Abstract
Cyclic diguanylate (c-di-GMP) is a near universal signaling molecule produced by diguanylate cyclases that can direct a variety of bacterial behaviors. A major area of research over the last several years has been aimed at understanding how a cell with dozens of diguanylate cyclases can deploy a given subset of them to produce a desired phenotypic outcome without undesired cross talk between c-di-GMP-dependent systems. Several models have been put forward to address this question, including specificity of cyclase activation, tuned binding constants of effector proteins, and physical interaction between cyclases and effectors. Additionally, recent evidence has suggested that there may be a link between the catalytic state of a cyclase and its physical contact with an effector. This review highlights several key studies, examines the proposed global and local models of c-di-GMP signaling specificity in bacteria, and attempts to identify the most fruitful steps that can be taken to better understand how dynamic networks of sibling cyclases and effector proteins result in sensible outputs that govern cellular behavior.
Keywords: cyclic diguanylate, c-di-GMP, diguanylate cyclase, signaling specificity, biofilms
INTRODUCTION
Over the last several decades, cyclic diguanylate (c-di-GMP) has been recognized as a major secondary signaling molecule in the bacterial domain. It is perhaps one of the better-studied examples of how bacteria gather environmental information and process that information into actionable cellular operations. The story of its discovery and involvement in a staggering number of cellular processes has been well reviewed (28, 52, 57). Briefly, the current paradigm involves proteins called diguanylate cyclases (DGCs) cyclizing two molecules of GTP to produce c-di-GMP, which is then free to find an effector. This effector, in turn, may bind c-di-GMP and initiate some downstream cellular action as a result. c-di-GMP may be degraded by proteins called phosphodiesterases (PDEs). DGCs and PDEs work in opposition to produce a level of c-di-GMP that may activate effector proteins that can influence cellular processes, including biofilm formation, motility, virulence activation, transcription, macromolecular synthesis, stress responses, and many others (34, 39, 61).
The focus of this review is the key step of c-di-GMP from a given cyclase finding its effector protein. As this paradigm was examined in a progressively larger number of bacteria, many microbes were found to contain a surprising number of DGCs and PDEs, numbering past 60 in some cases (23). Critically, despite the sheer number of DGCs at work, many studies have demonstrated that most DGCs appear to specifically signal for one or two c-di-GMP-dependent processes, indicating some kind of mechanism(s) that allows a given DGC to activate a specific effector (25, 43, 47, 66, 67). These observations gave rise to tension between the idea that effectors can sense the c-di-GMP signal from a specific DGC, and the idea that any one DGC is making the same small, diffusible molecule as its sibling enzymes in the open space of the bacterial cytoplasm. Evidence from several bacterial systems has given rise to varying interpretations of this phenomenon, leading to several possible models for conferring signaling specificity (Figure 1). Here, we explore these models in terms of what observations each is able to describe and how they might ultimately fit together. We also examine specific cases of DGC specificity and emerging work suggesting future paths that will usher in a more complete understanding of how bacteria use c-di-GMP-based networks to modulate many aspects of their behavior.
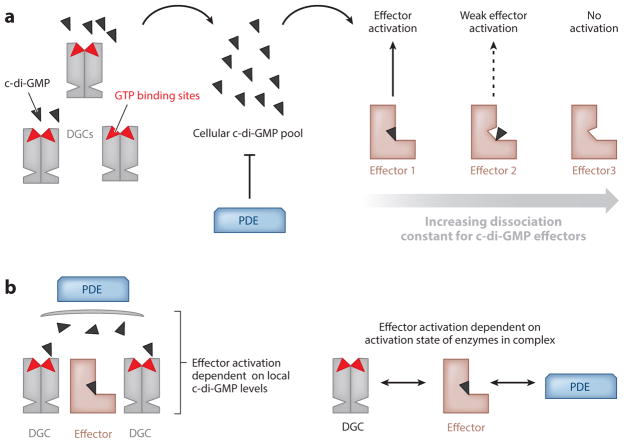
Figure 1.
Modes of c-di-GMP signaling specificity. A variety of models have been proposed to explain the signaling specificity of a given DGC for a specific cellular process. (a) In a global model of signaling specificity, different DGCs produce varying amounts of c-di-GMP that contribute to a global pool, and PDEs can reduce the amount of the global pool. Effectors with the lowest dissociation constant are activated first. As the global pool of c-di-GMP increases, effectors with higher dissociation constants are activated. (b) Local models of c-di-GMP signaling can operate on several different principles. DGCs that are in proximity of—but not necessarily interacting with—effectors can create a local pool of c-di-GMP that may activate target proteins, whereas PDEs may be responsible for keeping the local c-di-GMP pool from affecting other effectors (left). Alternatively, direct interaction between DGCs, effectors, and PDEs can result in functional complexes. The state of the effector depends on the activation state of the DGC or PDE enzymes. Additionally, the DGC catalytic rate may be affected by physical interaction with its partners (right).
THE COMPONENTS OF C-DI-GMP SIGNALING
DGCs operate as dimers, using their GGDEF domains to produce c-di-GMP. The domain’s namesake represents the catalytic residues they use to accomplish c-di-GMP synthesis, but in addition to GGDEF, active cyclases have also been found that make use of GGEEF, AGDEF, and GGDEM motifs. DGCs using SGDEF have also been discovered in eukaryotes (12). In addition to their catalytic residues, approximately half of GGDEF domains also contain residues linked to an autoinhibitory site (I-site). These residues include RXXD, called the primary I-site, which acts to bind c-di-GMP and quell the enzyme’s catalytic activity while bound (Figure 2). The GGDEF domain may exist independently, or it may be fused to a variety of other domains, including PAS, CACHE, CHASE, MASE, and response regulator domains (1, 18, 23). PDEs that break down c-di-GMP come in two varieties. Many PDEs make use of EAL domains to degrade c-di-GMP, while others contain HD-GYP domains (8, 16, 55, 63). A protein that has both GGDEF and EAL domains may act as a DGC, a PDE, or both depending on the circumstances. Additionally, not all GGDEF and EAL domains are catalytically active, a point we will return to in the next section. Finally, DGCs or PDEs either may be found in the cytoplasm or may contain transmembrane domains anchoring them to the inner membrane.
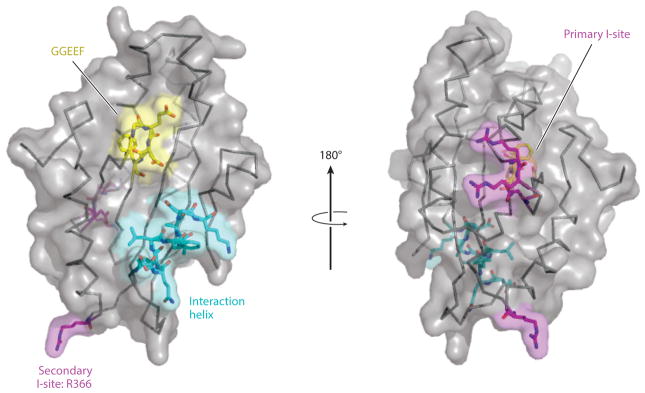
Figure 2.
Components of GGDEF domain of DGCs. Crystal structure of the GcbC GGDEF domain. The amino acid backbone is shown as a ribbon and is overlaid on the electrostatic surface. So-called GGDEF domains (which can also have a GGEEF motif) catalyze the cyclization of two molecules of GTP into c-di-GMP using the catalytically active pocket (GGEEF residues shown in yellow, left). The GGDEF domain makes contact with its effector LapD using the N-terminal portion of the α5GGDEF helix shown in cyan (left). The primary I-site, shown at right in purple, is used to quench catalytic activity when c-di-GMP binds to the I-site pocket. GcbC has an RRxxD motif as its primary I-site, and the R366 residue makes up the secondary I-site (left), which works in conjunction with the primary I-site to bind c-di-GMP.
The centerpieces of c-di-GMP signaling are those effectors, both proteins and RNA, that can bind c-di-GMP and direct the cell to take some action as a result. These c-di-GMP sensors provide a way for environmental signals converted into second messengers to be integrated into a sensible output. The first cellular process recognized to be allosterically activated by c-di-GMP was the synthesis of cellulose via the cellulose synthase from Komagataeibacter xylinus (53, 54), but it took a large-scale, multiple-bacteria analysis to recognize that the c-di-GMP binding property of the cellulose synthase complex resided in a PilZ domain (3). Since that period, PilZ domains have been found throughout the bacterial domain, and they are also known to regulate motility in various bacterial species including select examples in several enterobacteria (56), Caulobacter crescentus (15), and Pseudomonas aeruginosa (2, 40), in addition to virulence in Vibrio cholerae (49). The structure of the PilZ domain was later solved, and was revealed to bind c-di-GMP using RxxxR and DxSxxG motifs (26, 51), with further insight showing the PilZ domain may be undergoing structural rearrangements in order to transmit the c-di-GMP binding event to downstream proteins (58).
Importantly, the finding of widespread cyclases and PDEs encoded throughout the bacterial domain, coupled with the realization that not all c-di-GMP-dependent processes could be accounted for by PilZ-domain-containing proteins, resulted in the search for effector proteins that could bind c-di-GMP but were not related to the PilZ domain. Shortly after the PilZ domain became understood at the structural level, it was discovered that a second class of proteins could bind to c-di-GMP. One prominent example of this class was the PelD protein from P. aeruginosa. PelD, required for pellicle formation, contains a degenerate GGDEF domain but makes use of an intact inhibitory site to bind c-di-GMP and acts as an effector (37). It is currently speculated that conformational shifts in PelD upon binding c-di-GMP are largely responsible for this protein’s signaling abilities (57).
Concurrent to the discovery that degenerate c-di-GMP-producing enzymes may act as effectors, Newell et al. (46) identified a degenerate EAL-domain-containing protein, LapD, as a c-di-GMP receptor in a 2009 study. Given that the first step a PDE must take is to bind c-di-GMP, it is perhaps not surprising that a degenerate EAL domain could participate in c-di-GMP binding. Since then, LapD from Pseudomonas fluorescens (discussed below) has become the prototype example of a degenerate dual GGDEF/EAL-domain-containing protein that can bind c-di-GMP via its defunct EAL domain (45, 46).
More recently, several unique kinds of c-di-GMP binding proteins have been recognized that are neither PilZ domains nor degenerate versions of c-di-GMP enzymes. The transcription factor VpsT from V. cholerae appears to bind c-di-GMP through a W[F/L/M][T/S]R motif resulting in a change in oligomerization that allows this transcription factor to regulate biofilm formation and motility (32). Two more examples of c-di-GMP binding effectors that appear to regulate biofilm formation via transcription are FleQ from P. aeruginosa and Bcam1349 from Burkholderia cenocepacia (22, 29). While the mode of FleQ binding c-di-GMP has recently been described, the motif used to bind c-di-GMP in Bcam1349 is still unknown, although c-di-GMP does appear to enhance the binding of Bcam1349 to DNA (22, 41). Finally, RNA itself has been recognized as an effector that can bind c-di-GMP, providing another target to study in organisms that seem to have abundant DGCs and PDEs encoded in their genomes, but which previously had fewer identified effectors. Sudarsan et al. (62) first recognized an RNA domain they termed GEMM upstream of the open reading frames of some DGCs, PDEs, and genes regulated by c-di-GMP in diverse bacteria. This discovery was quickly followed by the discovery of a second class of riboswitch that made use of a group I self-spicing ribozyme to bind c-di-GMP in a pseudoknot to regulate virulence in Clostridium difficile (36, 60). Whether and how specificity occurs between DGCs and riboswitches remains unknown.
SPECIFICITY AS DERIVED FROM CYCLASE ACTIVITY
Perhaps the most straightforward way specificity between a given cyclase and cellular process could be achieved is through activation of a given cyclase(s) at the desired time. Although many DGCs have putative sensory domains attached to the GGDEF domain, relatively few substrates have been identified. Two prime examples of DGCs where this step has been resolved are a DGC called DosC and a PDE called DosP in E. coli; each senses oxygen, the former through a modified globin domain and the latter a heme-binding PAS domain (65). Together, these enzymes modulate the level of c-di-GMP produced in response to molecular oxygen. Conversely, zinc has been found to bind to a previously uncharacterized regulatory domain of DgcZ in E. coli, causing the GGDEF domains to come out of alignment and inhibit GTP cyclization (69). Another intriguing example of this kind of regulation comes from a 2015 study in V. cholerae A1552. This strain forms a biofilm at 15°C and has 31 predicted GGDEF-domain-containing proteins. When in-frame deletions of all 31 genes were tested, only 6 DGCs were found to be additively responsible for the temperature-dependent increase in c-di-GMP observed at 15°C compared to 37°C, although it is still unknown why these 6 enzymes, and not the other 25, appear to sense the temperature shift (64). Together, these findings help explain how a given DGC may be associated with a particular phenotype, but they do not fully answer the question of how only a given effector protein is engaged.
SPECIFICITY AS DERIVED FROM EFFECTOR BINDING AFFINITY
Many studies have been focused on understanding the organization and integration of the c-di-GMP signal generated by DGCs to the intended effectors as a function of a DGC catalytic rate and effector binding affinity. Such an idea would allow specificity between a given DGC and effector at a distance and creates what we refer to here as the global signaling model (Figure 1a). In 2012, Pultz et al. (50) noted that in Salmonella enterica serovar Typhimurium there are 3 known c-di-GMP-dependent phenotypes, 2 PilZ-domain-containing proteins, and 20 possible DGC/PDEs. They found a more than 40-fold difference in binding affinity to c-di-GMP between YcgR and BcsA, the effectors for halting motility and synthesizing cellulose, respectively. They were further able to demonstrate that the difference in c-di-GMP binding was in part due to the residues near the RxxxR binding motif, and that making substitutions to these residues could increase or decrease their affinity for c-di-GMP with concomitant phenotypic consequences. These results prompted the proposal of a global signaling model where some DGCs are responsible for the first, baseline level of c-di-GMP production, resulting in an inhibition of flagellar torque generation via YcgR (and thus reduced motility), followed by a higher burst of c-di-GMP production to the point of BcsA activation and subsequent cellulose synthesis.
Likewise, binding constants of PilZ domains in P. aeruginosa showed a wide range of binding affinities for c-di-GMP within this species, some differing by a factor of more than 140, providing evidence that global levels of c-di-GMP may be partially responsible for specificity of cellular output by virtue of the ability of various effectors to have differential sensitivity to the molecule (17, 21, 42, 50). In addition to these observations, Pultz et al. (50) put forth ideas for how the cell might modulate which activated effectors function under high global levels of c-di-GMP. For example, some PilZ domains have been found to have multiple binding sites for c-di-GMP, and it has been hypothesized that they may use the binding site as a bandpass, which might allow the effector to operate within a specific range of c-di-GMP and not above or below this range (50). Although this concept has never been experimentally demonstrated, it is worth investigation.
Further candidates for global c-di-GMP signaling include organisms with relatively few DGCs, such as Ehrlichia chaffeensis, which has a single known DGC responsible for regulating invasion of monocytes (35). Likewise, a recent study of Listeria monocytogenes identified three DGCs that appear to work synergistically to induce virulence, and three PDEs that each have different rates of c-di-GMP degradation (11). The single known DGC of Borrelia burgdorferi is required for survival through the tick vector, although dispensable for infection in humans (27). It has been noted that obligate bacterial pathogens tend to have fewer DGCs than free-living organisms (4). While this pattern is not without exception, it may help explain the need of some bacteria for a greater or lesser number of enzymes in their c-di-GMP signaling machinery, with organisms that see more varied environments requiring a greater degree of regulation.
SPECIFICITY AS DERIVED BY PHYSICAL INTERACTION
In contrast to the global signaling model discussed above, other studies have focused on the possibility that a given cyclase and effector may physically interact, where the cyclase would essentially be delivering c-di-GMP to the desired effector protein directly (Figure 1b). We refer to this possibility as the local signaling model. Several observations have emerged in recent years to support such a concept. The P. aeruginosa DGC WspR has been shown to form clustered patches in the cytoplasm when activated by phosphorylation, providing one of the first observations that suggested compartmentalization of c-di-GMP signaling may be occurring (24). Other reports have demonstrated physical interaction among the components of c-di-GMP signaling. For example, it has been noted that the c-di-GMP-bound YcgR effector in Escherichia coli interacts with FliG and FliM to control flagellar braking (48). Further, the DGC DosC and PDE DosP discussed above have been shown to form a complex with one another, opening up the possibility that a DGC and PDE may work together as a complex to regulate how much c-di-GMP is available for effector activation (65).
While the examples above demonstrate that members of the c-di-GMP network in bacteria can interact, they do not resolve how such interactions might result in signaling specificity. One sterling attempt to address this question came from a study conducted by Lindenberg et al. in 2013 (38) that showed that c-di-GMP signaling modules can work as a cascade (Figure 3). These investigators demonstrated that one DGC/PDE pair in E. coli, YegE and YhjH, could regulate the next DGC/PDE pair in the cascade, YdaM and YciR (38). In this manner, the PDE YciR acts as a so-called trigger enzyme, repressing the YdaM DGC until c-di-GMP from the upstream YegE DGC is bound and degraded, leaving YdaM free to act on downstream pathways, including MlrA activation and csgD transcription, resulting in the promotion of biofilm formation. Importantly, the second half of this cascade was found to rely on multiple physical interactions between the DGC, PDE, and transcription factor effector. The PDE YciR was found to make several strong interactions via its EAL domain to the DGC YdaM and effector MlrA, making it possible that physical interaction itself blocked further activation of the pathway until YciR bound and degraded c-di-GMP from YegE. Further, only in the absence of YciR were strong interactions observed between the YdaM GGDEF domain and MlrA (38). This study underlines the importance of physical interaction in target specificity, demonstrating that physical interaction can be important for both inhibition and activation of downstream targets. YegE produces c-di-GMP that apparently comes into contact with the inhibited downstream complex, but c-di-GMP is insufficient to fully activate MlrA. YdaM, in contrast, is only able to strongly interact with MlrA when YciR is otherwise disposed, at which point signaling is mediated by physical interaction.
Figure 3.
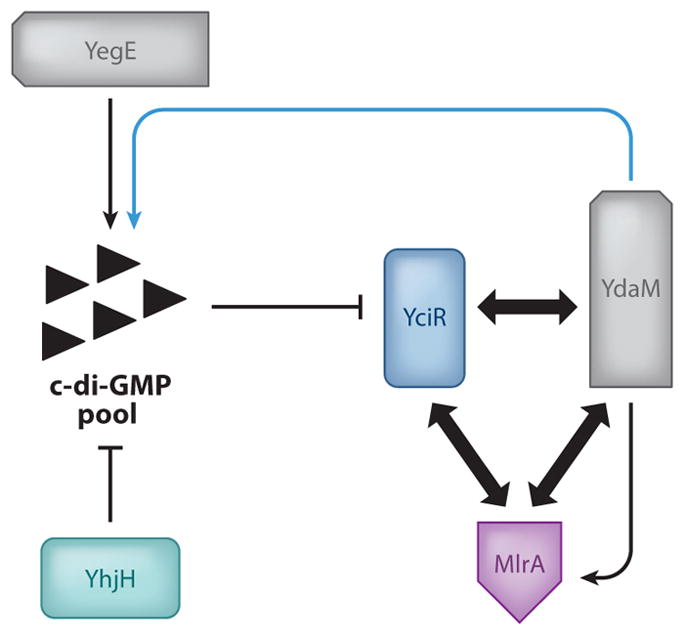
The trigger enzyme system of Escherichia coli. The global c-di-GMP pool is partially controlled by the DGC YegE and the PDE YhjH. When the c-di-GMP pool is low, the trigger enzyme and PDE YciR bind the DGC YdaM and its target, the transcription factor MlrA (bold arrows), preventing YdaM activation of MlrA. When the c-di-GMP pool is high, YciR degrades c-di-GMP and releases YdaM and MlrA in the process, leading to YdaM activation of MlrA and further contribution to the global c-di-GMP pool (blue arrow). Adapted from Lindenberg et al. (38).
While this study illuminated the ways DGCs, PDEs, and effectors can use physical interaction to regulate c-di-GMP signaling, it opened the door to several more important questions. First, it is not clear how interactions among the proteins examined are occurring, and as discussed by the Hengge group (38), it is especially confusing that multiple domains in isolation interact with multiple, isolated domains of their partners. Can a more discrete interface be detected? Second, it was determined that while physical interaction of the DGC YdaM was necessary for activation of the transcription factor MlrA, the catalytic activity of YdaM appeared to contribute only a minor extent to MlrA activation. It was further determined that MlrA does not appear to bind c-di-GMP in a specific manner (38). As a result, it was still unclear whether physical interaction mattered in terms of delivering c-di-GMP to a specific effector over other targets.
The discussion of the YegE/YhjH/YdaM/YciR/MlrA system raised the following question: Can signaling specificity be achieved through c-di-GMP delivery to an effector as a direct result of physical interaction between a DGC and said effector? The Lap system of P. fluorescens is a well-documented case of such a possible direct handoff. The Lap system is one of the best-studied examples of a c-di-GMP effector circuit that is understood from the level of c-di-GMP production to effector activation to downstream proteins exerting their impacts on cellular processes (Figure 4). The Lap genes making up the control switch for biofilm formation include the large, outer membrane adhesin LapA; the periplasmic protease LapG; the effector protein LapD; and the putative ABC transporter responsible for secretion of LapA. The ABC transporter comprises LapB, LapC, and LapE, which correspond to a cytoplasmic membrane ATPase, an inner membrane transporter, and an outer membrane fusion protein, respectively (30). Responsible for controlling biofilm formation in P. fluorescens, LapD is an inner-membrane dual-domain effector with defunct GGDEF and EAL domains (46). When c-di-GMP binds to the EAL domain, LapD undergoes a structural change translated through its cytoplasmic HAMP domain into its periplasmic domain (45). This structural change controls the development of biofilm formation via sequestration of the periplas-mic protease LapG. LapG is a cysteine protease that can bind Ca2+ via residues D134 and E136. Binding of Ca2+ by LapG is required both for its activity as a protease, but also for its binding to the activated LapD (5). A crystal structure of the Legionella pneumophila LapG was the first to shed mechanistic insight on the DUF920 domain conserved in LapG homologs. Critically, a conserved catalytic triad of C-H-D was identified and found throughout many other species containing LapG homologues (9). Cleavage of LapA by LapG requires a conserved double alanine motif at positions 108–109 in the N terminus of LapA. The binding of LapG to LapD is dependent on a highly conserved tryptophan residue on LapD that inserts into a hydrophobic pocket on LapG, relying primarily on shape complementarity (10).
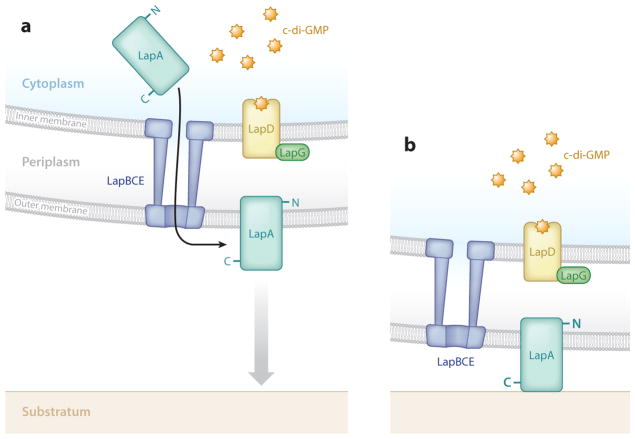
Figure 4.
The Lap system of Pseudomonas fluorescens. (a) A simplified model of the activated Lap system. The large adhesin LapA is translocated through the type I secretion apparatus composed of LapBCE. As long as LapD remains c-di-GMP bound, it sequesters the periplasmic protease LapG. (b) This LapD-LapG binding prevents LapG cleavage of the N terminus of LapA, and biofilm formation may commence via LapA adherence to a substratum. Low c-di-GMP levels result in a LapD protein unable to sequester LapG, LapG-mediated cleaving of the N-terminal 107 amino acids of LapA, loss of LapA from the cell surface, and a reduction in biofilm formation. From Boyd & O’Toole (6).
The Lap system forms a well-researched circuit, first elucidated by studying biofilm formation in P. fluorescens in the presence of high and low levels of inorganic phosphate (44). When phosphate concentrations are high, c-di-GMP is able to bind LapD, causing LapD to adopt its active conformation and bind LapG via its periplasmic domain. When sequestered, LapG is unable to cleave the large adhesin LapA from the outer membrane. LapA is then allowed to accumulate on the cellular surface; LapA adheres to a diverse array of surfaces partly through the use of its central repetitive region (7). As long as LapA remains on the surface of the bacterium, this adhesin may participate in irreversible attachment to a substrate. Conversely, under conditions of low inorganic phosphate, the rapA gene, encoding a PDE, is transcriptionally upregulated, degrading c-di-GMP and causing LapD to adopt its inactive state, releasing LapG and allowing for proteolysis of the LapA adhesin. While it is unclear if this system is found in other genera of bacteria, operons containing LapD and LapG homologs have been discovered in numerous bacteria, and studies have demonstrated that LapG from L. pneumophila can cleave LapA and be bound by LapD of P. fluorescens (9, 10).
While the downstream mechanism of LapD’s impact on biofilm formation is reasonably well characterized, the operation of the upstream c-di-GMP signaling machinery remained somewhat elusive until recently. In addition to not knowing any environmental inputs triggering c-di-GMP signaling aside from phosphate, it was not clear which of the GGDEF-domain-containing proteins and EAL-domain-containing proteins were involved in signaling until 2011, when Newell et al. (47) created mutations of each canonical c-di-GMP-synthesizing or -degrading enzyme predicted in the genome of P. fluorescens. This study revealed four DGCs that appeared to affect biofilm formation in a glycerol- and tryptone-containing minimal medium, two of which did so in a LapD-dependent manner (47). These DGCs, GcbB and GcbC, were shown to signal through LapD by monitoring the fractions of LapA retained or cleaved from the cell’s surface in response to GcbB and GcbC activity. The same study also identified another DGC that affects biofilm formation by virtue of regulating motility, and five PDEs whose disruption causes increased biofilm formation. Given that there are approximately 42 predicted c-di-GMP enzymes in P. fluorescens, these results raised two important questions: How do these DGCs and PDEs act independently of their sibling enzymes to specify a target, and what is the role of the larger network during these signaling events?
In two recent studies from our lab, we attempted to address the question of how a DGC can specifically signal to LapD. In the first study, we demonstrated that GcbC physically interacts with LapD and that this physical interaction is directly linked to signaling ability (19). This finding was significant because it provides an example of a DGC that uses physical interaction to deliver c-di-GMP to a desired target (Figure 5). In the process of determining that these proteins interact, we attempted to do a domain-level analysis to find out which domains of GcbC and LapD were responsible for the interaction. We found only the full-length proteins of each competent to interact in our assays. We viewed this result as indicating one of two possibilities. First, it could mean that there are multiple interaction surfaces spread across the proteins, a hypothesis supported by evidence discussed below. Second, it is also possible that the proteins need to take on specific structural conformations in order to be able to interact with one another, which requires the full-length protein. This latter idea draws attention to the HAMP domain present in LapD, which is known to mediate conformational shifts when LapD binds c-di-GMP.
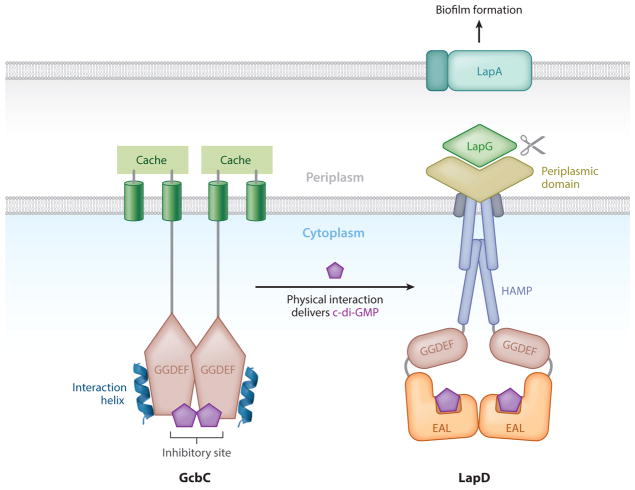
Figure 5.
GcbC physically delivers c-di-GMP to LapD. The inner membrane DGC requires physical contact to fully deliver its c-di-GMP signal to LapD. The interaction α5GGDEF helix of GcbC is depicted in blue. The inhibitory site of GcbC has also been shown to be a necessary element for interaction to occur and may help facilitate contact between these two proteins through an as yet undefined mechanism. c-di-GMP is depicted as purple pentagons located at the GcbC inhibitory site and bound at the LapD active site. Each protein functions as a dimer. GcbC has a cache domain that presumably allows enzymatic activity in response to an unidentified signal. The HAMP, GGDEF, EAL, and periplasmic domains of LapD are also shown. When LapD is activated by c-di-GMP, the periplasmic protease LapG is sequestered and the large adhesin LapA accumulates on the surface of the cell, allowing biofilm formation to commence. Adapted from Navarro et al. (45).
Mutational analysis of GcbC (Figure 2) and LapD revealed a discrete interaction surface made up of the N terminus of the α5GGDEF helix on the GGDEF domain of GcbC that makes contact with the complementary α helix on the EAL domain of LapD. Mutation of four of these residues found on the GcbC α helix resulted in partial loss of interaction with LapD, and a commensurate loss of biofilm formation, with no loss of catalytic ability observed. Intriguingly, mutation of a fifth residue on this helix, D484, was also found to contribute to interaction with LapD but demonstrated a partial loss of catalysis. We speculate this loss of catalysis is due to D484 coordinating a water molecule, which in turn stabilizes the product-bound state in the active site of GcbC, as is observed in the crystal structure of a DGC from Xanthomonas campestris (68). While transplanting residues from this α helix onto another DGC conferred some of GcbC’s functionality, wholesale helix transplants failed to alter the function of several other DGCs, indicating a multivariate system of physical and functional interaction.
In a second study, it was discovered that the autoinhibitory site of GcbC is a required element for interaction with LapD (20). Indeed, several disruptions of the I-site resulted in complete loss of interaction with LapD, an observation that was beyond the effect caused by stacked mutations from the interacting α helix. Disruptions to the I-site of a DGC traditionally result in increased c-di-GMP production and a commensurate increase in the dependent phenotype (14, 31). While increased c-di-GMP production was true for some of the I-site mutants observed in GcbC, other mutations demonstrated elevated levels of c-di-GMP production yet lower biofilm production—a consequence likely associated with loss of interaction between GcbC and LapD (Figure 5).
We note that the I-site marks the second example from GcbC where residues that can control the rate of catalysis have been linked as either a direct or indirect requirement for interaction with LapD, in conjunction with the D484 residue discussed above. These observations suggest that an important theme to physical interaction between a DGC and its effector may be the effector directly affecting the catalytic rate of the partner DGC when in complex.
MERGING THE MODELS
Important questions remain for unraveling signaling specificity in large c-di-GMP networks. How often is physical interaction used to localize signaling compared to the process of affecting global c-di-GMP levels? With the puzzle pieces of signaling specificity before us, it is worth reflecting on what experimental avenues are now available to the field and which might be most fruitful to pursue to create more complete models.
Understanding the Relationship Between DGC-Effector Contact and Catalysis
In studying a single DGC-effector interaction, we have observed two instances where DGC residues that may be involved in the rate control of catalysis appear to be required for interaction with their effector, LapD (19, 20). Demonstrating or disproving the hypothesis that effector contact can affect DGC catalysis is a likely next step from this work. It would also be beneficial to determine how often it is the case in DGC-effector complexes that residues that modulate catalysis are necessary for the interaction. Such a model would suggest a close interplay between function and/or regulation of the enzyme and receptor with which it specifically communicates.
Accurately Measuring c-di-GMP Production
If a given DGC may be involved in local signaling via physical interaction with its effector, it may not contribute to the global c-di-GMP pool. Such a situation could make quantitative measurement through whole-cell organic extractions followed by mass spectrometry misleading, as the global pool of c-di-GMP may not reflect local signaling events. The task becomes even more difficult if an effector protein can in any way modulate the rate of catalysis for its partner DGC as discussed above. In this case, dysfunction through loss of interaction may actually result in an increase in global c-di-GMP levels. Qualitative tests such as Congo red binding as a surrogate for measuring c-di-GMP levels when a given DGC is expressed heterologously in another organism may also prove inadequate in some cases, particularly if the c-di-GMP-dependent phenotype being assayed is not the result of global c-di-GMP levels, or if the DGC being tested happens to physically interact with host effectors (see the sidebar titled “Congo Red”). New in vitro and in vivo methods of c-di-GMP quantification at a local level may be required to analyze a given DGC, both with and without its cognate effector present.
CONGO RED.
Many DGCs are tested for catalytic activity through heterologous expression in P. aeruginosa. The more c-di-GMP produced by the cell, the more Pel polysaccharide is produced. Congo red is a dye capable of binding the Pel polysaccharide (as well as other polysaccharides). A qualitative correlation can be drawn between the redness of the P. aeruginosa colony due to binding of the Congo red dye and the amount of c-di-GMP being produced.
Mapping the Interaction Surfaces
Both the work we discuss in this review that found that multiple domains of interaction partners were competent to interact with one another (38) and our own work where we found that disruption of no one interaction face was enough to fully negate interaction (19, 20) suggest that there may be multiple interaction surfaces at play in any given signaling complex. In the case of the Lap system, we have defined interaction interfaces for LapD and GcbC. Are there additional such interfaces to define? And how conserved are these interfaces? That is, might LapD interact with some or all of the other DGCs in the cell in a manner analogous to GcbC? As it stands, it is also unclear if we can eventually define interaction interfaces to the point where we might even predict interaction, analogous to what has been done for two component systems (13, 59).
Distinguishing Interaction Versus Signaling
As described above, assuming there are several interaction surfaces involved between a DGC and effector, it also becomes necessary to tease apart the difference between physical association whereby proteins colocalize and functional interaction where they purposefully exchange signaling information. It is entirely possible that these two phenomena occur as discrete steps whereby physical interaction does not immediately imply active signaling, as a variety of signaling-defective mutants of both GcbC and LapD were still found to be competent to interact (19). Additionally, it is worthwhile to determine if there may be a step between association and signaling—some type of modulation that may be occurring after association that can either allow or disallow productive signaling to take place. Each of these steps may utilize different interaction surfaces. What these interaction sites are, and how they are used will be major topics to address going forward.
Characterizing the Dynamics of a Signaling Complex
If complexes of DGCs and effectors do commonly form, the next question to ask may be, how are they turned off when signaling is not desired? While transcriptional and translational regulation may play a role, it is unlikely every active signaling complex must wait for protein turnover before being switched off. The answer may lie in how frequently the DGC-effector pair makes contact, and how enduring this interaction is—another possible role for disparate interaction surfaces. Further possibilities include PDEs joining these complexes to help regulate them, other DGC members competitively binding the effector, and the cell preventing productive interaction through other means. Finally, given that most DGCs have an associated regulatory domain, regulation may require a combination of interactions and environmental inputs. Indeed, such inputs could regulate activity of the DGC, or perhaps its ability to interact with its receptor.
Examining the Relationship Between Local and Global Signaling
For organisms that rely on both local and global c-di-GMP signaling, it is possible both systems are at times employed to control a given cellular response. Of note is the P. fluorescens DGC called GcbB. Like GcbC, GcbB was found to signal in a LapD-dependent manner to control LapA accumulation on the cell surface. Despite this role in signaling, we have failed to demonstrate physical interaction between LapD and GcbB by bacterial two-hybrid and coprecipitation methods (unpublished data). While it is still possible that these proteins do in fact interact, it is worth considering how the global pool of c-di-GMP may be affecting LapD or similar effectors in tandem with local partners. What strategies bacteria can use to either insulate an effector against the global pool of c-di-GMP, or alternatively what strategies exist to integrate the global and local pool of c-di-GMP is an open question.
It is likely that PDEs will also need to become part of this analysis. There is evidence both for the idea that PDEs can physically interact with effectors and that PDEs can act globally to lower c-di-GMP levels (33, 38). How bacteria manage individual c-di-GMP signaling events may well depend on how many PDEs operate to keep the cytoplasm clear of c-di-GMP versus how many PDEs form complexes with effectors, shielding these receptors from the global c-di-GMP pool. And as always, the answer may lie somewhere in between.
CONCLUSIONS AND PERSPECTIVES
Owing to multiple prominent efforts around the world, the field of c-di-GMP research has recently entered into an era when we may ask how specificity in signaling is achieved in the cell. Importantly, it appears that both the global and the local c-di-GMP signaling models are used in nature. These distinct mechanisms of specificity provide large opportunities to understand DGC, PDE, and effector operations at a level of detail that may make prediction of functional modules in c-di-GMP networks easier and provide ways to target c-di-GMP signaling systems of various bacteria for therapeutic purposes. It is also noteworthy that a large signaling system with points of potential cross talk may provide a powerful model for bioengineers to study and develop more sophisticated bacterial circuitry than is currently possible.
There are also many major questions of basic biology left to answer. In many organisms, there are more identifiable cyclases than effectors. This may be partly due to the relative ease of identifying a putative DGC domain based on its sequence compared to the more diverse and still growing number of effector archetypes. On the other hand, it is possible that in some organisms several enzymes signal to the same effector. How a signal is integrated from multiple DGCs into one effector remains to be elucidated. Further, how the precise roles and responsibilities of DGCs are divided based upon timing and circumstance also remains a mystery. Finally, perhaps the most perplexing question is more thematic than scientific: Why do some organisms have so many of these enzymes? That so many DGCs and effectors in the literature appear to have no observable phenotype suggests that many environmentally relevant conditions are missing from laboratory tests that future studies are sure to identify. But it also suggests a requirement of many specialized enzymes when an organism must determine in short order if it will make a biofilm under a wide and disparate set of circumstances. The next large question must therefore move beyond understanding individual DGCs and PDEs, and toward understanding how the cell performs the computation to integrate the signal(s) across the entire DGC network for each c-di-GMP-driven phenotype.
Research into c-di-GMP signaling specificity is at a critical juncture. Many mechanisms used by individual DGCs and effectors are being discovered, and these insights must necessarily form the basis for the next set of hypotheses to test. A natural consequence of this period of productivity is that many competing models are being created in an attempt to explain wider and wider swaths of the observed systems. Accommodating disparate models in the literature and allowing them to be developed in their respective systems is an important step toward more fully describing a very complex mode of signaling. And when these models mature it is likely, as is often the case in science, that many will turn out not to be as mutually exclusive as they initially appeared.
Acknowledgments
This work was supported by NIH grant R01-AI097307, and grant T32-GM08704 (supporting K.M.D.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Agostoni M, Koestler BJ, Waters CM, Williams BL, Montgomery BL. Occurrence of cyclic di-GMP-modulating output domains in cyanobacteria: an illuminating perspective. mBio. 2013;4(4):e00451–13. doi: 10.1128/mBio.00451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm RA, Bodero AJ, Free PD, Mattick JS. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178:46–53. doi: 10.1128/jb.178.1.46-53.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 4.Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, et al. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol Microbiol. 2011;79:533–51. doi: 10.1111/j.1365-2958.2010.07470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd CD, Chatterjee D, Sondermann H, O’Toole GA. LapG, required for modulating biofilm formation by Pseudomonas fluorescens Pf0–1, is a calcium-dependent protease. J Bacteriol. 2012;194:4406–14. doi: 10.1128/JB.00642-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd CD, O’Toole GA. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol. 2012;28:439–62. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd CD, Smith TJ, El-Kirat-Chatel S, Newell PD, Dufrene YF, O’Toole GA. Structural features of the Pseudomonas fluorescens biofilm adhesin LapA required for LapG-dependent cleavage, biofilm formation, and cell surface localization. J Bacteriol. 2014;196:2775–88. doi: 10.1128/JB.01629-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang AL, Tuckerman JR, Gonzalez G, Mayer R, Weinhouse H, et al. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry. 2001;40:3420–26. doi: 10.1021/bi0100236. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee D, Boyd CD, O’Toole GA, Sondermann H. Structural characterization of a conserved, calcium-dependent periplasmic protease from Legionella pneumophila. J Bacteriol. 2012;194:4415–25. doi: 10.1128/JB.00640-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee D, Cooley RB, Boyd CD, Mehl RA, O’Toole GA, Sondermann H. Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. eLife. 2014;3:e03650. doi: 10.7554/eLife.03650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen LH, Koseoglu VK, Guvener ZT, Myers-Morales T, Reed JM, et al. Cyclic di-GMP-dependent signaling pathways in the pathogenic Firmicute Listeria monocytogenes. PLOS Pathog. 2014;10:e1004301. doi: 10.1371/journal.ppat.1004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488:680–83. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng RR, Morcos F, Levine H, Onuchic JN. Toward rationally redesigning bacterial two-component signaling systems using coevolutionary information. PNAS. 2014;111:E563–71. doi: 10.1073/pnas.1323734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christen B, Christen M, Paul R, Schmid F, Folcher M, et al. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281:32015–24. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 15.Christen M, Christen B, Allan MG, Folcher M, Jeno P, et al. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. PNAS. 2007;104:4112–17. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280:30829–37. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 17.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science. 2010;328:1295–97. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz DP, Huertas MG, Lozano M, Zarate L, Zambrano MM. Comparative analysis of diguanylate cyclase and phosphodiesterase genes in Klebsiella pneumoniae. BMC Microbiol. 2012;12:139. doi: 10.1186/1471-2180-12-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahlstrom KM, Giglio KM, Collins AJ, Sondermann H, O’Toole GA. Contribution of physical interactions to signaling specificity between a diguanylate cyclase and its effector. mBio. 2015;6(6):e01978–15. doi: 10.1128/mBio.01978-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlstrom KM, Giglio KM, Sondermann H, O’Toole GA. The inhibitory site of a diguanylate cyclase is a necessary element for interaction and signaling with an effector protein. J Bacteriol. 2016;198:1595–603. doi: 10.1128/JB.00090-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duvel J, Bertinetti D, Moller S, Schwede F, Morr M, et al. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods. 2012;88:229–36. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Fazli M, O’Connell A, Nilsson M, Niehaus K, Dow JM, et al. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol. 2011;82:327–41. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 23.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 24.Guvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–73. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ha DG, Richman ME, O’Toole GA. Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa PA14. Appl Environ Microbiol. 2014;80:3384–93. doi: 10.1128/AEM.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habazettl J, Allan MG, Jenal U, Grzesiek S. Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J Biol Chem. 2011;286:14304–14. doi: 10.1074/jbc.M110.209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He M, Ouyang ZM, Troxell B, Xu HJ, Moh A, et al. Cyclic di-GMP is essential for the survival of the Lyme disease spirochete in ticks. PLOS Pathog. 2011;7:e1002133. doi: 10.1371/journal.ppat.1002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–73. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 29.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–89. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49:905–18. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 31.Hunter JL, Severin GB, Koestler BJ, Waters CM. The Vibrio cholerae diguanylate cyclase VCA0965 has an AGDEF active site and synthesizes cyclic di-GMP. BMC Microbiol. 2014;14:22. doi: 10.1186/1471-2180-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–68. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:8165–78. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′–5′)-cyclic-GMP in virulence. PNAS. 2006;103:2839–44. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumagai Y, Matsuo J, Hayakawa Y, Rikihisa Y. Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J Bacteriol. 2010;192:4122–33. doi: 10.1128/JB.00132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–48. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Chen JH, Hao Y, Nair SK. Structures of the PelD cyclic diguanylate effector involved in pellicle formation in Pseudomonas aeruginosa PAO1. J Biol Chem. 2012;287:30191–204. doi: 10.1074/jbc.M112.378273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 2013;32:2001–14. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Gil M, Ramos-Gonzalez MI, Espinosa-Urgel M. Roles of cyclic di-GMP and the Gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J Bacteriol. 2014;196:1484–95. doi: 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Granero F, Navazo A, Barahona E, Redondo-Nieto M, Gonzalez de Heredia E, et al. Identification of flgZ as a flagellar gene encoding a PilZ domain protein that regulates swimming motility and biofilm formation in Pseudomonas. PLOS ONE. 2014;9:e87608. doi: 10.1371/journal.pone.0087608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuyama BY, Krasteva PV, Baraquet C, Harwood CS, Sondermann H, Navarro MV. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. PNAS. 2016;113:E209–18. doi: 10.1073/pnas.1523148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′–5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–95. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 43.Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, et al. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio. 2010;1(4):e00183–10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monds RD, Newell PD, Gross RH, O’Toole GA. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0–1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol. 2007;63:656–79. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- 45.Navarro MV, Newell PD, Krasteva PV, Chatterjee D, Madden DR, et al. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLOS Biol. 2011;9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newell PD, Monds RD, O’Toole GA. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0–1. PNAS. 2009;106:3461–66. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O’Toole GA. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0–1. J Bacteriol. 2011;193:4685–98. doi: 10.1128/JB.05483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell. 2010;38:128–39. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 2007;282:12860–70. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B, Miller SI. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol. 2012;86:1424–40. doi: 10.1111/mmi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramelot TA, Yee A, Cort JR, Semesi A, Arrowsmith CH, Kennedy MA. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins. 2007;66:266–71. doi: 10.1002/prot.21199. [DOI] [PubMed] [Google Scholar]
- 52.Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross P, Mayer R, Weinhouse H, Amikam D, Huggirat Y, et al. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum: chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J Biol Chem. 1990;265:18933–43. [PubMed] [Google Scholar]
- 54.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–81. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 55.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. PNAS. 2006;103:6712–17. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–14. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 57.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–35. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 58.Shin JS, Ryu KS, Ko J, Lee A, Choi BS. Structural characterization reveals that a PilZ domain protein undergoes substantial conformational change upon binding to cyclic dimeric guanosine monophosphate. Protein Sci. 2011;20:270–77. doi: 10.1002/pro.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–54. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith KD, Shanahan CA, Moore EL, Simon AC, Strobel SA. Structural basis of differential ligand recognition by two classes of bis-(3′–5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. PNAS. 2011;108:7757–62. doi: 10.1073/pnas.1018857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srivastava D, Hsieh ML, Khataokar A, Neiditch MB, Waters CM. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol. 2013;90:1262–76. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–13. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sultan SZ, Pitzer JE, Boquoi T, Hobbs G, Miller MR, Motaleb MA. Analysis of the HD-GYP domain cyclic dimeric GMP phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infect Immun. 2011;79:3273–83. doi: 10.1128/IAI.05153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Townsley L, Yildiz FH. Temperature affects c-di-GMP signalling and biofilm formation in Vibrio cholerae. Environ Microbiol. 2015;17:4290–305. doi: 10.1111/1462-2920.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tuckerman JR, Gonzalez G, Sousa EHS, Wan XH, Saito JA, et al. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry. 2009;48:9764–74. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- 66.Wei C, Jiang WD, Zhao MR, Ling JJ, Zeng X, et al. A systematic analysis of the role of GGDEF-EAL domain proteins in virulence and motility in Xanthomonas oryzae pv. oryzicola. Sci Rep. 2016;6:23769. doi: 10.1038/srep23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, et al. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416:207–11. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- 68.Yang CY, Chin KH, Chuah ML, Liang ZX, Wang AH, Chou SH. The structure and inhibition of a GGDEF diguanylate cyclase complexed with (c-di-GMP)2 at the active site. Acta Crystallogr D Biol Crystallogr. 2011;67:997–1008. doi: 10.1107/S090744491104039X. [DOI] [PubMed] [Google Scholar]
- 69.Zahringer F, Lacanna E, Jenal U, Schirmer T, Boehm A. Structure and signaling mechanism of a zinc-sensory diguanylate cyclase. Structure. 2013;21:1149–57. doi: 10.1016/j.str.2013.04.026. [DOI] [PubMed] [Google Scholar]