ABSTRACT
In Drosophila, formation of the cardiac extracellular matrix (ECM) starts during embryogenesis. Assembly and incorporation of structural proteins such as Collagen IV, Pericardin, and Laminin A, B1, and B2 into the cardiac ECM is critical to the maintenance of heart integrity and functionality and, therefore, to longevity of the animal. The cardiac ECM connects the heart tube with the alary muscles; thus, the ECM contributes to a flexible positioning of the heart within the animal's body. Moreover, the cardiac ECM holds the larval pericardial nephrocytes in close proximity to the heart tube and the inflow tract, which is assumed to be critical to efficient haemolymph clearance. Mutations in either structural ECM constituents or ECM receptors cause breakdown of the ECM network upon ageing, with disconnection of the heart tube from alary muscles becoming apparent at larval stages. Finally, the heart becomes non-functional. Here, we characterised existing and new pericardin mutants and investigated biosynthesis, secretion, and assembly of Pericardin in matrices. We identified two new pericardin alleles, which turned out to be a null (pericardin3-548) and a hypomorphic allele (pericardin3-21). Both mutants could be rescued with a genomic duplication of a fosmid coding for the pericardin locus. Biochemical analysis revealed that Pericardin is highly glycosylated and forms redox-dependent multimers. Multimer formation is remarkably reduced in animals deficient for the prolyl-4 hydroxylase cluster at 75D3-4.
KEY WORDS: Cellular biology, Developmental biology, Pericardin, Extracellular matrix, Cardiac matrix, Cardiogenesis, Dorsal vessel, Heart
Summary: We identified two new pericardin alleles. Both mutants could be rescued with a genomic duplication of a fosmid coding for the pericardin locus. Biochemical analysis revealed that Pericardin is highly glycosylated and forms redox-dependent multimers.
INTRODUCTION
By underlying or encasing a multitude of cells or tissues, extracellular matrices (ECMs) are essential to several physiological processes including tissue protection, tissue scaffolding, and cell signalling. Biochemical analysis, which is generally impeded by the insoluble and frequently cross-linked nature of the ECM, has shown that the complexity of matrices is much higher than previously expected. It has been reported that the ‘matrisome’, which collectively encompasses the proteins that constitute the ECM, comprises more than 300 proteins in mammals, including collagens, proteoglycans, growth factors, and receptors (Hynes and Naba, 2012). The complexity of matrices is not only reflected by the number of proteins that constitute the matrix, but also by the different ratio with which the various components contribute and by the appearance of unique components in matrices of specific tissues. For example, while a high amount of Collagen I is characteristic of tendons, basement membranes (BMs) contain large amounts of Collagen IV, Laminins, Perlecan, and other proteins. Due to its diverse physiological function, the ECM is more than a homogeneous mass of proteins and carbohydrates. Within the meshwork of its structural components, the ECM is spatially patterned and thereby provides locally restricted reaction environments and structural micro-compartments.
The Drosophila heart is considered as a model for a specialised ECM composition that ensures proper tissue integrity, functionality, and organ performance (Rotstein and Paululat, 2016). In Drosophila, at present, only four collagens or collagen-like proteins have been identified. One of these proteins is Pericardin (Prc) (Chartier et al., 2002); the others are Collagen IV alpha2 (Viking, Vkg) (Yasothornsrikul et al., 1997), Cg25c (Dcg1) (Cecchini et al., 1987; Natzle et al., 1982; Yasothornsrikul et al., 1997), and Multiplexin (Adams et al., 2003; Blumberg et al., 1988; Kelemen-Valkony et al., 2012; Monson et al., 1982; Volk et al., 2014; Yasothornsrikul et al., 1997). The Pericardin precursor protein consists of 1713 amino acids and harbours an N-terminal signal peptide as well as a long repeat region separated into a collagen-like domain and a non-collagen-like domain, with the former containing 26 atypical and several typical (Gly-X-Y)n repeats (Katti et al., 2000). In addition, a single potential Integrin-binding site (RGD) is present at the C-terminus (Chartier et al., 2002; Drechsler et al., 2013; Volk et al., 2014). In contrast to the ubiquitously distributed Collagen IV, Pericardin assembles specifically within distinct matrices: these include the matrix of the heart tube, the surface of pericardial cells and oenocytes, and the cap cells of chordotonal organs. Lack of Pericardin, or its ECM adapter protein Lonely heart (Loh), causes heart failure upon ageing (Drechsler et al., 2013; Rotstein et al., in revision). During development, Pericardin is synthesised and secreted by different tissues: first, during embryogenesis, the pericardial cells secrete Pericardin; later, in first and second instar larvae, the main source of Pericardin secretion is the adipocytes. After biosynthesis, secretion, and release into the haemolymph, Pericardin specifically assembles at the outer surface of the cardiac tube and incorporates into the meshwork formed by typical structural components of basement membranes such as Collagen IV, Perlecan, and Nidogen. Adipocyte-specific knock-down of Sar1 expression inhibits Pericardin secretion and thereby affects the formation of a proper heart ECM in Drosophila (Drechsler et al., 2013; this work). When Pericardin is not expressed, not secreted, or mislocalised, heart integrity is lost, which ultimately results in heart failure and heart collapse. These findings demonstrate that the assembly of a single structural protein, such as Pericardin, in the larval heart is essential for organ integrity and that adipocytes are the major source of distinct ECM components delivered to the heart tube.
Aiming to extend the current knowledge on how the specific meshwork of structural ECM constituents that characterise the heart matrix is established, we investigated aspects of the biosynthesis, secretion and deposition of Pericardin in the cardiac matrix in more detail. The Pericardin protein displays collagen-like features that led to the assumption that Pericardin forms, like Collagen IV, trimeric helices that incorporate into matrices (Chartier et al., 2002; Drechsler et al., 2013).
Thus, we focused particularly on components that are known to play an important role in Collagen IV processing, asking whether these enzymes also process Pericardin. Hydroxylation of proline and lysine residues of collagen proteins, taking place within the ER of the collagen-synthesising cells, leads to dimer- and trimerisation by converting proline or lysine into hydroxyproline or hydroxylysine (Gorres and Raines, 2010; Myllyharju, 2003). This reaction is catalysed by various proteins such as Prolyl 4-hydroxlases (PH4), which map to different loci within the genome. Lysine hydroxylation is performed by Lysyl-hydroxylases of which only one, dPlod, is present in the fly genome. Prolyl 4-hydroxylases are comprised of an α2β2 tetramer; the β-subunit is encoded, in Drosophila, by the pdi gene (Pdi, Protein-disulfide isomerase).
We found that Pericardin processing, i.e. multimerisation, is not blocked in mutants for pdi and dplod, and – to some extent – is inhibited in deficiencies that delete a cluster of PH4-encoding genes, which is in contrast to Collagen IV processing phenotypes seen in mutants for pdi, dplod, or PH4 genes (Abrams and Andrew, 2002; Bunt et al., 2011; Kelemen-Valkony et al., 2012; Molnar et al., 2005; Myllyharju and Kivirikko, 2004; Pastor-Pareja and Xu, 2011; Yasothornsrikul et al., 1997). Possibly, redundant or residual activity of the enzymes is sufficient for Pericardin (but not for Collagen IV) maturation and cardiac assembly. Furthermore, our recent results show that Pericardin deposition at the embryonic cardiac matrix is, unlike deposition of Collagen IV (Pastor-Pareja and Xu, 2011), not necessary for the recruitment and incorporation of additional structural ECM proteins such as Laminin, Nidogen, or Perlecan. Our Western blot analyses provide initial evidence that Pericardin forms intermediate dimers as well as multimeres. Like many other secreted matrix proteins, Pericardin is extensively glycosylated, indicating cross-linking of Pericardin with other ECM proteins via carbohydrate chains. Finally, we extended previous analyses of pericardin mutant phenotypes by characterising two new EMS-induced pericardin alleles, which we identified in a genetic screen for mutants displaying post-embryonic heart malformations. One of the new pericardin alleles turned out to be a protein null allele, whereas the other one represents most likely a hypomorphic allele with Pericardin being expressed but misassembled.
RESULTS
Identification of two new pericardin mutants
For screening a collection of pupal lethal EMS-induced mutants (Koundakjian et al., 2004), we made use of our previously established cardiac hand-GFP reporter to monitor age-related cardiac integrity defects in living animals without the need of dissection (Paululat and Heinisch, 2012; Sellin et al., 2006) (Fig. 1A). This led to the identification of two new mutant alleles carrying EMS-induced mutations on the third chromosome, PMM3-21 (prc3-21) and PMM3-548 (prc3-548), with both displaying an irreversible detachment of the pericardial nephrocytes from the heart tube eminent in third instar larvae (Fig. 1B,C). Age-dependent displacement of pericardial nephrocytes is accompanied by structural defects in the cardiac extracellular matrix, a phenotype that resembles the malformations previously observed in mutants for the Pericardin adapter protein Lonely heart, as well as in pericardin mutants (Chartier et al., 2002; Drechsler et al., 2013). In addition, we observed severely misguided myofibers in the cardiomyocytes of mutant larvae (Fig. 1B-G), a phenotype previously found in cardiac ECM mutants (Drechsler et al., 2013). In this context, the connection between sarcomeres and the ECM, which is established by Integrins and Dystroglycans, might be lost in ECM mutants, which in turn affects regular organisation and orientation of myofibers (Fig. 1).
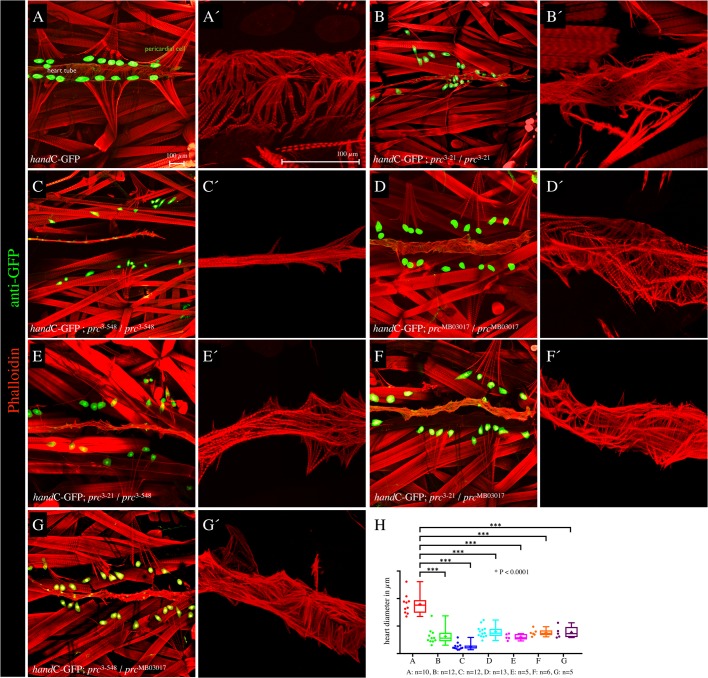
Fig. 1.
Cardiac phenotype of pericardin alleles. (A-G′) Dissected third instar larvae were stained with anti-GFP (green channel) to visualise the GFP reporter expression in cardiac cells driven by hand-GFP (Sellin et al., 2006) and counterstained with Phalloidin-TRITC to label the actin fibres in cardiomyoblasts and somatic muscles (red channel). Pictures show the heart of dissected third instar larvae with the genotype handC-GFP (A,A′), handC-GFP; prc3-21/prc3-21 (B,B′), handC-GFP; prc3-548/prc3-548 (C,C′), handC-GFP; prcMB03017/prcMB03017 (D,D′), handC-GFP; prc3-21/prc3-548 (E,E′), handC-GFP; prc3-21/prcMB03017 (F,F′) and handC-GFP; prc3-548/prcMB03017 (G,G′). The higher magnifications (red channel) illustrate the orientation of F-actin fibres in the heart tube. (H) For quantification of the observed reduction of the heart diameter, five to 13 individual animals were used to measure the width along the heart chamber (with 10 µm distance between the individual reading points). The heart diameter is significantly reduced compared to wild type (***P≤0.0001, unpaired t-test). Higher-magnification images show representative areas of the dorsal vessel with the respective genotype, either taken from the same animal shown on the right side or a different animal with the same genotype. H shows box-and-whisker plots with lower and upper quartiles and the median. Whiskers indicate variability outside the upper and lower quartiles. In addition, scatter plots are shown.
Genetic and phenotypic analysis of PMM3-21/PMM3-548 transheterozygous animals revealed that the two alleles fail to complement each other (Fig. 1E). This observation indicates that the same gene is mutated in both lines. A genetic analysis showed that the newly discovered mutants displaying a cardiac ECM phenotype failed to complement the previously characterised pericardin transposon allele pericardinMB03017 (Fig. 1F,G) as well as the pericardin deficiency Df(3L)vin6. The new pericardin alleles were named pericardin3-21 and pericardin3-548. Homozygous pericardinMB03017, pericardin3-21, pericardin3-548, and transheterozygous animals with the genotype pericardin3-21/pericardinMB03017, pericardin3-548/pericardinMB03017, and pericardin3-21/pericardin3-548 showed similar phenotypes when third instar larvae were stained for hand-GFP to illustrate dissociation of pericardial cells from the heart tube and for F-actin to visualise heart tube collapse (Fig. 1). The latter phenotype was systematically quantified by measuring the diameter of the heart tube every 10 µm along the anterior-posterior axis (heart chamber proper) in control and in mutant animals (Fig. 1H). All pericardin alleles (homozygous or transheterozygous conditions) showed a severe heart tube luminal collapse resulting in a significantly reduced heart diameter and a severe F-actin disarrangement, in addition to the detachment of the pericardial nephrocytes from the heart tube.
Next, we asked whether the pericardin transcript level is altered in mutant animals and we tested the three alleles pericardinMB03017, pericardin3-21, and pericardin3-358 by FISH (Fig. 2A-F). pericardin transcripts were not detectable in homozygous pericardinMB03017 and pericardin3-358 animals (Fig. 2B,D), which suggested that the EMS-induced mutation in pericardin3-358 affects either transcription or transcript stability. By contrast, pericardin transcripts were clearly detectable in the allele pericardin3-21 (Fig. 2F). The latter result indicated an EMS-induced mutation in the pericardin locus that does not significantly affect transcription, but rather severely impairs protein functionality (Fig. 1).
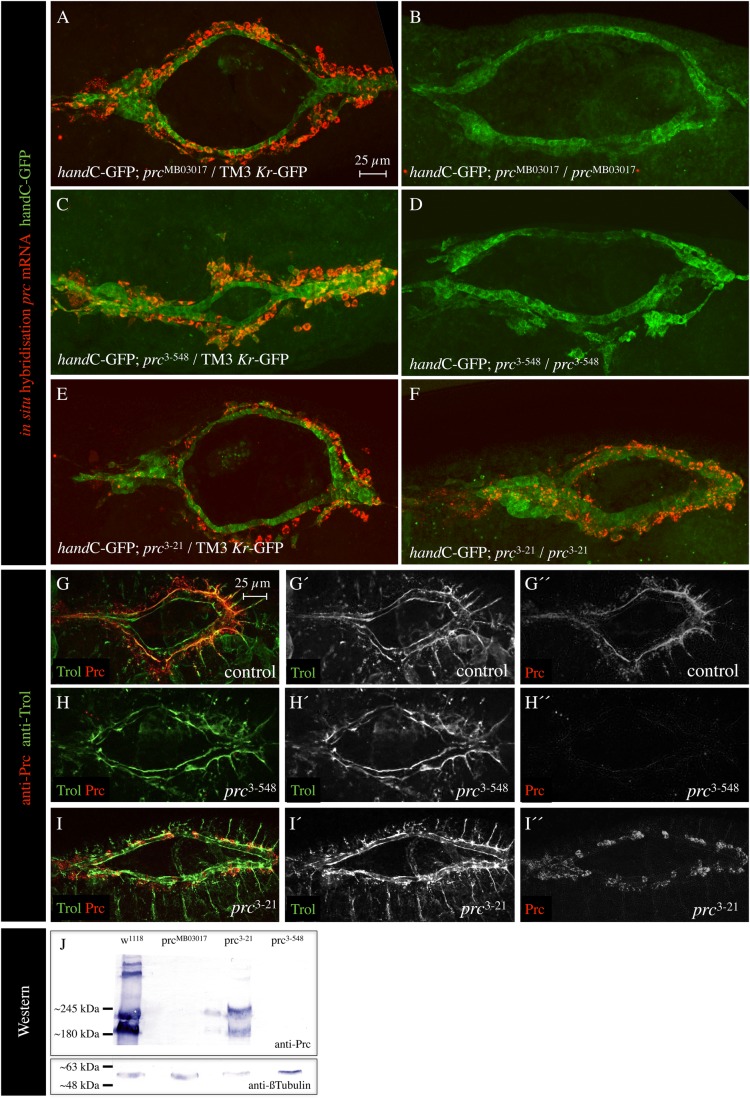
Fig. 2.
Characterisation of pericardin alleles. (A-F) Stage 15-16 embryos stained for prc transcripts (red channel) and handC-GFP (heart cells, green channel). The pericardin mutants with the genotype handC-GFP; prcMB03017/prcMB03017 (B), handC-GFP; prc3-548/prc3-548 (D) and handC-GFP; prc3-21/prc3-21 (F) were probed and compared to the control animals (A,C,E). (G-I) Stage 15-16 embryos stained for Trol/Perlecan (green channel) and Prc protein (red channel). Beside the merged images, single channels are shown individually, as well. (J) Western blot analysis of the respective prc mutants is depicted. As a loading control, the blot was probed for anti-β-Tubulin. All protein extracts contain sufficient amounts of proteins.
Anti-Pericardin staining of embryos or larvae furthermore revealed that in homozygous pericardin3-358 animals (same as pericardin3-358/pericardinMB03017, pericardin3-358/Df(3L)vin6, and pericardinMB03017/pericardinMB03017 individuals) the amount of Pericardin protein is reduced below detection limits (Fig. 2H). By contrast, we found Pericardin being present in homozygous pericardin3-21 animals (Fig. 2I). As shown previously (Drechsler et al., 2013), localisation of additional ECM constituents such as Nidogen and Laminin was not affected in pericardin mutants. This is further corroborated by staining pericardin3-21 and pericardin3-358 for Trol/Perlecan (Fig. 2G-I), which showed that Trol is successfully assembled at the cardiac matrix. Western blot analysis (Fig. 2J) confirmed our observation that Prc (mutated form) is present in pericardin3-21 and absent in homozygous pericardin3-358 animals. Thus, we consider pericardin3-21 as a new hypomorphic, and pericardin3-358 as a new amorphic, pericardin allele. Unfortunately, our sequence analysis of the gene region did not result in the identification of the EMS-induced mutations. Due to the fact that the mutant chromosomes display numerous polymorphisms within the pericardin locus, it was not possible to differentiate between variation and a real point mutation in an unambiguous manner.
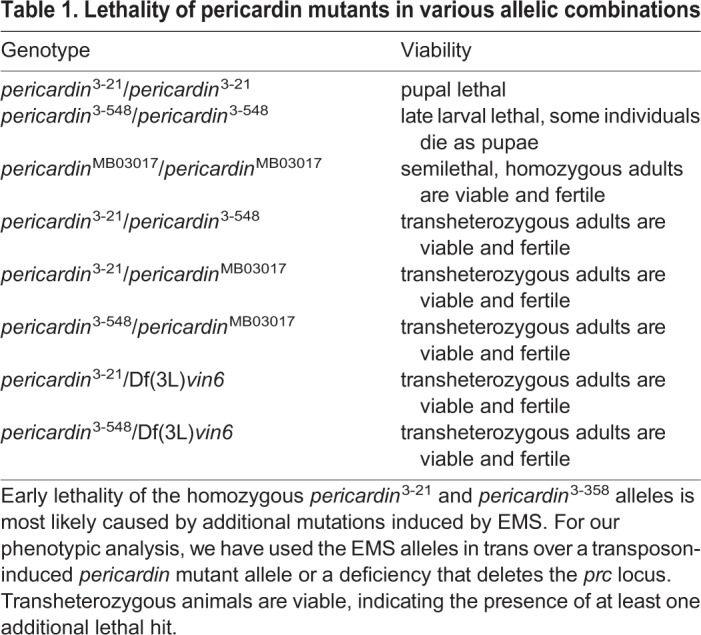
Lack of pericardin caused severe cardiac phenotypes (Fig. 1) and eventually results in reduced function or total block of cardiac performance (Drechsler et al., 2013). As shown previously, absence of a functional heart reduces fitness and lifespan but – under laboratory breeding conditions – does not cause immediate death of the fly. In agreement with this previous result, we found that all transheterozygous allelic pericardin combinations give rise to viable and fertile adult flies (Table 1).
Table 1.
Lethality of pericardin mutants in various allelic combinations
C-terminal GFP fusion proteins mimic endogenous Prc localisation but fail to rescue the pericardin phenotype, whereas an untagged version of Pericardin rescues null alleles
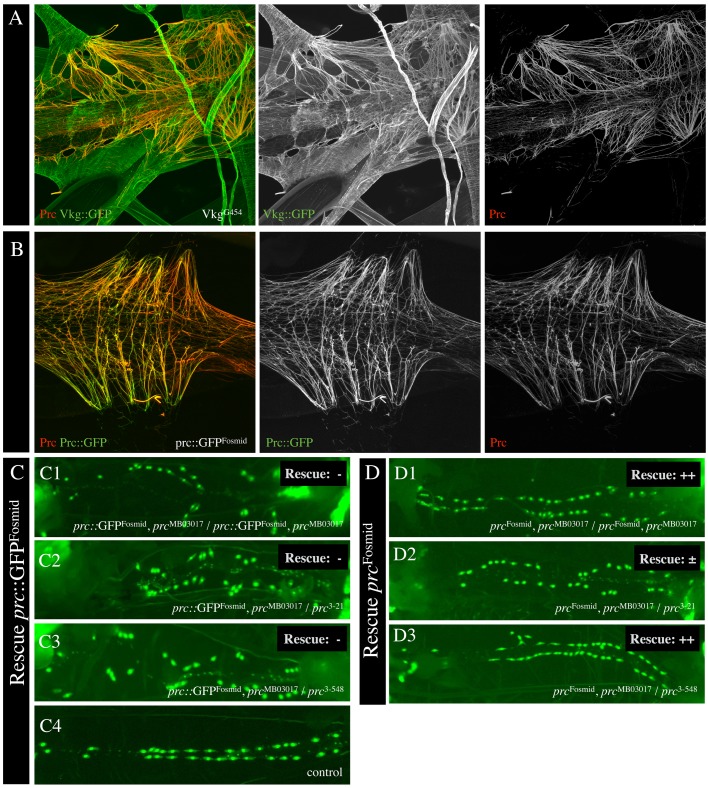
For rescue experiments, we used the fosmid insertion line fTRG497 (pericardin::GFPFosmid) (Ejsmont et al., 2009; Sarov et al., 2016). The transgenic fosmid line (fTRG497) duplicates the genomic region 11,945,299 to 11,977,577 that spans the pericardin locus. In addition, several epitopes have been fused to the C-terminus of Pericardin including an in-frame sGFP as well as V5, preTEV, BLRP, and 3x FLAG tags. We checked this line for Pericardin production and found that Pericardin is synthesised, secreted (Fig. 3A,B) and incorporates into the ECM of the target tissue (Fig. 3B). The fosmid insertion was recombined with pericardinMB03017 to generate a pericardin::GFPFosmid:: pericardinMB03017 chromosome. Rescue capability was checked in the homozygous condition and by crossing pericardin::GFPFosmid:: pericardinMB03017 carrying flies to pericardin3-21 or pericardin3-358 alleles to generate genetically defined transheterozygous animals with the fosmid rescue construct in the background (Fig. 3C). Presence of the handC-GFP transgene in all genotypes allowed us to stain for the heart and to check for rescue capability (Fig. 3C,D). In addition, we checked various genetic combinations for expression of Pericardin via Western blot analysis (not shown). Animals with the genotype pericardin::GFPFosmid, pericardinMB03017 (Fig. 3C1), pericardin::GFPFosmid, pericardinMB03017/pericardin3-21 (Fig. 3C2), and pericardin::GFPFosmid, pericardinMB03017/pericardin3-548 (Fig. 3C3) developed a heart that displayed matrix or disintegration defects, indicating that the Pericardin::GFP encoded by the gene copy present on the fosmid is not able to fully rescue the pericardin phenotype. Although the localisation of the pericardin::GFP construct appears to be normal, we speculated that the C-terminal tag renders Pericardin non-functional. To test this hypothesis, we generated a new transgenic fly line (pericardinFosmid) carrying the identical fosmid at the same landing site as fTRG497, but this time with an untagged version of pericardin (fosmid 028051). The VK00033 line (BL9750, Flybase ID FBst0009750) was used for injection, and two independent fly lines were obtained. Line pericardinFosmid was recombined with pericardinMB03017. The resulting line was used for rescue experiments as described above (Fig. 3D1-D3). We found that – in contrast to the Pericardin::GFP fusion protein – the untagged form of Pericardin rescues the cardiac phenotype of pericardin mutants. These experiments confirm that pericardin3-21 and pericardin3-548 represent new pericardin alleles. The C-terminal GFP-tagged Pericardin fusion protein expressed from the fosmid fTRG497 is properly secreted and assembles into the cardiac matrix, but turned out to be non-functional due to the fused epitope tag.
Fig. 3.
Prc::GFP localises to the cardiac ECM but fails to rescue the prc mutations. (A) The cardiac extracellular matrix of a dissected third instar wild-type larva (heart chamber region) stained for the Viking::GFP (anti-GFP, green channel) and endogenously expressed Pericardin protein (anti-Prc, red channel). Viking (Collagen IV) stains all basal laminae whereas Pericardin is restricted to the cardiac matrix. (B) The cardiac extracellular matrix of a dissected third instar wild-type larva (heart chamber region) stained for the Pericardin::GFP fusion protein expressed from the fosmid insertion (anti-GFP, green channel) and endogenously expressed Pericardin protein (anti-Prc, red channel). By staining of the endogenous Prc with anti-Prc, the antibody recognises the Prc::GFP protein as well, which indicates complete overlap. (C) Recombinant prc::GFPFosmid, prcMB03017 animals (C1) were mated with prc3-21 and prc3-548. F1 larvae at the third instar larval stage were analysed for partial or complete rescue of the cardiac phenotype (dissociation of pericardial cells as a read out for cardiac ECM disassembly). The analysis was carried out in homozygous animals with the genotype prc::GFPFosmid, prcMB03017/prc::GFPFosmid, prcMB03017 (C1) and animals with the genotypes prc::GFPFosmid, prcMB03017/prc3-21 (C2) and prc::GFPFosmid, prcMB03017/prc3-548 (C3). (D) prcFosmid was used instead of prc::GFPFosmid for the rescue experiment (D1-D3).
Pericardin is highly glycosylated and forms redox-dependent multimers
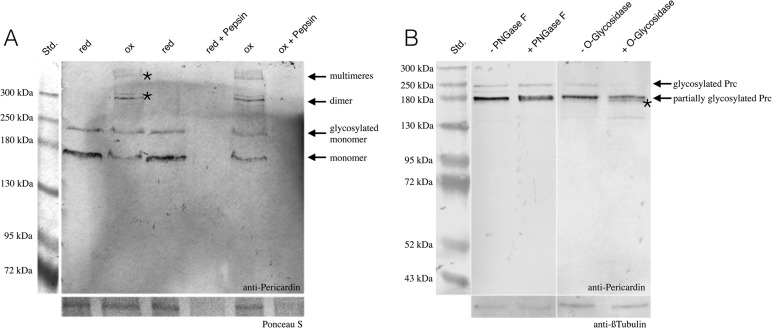
Pericardin encodes a 1713 aa protein with a predicted molecular mass of 163.5 kDa. Only one single isoform has been described (Chartier et al., 2002; Drechsler et al., 2013). Interestingly, the Pericardin-specific antibody EC11 (Zaffran et al., 1995) detected several bands in protein extracts isolated from wild-type embryos or larvae (Drechsler et al., 2013; Zaffran et al., 1995; this work). To assess the identity of these bands in more detail, we extracted proteins from precisely staged third instar wandering larvae and analysed the defined protein samples under reducing and non-reducing conditions. The defined redox conditions should allow identification of disulfide-bonded multimers, which are characteristic to collagens (Lamberg et al., 1996). As depicted in Fig. 4A, Western blot analysis under reducing conditions with anti-Prc antibodies detected two protein species of approximately 170 kDa and 235 kDa, respectively. Regarding the 170 kDa protein, it appeared likely that this band corresponds to the non-modified monomeric form of Pericardin (predicted molecular mass: 163.5 kDa), whereas the higher migrating protein could represent a glycosylated form of monomeric Pericardin. To address this indication we incubated larval protein extracts with deglycosylating enzymes that are specific to either N-linked (PNGase F) or O-linked glycans (O-glycosidase). Interestingly, application of PNGase F resulted in a blurred appearance of the 170 kDa band, concomitant with a slight reduction of the apparent molecular mass (Fig. 4B). While the blurry appearance may be due to an incomplete deglycosylation, the mass shift clearly indicates that the 170 kDa band corresponds to N-glycosylated monomeric Pericardin. Analysis of the Prc primary sequence identified two putative N-glycosylation sites at positions 72-75 and 1554-1557 (Motif Scan), which suggests a maximum mass shift of 2-3 kDa upon deglycosylation, provided that both predicted sites are functional. The observed shift in molecular mass indicates that at least one of them is glycosylated in vivo. Incubation with O-glycosidase in combination with Neuraminidase resulted in a considerable reduction of the 235 kDa band intensity, which suggests that this band corresponds to fully glycosylated Prc, holding both, N- as well as O-linked glycans. In addition, a band slightly below the 170 kDa band appeared as a result of O-glycosidase treatment (Fig. 4B, asterisk), which indicates that also the 170 kDa Prc species holds a limited number of O-linked glycans. Of note, in silico analysis predicts 231 O-glycosylation sites within the Prc primary sequence (NetOGlyc 4.0, http://www.cbs.dtu.dk/services/NetOGlyc). The considerable discrepancy between the predicted molecular mass of Prc (163.5 kDa) and the apparent mass of the fully glycosylated protein species (∼235 kDa) indicates that a high portion of the predicted sites are glycosylated in vivo. As shown previously, collagens form trimers and these trimers are known to be redox sensitive (Lamberg et al., 1996). Interestingly, we found that also Pericardin forms multimers of high molecular weight (approximately 300 kDa and above 500 kDa) under oxidising conditions, indicating the formation of redox-dependent dimers, trimers, or even higher order multimers, probably stabilised by disulfide bonds (Fig. 4A; Fig. S1). Of note, the formation of disulfide-bonded trimers represents a characteristic feature of all collagens (Fessler et al., 1993). However, unlike collagens, both, monomeric as well as multimeric Prc were susceptible to proteolytic cleavage, as confirmed by Pepsin digestion of reduced and non-reduced protein preparations (Fig. 4A). This result is significant since in most cases multimerisation protects the respective collagen helices from proteolytic digestion (Lamberg et al., 1996). The fact that this protective effect did not apply to Prc multimers may indicate that they are more flexible and thus more accessible to proteases than Collagen IV trimers. Of note, our data suggest that the epitope recognized by the anti-Prc antibody is located within the atypical repeats of the Prc collagen-like domain (Fig. 8). Since this domain is presumably part of the mature triple helix (Chartier et al., 2002), the possibility that only the epitope is cleaved, while the triple helix is still resistant, appears unlikely. We concluded from our analysis that Pericardin is post-translationally modified by glycosylation and that the protein forms redox-dependent dimers and trimers in vivo.
Fig. 4.
Mature Pericardin is glycosylated and assembles in redox-dependent multimers. (A) Protein extracts from wandering third instar larvae (w1118) were probed with anti-Pericardin antibodies. Under reducing buffer conditions (red), two prominent bands with a molecular mass of approximately 170 kDa and 235 kDa are apparent. Under oxidising conditions (ox), additional high molecular weight bands are detected (asterisks). Pepsin incubation of reduced (red+Pepsin) as well as non-reduced protein preparations (ox+Pepsin) results in absence of any protein detection. (B) Application of PNGase F results in a blurred appearance of the 170 kDa band, concomitant with a slight reduction of the apparent molecular mass. Incubation with O-glycosidase in combination with Neuraminidase results in reduced abundance of the 235 kDa band, which suggests that this band corresponds to fully glycosylated monomeric Prc. In addition, a band slightly below the 170 kDa band appears as a result of O-glycosidase treatment (asterisk).
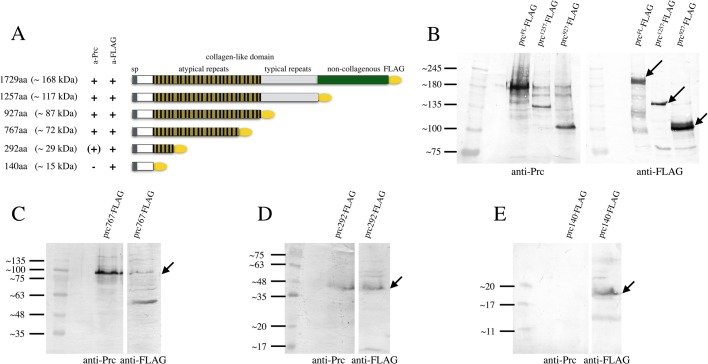
Fig. 8.
Expression of Pericardin constructs in H5 cells uncovers the epitope binding region of EC11 antibody. (A) Graphical presentation illustrating the FLAG-tagged Prc constructs used for expression in H5-cells. (B-E) A series of C-terminal deletions was generated and expressed, and protein extracts from H5 cell cultures were probed each for anti-Prc and for anti-FLAG.
Pericardin modification
Up to now, the biosynthesis and protein maturation pathway of Pericardin has not been well understood. At the sequence level, Pericardin shares a number of conserved sequence motifs with collagens in general, and, in particular, with the three other collagen genes present in the Drosophila genome: Viking, Cg25c, and Multiplexin (Volk et al., 2014). Among these motifs present in Pericardin, typical Collagen repeat domains appear to be most characteristic. Collagen repeats are highly enriched in relatively small amino acids, such as glycine, proline, or lysine, which allow the formation of trimers with a compact triple helical structure. Helix formation of Collagen IV, for example, is fostered by hydroxylation of proline and lysine residues, which is mediated by the enzymatic activity of Prolyl4-hydroxylases (PH4) and Lysyl-hydroxylases (LH) in the endoplasmic reticulum (ER). Proline represents the most abundant amino acid in the repeat region of Pericardin and accounts for more than 9% of all amino acids of the full-length protein (lysins, which are also known to be hydroxylated in collagens, account for about 2%). The PH4s in vertebrates and in Drosophila consist of α2β2-tetrameres, with the β-subunit representing the Protein disulfide isomerase (Pdi) (Annunen et al., 1999; Kivirikko and Myllyharju, 1998). In contrast, the Lysyl-hydroxylases form dimers that display enzymatic activity without the necessity of the presence of additional protein-complex components (Kellokumpu et al., 1994). Cross-linking of Collagen or Elastin fibres in the matrix, which ultimately leads to the formation of a ramified meshwork of ECM megafibres, is mediated by the extracellular enzymatic activity of Lysyl oxidases (Lox) (Kagan and Li, 2003). Two lox genes are present in the Drosophila genome (Molnar et al., 2005), and it has been shown that they regulate ECM stiffness in the nervous system of the fly (Kim et al., 2014). Thus, we tested whether one or more of the known enzymatic activities involved in Collagen biosynthesis and processing, such as PH4, LH or Pdi play a major role in Pericardin processing.
Pdi
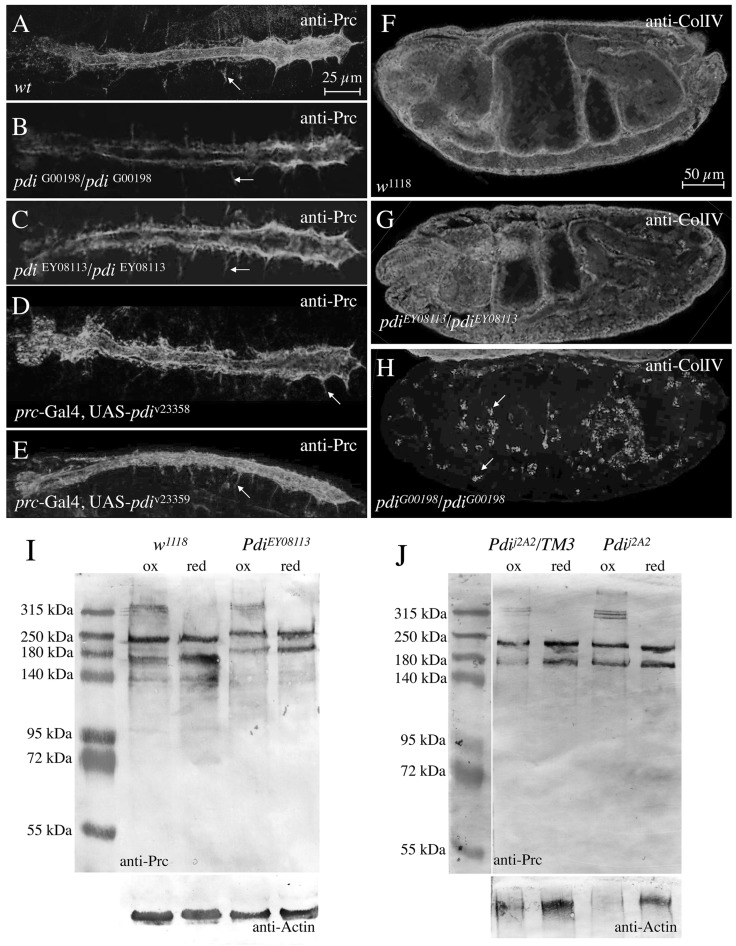
First, we analysed whether the Protein disulfide isomerase in Drosophila, encoded by the single gene pdi (dpdi), plays a role in cardiac matrix formation and Pericardin processing (Fig. 5). We tested three classical Pdi alleles and two RNAi lines. Two alleles, pdiG00198 and pdiJ2A2, were induced by the insertion of transposons, which are located at very similar positions within the first intron of Pdi (Flybase, 1999; Morin et al., 2001; Spradling et al., 1999). Both alleles are described as being capable of inducing lethality. The third allele, pdiEY08113, caused by a P-insertion in the 5′prime UTR region of pdi, is homozygous viable, and thus can be considered as a potentially hypomorphic allele (Bellen et al., 2004). Neither the three alleles induced by P-element insertions nor the two RNAi-mediated down-regulation instances of pdi, in which we used prc-Gal4 for inducing expression of the respective RNAi construct (v23358, Fig. 5D and v23359, Fig. 5E), caused severe abnormalities with respect to Pericardin deposition at the embryonic cardiac matrix (Fig. 5A-E). As control, we stained for Collagen IV (Fig. 5F-H), which is thought to be retained in collagen-producing cells in strong pdi mutants. Indeed, in contrast to wild type (Fig. 5A) or the hypomorphic allele pdiEY08113 (Fig. 5G), Collagen IV was retained in pdiG00198 embryonic haemocytes (Fig. 5H). Western blot analysis of proteins isolated from hypomorphic or amorphic embryos (Fig. 5I,J) confirmed presence of Pericardin in its monomeric, glycosylated, and multimeric forms in all cases tested.
Fig. 5.
Pdi has no visible effect on Pericardin (Prc) deposition or multimerisation. (A-H) We tested the potential role of Pdi on Prc secretion and deposition using pdi mutants stained for anti-Prc (A-E) or anti-ColIV (F-H). (I-J) To verify a potential role of Pdi on Prc multimerisation, we extracted protein from control and pdi mutants and probed the extracts by Western blot analysis. We tested RNAi-mediated down-regulation of pdi in adipocytes and pericardial cells, the main resources for structural ECM proteins (D,E). RNAi constructs (v23358 and v23359) were driven by prc-Gal4. Protein extracts isolated from PdiEY08113 (I) and Pdij2A2 (J) specimens were probed under oxidising and reducing conditions. Arrows in A-E highlight Pericardin-positive segmental ECM structures (compare to Fig. 6C). Arrows in H indicate exemplary hemocytes accumulating ColIV.
Possible role of Prolyl-hydroxylases for cardiac ECM assembly and maintenance
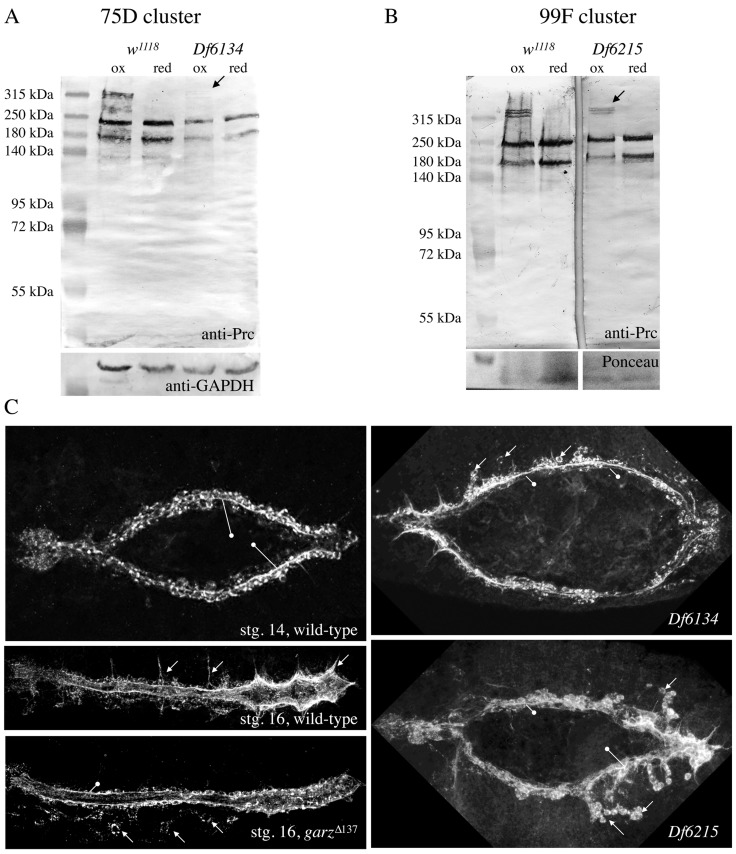
At least 19 genes encoding Prolyl4-hydroxylase α-subunits are present in the Drosophila genome; ten map to 99F8-9, six map to a cluster at 75D3-4, and three are dispersed to other chromosomal regions (Abrams and Andrew, 2002). Together with Pdi, they form the enzymatically active tetrameric Prolyl-hydroxylase complex, processing collagens in the ER of the cell (Annunen et al., 1999). Whether the PH4 proteins display a rigid substrate specificity or whether they process a broader spectrum of target proteins in a tissue-specific manner is yet unknown. However, it has been shown previously that one of the PH4 proteins at 99F, PH4αEFB (CG31022), affects Drosophila Collagen IV (Viking) processing efficiently. Thereby, PH4αEFB RNAi-mediated knock-down in adipocytes results in larvae in which Viking is secreted in its monomeric form. Under these conditions, Viking accumulates in the haemolymph but fails to become incorporated into BMs (Pastor-Pareja and Xu, 2011). Therefore, we tested PH4αEFB mutants as well as PH4αEFB RNAi-mediated knock-down in adipocytes for a possible influence on Pericardin secretion and multimerisation. It should be noted that PH4αEFB is the only PH4 protein, according to the expression data published in FlyAtlas and elsewhere, which is strongly co-expressed with Pericardin in larval adipocytes. In any case, Pericardin production appeared normal and unaltered under the experimental conditions we selected (data not shown). Therefore, we tested deficiencies that remove several Prolyl4-hydroxylase α-subunits at the same time, for instance the PH4 clusters at 75D and 99F with six and ten PH4 genes, respectively (Fig. 6). The deficiency Df(3L)Exel6134 deletes 16 genes including all six PH4 genes at 75D3 (Fig. 6A). The ten Prolyl4-hydroxylases clustered at 99F are removed by the deficiency Df(3R)Exel6215, which removes 15 genes in total (Fig. 6B). Homozygous mutants die during embryogenesis at stage 14, or 15 at the latest, before heart formation is completed and a ramified pericardin network is formed (Fig. 6C). However, we found that Prc is synthesized and secreted as indicated by the formation of Prc sheets and layers around the malformed heart tube in both mutants. For comparison we present garzΔ137 mutant embryos stained for Pericardin (Fig. 6C). It has been shown previously that secretion of Pericardin protein is inhibited in garz mutant embryos (Wang et al., 2012). However, our Western blot analysis of wild-type and PH4 mutants indicated a possible effects on Pericardin multimerisation (Fig. 6A,B). The upper-most band, corresponding to Prc multimeres, is severely reduced in homozygous Df(3L)Exel6134 mutant embryos (deleting the 75D PH4 gene cluster) and slightly reduced in homozygous Df(3R)Exel6215 mutant embryos (deleting the 99F PH4 gene cluster) (Fig. 6A,B). Testing individual PH4 genes from the 75D cluster by utilising RNAi-mediated down-regulation did not result in the identification of one single PH4 protein responsible for Pericardin processing. Possible explanations might be the insufficient efficiency of RNAi-mediated down-regulation of the respective genes or a redundant functionality of PH4s from the cluster, an issue that needs to be analysed further in future studies. However, our results indicated that hydroxylation of Pericardin might take place and insufficiently hydroxylated forms of Pericardin might fail to form multimers. It has been shown for vertebrate collagens that less hydroxylated proteins are less stable than the fully processed proteins and become degraded by the cell, a hypothesis that is corroborated by data on other collagens, which are degraded in the absence of ascorbic acid (vitamin C), a key cofactor of PH4 in mammals (Murad et al., 1981), as well as the fact that lack of functional PH4 keeps collagens retained in the ER of collagen-producing cells (Walmsley et al., 1999). Whether this applies to Pericardin as well remains to be elucidated in future experiments.
Fig. 6.
Deletion of the PH4 gene cluster at 75D inhibits efficient higher-order multimerisation of Pericardin. (A,B) Protein extracts isolated from homozygous animals with the genotype Df(3L)Exel6134 (deleting the PH4 gene cluster at 75D) (A) and Df(3R)Exel6215 (deleting the PH4 gene cluster at 99F) (B) were probed for anti-Pericardin under oxidising and reducing conditions. As loading control Western blots were probed for GAPDH or membranes were stained for Ponceau. (C) Wild-type, homozygous Df(3L)Exel6134 and Df(3R)Exel6215 and homozygous garzΔ137 mutant embryos (stg. 16/17) were stained with anti-Prc.
Lysyl-hydroxylase (dPlod)
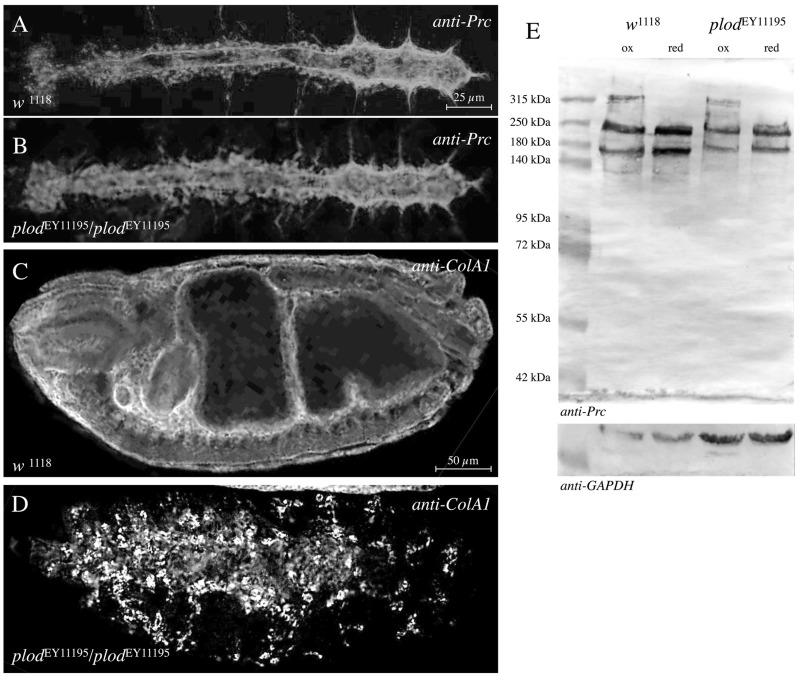
The Drosophila genome harbours a single orthologue of the vertebrate Lysyl hydroxylase 3 named dPlod (Bunt et al., 2011, 2010). This enzyme catalyses hydroxylation of lysine residues in collagens. In agreement with its expected function, dPlod is highly expressed in type-IV Collagen-producing cells that are haemocytes and adipocytes. According to FlyAtlas, dPlod shows an even broader tissue and temporal expression including embryonic and larval heart and fat body tissue; thereby, dPlod is also co-expressed with pericardin. Bunt and colleagues showed that Collagen IV is retained in embryonic haemocytes in dPlod mutants using a deficiency that removes the dPlod locus and eight additional genes (Bunt et al., 2011). Animals carrying the deficiency die during embryogenesis when homozygous for the deletion. Until now, FlyBase has annotated six P-element insertions associated with the dPlod locus; all of them locate within the 5′prime UTR of dPlod and none of them cause lethality (Flybase, 1999). We tested one of the published lines, dPlodEY11195, for a possible impact on Pericardin biosynthesis and found no distinct effect on the secretion of Pericardin or its incorporation into the embryonic cardiac ECM (Fig. 7A,B). Western blot analysis confirmed that multimerisation of Pericardin is not affected in this dPlod allele when compared to wild-type (Fig. 7E). As control, we used the identical line to stain for Collagen IV (Fig. 7C,D). We observed that Collagen IV is retained in the embryonic haemocytes, a result that confirmed previously made observations (Bunt et al., 2011) and also showed that this gene-specific P-insertion affects dPlod function. Our result indicates that Lysyl-hydroxylation plays no major role in Pericardin biosynthesis and maturation.
Fig. 7.
Plod has no visible effect on Pericardin deposition or multimerisation. (A-D) We tested the potential role of dPlod on Pericardin secretion and deposition using the molecularly characterised dplod mutant allele dplodEY11195 (B,D). Stage 17 embryos were stained for anti-Prc (A,B). The same allele was probed for anti-ColIV (C,D). (E) To check for a potential role of Plod on Pericardin multimerisation, we extracted protein from control and dPlod mutants and probed the extracts by Western blot analysis. As a loading control Western blots were probed for GAPDH.
Expression of Pericardin in H5 cells
To produce larger amounts of the protein in cultured cells for future biochemical and structural characterisation, we checked for possible expression of Pericardin in High-Five (H5) cells. Furthermore, we considered a cell culture system for expressing Pericardin as being potentially helpful to elucidate the function of protein domains present in the primary sequence of Pericardin, such as non-typical Collagen IV repeats or one RGD site close to the C-terminus of the protein. The latter motif might account for anchoring Pericardin to Integrin receptors at the cell surface. Therefore, we induced expression of a full-length FLAG-tagged version of Pericardin in H5 cells (Fig. 8). Interestingly, we found that H5 cells synthesise predominantly the monomeric, partially glycosylated form of Pericardin (Fig. 8B), whereas the fully glycosylated monomer or dimers and trimers are almost undetectable. Of note, earlier experiments that aimed to express human collagens in H5 cells demonstrated that multimerisation of the proteins fails due to the absence of the appropriate PH4 enzymes in the cultured cells (Lamberg et al., 1996). In line with these data, we have shown that a yet unknown PH4 (or a combination of several) at cluster 75D seems to be important, at least to some degree, to proper Prc multimer formation (Fig. 6A). Thus, lack of proper PH4 enzymes in H5 cells might explain the absence of higher-order multimers of Pericardin when expressed in this cell type.
Nevertheless, we used our full-length FLAG-tagged Pericardin construct as a starting plasmid to generate a series of truncated versions and expressed all of them in H5 cells to map the epitope recognised by EC11 (Fig. 8A). Since the antibody detects both monomeric, as well as multimeric Pericardin, lack of proper multimerisation is considered insignificant in this context. Our Western blot analysis showed that the EC11 antibody binds to an epitope within the N-terminal third of Pericardin, mapped between amino acids 140 to 292. Our data are in conflict with a previous report that showed that the EC11 antibody recognises a short sequence at the C-terminus of Pericardin (Chartier et al., 2002). The authors of the respective study used a trypsin-digested ECM preparation that was probed with EC11 as a basis for immuno-precipitation. Subsequently, a Pericardin antigen peptide with the sequence NFQSTYYTK was purified. This sequence is identical to a short stretch located at the C-terminus of Pericardin. However, the EC11 anti-Pericardin antibody clearly recognises Pericardin forms with truncations at the C-terminus (Fig. 8); therefore, we speculate that the previous results may have been impaired by off-target binding of the antibody.
DISCUSSION
The ECM protein Pericardin plays a fundamental role in supporting the structural integrity of the cardiac matrix in the developing Drosophila embryo and larvae (Chartier et al., 2002; Drechsler et al., 2013). Lack of Pericardin or inhibition of Pericardin recruitment to the cardiac matrix results in destabilisation of the larval cardiac ECM meshwork and loss of the alary muscles-pericardial nephrocytes-heart tube connection upon initiation of heart beat activity. Finally, upon ageing, this leads to luminal heart collapse and renders the heart nonfunctional (Fig. 1). Here, we introduce two new EMS-induced pericardin mutants that both display the characteristic cardiac phenotypes; one of the alleles turned out to be a null allele characterised by complete absence of the protein (Fig. 2). All pericardin alleles and transheterozygous combinations of pericardin alleles not only show ECM disintegration upon ageing but also heart collapse associated with disorientation of the cardiomyocyte sarcomeres. In wild-type animals the sarcomeres are highly organised and show a helical orientation (Lammers et al., 2017; Lehmacher et al., 2012). In pericardin mutants this orientation is lost, presumably due to the fact that the costameres lose their link – via integrins – to the extracellular matrix upon ECM disintegration.
Pericardin co-localises with type IV Collagen (Viking) (Fig. 3A). However, in contrast to Viking, which assembles into the basal lamina of virtually all tissues within the animal, Pericardin is highly restricted to the cardiac ECM. ECM fibres harbouring Viking and Pericardin connect the heart tube to the alary muscles (Fig. 3A). A Pericardin::GFP fusion protein expressed from an engineered fosmid carrying an approximately 40 kb genomic region including the pericardin locus with the pericardin gene tagged with GFP, is synthesised, secreted, distributed by haemolymph flow and assembles at the cardiac matrix (Fig. 3B). Co-staining for Prc::GFP and endogenous Pericardin shows a complete overlap. However, the Prc::GFP fusion protein fails to rescue the cardiac phenotype of pericardin mutants (Fig. 3C). By contrast, Pericardin, expressed from an identical fosmid but lacking the C-terminal GFP tag, harbours rescue capability (Fig. 3D). This demonstrates the importance of the C-terminus of Pericardin for full functionality. While future studies are needed to analyse why Prc::GFP fails to rescue, it appears likely that the C-terminal tag affects accessibility of the RGD-site, which is located close to the C-terminus and which might play a role in anchoring Pericardin to the cell surface via Integrin interaction.
Based on distinct sequence similarities, including a central Collagen-like repeat domain with typical (Gly-X-Y)n repeats, Pericardin was classified as a type IV Collagen-like protein. In addition, it has been speculated that, analogous to collagens, Pericardin has the ability to form triple helices (Chartier et al., 2002). However, experimental evidence for dimer-, trimer- or multimerisation of Pericardin has not been provided yet. By analysing protein samples under defined redox conditions, we found that non-reducing conditions result in formation of high molecular weight Prc multimers. Based on the apparent molecular mass (>500 kDa), the largest multimers most likely correspond to trimeric or even higher order multimeric Prc (Fig. 4A; Fig. S1). Thus, similar to collagens (Lamberg et al., 1996), Prc appears to form redox-dependent multimers, probably disulfide-bonded. Considering the fact that Prc is embedded into the cardiac extracellular matrix, which resides in an oxidising environment, multimeric Prc likely represent the mature, functional form, while the monomeric species presumably constitute biosynthesis intermediates. In addition to confirming Prc multimerisation, we also found that the protein is extensively glycosylated. Application of both, N- as well as O-glycosidic bond-specific enzymes resulted in distinct mass shifts (Fig. 4B). While the apparent shift of about 2-3 kDa resulting from PNGase F incubation suggests presence of 1-2 N-linked glycans, the huge mass shift that is obvious upon O-glycosidase incubation (∼65 kDa) indicates substantial O-glycosylation of the protein in the Golgi. This indication is supported by sequence analysis, which predicts only two N-glycosylation (MotifScan) but 231 O-glycosylation sites (NetOGlyc 4.0). Glycosylation represents a highly prevalent post-translational modification of ECM proteins and accounts for cell-cell and cell-matrix attachment by promoting the formation of ramified networks between the glycosylated proteins present in the matrix. Considering this, as well as the severe effects of Prc knock out (Drechsler et al., 2013), glycosylated Pericardin appears to be a core component of the ECM network present at the heart.
Trimerisation of type IV Collagen has been shown to depend on the enzymatic activity of Prolyl (PH4)- and Lysyl (LH)-Hydroxylases. Prolyl-Hydroxylases form a tetrameric complex, with Proteindisulfide-Isomerase (Pdi) being present in the complex. Hydroxylation occurs in the Collagen producing cells in the lumen of the ER prior to secretion of Collagen IV molecules (Stephens, 2012). Interestingly, the primary sequence of Pericardin contains a high number of prolines (158, 9.2% of total) as well as numerous type IV Collagen-like repeats, which indicates that Pericardin may undergo a similar biosynthesis pathway as collagens. Therefore, we analysed whether formation of high molecular mass forms of Pericardin (dimers and trimers) is affected in Pdi (Fig. 5), PH4 (Fig. 6) or LH (Fig. 7) mutants. Only in PH4 mutants we observed the absence of these multimers in Western blot analysis. The PH4 mutants we used harbour deletions that remove six annotated Prolyl-Hydroxylases at once, the so-called 75D cluster. Single mutant lines for each of these PH4s are not available; therefore we tested whether RNAi-mediated down-regulation of the individual PH4 genes in the cluster results in an inhibition of the formation of high molecular mass forms of Pericardin, which was not the case. This is either caused by inefficient down-regulation of the target gene or by a redundant function of more than one of the PH4s in the 75D cluster. Pdi and LH seem to play no major role in Pericardin biosynthesis (Figs 5 and 7).
In H5 cells, which are widely used to express high amounts of recombinant protein for further biochemical characterisation, Pericardin is not expressed endogenously at detectable levels (data not shown). Using a full-length FLAG-tagged version of Pericardin we found that, after the cells were transfected with the construct, only the monomeric form of Pericardin is produced (Fig. 8). We conclude that H5 cells derived from the cabbage looper Trichoplusia ni lack activity of certain enzymes critical to the biosynthesis of multimeric forms of Pericardin. Interestingly, it has been noticed earlier that the expression of human Collagens is highly efficient in H5 cells, but multimerisation fails due to absence of the appropriate PH4 enzymes in the cultured cells (Lamberg et al., 1996). Our result that a yet unidentified PH, or a combination of several PHs from the 75D cluster, appears to be essential to proper formation of Pericardin multimers indicates that, like in Collagens, Pericardin multimer formation fails due to absence of the required PH in the H5 cells. However, we successfully used the cell culture system to determine the epitope recognised by the widely used EC11 antibody. EC11 recognises Pericardin in the native and the denatured state and our experiments indicate that the epitope bound by this antibody locates to the N-terminus of Pericardin (Fig. 8).
MATERIALS AND METHODS
Drosophila stocks
Fly strains used in this study were w1118 vkg::GFP-454 (Collagen IV α2) (Morin et al., 2001), and handC-GFP (Hallier et al., 2015; Paululat and Heinisch, 2012; Sellin et al., 2006). The following strains were obtained from the Bloomington Drosophila Stock Center: pdiG00198 (BL110624, FlyBase ID FBst0307666), pdiEY08113 (BL19868, FlyBase ID FBst0019868), pdij2a2 (BL12090, FlyBase ID FBst0012090) dplodEY11195 (BL20273, FlyBase ID FBst0020273), Df(3R)6134 (BL7613, FlyBase ID FBst0007613) (deletes the PH4 cluster at 75D1), and Df(3R)Exel6215 (BL7693, FlyBase ID FBst0007693) (deletes the PH4 cluster at 99F8). The RNAi lines v23358 (FlyBase ID FBst0454967) and v23359 (FlyBase ID FBst0454968) were obtained from the Vienna Stock Collection.
Mapping of PMM3-21 and PMM3-548
To map the mutation, we generated transheterozygous flies with PMM3-21 and PMM3-548, and approximately 160 deficiency lines from the third chromosome deficiency kit (FlyBase, Bloomington, USA), and scored the progeny for the presence of the cardiac phenotype. Subsequently, fine mapping was performed with smaller deletions, transposon and EMS mutants that affect individual genes.
Rescue of prc-mutants
For rescue experiments, we used the fosmid line fTRG947 (pericardin::GFPFosmid) (Ejsmont et al., 2009; Sarov et al., 2016) (gift from F. Schnorrer, IBDM - Institute de Biologie du Developpement de Marseille, Marseille, France) and pericardinFosmid, which duplicate the genome region 11,945,299 to 11,977,577. Both lines were recombined with pericardinMB03017 and used in trans to pericardin3-21 and pericardin3-548. For generating transgenic Drosophila lines, a commercial service was used (BestGene, Chino Hills, USA). The Drosophila line carrying the landing side was BL9750 (FlyBase ID FBst0009750).
Immunohistochemistry
Wandering third instar larvae were dissected and stained as previously described (Drechsler et al., 2013). The following antibodies were used: rabbit anti-Nidogen (1:1000, gift from S. Baumgartner, Faculty of Medicine/Department of Experimental Medical Science, Lund University, Lund, Sweden), mouse anti-Pericardin [1:5, Developmental Studies Hybridoma Bank (DSHB), Iowa City, USA], rabbit anti-Perlecan/Trol (1:1000, gift from S. Baumgartner), mouse anti-Col4A1 (1:50, gift from M. Mink, Department of Genetics, University of Szeged, Szeged, Hungary) and rabbit anti-GFP (1:2000, Abcam). Secondary antibodies used were anti-mouse Cy2/Cy5 and anti-rabbit Cy3/Cy5 (1:100, Dianova /Abcam).
Deglycosylation assay
Animals were collected and anaesthetised on ice. Per reaction, five wandering third instar larvae were homogenised in 100 µl Glycoprotein Denaturing Buffer (0.5% SDS, 40 mM DTT) and boiled for 10 min. PNGase F and O-Glycosidase/Neuraminidase treatments (12 h, 37°C) were done according to the manufacturer′s instructions (New England Biolabs, Ipswich, USA). Subsequently, samples were boiled in Laemmli buffer and subjected to SDS-PAGE and Western blot analysis.
H5 cells, transfection, and protein extraction from H5 cells
Cell culture and transfection was performed as described in (Hallier et al., 2016). For heterologous expression in H5 cells, full-length Prc was cloned into the pFastBacDual vector (Life Technologies). The endogenous stop codon of Prc was removed and a C-terminal FLAG tag was inserted by appropriate primer design. Truncated versions of Pericardin were established using the respective FLAG tagged construct as a template.
Supplementary Material
Acknowledgements
We thank Stefan Baumgartner and Mátyás Mink for providing antibodies, and Frank Schnorrer for providing fTRG497. We thank Lea Weber and Nicole Kasselmann for their phenotypic analysis on pericardin alleles as well as Yvonne Peters for her assistance with the epitope mapping. Furthermore, we thank Mechtild Krabusch and Martina Biedermann very much for technical assistance and taking care of the fly stocks. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Transgenic fly stocks were obtained from the Vienna Drosophila Resource Center (VDRC, www.vdrc.at). The EC11 anti-Pericardin developed by Laboratoire de Génétique et Physiologie du Développement (LGPD)/CNRS was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: B.R., H.M., A.P.; Validation: H.M.; Investigation: A.C.W., N.K., H.M.; Writing - original draft: A.P.; Writing - review & editing: A.C.W., H.M.; Visualization: A.P.; Supervision: B.R., A.P.; Project administration: A.P.; Funding acquisition: A.P.
Funding
This work was supported by grants from Deutsche Forschungsgemeinschaft to A.P. (DFG PA 13-1, SFB 944 TP7, SFB 944 Z-Project), Deutscher Akademischer Austauschdienst (ProCope program), the State of Lower-Saxony (ZN2832) and Osnabrück University (Open Access Publishing Fund).
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.030361.supplemental
References
- Abrams E. W. and Andrew D. J. (2002). Prolyl 4-hydroxylase alpha-related proteins in Drosophila melanogaster: tissue-specific embryonic expression of the 99F8-9 cluster. Mech. Dev. 112, 165-171. 10.1016/S0925-4773(01)00636-0 [DOI] [PubMed] [Google Scholar]
- Adams J. C., Monk R., Taylor A. L., Ozbek S., Fascetti N., Baumgartner S. and Engel J. (2003). Characterisation of Drosophila thrombospondin defines an early origin of pentameric thrombospondins. J. Mol. Biol. 328, 479-494. 10.1016/S0022-2836(03)00248-1 [DOI] [PubMed] [Google Scholar]
- Annunen P., Koivunen P. and Kivirikko K. I. (1999). Cloning of the alpha subunit of prolyl 4-hydroxylase from Drosophila and expression and characterization of the corresponding enzyme tetramer with some unique properties. J. Biol. Chem. 274, 6790-6796. 10.1074/jbc.274.10.6790 [DOI] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., Tsang G., Evans-Holm M., Hiesinger P. R., Schulze K. L., Rubin G. M. et al. (2004). The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761-781. 10.1534/genetics.104.026427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B., MacKrell A. J. and Fessler J. H. (1988). Drosophila basement membrane procollagen alpha 1(IV). II. Complete cDNA sequence, genomic structure, and general implications for supramolecular assemblies. J. Biol. Chem. 263, 18328-18337. [PubMed] [Google Scholar]
- Bunt S., Hooley C., Hu N., Scahill C., Weavers H. and Skaer H. (2010). Hemocyte-secreted type IV Collagen enhances BMP signaling to guide renal tubule morphogenesis in Drosophila. Dev. Cell 19, 296-306. 10.1016/j.devcel.2010.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt S., Denholm B. and Skaer H. (2011). Characterisation of the Drosophila procollagen lysyl hydroxylase, dPlod. Gene Expr. Patterns 11, 72-78. 10.1016/j.gep.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini J.-P., Knibiehler B., Mirre C. and Le Parco Y. (1987). Evidence for a type-IV-related Collagen in Drosophila melanogaster. Evolutionary constancy of the carboxyl-terminal noncollagenous domain. Eur. J. Biochem. 165, 587-593. 10.1111/j.1432-1033.1987.tb11480.x [DOI] [PubMed] [Google Scholar]
- Chartier A., Zaffran S., Astier M., Sémériva M. and Gratecos D. (2002). Pericardin, a Drosophila type IV Collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development 129, 3241-3253. [DOI] [PubMed] [Google Scholar]
- Drechsler M., Schmidt A. C. and Paululat A. (2013). The conserved ADAMTS-like protein Lonely heart mediates matrix formation and cardiac tissue integrity. PLoS Gent. 9, e1003616 10.1371/journal.pgen.1003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsmont R. K., Sarov M., Winkler S., Lipinski K. A. and Tomancak P. (2009). A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods 6, 435-437. 10.1038/nmeth.1334 [DOI] [PubMed] [Google Scholar]
- Fessler L. I., Condic M. L., Nelson R. E., Fessler J. H. and Fristrom J. W. (1993). Site-specific cleavage of basement membrane Collagen IV during Drosophila metamorphosis. Development 117, 1061-1069. [DOI] [PubMed] [Google Scholar]
- Flybase (1999). The FlyBase database of the Drosophila genome projects and community literature. Nuclei Acids Res. 27, 85-88. 10.1093/nar/27.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorres K. L. and Raines R. T. (2010). Prolyl 4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 45, 106-124. 10.3109/10409231003627991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallier B., Hoffmann J., Roeder T., Tögel M., Meyer H. and Paululat A. (2015). The bHLH transcription factor Hand regulates the expression of genes critical to heart and muscle function in Drosophila melanogaster. PLoS ONE 10, e0134204 10.1371/journal.pone.0134204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallier B., Schiemann R., Cordes E., Vitos-Faleato J., Walter S., Heinisch J. J., Malmendal A., Paululat A. and Meyer H. (2016). Drosophila neprilysins control insulin signaling and food intake via cleavage of regulatory peptides. Elife 5, e19430 10.7554/eLife.19430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. and Naba A. (2012). Overview of the Matrisome-An inventory of extracellular matrix constituents and functions. In Extracellular Matrix Biology (ed. Hynes R. O. and Yamada K. M.), pp. 1-16. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan H. M. and Li W. (2003). Lysyl oxidase: Properties, specificity, and biological roles inside and outside of the cell. J. Cell. Biochem. 88, 660-672. 10.1002/jcb.10413 [DOI] [PubMed] [Google Scholar]
- Katti M. V., Sami-Subbu R., Ranjekar P. K. and Gupta V. S. (2000). Amino acid repeat patterns in protein sequences: their diversity and structural-functional implications. Protein Sci. 9, 1203-1209. 10.1110/ps.9.6.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen-Valkony I., Kiss M., Csiha J., Kiss A., Bircher U., Szidonya J., Maróy P., Juhász G., Komonyi O., Csiszár K. et al. (2012). Drosophila basement membrane Collagen col4a1 mutations cause severe myopathy. Matrix Biol. 31, 29-37. 10.1016/j.matbio.2011.09.004 [DOI] [PubMed] [Google Scholar]
- Kellokumpu S., Sormunen R., Heikkinen J. and Myllyla R. (1994). Lysyl hydroxylase, a collagen processing enzyme, exemplifies a novel class of luminally-oriented peripheral membrane-proteins in the endoplasmic-reticulum. J. Biol. Chem. 269, 30524-30529. [PubMed] [Google Scholar]
- Kim S. N., Jeibmann A., Halama K., Witte H. T., Walte M., Matzat T., Schillers H., Faber C., Senner V., Paulus W. et al. (2014). ECM stiffness regulates glial migration in Drosophila and mammalian glioma models. Development 141, 3233-3242. 10.1242/dev.106039 [DOI] [PubMed] [Google Scholar]
- Kivirikko K. I. and Myllyharju J. (1998). Prolyl 4-hydroxylases and their protein disulfide isomerase subunit. Matrix Biol. 16, 357-368. 10.1016/S0945-053X(98)90009-9 [DOI] [PubMed] [Google Scholar]
- Koundakjian E. J., Cowan D. M., Hardy R. W. and Becker A. H. (2004). The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167, 203-206. 10.1534/genetics.167.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberg A., Helaakoski T., Myllyharju J., Peltonen S., Notbohm H., Pihlajaniemi T. and Kivirikko K. I. (1996). Characterization of human type III Collagen expressed in a baculovirus system. Production of a protein with a stable triple helix requires coexpression with the two types of recombinant prolyl 4-hydroxylase subunit. J. Biol. Chem. 271, 11988-11995. 10.1074/jbc.271.20.11988 [DOI] [PubMed] [Google Scholar]
- Lammers K., Abeln B., Hüsken M., Lehmacher C., Psathaki O. E., Alcorta E., Meyer H. and Paululat A. (2017). Formation and function of intracardiac valve cells in the Drosophila heart. J. Exp. Biol. 220, 1852-1863. 10.1242/jeb.156265 [DOI] [PubMed] [Google Scholar]
- Lehmacher C., Abeln B. and Paululat A. (2012). The ultrastructure of Drosophila heart cells. Arthr. Struct. Dev. 41, 459-474. 10.1016/j.asd.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Molnar J., Ujfaludi Z., Fong S. F. T., Bollinger J. A., Waro G., Fogelgren B., Dooley D. M., Mink M. and Csiszar K. (2005). Drosophila Lysyl Oxidases Dmloxl-1 and Dmloxl-2 Are differentially expressed and the active DmLOXL-1 influences gene expression and development. J. Biol. Chem. 280, 22977-22985. 10.1074/jbc.M503006200 [DOI] [PubMed] [Google Scholar]
- Monson J. M., Natzle J., Friedman J. and McCarthy B. J. (1982). Expression and novel structure of a Collagen gene in Drosophila. Proc. Natl. Acad. Sci. USA 79, 1761-1765. 10.1073/pnas.79.6.1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M. and Chia W. (2001). A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98, 15050-15055. 10.1073/pnas.261408198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad S., Grove D., Lindberg K. A., Reynolds G., Sivarajah A. and Pinnell S. R. (1981). Regulation of Collagen synthesis by ascorbic acid. Proc. Natl. Acad. Sci. USA 78, 2879-2882. 10.1073/pnas.78.5.2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J. (2003). Prolyl 4-hydroxylases, the key enzymes of Collagen biosynthesis. Matrix Biol. 22, 15-24. 10.1016/S0945-053X(03)00006-4 [DOI] [PubMed] [Google Scholar]
- Myllyharju J. and Kivirikko K. I. (2004). Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 20, 33-43. 10.1016/j.tig.2003.11.004 [DOI] [PubMed] [Google Scholar]
- Natzle J. E., Monon K. M. and McCarthy B. J. (1982). Cytogenetic location and expression of Collagen-like genes in Drosophila. Nature 296, 368-371. 10.1038/296368a0 [DOI] [PubMed] [Google Scholar]
- Pastor-Pareja J. C. and Xu T. (2011). Shaping cells and organs in Drosophila by opposing roles of fat body-secreted Collagen IV and Perlecan. Dev. Cell 21, 245-256. 10.1016/j.devcel.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paululat A. and Heinisch J. J. (2012). New yeast/E. coli/Drosophila triple shuttle vectors for efficient generation of Drosophila P element transformation constructs . Gene 511, 300-305. 10.1016/j.gene.2012.09.058 [DOI] [PubMed] [Google Scholar]
- Rotstein B. and Paululat A. (2016). On the morphology of the Drosophila heart. J. Cardiovasc. Dev. Dis. 3, 1-13. 10.3390/jcdd3020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotstein B., Post Y., Reinhardt M., Lammers K., Buhr A., Heinisch J. J., Meyer H. and Paululat A. (2018). Distinct domains in the matricellular protein Lonely heart are crucial for cardiac extracellular matrix formation and heart function in Drosophila. J. Biol. Chem. jbc.M117.817940 10.1074/jbc.M117.817940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M., Barz C., Jambor H., Hein M. Y., Schmied C., Suchold D., Stender B., Janosch S., Kj V. V., Krishnan R. T. et al. (2016). A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 5, 1-38. 10.7554/eLife.12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin J., Albrecht S., Kölsch V. and Paululat A. (2006). Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand. Gene Expr. Patterns 6, 360-375. 10.1016/j.modgep.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Stern D., Beaton A., Rhem E. J., Laverty T., Mozden N., Misra S. and Rubin G. M. (1999). The Berkeley Drosophila genome project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens D. J. (2012). Cell biology: Collagen secretion explained. Nature 482, 474-475. 10.1038/482474a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T., Wang S., Rotstein B. and Paululat A. (2014). Matricellular proteins in development: perspectives from the Drosophila heart. Matrix Biol. 37, 162-166. 10.1016/j.matbio.2014.03.006 [DOI] [PubMed] [Google Scholar]
- Walmsley A. R., Batten M. R., Lad U. and Bulleid N. J. (1999). Intracellular retention of procollagen within the endoplasmic reticulum is mediated by prolyl 4-hydroxylase. J. Biol. Chem. 274, 14884-14892. 10.1074/jbc.274.21.14884 [DOI] [PubMed] [Google Scholar]
- Wang S., Meyer H., Ochoa-Espinosa A., Buchwald U., Onel S., Altenhein B., Heinisch J. J., Affolter M. and Paululat A. (2012). GBF1 (Gartenzwerg)-dependent secretion is required for Drosophila tubulogenesis. J. Cell Sci. 125, 461-472. 10.1242/jcs.092551 [DOI] [PubMed] [Google Scholar]
- Yasothornsrikul S., Davis W. J., Cramer G., Kimbrell D. A. and Dearolf C. R. (1997). viking: identification and characterization of a second type IV Collagen in Drosophila. Gene 198, 17-25. 10.1016/S0378-1119(97)00274-6 [DOI] [PubMed] [Google Scholar]
- Zaffran S., Astier M., Gratecos D., Guillen A. and Sémériva M. (1995). Cellular interactions during heart morphogenesis in the Drosophila embryo. Biol. Cell 84, 13-24. 10.1016/0248-4900(96)81314-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.