted by miR156 (HvSPL3, 11, 13, 16, 17 and 23) were highly expressed and displayed tissue-specific patterns of expression. Interestingly, expression of miR156 targeted HvSPL18 and miR156 non-targeted HvSPL7 and HvSPL20 were unique to INF2 tissue. In contrast, the others (HvSPL7A, 8, 21, and 22) showed very low expression in all tissues. Most of the HvSPLs genes (except HvSPL8 and 21) were highly expressed in inflorescence 2, whereas expression of only HvSPL17, 20, 21 and 22, was higher in inflorescence 1, suggesting their involvement in barley inflorescence development. HvSPL13 was expressed mainly in the inflorescence, and its expression at the NOD and CAR5 stages was negligible. HvSPL6 and HvSPL15 were constitutively expressed at high levels in all tissues.
Figure 4.
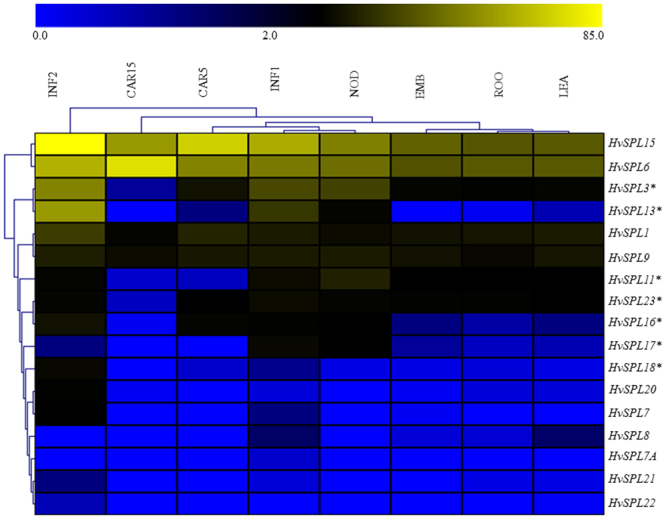
Spatio-temporal expression patterns of HvSPL genes in eight different tissues. The colour scale bar at the top of heat map represents FPKM normalized log2 transformed values based on “Morex” RNA-seq data, and represents high and low expression, respectively. EMB, ROO, NOD, LEA, INF1, INF2, CAR5 and CAR15tissues were used for expression profiling and indicated at the top of the heat map. Details about these tissues have been explained in material and method section. HvSPL genes that contain miR156 target sites are indicated by (*) asterisks.
Vegetative to Reproductive Phase in Barley: Expression of miR156, miR172 and Specific SPL Genes
The timing of juvenile to adult phase transition in A. thaliana is known to be regulated by miR156 and miR172, along with several members of the SPL family38. We examined their expression patterns in barley during the vegetative to reproductive phase change. The sequences of three miR172 family members (miR172a/b/c) in barley were retrieved from the mirex2.0 database (Fig. 5A,B). The expression patterns of miR156b/c/d and miR172a/b/c in barley tissues collected at 11, 13, 20–21, 32–36 and 75–77 days old plants were examined46 (Fig. 5C). As expected, expression of miR156 family members was higher in 11-d-old seedlings stage (vegetative phase) and lower in 70–75 days old plants (reproductive phase). Interestingly, the expression of only miR172b was lower in 11-d-old seedlings and higher in 70–75-d-old-plants. However, the expression of miR172a and of miR172c was lower only in 11, 20–21 and 75–77 day old plants. Thus, miR156b/c/d and only miR172b showed an inverse expression relationship during the vegetative to reproductive phase change, suggesting their contribution in growth phase transition of barley.
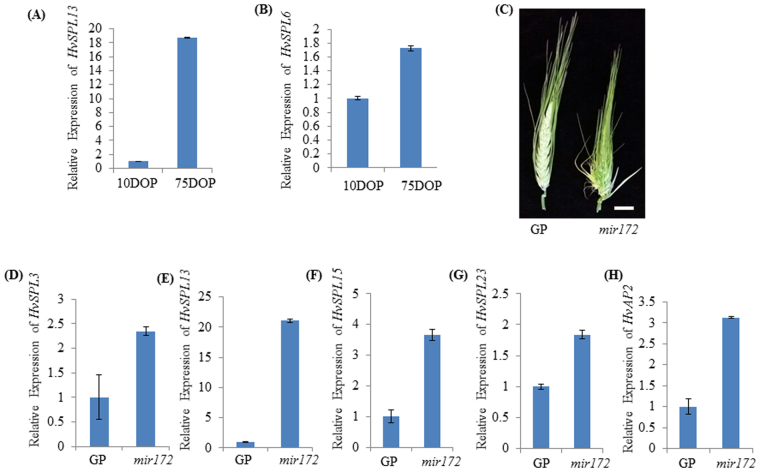
To validate the expression patterns of selected HvSPLs, barley seedlings were harvested at 10, 16, and 75 days old plants (DOP) and expression of HvSPLs was measured by RT-PCR and qRT-PCR (Figs 6A,B and S4). Five HvSPLs (HvSPL3, 6, 13, 15, and 23) were selected based on their differential expression in silico. All these genes were expressed at low levels during the vegetative phase (10 DAP) and higher levels at the reproductive phase (75 DAP), implying their association with reproductive phase development (Fig. S4). Nevertheless, expression of HvSPL15 remained stable throughout the development. Further investigation by qRT-PCR showed that the transcript abundance of HvSPL13 and HvSPL6 in reproductive phase vs. vegetative phase was 18- and 1.7-fold higher, respectively (Fig. 6A,B).
Figure 6.
Expression of HvSPL genes during growth phase transition in barley. (A) Transcript abundance of HvSPL13 and (B) HvSPL6 at vegetative (10 days) and reproductive (75 days) phases. (C) Spike architecture of barley mir172 mutant and wild type GP. A single 3.6 kb Ds insertion in mir172 mutant was previously identified by Brown and Bregitzer, 2011. This mutant possesses abnormal spike at both apical region (glumes were changed to florets) and basal region (abnormal branched phenotype). (D–H) Expression of HvSPL3, 13, 15, 23 and AP2 genes in the spike of barley mir172 mutant and wild type GP. Transcript abundance was measured by qRT-PCR.
Interaction Analysis of miR172, HvSPL & HvAP2 Genes
Previously, a barley mir172 mutant line was developed through transposon tagging system in which a 3.6 kb Ds sequence was inserted into the mature sequence of miR17243. This insertion produced abnormal spikelet development in that the apical region of spike glumes were converted to partially developed florets and basal region showed abnormal branched phenotype. Comparison of miR172a/b/c precursor sequence with Ds flanking sequence suggested us that the Ds was inserted in the mature miR172c sequence (Fig. 5A). Since SPL/miR156 module control panicle branching by directly regulating the miR172/AP2 module in rice30,47, bract and ear glume development in maize48,49 and floral meristem identity in A. majus2,28, expression of HvSPL genes in the mir172 barley mutant was analysed. The expression of HvSPL3, 6, 13, 15 and 23 was initially investigated through semi-quantitative RT-PCR and followed by qRT-PCR to investigate expression in the immature spikes the mir172 mutant and wild-type barley plants (Figs S5 and 6C–G). Expression of HvSPL3, 13, 15 and 23 was higher in the mir172 mutant than in the wild-type counterpart. Expression of HvSPL3, 13, 15 and 23 was further investigated through qRT-PCR which showed 2.3 to 21 fold higher expression in the mir172 mutant. As expression of HvAP2 is regulated by miR172, we also examined the expression of HvAP2 in the spike of wild type and mir172 mutant lines (Figs S5 and 6H). As expected, expression of HvAP2 was higher in the mir172 mutant spike as compared to its wild type counterparts. These results suggest the possible indirect feedback regulation of the AP2/miR172 module on HvSPL genes in barley.
Discussion
SPL family genes are plant specific transcription factors and have been identified in many plant species, namely A. thaliana, rice, wheat, maize, tomato, populus, chlamydomonas, silver birch, Brassica napus, and soybean7,9,10,13,14,18,19,45,50,51. The present study represents the first comprehensive analysis of the miR156/SPL/miR172 regulatory hub in barley. Phylogenetic analysis based on amino acid sequences of conserved SBP domain from wheat, rice, and barley (monocot) and from A. thaliana (dicot) classified HvSPLs into eight different groups (Fig. 1). SPL proteins belonging to the same group appeared to be more closely related to each other than to those of other groups within a species. Differences in exon/intron structures and SBP domains of HvSPLs suggest functional diversity in plant development (Fig. S1A,B). The SBP domain binds to consensus nucleotide sequences TNCGTACAA17,50, with GTAC being an essential core sequence present on the promoter of its target genes. The zinc finger motif with two Zn ions binding sites in the SBP domain in barley was Cys3His1 and Cys2His1Cys1, and there was a NLS signal at C-Terminus (Fig. S1B). The NLS signal present at the C-terminus partly overlaps with the second Zn ion binding structure3. The size of A. thaliana SPL proteins range from 131 amino acids (SPL3) to 927 amino acids (SPL12)17. In contrast, the size of barley SPL proteins ranged from 112aa (SPL22) to 1130aa (SPL15) (Table 1). Different SPLs may have various numbers of exons/introns even though SBP domain of all land plants is determined by the first and second exons52.
Categorization of HvSPLs by phylogenetic analysis was supported by classification based on motif composition. The majority of SPL proteins belonging to the same group contained similar motif distributions (Fig. S2) vs greater dissimilarity compared to other phylogenetic groups, a finding that is similar to those reported for soybean13, wheat14, and maize45, suggesting that these differences could be important for the function of SPLs. We found that motifs 1, 2,7 and 8 were highly conserved and present in most of the HvSPL proteins, a finding similar to that found in wheat14. The diversity in motif composition and variation in the amino acid sequences in the SBP domain reveals that SPL is a diverse gene family.
miRNAs play key functions in controlling the transcription of target genes. We identified 4 members of miR156 family in barley and target prediction showed that 7 of the 17 SPL genes contained a complementary site for this miRNA (Fig. 2A–C; Table S5). The miR156-complementary site was present in coding regions of HvSPL3, 11, 16, 17, 18, 23, and in the 3′UTR of HvSPL13. In A. thaliana, 10 of 17 SPL genes are targeted by miR156, suggesting that miR156 complementary sites in SPL genes are conserved across plant species.
Alternative splicing (AS) acts as a “molecular thermometer” that allows plants to generate efficient transcripts to cope up with environmental perturbations53. Recently, AS was identified in barley and its possible function was explored by network analysis54. FLOWERING LOCUS T (FT) in Brachypodium undergoes age dependent AS and produces two (FT2α and FT2β) splice variants55 but, only FT2β was found to be involved in the regulation of flowering. Similarly, 16 of 17 HvSPLs undergo AS and generate diverse transcript and protein sequences (Fig. 3, Table S6). Most of the splice variants of miR156-targeted HvSPLs exhibited miR156 complementary sites, implying the existence of alternate splicing-mediated regulation of biological processes in barley (Fig. 3, Table S7). Splice variants of HvSPL8, 11, and 20 displayed age-dependent differential expressions, implying their role in barley growth phase transition (Fig. 3B–G). The different splice variants of HvSPL8, 11, and 20 were expressed less in the vegetative phase and more in the reproductive phase, revealing their possible association with reproductive development. This is consistent with the previous results for phase transition in A. thaliana by SPL genes5,38. In A. thaliana, SPL4 promotes vegetative phase change and flowering5. Similarly, the male fertility and gynoecium differentiation in A. thaliana was shown to be regulated by SPL856,57. Exploration of cis-regulatory elements in the promoter regions of HvSPLs exhibited both conservation and divergence (Table 2). SPL genes have been shown to be responsive to light, hormone and abiotic stresses45,58. The majority of the cis-regulatory elements were involved in response to light signalling, plant growth and development, stresses and hormones. The presence of plant_AP-2-like motif in HvSPL9, HvSPL17 and HvSPL18 promoter region suggest the possible feedback regulation of HvSPL genes by AP2 like transcription factor. The elements for abscisic acid (ABA), gibberellic acid (GA), salicylic acid (SA), methyl jasmonate (MeJA) and auxin were enriched. Earlier studies in A. thaliana indicated that floral transition was regulated by gibberellin guided miR156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors58. The hormonal pathways phenomenon awaits investigation which has not thus far been reported for the HvSPL genes. This result was consistent with previous studies reported in wheat59 and A. thaliana60 which suggested the diverse role of SPL gene family on the life cycle of plants.
Upregulated expression of HvSPL genes in barley inflorescence revealed their major role in inflorescence development (Fig. 4). Importantly, tissue-specific differential expression of miR156-targeted HvSPL genes also suggests that they have possible key role in barley growth and development. Fully developed plants progressed through juvenile to adult (vegetative phase) and reproductive phases that are certainly under precise genetic regulation. In A. thaliana, these phases are regulated by miR156 and miR17238 via SPL genes. The expression pattern of miR156 family members and miR172b in barley vegetative to reproductive phase was also antagonistically related (Fig. 5A–C). Expression analyses of HvSPL3, 6, 13, and 23 in our study are aligned with the expression pattern of miR156 and miR172b during vegetative and reproductive phases suggesting the similar role of miR156-HvSPL-miR172b module in growth phase modifications in barley as observed in A. thaliana38 and maize61 (Figs 6A,B and S5).
The regulation of different aspects of plant developments is contributed by the diversity of SPL genes, for instance in bract and ear glume development48,49, fruit ripening and grain yield10,62, juvenile to adult phase transition and flowering63,64, fertility16, and embryonic development65. To see the effect of AP2/miR172 module on HvSPL genes, we examined the expression patterns of HvSPLs in the spikes of a mir172 mutant and its wild type counterpart, which produce indeterminate and normal spikes, respectively (Fig. 6C–G). In our study, HvSPL3, 13, 15, and 23 were differentially expressed in the mir172 mutant when compared to control spikes, suggesting a possible feedback regulation of the miR172/AP2 module. This possibility is in need of further investigation. Down-regulation of AP2 genes is also mediated by miR17239. The up-regulation of HvAP2 in mir172 mutant spikes as compared to their wild type counterparts proved its negative regulation by miR172 (Figs S5 and 5H). Recently we have observed that Ago4_9 genes in barley and wheat are differentially expressed during the reproductive phase66,67. As Ago4_9 is part of the RdDM pathway machinery, it remains to be seen if reproductive phase-specific SPLs are also epigenetically controlled via the RdDM pathway. We are currently elucidating these scenarios in reproductive phase development of cereals. We conclude that when inflorescence architecture is altered by down-regulation of miR172, the SPL gene expression may be altered as a consequence of that or due to up-regulation of AP2-like gene. The results of the current study revealed that the miR156/HvSPL/miR172 module functions as key molecular integrators that affected developmental phase transitions and spike development in barley.
Materials and Methods
Identification and Annotation of SPL Genes in Barley
The DNA coding sequences (Table S1), protein sequences (Table S2), and DNA genomic sequences (Table S3) of HvSPLs were obtained from the Ensemble H. vulgare database (http://plants.ensembl.org/Hordeum_vulgare/Info/Index) database. The pHMMER search function was used, with the A. thaliana SBP domain (Pfam: PF03110) sequence as the query. The IPK Barley BLAST server (http://webblast.ipk-gatersleben.de/barley_ibsc/) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Hvulgare_er) databases were also searched by performing TBLASTN using SBP domain sequence as a query. The HvSPL gene accession numbers were extracted. The nomenclature of putative SPL genes in H. vulgare was based on rice and wheat orthologs.
Gene Structure and Phylogenetic Analysis of HvSPL Genes
The Gene Structure Display Server program (http://gsds.cbi.pku.edu.cn/index.php) was used to predict the exon/intron structure of each HvSPL gene by comparing their coding and genomic sequences. SPL sequences of A. thaliana were obtained from TAIR (http://www.arabidopsis.org/index.jsp)17. SPL sequences of rice were obtained from the rice genome annotation project database. Wheat SPL sequences were taken from14. The amino acid sequences of conserved SBP domains were selected for phylogenetic analysis. The SBP domain sequences of SPL proteins from A. thaliana, rice, wheat, and barley were identified by the SMART tool68 and are presented in Table S4. The phylogenetic tree was constructed using the maximum likelihood method using JTT + G as a best model. SBP domain amino acid sequences were converted to the PHYLIP format and analysed with the PhyML3.0 software (http://www.atgc-montpellier.fr/phyml/)69, which uses the approximate likelihood-ratio test (aLRT) and depends on a non-parametric Shimodaira-Hasegawa-like (SH-like) approach.
Conserved Motif Identification, Cis-Regulatory Elements, miR156 Target Site Prediction and Alternative Splicing Event Analysis
A search for conserved motifs within HvSPL proteins was performed by using the MEME 4.11.0 tool (http://meme-suite.org/tools/meme)70,71 using default settings, except that the maximum width was 50, the minimum width was 6, and the maximum number of motifs to find was 10. The online WebLogo3 platform (http://weblogo.threeplusone.com/) was used to create the sequence logo of the barley SBP domain. The genomic and cDNA sequences of HvSPLs were analysed to predict the putative target sites of miR156 using psRNATarget tool (http://plantgrn.noble.org/psRNATarget/?function). Information on alternative splice events for each HvSPL gene was obtained from the Ensemble database (http://plants.ensembl.org/Hordeum_vulgare/Info/Index). Promoter regions, defined as the 1000-bp sequences upstream of start codons were searched for cis-regulatory elements using the PlantCARE database72.
In Silico Gene Expression Analysis of HvSPLs
‘Morex’ RNA-seq data was obtained from plant expression ATLAS (https://www.ebi.ac.uk/gxa/plant/experiments) which was generated by International Barley Sequencing Consortium (https://ics.hutton.ac.uk/morexGenes/), and the log2-transformed fragments per kilobase per million fragments measured (FPKM) values were used to study the expression of HvSPLs in eight tissues: 4-day-old embryos dissected from germinating grains (EMB); roots (Roo) and shoots (LEA) collected from seedlings (10-cm shoot stage); developing inflorescences (5 mm; INF1 (5 mm) and INF2 (1–1.5 cm); developing tillers at the six-leaf stage (3rd internode, NOD); developing grain 5 days post anthesis (DPA); spikelets with bracts removed at 5 DPA (CAR5) and 15 DPA (CAR15). A heat map of the expression of HvSPLs was generated by the average hierarchical clustering method73 using the MeV tool (http://www.tm4.org/mev.html).
Expression Analysis of Barley miR156 and miR172 Family Members
The mirEX2.0 web portal (http://www.combio.pl/mirex) provides a comprehensive platform for the examination of microRNA expression data based on next generation sequencing (NGS) experiments. For barley, data from the two-rowed cultivar Rolap was obtained for five developmental stages: 1-wk-old and 2-wk-old seedlings, whole plants at the beginning of tillering and stem elongation, and at the milk development stage of the kernel46. Data are expressed as RPM (reads per million) for the miR172 and miR156 members normalized to all miRNAs identified in the sample. Heat map based expression pattern was generated using the MeV tool.
Plant Material, Sample Preparation and RNA Isolation
Barley plants were grown on a 14/10hrs-day/night cycle in a controlled growth room, with a day temperature of 25 °C and a night temperature of 20 °C. Tissue samples were collected from 10, 16, and 75 days old plants (10, 16, and 75 DOP, respectively) and immature spikes. The samples were frozen immediately after harvest by immersion in liquid nitrogen, and stored at −80 °C prior to RNA isolation.Total RNA was extracted using the spectrum plant total RNA Kit (Sigma- Aldrich, St. Louis, MO, USA) according to the manufacturer’s protocol. All samples were quantified for RNA concentration on a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA) and electrophoresed on 1% agarose gel to test the integrity and purity. Each sample was treated with DNase I to remove genomic DNA contamination (Invitrogen, USA). The samples were incubated at 23 °C for 15 minutes, followed by the addition of, 1 µl of 25 mM EDTA to each sample, and further incubation at 65 °C for 10 minutes to terminate the reaction.
First Strand cDNA Synthesis and Quantitative Real-Time PCR (qRT-PCR) Analysis
For each sample, first strand cDNA was synthesized from 1 µg total RNA sample using the AffinityScript QPCR cDNA Synthesis Kit (Agilent technology, Canada). Analysis via qRT-PCR was performed in optical strip tubes using the Mx3000 qPCR system (Stratagene, USA). Each reaction was carried out in a 20-μl volume containing 1 μl diluted cDNA, 5 µM gene specific primers, and 10 μl Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix (Agilent, USA) with the following conditions: 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 30 s at 60 °C. The expression of actin and GAPDH were used as internal control67. Three technical and two biological replicates were used. The relative level of gene expression was analysed by the 2−ΔΔCq method (Livak and Schmittgen 2001). Barley actin or GAPDH transcript was used to adjust the relative transcript level for semi-quantitative RT-PCR. The gene‐specific primers used in semi-quantitative RT‐PCR and qRT-PCR for barley SPL genes are presented in Table S8. The primers for barley SPL genes were designed based on their cDNA sequences. PCR was performed using GoTaq® Green master mix (Promega, USA). PCR for barley actin or GAPDH was run for 30 cycles, whereas PCR cycles for barley SPL genes was run for 34 to 35 cycles. Twelve μl of the RT-PCR products were analysed by 1.2% agarose gel electrophoresis.
Electronic supplementary material
Acknowledgements
The project was supported by a grant to Jaswinder Singh from the Natural Sciences and Engineering Research Council of Canada through discovery program (NSERC-Discovery).
Author Contributions
R.K.T. and J.S. Conceived and designed the experiments. R.K.T. Performed the experiments. R.K.T. and J.S. Analysed the data. R.K.T. and P.B., J.S. wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25349-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Clifford, H.T. (Soderstrom, T. R., and others ed (s). Grass systematics and evolution: an International Symposium held at the Smithsonian Institution, Washington, DC, 27–31 July 1986. Washington, DC London, Smithsonian Institution Press, 1987).
- 2.Klein J, Saedler H, Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of theAntirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996;250:7–16. doi: 10.1007/BF02191820. [DOI] [PubMed] [Google Scholar]
- 3.Birkenbihl RP, Jach G, Saedler H, Huijser P. Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J. mol. biol. 2005;352:585–596. doi: 10.1016/j.jmb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki K, et al. A Novel Zinc-binding Motif Revealed by Solution Structures of DNA-binding Domains of Arabidopsis SBP-family Transcription Factors. J. mol. biol. 2004;337:49–63. doi: 10.1016/j.jmb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eriksson M, et al. Genetic dissection of nutritional copper signaling in Chlamydomonas distinguishes regulatory and target genes. Genetics. 2004;168:795–807. doi: 10.1534/genetics.104.030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kropat J, et al. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arazi T, et al. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 9.Lannenpaa M, et al. A new SBP-box gene BpSPL1 in silver birch (Betula pendula) Physiol. Plant. 2004;120:491–500. doi: 10.1111/j.0031-9317.2004.00254.x. [DOI] [PubMed] [Google Scholar]
- 10.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 11.Shao CX, Takeda Y, Hatano S, Matsuoka M, Hirano HY. Rice genes encoding the SBP domain protein, which is a new type of transcription factor controlling plant development. Rice Genet. News. 1999;16:114. [Google Scholar]
- 12.Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes & Dev. 1997;11:616–628. doi: 10.1101/gad.11.5.616. [DOI] [PubMed] [Google Scholar]
- 13.Tripathi RK, Goel R, Kumari S, Dahuja A. Genomic organization, phylogenetic comparison, and expression profiles of the SPL family genes and their regulation in soybean. Dev. Genes Evol. 2017;227:101. doi: 10.1007/s00427-017-0574-7. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B, et al. Molecular characterization and expression analysis of Triticum aestivum squamosa-promoter binding protein-box genes involved in ear development. J. Integr. Plant Biol. 2014;56:571–581. doi: 10.1111/jipb.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Genomic organization, differential expression, and functional analysis of the SPL gene family in Gossypium hirsutum. Mol. Genet. Genomics. 2015;290:115–126. doi: 10.1007/s00438-014-0901-x. [DOI] [PubMed] [Google Scholar]
- 16.Xing S, Salinas M, Hohmann S, Berndtgen R, Huijser P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 2010;22:3935–3950. doi: 10.1105/tpc.110.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardon G, et al. Molecular characterisation of the Arabidopsis SBP-box genes. Gene. 1999;237:91–104. doi: 10.1016/S0378-1119(99)00308-X. [DOI] [PubMed] [Google Scholar]
- 18.Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006;142:280–293. doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Lu S. Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 2014;14:131. doi: 10.1186/1471-2229-14-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandikota M, et al. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- 21.Unte US, et al. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell. 2003;15:1009–1019. doi: 10.1105/tpc.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Schwarz S, Saedler H, Huijser P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007;63:429–439. doi: 10.1007/s11103-006-9099-6. [DOI] [PubMed] [Google Scholar]
- 23.Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009;50:2133–2145. doi: 10.1093/pcp/pcp148. [DOI] [PubMed] [Google Scholar]
- 24.Xu M, et al. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2017;12:e1006263. doi: 10.1371/journal.pgen.1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell. 2009;21:347–361. doi: 10.1105/tpc.108.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Z-X, et al. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and Patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Mol. Plant. 2015;8:98–110. doi: 10.1016/j.molp.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Gou J-Y, Felippes FF, Liu C-J, Weigel D, Wang J-W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preston JC, Hileman LC. SQUAMOSA PROMOTER BINDING PROTEIN 1 initiates flowering in Antirrhinum majus through the activation of meristem identity genes. Plant J. 2010;62:704–712. doi: 10.1111/j.1365-313X.2010.04184.x. [DOI] [PubMed] [Google Scholar]
- 29.Preston JC, Jorgensen SA, Orozco R, Hileman LC. Paralogous SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes differentially regulate leaf initiation and reproductive phase change in petunia. Planta. 2016;243:429–440. doi: 10.1007/s00425-015-2413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nature Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 32.Si L, et al. OsSPL13 controls grain size in cultivated rice. Nature Genet. 2016;48:447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, et al. Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res. 2017;27:1142. doi: 10.1038/cr.2017.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Cheng X, Liu P, Sun J. miR156-targeted SBP-Box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 2017;174:1931–1948. doi: 10.1104/pp.17.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chuck GS, Brown PJ, Meeley R, Hake S. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc. Natl. Acad. Sci. USA. 2014;111:18775–18780. doi: 10.1073/pnas.1407401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gou J, et al. The miR156-SPL4 module predominantly regulates aerial axillary bud formation and controls shoot architecture. New Phytol. 2017;216:829–840. doi: 10.1111/nph.14758. [DOI] [PubMed] [Google Scholar]
- 37.Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu G, et al. The Sequential Action of miR156 and miR172 Regulates Developmental Timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair SK, et al. Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc. Natl. Acad. Sci. USA. 2010;107:490–495. doi: 10.1073/pnas.0909097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown RH, Bregitzer P. A insertional mutant of a barley gene results in indeterminate spikelet development. Crop Sci. 2011;51:1664–1672. doi: 10.2135/cropsci2010.09.0532. [DOI] [Google Scholar]
- 44.Houston K, et al. Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc. Natl. Acad. Sci. USA. 2013;110:16675–16680. doi: 10.1073/pnas.1311681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao H-D, et al. Genome-wide analysis of the SPL family transcription factors and their responses to abiotic stresses in maize. Plant Gene. 2016;6:1–12. doi: 10.1016/j.plgene.2016.03.003. [DOI] [Google Scholar]
- 46.Zielezinski A, et al. mirEX 2.0-an integrated environment for expression profiling of plant microRNAs. BMC Plant Biol. 2015;15:144. doi: 10.1186/s12870-015-0533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, et al. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA. 2015;112:15504–15509. doi: 10.1073/pnas.1521949112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, et al. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chuck G, Whipple C, Jackson D, Hake S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development. 2010;137:1243–1250. doi: 10.1242/dev.048348. [DOI] [PubMed] [Google Scholar]
- 50.Cardon GH, Hohmann S, Nettesheim K, Saedler H, Huijser P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 1997;12:367–377. doi: 10.1046/j.1365-313X.1997.12020367.x. [DOI] [PubMed] [Google Scholar]
- 51.Cheng H, et al. Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Biol. 2016;16:196. doi: 10.1186/s12870-016-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo A-Y, et al. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 53.Capovilla G, Pajoro A, Immink RGH, Schmid M. Role of alternative pre-mRNA splicing in temperature signaling. Curr. Opin. Plant Biol. 2015;27:97–103. doi: 10.1016/j.pbi.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 54.Panahi B, Mohammadi SA, Khaksefidi RE, Fallah Mehrabadi J, Ebrahimie E. Genome-wide analysis of alternative splicing events in Hordeum vulgare: Highlighting retention of intron-based splicing and its possible function through network analysis. FEBS Lett. 2015;589:3564–3575. doi: 10.1016/j.febslet.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 55.Qin, Z. et al. Regulation of FT splicing by an endogenous cue in temperate grasses. Nat. Commun. 8 (2017). [DOI] [PMC free article] [PubMed]
- 56.Xing S, et al. SPL8 and miR156 targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J. 2013;75:566–577. doi: 10.1111/tpj.12221. [DOI] [PubMed] [Google Scholar]
- 57.Xing S, Salinas M, Hohmann S, Berndtgen R, Huijser P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 2010;22:3935–3950. doi: 10.1105/tpc.110.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu S, et al. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING LIKE transcription factors. Plant Cell. 2012;24:3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang B, et al. Characterization of Squamosa Promoter Binding Protein-LIKE. J. Plant Biol. 2015;58:220–229. doi: 10.1007/s12374-015-0105-x. [DOI] [Google Scholar]
- 60.Bernal M, et al. Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell. 2012;24:738–761. doi: 10.1105/tpc.111.090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nature Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 62.Jiao Y, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 63.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J-W, Czech B, Weigel D. miR156-Regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 65.Nodine MD, Bartel DP. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010;24:2678–2692. doi: 10.1101/gad.1986710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh M, Singh J. Seed development-related expression of ARGONAUTE4_9 class of genes in barley: possible role in seed dormancy. Euphytica. 2012;188:123–129. doi: 10.1007/s10681-012-0624-1. [DOI] [Google Scholar]
- 67.Singh M, Singh S, Randhawa H, Singh J. Polymorphic homoeolog of key gene of RdDM pathway, ARGONAUTE4_9 class is associated with pre-harvest sprouting in wheat (Triticum aestivum L.) PloS One. 2013;8:e77009. doi: 10.1371/journal.pone.0077009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaur R, Singh K, Singh J. A root-specific wall-associated kinase gene, HvWAK1, regulates root growth and is highly divergent in barley and other cereals. Funct. Integr. Genomics. 2013;13:167–177. doi: 10.1007/s10142-013-0310-y. [DOI] [PubMed] [Google Scholar]
- 69.Guindon Sp, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 70.Bailey, T.L. et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. gkp335 (2009). [DOI] [PMC free article] [PubMed]
- 71.Dhaliwal AK, Mohan A, Gill KS. Comparative analysis of ABCB1 reveals novel structural and functional conservation between monocots and dicots. Front. Plant Sci. 2014;5:657–667. doi: 10.3389/fpls.2014.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh S, Tripathi RK, Lemaux PG, Buchanan BB, Singh J. Redox- dependent interaction between thaumatin-like protein and β-glucan influences malting quality of barley. Proc. Natl. Acad. Sci. USA. 2017;114:7725–7730. doi: 10.1073/pnas.1701824114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.