Abstract
We describe a novel mutation in the Chlamydomonas reinhardtii STA11 gene, which results in significantly reduced granular starch deposition and major modifications in amylopectin structure and granule shape. This defect simultaneously leads to the accumulation of linear malto-oligosaccharides. The sta11-1 mutation causes the absence of an α-1,4 glucanotransferase known as disproportionating enzyme (D-enzyme). D-enzyme activity was found to be correlated with the amount of wild-type allele doses in gene dosage experiments. All other enzymes involved in starch biosynthesis, including ADP-glucose pyrophosphorylase, debranching enzymes, soluble and granule-bound starch synthases, branching enzymes, phosphorylases, α-glucosidases (maltases), and amylases, were unaffected by the mutation. These data indicate that the D-enzyme is required for normal starch granule biogenesis in the monocellular alga C. reinhardtii.
Starch is one of the most abundant biological polymers present in the earth's biosphere and remains the major supply of calories in both human and animal diets. As is the case for animal, fungal, and bacterial glycogen, starch is made solely of Glc residues linked in α-1,4 positions and branched in α-1,6 positions. Unlike glycogen, starch granules consist of complex, semicrytalline structures of unlimited size (for review, see Buléon et al., 1998). Amylopectin, the major polysaccharide fraction of starch, is considered to be the only molecular fraction required to generate normal granules. The building of the polymer structure depends on the transfer by SS of Glc in the α-1,4 position from ADP-Glc to the nonreducing end of growing chains. Introduction of the α-1,6 branch proceeds through the cleavage (by branching enzyme) of a pre-existing α-1,4-linked glucan and the transfer of the cleaved glucan in the α-1,6 position. It was previously thought that the specific features of starch structure depended solely on the concerted action of multiple forms of SS and branching enzyme; however, other enzymes are likely to be involved in the process.
Our strategy consisted of isolating mutants defective in amylopectin synthesis, thereby defining the functions involved in starch granule biogenesis. From the analysis of mutants of the monocellular green alga Chlamydomonas reinhardtii (Mouille et al., 1996), maize (James et al., 1995), rice (Nakamura et al., 1996), and Arabidopsis (Zeeman et al., 1998b), it was determined that debranching enzymes were required to trim α-1,6 linkages from a precursor (pre-amylopectin) into a mature amylopectin molecule (Ball et al., 1996) or that they were required to prevent glycogen production by the starch synthesis machinery (Zeeman et al., 1998b). Continued genetic analysis is likely to identify additional enzymes needed for starch biosynthesis that would not have been foreseen from the basic biosynthetic steps (Mouille et al., 1996).
Eukaryotic algae are of particular relevance for studies dealing with starch synthesis. Starch polysaccharides are not found in bacteria or fungi. Because C. reinhardtii is the only starch-storing unicellular organism intensively studied by geneticists, it offers a unique opportunity to understand the basic mechanisms of starch granule biogenesis. Indeed, growth-arrested C. reinhardtii cells accumulate glucopolysaccharides that are very similar to cereal endosperm storage starch, so this organism serves as a highly useful model for starch synthesis in crop plants. C. reinhardtii cells contain the same set of starch biosynthesis enzymes and, most importantly, respond in an analogous fashion to mutations affecting these activities (for review see Ball, 1998).
D-enzymes define α-glucanotransferases that transfer unbranched malto-oligosaccharide groups from a donor α-1,4 glucan of at least three Glc residues (maltotriose) to a recipient oligosaccharide at the final expense of Glc formation (Peat et al., 1956). It is thought that this activity disproportionates short glucans into longer oligosaccharides to facilitate their degradation through phosphorylases or hydrolases (Boos and Schuman, 1998).
This study characterized a novel C. reinhardtii mutation, sta11-1, which causes decreased levels of starch, modification of amylopectin structure, increased amylose content, modification of granule size and shape, and accumulation of unbranched oligosaccharides. All enzymes previously suspected to be involved in starch biosynthesis are unaffected by the presence of the sta11-1 mutation; however, sta11-1 mutants lack an enzyme required for maltotriose hydrolysis. Zymogram analysis established that the missing maltotriose-metabolizing enzyme, as described for higher plants, is a D-enzyme.
These data indicate that α-1,4 glucanotransferases are important components of the amylopectin synthesis machinery in C. reinhardtii. The results are generally similar to those regarding debranching enzyme function, because in both instances enzymes thought to be involved in the breakdown of glucopolysaccharides are apparently involved in starch biosynthesis.
MATERIALS AND METHODS
Materials
U-14C Glc-1-P and [d-Glc-U-14C]ADP-Glc were purchased from Amersham. ADP-Glc and maize amylopectin were from Sigma. Pseudomonas amyloderamosa isoamylase was from Megazyme (Sydney, Australia). Glc-1-P, rabbit muscle glycogen, yeast hexokinase, yeast Glc-6-P dehydrogenase, and rabbit muscle phosphorylase were obtained from Boehringer Mannheim.
Chlamydomonas reinhardtii Strains, UV Mutagenesis, Growth Conditions, Cytological Observations, and Media
The wild-type reference Chlamydomonas reinhardtii strains used in this study were 330 (mt+ arg7 cw15 nit1 nit2) and 137C (mt− nit1 nit2). According to the genotypes of the mutants or segregants tested, diploids were selected by complementation on minimal medium after crossing either with strain A35 (mt+ pab2 ac14), 37 (mt− pab2 ac14), NV314 (mt− pab2 ac14 sta1-1), strain 37E-17 (mt− pab2 ac14 sta3-1), GST− (mt− nit1 nit2 sta5-1), BAFR1 (mt+ nit1 nit2 cw15 arg7-7 sta2–29::ARG7), or 18B (mt− nit1 nit2 sta2-1). The sta11-1 strains most commonly used for complementation were CO 214 (mt+ nit1 nit2 sta11-1), CO 29 (mt+ pab2 ac14 sta11-1), and JV45J (mt− nit1 nit2 sta11-1).
UV mutagenesis was performed by irradiating cells at 20% survival using a 254-nm transilluminator (model TS-15, Ultra-Violet Products, San Gabriel, CA) displaying a peak intensity of 7.0 mW cm−2. Irradiation was followed by overnight incubation in high-salt medium in darkness.
All experiments were carried out in continuous light (80 μE/m2 s−1) in the presence of acetate at 24°C in liquid cultures that were shaken vigorously without air or CO2 bubbling. Late-log-phase cultures were inoculated at 105 cells mL−1 and harvested at 2 × 106 cells mL−1. Nitrogen-starved cultures were inoculated at 5.105 cells mL−1 and harvested after 4 d at a final density of 1 to 2 × 106 cells mL−1. Genetic techniques were as described by Harris (1989a). Standard TAP (Tris acetate phosphate) medium was fully detailed in Harris (1989b), while nitrogen-starved medium (TAP-N) and diploid clone selection were described in Ball et al. (1990, 1991) and Delrue et al. (1992). Fixation and embedding protocols were as described in Dauvillée et al. (1999).
Structural Analysis of Polysaccharides
Wide-angle x-ray diffraction and TEM and SEM analyses were as detailed in Buléon et al. (1997). Gel permeation chromatography of delipidated starch fractions dispersed in 10 mm NaOH was as described in Delrue et al. (1992) and Libessart et al. (1995), and was performed on Sepharose CL2B (Pharmacia). Gel permeation chromatography-purified amylose and amylopectin were debranched with isoamylase. 1H-NMR of gel permeation chromatography-purified starch fractions were as described previously (Fontaine et al., 1993). The APTS-tagged chains produced by isoamylase-mediated debranching were separated by high-resolution slab gel electrophoresis on a DNA sequencer (O'Shea and Morell, 1996). The results obtained were confirmed by capillary electrophoresis analysis of debranched amylopectins obtained from two mutant and two wild-type meiotic offspring. Capillary electrophoresis was carried out as previously described (O'Shea et al., 1998).
Measures of Starch Levels, Starch Purification, and Spectral Properties of the Iodine-Starch Complex
A full account of amyloglucosidase assays, starch purification on Percoll gradients, and λmax, the wavelength of the maximal absorbance of the iodine-polysaccharide complex, can be found in Delrue et al. (1992).
Crude Extract Preparation, Enzyme Assays, Partial Purification of Enzyme Activities, and Zymograms
Soluble crude extracts were always prepared from late- log-phase cells (2 × 106 cells mL−1) grown in high-salt acetate medium under continuous light (80 μE m−2 s−1). All assays were conducted in conditions of linearity with respect to time and amount of crude extract. Phosphoglucomutase, ADP-Glc pyrophosphorylase, and phosphorylase activities were monitored by using the standard assays described in Ball et al. (1991) and Van den Koornhuyse et al. (1996). For SS and branching enzymes, the assays used were those described by Fontaine et al. (1993) and Libessart et al. (1995). GBSS I was monitored as previously described (Delrue et al., 1992) from the starch purified from nitrogen-supplied cultures. α-Glucosidase was monitored by measuring the Glc produced from maltose hydrolysis in sodium acetate, pH 6.5, by the standard Glc-6-P dehydrogenase assay described in Ball et al. (1991). The analysis was completed by zymograms as detailed in Buléon et al. (1997) and in Mouille et al. (1996). For partial purification, a 10% (w/v) protamine sulfate precipitation was applied to 5 ×109 cells in 4 mL of 50 mm sodium acetate, pH 6.0, and 1 mm DTT, loaded on a FPLC anion-exchange column (1.6 × 10 cm, 1 mL min−1, 0–0.5 m NaCl gradient; DEAE Mono-Q, Pharmacia), and 1-mL fractions were collected. Aliquots (300 μL) corresponding to each fraction were stored at −80°C, while the different enzyme activities were detected by our standard enzyme assays or zymogram procedures.
D-enzyme activity was first monitored by measuring the production of Glc from maltotriose as follows. Amounts of crude extracts corresponding to 20 to 200 μg of total protein buffered in sodium acetate, pH 6.5, in a 1-mL final volume were incubated from 5 to 30 min at 30°C with 50 mm maltotriose. Glc was monitored by the Glc-6-P dehydrogenase assay as previously described (Ball et al., 1991).
The activity was followed on zymograms under either denaturing or native migration conditions. Denaturation of the extract was as previously described (Mouille et al., 1996). The equivalent of 50 to 200 μg of undenatured or denatured crude extract protein was loaded on a 30:1 (acry:bis), 7.5% (w/v) acrylamide, 1.5-mm-thick polyacrylamide gel (native conditions) or in a similar gel containing 0.1% (w/v) SDS (denaturing conditions). Migration at 15 V cM−1 was performed for 90 min at 4°C. At the end of the run, the denaturing gels were first subjected to renaturation as described in Mouille et al. (1996). The renatured and native gels were incubated overnight in the dark at room temperature in 30 mL of 3 mg mL−1 maltotriose; 200 mm Tris/HCl pH 8.0, 1 mm EDTA, 42 mm MgCl2, 0.014% (w/v) NADP, 0.027% (w/v) NAD, 0.027% (w/v) MTT, 0.015% (w/v) PMS, 1.5 mm ATP, 1 unit mL−1 hexokinase, and 0.5 unit mL−1 Glc-6-PDH. The gel was subsequently rinsed with distilled water and photographed.
To ascertain that the 62-kD Glc-producing activity displayed D-enzyme activity, we cut off 5-mm-wide strips of gels containing the renatured activity and incubated them in 0.5 mL of buffer containing 2 mm Glc, maltose, maltotriose, maltotetraose, maltopentaose, maltohexaose, and maltoheptaose. After overnight incubation, the gel strip was discarded and the buffer was lyophylized and redissolved in 50 μL of distilled water to be spotted immediately on a TLC plate (Silica Gel 60, Merck) in butanol:ethanol:water (5:5:4), and revealed by orcinol sulfuric staining.
Oligosaccharide Purification
Malto-oligosaccharides were prepared from 3 L of nitrogen-starved culture, inoculated at 105 cells mL−1, and harvested after 7 d of growth under continuous light (80 μE m−2 s−1) on high-salt acetate medium (Harris, 1989b) with gentle shaking. Algae were ruptured by passage in a French press cell (10,000 p.s.i.) at a density of 108 cells mL−1 in water. The crude extract was immediately frozen at −80°C. After thawing, cell debris were discarded by centrifugation at 10,000g for 15 min at 4°C. The supernatant was boiled for 5 min to inactivate enzymatic activities, and centrifuged at 10,000g for 15 min. The supernatant was then lyophilized, and the lyophilized material was resuspended in 1 mL of distilled water and loaded onto a Dowex 50:2 (1- × 6-cm) column immediately coupled to a Dowex 1:2 (1- × 6-cm) column equilibrated with water. Four microliters of each fraction (1 mL) was subjected to TLC (Silica Gel 60 column). Malto-oligosaccharides were revealed by spraying with orcinol (2 g L−1) dissolved in 20% (v/v) sulfuric acid. The TLC plate was subsequently incubated at 80°C for 10 min. After pooling and neutralization with one drop of 30% ammonium hydroxide, the pool was concentrated by rotary evaporation and desalted by gel filtration chromatography on a column (TSK HW-40, Merck) equilibrated in 0.5% (w/v) acetic acid. The desalted material was concentrated by rotary evaporation and lyophilized. The sample was kept at room temperature until it was subjected to NMR analysis.
Gene Dosage Experiments
Diploid and triploid strains were constructed as follows. To obtain the homozygous mutant diploid, we crossed CO 216 (mt− ac14 nit1 nit2 sta11-1) and CO 27 (mt+ pab2 nit1 nit2 sta11-1) and selected the diploid after 4 d of growth on minimal medium supplied with ammonium. Vegetative diploid strains heterozygous for mating type display an mt− mating type. After checking the phenotype, cellular volume, protein content, and mating type, we crossed the homozygous mutant either with CO 29 to obtain the homozygous triploid mutant or with strain 37 to obtain the sta11-1/sta11-1/+ triploid. To obtain the homozygous wild-type diploid, we crossed CO 218 (mt− ac14 nit1 nit2) and CO 42 (mt+ pab2 ac14). The colonies were selected on minimal medium supplied with acetate using nitrate as the nitrogen source (Ball et al., 1991).
The homozygous wild-type diploid was crossed with CO 35 (mt+pab2 nit1 nit2) to obtain the triploid wild-type (+/+/+) and with CO 26 (mt+pab2 nit1 nit2 sta11-1) to obtain +/+/sta11-1. The triploid strains were selected on minimal medium with nitrate as the nitrogen source. The heterozygous diploid strain was obtained by crossing JV45J and 37, and was selected on minimal medium with nitrate as the nitrogen source. After selection on the appropriate medium, the haploid, diploid, or triploid nature of the clones was confirmed by retesting the phenotypes and measuring both the average cell volume distribution and the cell protein content from unsynchronized cultures. We also confirmed the mating types of the clones. For each construct we selected three independent clones. Gene dosages are thus averages from three separate colonies for each construct. Phosphoglucomutase activity was assayed (Ball et al., 1991) and used as an internal standard during these experiments.
RESULTS
Isolation and Characterization of Low-Starch Mutants
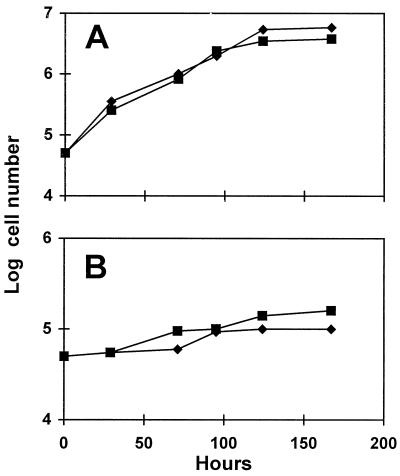
Among 5 × 104 colonies screened by the standard iodine-staining technique after UV mutagenesis of a wild-type C. reinhardtii strain (137C), we isolated a low-starch mutant (JV45J) that accumulated 8% of the normal amount of starch under conditions of maximum synthesis (Fig. 1). The defect behaved as a standard single-recessive Mendelian trait upon crossing. In addition, all diploids generated by crossing with reference mutant strains carrying a defect in the STA1, STA2, STA3, STA4, STA5, STA6, STA7, or STA8 genes proved to be wild type for starch amount and structure. Therefore, we defined a novel genetic locus, which we named STA11. In addition to starch, the sta11-1 mutants accumulated a significant amount (2% of the amount of starch in a wild-type strain) of water-soluble, amyloglucosidase-digestible material. No growth defects could be detected upon growing the mutants for over a week in continuous light with or without acetate or under a 12-h day/12-h night cycle with or without acetate (Fig. 2).
Figure 1.
Wild-type and mutant iodine-staining phenotype. Iodine stain of cell patches incubated for 7 d on solid nitrogen-deprived medium. Genotypes with respect to starch are indicated for our reference strains. sta2–27::ARG7, sta3–1, and sta7–5::ARG7 correspond to highly specific defects respectively in GBSS, SS, and debranching enzyme (Delrue et al., 1992; Fontaine et al., 1993, Mouille et al., 1996). +, Wild type. The original mutant strain JV45J (sta11-1) and two recombinants, CO199 and CO180, display the typical yellow stain of low-starch mutants, while the wild-type recombinant CO23 shows the typical dark-blue color of the wild-type reference strain.
Figure 2.
Growth curves of wild-type and mutant sta11-1 strains. Unsynchronized precultures of three mutant and three wild-type progeny from a cross involving the mutant JV45J and the wild-type strain 37 were grown in TAP medium (Harris, 1989b) to late log phase at 2 × 106 cells mL−1. The cultures were inoculated at 5 × 104 cells mL−1 and subjected to a 12-h d (80 μE m−2 s−1)/12-h night cycle of culture at 20°C with vigorous shaking. A, TAP medium (with acetate). Average mutant cell counts (▪) and wild-type (♦) cell counts are displayed as log of cell number versus time. B, TP medium (without acetate). Average mutant cell counts (▪) and wild-type (♦) cell counts are displayed as log of cell number versus time. The cultures are severely CO2 limited under these conditions and therefore grew at a very slow but comparable rate.
Characterization of the Water-Soluble, Amyloglucosidase-Digestible Material
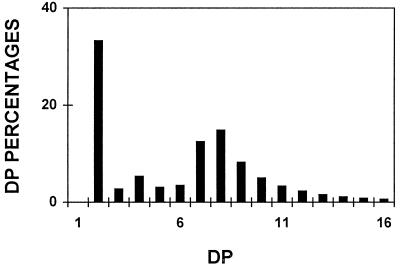
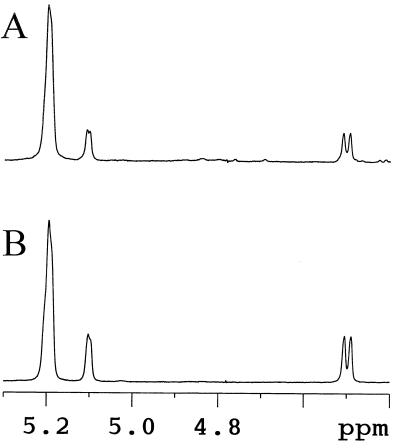
We found no evidence for the presence of high-mass, water-soluble polysaccharides. Indeed, the amount of water-soluble amyloglucosidase-digestible material excluded from the gel permeation chromatography column was less than 1% of the total water-soluble polysaccharide found in crude extracts. The water-soluble polysaccharide thus consisted solely of α-1,4-linked glucans, the size distribution of which is displayed in Figure 3. These glucans were further purified (Table I) and subjected to a detailed structural characterization. The size distribution before and after purification was identical. The mix of oligosaccharides generated an averaged 1H-NMR spectrum very similar to that of maltotriose standards (Fig. 4). As demonstrated by 1H-NMR analysis, branches, if present, fell below our detection level (1%). Co-segregation between the oligosaccharide fraction and the low-starch phenotype was found among all sta11-1-carrying strains tested (n = 50).
Figure 3.
Relative frequency distributions of oligosaccharides. Undebranched malto-oligosaccharides accumulated by the sta11-1 reference strain JV45J were separated according to length after APTS fluorescence labeling and separation on a DNA sequencer. Percentages of chains ranging between a DP of 1 to 16 (chains containing 1–16 Glc residues) are scaled on the y axis.
Table I.
Malto-oligosaccharide purification table
| Extract | Polysaccharide Quantitya | Yield |
|---|---|---|
| mg | % | |
| Crude extract | 3.1 | 100 |
| Lyophilizate | 2 | 65 |
| Dowex 50:2, 1:2 | 1.5 | 50 |
| TSK HW40 | 1.43 | 46 |
Polysaccharide amounts were measured by the standard amyloglucosidase assay. The purification reported here was from 3-L cultures of nitrogen-starved mutant strain JV45J.
Figure 4.
1H-NMR analysis of malto-oligosaccharides. Part of the 1H-NMR spectra of amylopectin in dimethyl-sulfoxyde-d6/2H2O (80:20) at 80°C is displayed. The chemical shifts for the α- and β-anomers of the reducing end are at 5.1 and 4.5 ppm, respectively. If present, α-1,6-linkage anomeric protons should be found at 4.85 ppm. The integration area of the α- and β-anomers reducing end signals divided by that of the α-1,4-linkage anomeric proton at 5.2 ppm yields the average DP of the sample. NMR conditions were similar to those described in Delrue et al. (1992). A, Malto-oligosaccharides purified from strain JV45J. B, Maltotriose reference.
Characterization of the Residual Mutant Starch
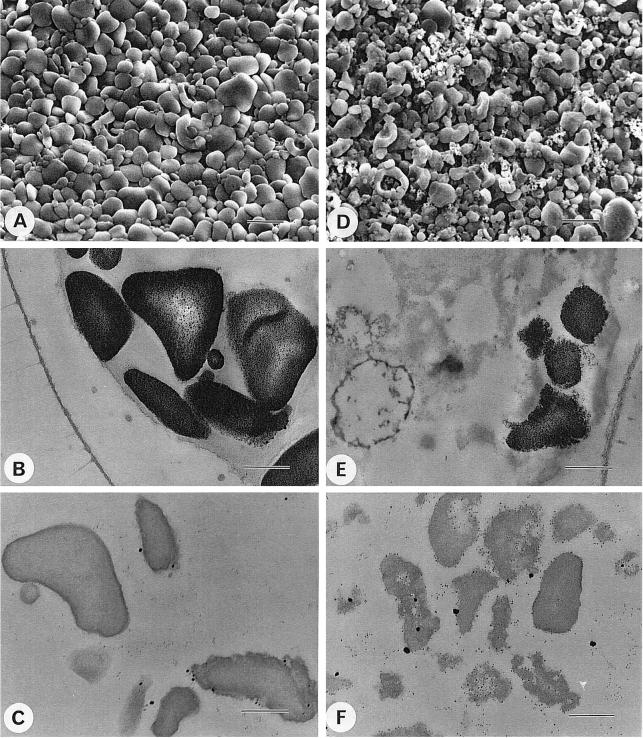
The residual starch structure was monitored through a variety of techniques, including wide-angle x-ray diffraction analysis (not shown), TEM and SEM (Fig. 5), separation of amylose and amylopectin through gel permeation chromatography (Fig. 6), and enzymatic debranching of the purified amylose and amylopectin (Fig. 7). The starch granules showed a significant relative increase in amylose (Table II). The distributions displayed in Figure 6, A and B, suggested a decrease in the mass of the amylose fraction. This was confirmed by mixing a low amount of labeled mutant starch with a high amount of wild-type reference polysaccharide on the same column. The x-ray diffractograms switched from the wild-type A-type lattice with high crystallinity to a mix of A- and B-type lattices with very low crystallinities. The shape of the granules was particularly affected, as illustrated in Figure 5.
Figure 5.
SEM and TEM of starches from wild-type and mutant sta11-1 strains. Electron micrographs of purified starches and in cells from nitrogen-starved wild-type (137C) and mutant sta11-1 (JV45J) C. reinhardtii strains. A to C, Wild-type strain 137C; D to F, mutant sta11-1 (JV45J) strain. A and D, SEM of purified starches (bar = 2 μm); B and E, TEM of starch-containing cells after PATAg staining (bar = 0.5 μm); C and F, TEM of purified starches after PATAg staining (bar = 0.5 μm).
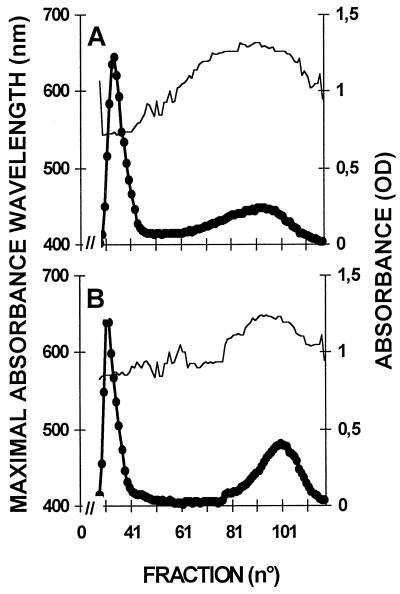
Figure 6.
Separation of amylopectin and amylose by CL2B-Sepharose chromatography. The optical density (•) was measured for each 2-mL fraction at λmax (unbroken thin line). All samples were loaded on the same column setup described by Delrue et al. (1992). The wild-type haploid 137C strain starch extracted from nitrogen-starved cultures (storage starch) (A) displays both amylopectin and low-Mr amylose. B, Starch from the mutant strain JV45J carrying the sta11-1 mutation. Starch was also extracted under nitrogen starvation (storage starch). Quantification of amylose and amylopectin ratios was obtained by pooling amylopectin and amylose fractions separately and measuring the amount of Glc through the standard amyloglucosidase assay.
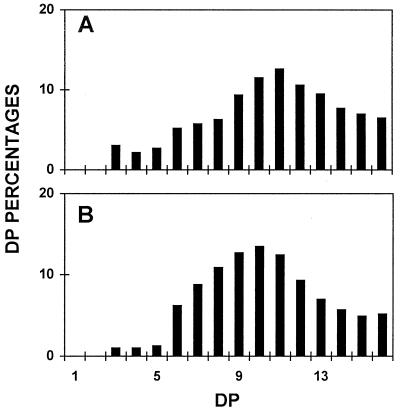
Figure 7.
Chain-length distributions of wild-type and mutant amylopectin. Isoamylase-debranched chains were separated according to length after APTS fluorescence labeling and separation on a DNA sequencer. Percentages of chains ranging between DP 1 to 16 (chains containing 1–16 Glc residues) are scaled on the y axis. A, Debranched chains of gel permeation chromatography-purified amylopectin from the mutant JV45J. B, Debranched chains from gel permeation chromatography-purified C. reinhardtii reference amylopectin (extracted from the amylose-free BAFR1 strain).
Table II.
Phenotype of wild-type and mutant strains during storage or transitory starch synthesis
| Strain | Genotype | λmaxa
|
Starchb
|
MOSc
|
Amd
|
||||
|---|---|---|---|---|---|---|---|---|---|
| +N | −N | +N | −N | +N | −N | +N | −N | ||
| % | |||||||||
| CO23 | + | 566 | 560 | 1.2 | 13 | 0.011 | 0.018 | 1 | 14 |
| CO65 | + | 570 | 542 | 0.4 | 24.7 | 0.007 | 0.001 | 2 | 15 |
| CO35 | + | 564 | 554 | 0.91 | 22.8 | 0.012 | 0.017 | 5 | 25 |
| CO29 | sta11-1 | 576 | 570 | 1 | 1.7 | 0.45 | 0.45 | 7 | 38 |
| CO137 | sta11-1 | 572 | 564 | 2 | 0.83 | 0.3 | 0.17 | 10 | 25 |
| CO214 | sta11-1 | 575 | 562 | 0.31 | 0.78 | 0.2 | 0.2 | 12 | 24 |
Values listed are average of three separate measures in a single experiment.
Wavelength of maximal absorbance of the iodine-polysaccharide complex of amylopectin purified by gel filtration.
Amount of insoluble polysaccharide, expressed in μg 10−6 cells, purified through sedimentation as measured by the standard amyloglucosidase assay.
Amount of soluble malto-oligosaccharides expressed in μg 10−6 cells. 0.5 μg 10−6 cells corresponds to a plastidial concentration of 10 mm if all MOS was to be considered as maltotriose.
The percentage of amylose in the purified starch was calculated after gel filtration of the dispersed polysaccharides.
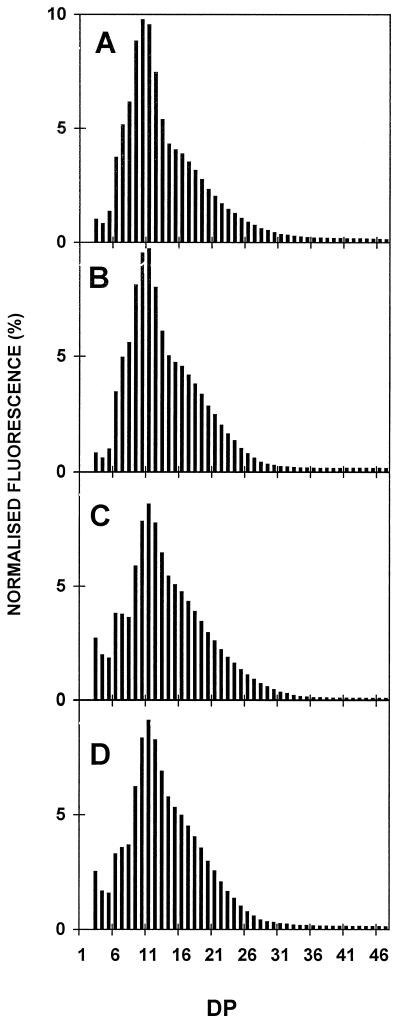
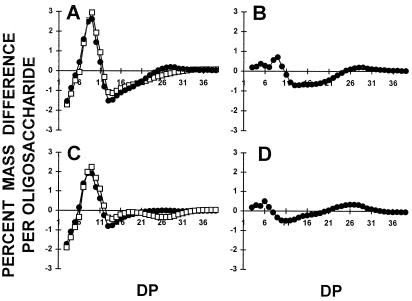
In addition, the starch granules decreased their overall mean size and the regularly shaped smooth surface became rugged and highly irregular. The purified amylopectin and amylose were subjected to enzymatic debranching, labeling of the reducing ends by APTS, and separation of the chains according to length on DNA sequencing gels. While no changes were detected in the amylose chain-length distributions, a significant modification of the very small chain-length distribution (increase of DP 3–5 and, to a lesser extent, DP 12–16 at the expense of DP 6–11) could be detected (compare Fig. 7A with Fig. 7B). Capillary electrophoresis was performed to confirm those differences between four wild-type and four mutant strains. An example is shown for four of these strains (Fig. 8), and results are summarized by subtractive analysis in Figure 9. Figure 9 clearly confirms the enrichment found in the mutants in the extra-short glucan range (DP 3–5 at the expense of DP 6–11). The increase previously noted for DP 12 to 14, while significant, is not far from the natural variation range for each genotype class. These results were confirmed on the two other pairs of wild-type and mutant recombinants.
Figure 8.
Chain-length distributions of amylopectin from wild-type and mutant strains. Distribution of chain lengths of wild-type and mutant amylopectin after isoamylase-mediated debranching were confirmed by capillary electrophoresis of APTS-labeled glucans following a procedure previously described (O'Shea et al., 1998). The relative amount of chains corresponding to each DP is strictly equivalent to the normalized fluorescence percentage. A and B correspond to wild-type strains 137C (A) and CO65 (B), while C and D correspond to sta11-1-carrying strains JV45J (C) and CO29 (D).
Figure 9.
Comparison of the normalized masses of APTS-labeled oligosaccharides from isoamylase debranched amylopectin. The percentage difference of the total mass present in each individual oligosaccharide has been obtained by subtracting the chain-length distribution from debranched amylopectins. A, Subtractive analysis from the wild-type reference strain 137C minus that of the mutant sta11-1 JV45J (□) and from the wild-type strain 137C minus that of the mutant sta11-1 CO29 (•). B, Subtractive analysis from the wild-type reference strain 137C minus that of the wild-type strain CO65 (•). C, Subtractive analysis from the wild-type reference strain CO65 minus that of the mutant sta11-1 JV45J (□) and from the wild-type strain CO65 minus that of the mutant sta11-1 CO29 (•). D, Subtractive analysis from the mutant reference strain JV45J minus that of the mutant CO29 (•).
Finding the Enzymatic Defect
A detailed enzymological analysis was done in crude and partially purified extracts for all major enzymes reported to be involved in starch biosynthesis. This study involved quantitative and qualitative measures of enzyme activities, together with kinetic characterizations and investigations of the elution behavior during FPLC on anion-exchange columns. The enzymes tested include ADP-Glc pyrophosphorylase, phosphoglucomutase, SS I, SS II, GBSS, both types of branching enzymes, debranching enzymes (limit-dextrinase and isoamylase), phosphorylases, maltase (α-glucosidase), and all starch hydrolases that could be detected in starch-containing zymogram gels (Table III).
Table III.
Enzyme activities in wild-type and mutant sta11-1 progeny
| Enzyme | Wild-Type Strains | Mutant Strains | Zymogram |
|---|---|---|---|
| ADP-Glc pyrophosphorylase | 3.2 ± 0.2 (4)a | 4.2 ± 0.2 (6) | No |
| Starch phosphorylase | 20 ± 4 (6) | 20 ± 2 (4) | Yes (7, 15)b |
| Phosphoglucomutase | 6.7 ± 2.9 (6) | 6 ± 2 (4) | Yes (10, 8) |
| SS | 2.4 ± 1 (5) | 2.3 ± 0.7 (4) | Yes (9, 9) |
| Branching enzyme | 0.6 ± 0.2 (6) | 0.5 ± 0.2 (6) | Yes (6, 4) |
| GBSS | 45 ± 10 (2) | 42 ± 8 (3) | No |
| α-Glucosidase | 5.8 ± 0.8 (9) | 4.6 ± 1 (7) | No |
| Limit-dextrinase | 0.37 ± 0.1 (6) | 0.34 ± 0.1 (6) | No |
| α-Amylase | N/A | N/A | Yes (22, 20) |
| Isoamylase | N/A | N/A | Yes (22, 20) |
| D-enzyme | 83 ± 4 (10) | 1.3 ± 0.5 (10) | Yes (36, 39) |
AGPase (assayed in direction of pyrophosphorolysis in the presence of 1.5 mm 3-PGA), starch phosphorylase, and phosphoglucomutase units are expressed in nmol Glc-1-P produced min−1 mg−1 protein. SS and GBSS are expressed in nmol ADP-Glc incorporated into polysaccharide min−1 mg−1 protein (SS) or mg starch (GBSSI). Branching enzyme is expressed as nmol Glc-1-P incorporated into polysaccharide min−1 mg−1 protein (phosphorylase amplification assay). Limit-dextrinase and D-enzyme are expressed in nmol maltotriose formed from pullulan min−1 mg−1 protein and nmoles Glc formed from maltotriose min−1 mg−1 protein, respectively. α-Glucosidase activities are expressed in nmol Glc formed from maltose min−1 mg−1 protein. N/A, Not applicable.
The numbers in parentheses correspond to the number of different strains examined.
The two numbers in parentheses correspond to the number of wild-type and mutant strains examined, respectively.
No qualitative nor quantitative differences in these enzyme activities were found to co-segregate with the mutant gene. However, when we applied the standard assay used in plants to measure D-enzyme activity, we were surprised to find a 99% decrease in the rate of Glc production from maltotriose (Table III). The mutants were also clearly defective in the production of Glc from maltotetraose, maltopentaose, maltohexaose, and maltoheptaose. However, the production of Glc from maltose was comparable in mutant and wild-type strains. This production amounted to less than 5% of that measured with maltotriose in the wild-type progeny. We therefore assume that the production of Glc from maltoriose by α-glucosidase or other hydrolases is negligible in face of the amount of D-enzyme activity present in our extracts. This result is similar to that reported for higher plant leaf extracts (Zeeman et al., 1998a).
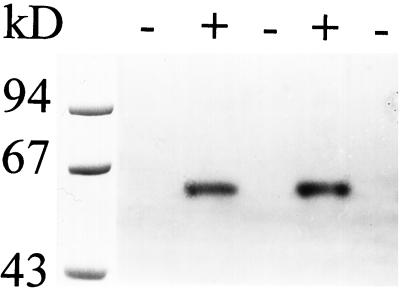
Glc production from maltotriose thus defines a quantitative and specific assay for D-enzyme activity in C. reinhardtii extracts. We adapted the zymogram techniques used to detect Glc production to assay the action of D-enzyme on maltotriose (Fig. 10). We were able to detect the activity after denaturation and renaturation, which allowed us to estimate the mass of the enzyme. This 62-kD band was absent in all meiotic progeny bearing the mutation (n = 75) and was systematically present in wild-type strains. To make sure that the 62-kD band contained oligosaccharide-disproportionating activity, we incubated gel slices in buffer containing Glc, maltose, maltotriose, maltotetraose, maltopentaose, maltohexaose, or maltoheptaose (not shown). The lyophilized incubation buffers were then spotted onto TLC plates and the oligosaccharides revealed by the orcinol-sulfuric acid method (Mouille et al., 1996). The 62-kD band obeyed the rules set years ago by Peat et al. (1956) to define D-enzymes: it could not use Glc or maltose as sole substrates, but could disproportionate α-1,4-linked oligosaccharides of at least three Glc residues into longer oligosaccharides.
Figure 10.
The enzymatic defect of sta11-1 mutant strains. Denatured crude extracts (100 μg of protein) from two wild-type (+) and three sta11-1 (−) were loaded on denaturing polyacrylamide gels. The proteins were renatured after electrophoresis and incubated overnight. Glc production was revealed after overnight incubation of the gel with 3 mg mL−1 maltotriose. The 62-kD blue-staining band can be easily distinguished in the wild-type cells. This band contained the D-enzyme activity.
Gene Dosage Experiments
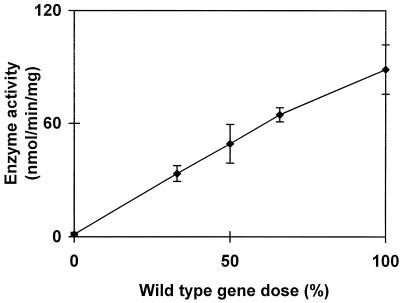
We were able to monitor the amount of enzyme activity present within the plastids (8.7 μmol of Glc formed from maltotriose min−1 mL−1) or to perform gene dosage experiments in diploid and triploid zygotes. The enzyme activity correlated with the relative amount of wild-type alleles, as would be expected if STA11 encoded D-enzyme (Fig. 11). Knowing the cell and organelle volumes of the strains used in this work, we were able to calculate the physiological enzyme concentrations (Schötz et al., 1972). We were also able to estimate the amount of malto-oligosaccharides in wild-type (50–200 μg mL−1 of chloroplast) and mutant (0.5–1 mg mL−1) strains.
Figure 11.
Gene dosages expressed from 0% (homozygous mutant) to 100% (homozygous wild-type); 50% corresponds to the heterozygous diploids, while 33% and 66% correspond to a sta11-1/sta11-1/+ and a sta11-1/+/+ triploid respectively. Haploid, diploid, or triploid homozygous combinations gave identical results when the activity was expressed per milligram of protein. Means (♦) and sds of measures from three different diploid or triploid constructs were calculated for each gene dose. Total phosphoglucomutase-specific activities were monitored as internal controls and proved similar in all constructs (2.5 ± 0.4 nmol Glc-6-P formed from Glc-1-P min−1 mg−1 protein).
The Expression of the Mutant Phenotype Is Partly Conditional
We have previously noted that expressivity of mutant phenotypes can vary when transitory starch is compared with the more classical storage form of the polysaccharide (Libessart et al., 1995). In C. reinhardtii, storage starch synthesis is obtained by using nutrient starvation conditions, leading to the arrest of cell division and to the accumulation of starch. On the other hand, transitory starch synthesis is obtained under conditions of active photosynthesis and cell division. Mutant phenotypes of C. reinhardtii fall into three classes. The first class concerns mutations whose phenotypes express themselves equally in both physiological conditions. Mutations affecting the small and large subunits of ADP-Glc pyrophosphorylase, GBSS, and debranching enzyme fall within this class (Ball et al., 1991; Delrue et al., 1992; Libessart et al., 1995; Mouille et al., 1996; Dauvillée et al., 1999). The functions thereby defined are said to be mandatory for either amylose or starch synthesis.
The second class contains mutations expressed in both conditions but with lesser expressivity upon transitory starch. These include mutations affecting the major SS (Libessart et al., 1995). These functions are thus involved in the process of normal starch synthesis, but cannot be defined as mandatory in C. reinhardtii. We believe this to be the result of some level of functional redundancy due either to the presence of multiple enzyme forms belonging to same family or to the presence of alternative pathways such as those defined by hydrolysis or phosphorolysis. Finally, some loci, when defective, lead to a detectable phenotype only in storage conditions. Their expression is said to be conditional. This applies to the high-amylose sta4 mutants, for which no enzymatic defect has yet been reported (Libessart et al., 1995).
We compared the phenotypes recorded on the wild-type and mutant sta11-1 offspring during nitrogen starvation with those obtained during normal growth. An example of such an analysis is displayed in Table II. The results clearly define the sta11-1 mutant defect as partly conditional, and therefore it belongs to the second class. Maximal expression of the defect is obtained under nitrogen starvation, where decreases in starch exceeding 90%, together with increases in amylose percentage and malto-oligosaccharide content, are obtained. However, only the increases in malto-oligosaccharide and amylose percentages are systematically recorded in undepleted medium. We believe this defines STA11 as being required but not mandatory for normal starch biosynthesis.
DISCUSSION
It was recently reported that a 98% reduction in D-enzyme activity did not affect starch biosynthesis or structure in potato (Takaha et al., 1998). However, there are several known instances in which potato antisense RNA technology has yet to confirm phenotypes of mutants obtained in a wide variety of starch-synthesizing species. We believe this to be due to the presence in many cases of a sufficient amount of residual wild-type activity to carry out the steps, which could be far from rate controlling. This study establishes a correlation between the disappearance of D-enzyme and major alterations in starch structure and accumulation, and thus raises the possibility that this enzyme is a constituent of the starch biosynthetic pathway.
In the progeny of a cross between wild type and sta11-1 mutant strains, co-segregation was observed for the phenotypes of D-enzyme deficiency, malto-oligosaccharide accumulation, and specific starch structural defects. That unbranched malto-oligosaccharides accumulated in the mutant progeny was to be expected, because these are the normal substrates of D-enzymes. The relative abundance of maltose to that of longer oligosaccharides fluctuates because of the presence of other starch hydrolases. Maltose production probably results from the action of α-amylase on longer oligosaccharides that would otherwise accumulate in the mutants. However, the major changes in starch biosynthesis in conditions of nitrogen starvation were unexpected in light of what is currently known about the functions of D-enzyme, in particular because these enzymes are usually assumed to be utilized for catabolism of unbranched malto-oligosaccharides. The D-enzyme deficiency co-segregated not only with a reduction in total glucopolysaccharide accumulation, but also with abnormal starch granule shape and size, an altered amylose/amylopectin ratio in the mutant granules, and changes in the chain-length distribution of amylopectin in those granules. To explain these observations we propose that D-enzyme has a specific function in the process of starch biosynthesis.
If the absence of D-enzyme is not responsible for the starch biosynthesis defects, then the alternative explanation requires that sta11-1, through pleiotropic effects, causes a deficiency of more than one enzyme. This situation could result from, for instance, the collapse of a specific multienzyme complex or from some complex feedback-regulatory mechanism acting on enzymes that have not yet been characterized. In this view an unidentified enzyme other than D-enzyme is also affected in the sta11-1 mutant strains, and is required for normal starch biosynthesis. We consider this possibility unlikely because all known enzyme activities in the starch biosynthesis pathway have been monitored very carefully in the mutant strains.
Some minor quantitative and qualitative fluctuations were observed, however, and were found equally in the mutant and nonmutant progeny in the segregating population; therefore, they cannot be relevant to the observed starch biosynthesis phenotype. Furthermore, complementation tests established that STA11 is distinct from any genetic element known in C. reinhardtii to code for or determine activity of an enzyme involved in starch biogenesis, including the genes required for GBSS I, SS II, phosphoglucomutase, the ADP-Glc pyrophosphorylase large and small subunits, and a starch-debranching enzyme. Although this alternative explanation cannot be definitively excluded, the most straightforward explanation of the phenotype caused by sta11-1 is that D-enzyme itself is required for normal starch biosynthesis.
Gene dosage experiments in diploid and triploid genetic backgrounds established a very good correlation between the amount of D-enzyme activity and the STA11 wild-type allele dose. These data suggest that STA11 codes for D-enzyme, although further analysis is required to characterize definitively the nature of this gene. Even if STA11 is proven to code for D-enzyme, however, the possibility of a pleiotropic effect causing the starch structural defects in sta11-1 mutants cannot be ruled out definitively. Complementation of the sta11-1 defect by transgenesis of a wild-type D-enzyme gene is entailed with similar limitations. Gene cloning, however, is inherently difficult in C. reinhardtii owing to technical limitations such as the absence of powerful yeast-type shuttle vectors precluding complementation cloning and the absence of complete banks of tagged mutants.
We expect, however, that application of this powerful genetic screening system to identify a previously unsuspected enzyme likely involved in starch biosynthesis enables analysis in plants such as maize or Arabidopsis. In such an application, specific structural gene mutations could be identified by methods such as transposon insertion screening. What is needed at present, before any discussion is made on the mutant phenotype and its relevance to our understanding of starch biosynthesis, is to investigate the D-enzyme wild-type activity in greater detail. In particular, we would like to determine if this enzyme could be directly involved in the building of the wild-type amylopectin structure. D-enzyme has been documented as working essentially on soluble malto-oligosaccharides, and we have thus proceeded to further investigate its activity on amylopectin structure.
ACKNOWLEDGMENTS
We thank André Decq and Jocelyn Celen for their excellent technical assistance.
Abbreviations:
- APTS
8-amino-1,3,6-pyrenetrisulfonic acid
- D-enzyme
disproportionating enzyme
- DP
degree of polymerization
- GBSS
granule-bound starch synthase
- SEM
scanning electron microscopy
- SS
soluble starch synthase
- TEM
transmission electron microscopy
Footnotes
This work was supported by grants from the Ministère de l'Education Nationale, by the Centre National de la Recherche Scientifique (Unité Mixte de Recherche du Centre National de la Recherche Scientifique no. 8576, Director André Verbert), by the University of Lille, by the University of Reims, and by a grant from Biogemma (Cambridge, UK).
LITERATURE CITED
- Ball S. Regulation of starch biosynthesis. In: Rochaix J-D, Goldschmidt-Clermont M, Merchant S, Govindjee, editors. The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Advances in Photosynthesis, Vol 7. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 549–567. [Google Scholar]
- Ball S, Dirick L, Decq A, Martiat JC, Matagne RF. Plant Sci. 1990;66:1–9. [Google Scholar]
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J. From glycogen to amylopectin: a model explaining the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- Ball S, Marianne T, Dirick L, Fresnoy M, Delrue B, Decq A. A Chlamydomonas reinhardtii low-starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta. 1991;185:17–26. doi: 10.1007/BF00194509. [DOI] [PubMed] [Google Scholar]
- Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buléon A, Colonna P, Planchot V, Ball S. Starch granules: structure and biosynthesis. Int J Biol Macromol. 1998;23:85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- Buléon A, Gallant DJ, Bouchet B, Mouille G, D'Hulst C, Kossman J, Ball SG. Starches from A to C: Chlamydomonas reinhardtii as a model microbial system to investigate the biosynthesis of the plant amylopectin crystal. Plant Physiol. 1997;115:949–957. doi: 10.1104/pp.115.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D, Colleoni C, Shaw E, Mouille G, D'Hulst C, Morell M, Samuel MS, Bouchet B, Gallant DJ, Sinskey A. Plant Physiol. 1999;119:321–330. doi: 10.1104/pp.119.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski JM, Van Den Koornhuyse N, Maddelein M-L, Fournet B, Ball S. Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase accumulate a structurally modified amylopectin. J Bacteriol. 1992;174:3612–3620. doi: 10.1128/jb.174.11.3612-3620.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine T, D'Hulst C, Maddelein M-L, Routier F, Marianne-Pepin T, Decq A, Wieruszeski JM, Delrue B, Van Den Koornhuyse N, Bossu JP. Toward an understanding of the biogenesis of the starch granule: evidence that Chlamydomonas soluble starch starch synthase II controls the synthesis of intermediate size glucans of amylopectin. J Biol Chem. 1993;268:16223–16230. [PubMed] [Google Scholar]
- Harris EH. Genetic analysis. In: Harris E, editor. The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989a. pp. 399–446. [DOI] [PubMed] [Google Scholar]
- Harris EH. Culture and storage methods. In: Harris E, editor. The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989b. pp. 25–63. [DOI] [PubMed] [Google Scholar]
- James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libessart N, Maddelein M-L, Van Den Koornhuyse N, Decq A, Delrue B, Ball SG. Storage, photosynthesis and growth: the conditional nature of mutations affecting starch synthesis and structure in Chlamydomonas reinhardtii. Plant Cell. 1995;7:1117–1127. doi: 10.1105/tpc.7.8.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Maddelein M-L, Libessart N, Talaga P, Decq A, Delrue B, Ball S. Phytoglycogen processing: a mandatory step for starch biosynthesis in plants. Plant Cell. 1996;8:1353–1366. doi: 10.1105/tpc.8.8.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Umemoto T, Takahata Y, Komae K, Amano E, Satoh H. Changes in structure of starch and enzyme activities affected by sugary mutations in developing rice endosperm: possible role of starch debranching enzyme in amylopectin biosynthesis. Physiol Plant. 1996;97:491–498. [Google Scholar]
- O'Shea MG, Morell MK. High resolution slab gel electrophoresis of 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis. 1996;17:681–688. doi: 10.1002/elps.1150170410. [DOI] [PubMed] [Google Scholar]
- O'Shea MG, Samuel MS, Konik CM, Morell MK. Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labelling and high-resolution separation. Carbohydr Res. 1998;307:1–12. [Google Scholar]
- Peat S, Whelan WJ, Rees WR. The enzymic synthesis and degradation of starch: the disproportionating enzyme of potato. J Chem Soc. 1956;1956:44–53. [Google Scholar]
- Schötz F, Bathelt H, Arnold CG, Schimmer Die architektur und organisation der Chlamydomonas-zelle: ergebnisse der elektronenmikroskopie von seriensschnitten und der daraus resultierenden dreidimensionalen rekonstruktion. Protoplasma. 1972;75:229–254. doi: 10.1007/BF01279818. [DOI] [PubMed] [Google Scholar]
- Takaha T, Critchley J, Okada S, Smith SM. -glucanotransferase) Planta. 1998;205:445–451. [Google Scholar]
- Van den Koornhuyse N, Libessart N, Delrue B, Zabawinski C, Decq A, Iglesias A, Preiss J, Ball S. Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J Biol Chem. 1996;271:16281–16288. doi: 10.1074/jbc.271.27.16281. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, Rees T. A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch hydrolyzing enzyme. Plant J. 1998a;15:357–365. doi: 10.1046/j.1365-313x.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Umemoto T, Lue WL, Au-Yeung P, Martin C, Smith AM, Chen J. A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell. 1998b;10:1699–1712. doi: 10.1105/tpc.10.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]