Abstract
Constitutive activation of the non-receptor tyrosine kinase c-Abl (Abl1) in the Bcr-Abl1 fusion oncoprotein is the molecular cause of chronic myeloid leukemia. Recent studies have indicated that an interaction between the SH2 domain and the N-lobe of the c-Abl kinase domain has a critical role in leukemogenesis. To dissect the structural basis of this phenomenon we studied c-Abl constructs comprising the SH2 and kinase domains in vitro. We present a crystal structure of an SH2-kinase domain construct bound to dasatinib, which contains the relevant interface between the SH2 domain and the N-lobe of the kinase domain. We show that the presence of the SH2 domain enhances kinase activity moderately and that this effect depends on contacts in the SH2-N-lobe interface and is abrogated by specific mutations. Consistently, formation of the interface decreases slightly the association rate of imatinib with the kinase domain. That the effects are small compared to the dramatic in vivo consequences suggests an important function of the SH2-N-lobe interaction might be to help disassemble the autoinhibited conformation of c-Abl and promote processive phosphorylation, rather than substantially stimulate kinase activity.
Keywords: Abl, tyrosine kinase, SH2 domain, imatinib, dasatinib, CML
INTRODUCTION
Under normal conditions the catalytic activity of the non-receptor tyrosine kinase c-Abl (cellular Abelson tyrosine protein kinase 1, Abl1) is tightly regulated by various autoinhibitory mechanisms (for review, see [1]). In chronic myeloid leukemia (CML) c-Abl is constitutively active through genetic fusion with the breakpoint cluster region (BCR), resulting in the expression of the fusion protein Bcr-Abl, which leads to Abl activation through dimerization and also due to the loss of an N-terminal regulatory interaction (for review, see [1]). In the following, we use ‘Abl’ when referring to common features of c-Abl and Bcr-Abl.
c-Abl has a modular architecture. Like Src family kinases, the N-terminal part of c-Abl contains an SH3, an SH2, and a kinase domain (Figure 1A). The C-terminal region consists of an F-actin binding domain and various short interaction motifs. While the structural basis for the autoinhibition of the N-terminal half of c-Abl [2] and various states in the conformational equilibrium of the catalytic domain have been described [3–5], it is incompletely understood how Abl gains full catalytic activity.
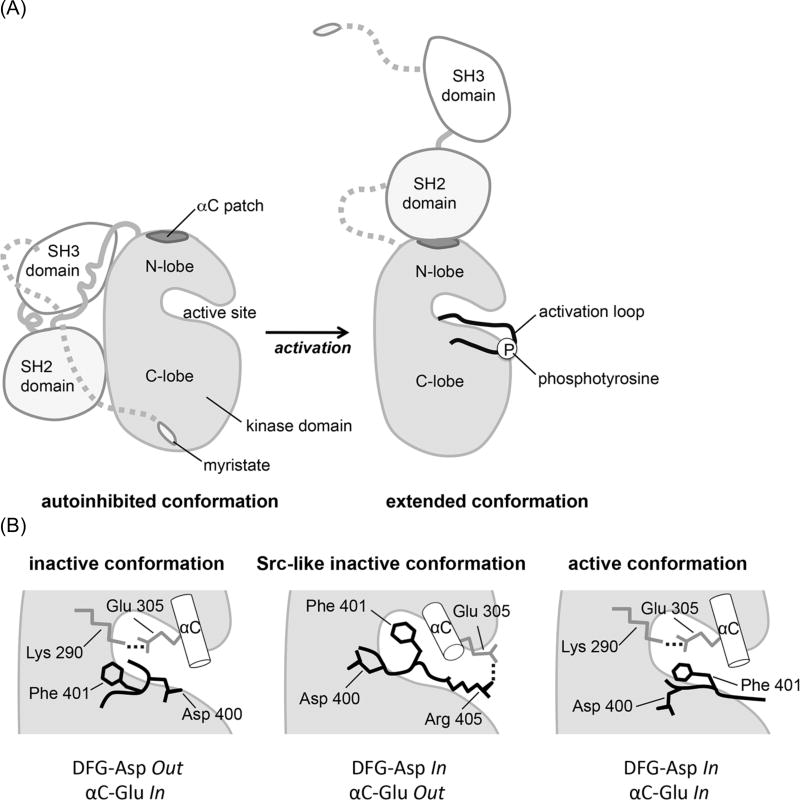
Figure 1. Conformational states of c-Abl.
(A) Schematic depicting two distinct domain arrangements in the autoinhibited and active states of c-Abl. In the autoinhibited, assembled state the N-terminal myristoyl group binds to the kinase domain and allows the SH2 and SH3 domains to dock onto it [2,40]. Open autoinhibited conformations also exist [5]. In the extended, active conformation, the SH2 domain forms an interface with the N-lobe of the kinase domain, involving the αC patch and Tyr 412 in the activation loop undergoes rapid autophosphorylation.
(B) Schematic representation of critical components of the active site in various states of the Abl kinase domain: the inactive conformation as seen in the complex with imatinib, the Src-like inactive conformation, and the active conformation. The DFG motif at the base of the activation loop and αC helix are shown. Ionic interactions are indicated as dotted lines.
Based on studies of Hck [6] and Btk [7], kinase active conformations were originally thought to resemble ‘beads on a string’. It has since become clear, however, that kinase active conformations are typically not fully disassembled and in many cases rely on distinct domain interactions to sustain activity. Likewise, the autoinhibited state has recently been shown to possess significant domain flexibility and to be in equilibrium with less compact conformations [5].
What do such alternative conformations that are distinct from the autoinhibited states look like and how do they stimulate activity? A recurrent docking site for allosteric effectors in various kinase families is a hydrophobic patch on the N-lobe of the kinase domain, termed ‘helix αC patch’ (for review see [8]). This hydrophobic region communicates stimulatory signals either intermolecularly, as seen for CDK/Cyclin, EGFR asymmetric dimer, and Aurora/Tpx2 among others or intramolecularly, as seen for PKA, PDK1, PKB/Akt, Rho-kinase and Fes. In the case of Abl, the helix αC patch can interact with the flanking SH2 domain to give rise to an extended conformation, as opposed to the assembled, autoinhibited state in which the SH2 domain docks onto the backside of the C-lobe (Figure 1A).
The extended conformation was originally observed in a crystal structure of a c-Abl three-domain construct containing the SH3, SH2 and kinase domains in complex with myristic acid and a small-molecule inhibitor, PD166326 [2]. In this crystal the extended, myristoyl-free conformation occurred fortuitously alongside the assembled, myristoyl-bound state, and its significance initially remained unclear. Small-angle X-ray scattering studies later revealed that an activated three-domain Abl construct, in which autoinhibitory constraints have been released through mutations in regulatory interfaces, has indeed an extended shape in solution that is in good agreement with the extended conformation found in the crystal [9].
Contacts in the SH2-N-lobe interface of the extended conformation have been shown to be important for kinase activation [10] and for the deleterious effects of the overactive forms of Abl, such as Bcr-Abl, in vivo [11,12]. Remarkably, perturbation of the interface by a single point mutation (I164E) inhibits downstream signaling events that are important for CML maintenance, in particular STAT5 phoshorylation, and abolishes leukemogenesis in a CML mouse model [11]. Furthermore, a mutation (T231R) that potentially stabilizes the SH2-N-lobe interface was identified in a CML patient and was associated with resistance to imatinib (Gleevec) treatment [12]. While the functional significance of the SH2-N-lobe interaction in vivo is thus undoubted, it remains unclear on a structural level how formation of this interface stimulates kinase activity.
It is possible, for instance, that the extended conformation of Abl facilitates processive substrate phosphorylation by having the SH2 domain in a suitable position to capture substrates upon phosphorylation and to present further tyrosine residues on these substrates to the active site on the kinase domain. In line with this idea, multi-site phosphorylation of substrates was found to be reduced upon mutational perturbation of the phosphate-binding site on the SH2 domain and upon mutation of the SH2-N-lobe interface [11]. Alternatively, the extended conformation of the SH2-kinase domain module may stimulate activity by enabling yet unknown intramolecular interactions in the context of full-length Abl. The extended conformation may also promote interactions with effector proteins to aid substrate phosphorylation or prevent substrate dephosphorylation in the cell. Finally, the SH2-N-lobe interaction may enhance kinase activity through allosteric effects that impact the structure of the kinase active site.
The active site is located between the N- and the C-lobe of the kinase domain and contains conserved structural features critical for the positioning of substrates and ATP (Figure 1A) (for review see [8]). One important element is the activation loop that adopts an open conformation in the kinase active conformation, thereby providing a docking site for the substrate. An acidic side chain, Asp 400, that is part of a conserved Asp-Phe-Gly (DFG) motif at the base of the activation loop, contributes to the binding of ATP through coordination of a magnesium ion (Figure 1B). This conformation is known as ‘DFG-Asp In’. ATP binding further involves contacts with a glycine-rich loop (commonly referred to as ‘P-loop’) and a specific orientation of helix αC relative to the rest of the N-lobe (‘αC-Glu In’), as characterized by a salt bridge between Glu 305 and the ATP-coordinating residue Lys 290. An intricate network of contacting residues near the active site have been suggested to act as regulatory and catalytic ‘spines’ that stretch across the two lobes of the kinase domain and sustain the catalytically competent conformation [13]. The important physiological role of the SH2-N-lobe interaction for Abl activity might indicate that these spines are part of an even more extended contact network or ‘skeleton’ that serves to propagate allosteric effects across domains boundaries to the active site. To investigate a possible structural communication between the SH2-N-lobe interface and the active site, in the absence of other potentially contributing factors, we studied an Abl fragment that contains only the SH2 and kinase domains in vitro.
EXPERIMENTAL
Protein expression and purification
Genes encoding human c-Abl 1b constructs (KD: residues 248–534, SH2-KD: residues 138–534) were cloned into pET21d (NheI/XhoI) and co-expressed with YopH phosphatase in E. coli BL21 DE3 cells, following established protocols [14]. Protein purification of the C-terminally hexa-His-tagged constructs included nickel affinity chromatography and anion exchange chromatography, as previously described [14], and an additional gel filtration (Superdex 75, Amersham Biosciences) in 100 mM NaCl, 20 mM Tris (pH 8.0), 5% glycerol, and 1 mM DTT.
Activity assays
Kinase activity measurements were performed with the “ADP Quest Assay” (DiscoveRx), which is an enzyme-coupled, fluorescence-based system for monitoring ADP production. The substrate peptide (“Abltide”) used in our study (sequence: EAIYAAPFAKKK) has previously been shown to be a good substrate for c-Abl [14,15]. To be pre-phosphorylated the Abl constructs were incubated with purified, autophosphorylated Hck as follows: 200 nM Hck was first incubated with 2 mM ATP in 100 mM Tris (pH 7.5), 10 mM MgCl2, and 1 mM vanadate for 30 minutes. Subsequently, c-Abl constructs were incubated at 10 µM concentration with 50 nM of pre-incubated Hck in 100 mM Tris (pH 7.5), 10 mM MgCl2, additional 500 µM ATP, and 1 mM vanadate for 30 minutes. Samples without pre-phosphorylation were prepared in the same way, but leaving out Hck. For all samples, the kinase activity of 2.5 nM Abl towards 300 µM substrate was monitored at 25°C for 1 hour. Fluorescence intensity (λexcitation = 530 nm, λemission = 590 nm) was measured in 4 min intervals and Vmax values were obtained by linear fitting of the steady-state region of the data, corrected for ATP-hydrolysis in the absence of peptide and referenced to the value obtained for the wildtype kinase domain construct. Finally, relative Vmax values were averaged from three replicate runs.
Drug binding assays
To measure the kinetics of imatinib binding, equal volumes of protein and drug were mixed at room temperature using a stopped-flow instrument (Applied Photophysics) following established protocols [16]. Imatinib was obtained from clinically available capsules (Novartis). Upon excitation at λ = 280 nm the decrease in fluorescence at λ = 348 nm was monitored with a Jobin Yvon FluoroMax-3 (Horiba) spectrofluorometer. After mixing, the protein concentration was 50 nM and the drug concentrations between 0.1 µM and 12.5 µM in 100 mM NaCl, 1 mM DTT, 50 mM Tris, pH 6.5. Data were fit to a single exponential with a sloping baseline to account for photobleaching. At low drug concentrations a slight deviation from the model was observed. However, this deviation was small enough (residuals < 5% of signal amplitude) to be ignored. 4 replicates of each run were fit individually and the results were averaged. The association rate constants were derived from a linear fit of the observed rate constants plotted over the drug concentration. The corresponding dissociation rate constants are not discussed here, for they are poorly defined in this experimental setup.
X-ray crystallography and structure determination
16 mg/ml of the wild SH2-KD construct in 100 mM NaCl, 20 mM Tris (pH 8.0), 5% glycerol, and 1 mM DTT was mixed with a two-fold molar excess of dasatinib (Symansis), and sitting drops were set up using a Phoenix crystallization robot (Art Robbins Instruments) using 100 nl + 100 nl drops. The SH2-KD construct crystallized readily as bundles of small needles unsuited for crystallographic handling. While extensive optimization efforts were unsuccessful in bringing about significant improvements, we eventually obtained a single sufficiently robust needle after a growth period of several weeks. The crystal grew at 20°C in 0.1 M sodium citrate (pH 5.0) and 8% PEG 8000, was cryoprotected in 20% PEG 8000, 25% glycerol, and 0.1 M sodium citrate (pH 5.0). Diffraction data were collected at the Advanced Light Source (Lawrence Berkeley National Laboratory), beamline 8.3.1 to 2.9 Å resolution and were processed with HKL2000 [17]. Molecular replacement was performed with Phaser [18], as implemented in the ccp4 suite [19]. Refinement was performed using Phenix [20] with NCS-restraints and group B-factors. TLS refinement was performed with individual domains as separate TLS objects. Manual model building was performed in Coot [21].
RESULTS AND DISCUSSION
The SH2-N-lobe interaction enhances kinase activity in vitro
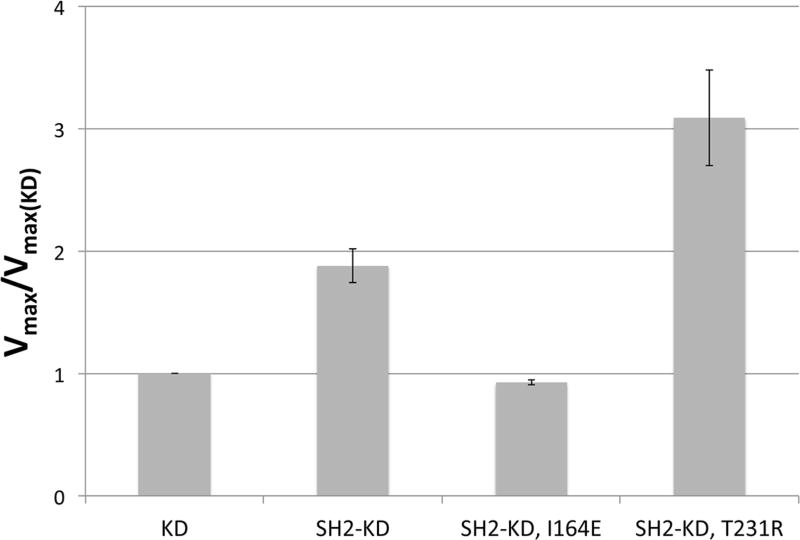
To determine the effect of the SH2-N-lobe interaction on Abl kinase activity in vitro, we compared the activities of the recombinantly expressed and purified kinase domain (KD) and of a construct comprising both the SH2 and kinase domains (SH2-KD) using a continuous enzyme-coupled assay. By co-expressing the constructs with the phosphatase YopH, we ensured that the kinase constructs were initially unphosphorylated, as confirmed by mass spectrometry (data not shown) [14]. All activity measurements were made using saturating concentrations of an Abl-optimized substrate (“Abltide”) [15], and so the measured rates are representative of the Vmax values (the Km for Abltide is under 10 µM for Abl as the kinase [22]). The presence of the SH2 domain was found to increase the activity of the kinase domain, measured as the Vmax-value, by a factor of ~ 2 (Figure 2). The activating effect of the SH2 domain is lost upon introducing the mutation I164E that should perturb the SH2-N-lobe interface. In contrast, a disease-relevant mutation, T231R, that presumably stabilizes the SH2-N-lobe interface [12], further increases the activity compared to the kinase domain alone (by a factor of ~ 3).
Figure 2. Effect of the SH2-N-lobe interaction on kinase activity.
Kinase activities, as measured by the Vmax values, of the SH2-KD construct and two mutanted variants thereof referenced to the activity of the isolated kinase domain. Perturbation of the SH2-N-lobe interface by the mutation I164E nullifies the stimulating effect of the SH2 domain, while the mutation T231R enhances it.
To test how phosphorylation of Abl impacts the relative activities of the constructs used here, we performed analogous activity assays after pre-incubation of Abl with the tyrosine kinase Hck. Hck treatment had no detectable effect on the activity of the isolated kinase domain, but was found to slightly activate all SH2-kinase domain constructs (by factors < 2). The relative activities of the SH2-KD constructs compared to the isolated kinase domain were qualitatively similar to what was observed without pre-phosphorylation (Supplementary Figure 1).
Taken together, these data confirm that the SH2-N-lobe interaction enhances Abl kinase activity in vitro, albeit to a rather modest extent. These results are qualitatively consistent with previous studies monitoring the activity of immunoprecipitated c-Abl and Bcr-Abl mutants, respectively, from human cancer cell extracts [10–12].
The SH2-N-lobe interaction decreases the rate of imatinib binding
Mutations at the SH2-N-lobe interface have been shown to modulate the sensitivity of Bcr-Abl towards active-site inhibitors [11,12]. To study how changes at the SH2-kinase domain interface are propagated to the active site of the kinase domain, we monitored imatinib binding rates by stopped-flow fluorimetry. Since imatinib specifically recognizes the inactive (DFG-Asp Out, αC-Glu In) state, its binding kinetics provide a read-out of which active site conformations are preferentially adopted [16,23].
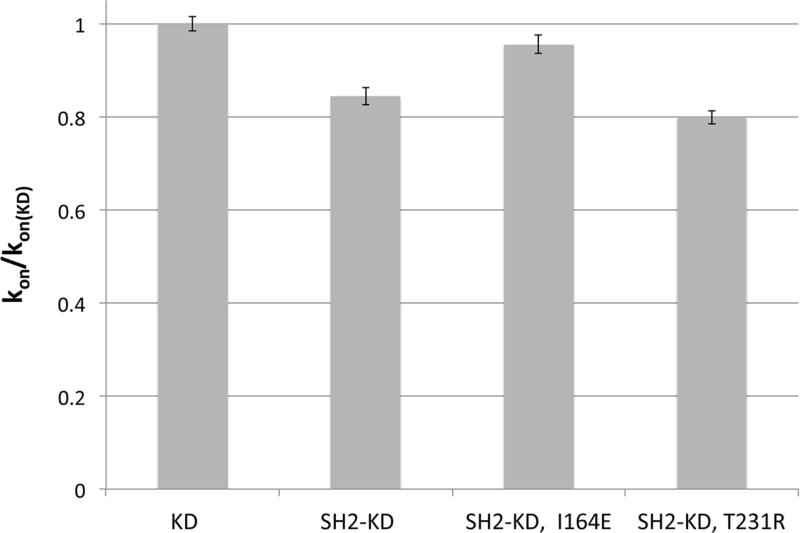
We found that the presence of the SH2 domain leads to a small (~1.2 fold), but significant, decrease in the rate of imatinib binding to the kinase domain from 0.82 +/− 0.01 µM−1 s−1 to 0.69 +/− 0.02 µM−1 s−1 (Figure 3). The T231R mutation slows down binding even further (0.65 +/− 0.01 µM−1 s−1; ~1.3 fold compared to the kinase domain alone). In contrast, disruption of the interface in the I164E mutant restores the rate of imatinib binding to nearly that of the isolated kinase domain (0.78 +/− 0.02 µM−1 s−1).
Figure 3. Effect of the SH2-N-lobe interaction on imatinib binding.
Imatinib binding rates (kon) of the SH2-KD construct and two mutants thereof referenced to that of the isolated kinase domain. Formation of the native SH2-N-lobe interface decreases the rate of drug binding and is reduced further by the mutation T231R. Mutational perturbation of the interface (I164E), however, enhances the binding rate compared to the wildtype.
These results show that changes at the SH2-kinase domain interface are reflected - to a small extent - in conformational features of the active site and impact drug binding kinetics. In line with our expectations, the native SH2-N-lobe interaction slows down imatinib binding to the kinase active site, indicating a preferential occupation of the active conformation.
Crystal structure of the Abl SH2-KD construct in complex with dasatinib
To obtain structural insights into how the SH2-N-lobe interaction may affect the kinase active site we crystallized the wild-type SH2-KD construct in complex with the multi-targeted Abl-and Src-family kinase inhibitor dasatinib (Sprycel) and solved the structure at 2.9 Å resolution. The rationale for choosing this particular drug was the notion that dasatinib preferentially binds to the active kinase conformation [4,24]. Crystallographic data and refinement statistics are given in Table 1.
Table 1.
Structure determination and refinement
| Data collection | ||
| wavelength (Å) | 1.116 | |
| space group | P 21 21 2 | |
|
| ||
| cell dimensions | ||
| a,b,c (Å) | 114.1, 125.4, 56.2 | |
|
| ||
| resolution (Å) | 46.8-2.90 (2.95-2.90) | |
| Rsym (%) | 14.5 (5.69) | |
| I/σI | 9.2 (2.1) | |
| completeness (%) | 99.6 (99.6) | |
| redundancy | 4.0 (4.0) | |
| Wilson B factor | 50.47 | |
|
| ||
| Refinement | ||
| resolution (Å) | 46.8-2.90 | |
| reflections used | 18657 | |
| Rfree reflections | 947 | |
| Rwork/Rfree | 0.231/0.259 | |
|
| ||
| no. atoms | ||
| protein | 5114 | |
| ligand | 66 | |
|
| ||
| average B-factors | ||
| protein | 51.1 | |
| ligand | 44.8 | |
|
| ||
| RMSD from ideality | ||
| bonds (Å) | 0.004 | |
| angles (°) | 0.81 | |
|
| ||
| Ramachandran statistics | ||
| favored (%) | 95 | |
| disallowed (%) | 0 | |
| MolProbity clash score | 5.77 | |
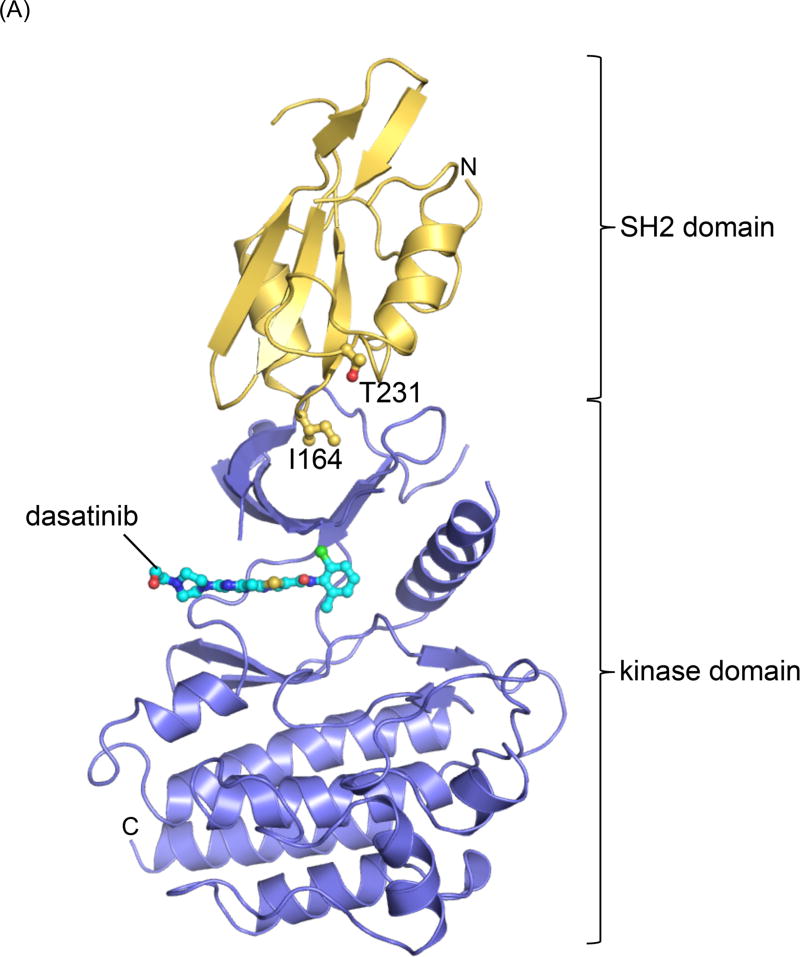
The crystal form has two copies of the kinase domain in the asymmetric unit, however only one SH2 domain could be localized in the electron density map, presumably due to a high degree of disorder in the second SH2 domain. Since the conformation of both kinase domains is very similar, we focus our interpretation on the molecule for which the SH2 domain could be built. The structure shows the SH2 and kinase domain in an extended conformation (Figure 4A), as was observed previously for the SH3-SH2-kinase domain construct bound to PD166326 (PDB ID: 1OPL) [2]. The previous crystal form, however, contained two different conformational states of the SH3-SH2-KD construct, (i) the compact autoinhibited conformation, in which the SH2 domain docks onto the C-lobe of the kinase domain and (ii) the extended conformation, for which only the SH2 and kinase domains could be modeled and which was characterized by very high temperature factors.
Figure 4. Crystal structure of the Abl SH2-KD construct.
(A) Cartoon representation of the crystal structure of the SH2-KD construct (chain B) in complex with dasatinib (cyan, ball-and-stick representation). The SH2 domain is colored yellow, the kinase domain blue. The side chains of two residues at the SH2-N-lobe interface, Ile 164 and Thr 231, whose roles were studied by mutagenesis are highlighted.
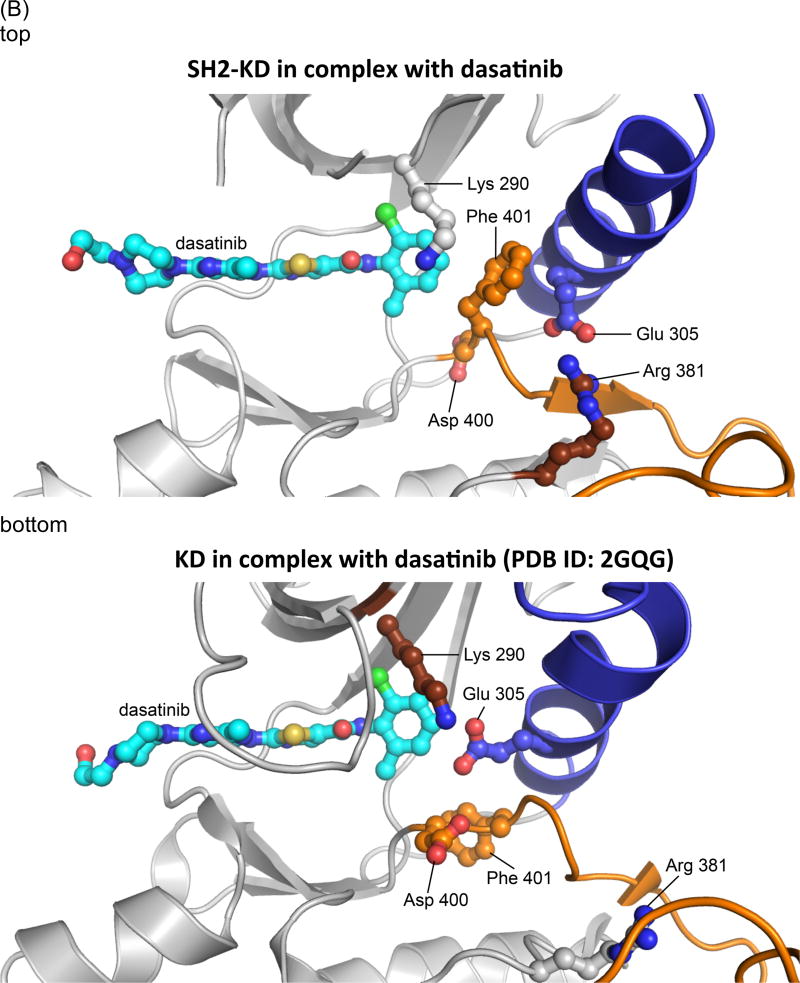
(B) Detailed view of the active site in the SH2-KD structure (top) compared to the published structure of the isolated kinase domain in complex with dasatinib, pdb ID: 2GQG (bottom) [24]. The activation loop is highlighted in orange and helix αC in blue. In contrast to the active state of the isolated kinase domain (DFG-Asp In/αC-Glu In), the SH2-KD construct adopts an intermediate state (DFG-Asp Out/αC-Glu Out).
The previously determined structure and our current structure both show a predominantly hydrophobic interface between the SH2 domain and the N-lobe of the kinase domain, surrounding Ile 164 and burying ~ 1000 Å2 of total surface area. Significant differences between the two structures are observed at the kinase active site: While the extended structure in complex with PD166326 showed an inactive conformation with a flipped DFG-motif (DFG-Asp Out, αC-Glu In), the active site in our SH2-KD construct, bound to dasatinib, adopts an intermediate state, with helix αC being partially swung out of the active site (DFG-Asp Out, αC-Glu Out) (Figure 4B, top). The conserved salt bridge between Glu 305 and Lys 290, characteristic of the inactive state, is disrupted due to rotation of the Glu 305 side chain, and instead forms an ion pair with Arg 381 of the conserved HRD-motif in the catalytic loop. Such an intermediate conformation of the Abl active site has also been observed in crystal structures of the isolated kinase domain in complex with PD166326 (PDB ID: 2G2H) and an ATP analog-peptide conjugate (PDB ID: 2G2F), respectively, and was suggested to represent an intermediate state on the transition pathway between a ‘Src-like inactive’ (DFG-Asp In, αC-Glu Out) and the active (DFG-Asp In, αC-Glu In) state of the Abl kinase domain [3]. Notably, our crystal form captured this intermediate state in the presence of dasatinib, a drug that had previously been crystallized in complex with the active state of the isolated c-Abl kinase domain (PDB ID: 2GQG) (Figure 4B, bottom) [24]. The differences in the conformation of the DFG motif and the Glu 305 side chain between the intermediate and the active state do not significantly affect the general binding mode of dasatinib, the P-loop being poorly ordered in both structures. Fo-Fc omit electron density for dasatinib and the DFG motif in our structure is shown in Supplementary figure 2.
This comparison shows that dasatinib can, in principle, recognize various conformational states of the kinase active site, which is in line with computational predictions [4,24,25] and might provide a rationale for its greater therapeutic potency compared to conformationally more selective inhibitors, such as imatinib. It should be noted, however, that dasatinib strongly favors the active ‘DFG-Asp In’ conformation in solution, as was determined by NMR studies of the isolated kinase domain [4]. Taken together, our structural analysis indicates that formation of the SH2-N-lobe interface – while functionally favoring the kinase active state - is compatible with various different conformations of the catalytic center.
An alternative SH2-kinase interface in the crystal lattice
When examining the crystal lattice, we noticed a crystallographic dimer in which the SH2 domain and parts of the flanking N-lobe of one molecule interact with the C-lobe of a neighboring kinase molecule (Supplementary figure 3A). The dimeric interface buries a total surface area of ~ 1200 Å2 and is centered on a key aromatic stacking interaction between Tyr 158 of the SH2 domain and Tyr 468 in helix αC of the C-lobe (Supplementary figure 3B). Surrounding polar contacts involve Ser 161 in the SH2 domain as well as Thr 262 and Lys 281 in the flanking N-lobe, which interact with Glu 472 in the C-lobe of the neighboring molecule. An additional hydrogen bond links Thr 259 in the SH2 domain to Tyr 432 in the neighboring C-lobe. Interestingly, this dimeric arrangement brings the phosphotyrosine binding site on the SH2 domain of one molecule in proximity of the active site of the other kinase molecule, which leads us to speculate that it may provide a mechanism for processive substrate phosphorylation.
Abl has long been known to efficiently phosphorylate substrates at multiple sites – a property that has been attributed to its SH2 domain [26–28]; the underlying molecular mechanism, however, has remained elusive. One possibility is that the SH2 domain binds phosphorylated substrates, increases their local concentration and thus the probability of further phosphorylation, as was suggested for Fes kinase [10]. Besides such a proximity-based mechanism, it is not immediately obvious how the SH2 domain in the active, extended conformation of Abl would be able to specifically present bound substrates to the kinase active site in cis. In the context of the crystallographic dimer observed here, however, binding of a pre-phosphorylated substrate to the SH2 domain of one Abl molecule may serve to position other tyrosine residues on the substrate for phosphorylation in trans. To illustrate such a mechanism we modeled two regions of a putative substrate onto the crystallographic Abl dimer, based on the crystal structures of the Abl kinase domain bound to an ATP-peptide conjugate (PDB ID: 2G2I [3]) and the SH2 domain of Src in complex with a phosphotyrosine-containing peptide (PDB ID: 1SPS) (Supplementary figure 3B) [29]. Our model is consistent with the idea that a substrate containing multiple tyrosine residues might be captured through one phosphotyrosine interacting with the SH2 domain of one Abl subunit, while spanning across the dimer and presenting another tyrosine residue to the active site of the other Abl subunit for phosphorylation.
Conclusions
We set out to dissect the structural mechanism by which the SH2 domain of Abl activates the kinase domain by studying these two domains under defined conditions in vitro. While our functional data monitoring kinase activity and drug binding rates are qualitatively consistent with the observation that the SH2-N-lobe interaction in the extended conformation stimulates kinase activity, the effects of this physiologically critical interaction are rather small in our experimental set-up. Nevertheless, our studies validate the N-lobe docking of the SH2 domain as a relevant conformation of c-Abl, which is in line with previous functional data that were obtained using different constructs and experimental approaches [10–12]. For instance, when testing the activity of immunoprecipitated full-length Bcr-Abl from human cancer cells towards a peptide substrate, the T231R mutation was found to increase activity (Vmax) by a factor of ~1.4 [12], while the disruptive I164E mutation decreased Vmax by a factor of ~ 3 [11]. The presence of the SH2 domain in the context of the SH2-kinase domain construct compared to the kinase domain alone was found to enhance Vmax ~ 3.5 fold using immunoprecipitated material [10]. In contrast, Nagar et al. did not detect a significant difference in activity when comparing the isolated kinase domain to an activated SH3-SH2-KD construct (containing the mutations P242E and P249E), both of which had been bacterially expressed and purified [9]. In the same study, however, the SH3-SH2-KD construct showed a significantly reduced activity below the level of the isolated kinase domain upon the simultaneous introduction of three mutations (I164E, T291E, and Y331A) at the SH2-N-lobe interface.
Taken together, these various studies consistently report stimulatory effects of the SH2-N-lobe interaction on kinase activity. However, the measured effects are all rather small compared to the biological consequences brought about by mutations in the SH2-N-lobe interface. It is thus relevant to speculate what factors contribute to the important physiological impact of this interaction.
Naturally, moderate changes in velocity or Km can result in steeper, threshold responses in a cellular context, due to downstream signaling molecules or changes in the activity of counteracting enzymes (phosphatases). Such “extrinsic” signal amplification is certainly an important factor [11], but structural mechanisms intrinsic to Abl may also have a role [30]. One such possibility is that the SH2-N-lobe interaction provides an alternative docking site for the kinase domain, which might help destabilize the fully assembled and autoinhibited conformation. Little is known about how the C-terminal half of c-Abl interacts with the N-terminal half in the autoinhibited state, in which the latter adopts an inactive Src-like conformation. Posttranslational modifications of Abl may also be a deciding factor, since Bcr-Abl is multi-phosphorylated in the cell [31,32]. The functional characterization of native phosphorylation sites on Abl will, therefore, be an important area for future studies. It has also been hypothesized that the formation of the SH2-N-lobe interface may stimulate autophosphorylation on Tyr245 in the SH2 kinase linker, thereby prolonging the activated state [9]. This idea is consistent with recent data demonstrating that the SH2-N-lobe interface impacts strongly autophosphorylation in the SH2-kinase linker and in the activation loop [11,33]. Furthermore, binding partners, as identified in extensive proteomic analyses [34], may modulate Abl activity in the cell. It is also possible that domains other than the SH2 domain provide intramolecular docking sites in the context of the full-length protein, thereby stabilizing the active state. Hence, it will be interesting to investigate if and how the effects of mutations that simultaneously occur in relapsed CML patients and may impact domain interactions cooperate to confer drug resistance. The T231R mutation at the SH2-N-lobe interface, for instance, was found to coincide with a second SH2 domain mutation, S173N, in 50% of the Bcr-Abl clones tested [12].
We further speculate that oligomerization of Abl, which is known to occur in the cell may have a role in rendering the phosphorylation of Abl substrates processive and thereby increasing activity [35–38]. The crystallographic dimer that we identified might provide a mechanism for how dimerization and efficient multi-site phosphorylation may be linked. However, it is currently unclear if such a dimer exists and what factors would stabilize it. While the N-terminal region of Abl containing the SH3, SH2 and kinase domains has been reported to mediate the association with full-length Abl in the cell [35], the shorter SH2-kinase domain construct used by us does not dimerize to a detectable degree in vitro (data not shown). The model that we have constructed (Supplementary figure 3A) thus requires thorough experimental testing, which is beyond the scope of this paper and the subject of ongoing studies.
A detailed understanding of the various mechanisms that cooperate in Abl activation is expected to uncover novel targets for therapeutic intervention in CML that could complement traditional ATP-competitive inhibitors. The potential of such approaches has been demonstrated by the discovery of GNF-2, a selective allosteric inhibitor that occupies the myristate binding pocket and displayed in vivo efficacy in murine bone marrow transplantation models [39].
Supplementary Material
Summary statement.
Constitutive activation of the tyrosine kinase c-Abl in the cell involves interactions between the SH2 and kinase domains. We present a crystal structure of a c-Abl construct comprising these domains and analyze the functional role of their interface in vitro.
Acknowledgments
We thank the staff at beamline 8.3.1 of the Advanced Light Source at the Lawrence Berkeley National Laboratory for technical support, David King for peptide synthesis and mass spectrometry, and members of the Kuriyan lab for advice, particularly Brian Kelch, Tiago Barros, Jonathan Winger, and Qi Wang. We would also like to acknowledge Markus Seeliger, who kindly shared experimental protocols and provided assistance with data analysis.
FUNDING
This work was supported in part by a grant from the Leukemia and Lymphoma Society (LLS 7393-06, J. Kuriyan/ B. Druker) and by institutional funds from the Howard Hughes Medical Institute. Sonja Lorenz held a postdoctoral fellowship by the Leukemia & Lymphoma Society and is now an Emmy Noether fellow of the German Research Foundation. Oliver Hantschel is supported by the ISREC Foundation.
Abbreviations footnote
- Akt
AKT8 virus oncogene cellular homolog
- Bcr
breakpoint cluster region
- Btk
Bruton tyrosine kinase
- CDK
cyclin-dependent kinase
- C-lobe
carboxyl-terminal lobe
- EGFR
epidermal growth factor receptor
- Hck
hemopoietic cell kinase
- N-lobe
amino-terminal lobe
- NCS
non-crystallographic symmetry
- NMR
nuclear magnetic resonance
- PKA
protein kinase A
- PKB
protein kinase B
- PDK1
phosphoinositide-dependent protein kinase 1
- SH2
Src-homology 2
- SH3
Src-homology 3
- STAT
signal transducer and activator of transcription
- TLS
translation/libration/screw
- Tpx2
targeting protein for Xklp2
Footnotes
AUTHOR CONTRIBUTIONS
Sonja Lorenz and Tricia Deng prepared all proteins, performed activity assays, and drug-binding assays, crystallized the SH2-KD construct, and collected diffraction data. Sonja Lorenz solved the crystal structure and prepared the manuscript. Oliver Hantschel provided the DNA constructs and helpful discussions. Giulio Superti-Furga and John Kuriyan conceived the work. John Kuriyan provided critical assistance in experimental design, data interpretation, structure analysis, and manuscript preparation. All authors read and approved the final paper.
References
- 1.Hantschel O, Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- 2.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 3.Levinson NM, Kuchment O, Shen K, Young MA, Koldobskiy M, Karplus M, Cole PA, Kuriyan J. A Src-like inactive conformation in the abl tyrosine kinase domain. PLos Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vajpai N, Strauss A, Fendrich G, Cowan-Jacob SW, Manley PW, Grzesiek S, Jahnke W. Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of imatinib/nilotinib and dasatinib. J. Biol. Chem. 2008;283:18292–18302. doi: 10.1074/jbc.M801337200. [DOI] [PubMed] [Google Scholar]
- 5.Skora L, Mestan J, Fabbro D, Jahnke W, Grzesiek S. NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc. Natl. Acad. Sci. USA. 2013;110:E4437–45. doi: 10.1073/pnas.1314712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell. 2001;105:115–126. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 7.Márquez JA, Smith CIE, Petoukhov MV, Surdo, Lo P, Mattsson PT, Knekt M, Westlund A, Scheffzek K, Saraste M, Svergun DI. Conformation of full-length Bruton tyrosine kinase (Btk) from synchrotron X-ray solution scattering. EMBO J. 2003;22:4616–4624. doi: 10.1093/emboj/cdg448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagar B, Hantschel O, Seeliger M, Davies JM, Weis WI, Superti-Furga G, Kuriyan J. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol. Cell. 2006;21:787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 10.Filippakopoulos P, Kofler M, Hantschel O, Gish GD, Grebien F, Salah E, Neudecker P, Kay LE, Turk BE, Superti-Furga G, et al. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell. 2008;134:793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grebien F, Hantschel O, Wojcik J, Kaupe I, Kovacic B, Wyrzucki AM, Gish GD, Cerny-Reiterer S, Koide A, Beug H, et al. Targeting the SH2-kinase interface in Bcr-Abl inhibits leukemogenesis. Cell. 2011;147:306–319. doi: 10.1016/j.cell.2011.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherbenou DW, Hantschel O, Kaupe I, Willis S, Bumm T, Turaga LP, Lange T, Dao K-H, Press RD, Druker BJ, et al. BCR-ABL SH3-SH2 domain mutations in chronic myeloid leukemia patients on imatinib. Blood. 2010;116:3278–3285. doi: 10.1182/blood-2008-10-183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornev AP, Taylor SS. Defining the conserved internal architecture of a protein kinase. Biochim. Biophys. Acta. 2010;1804:440–444. doi: 10.1016/j.bbapap.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeliger MA, Young M, Henderson MN, Pellicena P, King DS, Falick AM, Kuriyan J. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Songyang Z, Carraway KL, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 16.Seeliger MA, Nagar B, Frank F, Cao X, Henderson MN, Kuriyan J. c-Src Binds to the Cancer Drug Imatinib with an Inactive Abl/c-Kit Conformation and a Distributed Thermodynamic Penalty. Structure. 2007;15:299–311. doi: 10.1016/j.str.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, Sweet RM, editors. Meth. Enzymol. Vol. 276. 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 18.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Diculescu VC, Enache TA. Electrochemical evaluation of Abelson tyrosine-protein kinase 1 activity and inhibition by imatinib mesylate and danusertib. Anal. Chim. Acta. 2014;845:23–29. doi: 10.1016/j.aca.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 23.Schindler T. Structural Mechanism for STI-571 Inhibition of Abelson Tyrosine Kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 24.Tokarski JS, Newitt JA, Chang CYJ, Cheng JD, Wittekind M, Kiefer SE, Kish K, Lee FYF, Borzillerri R, Lombardo LJ, et al. The structure of Dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 25.Verkhivker GM. In silico profiling of tyrosine kinases binding specificity and drug resistance using Monte Carlo simulations with the ensembles of protein kinase crystal structures. Biopolymers. 2007;85:333–348. doi: 10.1002/bip.20656. [DOI] [PubMed] [Google Scholar]
- 26.Baskaran R, Dahmus ME, Wang JY. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc. Natl. Acad. Sci. USA. 1993;90:11167–11171. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duyster J, Baskaran R, Wang JY. Src homology 2 domain as a specificity determinant in the c-Abl-mediated tyrosine phosphorylation of the RNA polymerase II carboxyl-terminal repeated domain. Proc. Natl. Acad. Sci. USA. 1995;92:1555–1559. doi: 10.1073/pnas.92.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer BJ, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr. Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- 29.Waksman G, Shoelson SE, Pant N, Cowburn D, Kuriyan J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
- 30.Dölker N, Górna MW, Sutto L, Torralba AS, Superti-Furga G, Gervasio FL. The SH2 domain regulates c-Abl kinase activation by a cyclin-like mechanism and remodulation of the hinge motion. PLoS Comput. Biol. 2014;10:e1003863. doi: 10.1371/journal.pcbi.1003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyn MA, Wilson MB, Abdi FA, Fahey N, Schiavone AP, Wu J, Hochrein JM, Engen JR, Smithgall TE. Src family kinases phosphorylate the Bcr-Abl SH3-SH2 region and modulate Bcr-Abl transforming activity. J. Biol. Chem. 2006;281:30907–30916. doi: 10.1074/jbc.M605902200. [DOI] [PubMed] [Google Scholar]
- 32.Salomon AR, Ficarro SB, Brill LM, Brinker A, Phung QT, Ericson C, Sauer K, Brock A, Horn DM, Schultz PG, et al. Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc. Natl. Acad. Sci. USA. 2003;100:443–448. doi: 10.1073/pnas.2436191100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamontanara AJ, Georgeon S, Tria G, Svergun DI, Hantschel O. The SH2 domain of Abl kinases regulates kinase autophosphorylation by controlling activation loop accessibility. Nat. Commun. 2014;5:5470. doi: 10.1038/ncomms6470. [DOI] [PubMed] [Google Scholar]
- 34.Brehme M, Hantschel O, Colinge J, Kaupe I, Planyavsky M, Köcher T, Mechtler K, Bennett KL, Superti-Furga G. Charting the molecular network of the drug target Bcr-Abl. Proceedings of the National Academy of Sciences. 2009;106:7414–7419. doi: 10.1073/pnas.0900653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan P-D, Cong F, Goff SP. Homo- and hetero-oligomerization of the c-Abl kinase and Abelson-interactor-1. Cancer Res. 2003;63:873–877. [PubMed] [Google Scholar]
- 36.McWhirter JR, Galasso DL, Wang JY. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol. Cell Biol. 1993;13:7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golub TR, Goga A, Barker GF, Afar DE, McLaughlin J, Bohlander SK, Rowley JD, Witte ON, Gilliland DG. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol. Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith KM, Van Etten RA. Activation of c-Abl kinase activity and transformation by a chemical inducer of dimerization. J Biol Chem. 2001;276:24372–24379. doi: 10.1074/jbc.M100786200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Adrián FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.