Abstract
Background
This study aimed to establish an effective predictive nomogram for non-small cell lung cancer (NSCLC) patients with chronic hepatitis B viral (HBV) infection.
Methods
The nomogram was based on a retrospective study of 230 NSCLC patients with chronic HBV infection. The predictive accuracy and discriminative ability of the nomogram were determined by a concordance index (C-index), calibration plot and decision curve analysis and were compared with the current tumor, node, and metastasis (TNM) staging system.
Results
Independent factors derived from Kaplan–Meier analysis of the primary cohort to predict overall survival (OS) were all assembled into a Cox proportional hazards regression model to build the nomogram model. The final model included age, tumor size, TNM stage, treatment, apolipoprotein A-I, apolipoprotein B, glutamyl transpeptidase and lactate dehydrogenase. The calibration curve for the probability of OS showed that the nomogram-based predictions were in good agreement with the actual observations. The C-index of the model for predicting OS had a superior discrimination power compared with the TNM staging system [0.780 (95% CI 0.733–0.827) vs. 0.693 (95% CI 0.640–0.746), P < 0.01], and the decision curve analyses showed that the nomogram model had a higher overall net benefit than did the TNM stage. Based on the total prognostic scores (TPS) of the nomogram, we further subdivided the study cohort into three groups: low risk (TPS ≤ 13.5), intermediate risk (13.5 < TPS ≤ 20.0) and high risk (TPS > 20.0).
Conclusion
The proposed nomogram model resulted in more accurate prognostic prediction for NSCLC patients with chronic HBV infection.
Keywords: Hepatitis B viral, Liver function, Nomogram, Non-small cell lung cancer, Prognosis
Background
Lung cancer remains the most common cause of cancer deaths worldwide. The recent literature reported that 5-year survival rate of lung cancer was 18% [1]. More than 80% of total lung cancer cases are non-small cell lung cancer (NSCLC) [2]. The low survival rates of NSCLC patients are affected by many factors, including poor early diagnosis, tumor recurrence, and distant metastasis.
Chronic hepatitis B viral (HBV) infection is also a serious global public problem. Two billion people are estimated to be infected with HBV worldwide [3]. In particular, China has a high prevalence of HBV infections, and HBV patients in China account for approximately 38% of all patients worldwide [4]. A national epidemiology survey announced in April 2008 by the Ministry of Health showed that 93 million people in China had been infected with HBV.
Numerous studies have reported that many prognostic factors are correlated with the prognosis of NSCLC patients. The prognostic factors include liver function indicators (LFIs), such as aspartate aminotransferase (AST) [5], lactate dehydrogenase (LDH) [6], and alkaline phosphatase (ALP) [7], among others.
The risk of liver injury may increase when HBV infects patients. Most studies suggest that liver injury in viral hepatitis is not caused by the direct cytopathic effects of viruses but by the host immune response to viral proteins expressed by infected hepatocytes [8], which causes liver dysfunction. Research has shown that chronic HBV infection is an independent prognostic factor in patients with nasopharyngeal carcinoma [9], pancreatic cancer [10] and NSCLC [11]. These results suggest that NSCLC patients with HBV infection should be distinguished from those without HBV infection because they have different clinicopathological characteristics, prognostic factors, and outcomes after treatment, which require a distinct prognostic, predictive model.
Nomogram is currently widely applied as graphical representations of complex mathematical formulas. They can integrate essential factors to build a statistical prognostic model for estimating prognosis in the outcomes of many cancers [12, 13]. Furthermore, nomogram has been shown to make more precise predictions than do the traditional staging systems used in many cancers [14, 15]. However, no study has established a prognostic nomogram for NSCLC patients with HBV. Therefore, our study aimed to develop a practical clinical tool by combining clinicopathologic factors and markers of liver function tests. We also tested whether the nomogram model provides a more accurate prediction of patient survival than does the 7th edition of the American Joint Committee on Cancer (AJCC) TNM Staging.
Methods
Patients and study design
A retrospective observational study was performed including a total of 230 NSCLC patients with chronic HBV infection, and the patients first visited Sun Yat-sen University Cancer Center (Guangzhou, China) between January 2008 and December 2010. The inclusion criteria were as follows: (1) patients with a confirmed pathological diagnosis of NSCLC; (2) patients who were positive for the hepatitis B surface antigen (HBsAg), excluding acute hepatitis; (3) patients without co-infection of other types of hepatitis viruses; (4) patients with complete clinical data; (5) patients without secondary carcinomas as assessed by clinical history, computed tomography (CT), ultrasonographic examination and routine laboratory tests; and (6) patients without liver fibrosis, steatosis, and cirrhosis as detected by CT or ultrasonographic examination.
All the samples were collected at the time of diagnosis before any treatment. We obtained the patients’ clinicopathologic parameters including gender, age, family history, smoking history, body mass index (BMI), pathologic TNM stage [16], tumor size, and treatment history. LFIs including alanine transaminase (ALT), aspartate aminotransferase (AST), the AST-to-ALT ratio (SLR), apolipoprotein A-I (APOAI), apolipoprotein B (APOB), alkaline phosphatase (ALP), albumin (ALB), glutamyl transpeptidase (GGT), lactate dehydrogenase (LDH), total bilirubin (TBIL) and direct bilirubin (DBIL) and HBV infection markers including HbsAg, hepatitis B surface antibody (HbsAb), hepatitis B e antigen (HbeAg), hepatitis B e antibody (HbeAb) and hepatitis B core antibody (HbcAb) were recorded.
We randomly divided the patients into a primary cohort and a validation cohort. Computer-generated random numbers were used to assign 141 of the patients to the primary cohort and 89 patients to the validation cohort. The overall survival (OS) of the patients was recorded based on a follow-up clinical visit or a telephone call. The OS was calculated from the time of initial diagnosis until the time of death from any cause, or until the last follow-up. All the patients were followed up until death or January 2016, if still alive. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (http://www.researchdata.org.cn), with the approval RDD Number as RDDA2018000554.
Laboratory measurements
ALT, AST, APOAI, APOB, ALP, ALB, GGT, LDH, TBIL, and DBIL were measured using a Hitachi 7600 Automatic Analyzer (Tokyo, Japan). HBsAg, HbsAb, HbeAg, HbeAB, and HbcAb were detected by enzyme-linked immunosorbent assay (ELISA) technology. The values of all the variables tested a few days before pretreatment were recorded. The SLR was defined as the serum AST level divided by the serum ALT level. The coefficient of variation for these methods over the range of measurements was < 5% as established by routine quality control procedures.
Statistical analysis
Categorical variables were classified based on clinical findings, and continuous variables were transformed into categorical variables based on cutoff points, which were determined by the minimum P value from log-rank ×2 statistics using the X-tile program [17]. Survival curves were depicted using the Kaplan–Meier method and compared with a log-rank test stratified according to the prognostic factors. The P values of variables less than or equal to 0.05 in the univariate analyses were incorporated into the Cox’s proportional hazards regression. A predictive nomogram model was built based on the Cox model parameter estimates in the primary cohort, and the selection of the final prediction model was performed with a backward step-down selection process using the Akaike information criterion [18]. The accuracy of the nomogram model was estimated by the Harrell’s C-index (C-index). The value of the C-index ranges from 0.5 to 1.0, with 0.5 indicating random chance and 1.0 indicating a perfect ability to correctly discriminate the outcome with the model. Validation was performed using a bootstrap method to quantify our modeling strategy. Finally, a calibration curve of the nomogram model for the 1-, 3-, and 5-year OS and decision curve analyses [19] was plotted to assess the predictive value of the model. The nomogram model was divided into three groups (low-risk prognosis, intermediate-risk prognosis, and high-risk prognosis) based on the total prognostic scores (TPS) in the primary cohort and validation cohort. Correlation analysis was adopted using Pearson’s correlation. All the statistical analyses and graphics were performed using the SPSS 20.0 statistical package (SPSS Inc., Chicago, IL, USA) and R version 3.3.2 (http://www.r-project.org/). P values less than 0.05 were considered statistically significant.
Results
Patient characteristics
The study enrolled a total of 230 patients. We randomly divided the patients into a primary cohort and a validation cohort. The patients’ demographic and clinical characteristics are listed in Table 1. There were 141 patients in the primary cohort, comprising 72 male patients (51.1%) and 69 female patients (48.9%), and the age of the patients ranged from 27 to 82 years. The median follow-up time was 33 months. The validation cohort included 50 male patients (56.2%) and 39 female patients (43.8%), and the age of the patients ranged from 33 to 79 years. The median follow-up time was 35 months. The 5-year OS rates for the primary cohort and the validation cohort were 33.3 and 33.7%, respectively.
Table 1.
Patient demographics and clinical characteristics
| Primary cohort | Validation cohort | |
|---|---|---|
| n = (141) | n = (89) | |
| Characteristic | No. (%) | No. (%) |
| Gender | ||
| Male | 72 (51.1%) | 50 (56.2%) |
| Female | 69 (48.9%) | 39 (43.8%) |
| Age (years) | ||
| ≤ 42 | 19 (13.5%) | 11 (12.4%) |
| > 42 | 122 (86.5%) | 78 (87.6%) |
| Family history | ||
| Yes | 36 (25.5%) | 18 (20.2%) |
| No | 105 (74.5%) | 71 (79.8%) |
| Smoking behaviour | ||
| Yes | 84 (59.6%) | 50 (56.2%) |
| No | 57 (40.4%) | 39 (43.8%) |
| BMI | ||
| ≥ 25 | 30 (21.3%) | 14 (15.7%) |
| 18.5 ≤ BMI < 25 | 103 (73.0%) | 65 (73.0%) |
| < 18.5 | 8 (5.7%) | 10 (11.3%) |
| TNM stagea | ||
| I | 31 (22.0%) | 23 (25.8%) |
| II | 17 (12.1%) | 6 (6.7%) |
| III | 53 (37.6%) | 34 (38.2%) |
| IV | 40 (28.3%) | 26 (29.3%) |
| Size (cm)b | ||
| ≤ 6.3 | 122 (86.5%) | 70 (78.7%) |
| > 6.3 | 19 (13.5%) | 19 (21.3%) |
| Treatment | ||
| Surgery | 37 (26.2%) | 21 (23.6%) |
| Radiotherapy/chemotherapy | 47 (33.3%) | 31 (34.8%) |
| Surgery and radiotherapy/chemotherapy | 49 (34.8%) | 25 (28.1%) |
| Other | 8 (5.7%) | 12 (13.5%) |
| ALT (U/L) | ||
| ≤ 12.5 | 15 (10.6%) | 11 (12.4%) |
| > 12.5 | 126 (89.4%) | 78 (87.6%) |
| AST (U/L) | ||
| ≤ 22.0 | 73 (51.8%) | 45 (50.1%) |
| > 22.0 | 68 (48.2%) | 44 (49.9%) |
| SLR | ||
| ≤ 1.24 | 102 (72.3%) | 61 (68.5%) |
| > 1.24 | 39 (27.7%) | 28 (31.5%) |
| APOAI (g/L) | ||
| ≤ 1.17 | 56 (39.7%) | 39 (43.8%) |
| > 1.17 | 85 (60.3%) | 50 (56.2%) |
| APOB (g/L) | ||
| ≤ 0.99 | 92 (65.2%) | 50 (56.2%) |
| > 0.99 | 49 (34.8%) | 39 (43.8%) |
| ALP (U/L) | ||
| ≤ 82.9 | 96 (68.1%) | 51 (57.3%) |
| > 82.9 | 45 (31.9%) | 38 (42.7%) |
| ALB (g/L) | ||
| ≤ 40.1 | 108 (76.6%) | 62 (69.7%) |
| > 40.1 | 33 (23.4%) | 27 (30.3%) |
| GGT (U/L) | ||
| ≤ 44.2 | 117 (83.0%) | 72 (80.9%) |
| > 44.2 | 24 (17.0%) | 17 (19.1%) |
| LDH (U/L) | ||
| ≤ 245.6 | 125 (88.7%) | 77 (86.5%) |
| > 245.6 | 16 (11.2%) | 12 (13.5%) |
| TBIL (μmol/L) | ||
| ≤ 13.6 | 113 (80.1%) | 61 (68.5%) |
| > 13.6 | 28 (19.9%) | 28 (31.5%) |
| DBIL (μmol/L) | ||
| ≤ 3.3 | 67 (47.5%) | 38 (42.7%) |
| > 3.3 | 74 (52.5%) | 51 (57.2%) |
BMI body mass index, TNM pathological tumour node metastasis stage, ALT alanine transaminase, AST aspartate aminotransferase, SLR AST-to-ALT ratio, APOAI apolipoprotein AI, APOB apolipoprotein B, ALP alkaline phosphatase, ALB albumin, GGT glutamyl transpeptidase, LDH lactic dehydrogenase, TBIL total bilirubin, DBIL direct bilirubin
aTNM stage was classified according to the AJCC 7th TNM staging system
bThe tumor maximum diameter
Univariate analysis of the OS in the primary cohort
In the univariate analysis (Table 2), Kaplan–Meier survival analysis and log-rank tests were performed to predict patients’ OS. Univariate analysis indicated that age (P < 0.001), TNM stage (P < 0.001), tumor size (P = 0.001), treatment (P < 0.001), SLR (P = 0.044), APOAI (P < 0.001), APOB (P = 0.024), ALP (P = 0.018), GGT (P = 0.003) and LDH (P < 0.001) were significantly associated with OS in NSCLC patients with HBV infection.
Table 2.
Univariate analysis of OS in the primary cohorts
| Variable | Univariate analysis | |
|---|---|---|
| HR (95% CI) | P | |
| Gender | ||
| Male | Reference | |
| Female | 1.088 (0.678–1.746) | 0.726 |
| Age (years) | ||
| ≤ 42 | Reference | |
| > 42 | 0.316 (0.171–0.587) | < 0.001 |
| Family history | ||
| Yes | Reference | |
| No | 0.877 (0.512–1.501) | 0.631 |
| Smoking behaviour | ||
| Yes | Reference | |
| No | 0.965 (0.595–1.564) | 0.884 |
| BMI | ||
| ≥ 25 | Reference | |
| 18.5 ≤ BMI < 25 | 1.419 (0.757–2.660) | 0.275 |
| < 18.5 | 1.489 (0.480–4.618) | 0.491 |
| TNM stage | ||
| I | Reference | |
| II | 1.058 (0.318–3.517) | 0.927 |
| III | 3.256 (1.477–7.181) | 0.003 |
| IV | 6.575 (2.939–14.711) | < 0.001 |
| Size (cm)d | ||
| ≤ 6.3 | Reference | |
| > 6.3 | 2.769 (1.503–5.101) | 0.001 |
| Treatment | ||
| Surgery | Reference | |
| Radiotherapy/chemotherapy | 3.737 (1.891–7.387) | < 0.001 |
| Surgery and radiotherapy/chemotherapy | 1.049 (0.517–2.126) | 0.895 |
| Other | 6.845 (2.630–17.818) | < 0.001 |
| ALT (U/L) | ||
| ≤ 12.5 | Reference | |
| > 12.5 | 0.630 (0.311–1.278) | 0.200 |
| AST (U/L) | ||
| ≤ 22.0 | Reference | |
| > 22.0 | 1.366 (0.849–2.197) | 0.198 |
| SLR | ||
| ≤ 1.24 | Reference | |
| > 1.24 | 1.673 (1.015–2.759) | 0.044 |
| APOAI (g/L) | ||
| ≤ 1.17 | Reference | |
| > 1.17 | 0.393 (0.244–0.633) | < 0.001 |
| APOB (g/L) | ||
| ≤ 0.99 | Reference | |
| > 0.99 | 0.533 (0.308–0.922) | 0.024 |
| ALP (U/L) | ||
| ≤ 82.9 | Reference | |
| > 82.9 | 1.800 (1.106–2.929) | 0.018 |
| ALB (g/L) | ||
| ≤ 40.1 | Reference | |
| > 40.1 | 0.693 (0.385–1.246) | 0.220 |
| GGT (U/L) | ||
| ≤ 44.2 | Reference | |
| > 44.2 | 2.371 (1.348–4.169) | 0.003 |
| LDH (U/L) | ||
| ≤ 245.6 | Reference | |
| > 245.6 | 3.423 (1.847–6.345) | < 0.001 |
| TBIL (μmol/L) | ||
| ≤ 13.6 | Reference | |
| > 13.6 | 0.541 (0.276–1.059) | 0.073 |
| DBIL (μmol/L) | ||
| ≤ 3.3 | Reference | |
| > 3.3 | 1.583 (0.974–2.572) | 0.064 |
BMI body mass index, TNM pathological tumour node metastasis stage, ALT alanine transaminase, AST aspartate aminotransferase, SLR AST-to-ALT ratio, APOAI apolipoprotein AI, APOB apolipoprotein B, ALP alkaline phosphatase, ALB albumin, GGT glutamyl transpeptidase, LDH lactic dehydrogenase, TBIL total bilirubin, DBIL direct bilirubin
Prognostic nomogram model for OS
The variables age, TNM stage, tumor size, treatment, SLR, APOAI, APOB, ALP, GGT and LDH were identified as predictors of OS in the univariate analysis and were entered into the Cox’s proportional hazards regression. The selection of the final model was performed using a backward step-down selection process with the Akaike information criterion (AIC). We selected the optimal model based on the smallest AIC. The factors in our final model included age, TNM stage, tumor size, treatment, APOAI, APOB, GGT and LDH, and the AIC of the final model was 559.41. The nomogram model was constructed based on the selected factors, and Fig. 1 shows the nomogram model predicting the 1-, 3- and 5-year OS in the primary cohort. Patients with higher scores corresponded to worse prognoses.
Fig. 1.
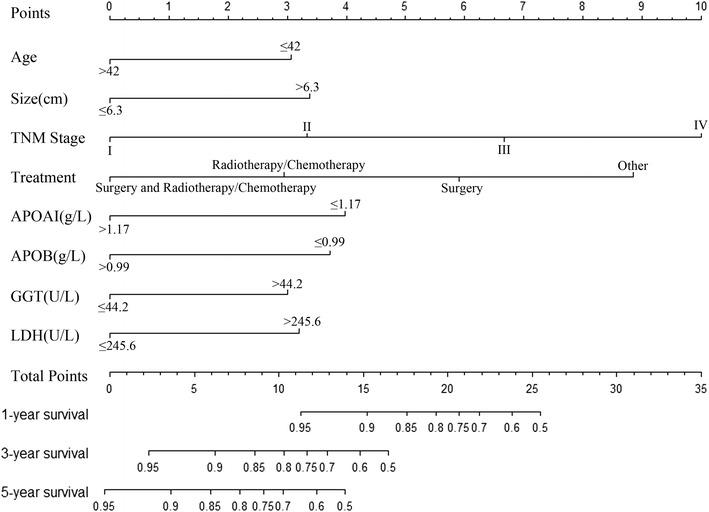
Nomogram model predicting the 1-, 3- and 5-year OS in NSCLC patients with chronic HBV infection. The nomogram was used summing the points identified on the points scale for each variable. The total points projected on the bottom scales indicate the probability of 1-, 3- and 5-year survival
Internal and external validation of the nomogram model
The predictive accuracies for OS in NSCLC patients with chronic HBV infection between the nomogram model and conventional TNM staging systems were compared by calculating the Harrell’s C-index (Table 3). In the primary cohort, the C-index was 0.780 (95% confidence interval (CI) 0.733–0.827), which was significantly higher than that of the TNM staging system, with a value of 0.693 (95% CI 0.640–0.746, P < 0.01). This result was also confirmed in the validation cohort, where the C-index of the nomogram model (0.786, 95% CI 0.731–0.841) was higher than the C-index of the TNM staging system (0.704, 95% CI 0.642–0.766, P < 0.01). Calibration curves at 1, 3 and 5 years were then used to assess the agreement between the predicted and actual outcomes (Fig. 2). The diagonal gray line represents the actual OS probability, while the solid black line represents the performance of the nomogram model in predicting the OS probability. The two lines overlap closely, indicating that the nomogram model provided better estimations in the patient cohort.
Table 3.
The C-index of nomogram model and TNM stage for prediction of OS in the primary cohort and validation cohort
| Factor | Primary cohort | Validation cohort | ||
|---|---|---|---|---|
| C-index (95% CI) | p | C-index (95% CI) | p | |
| Nomogram model | 0.780 (0.733–0.827) | 0.786 (0.731–0.841) | ||
| TNM stage | 0.693 (0.640–0.746) | 0.704 (0.642–0.766) | ||
| Nomogram model vs TNM stage | < 0.01 | < 0.01 | ||
Nomogram Model: including eight risk factors (age, size, pTNM, treatment, APOAI, APOB GGT and LDH)
C-index concordance index, CI confidence interval
Fig. 2.
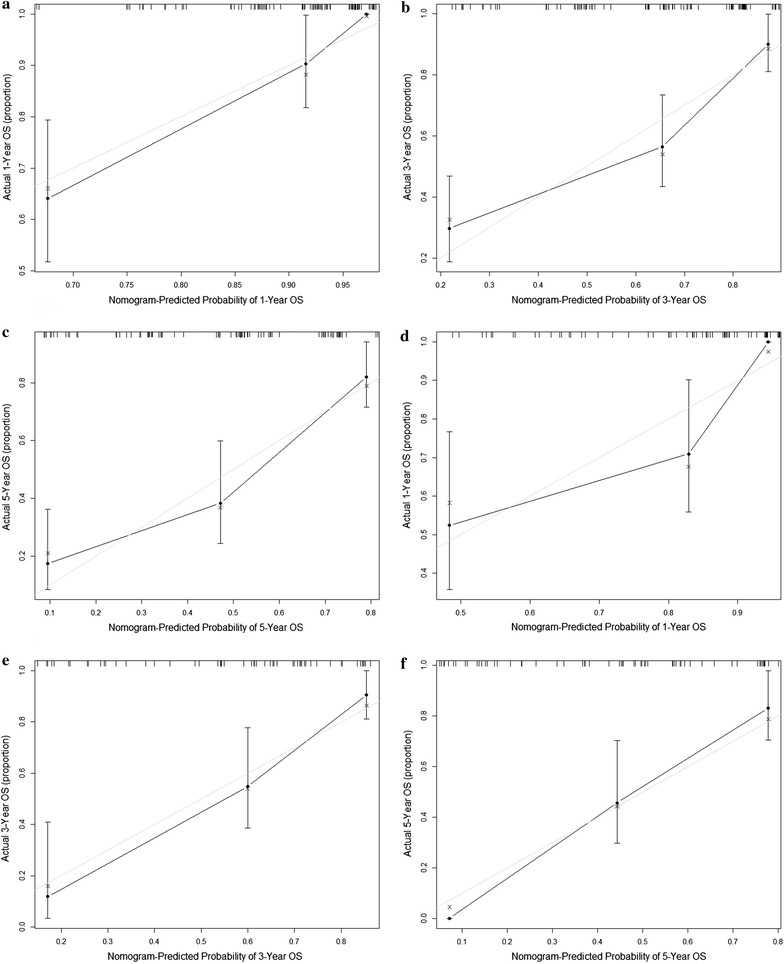
The calibration curves for predicting patient OS at (a) 1 year, (b) 3 years and (c) 5 years in the primary cohort and at (d) 1 year, (e) 3 years and (f) 5 years in the validation cohort. The nomogram model predicted OS is plotted on the x-axis, and the actual OS is plotted on the y-axis. Solid black line = performance of the nomogram model; closer alignment with the diagonal gray line represents a better estimation
Performance of the nomogram model in stratifying patient risk
Next, we determined the cutoff values using the X-tile program by grouping the patients in the primary cohort evenly into the following 3 groups based on the predictions of the nomogram model: low-risk prognosis (TPS ≤ 13.5, 74 patients), intermediate-risk prognosis (13.5 < TPS ≤ 20.0, 43 patients) and high-risk prognosis (TPS > 20.0, 24 patients). The low-risk group had the highest probability of survival (95.9% for 1 year, 68.9% for 3 years and 55.4% for 5 years), followed by the intermediate-risk group with survival probabilities of 76.7, 37.2 and 14.0% for 1, 3 and 5 years, respectively. The high-risk group had survival probabilities of 45.8, 8.3 and 0% for 1, 3 and 5 years, respectively. The differences in survival could be used to discriminate these three groups (Table 4). We then applied the cutoff values to plot Kaplan–Meier curves in the primary cohort and the validation cohort (Fig. 3). Both were significantly associated with OS outcomes within the three groups (P < 0.001).
Table 4.
Cox regression analysis for groups based on the model in the primary cohorts
| Groups | OS mean | 1-year (%) | 3-years (%) | 5-years (%) | Sig. | HR | 95% CI for HR | |
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| Low | 75.976 | 95.9 | 68.9 | 55.4 | – | – | – | – |
| Intermediate | 38.839 | 76.7 | 37.2 | 14.0 | < 0.001 | 4.098 | 2.274 | 7.385 |
| High | 13.852 | 45.8 | 8.3 | 0.0 | < 0.001 | 15.318 | 7.739 | 30.320 |
Groups were divided by cutoff values of total prognostic scores (TPS) cumulated from nomogram we designed. (TPS ≤ 13.5, 13.5 < TPS ≤ 20.0, TPS > 20.0) (group lowest risk, intermediate risk and high risk with 74, 43 and 24 patients, respectively.). CI confidence interval, HR hazard ratio, OS overall survival
Fig. 3.
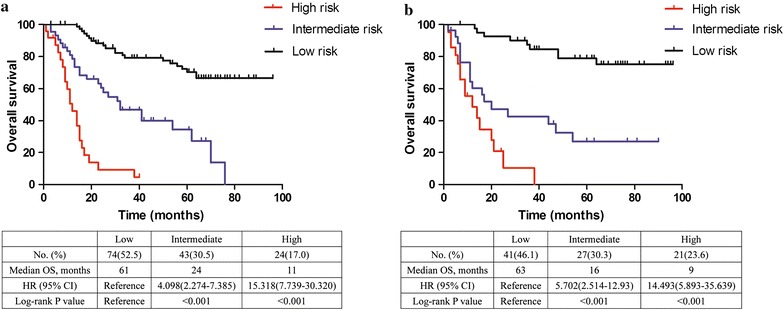
Graphs showing the results of the Kaplan–Meier curves for all three groups based on the prediction from the nomogram model in the primary cohort (left a) and in the validation dataset (right b). A significant association of the radiomics signature with the OS was shown in the training dataset, which was then confirmed in the validation dataset
Decision curve analysis for 5-year survival predictions
Figure 4 presents the results of the decision curve analysis at 5 years. The results showed that our nomogram model had a higher overall net benefit than did the traditional TNM staging system across a wide range of threshold probabilities.
Fig. 4.
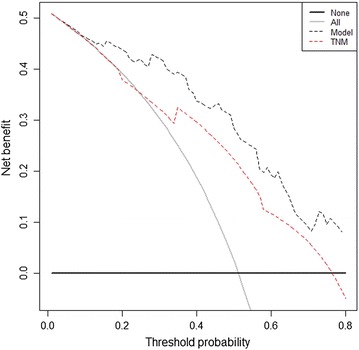
Decision curve analysis for the 5-year survival predictions. In the decision curve analysis, the y-axis indicates net benefit, calculated by summing the benefits (true positives) and subtracting the harms (false positives). The nomogram model (black dotted line) had the highest net benefit compared with the TNM staging system (red dotted line). The straight line represents the assumption that all the patients will die, and the horizontal line represents the assumption that none of the patients will die
Correlations among the variables of the nomogram model
Figure 5 shows the correlations among the different variables of the nomogram model. In this plot, positive correlations are displayed in blue and negative correlations in red. The color intensity and the size of the circle are proportional to the correlation coefficients. Additionally, the numbers in the graph show the Pearson’s correlation coefficient between the different variables. The results revealed that there was not a significant correlation among the various variables.
Fig. 5.
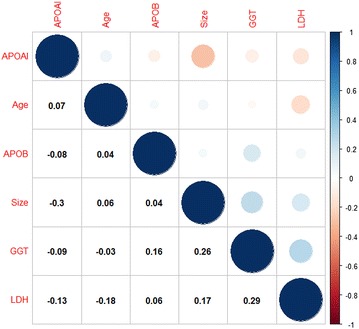
The correlations among the various variables of the nomogram model
Discussion
In this study, we evaluated the prognostic power of LFIs in NSCLC patients with HBV infection, taking advantage of the ability of a nomogram model that combined LFIs with clinicopathological characteristics to establish an effective predictive nomogram model for NSCLC patients with HBV infection. To our knowledge, this study is the first to establish a prognostic nomogram model for NSCLC patients with HBV infection based on the clinicopathologic data of 230 patients. The model included age, TNM stage, tumor size, treatment, APOAI, APOB, GGT, and LDH. Our nomogram model had better discriminatory ability than the current AJCC TNM classification system. The nomogram model also had a higher overall net benefit than the TNM staging system at 5 years.
Many studies have shown that LFIs are correlated with cancer prognosis. Our model included APOAI, APOB, GGT and LDH, which are good prognostic markers in different types of cancers. APOAI has been shown to have cardioprotective, anti-inflammatory, anti-viral, anti-parasitic, anti-bacterial and anti-tumor activity functions [20]. APOAI is a useful prognostic factor in breast cancer [21], renal cell carcinoma [22], nasopharyngeal carcinoma [23] and lung cancer [24]. APOB is a major structural protein for atherogenic APOB-containing lipoproteins [25]. The levels of APOB are positively associated with the risk of colorectal cancer, breast cancer, lung cancer [26, 27]. In addition, our study is the first to report that APOB is correlated with lung cancer prognosis. GGT is a membrane-bound enzyme involved in the metabolism of glutathione. Several previous studies revealed that GGT is related to tumor development, progression, invasion, drug resistance and prognosis [28, 29]. Elevated serum levels of GGT were also found to be associated with poorer prognosis in several human cancers. LDH, a hypoxia regulator, plays a vital role in alternative metabolic pathways of cancer cells [30]. Serum LDH levels could be a low-cost and useful prognostic factor in patients with lung cancer [31, 32].
Compared with previous studies, our results differed slightly in that our model excluded ALP and ALB. Serum ALP is used as an indicator of hepatic and bone diseases as it is convenient to measure. Arife et al. [7] showed that the risk of progression with normal levels of ALP was significantly higher than the risk with high ALP levels among advanced NSCLC patients. ALP is an independent prognostic factor related to OS and progression-free survival in NSCLC patients. Serum ALB is used to assess nutritional status. Many studies have shown that serum albumin as an independent prognosticator of survival in lung cancer [33, 34]. These results may be explained by the different prognostic outcomes between NSCLC patients with HBV infection and those of patients without HBV infection. Furthermore, the cutoff value of ALB was different from that of previous reports. Here, we adopted the X-tile program to choose the optimal cutoff points, which may have led to the different results.
HBV is a noncytopathic virus that does not cause direct damage to liver cells. Instead, it is the immune system’s aggressive response to the virus that leads to inflammation and damage to the liver [35, 36]. Thereby, inflammation and biochemical indicators of liver function are correlated with cancer prognosis and influence the prognosis of cancer patients. Therefore, we developed an effective nomogram model to predict OS in NSCLC patients with HBV infection.
In addition to these strengths, our study has various limitations. First, due to the retrospective nature of our study, we cannot avoid potential biases, and we enrolled relatively few patients. Second, the data were obtained from a single center and represent a small sample size. Therefore, further multi-center studies using higher sample sizes are needed to externally validate the nomogram model to verify whether our findings are universally applicable. Third, we only analyzed the impact of the biochemical indicators of liver function on the prognosis of NSCLC patients with HBV infection. Other prognostic factors such as inflammatory factors, HBV DNA, serum carcinoembryonic antigen (CEA) [37] and oncogenic mutations [38] were not included. These factors should be considered in future studies. Despite these limitations, the nomogram model was effective and may be useful in predicting the outcomes of NSCLC patients with HBV infection.
Conclusion
In summary, we developed and validated an effective nomogram model predicting the 1-, 3- and 5-year OS in NSCLC patients with HBV infection. The proposed nomogram in this study showed a better level of discrimination and accuracy than the current AJCC TNM classification system. Furthermore, it could be useful for patient counseling and individualized prediction of survival for NSCLC patients with HBV infection.
Authors’ contributions
All the authors contributed to this manuscript, including the conception and design (SLC, YZL, ZQH), the acquisition of data (JPL, XH), the analysis and interpretation of the data (RS), material support (QYD), study supervision (SGP, HC, WLL), and the writing, review and revision of the manuscript (SLC, YZL, ZQH, JPL, XH, SGP, HC, WLL). All authors read and approved the final manuscript.
Acknowledgements
We thank the staff at the Director of Clinical Laboratories, Sun Yat-sen University Cancer Center, for providing support for the research conditions in this study.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All the patients provided written informed consent for their medical information to be stored and used in the hospital database at the first visit to our center. This study was approved by the Clinical Research Ethics Committee of the Sun Yat-sen University Cancer Center.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 81472008).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Shulin Chen, Yanzhen Lai and Zhengqiang He contributed equally to this work
Hao Chen, Songguo Peng and Wanli Liu contributed equally to this work
Contributor Information
Shulin Chen, Email: chenshl@sysucc.org.cn.
Yanzhen Lai, Email: laiyzh@sysucc.org.cn.
Zhengqiang He, Email: hezhengq@sysucc.org.cn.
Jianpei Li, Email: lijp@sysucc.org.cn.
Xia He, Email: hexia@sysucc.org.cn.
Rui Shen, Email: 81219359@qq.com.
Qiuying Ding, Email: dingqy@sysucc.org.cn.
Hao Chen, Email: chenhao@sysucc.org.cn.
Songguo Peng, Email: pengsg@sysucc.org.cn.
Wanli Liu, Phone: +86-20-8734-3438, Email: liuwlsysucc@163.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:5. doi: 10.3322/caac.21360. [DOI] [PubMed] [Google Scholar]
- 2.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584. doi: 10.1016/S0025-6196(11)60735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 4.Zheng X, Wang J, Yang D. Antiviral therapy for chronic hepatitis B in China. Med Microbiol Immunol. 2015;204:115. doi: 10.1007/s00430-014-0380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen SL, Xue N, Wu MT, Chen H, He X, Li JP, Liu WL, Dai SQ. Influence of preoperative serum aspartate aminotransferase (AST) level on the prognosis of patients with non-small cell lung cancer. Int J Mol Sci. 2016;17(9):1474. doi: 10.3390/ijms17091474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhuo Y, Lin L, Wei S, Zhang M. Pretreatment elevated serum lactate dehydrogenase as a significant prognostic factor in malignant mesothelioma: a meta-analysis. Medicine. 2016;95:e5706. doi: 10.1097/MD.0000000000005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulas A, Turkoz FP, Silay K, Tokluoglu S, Avci N, Oksuzoglu B, Alkis N. A laboratory prognostic index model for patients with advanced non-small cell lung cancer. PLoS ONE. 2014;9:e114471. doi: 10.1371/journal.pone.0114471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamoto Y, Kaneko S. Mechanisms of viral hepatitis induced liver injury. Curr Mol Med. 2003;3:537. doi: 10.2174/1566524033479591. [DOI] [PubMed] [Google Scholar]
- 9.Xu LM, Xing LM, Ning J, Ying LM, Ling-Long Tang MD, Lei CM, Guan-Qun Zhou MD, PhD YSM, Dan YM, Rui GM. Prognostic value of chronic hepatitis B virus infection in patients with nasopharyngeal carcinoma: analysis of 1301 patients from an endemic area in China. Cancer. 2014;120:68–76. doi: 10.1002/cncr.28377. [DOI] [PubMed] [Google Scholar]
- 10.Wei XL, Qiu MZ, Chen WW, Ying J, Chao R, Feng W, Luo HY, Wang ZQ, Zhang DS, Wang FH. The status of HBV infection influences metastatic pattern and survival in Chinese patients with pancreatic cancer. J Transl Med. 2013;11:249. doi: 10.1186/1479-5876-11-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng JW, Liu DY, Lin GN, Xiao JJ, Xia ZJ. Hepatitis B virus infection is associated with poor prognosis in patients with advanced non small cell lung cancer. APJCP. 2015;16:5285–5288. doi: 10.7314/apjcp.2015.16.13.5285. [DOI] [PubMed] [Google Scholar]
- 12.See W. Postoperative nomogram predicting risk of recurrence after radical cystectomy for bladder cancer. Urol Oncol Semin Orig Invest. 2007;25:3967. doi: 10.1200/JCO.2005.05.3884. [DOI] [PubMed] [Google Scholar]
- 13.Karakiewicz PI, Briganti A, Chun FK, Trinh QD, Perrotte P, Ficarra V, Cindolo L, De La Taille A, Tostain J, Mulders PF. Multi-institutional validation of a new renal cancer-specific survival nomogram. J Clin Oncol. 2007;25:1316. doi: 10.1200/JCO.2006.06.1218. [DOI] [PubMed] [Google Scholar]
- 14.Mariani L, Miceli R, Kattan MW, Brennan MF, Colecchia M, Fiore M, Casali PG, Gronchi A. Validation and adaptation of a nomogram for predicting the survival of patients with extremity soft tissue sarcoma using a three-grade system. Cancer. 2005;103:402–408. doi: 10.1002/cncr.20778. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Hricak H, Kattan MW, Chen HN, Scardino PT, Kuroiwa K. Prediction of organ-confined prostate cancer: incremental value of MR imaging and MR spectroscopic imaging to staging nomograms. Radiology. 2006;238:597–603. doi: 10.1148/radiol.2382041905. [DOI] [PubMed] [Google Scholar]
- 16.Ramiporta R, Crowley JJ, Goldstraw P. The revised TNM staging system for lung cancer. Ann Thorac Cardiovasc Surg. 2009;15:4–9. [PubMed] [Google Scholar]
- 17.Camp RL, Dolledfilhart M, Rimm DL. X-Tile a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 18.Jr HF, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangaraj M, Nanda R, Panda S. Apolipoprotein A-I: a molecule of diverse function. Indian J Clin Biochem. 2016;31:253. doi: 10.1007/s12291-015-0513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pakzad R, Safiri S. The effect of preoperative serum triglycerides and high-density lipoprotein-cholesterol levels on the prognosis of breast cancer: methodological issue. Breast. 2017;36:103–104. doi: 10.1016/j.breast.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 22.Guo S, He X, Qian C, Yang G, Kai Y, Pei D, Ye Y, Dong C, Zhang Z, Qin Z. The effect of preoperative apolipoprotein A-I on the prognosis of surgical renal cell carcinoma: a retrospective large sample study. Medicine. 2016;95:e3147. doi: 10.1097/MD.0000000000003147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo XL, Zhong GZ, Hu LY, Jie C, Ying L, Chen QY, Liu Q, Rao HL, Chen KL, Cai QQ. Serum apolipoprotein A-I is a novel prognostic indicator for non-metastatic nasopharyngeal carcinoma. Oncotarget. 2011;6:44037–44048. doi: 10.18632/oncotarget.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charet JC, Watine J, Marre A, Charet P. Prognostic value of serum levels of cholesterol and apolipoprotein A1 in pulmonary cancer. Ann Biol Clin. 1997;55:52. [PubMed] [Google Scholar]
- 25.Kane JP. Apolipoprotein B: structural and metabolic heterogeneity. Annu Rev Physiol. 1983;45:637. doi: 10.1146/annurev.ph.45.030183.003225. [DOI] [PubMed] [Google Scholar]
- 26.Borgquist S, Butt T, Almgren P, Shiffman D, Stocks T, Orhomelander M, Manjer J, Melander O. Apo-lipoproteins, lipids and risk of cancer. Int J Cancer. 2016;138:2648–2656. doi: 10.1002/ijc.30013. [DOI] [PubMed] [Google Scholar]
- 27.Chandler PD, Song Y, Lin J, Zhang S, Sesso HD, Mora S, Giovannucci EL, Rexrode KE, Moorthy MV, Li C. Lipid biomarkers and long-term risk of cancer in the Women’s Health Study. Am J Clin Nutr. 2016;103:1397. doi: 10.3945/ajcn.115.124321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield JB. Gamma glutamyl transferase. Crit Rev Clin Lab Sci. 2001;38:263–355. doi: 10.1080/20014091084227. [DOI] [PubMed] [Google Scholar]
- 29.Corti A, Franzini M, Paolicchi A, Pompella A. Gamma-glutamyltransferase of cancer cells at the crossroads of tumor progression, drug resistance and drug targeting. Anticancer Res. 2010;30:1169–1181. [PubMed] [Google Scholar]
- 30.Serganova I, Rizwan A, Ni X, Thakur SB, Vider J, Russell J, Blasberg R, Koutcher JA. Metabolic imaging: a link between lactate dehydrogenase A, lactate, and tumor phenotype. Clin Cancer Res. 2011;17:6250–6261. doi: 10.1158/1078-0432.CCR-11-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inomata M, Hayashi R, Tokui K, Taka C, Okazawa S, Kambara K, Ichikawa T, Yamada T, Miwa T, Kashii T. Lactate dehydrogenase and body mass index are prognostic factors in patients with recurrent small cell lung cancer receiving amrubicin. Tumori. 2015;102:606–609. doi: 10.5301/tj.5000435. [DOI] [PubMed] [Google Scholar]
- 32.Zhu L, Li X, Shen Y, Ying C, Fang X, Chen J, Ying Y. A new prognostic score based on the systemic inflammatory response in patients with inoperable non-small-cell lung cancer. Oncotargets Ther. 2016;9:4879–4886. doi: 10.2147/OTT.S107279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam PT, Leung MW, Tse CY. Identifying prognostic factors for survival in advanced cancer patients: a prospective study. Hong Kong Med J. 2007;13:453. [PubMed] [Google Scholar]
- 34.Bagan P, Berna P, De DF, Jc DNP, Mordant P, De LTB, Le PBF, Riquet M. Nutritional status and postoperative outcome after pneumonectomy for lung cancer. Ann Thorac Surg. 2013;95:392–396. doi: 10.1016/j.athoracsur.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 35.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 36.Lamontagne RJ, Bagga S, Bouchard MJ. Hepatitis B virus molecular biology and pathogenesis. Hepatoma Res. 2016;2:163. doi: 10.20517/2394-5079.2016.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda R, Suda T, Hachimaru A, Tochii D, Tochii S, Takagi Y. Clinical significance of preoperative carcinoembryonic antigen level in patients with clinical stage IA non-small cell lung cancer. J Thorac Dis. 2017;9:176. doi: 10.21037/jtd.2017.01.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, Sun T, Kang P, Qian K, Deng B, Zhou J, Wang R, Jiang B, Li K, Liu F. Combined analysis of rearrangement of ALK, ROS1, somatic mutation of EGFR, KRAS, BRAF, PIK3CA, and mRNA expression of ERCC1, TYMS, RRM1, TUBB3, EGFR in patients with non-small cell lung cancer and their clinical significance. Cancer Chemother Pharmacol. 2016;77:583. doi: 10.1007/s00280-016-2969-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.


