Summary
Embryonic stem cells (ESCs) cultured in leukemia inhibitory factor (LIF) plus fetal bovine serum (FBS) exhibit heterogeneity in the expression of naive and primed transcription factors. This heterogeneity reflects the dynamic condition of ESCs and their versatility to promptly respond to signaling effectors promoting naive or primed pluripotency. Here, we report that ESCs lacking Nanog or overexpressing Otx2 exhibit an early primed identity in LIF + FBS and fail to convert into 2i-induced naive state. Conversely, Otx2-null ESCs possess naive identity features in LIF + FBS similar to Nanog-overexpressing ESCs and convert poorly into FGF-induced early primed state. When both Nanog and Otx2 are inactivated, ESCs cultured in LIF + FBS exhibit primed identity and weakened ability to convert into naive state. These data suggest that, through mutual antagonism, NANOG and OTX2 specify the heterogeneous identity of ESCs cultured in LIF + FBS and individually predispose them for optimal response to naive or primed inducing factors.
Keywords: embryonic stem cell heterogeneity, OTX2, NANOG, LIF signaling, pluripotency
Graphical Abstract
Highlights
-
•
Loss of Nanog or Otx2 overexpression induces primed identity in ESCs cultured in LIF
-
•
Loss of Nanog or Otx2 overexpression affects naive conversion
-
•
Otx2 null ESCs in LIF show naive identity and abnormal conversion into primed state
-
•
ESCs lacking Otx2 and Nanog show primed identity and convert into naive and primed states
In this article, Simeone and colleagues study the molecular mechanism controlling the heterogeneous and flexible identity of ESCs cultured in LIF + FBS. They analyze a series of ESC lines carrying different dosages of OTX2 and/or NANOG. They find that synergism between the OTX2 and NANOG antagonistic regulatory networks specifies the identity of ESCs in LIF + FBS.
Introduction
Pluripotency is the capability of a single cell to generate all embryonic and adult cell types. In vivo this ability is exhibited by the epiblast, and in vitro by pluripotent stem cells (Nichols and Smith, 2009, Rossant and Tam, 2009, Gardner and Beddington, 1988). Mouse ESCs may be derived from both the inner cell mass and early preimplantation epiblast; they can be indefinitely propagated in culture by ensuring provision of leukemia inhibitory factor (LIF) plus fetal bovine serum (FBS) and may efficiently integrate into host blastocysts and contribute to all body tissues (Nichols and Smith, 2009, Silva and Smith, 2008, Martin, 1981, Evans and Kaufman, 1981). However, their state depends strictly on a regulatory network controlled by core pluripotency transcription factors OCT4, SOX2, KLF2/4, NANOG, and ESRRB as well as LIF, WNT, and BMP4 signaling pathways (Kalkan and Smith, 2014, Festuccia et al., 2012, Martello et al., 2012, ten Berge et al., 2011, Silva et al., 2009, Ying et al., 2008). ESCs cultured in LIF + FBS are characterized by cell heterogeneity in both expression of specific transcription factors and sensitivity to signaling molecules, which together define a state ensuring self-renewal and opportunity to convert into naive or primed pluripotency. This cell heterogeneity is exemplified by the fluctuating expression of Nanog and by the detection of naive and primed markers in specific ESC sub-type compartments (Smith, 2017, Acampora et al., 2013, Acampora et al., 2016, Torres-Padilla and Chambers, 2014, Cahan and Daley, 2013, Martinez Arias et al., 2013, Muñoz Descalzo et al., 2012, Nichols and Smith, 2011, Kalmar et al., 2009, Hayashi et al., 2008, Chambers et al., 2007). A similar heterogeneity exists in the preimplantation mouse embryo at E4.5–E4.7 when the epiblast gradually loses naive identity and begins to induce early primed pluripotency (Acampora et al., 2016). Recently, the state of the early primed epiblast has been discussed as representing a new phase of pluripotency, named formative, which is interposed between naive and primed pluripotency (Smith, 2017). Formative pluripotency is hypothesized to represent an essential staging post required to enable naive cells to successfully remodel transcriptional, epigenetic, signaling, and metabolic networks in preparation for transit into a mature primed state responsive to differentiation cues (Smith, 2017). ESCs cultured in LIF + FBS may be committed to naive or primed pluripotency if adequately stimulated. For example, ESCs cultured in LIF may convert into a naive state of pluripotency if provided with the two inhibitor molecules (2i), which respectively inhibit FGF signaling and activate WNT signaling (Marks et al., 2012, Nichols et al., 2009, Ying et al., 2008); alternatively ESCs may also convert to a primed state of pluripotency if LIF is replaced with FGF and Activin A (Kunath, 2011, Lanner and Rossant, 2010, Brons et al., 2007, Tesar et al., 2007). Signaling-pathway-mediated modification of the pluripotent state is associated with a response in the expression of specific genes, which ultimately determine the state of pluripotency. This implies that the precise dosage and relationship between pluripotency factors should determine optimal functioning of the entire circuitry (Smith, 2017, Torres-Padilla and Chambers, 2014, Karwacki-Neisius et al., 2013, Muñoz Descalzo et al., 2012, Takahashi and Yamanaka, 2006, Niwa et al., 2000). For example, Nanog overexpression is sufficient to drive LIF-independent self-renewal, and the gene dosage of Nanog determines the efficiency with which ESCs can self-renew (Chambers et al., 2007). Nanog null ESCs, although predisposed to differentiate, retain an undifferentiated phenotype and ability to self-renew (Theunissen et al., 2011, Silva et al., 2009, Chambers et al., 2003, Chambers et al., 2007, Mitsui et al., 2003). On the other hand, OTX2 opposes LIF-independent self-renewal, is required for transition into early primed pluripotency, and stabilizes the EpiSC state (Buecker et al., 2014, Yang et al., 2014, Acampora et al., 2013). OTX2 may also bind to the Nanog promoter and positively sustain Nanog expression to optimize in vivo the specification of the epiblast and in vitro the integrity of ESC sub-type compartments (Acampora et al., 2016). We hypothesize that integration of the OTX2 and NANOG antagonistic networks specifies the heterogeneous identity of ESCs cultured in LIF + FBS. To gain insights into this mechanism, we studied whether different dosages of OTX2 and/or NANOG may intrinsically modify the pluripotent state of ESCs cultured in LIF + FBS without the contribution of signaling effectors. Our data suggest that OTX2 and NANOG cooperate to deliver the heterogeneous and flexible identity of ESCs cultured in LIF + FBS from their antagonistic regulatory networks.
Results
OTX2 and NANOG Antagonism Controls the Specification of ESC Sub-types Cultured in LIF + FBS
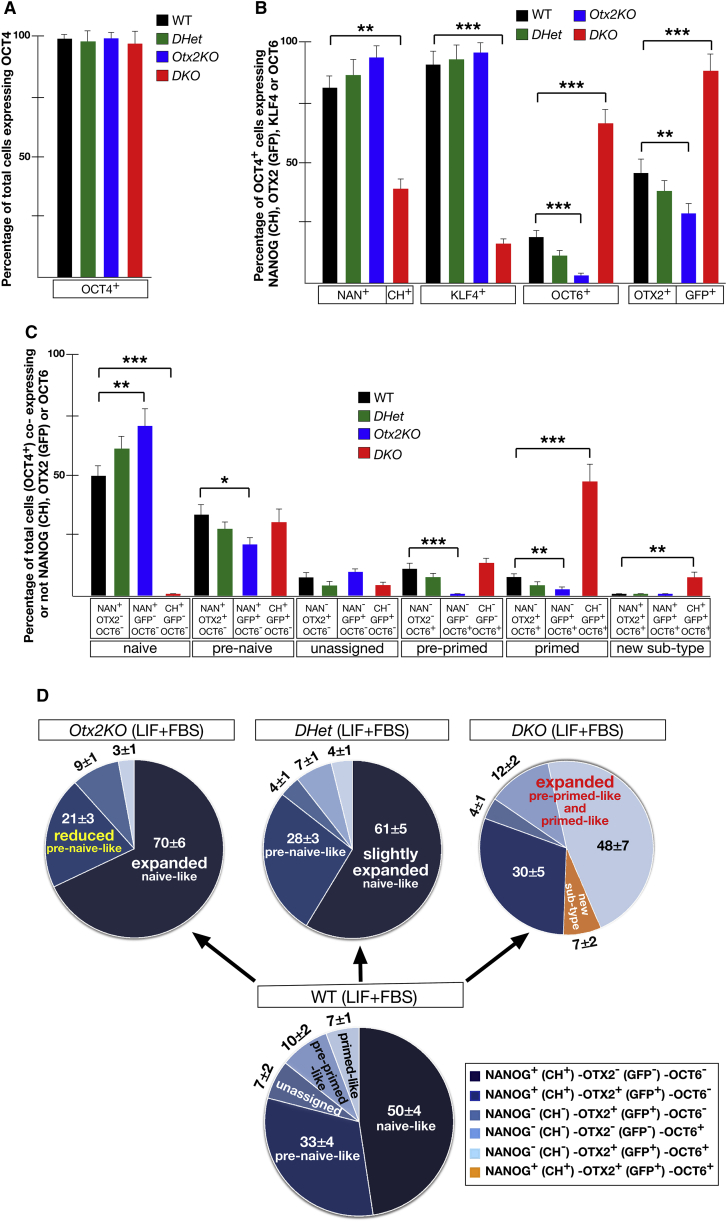
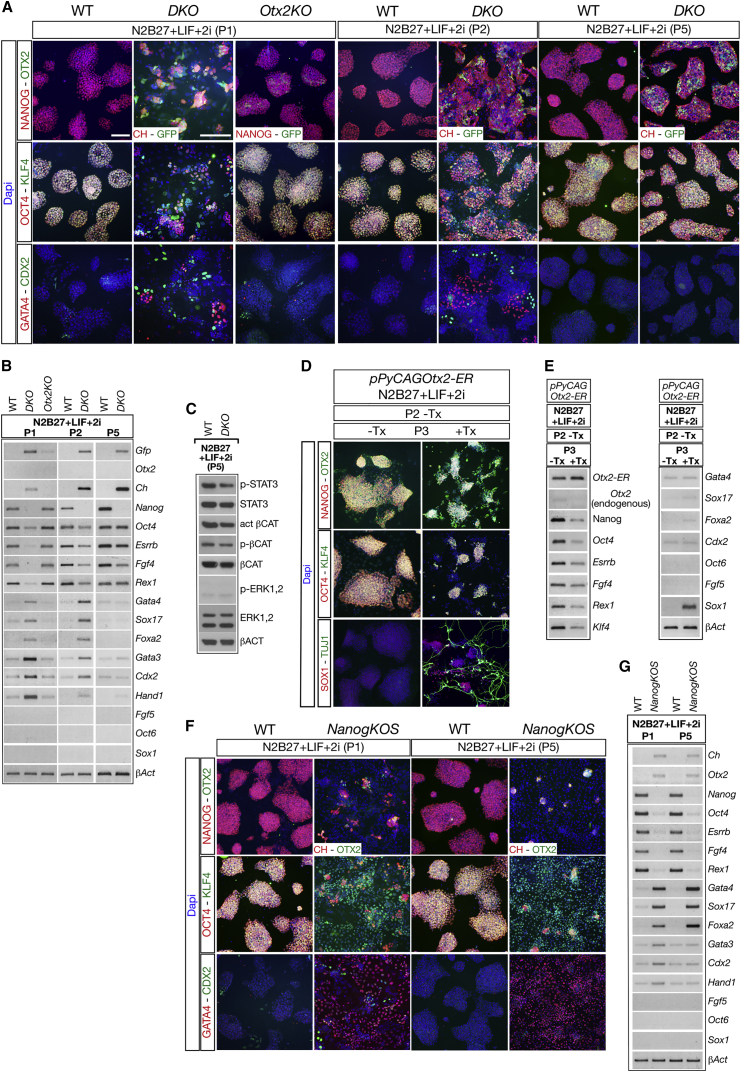
We recently reported that the combined analysis of NANOG, OCT6, and OTX2 protein expression identified five OCT4+ compartments in ESCs cultured in LIF + FBS. In addition to a large naive-like (NANOG+-OTX2−-OCT6−) and a small primed-like (NANOG−-OTX2+-OCT6+) compartment, three potentially transitional compartments were identified that we refer to as pre-naive-like (NANOG+-OTX2+-OCT6−), pre-primed-like (NANOG−-OTX2−-OCT6+), and unassigned (NANOG−-OTX2+-OCT6−) (Acampora et al., 2016). To test our hypothesis that the antagonism between the OTX2 and NANOG regulatory networks coordinates the specification of ESC compartments, we analyzed the expression of these transcription factors in a new allelic series of ESC mutant lines, which have different gene dosages of Otx2 and/or Nanog, including Nanog null and Otx2 null ESC lines.
We first targeted CreER into the Rosa26 locus of wild-type (WT) ESCs (R26CreER/+) and used this ESC line to generate all mutant ESCs (Figure S1). To monitor expression from the Nanog locus in Nanog null ESCs (NanogCh/−; R26CreER/+ referred to as NanogKO) mCherry (Ch) was inserted into one of the Nanog alleles. To generate the NanogKO mutant, Nanogflox/Ch; R26CreER/+ (referred to as NanogCKO) ESCs (Figures S1A–S1C) were plated at clonal density and treated with 4-OH-tamoxifen (Tx) 3 hr later. Only very few NanogKO clones could be cultured (Figure S2A) for more than 12 passages (P12). Although these clones generated abundant OCT4− cells with primitive endoderm (PE)-like identity, they also exhibited numerous small and flat OCT4+ colonies (Figures S2B and S2C) (Mitsui et al., 2003). These OCT4+ cells accounted for 13% of total cells (Figures S2E and S3A; Table S1). We next analyzed the identity of OCT4+ cells by monitoring the expression of CH, KLF4, OTX2, and OCT6. Cell-counting experiments showed that compared with WT, in NanogKO ESCs the number of OCT4+ cells expressing CH or KLF4 was severely decreased, while that of OTX2 and OCT6 increased (Figures S2D, S2F, and S3A; Table S1). OCT4+ ESCs were then analyzed for co-expression of OCT6, CH, and OTX2. Compared with WT, in NanogKO ESCs the naive-like and the pre-naive-like compartments were heavily reduced, and the primed-like compartment markedly expanded (Figures S2G and S3B; Table S1). Noteworthy, the sum of all compartments accounted for the expected percentage of OCT4+ ESCs (Figure S2H). This indicates that the cell-counting analysis is not excluding OCT4+ ESC sub-types negative for CH, OTX2, and OCT6 expression. These data revealed that loss of Nanog causes a marked expansion of the primed-like compartment at the expense of the naive-like and pre-naive-like compartments (Figure S2I). Based on this, we reasoned that, despite the large expansion of the primed-like sub-type, a fraction of NanogKO ESCs retained naive-like and pre-naive-like identity, suggesting that CH+ ESCs may include a fraction of cells protected from conversion into primed-like identity. To test this possibility, we sorted CH+ ESCs (Figure 1A) from NanogKO ESCs and studied whether these NanogKO sorted (NanogKOS) ESCs generated colonies with a different identity. Alkaline phosphatase (ALP) assay showed that none of the NanogKOS ESC colonies was uniformly ALP+, but most of them retained an evident mixed staining (Figure 1B). Immunohistochemistry assays and cell counting showed that compared with NanogKO ESCs, NanogKOS ESCs generated much less PE-like OCT4− cells (Figures 1C, 1D, S2B, S2C, and S4A) and showed a remarkable increase in the percentage of total cells expressing OCT4 (Figures 1F and S2E; Table S1). Cell counting showed that in NanogKOS ESCs, the number of CH+ and KLF4+ cells was decreased compared with WT ESCs but increased compared with NanogKO ESCs; in a complementary trend, the number of OCT6+ and OTX2+ cells in NanogKOS ESCs was increased compared with WT ESCs but diminished compared with NanogKO ESCs (Figures 1G, S2F, and S3A; Table S1). Analysis of NanogKOS sub-types showed that, compared with WT, the naive-like compartment was severely reduced, the pre-naive-like compartment was unaffected, and the unassigned, pre-primed-like, and primed-like compartments were expanded (Figures 1H and 1K). Compared with NanogKO, the size of the naive-like, pre-naive-like and primed-like compartments of NanogKOS ESCs was partially restored (Figures 1K and S2I). Coherently, compared with NanogKO, in NanogKOS ESCs the expression of Fgf4, Rex1, and Esrrb was moderately increased and that of Oct6 and Fgf5 decreased (Figures 1J and S4B) (Chambers et al., 2007). These data suggest that the inactivation of Nanog has a remarkable impact on the identity of OCT4+ ESCs, which become shifted toward the primed-like identity (Figure S2I). Our analysis also reveals that the CH+ ESC fraction is protected from complete conversion into primed-like identity (Figure 1K).
Figure 1.
NanogKOS ESCs Exhibit Partial Recovery of Naive-like and Pre-naive-like ESC Sub-types
(A) Flow cytometry plot of NanogKO ESCs showing CH+ cells versus side scatter (SSC).
(B) Number of WT and NanogKOS colonies uniformly ALP+. Data are the means ± SD from four independent experiments.
(C) Representative images of WT and P4 NanogKOS ESCs immunostained with OCT4 and KLF4 and with CDX2, GATA4, and OCT4. Scale bar, 100 μm.
(D) RT-PCR assays with Oct4 and PE (Gata6, Gata4, Sox17, Foxa2, and Igf2) markers on WT and NanogKOS RNAs normalized by β-Actin (n = 3 independent experiments).
(E) Representative images of WT and NanogKOS ESCs immunostained with OCT4 and OCT6, with NANOG or CH, OTX2, and OCT4 and with NANOG or CH, OTX2, and OCT6; ESCs are also stained with DAPI. Scale bar, 100 μm.
(F–I) Cell-counting analysis showing in WT and NanogKOS ESCs the percentage of total cells expressing OCT4 (F), the percentage of OCT4+ cells expressing NANOG or CH, KLF4, OCT6, and OTX2 (G), the percentage of total cell sub-types showing naive-like, pre-naive-like, unassigned, pre-primed-like, and primed-like identity (H), and the percentage of total cells expressing the sum of all cell sub-types (I). Data are reported as means ± SD from four independent experiments. ∗∗∗p ≪ 0.001; ∗∗p < 0.001; ∗p value is between 0.005 and 0.001.
(J) RT-PCR assays showing the expression of Nanog, Ch, Otx2, Fgf5, Oct6, Fgf4 Rex1, and Esrrb in WT, NanogKO, and NanogKOS RNAs normalized by Oct4 (n = 3 independent experiments).
(K) Schematic representation of ESC sub-type compartments showing that NanogKOS ESCs exhibit a reduction of the naive-like compartment and moderate expansion of the unassigned, pre-primed-like, and primed-like compartments. A color code of the different compartments is also shown.
See also Figures S1–S4 and Table S1.
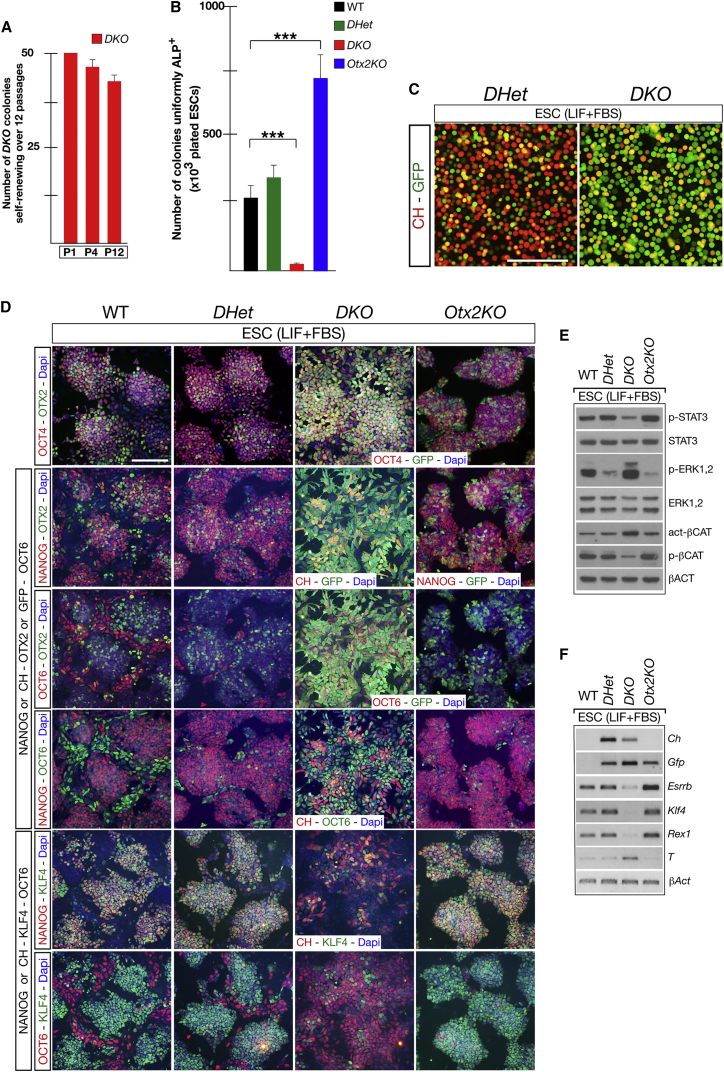
Since we initially hypothesized that NANOG and OTX2 act through reciprocal antagonism to control the state of ESCs cultured in LIF + FBS, we next studied mutant ESCs lacking Nanog in an Otx2 null background (NanogCh/−; Otx2Gfp/−; R26CreER/+ referred to as DKO) in comparison with NanogCh/+; Otx2Gfp/+; R26CreER/+ (referred to as DHet) and Otx2Gfp/−; R26CreER/+ (referred to as Otx2KO) ESCs (Figure S1). First, Nanogflox/Ch; Otx2Gfp/−; Rosa26CreER/+ (referred to as NanogCKO; Otx2KO) (Figure S1D) ESCs were plated at clonal density and with Tx to select DKO ESCs. Importantly, about 80% of the DKO ESC clones could be extensively and stably passaged (Figure 2A). Analysis of ten of these clones showed flat colonies and no sign of differentiation into PE-like cells (data not shown). This indicates that loss of Nanog in an Otx2 null background prevents both loss of efficient self-renewal and differentiation into PE-like cells. We next monitored the ability of mutant ESC lines to generate colonies uniformly stained with ALP. Compared with WT and DHet, DKO ESCs similar to NanogKOS ESCs produced very few uniformly ALP+ colonies, whereas Otx2KO ESCs showed a higher number of fully stained ALP+ colonies (Figure 2B) (Acampora et al., 2013). Analysis of trypsinized living cells showed that compared with DHet, the ratio between CH+ and GFP+ cells is reversed in DKO ESCs (Figure 2C). Of note, the CH+-GFP− ESC subset was absent in DKO mutant (Figure 2C). Cell counting of immunohistochemistry experiments showed that OCT4 was ubiquitously expressed in all mutant ESCs (Figures 2D and 3A). The number of NANOG+ or CH+ and KLF4+ cells was similar in WT and DHet and reduced in DKO ESCs, and the number of OTX2+ or GFP+ and OCT6+ ESCs was severely diminished in Otx2KO, moderately reduced in DHet, and massively increased in DKO ESCs (Figures 2D and 3B; Table S2). We next analyzed how these abnormalities in cell number affected the size of ESC sub-type compartments. Compared with WT, DHet ESCs exhibited a moderate expansion of the naive-like compartment and a corresponding contraction of the pre-naive-like sub-type; Otx2KO ESCs lacked the pre-primed-like sub-type and displayed a significant reduction of the primed-like and pre-naive-like compartments, which was counterbalanced by the marked expansion of the naive-like sub-type; and DKO ESCs lacked the naive-like sub-type and heavily expanded the size of the primed-like compartment (Figures 3C, 3D, and S3C; Table S2). Noteworthy, DKO ESCs, like Nanog null ESCs (Figures 1K and S2J), showed a small new sub-type co-expressing CH, GFP, and OCT6 (Figures 3C and 3D). These data were further corroborated by the expression of additional markers and signaling effectors. Compared with WT, naive markers such as Klf4, Esrrb, Rex1, the phosphorylated form of the LIF signaling effector STAT3 (p-STAT3) and the phosphorylated form of the βCATENIN (p-βCAT) were almost unaffected in DHet, downregulated in DKO, and several of them such as p-STAT3, Esrrb, Klf4, and Rex1, were upregulated in Otx2KO ESCs (Figures 2E, 2F, and S4C). Primed markers such as T and p-ERK1,2 were upregulated in DKO, while in Otx2KO and DHet ESCs, p-ERK1,2 was downregulated (Figures 2E, 2F, and S4C). These data indicate that (1) in contrast to Nanog null ESCs, loss of Nanog in an Otx2 null background generates a compensatory effect resulting in a restored LIF-dependent self-renewal and suppression of PE-like differentiation, and (2) in contrast to Otx2KO, the identity of DKO ESCs exhibits a marked expansion of the primed-like compartment. Overall, these data suggest that NANOG and OTX2 cooperate through their antagonistic transcriptional networks to specify and maintain the identity of ESCs cultured in LIF + FBS.
Figure 2.
NANOG and OTX2 Antagonism Controls the Identity and Size of ESC Sub-type Compartments and Influences Self-Renewal
(A and B) Number of DKO colonies with efficient self-renewal (A) and number of ESC colonies uniformly stained with ALP in WT and mutant ESCs (B). Data are reported as means ± SD of four independent experiments. ∗∗∗p ≪ 0.001.
(C) Representative images showing that DHet and DKO trypsinized ESCs exhibit mirror distribution of GFP+ and CH+ cells. Scale bar, 100 μm.
(D) Representative immunohistochemistry assays performed in WT, DHet, DKO, and Otx2KO ESCs with OTX2 or GFP and OCT4, with NANOG or CH, OTX2 or GFP and OCT6, and with NANOG or CH, KLF4 and OCT6; ESCs are also stained with DAPI. Scale bar, 100 μm.
(E and F) Western blots (E) and RT-PCR assays (F) performed on WT, DHet, Otx2KO, and DKO ESCs to assess the expression level of p-STAT3, STAT3, p-ERK1,2, ERK1,2, activated-βCAT and p-βCAT (E) as well as Ch, Gfp, Esrrb, Klf4, Rex1, and T (F). β-Actin is used as quantitative internal standard (n = 3 independent experiments).
See also Figures S1 and S4.
Figure 3.
Cell-Counting Analysis of Mutant ESC Sub-type Compartments
(A–C) Cell-counting data showing for WT, DHet, DKO, and Otx2KO ESCs the percentage of total cells expressing OCT4 (A), the percentage of OCT4+ cells expressing NANOG or CH, KLF4, OCT6 and OTX2 or GFP (B), and the percentage of total cells (OCT4+), corresponding to the naive-like, pre-naive-like, unassigned, pre-primed-like, primed-like compartments (C); note that DKO ESCs show a new small sub-type (CH+-GFP+-OCT6+) (C). Data are reported as the means ± SD from four independent experiments. ∗∗∗p ≪ 0.001; ∗∗p < 0.001; ∗p value is between 0.005 and 0.001.
(D) Schematic representation of ESC compartments showing that Otx2KO ESCs exhibit a relevant expansion of the naive-like compartment counterbalanced by a significant contraction of the pre-naive-like and loss of the pre-primed-like compartments; and DKO ESCs show loss of the naive-like compartment and expansion of primed-like and pre-primed-like compartments.
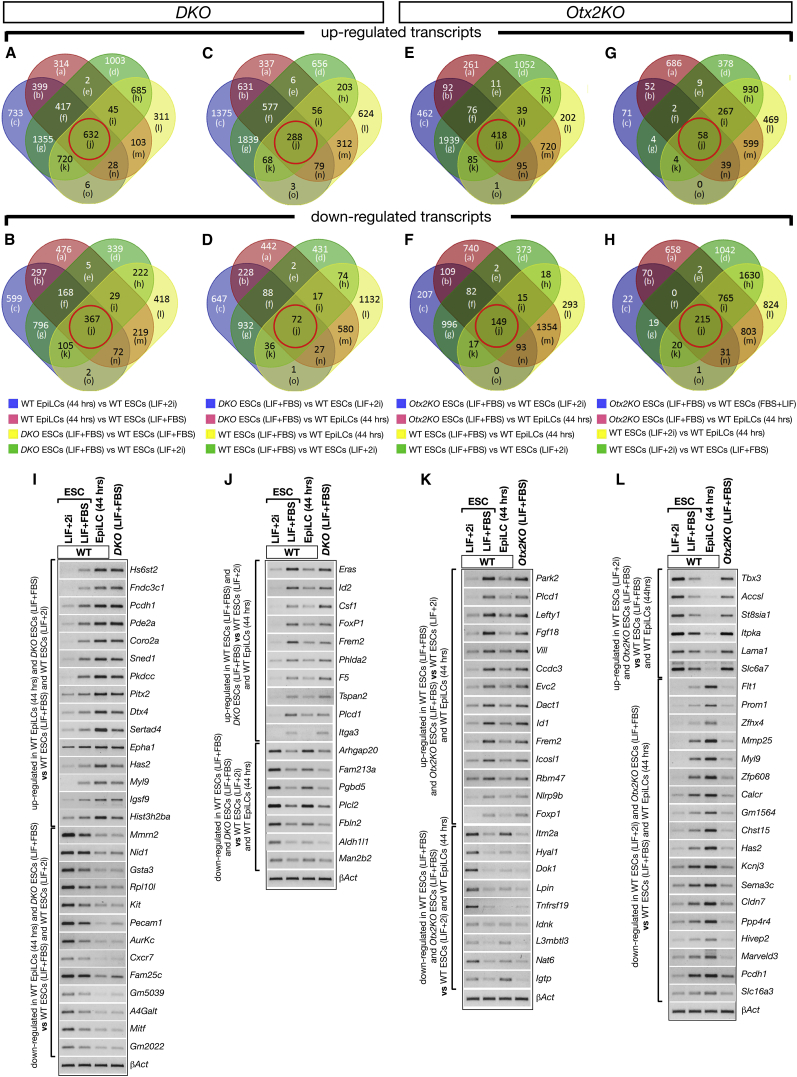
Identity Assignment of DKO and Otx2KO ESCs by Transcriptome Analysis
To evaluate the identity of DKO and Otx2KO cells, we used global transcriptome analyses. We compared DKO and Otx2KO ESCs cultured in LIF + FBS with WT ESCs cultured in LIF + 2i, LIF + FBS, or in FGF2 + Activin A for 44 hr to generate EpiLCs (44 hr) (Figures 4 and S5; Table S3. Up- and Downregulated Gene Transcripts Detected in All Comparisons, Related to Figures 4 and S5, Table S4. List of Up- and Downregulated Transcripts as They Appear within Each Box of the Venn Diagrams Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P, Table S5. List of Up and Downregulated Gene Transcripts Reported in Box (j) of Each Venn Diagram Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P). We first performed pairwise analysis of DKO ESCs (LIF + FBS) and WT EpiLCs (44 hr) both compared with WT ESCs (LIF + FBS) or WT ESCs (LIF + 2i) to identify DKO ESCs transcripts in common with or differentially expressed in WT EpiLCs (Figures S5A–S5D; Table S3. Up- and Downregulated Gene Transcripts Detected in All Comparisons, Related to Figures 4 and S5, Table S4. List of Up- and Downregulated Transcripts as They Appear within Each Box of the Venn Diagrams Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P, Table S5. List of Up and Downregulated Gene Transcripts Reported in Box (j) of Each Venn Diagram Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P). These data were integrated to generate comparative Venn diagrams showing that DKO exhibited 999 (632 upregulated and 367 downregulated) transcripts in common with those specifically expressed in WT EpiLCs (44 hr) (box [j] highlighted by red circle in Figures 4A and 4B; Table S3. Up- and Downregulated Gene Transcripts Detected in All Comparisons, Related to Figures 4 and S5, Table S4. List of Up- and Downregulated Transcripts as They Appear within Each Box of the Venn Diagrams Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P, Table S5. List of Up and Downregulated Gene Transcripts Reported in Box (j) of Each Venn Diagram Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P). In contrast, DKO shared only 360 and 208 differentially expressed genes with those specifically expressed in WT ESCs (LIF + FBS) and WT ESCs (LIF + 2i), respectively (box [j] highlighted by red circle in Figures 4C, 4D, S5M, and S5N; Table S3. Up- and Downregulated Gene Transcripts Detected in All Comparisons, Related to Figures 4 and S5, Table S4. List of Up- and Downregulated Transcripts as They Appear within Each Box of the Venn Diagrams Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P, Table S5. List of Up and Downregulated Gene Transcripts Reported in Box (j) of Each Venn Diagram Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P). RT-PCR assays validated a number of DKO ESC (LIF + FBS) transcripts selectively up- or downregulated in WT EpiLCs (44 hr) or WT ESCs (LIF + FBS) (Figures 4I, 4J, S5I, and S5J). These data suggest that DKO ESCs exhibit a transcriptome profile with a marked signature in common with WT EpiLCs (44 hr). A similar analysis was performed for the transcriptome of Otx2KO ESCs (LIF + FBS). Pairwise analysis of Otx2KO ESCs (LIF + FBS) and WT ESCs (LIF + 2i) both compared with WT EpiLCs (44 hr) (Figures S5E and S5F), and pairwise analysis of Otx2KO ESCs (LIF + FBS) and WT ESCs (LIF + FBS) both compared with WT ESCs (LIF + 2i) (Figures S5G and S5H) were performed to identify the Otx2KO ESCs (LIF + FBS) transcriptome fraction in common with or differentially expressed in WT ESCs (LIF + 2i) or WT ESCs (LIF + FBS). Comparative Venn diagrams showed that, although the highest overlap was with WT ESCs (LIF + FBS), specific transcripts (box [j] highlighted by red circle in Figures 4E and 4F), Otx2KO ESCs (LIF + FBS) also showed 215 gene transcripts in common with those specifically downregulated in WT ESCs (LIF + 2i) (box [j] highlighted by red circle in Figure 4H). Otx2KO ESCs (LIF + FBS) shared very few differentially expressed transcripts (n = 55) with WT EpiLCs (44 hr) (box [j] highlighted by red circle in Figures S5O and S5P). This suggests that OTX2 is required to activate genes repressed by LIF + 2i in WT ESCs. These data were also validated by RT-PCR assays (Figures 4K, 4L, S5K, and S5L). Then we compared the transcriptome of DKO ESCs (LIF + FBS) with that of Otx2KO ESCs (LIF + FBS). We identified 4,209 differentially expressed transcripts, which included 264 of the 500 genes reported as those more sensitive to NANOG (Festuccia et al., 2012) (Figure S5Q; Table S3, sheet 25). Based on this, we compared these 500 NANOG-sensitive genes (250 up- and 250 downregulated) with the OTX2-sensitive genes (230 up- and 378 downregulated) previously identified (Table S3, sheets 19 and 20). Pairwise analysis showed an overlap of 82 genes (Figure S5R), which included 18 genes downregulated in Nanog null and upregulated in Otx2KO ESC lines, 58 genes with the opposite expression profile and 6 genes downregulated in both Nanog and Otx2 null ESCs (Table S3, highlighted in sheet 26). Noteworthy, 17 of the 18 genes were in common with those selectively upregulated in WT ESCs (LIF + FBS) and included Klf4 and Tbx3, which are required to maintain pluripotency (Niwa et al., 2009); on the other hand, 23 out of the 58 genes were shared with those selectively upregulated in WT EpiLCs (44 hr). This suggests that the antagonism between OTX2 and NANOG may depend, at least in part, on opposing transcriptional regulation exerted directly or indirectly by these two transcription factors on a limited number of common target genes.
Figure 4.
Analysis of DKO and Otx2KO Transcriptomes
(A–H) Venn diagrams showing the different categories of gene transcripts (boxes from (a) to (o) in each Venn diagram) up- or downregulated in DKO (A–D) and Otx2KO (E–H) ESCs in LIF + FBS when compared with WT ESCs (LIF + FBS) or WT ESCs (LIF + 2i) or WT EpiLCs (44 hr); this analysis allows us to identify DKO (LIF + FBS) gene transcripts shared with those up- or downregulated specifically in WT EpiLCs (44 hr) or WT ESCs (LIF + FBS) (red circle in box [j] in A–D); the same analysis was performed to identify Otx2KO ESCs (LIF + FBS) transcripts shared with those specifically expressed in WT ESCs (LIF + FBS) and WT ESCs (LIF + 2i) (red circle in box [j] in E–H).
(I–L) RT-PCR assays of selected genes up- or downregulated specifically in DKO (LIF + FBS) and WT EpiLCs (44 hr) (I), or in DKO (LIF + FBS) and WT ESCs (LIF + FBS) (J), or in Otx2KO (LIF + FBS) and WT ESCs (LIF + FBS) (K), or in Otx2KO (LIF + FBS) and WT ESCs (LIF + 2i) (L).
The selected genes belong to the box (j) highlighted with red circles in the Venn diagrams (n = 3 independent experiments).
See also Figure S5 and Table S3. Up- and Downregulated Gene Transcripts Detected in All Comparisons, Related to Figures 4 and S5, Table S4. List of Up- and Downregulated Transcripts as They Appear within Each Box of the Venn Diagrams Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P, Table S5. List of Up and Downregulated Gene Transcripts Reported in Box (j) of Each Venn Diagram Shown in Figures 4 and S5M-P, Related to Figures 4 and S5M–S5P.
DKO ESCs Colonize Preimplantation Embryos with Reduced Efficiency
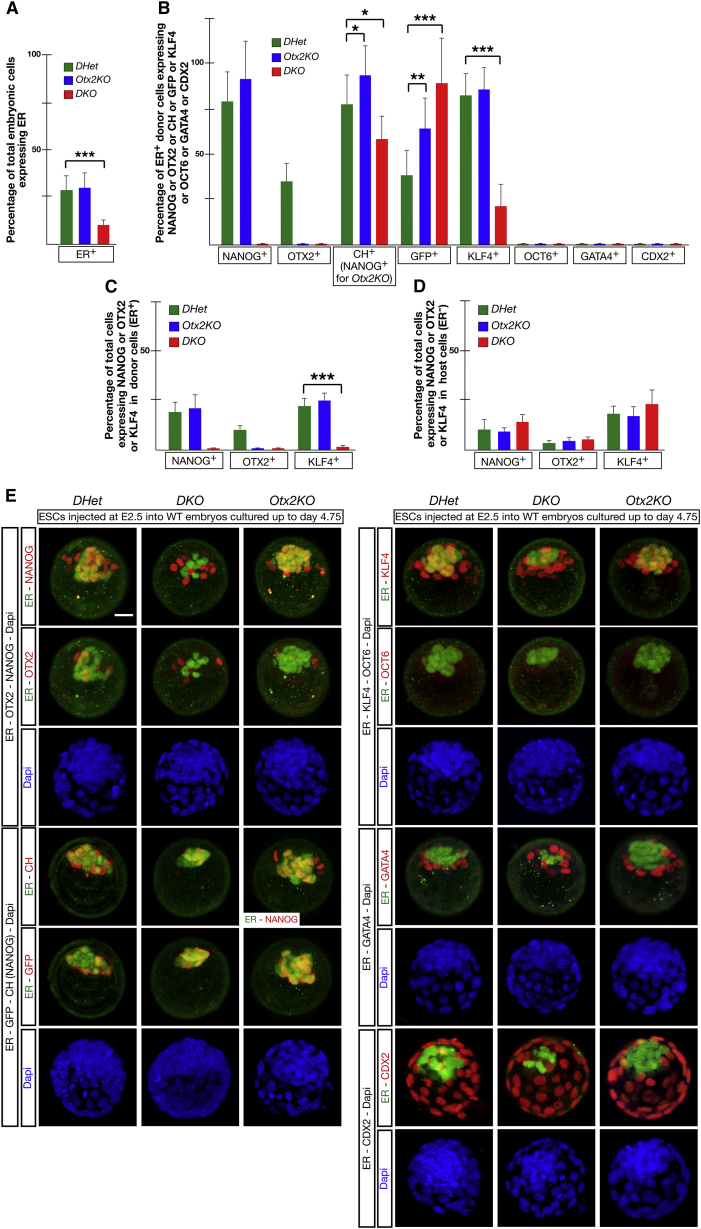
We next examined whether DKO ESCs may colonize host blastocysts and be maintained in postimplantation embryos and/or adult mice. The contribution from mutant ESCs was assessed by immunostaining with the ER antibody recognizing R26-driven constitutive expression of CreER (Figure S1), PCR assays, and coat-color inspection. DHet and DKO ESCs cultured in LIF + FBS or in LIF + 2i up to P5 were injected into E2.5 host embryos, which were allowed to develop in vitro for 24–30 hr before implantation. We found that, in contrast with DHet, DKO ESCs did not generate evident chimerism in E12.5 embryos and mice (Figure S6A). This may be contributed by intrinsic loss of their preimplantation colonizing ability and/or loss of pluripotency. To rule out the last possibility, we analyzed DKO teratomas and found that they generated derivatives of all germ layers (Figure S6B). Then, we studied the colonization efficiency of DKO ESCs in preimplantation embryos. Mutant ESCs were injected at E2.5 in WT embryos cultured up to day 4.75 and tracked using CreER immunofluorescence. Cell counting of ER+ ESCs versus DAPI+ total cells showed that, compared with DHet and Otx2KO, DKO ESCs were less efficiently integrated into host embryos (Figure 5A; Table S6). Then we studied the identity of both injected mutant ESCs and cells of the host embryos. We first determined the percentage of ER+ cells expressing NANOG or CH and OTX2 or GFP. We found that compared with DHet, the percentage of ER+ cells expressing NANOG or CH was increased in Otx2KO and diminished in DKO ESCs, while the percentage of ER+ cells expressing GFP was significantly increased in Otx2KO and DKO ESCs (Figures 5B and 5E; Table S6). When extended to KLF4, GATA4, OCT6, and CDX2, this analysis showed that compared with DHet ESCs, the percentage of ER+ cells expressing KLF4 was remarkably diminished in blastocysts injected with DKO ESCs; ER+ cells co-expressing OCT6 or GATA4 or CDX2 were not detected (Figures 5B and 5E; Table S6). This analysis suggests that the host environment slightly modifies the expression of NANOG or CH, OTX2 or GFP and KLF4 described in vitro for mutant ESCs, while it efficiently prevents/suppresses OCT6. These data were further confirmed by determining the percentage of total cells co-expressing ER with NANOG or OTX2 or KLF4 (Figure 5C; Table S6). Then, we studied whether the percentage of total cells expressing NANOG, OTX2, or KLF4 in host cells (ER−) was affected. For these markers, this percentage was slightly increased only in blastocysts injected with DKO ESCs (Figures 5D and 5E; Table S6), suggesting that it may be a consequence of the reduced colonizing ability of DKO ESCs.
Figure 5.
Abnormalities of Mutant ESCs in Chimeric Blastocysts
(A) Graphic representation showing that, compared with DHet and Otx2KO, DKO ESCs exhibit reduced efficiency of integration into host embryos. Data are the means ± SD from n = 10 blastocysts for each ESC line; ∗∗∗p ≪ 0.005.
(B–D) Cell counting showing the percentage of ER+ donor cells expressing NANOG, OTX2, CH, GFP, KLF4, OCT6, GATA4, and CDX2 (B) or the percentage of total cells (DAPI+) expressing NANOG, OTX2, or KLF4 in the donor (ER+) (C) or in the host cells (ER−) (D). Data are the means ± SD from n = 7 blastocysts for each combination of antibodies reported in (E) and for each ESC line. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005.
(E) Representative confocal images of embryos injected at E2.5 with DHet, DKO, and Otx2KO ESCs and immunostained at E4.75 with ER plus NANOG and OTX2, or CH and GFP, or KLF4 and OCT6, or GATA4, or CDX2. Note that the externalmost plane of acquisition is removed to allow a better view of the inner cell mass. DAPI staining is also shown. Scale bar, 30 μm.
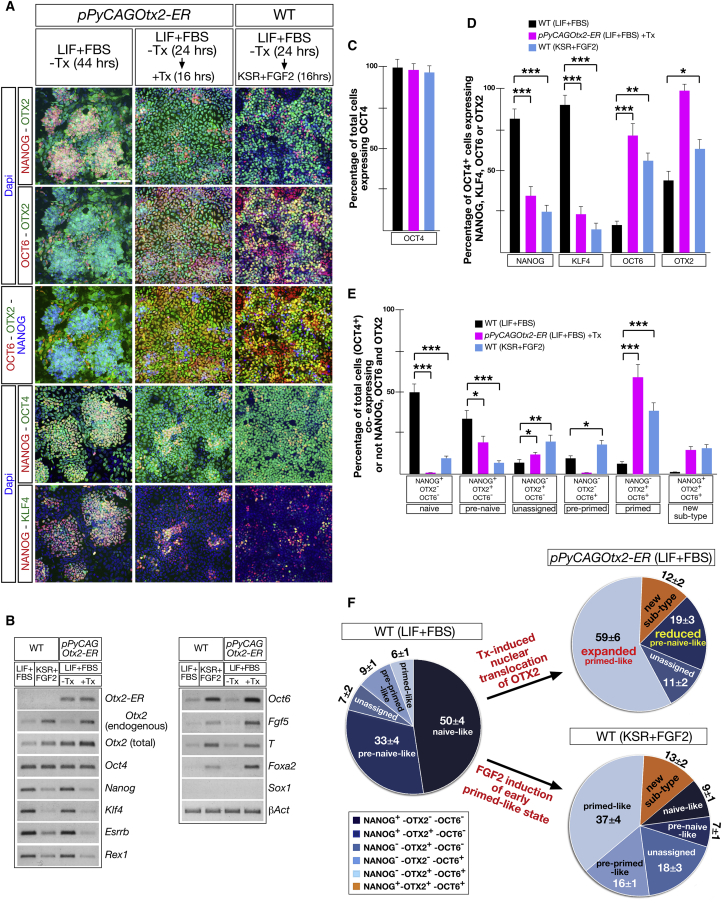
Otx2 Overexpression Is Sufficient to Induce Early Primed-like Identity in ESCs Cultured in LIF without the Contribution of FGF
Transition of ESCs into early primed pluripotency is characterized by rapid and wide activation of OTX2 and OCT6 (Acampora et al., 2016). Therefore, we investigated whether ubiquitous activation of OTX2 in ESCs cultured in LIF + FBS was sufficient to impart an early primed-like identity similar to that observed when FGF2 was administered to WT ESCs. To this aim, we generated a further mutant ESC line (pPyCAGOtx2-ER) showing constitutive expression of a fusion protein between OTX2 and ERT2 (OTX2-ER) (Figure S7A). Western blot analysis of the Tx-untreated pPyCAGOtx2-ER selected clone (red arrow in Figure S7B) showed that the amount of the OTX2-ER fusion protein was about four times that of the endogenous OTX2. Assuming that the endogenous OTX2 was expressed in about 40% of ESCs and that the OTX2-ER protein is present in all ESCs, we concluded that the OTX2-ER level/cell should be about two times that of the endogenous OTX2. Shortly after Tx administration, OTX2-ER was translocated to the nucleus (Figure S7C). Compared with WT, in pPyCAGOtx2-ER ESCs, the distribution and/or expression level of a number of markers including NANOG, OCT4, KLF4, OCT6, Esrrb, Rex1, Fgf5, T, Foxa2, and Sox1 was apparently not influenced by OTX2-ER when Tx was absent (Figures 6A, 6B, and S4D). However, when Tx was added to LIF + FBS-containing medium, nuclear translocation of OTX2-ER induced a profound change in the identity of ESCs, which exhibited marked similarity with WT ESCs primed with FGF2 (Figure 6A). Indeed, Tx-treated pPyCAGOtx2-ER ESCs (LIF + FBS) and WT ESCs exposed to FGF2 in KSR retained an ubiquitous distribution of OCT4 and exhibited a relevant contraction of NANOG+ and KLF4+ cells counterbalanced by the expansion of OCT6+ cells (Figures 6A, 6C, 6D, and S7D; Table S2). Analysis of ESC sub-types showed that in Tx-treated pPyCAGOtx2-ER ESCs, the naive-like sub-type was lost, the pre-naive-like sub-type was reduced, and the primed-like sub-type was expanded (Figures 6E and 6F; Table S2). Importantly, FGF2-mediated priming of WT ESCs induced a similar organization of sub-type compartments (Figures 6E and 6F; Table S2). Moreover, in Tx-treated pPyCAGOtx2-ER ESCs, additional naive factors such as Esrrb and Rex1 were downregulated, whereas primed markers such as Fgf5, T, and Foxa2 were upregulated (Figures 6B and S4D). Noteworthy, when Tx administration was interrupted at P1, the expression of NANOG, OTX2, OCT4, KLF4, and OCT6 reverted and, at P3, was very similar to that of WT ESCs (Figure S7E). These findings were confirmed in a second independent clone (black arrow in Figure S7B) (data not shown). These data indicate that in LIF + FBS medium, OTX2 is sufficient, without the contribution of FGF2, to induce an expansion of the primed-like ESC compartment as observed in FGF2-primed WT ESCs and NanogKO ESCs in LIF + FBS medium. Then, we analyzed the response to FGF2 of NanogKOS, DKO, and Otx2KO ESCs. Indeed, on the basis of their identity in LIF + FBS, the NanogKOS and DKO ESCs should be primed more efficiently than Otx2KO ESCs. Consistently, when compared with WT, NanogKOS and DKO cells exhibited higher induction of OCT6 and lower expression of CH, while Otx2KO cells induced OCT6 in fewer cells and retained a higher number of NANOG+ cells (Figure S7F). OTX2 or GFP were detected in most of the NanogKOS and Otx2KO cells and virtually in all DKO cells (Figure S7F). We next analyzed the identity of Tx-treated pPyCAGOtx2-ER ESCs in chimeric blastocysts. We found that the percentage of total cells expressing ER was not affected by Tx treatment (Figure S7G). Marker analysis showed that, compared with Tx-untreated blastocysts, in Tx-treated blastocysts, KLF4 and NANOG were detected in a reduced percentage of ER+ cells, OCT6 was activated in a small but significant percentage of ER+ cells, and GATA4 and CDX2 were detected only in ER− cells (Figures S7H and S7K; Table S6). These data were confirmed by determining the percentage of total cells expressing NANOG, KLF4, and OCT6 in ER+ donor cells (Figures S7I and S7K; Table S6). The complementary analysis showed that the percentage of total cells expressing NANOG or KLF4 in ER− host cells was not significantly affected (Figures S7J and S7K; Table S6). These findings suggest that the wide activation of OCT6 observed in cell-culture experiments is partially prevented by the host environment, which instead is unable to recover loss of naive markers.
Figure 6.
Otx2 Overexpression Is Sufficient to Prime ESCs in LIF-Containing Medium and without FGF2 Contribution
(A) Representative images of pPyCAGOtx2-ER ESCs administered or not with Tx and WT ESCs primed with FGF2 immunostained with NANOG, OTX2, and OCT6 and with NANOG, OCT4, and KLF4. Scale bar, 100 μm.
(B) Representative RT-PCR assays show that in WT and pPyCAGOtx2-ER ESCs, FGF2 treatment and Tx-induced nuclear translocation of OTX2-ER downregulate Nanog, Klf4, Esrrb, and Rex1 and upregulate endogenous Otx2, Fgf5, T, and Foxa2 (n = 3 independent experiments).
(C–E) Cell counting showing for WT ESCs (LIF + FBS), Tx-treated pPyCAGOtx2-ER in LIF + FBS, and WT ESCs in KSR plus FGF2 the percentage of total cells expressing OCT4 (C), the percentage of OCT4+ cells expressing NANOG or KLF4 or OCT6 or OTX2 (D), and the percentage of total cells (OCT4+) expressing the different sub-types identified by the combinatorial analysis of NANOG, OCT6, and OTX2 (E). Data are presented as means ± SD from four independent experiments. ∗∗∗p ≪ 0.001; ∗∗p < 0.001; ∗p value is between 0.005 and 0.001.
(F) Cell-counting data show that nuclear translocation of OTX2-ER is sufficient to induce a re-organization of sub-type compartments similar to that induced by FGF2 in WT ESCs.
See also Figures S4 and S7 and Table S2.
Loss of Otx2 and/or Nanog Affects the Responding Competence to 2i-mediated Conversion into Naive State
Finally, we tested whether the transition to the naive state was affected in mutant ESCs. In WT ESCs cultured in LIF + 2i, this transition was characterized by increased expression of naive markers such as OCT4, NANOG, KLF4, Esrrb, Fgf4, and Rex1 and loss of primed markers such as OTX2 and OCT6 (Figures 7A, 7B, and S4E). At P1, WT, DHet (data not shown), and Otx2KO ESCs exhibited a stable naive-like state while DKO ESCs showed small OCT4+ colonies expressing KLF4, CH and, unexpectedly, GFP (Figures 7A, 7B, and S4E). Moreover, most of the OCT4− DKO cells expressed PE (GATA4, KLF4, Foxa2, Sox17) or trophectoderm (TE) (CDX2, Hand1, Gata3) markers (Figures 7A, 7B, and S4E), which was reminiscent of NanogKO and NanogKOS ESCs in LIF + FBS. Nevertheless, under sustained culture in LIF + 2i, DKO ESCs gradually expanded the naive-like cell fraction and at P5 were similar to WT ESCs (Figures 7A–7C). We suggest that the phenotype of DKO ESCs may reflect the fact that, although NANOG is ablated, the simultaneous loss of OTX2 restrains but does not suppress naive conversion. To test this possibility, we first cultured pPyCAGOtx2-ER (-Tx) ESCs in LIF + 2i for two passages to allow conversion into the naive-like state and then administered Tx for a third passage. Compared with untreated pPyCAGOtx2-ER ESCs, Tx treatment generated hypomorphic OCT4+ colonies expressing NANOG and KLF4 and numerous OCT4− cells showing neural specific markers SOX1 and TUJ1 (Figure 7D). RT-PCR assays showed that Tx administration downregulated Esrrb, Rex1, and Fgf4 expression and activated strong Sox1 expression (Figures 7E and S4F). Then, we analyzed NanogKOS ESCs cultured in LIF + 2i. We found that at P1, NanogKOS ESCs exhibited small OCT4+-CH+ colonies expressing KLF4 only in a few OCT4+ cells (Figure 7F). Noteworthy, as observed for GFP in DKO ESCs, OTX2 was partially suppressed also in NanogKOS ESCs. Furthermore, NanogKOS ESCs generated a relevant number of PE-like OCT4− cells (Figures 7F, 7G, and S4G). At P5, in contrast with DKO, NanogKOS ESCs did not recover a naive-like phenotype (Figures 7F, 7G, and S4G). Together these and previous findings highlight the relevance of the antagonism between NANOG and OTX2 for the control of the heterogeneous identity of ESCs in LIF + FBS. Without integration of this mutual antagonism, the plasticity of ESCs is lost, and the capacity for bidirectional conversion into naive or primed pluripotency is forced in a more unidirectional manner either forward (primed) or reverse (naive), which is established by the dominance of the OTX2 regulatory network over that controlled by NANOG or vice versa. When both networks are abolished, ESCs retained a degree of plasticity sufficient to accomplish bidirectional conversion.
Figure 7.
Conversion of Mutant ESC Lines into Naive-like and Primed-like Pluripotent States
(A) Representative immunohistochemistry assays with OTX2 or GFP and NANOG or CH, OCT4 and KLF4, and with GATA4 and CDX2 performed at passage (P) 1, P2, and P5 in WT, Otx2KO, and DKO ESCs cultured in LIF + 2i show that DKO ESC colonies acquire a domed morphology, ubiquitous expression of OCT4, KLF4, and CH and stop generating OCT4− cells. Scale bars, 100 μm.
(B) RT-PCR assays show that in DKO ESCs, the expression of naive, PE, and TE markers progressively recovers a profile similar to that of WT ESCs (n = 3 independent experiments).
(C) Western blots show that at P5 also the level of p-STAT3, p-ERK1,2, act-βCAT, and p-βCAT is similar in WT and DKO ESCs (n = 3 independent experiments).
(D) Representative immunohistochemistry assays with OTX2 and NANOG, OCT4 and KLF4, and SOX1 and TUJ1 on pPyCAGOtx2-ER ESCs cultured in LIF + 2i up to P3 without Tx or cultured up to P2 without Tx and for an additional passage (P3) with Tx.
(E) RT-PCR assays of Tx-treated pPyCAGOtx2-ER ESCs show reduced expression of Esrrb, Fgf4, and Rex1 and Sox1 upregulation; expression of PE, TE, and primed markers appear unaffected (n = 3 independent experiments).
(F) Representative immunohistochemistry assays with OTX2 and NANOG or CH, OCT4, and KLF4 and with GATA4 and CDX2 show that NanogKOS ESCs cultured in LIF + 2i exhibit small and rare OCT4+ colonies, abundant differentiation of PE-like cells expressing KLF4 and GATA4, and do not recover a naive-like phenotype at P5.
(G) RT-PCR assays performed on WT and NanogKOS ESCs cultured in LIF + 2i show that naive markers are downregulated, PE markers are stably upregulated, TE markers are transiently upregulated, and primed markers are unaffected (n = 3 independent experiments).
See also Figure S4.
Discussion
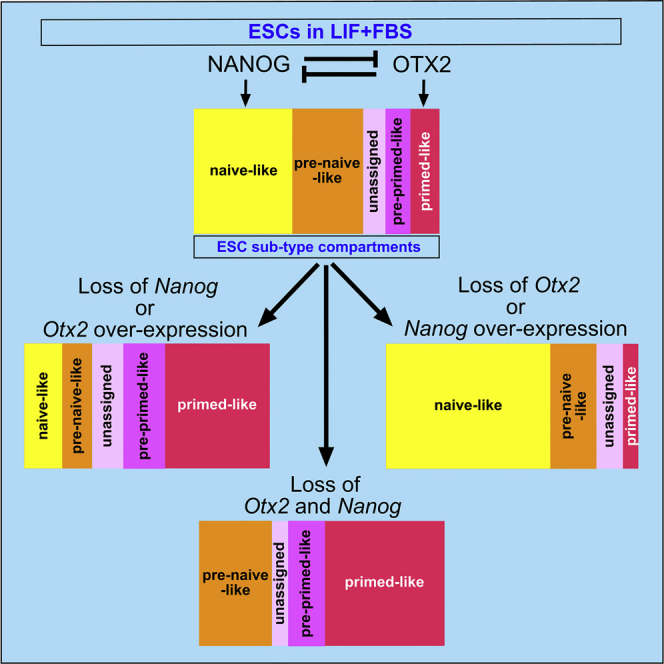
Heterogeneity in gene expression is a typical feature of ESCs cultured in LIF + FBS. Understanding the molecular basis controlling this heterogeneous condition is important to decipher the plasticity of ESCs (Smith, 2017, Torres-Padilla and Chambers, 2014, Martinez Arias et al., 2013, Nichols and Smith, 2009, Nichols and Smith, 2011, Chambers et al., 2007). Although different mechanisms may operate together to determine the final state of ESCs, the control exerted by specific transcription factors may have a prominent role. In this context, allelic control of NANOG dosage has been proposed to have a causative role for the heterogeneity of ESCs cultured in LIF + FBS (Torres-Padilla and Chambers, 2014, Miyanari and Torres-Padilla, 2012). On the other hand, OTX2 exhibits heterogeneous expression in ESCs cultured in LIF + FBS and is required to promote the transition into the early primed state (Acampora et al., 2013, Acampora et al., 2016, Buecker et al., 2014, Yang et al., 2014). These previous findings led us to investigate the possibility that OTX2 and NANOG may be antagonistic determinants that together specify the heterogeneity of ESCs cultured in LIF + FBS. We therefore studied how these two factors influence the identity and size of ESC sub-type compartments to assess whether they privilege or affect specific states of pluripotency co-existing in ESCs self-renewing in LIF + FBS. To this aim, we analyzed an allelic series of mutant ESCs carrying different dosages of OTX2 and/or NANOG. We have shown that, without the addition of exogenous factors that promote naive or primed pluripotency, loss of Otx2 causes expansion of the naive-like compartment, which is consistent with previous data on Nanog overexpression (Chambers et al., 2003). In contrast, loss of Nanog analyzed in NanogKO ESCs or Otx2 overexpression generate a strikingly similar phenotype characterized by remarkable contraction of the naive-like compartment and expansion of the primed-like sub-type. When both genes are deleted, ESCs acquire a pluripotent state exhibiting identity features in common with early primed-like cells. These data indicate that antagonism between NANOG and OTX2 is an essential requirement to specify and maintain ESC heterogeneity, which we visualize through the analysis of OCT4+ cells co-expressing different combinations of NANOG, OTX2, and OCT6 (Acampora et al., 2016). We envisage that between naive-like and primed-like compartments, there are at least three transitional and dynamic compartments—the pre-naive-like, the pre-primed-like, and the unassigned—ready to be converted and stabilized into naive or primed pluripotent states. Therefore, in addition to previous studies indicating that NANOG and OTX2 are important coordinators of naive and primed pluripotency, respectively (Acampora et al., 2013, Acampora et al., 2016, Buecker et al., 2014, Yang et al., 2014, Silva et al., 2009, Chambers et al., 2003, Chambers et al., 2007, Mitsui et al., 2003), our new data suggest that the combined action of these two transcription factors is instrumental in determining the heterogeneous identity of ESCs cultured in LIF + FBS. Importantly, a similar heterogeneity can be identified in vivo between E4.5 and E4.7 when the naive pluripotency circuit is being shut down but before the early primed state becomes dominant. Most of the ESC sub-types detected in LIF + FBS cultures are present in the E4.5–4.7 epiblast (Acampora et al., 2016). We suggest that at this stage, the transitional heterogeneity of the epiblast is captured in cultured ESCs by LIF + FBS and is maintained by antagonism between OTX2 and NANOG. These observations are relevant to the recently elaborated concept of formative pluripotency, a state hypothesized to interpose between naive and primed pluripotency and suggested to occur abruptly (Smith, 2017). However, the finding that the epiblast of E4.5–4.7 embryos and ESCs in LIF + FBS exhibit a spectrum of distinct and precise combinations of naive and primed markers leads us to suggest that this transition may not necessarily occur abruptly. In this context, an imbalance in favor of the OTX2-dependent regulatory network due to downregulation of Nanog expression and upregulation of Otx2 might trigger conversion of pluripotency from a metastable and heterogeneous condition toward a formative state.
The co-existence of different pluripotent sub-types is an intermediate gate preceding the unidirectional transition of the epiblast into the early primed state in vivo, and allowing bidirectional conversion into naive or early primed state in vitro (Smith, 2017, Torres-Padilla and Chambers, 2014, Martinez Arias et al., 2013, Nichols and Smith, 2011, Silva et al., 2009, Ying et al., 2008). The capability of NANOG to impose a unimodal naive-like identity is counterbalanced by OTX2, which, in turn, promotes primed-like identity. We propose that integration of these opposing regulatory circuits underlies the mechanism that specifies heterogeneity in ESCs. This identity can be established only in LIF-containing medium where exogenous signal constraints (FGF or 2i) are minimized and ESCs are allowed to manifest their flexible nature. Signaling pathway activation perturbs this state and determines the dominance of one of the two regulatory networks. This process requires the contemporary and co-ordinated response by both NANOG and OTX2. Indeed, 2i-mediated induction of naive state could not be efficiently accomplished without Nanog activation and Otx2 repression; similarly, FGF-mediated induction of primed state is incomplete without Otx2 activation and Nanog downregulation. Specification and maintenance of this mechanism require that the dosage and expression pattern of OTX2 and NANOG should be established within precise parameters, which ultimately determine the degree of integration between their antagonistic transcriptional networks. According to a hierarchically high role of OTX2 and NANOG in interpreting and responding to primed and naive inducers, we report that transitions between naive and primed compartments can be coherently mimicked in LIF + FBS only by genetic manipulations of OTX2 and/or NANOG dosage.
Experimental Procedures
Generation of ESC Lines
All mutant ESC lines (Figure S1) were generated in an E14Tg2a-derived ESC line, whose Rosa26 locus was previously targeted with a CreER-pac gene cassette (Acampora et al., 2013). The sequential steps for the generation of mutant ESCs are described in the Supplemental Experimental Procedures.
Cell-Culture Experiments
ESC culture procedures are described in the Supplemental Experimental Procedures.
RT-PCR and Western Blotting
RT-PCRs were performed in non-saturating conditions using the primers and cycles listed in Table S7. Western blots were probed with antibodies and at the dilutions listed in Table S7.
FACS Analysis
NanogKO ESCs were trypsinized, centrifuged, and resuspended in ESC medium containing 2% FBS and 2 mM EDTA. The CH+ fraction was sorted using a BD FACSAria III (Becton Dickinson) into a 6-well plate. The data were analyzed using the BD FACSDiva software.
Immunohistochemistry
Immunohistochemistry assays on ESCs and embryos and ALP assays were performed as described (Acampora et al., 2013). Antibodies and dilutions used in this study are listed in Table S7.
RNA-Seq Experiments, Analysis of RNA-Seq Data, and RNA-Seq Validation Assays
RNA-seq experiments were performed on DKO ESCs (LIF + FBS), Otx2KO ESCs (LIF + FBS), WT ESCs (LIF + FBS), WT ESCs (LIF + 2i), and WT EpiLCs (44 hr). Indexed sequencing libraries were generated and analyzed as described in the Supplemental Experimental Procedures.
Chimerism Experiments
Chimerism experiments are described in the Supplemental Experimental Procedures. Animals were handled in accordance with the authorization 1196/2015-PR released by the Italian Ministry of Health.
Cell-Counting Experiments
Cell counting of ESC sub-types was manually performed on immunohistochemistry images printed in A4 format (Acampora et al., 2013, Acampora et al., 2016). SD was calculated from the analysis of four independent experiments. p values were determined using the one-tailed Student’s t test. Cell counting of chimeric blastocysts was performed on n = 10 blastocysts for colonization efficiency experiments and on n = 7 blastocysts for each combination of antibodies and for each injected ESC line (Table S6). Images were printed in A4 format for manual cell counting. Data are reported as means ± SD; p values were determined using the one-tailed Student’s t test. For details see Supplemental Experimental Procedures.
Author Contributions
D.A. performed and supervised most of the in vitro and in vivo experiments. L.G.D.G. generated targeting molecules and contributed to the generation and analysis of mutant ESCs. A.G. and V.N. performed and analyzed RNA-seq experiments. D.O. generated the pPyCAGOtx2-ER ESC line. J.Z. generated the pPyCAGOtx2-ER plasmid. A.L. contributed to the generation of mutant ESCs. I.C. contributed to the preparation of the manuscript. A.S. conceived the experiments and wrote the manuscript.
Acknowledgments
We thank the staff of the IGB animal house facility, the IGB integrated microscopy facility, the IGB FACS facility, and the Tigem NGS facility. We also thank D. Graniero for typing and formatting the manuscript and S. Arbucci and V. Mercadante for technical support. This work was supported by the Italian Association for Cancer Research (AIRC) (Project IG-14152), the POR MOVIE of Regione Campania (B25C13000240007), and the PRIN project (20157JF8P5_004) to A.S. and the Medical Research Council to I.C.
Published: October 19, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.09.019.
Accession Numbers
RNA-seq data are available in European Nucleotide Archive database (https://www.ebi.ac.uk/ena/submit/sra/#home) under accession number ENA: PRJEB19241/ERP021225.
Supplemental Information
Table S3 contains data from RNA-seq experiments.
Table S4 contains data from RNA-seq experiments.
Table S5 contains data from RNA-seq experiments.
Table S7 includes the list of oligos and antibodies employed in this study and is.
References
- Acampora D., Di Giovannantonio L.G., Simeone A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development. 2013;140:43–55. doi: 10.1242/dev.085290. [DOI] [PubMed] [Google Scholar]
- Acampora D., Omodei D., Petrosino G., Garofalo A., Savarese M., Nigro V., Di Giovannantonio L.G., Simeone A. Loss of the Otx2-binding site in the Nanog promoter affects the integrity of embryonic stem cell subtypes and specification of inner cell mass-derived epiblast. Cell Rep. 2016;15:2651–2664. doi: 10.1016/j.celrep.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., Faial T., Simeone A., Tan M., Swigut T., Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., Daley G.Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Festuccia N., Osorno R., Halbritter F., Karwacki-Neisius V., Navarro P., Colby D., Wong F., Yates A., Tomlinson S.R., Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R.L., Beddington R.S. Multi-lineage 'stem' cells in the mammalian embryo. J. Cell Sci. Suppl. 1988;10:11–27. doi: 10.1242/jcs.1988.supplement_10.2. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T., Smith A. Mapping the route from naive pluripotency to lineage specification. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T., Lim C., Hayward P., Munoz-Descalzo S., Nichols J., Garcia-Ojalvo J., Martinez Arias A. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwacki-Neisius V., Göke J., Osorno R., Halbritter F., Ng J.H., Weiße A.Y., Wong F.C., Gagliardi A., Mullin N.P., Festuccia N. Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell. 2013;12:531–545. doi: 10.1016/j.stem.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T. Primed for pluripotency. Cell Stem Cell. 2011;8:241–242. doi: 10.1016/j.stem.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Lanner F., Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Gottgens B., Niwa H., Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A., Nichols J., Schroter C. A molecular basis for developmental plasticity in early mammalian embryos. Development. 2013;140:3499–3510. doi: 10.1242/dev.091959. [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Miyanari Y., Torres-Padilla M.E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- Muñoz Descalzo S., Rué P., Garcia-Ojalvo J., Martinez Arias A. Correlations between the levels of Oct4 and Nanog as a signature for naïve pluripotency in mouse embryonic stem cells. Stem Cells. 2012;30:2683–2691. doi: 10.1002/stem.1230. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. The origin and identity of embryonic stem cells. Development. 2011;138:3–8. doi: 10.1242/dev.050831. [DOI] [PubMed] [Google Scholar]
- Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Rossant J., Tam P.P. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- Silva J., Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. Formative pluripotency: the executive phase in a developmental continuum. Development. 2017;144:365–373. doi: 10.1242/dev.142679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R.K., Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen T.W., van Oosten A.L., Castelo-Branco G., Hall J., Smith A., Silva J.C. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr. Biol. 2011;21:65–71. doi: 10.1016/j.cub.2010.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla M.E., Chambers I. Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- Yang S.H., Kalkan T., Morissroe C., Marks H., Stunnenberg H., Smith A., Sharrocks A.D. Otx2 and Oct4 drive early enhancer activation during embryonic stem cell transition from naive pluripotency. Cell Rep. 2014;7:1968–1981. doi: 10.1016/j.celrep.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3 contains data from RNA-seq experiments.
Table S4 contains data from RNA-seq experiments.
Table S5 contains data from RNA-seq experiments.
Table S7 includes the list of oligos and antibodies employed in this study and is.